Abstract
Childhood stroke occurs from birth to 18 years of age, ranks among the top ten childhood causes of death, and leaves lifelong neurological impairments. Arterial ischemic stroke in infancy and childhood occurs due to arterial occlusion in the brain, resulting in a focal lesion. Our understanding of mechanisms of injury and repair associated with focal injury in the developing brain remains rudimentary. Neuroimaging can reveal important insights into these mechanisms. In adult stroke population, multi-center neuroimaging studies are common and have accelerated the translation process leading to improvements in treatment and outcome. These studies are centered on the growing evidence that neuroimaging measures and other biomarkers (e.g., from blood and cerebrospinal fluid) can enhance our understanding of mechanisms of risk and injury and be used as complementary outcome markers. These factors have yet to be studied in pediatric stroke because most neuroimaging studies in this population have been conducted in single-centred, small cohorts. By pooling neuroimaging data across multiple sites, larger cohorts of patients can significantly boost study feasibility and power in elucidating mechanisms of brain injury, repair and outcomes. These aims are particularly relevant in pediatric stroke because of the decreased incidence rates and the lack of mechanism-targeted trials.
Toward these aims, we developed the Pediatric Stroke Neuroimaging Platform (PEDSNIP) in 2015, funded by The Brain Canada Platform Support Grant, to focus on three identified neuroimaging priorities. These were: developing and harmonizing multisite clinical protocols, creating the infrastructure and methods to import, store and organize the large clinical neuroimaging dataset from multiple sites through the International Pediatric Stroke Study (IPSS), and enabling central searchability. To do this, developed a two-pronged approach that included building 1) A Clinical-MRI Data Repository (standard of care imaging) linked to clinical data and longitudinal outcomes and 2) A Research-MRI neuroimaging data set acquired through our extensive collaborative, multi-center, multidisciplinary network. This dataset was collected prospectively in eight North American centers to test the feasibility and implementation of harmonized advanced Research-MRI, with the addition of clinical information, genetic and proteomic studies, in a cohort of children presenting with acute ischemic stroke.
Here we describe the process that enabled the development of PEDSNIP built to provide the infrastructure to support neuroimaging research priorities in pediatric stroke. Having built this Platform, we are now able to utilize the largest neuroimaging and clinical data pool on pediatric stroke data worldwide to conduct hypothesis-driven research. We are actively working on a bioinformatics approach to develop predictive models of risk, injury and repair and accelerate breakthrough discoveries leading to mechanism-targeted treatments that improve outcomes and minimize the burden following childhood stroke. This unique transformational resource for scientists and researchers has the potential to result in a paradigm shift in the management, outcomes and quality of life in children with stroke and their families, with far-reaching benefits for other brain conditions of people across the lifespan.
Keywords: Neuroimaging, MRI, Pediatricstroke, Data platform, Multi-center study
1. Introduction
Pediatric arterial ischemic stroke affects 1 per 2500 newborns and 6 per 100,000 children annually. Stroke is among the ten most common causes of childhood death (deVeber et al., 2017, Goeggel Simonetti et al., 2015, Greenham et al., 2016). In addition, most survivors face cognitive, mental or physical disabilities impacting them for many decades (deVeber et al., 2000, Fullerton et al., 2002, Ganesan et al., 2000). Since 1995, the Children’s Stroke Program at the Hospital for Sick Children in Toronto, Canada, has led a Canadian clinical research network which has been instrumental in advancing our understanding of pediatric stroke outcomes (deVeber et al., 2017). This clinical research network was developed originally by the Canadian Pediatric Ischemic Stroke Registry (1992–2002), which grew into the International Pediatric Stroke Study (IPSS) (2003-current). A substantial achievement of the IPSS has been the enrollment of > 7,000 patients at 110 national and international sites.
Data collected through these networks employed standardized clinical data tools and validated clinical neurological outcomes (Kitchen et al., 2012, Slim et al., 2020). This data is stored in a secure web-based system accessible to all collaborators for analysis (details in Data Sharing section below). Analysis of this clinical data has produced significant discoveries in pediatric stroke incidences, treatment practices, risk factors and outcomes (deVeber et al., 2017, Goeggel Simonetti et al., 2015, Greenham et al., 2016, deVeber et al., 2000, Fullerton et al., 2002, Ganesan et al., 2000). However, our understanding of mechanisms of injury and repair associated with focal injury to the brain caused by pediatric stroke remains rudimentary.
Neuroimaging can reveal important insights about these mechanisms. In adult stroke, multicenter neuroimaging studies are common and offer advantages over single-center studies. They have accelerated the translation process in diseases such as Alzheimer's and mild cognitive impairment (Duchesne et al., 2019, Mueller et al., 2005) and, more recently, adult stroke (Liew et al., 2022). These studies are centered on the growing evidence that neuroimaging measures and other biomarkers (e.g., from blood and cerebrospinal fluid) can improve understanding mechanisms of risk and injury and be used as complementary outcome markers. To date, most pediatric neuroimaging studies on stroke have been conducted in single-centred small cohorts. By pooling neuroimaging data across multiple sites, larger cohorts of patients can significantly boost study feasibility and power in elucidating mechanisms of brain injury, repair and outcomes. These aims are particularly relevant in pediatric stroke because of the decreased incidence rates and the lack of mechanism-targeted trials. The major challenge with pooling multicentre data is the lack of standardized imaging protocols leading to sustained non-uniformity across institutions.
The inherent variability of image acquisition per site relates to non-standardized imaging protocols across sites. due to differences in the MRI vendors and models, acquisition parameters, staffing resources and expertise, to name a few. In addition, this variability exists in both the commonly available clinical images (i.e. 2D MRI acquired in the clinical setting to diagnose stroke) and the less commonly available images acquired in research protocols (i.e. high resolution 3D images). The added challenges with research acquisitions are the extended time for acquisition and the difficulty in standardizing protocols due to dynamic changes in the developing brain. For example, diffusion tensor imaging (DI) measures the direction of water diffusion in the brain, reflecting the integrity of fiber tracts. Due to myelination changes in the fiber tracks in the developmental brain at various ages, standardization of research protocols is difficult. There has been no research to address these issues in pediatric stroke. Therefore, the rich data set of neuroimaging on these patients remains underutilized.
Towards these aims, we identified the need for a two-pronged approach to advance the field by leveraging and pooling extensive clinically-acquired imaging data and collecting research-acquired neuroimaging data. Three neuroimaging-focused priorities were identified in 2015, including developing and harmonizing multisite clinical protocols and pathways and developing the infrastructure and methods among sites to import, store and organize the large accrued clinical neuroimaging dataset that enables central searchability, and develop, apply and test the feasibility of implementing prospective, research-based pediatric neuroimaging protocols in a multisite study. To provide the infrastructure to support these priorities, PEDSNIP was initiated, and funding from Brain Canada in 2016 enabled the development of the Platform, a conduit to address these challenges in pediatric stroke (Dlamini et al., 2017, Domi et al., 2017, Lee et al., 2017, Mirsky et al., 2017). Having built this Platform, we are in a position to utilize the largest neuroimaging and clinical data pool on pediatric stroke data worldwide. In addition, this Platform will facilitate hypothesis-driven research using neuroimaging in clinical and research-acquired MRI modalities. This purpose of this paper is to describe the process that enabled the development of this Platform to mobilize the knowledge generated and optimize impact.
2. Development of consensus clinical and research neuroimaging protocols and platform
To facilitate the use of the accrued clinical data collected in the International Pediatric Stroke Study (IPSS), a process to build the capacity of the existing data Platform so that clinical imaging data could be stored, analyzed and shared was needed. In 2013 the Children’s Stroke Program launched the Stroke Imaging Laboratory for Children (SILC). The vision of SILC is to develop and apply neuroimaging techniques to unravel the neurobiological processes underlying focal ischemic infarction, and the neural substrates of plasticity and recovery in children. SILC houses the clinical MRI, clinical and demographic data from the IPSS sub-studies, including VIPS (Vascular Effects of Infection in Pediatric Stroke), SPORT (Stimulation for Perinatal Stroke Optimizing Recovery Trajectories), and others (i.e. Ontario Cerebral Palsy Network study, Ontario Brain Institute). The neuroimaging data collected within initial SILC infrastructure was stored on a separate framework with limited capability to support optimized data collection, storage, organization and sharing.
Following this, we published a roadmap for how to utilize neuroimaging to advance pediatric stroke (Dlamini et al., 2017), consensus-based standardized neuroimaging protocols for acute and follow-up management of neonatal (Lee et al., 2017), and childhood stroke (Mirsky et al., 2017). Key Opinion Leaders and site-specific knowledge of resources and practices of the network informed these neuroimaging protocols. In addition, we reported the potential of advanced research imaging techniques in pediatric stroke (Domi et al., 2017). A follow-up survey collected through the IPSS Neuroimaging network in 2019 revealing an adherence of over 80% to our published neuroimaging protocols.
2.1. Study 1: The Retrospective Clinical-MRI Data Repository
Materials
To house neuroimaging data for participants enrolled in the IPSS, an Imaging Repository was developed to establish a standardized dataset by stroke type. The development of the Repository was implemented following the Brain Canada Neuroimaging Interest Group meeting held at the Child Neurology Society conference in October 2018. At this meeting, we hosted survey-informed discussions regarding each site’s capacity, resources, feasibility, data governance, data intake workflow and potential analyses.
Funded by the Brain Canada Platform Support Grant, the Repository, hosted in a high-performance computing environment, is supported with technology expertise by the Centre for Computational Medicine (CCM) at SickKids, the SILC leadership team, and the Research Information Technology department.
Methods
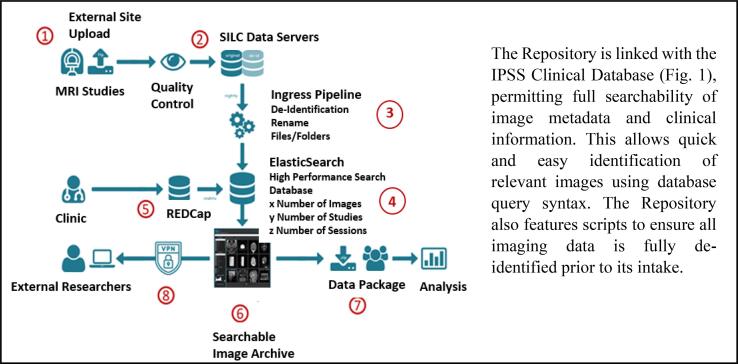
The Repository operates within the CCM’s high-performance computing (HPF) environment and is linked to the IPSS Clinical Database (Fig. 1). This enables powerful processing, large capacity storage and high-speed network infrastructure that permits searchability of image metadata clinical information. Imaging data analysis is also supported. Scripts were developed that ensure all imaging data is fully de-identified before intake. In addition, a Manual of Operations was supplied to collaborating sites to assist with transferring imaging data to SickKids through a secure file transfer portal (FTP). This manual details standard data requirements by stroke type and standardized time points.
Fig. 1.
Imaging intake workflow for clinical images for the IPSS-SILC Imaging Repository.
Clinically-indicated imaging is acquired from external sites (see Table 1), and sent over secure file transfer portal onto SILC servers. All incoming data is run through de-identification pipelines to scrub dicom headers, visually checked for quality control, renamed to match our naming conventions, and sorted into appropriate folders. Simultaneously, clinical data is collected at all sites, and is entered into the clinical IPSS Database, and stored on Research Electronic Data Capture (REDCap). Using the ElasticSearch on REDCap, syntax is created that is required to search the Imaging Archive for variables saved within the Clinical Database (for example, how many imaging studies do we have by stroke type and age at stroke?). External researchers are currently able to request queries using syntax to identify the number of images that fit their research criteria. SILC is then able to create data packages that can be sent to external collaborators for analysis (See Fig. 1).
Table 1.
Retrospective Clinical-MRI Data Repository: Institutions, Locations, Scanner Types
| Institution Name | City | Country | Vendor Field Combinations |
|---|---|---|---|
| Alberta Children's Hospital | Calgary | CANADA | 1.5T Siemens, 3T GE, 1.5T GE, 3T Siemens |
| Cook Children's Hospital | Forth Worth | USA | 3T Siemens, 1.5T Siemens, 1.5T GE, 1.493806T Siemens, 3T GE, 1.5T Philips, 3T Philips |
| Boston Children's Hospital | Boston | USA | 3T Siemens, 1.5T GE, 1.5T Siemens, 1.5T Philips, 3T GE |
| Nationwide Children's Hospital | Columbus | USA | 3T GE, 3T Siemens, 1.5T GE, 1.5T Siemens |
| University of Texas Southwestern Medical Center | Dallas | USA | 3T Philips, 1.5T Philips, 3T Siemens, 1.5T Siemens, 1.5T GE |
| University of California San Francisco | San Francisco | USA | 3T GE, 1.5T GE, 1.5T Siemens, 3T Philips, 1.5T Philips |
| Children's Central Hospital | Tbilisi | GEORGIA | 1.5T Siemens, 3T Siemens, 1T Siemens |
| Children's Hospital Colorado | Aurora | USA | 3T Philips, 1.5T Philips, 1.5T Siemens, 3T GE, 1.5T GE, 3T Siemens |
| Children's Hospital of Eastern Ontario | Ottawa | CANADA | 3T Siemens, 1.5T GE |
| Seattle Children's Hospital | Seattle | USA | 3T Siemens, 1.5T Siemens, 1.5T GE |
| Children's National Medical Center | Washington D.C. | USA | 3T GE, 1.5T GE, 1.5T Siemens |
| Royal Children's Hospital | Parkville | AUSTRALIA | 1.5T Siemens, 3T Siemens, 3T GE |
| St. Louis Children's Hospital | St. Louis | USA | 2.893620014T Siemens, 3T Siemens, 1.5T Siemens, 1.5T GE |
| Monroe Carell Jr. Children's Hospital at Vanderbilt | Nashville | USA | 3T Philips, 1.5T Philips |
| Winnipeg Children's Hospital | Winnipeg | CANADA | 3T Siemens, 1.5T GE, 1.5T Siemens |
| Ain Shams University | Cairo | EGYPT | 1.5T Philips |
| Oregon Health & Science University | Portland | USA | 1.5T Philips, 3T Philips, 1.5T GE, 1T Philips, 3T Siemens |
| Children's Mercy Hospitals and Clinics | Kansas City | USA | 1.5T Philips, 1.5T GE, 1.5T Siemens, 3T Siemens, 3T GE |
| Ann & Robert H. Lurie Children's Hospital of Chicago | Chicago | USA | 1.5T Siemens |
| Le Bonheur Children's Hospital | Memphis | USA | 1.5T GE, 3T GE, 3T Siemens, 1.5T Siemens |
| The Hospital for Sick Children | Toronto | CANADA | 1.5T Philips, 3T Philips, 1.5T Siemens, 1.5T GE, 3T Siemens |
| Children's Hospital of Wisconsin | Wauwatosa | USA | 1.5T GE, 1.5T Siemens, 3T Siemens, 1.5T Philips, 3T GE |
| Pontificia Universidad Catolica de Chile | Santiago | CHILE | 1.5T Philips, 1.5T Siemens |
| Cleveland Clinic Foundation | Cleveland | USA | 1.5T Siemens, 3T Siemens, 1.5T Philips, 1.5T GE |
| SUNY Buffalo Children's Hospital | Buffalo | USA | 1.5T GE, 3T GE |
| Johns Hopkins Hospital | Baltimore | USA | 1.5T Siemens, 3T Siemens, 3T GE |
| Maimonides Medical Center | New York City | USA | 1.5T Siemens |
| McMaster University Medical Centre | Hamilton | CANADA | 1.5T Siemens |
| Mother and Child Health Care Institute | Belgrade | SERBIA | 1.5T Siemens |
| Children's Hospital of Philadelphia | Philadelphia | USA | 1.5T Siemens, 1.5T GE, 3T Siemens, 3T GE, 3.0T Siemens |
| The University of Utah and Primary Children's Medical Center | Salt Lake City | USA | 3T GE, 1.5T GE, 1.5T Siemens, 1.5T Philips |
| Children's Clinic of Tartu University Hospital | Tartu | ESTONIA | 1.5T Siemens |
| Chinese PLA General Hospital | Beijing | CHINA | 1.5T Philips |
| Stollery Children's Hospital (University of Alberta Hospital) | Edmonton | CANADA | 1.5T Siemens |
| West Virginia University | Morgantown | USA | 1.5T Siemens, 3T Siemens |
| Columbia University Medical Centre | New York City | USA | 1.5T GE, 1.5T Siemens, 3T GE, 1.5T Philips, 3T Philips |
| Stanford University Medical Center | Palo Alto | USA | 3T GE, 1.5T Philips, 1.5T GE |
| Medical University of Silesia | Katowice | POLAND | 1.5T Siemens |
| Aristotle University of Thessaloniki | Thessaloniki | GREECE | 1.5T Siemens |
| Queen Mary Hospital | Hong Kong | CHINA | 1.5T GE |
| Robert Debre Hospital | Paris | FRANCE | 1.5T Philips |
| Akron Children's Hospital | Akron | USA | 3T Siemens, 1.5T Philips, 1.5T Siemens |
| Cincinnati Children's Hospital Medical Center | Cincinnati | USA | 3T Philips, 1.5T GE, 1.5T Philips, 1.5T Siemens, 3T GE |
| Helen DeVos Children's Hospital | Grand Rapids | USA | 1.5T GE, 3T GE, 1.5T Siemens, 1.5T Philips |
| Phoenix Children's Hospital | Phoenix | USA | 3T Philips, 1.5T Philips |
| Children's Hospital of Pittsburgh | Pittsburgh | USA | 1.5T GE, 3T Siemens |
| Ben Gurion University | Beersheba | ISRAEL | 1.5T Philips, 3T Philips |
| Wolfson Children's Hospital | Jacksonville | USA | 3T GE, 1.5T GE |
Results
Regulatory compliance documents (i.e., Data Transfer Agreements, financial agreements) were executed in 26 IPSS sites. Standard of care, clinically-acquired MRI and CT studies of approximately 1700 patients and 3000 imaging sessions acquired at the time of stroke diagnosis and at standardized time points for follow-up visits have been transferred and stored in the Repository (see Table 1). These images have been linked to the clinical profile and serial outcome data collected using the Pediatric Stroke Outcome Measure and Recovery and Recurrence Questionnaire) (Kitchen et al., 2012). Missing data reports are generated quarterly for all sites to ensure a comprehensive clinical and imaging data set.
The clinical imaging protocol and acquisition parameters were designed by the lead site considering the published IPSS Recommended Clinical Imaging Protocol (designed by colleagues with expertise in pediatric neuroradiology) and input from local site collaborators. Efforts were made to harmonize the clinical imaging protocols and acquisition parameters across study sites.
Data sharing and standardization
The Platform Operations and Neuroimaging Database Committees established research MR protocols to ensure standardized methodology and uniform data collection across sites. Imaging data are stored in a high-performance computing facility whose management satisfies security and privacy policies for patient health information based on the Canadian Standards Association Privacy Principles (Personal Health Information Protection Act [PHIPA], 2004, SO 2004, c 3, Sch A, https://canlii.ca/t/55g5p). Ethical approval and a fully executed Data Sharing Agreement are required to send/receive and analyze imaging data. Sites transfer de-identified imaging via secure FTP services while complying with the Platform procedures and all applicable laws (i.e., PHIPA, Personal Information Protection and Electronic Documents Act [PIPEDA], and any applicable provincial, state or national legislation concerning the protection of privacy and personal health information [PHI]). The Operations Committee and the Neuroimaging Subcommittee oversaw adherence to standardized neuroimaging protocols and Standard Operating Procedures [SOPs] for clinical data and saliva sample collection/processing, ensuring uniform data collection is obtained across all sites.
Our consent forms with language to address open data access, partnership with industry, and permission to approach for future research and clinical trials. This extensive clinical neuroimaging data pool represents an unparalleled resource for research in acquired focal brain injury in children.
2.2. Study 2: The Prospective Research-MRI Study
Materials
In addition to the data acquired in the clinical-MRI Repository, prospective research-MRI protocols (see Supplementary Material, 1for MRI protocols) have been implemented in children presenting with acute ischemic stroke in seven North American centers (Table 2). These include The Hospital for Sick Children (Toronto, Canada), British Columbia Children’s Hospital (British Columbia, Canada), London Health Sciences Center (London, Canada), Winnipeg Children’s Hospital (Winnipeg, Canada), Children’s Hospital of Colorado (Colorado, USA), Alberta Children’s Hospital (Calgary, Canada), Boston Children’s Hospital (Boston, Massachusetts), the University of Texas South Western Medical Center (Dallas, Texas). Clinical data and neuroimaging were collected serially in patients and healthy neurotypical controls in these centers. This is the first prospective study in this patient population to utilize neuroimaging at serial time points from the acute to the chronic stage to study the evolution of injury and repair in the brain following stroke in childhood.
Table 2.
Prospective Research-MRI Study, Institutions, Locations, Scanner Types.
| Hospital | City, State/Province/Country | Scanner Type |
|---|---|---|
| Alberta Children’s Hospital | Calgary, Alberta, Canada | 3T Siemens Magnetom Vida, 3T GE, Discovery 750W |
| Boston Children’s Hospital | Boston, Massachusetts, United States | 3T Siemens Prisma |
| British Columbia Children’s Hospital | Vancouver, British Columbia, Canada | 3T GE 750 |
| Children’s Hospital of Colorado | Aurora, Colorado, United States | 3T Philips Ingenia |
| The Hospital for Sick Children | Toronto, Ontario, Canada | 3T Siemens Prisma |
| London Health Sciences Center | London, Ontario, Canada | 1.5T GE Signa |
| The University of Texas South Western Medical Center | Dallas, Texas, United States | 3T Siemens Skyra |
| Winnipeg Children’s Hospital | Winnipeg, Manitoba, Canada | 1.5T Siemens Aero |
Patient Inclusion
The age inclusion criteria were 7-18 years based on expected participant compliance. However, the age limit was lowered to include 6-year-olds at the discretion of site investigators if they identified children able to comply. In addition, minimum study requirements for inclusion were defined to ensure the collection of a base set of homogeneous data. Participants required two imaging time points, including an acute scan, at least one blood sample, and outcome measures (Table 3) to be considered eligible for reimbursement. These requirements for a blood draw were subsequently adjusted due to research ethics at specific sites; therefore, saliva samples became the minimum requirement for biosamples.
Table 3.
Study Procedures and Data Collection Timepoints.
| Procedure | Acute (Day 3–7) | Sub-Acute (Day 10–14) | Chronic (Month 3) | Chronic 12 (Month 12) |
|---|---|---|---|---|
| Clinical Imaging Protocol | ✓ | ✓ | ||
| Research Imaging Protocol | ✓ | ✓ | ✓ | ✓ |
| Clinical Assessment | ✓ | ✓ | ✓ | ✓ |
| Behavioural Assessment | ✓ | ✓ | ✓ | ✓ |
| Saliva Sample | ✓ | |||
| Blood Sample | ✓ | ✓ |
3. Methods
Study DesignConsiderations
In order to minimize participant time requirements, research imaging sequences were collected during clinical scanning sessions. In order to accommodate the imaging requirements for individual participants, we designated a minimum data set per session and acquired imaging to be counted as a research session. Imaging time points were chosen to align with clinical practice when most children with AIS are scanned to track recurrence and recovery, thus minimizing any burden on the participant. Patients were scanned in the acute and chronic recovery phase (i.e., 3-7 days, 10-14 days, 3 months and 12 months post-stroke)(see Table 3), however the first diagnostic hyperacute scan was not included as part of the research protocol. These time windows for imaging data collection were deemed suitable in collaboration with participating sites to facilitate and maximize recruitment and general coordination of the study. Other factors included MRI vendors, field strength and scanner availability for research scanning at an acute time point (see Table 3).
Quality Assurance and Harmonization
An essential aspect of onboarding is the collection of imaging protocols through exam cards and pilot data to compare data quality and optimize imaging parameters aiming towards a harmonized imaging acquisition protocol. In addition, the onboarding process included meetings with principal investigators, neuroradiologists, MR technologists and site coordinators to ensure compliance with the imaging protocol in clinical settings and reiterate research imaging priorities so that minimum requirements were met.
Following this, our Platform adopted cross-cutting harmonization procedures to counteract variability in neuroimages acquired in multisite, multi-vendor studies. To further confirm data quality, human phantom data was collected across sites to assess variations in signal-to-noise ratio (SNR) and phantom geometry (Fu et al., 2006). During site initiation and onboarding, the same individual visited each study centre and was scanned using the standardized study research protocol at all sites participating in the prospective research-neuroimaging arm. This also allowed for the comparison of BOLD data acquisition and ensured that imaging stored within the Repository meets data standards.
4. Data sharing
In this study, we partnered with the Ontario Brain Institute (OBI) to leverage their existing neuroimaging storage service, BrainCODE (https://www.braincode.ca/content/getting-started#toc-2). BrainCODE is a large-scale imaging repository that allows investigators access to study data and features quality assurance pipelines to ensure data quality throughout the study. Imaging collected in the study is de-identified, named according to OBI conventions, and uploaded to BrainCODE. It can then be analyzed for quality, run through several analysis pipelines, and downloaded for local analyses. Following the study's conclusion, imaging data stored on BrainCODE are made available to outside collaborators for inclusion in large-scale data analyses.
5. Site Capacity Considerations
Several requirements were considered to determine each site's capacity to collaborate. Ideal sites had staffing resources, including a dedicated coordinator supported by study funding and (neuro)radiologists, MRI technologists and physicists, and the principal investigator, who was on-site and invested in research. The recruitment and data collection (i.e., multiple time points for neuroimaging, biosample and outcome measures) is labour-intensive and best executed at sites with financial and personnel resources to allocate to the study. These requirements were evaluated at the study's outset to ensure data collection and quality. Other requirements included the existing infrastructure at each site, the ability to enroll patients, and the ability to acquire the research-MRI protocols. The latter entailed the type of scanner, availability of MR technologists, their experience with functional MRI paradigms (resting state, task-based and breath holds) and capability to execute the collection of this dataset.
6. Results
Significant scientific advances using research-MRI techniques by participating centers include using cerebrovascular reactivity (using Blood Oxygen Level Dependant imaging [BOLD]) to predict stroke risk in children with moyamoya and sickle cell disease (Dlamini et al., 2020, Leung et al., 2016). In addition, the development of educational materials (including a webinar) by British Columbia Children’s Hospital, Vancouver, at the Inaugural Brain Canada Functional Cerebrovascular Imaging Symposium hosted at SickKids in 2015. Knowledge has been furthered for the design of future neuroimaging studies in pediatric stroke particularly with regard to determining protocol lengths (i.e. limiting scanning sessions to <60 minutes to improve compliance); the use of contrast (i.e., the time required to place IV lines, patient discomfort, concerns regarding renal function and potential gadolinium deposition); specific neurovascular imaging sequences (e.g., establishing a suitable delay time for arterial spin labelling and vessel wall imaging), and the optimal timing of injecting contrast for vessel wall imaging (Gulani et al., 2017).
7. Quality Assurance and Harmonization
To maximize data quality and improve homogeneity, great care was taken to harmonize parameters of the sequences collected across sites (see Supplementary Material, Table 1 for acquistion protocols). This was achieved by comparing pilot data scans, and working discussions between site technologists, neuroradiologists and the MR physicist at the lead site. The final cohort of collaborating centers included imaging collected on Siemens, GE and Philips scanners, with six 3.0T and two 1.5T MR scanners. Imaging data were collected on research-dedicated scanners in 2 sites and clinical scanners in the remaining six. The capability of the 3T MRI scanner (PrismaFIT, Siemens Healthineers) at the lead site to collect advanced imaging sequences in a shorter time frame due to better hardware performance, was an unforeseen hurdle. This led to timing parameters that could not be replicated each subsite. Despite this, the data collected at each site was found to be analogous and suitable for cross-site comparison.
In the human phantom study, a single volunteer (same person) was scanned utilizing the SILC protocol modified at each of the five scanners: British Columbia Children’s Hospital (BCH; General Electric (GE) 3 T Discovery MR750), Hospital for Sick Children Toronto (HSC; Siemens 3 T Prisma), Alberta Children’s Hospital (UCA; Siemens 3 T Magnetom Vida), Health Sciences Centre Children’s Hospital of Winnipeg (UMB; Siemens 1.5 T Aera), University of Colorado (UCD; Philips 3 T Ingenia). The FBIRN comparison was done using the phantom data, comparing the resting state data collected from British Columbia, SickKids, Alberta, Winnipeg, and Denver.
Functional MRI data were processed through an fBIRN pipeline to produce five summary QA measures, including Mean Signal Intensity, Mean Signal to Noise Ratio, and mean voxel smoothness as revealed by the Full Width at Half Maximum (FWHM) measurements in the X, Y, and Z directions. Each site’s five mean values were plotted as vertical lines in their corresponding plots, overlaid on fBIRN summary results from an independent database of harmonized fMRI scans, including 893 fMRI scans from 6 3 T Siemens scanners, 2,064 fMRI scans from 5 GE 3 T scanners, and 566 fMRI scans from 2 3 T Philips scanners. Consistent with the database results for Siemens vs GE scanners, the Siemens scanners of HSC, UCA and UMB generated very similar mean signal intensity values to one another. The signal intensity values were significantly lower than those generated by the two fMRI scans from the GE scanner at BCH (Note, UCD’s Philip’s scanner is a newer Ingenia model, relative to the older Achieva models from the Philips database and therefore not as comparable). The FWHM results from each site replicate the inherent vendor smoothing differences observed in the independent database for Siemens versus GE scanners, and are thus suited for post-processing corrections that will further harmonize these multisite data.
8. Genomics and proteomics
Blood and saliva samples were collected for storage and future analysis of genetic biomarkers of stroke subtypes (phenotype) and to clarify the causes and mechanisms of stroke in conjunction with the neuroimaging data. Saliva samples were collected using Oragene 500 kits (DNA Genotek, Ottawa, ON). Saliva samples are economical, non-invasive and do not require a trained phlebotomist or clinical laboratory. They have been shown to provide an acceptable yield of genomic DNA suitable for the identification of both small- (single nucleotide, insertion/deletion) and large-scale (copy number variation, structural variation) events (Trost et al., 2019), Standardized operating procedures were developed supporting the regulatory, quality control and logistic processes to ensure timely site activation, reliable collection, processing, storage and shipping of biosamples. Sites received support to add biosample collection and shipping to their regulatory documents and REB protocols.
9. Discussion
The creation of PEDSNIP was driven by the need to harness large clinical neuroimaging data acquired at multiple sites, and to address and overcome the challenges associated with conducting multisite prospective research imaging in pediatric stroke. There was also a need for updated neuroimaging protocols to capture the scope of pediatric stroke, including patient-specific primary and secondary morbidities. To address these needs, we built a Platform that enables the acquisition, pooling and shared analysis of comprehensive imaging data needed to conduct multicenter imaging studies of the developing brain. The Platform includes two parallel imaging arms, one with clinically-acquired MRI sequences images, and the other with research-acquired MRI sequences, with corresponding clinical data on pediatric stroke patients.
Although our paper is centered on MRI, the authors acknowledge that the use of computed tomography (CT) in pediatric stroke in the hyper-acute is equally as important. The Save ChildS Study reported the safety of endovascular treatment in pediatric stroke patients and showed that a majority of patients underwent CT. This is understandeable given that rapid imaging with CT with or without MRI is the first priority in order to confirm a suspicion of ischemia and to rule out hemorrhage or nonvascular disorders. There may also be contraindications for the use of MRI in children suspected of stroke including those with cardiac comorbidities and artificial heart devices. In addition, MRI may not be available or accessible in all settings. In PEDSNIP, CT images are also imported and stored, however, the use of MRI beyond the acute setting is an ongoing focus in our lab.
The planning and execution of this multi-layer Platform have been crucial components to ensure ongoing success. For each participating site, this includes a commitment from collaborating neurologists and radiologists, determining the feasibility of participating in this collaboration and facilitating the collection of required imaging in the clinical and research protocols. In addition, as the lead site, the SILC team has been responsible for defining clear objectives and developing the study proposal, securing funding for the Platform (Brain Canada Platform Support Grant), site support, image transfer, storage, data analysis, and managing the logistics of all of these processes. The latter is critical to the success of PEDSNIP due to the complexity and number of considerations paramount to the successful execution of these processes in the Platform. Currently, data collected from the first phase of PEDSNIP is being analyzed. This dataset now populates the neuroimaging Repository in the Platform and includes neuroimaging and clinical details on 1700 pediatric stroke patients.
The success of PEDSNIP is due to the commitment and efforts of the collaborative network of pediatric stroke experts and investigators (>150) and a diverse team of neuroradiologists (n = 5), data engineers (n = 4), graduate trainees and post-docs (n = 4). We published ten peer-reviewed manuscripts and presented 20 scientific abstracts in key journals, meetings and conferences worldwide. These include four seminal publications (Dlamini et al., 2017, Domi et al., 2017, Lee et al., 2017, Mirsky et al., 2017) that defined consensus neuroimaging guidelines for the management of neonatal (Lee et al., 2017) and childhood (Mirsky et al., 2017) stroke. Finally, we have conducted the first-ever, multisite, harmonized research-acquired neuroimaging study in a prospective cohort of children (n = 30 patients presenting with acute stroke and n = 30 controls, followed across four-time points over one year (see Table 2).
10. Future directions
We have now begun the second phase of PEDSNIP, where we continue to collect clinical imaging to increase our dataset (to approximately 2100 patients) and expand the biorepository to allow for future genomic approaches, including whole genome sequencing, through existing collaborations within the Platform. However, there remain barriers to large-scale analysis of the clinically-acquired imaging due to inadequate processing tools not designed for images that deviate from standardized normative brain maps, heterogeneity associated with variability in individual lesion characteristics, and differences in neuroimaging protocols across centres. Moreover, clinically- acquired MR images are often low resolution, particularly in two-dimensional acquisitions with thick slices and interslice gaps. Consequently, it is not possible to extract reliable quantitative structural (i.e., lesion, cortical and subcortical volume measures) from these images. This second phase aims to further develop our dataset into a transformational neuroinformatics Platform. To do this, our large multi-parametric and multi-domain data Platform will be leveraged to apply artificial intelligence (AI) to build predictive models from which biomarkers of risk, recurrence and outcomes in childhood stroke can be derived.
In brief, the imaging repository dataset will be used to train a machine-learning algorithm to identify stroke subtypes. These results will then be validated against manual expert assessment by a centralized neuroradiology adjudication committee consisting of an expert panel of three subspecialized pediatric neuroradiologists with expertise in pediatric stroke (Ertl-Wagner and Krishnan [Toronto], Stence [Denver]) and a fully trained radiologist (Sheng [Toronto]), with expertise in image segmentation. This team will confirm index stroke diagnosis through a centralized, structural assessment of acute and follow-up imaging according to pre-defined published criteria. This classification will establish a basis for the labels used to classify lesion characteristics across the Imaging Repository. Imaging data will be segmented using ischemic/infarcted brain parenchyma to determine lesion volumes. Lesion characteristics will be scored using these manual lesion segmentations as a reference standard. Finally, deep learning algorithms will be trained based on these segmentations to perform automated segmentations on the remaining neuroimaging data in the Repository.
11. Conclusion
To date, PEDSNIP has developed the largest cohort of longitudinal clinical imaging data on childhood stroke worldwide. Our Platform now integrates our existing multidisciplinary experts with new members in artificial intelligence, genomics and bioinformatics in a unique collaboration. Our goal is that these data models will not only serve our pediatric stroke population but will be translatable to other lesion-based injuries in the developing brain. This will provide an opportunity to improve understanding of the complex relationships between the brain, behaviour and critical outcomes of interest.
CRediT authorship contribution statement
Trish Domi: Conceptualization, Methodology, Visualization, Writing – original draft, Writing – review & editing, Formal analysis, Supervision, Project administration, Funding acquisition. Amanda Robertson: Methodology, Validation, Formal analysis, Investigation, Data curation, Visualization, Writing – original draft, Writing – review & editing, Supervision, Project administration. Wayne Lee: Software, Validation, Formal analysis, Investigation, Resources, Data curation, Supervision, Project administration. Richard Wintle: Methodology, Writing – review & editing. Nicholas Stence: Conceptualization, Methodology, Investigation, Visualization, Writing – review & editing, Project administration. Timothy Bernard: Conceptualization, Methodology, Investigation, Writing – review & editing, Project administration. Adam Kirton: Conceptualization, Methodology, Investigation, Writing – review & editing. Helen Carlson: Investigation, Writing – review & editing. Andrea Andrade: Investigation, Writing – review & editing. Mubeen Rafay: Conceptualization, Methodology, Investigation, Writing – review & editing. Bruce Bjornson: Conceptualization, Methodology, Investigation, Project administration. Danny Kim: Validation, Investigation, Resources. Michael Dowling: Conceptualization, Methodology, Investigation, Project administration. Wilmot Bonnett: Investigation, Project administration. Michael Rivkin: Conceptualization, Methodology, Investigation. Pradeep Krishnan: Methodology, Writing – review & editing. Manohar Shroff: Conceptualization, Resources, Writing – review & editing. Birgit Ertl-Wagner: Methodology, Resources, Writing – review & editing, Project administration. Stephen Strother: Methodology, Software, Validation, Resources. Steven Arnott: Methodology, Software, Validation, Resources, Data curation, Writing – review & editing. Max Wintermark: Conceptualization, Methodology, Writing – review & editing. Andrea Kassner: Conceptualization, Methodology, Formal analysis, Resources, Writing – review & editing, Supervision, Project administration. Gabrielle deVeber: Conceptualization, Methodology, Investigation, Writing – review & editing, Visualization, Supervision, Project administration, Funding acquisition. Nomazulu Dlamini: Conceptualization, Methodology, Investigation, Writing – review & editing, Visualization, Supervision, Project administration.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
The Pediatric Stroke Neuroimaging Platform was supported by funding from The Brain Canada Platform, Support Grant, The Auxilium Foundation, and The Nolen Hicks Family Fund.
Funding acquisition
*Author Contributions described by Contributor Role Taxonomy (CRediT) by Allen, L., O’Connell, A. and Kiermer, V. (2019), How can we ensure visibility and diversity in research contributions? How the Contributor Role Taxonomy (CRediT) is helping the shift from authorship to contributorship. Learned Publishing, 32: 71-74. https://doi.org/10.1002/leap.1210.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nicl.2023.103438.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- deVeber G.A., MacGregor D., Curtis R., Mayank S. Neurologic outcome in survivors of childhood arterial ischemic stroke and sinovenous thrombosis. J. Child Neurol. 2000;15(5):316–324. doi: 10.1177/088307380001500508. [DOI] [PubMed] [Google Scholar]
- deVeber G.A., Kirton A., Booth F.A., Yager J.Y., Wirrell E.C., Wood E., Shevell M., Surmava A.-M., McCusker P., Massicotte M.P., MacGregor D., MacDonald E.A., Meaney B., Levin S., Lemieux B.G., Jardine L., Humphreys P., David M., Chan A.K.C., Buckley D.J., Bjornson B.H. Epidemiology and Outcomes of Arterial Ischemic Stroke in Children: The Canadian Pediatric Ischemic Stroke Registry. Pediatr. Neurol. 2017;69:58–70. doi: 10.1016/j.pediatrneurol.2017.01.016. [DOI] [PubMed] [Google Scholar]
- Dlamini N., Wintermark M., Fullerton H., Strother S., Lee W., Bjornson B., Guilliams K.P., Miller S., Kirton A., Filippi C.G., Linds A., Askalan R., deVeber G. Harnessing Neuroimaging Capability in Pediatric Stroke: Proceedings of the Stroke Imaging Laboratory for Children Workshop. Pediatr. Neurol. 2017;69:3–10. doi: 10.1016/j.pediatrneurol.2017.01.006. [DOI] [PubMed] [Google Scholar]
- Dlamini N., Slim M., Kirkham F., Shroff M., Dirks P., Moharir M., MacGregor D., Robertson A., Deveber G., Logan W. Predicting ischemic risk using blood oxygen level–dependent MRI in children with moyamoya. Am. J. Neuroradiol. 2020;41(1):160–166. doi: 10.3174/ajnr.A6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domi T., Vossough A., Stence N.V., Felling R.J., Leung J., Krishnan P., Watson C.G., Grant P.E., Kassner A. The Potential for Advanced Magnetic Resonance Neuroimaging Techniques in Pediatric Stroke Research. Pediatr. Neurol. 2017;69:24–36. doi: 10.1016/j.pediatrneurol.2016.12.015. [DOI] [PubMed] [Google Scholar]
- Duchesne S., Chouinard I., Potvin O., Fonov V.S., Khademi A., Bartha R., Bellec P., Collins D.L., Descoteaux M., Hoge R., McCreary C.R., Ramirez J., Scott C.J.M., Smith E.E., Strother S.C., Black S.E., CIMA-Q group and the CCNA group The Canadian Dementia Imaging Protocol: Harmonizing National Cohorts. J. Magn. Resonance Imaging: JMRI. 2019;49(2):456–465. doi: 10.1002/jmri.26197. [DOI] [PubMed] [Google Scholar]
- Fu L., Fonov V., Pike B., Evans A.C., Collins D.L. Automated analysis of multi-site MRI phantom data for the NIHPD project. Med. Image Comput. Comput.-Assisted Intervent. 2006;9(Pt 2):144–151. doi: 10.1007/11866763_18. [DOI] [PubMed] [Google Scholar]
- Fullerton H.J., Chetkovich D.M., Wu Y.W., Smith W.S., Johnston S.C. Deaths from stroke in US children, 1979 to 1998. Neurology. 2002;59(1):34–39. doi: 10.1212/wnl.59.1.34. [DOI] [PubMed] [Google Scholar]
- Ganesan V., Hogan A., Shack N., Gordon A., Isaacs E., Kirkham F.J. Outcome after ischaemic stroke in childhood. Dev. Med. Child Neurol. 2000;42(7):455–461. doi: 10.1017/s0012162200000852. [DOI] [PubMed] [Google Scholar]
- Goeggel Simonetti B., Cavelti A., Arnold M., Bigi S., Regényi M., Mattle H.P., Gralla J., Fluss J., Weber P., Hackenberg A., Steinlin M., Fischer U. Long-term outcome after arterial ischemic stroke in children and young adults. Neurology. 2015;84(19):1941–1947. doi: 10.1212/WNL.0000000000001555. [DOI] [PubMed] [Google Scholar]
- Greenham M., Gordon A., Anderson V., Mackay M.T. Outcome in Childhood Stroke. Stroke. 2016;47(4):1159–1164. doi: 10.1161/STROKEAHA.115.011622. [DOI] [PubMed] [Google Scholar]
- Gulani V., Calamante F., Shellock F.G., Kanal E., Reeder S.B. Gadolinium deposition in the brain: summary of evidence and recommendations. Lancet Neurol. 2017;16(7):564–570. doi: 10.1016/s1474-4422(17)30158-8. [DOI] [PubMed] [Google Scholar]
- Kitchen L., Westmacott R., Friefeld S., MacGregor D., Curtis R., Allen A., Yau I., Askalan R., Moharir M., Domi T., deVeber G. The pediatric stroke outcome measure: a validation and reliability study. Stroke. 2012;43(6):1602–1608. doi: 10.1161/STROKEAHA.111.639583. [DOI] [PubMed] [Google Scholar]
- Lee S., Mirsky D.M., Beslow L.A., Amlie-Lefond C., Danehy A.R., Lehman L., Stence N.V., Vossough A., Wintermark M., Rivkin M.J. Pathways for Neuroimaging of Neonatal Stroke. Pediatr. Neurol. 2017;69:37–48. doi: 10.1016/j.pediatrneurol.2016.12.008. [DOI] [PubMed] [Google Scholar]
- Leung J., Duffin J., Fisher J.A., Kassner A. MRI-based cerebrovascular reactivity using transfer function analysis reveals temporal group differences between patients with sickle cell disease and healthy controls. Neuroimage Clin. 2016;12:624–630. doi: 10.1016/j.nicl.2016.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew S.-L., Zavaliangos-Petropulu A., Jahanshad N., Lang C.E., Hayward K.S., Lohse K.R., Juliano J.M., Assogna F., Baugh L.A., Bhattacharya A.K., Bigjahan B., Borich M.R., Boyd L.A., Brodtmann A., Buetefisch C.M., Byblow W.D., Cassidy J.M., Conforto A.B., Craddock R.C., Thompson P.M. The ENIGMA Stroke Recovery Working Group: Big data neuroimaging to study brain-behavior relationships after stroke. Hum. Brain Mapp. 2022;43(1):129–148. doi: 10.1002/hbm.25015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirsky D.M., Beslow L.A., Amlie-Lefond C., Krishnan P., Laughlin S., Lee S., Lehman L., Rafay M., Shaw D., Rivkin M.J., Wintermark M. Pathways for Neuroimaging of Childhood Stroke. Pediatr. Neurol. 2017;69:11–23. doi: 10.1016/j.pediatrneurol.2016.12.004. [DOI] [PubMed] [Google Scholar]
- Mueller S.G., Weiner M.W., Thal L.J., Petersen R.C., Jack C., Jagust W., Trojanowski J.Q., Toga A.W., Beckett L. The Alzheimer’s Disease Neuroimaging Initiative. Neuroimaging Clin. N. Am. 2005;15(4):869–877. doi: 10.1016/j.nic.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slim M., Fox C.K., Friefeld S., Dlamini N., Westmacott R., Moharir M., MacGregor D., deVeber G., Investigators S.I.P.S. Validation of the pediatric stroke outcome measure for classifying overall neurological deficit. Pediatr. Res. 2020;88(2):234–242. doi: 10.1038/s41390-020-0842-5. [DOI] [PubMed] [Google Scholar]
- Trost B., Walker S., Haider S.A., Sung W.W.L., Pereira S., Phillips C.L., Higginbotham E.J., Strug L.J., Nguyen C., Raajkumar A., Szego M.J., Marshall C.R., Scherer S.W. Impact of DNA source on genetic variant detection from human whole-genome sequencing data. J. Med. Genet. 2019;56(12):809–817. doi: 10.1136/jmedgenet-2019-106281. [DOI] [PMC free article] [PubMed] [Google Scholar]
Further reading
- Sporns P.B., Sträter R., Minnerup J., Wiendl H., Hanning U., Chapot R., Henkes H., Henkes E., Grams A., Dorn F., Nikoubashman O., Wiesmann M., Bier G., Weber A., Broocks G., Fiehler J., Brehm A., Psychogios M., Kaiser D., Yilmaz U., Morotti A., Marik W., Nolz R., Jensen-Kondering U., Schmitz B., Schob S., Beuing O., Götz F., Trenkler J., Turowski B., Möhlenbruch M., Wendl C., Schramm P., Musolino P., Lee S., Schlamann M., Radbruch A., Rübsamen N., Karch A., Heindel W., Wildgruber M., Kemmling A. Feasibility, Safety, and Outcome of Endovascular Recanalization in Childhood Stroke: The Save ChildS Study. JAMA Neurol. 2020;77(1):25–34. doi: 10.1001/jamaneurol.2019.3403. PMID: 31609380; PMCID: PMC6802048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.