Abstract
Induced pluripotent stem cells (iPSCs) have enormous potential in producing human tissues endlessly. We previously reported that type V collagen (COL5), a pancreatic extracellular matrix protein, promotes islet development and maturation from iPSCs. In this study, we identified a bioactive peptide domain of COL5, WWASKS, through bioinformatic analysis of decellularized pancreatic ECM (dpECM)-derived collagens. RNA-sequencing suggests that WWASKS induces the formation of pancreatic endocrine progenitors while suppressing the development of other types of organs. The expressions of hypoxic genes were significantly downregulated in the endocrine progenitors formed under peptide stimulation. Furthermore, we unveiled an enhancement of iPSC-derived islets’ (i-islets) glucose sensitivity under peptide stimulation. These i-islets secrete insulin in a glucose responsive manner. They were comprised of α, β, δ, and γ cells and were assembled into a tissue architecture similar to that of human islets. Mechanistically, the peptide is able to activate the canonical Wnt signaling pathway, permitting the translocation of β-catenin from the cytoplasm to the nucleus for pancreatic progenitor development. Collectively, for the first time, we demonstrated that an ECM-derived peptide dictates iPSC fate toward the generation of endocrine progenitors and subsequent islet organoids.
Keywords: Short peptide, pancreatic endocrine progenitor, islet organoid, RNA-sequencing, iPSC differentiation, canonical Wnt signaling pathway
Introduction
The shortage of cadaveric human islets necessitates an alternative source for islets. Islets derived from differentiation of human pluripotent stem cells (hPSCs) are promising islet cell sources. 1 A tissue microenvironment plays a paramount role in in vitro islet generation.2,3 Tissue niches such as insoluble signals, including extracellular matrix (ECM), coordinate with soluble signals such as growth factors, to regulate cell proliferation, differentiation, and maturation.4,5 For example, Rezania et al. investigated a panel of small molecules and growth factors that can promote stepwise differentiation of hPSCs into pancreatic β cells. 6 Chmielowiec et al. reported that growth factors secreted from mesenchymal and endothelial cells facilitate β cell differentiation from human embryonic stem cells (hESCs). 4 We also revealed that porous membrane substrates, polyester and polyethylene terephthalate, facilitate hPSC metabolism, self-renewal, and differentiation.5,7 Furthermore, we reported that a three-dimensional collagen scaffold augments the generation of pancreatic endoderm and endocrine from hESCs. 8 Our recent study unveiled that angiopoietin-2, a secreted factor found in pancreatic tissue, endorses in vitro islet development from hPSCs. 9 While extensive effects have been made to understand tissue niches for in vitro pancreatic endocrine development, an understanding of key molecules involved remains incomplete. In our previous studies, we have discovered that type V collagen (COL5), a decellularized pancreatic ECM (dpECM) protein, induces islet organogenesis and morphogenesis from both human induced pluripotent stem cells (iPSCs) and hESCs. 3 Our study hinted at the existence of peptide motifs critical to islet development and maturation.
There have been only a few developmental peptides identified to date. These peptides have been found to be able to facilitate stem cell differentiation into varied tissues, including epidermis, mesenchyme, nervous, neurons, 10 lung cells, 11 and osteogenic lineages. 12 A short peptide with amino acid sequence KEDW known as pancragen has led to increased expression of pancreatic markers PDX1, FOXA2, and NKX2.2 for pancreatic acinar cells MIA PaCa-2. 13 Pancragen is a synthetic analog of a peptide obtained from cattle pancreas, which penetrates to the nucleus. 13 Treatment of pancreatic acinar cells with pancragen induces the trans-differentiation of insulin, glucagon, somatostatin, and pancreatic polypeptide producing cells. 13 Other molecules of interest, including glucagon-like peptide 1 (GLP-1) and its agonists, have been found to be able to enhance insulin production and secretion in rat islet cell lines. 14 Treatment of the rat AR42J cell line with GLP-1 agonists leads to an increase in proliferation and expression of insulin, glucagon, and somatostatin in the cells. 14 GLP-1 promotes differentiation of rat exocrine cells or immature islet progenitors toward a more differentiated β cell phenotype. 15 Previous studies also suggested that the peptides RGD, PDSGR, and LRE derived from laminins enhanced human islet viability and glucose induced insulin secretion when added to alginate-encapsulated islets. 16 However, dpECM protein-derived peptides that promote iPSC differentiation into pancreatic endocrine lineages and subsequently islet organoids have not been reported. In this study, we discovered a bioactive short peptide as a chemically defined niche for iPSC pancreatic differentiation. RNA-sequencing (RNA-seq) revealed that it suppresses the development of other types of organs while promoting the generation of pancreatic endocrine lineages. It is able to enhance iPSC-derived islet organoids (i-islets)’ glucose sensitivity and augment the insulin stimulation index. The formed i-islets comprised of α, β, δ, and γ cells. They were assembled into a tissue architecture similar to that of human islets. As these distinct endocrine cells are required synergistically to regulate islet functions, 17 techniques that permit the in vitro generation of islet organoids consisting of all the major islet cell types are highly desired. The ECM-derived peptide discovered in this study could help unlock the tissue niches indispensable to islet development and maturation, leading to the development of chemically defined differentiation media for producing clinically relevant islets for diabetes research and treatment.
Materials and methods
Identification of dpECM-derived peptide motifs
A Basic Local Alignment Search Tool (BLAST) alignment was carried out to compare the amino acid sequences of different collagen types. From these alignments, each group of three or more similar or identical amino acids was cross-referenced with the amino acid sequences of each subunit for collagens type I through V. Each amino acid sequence used in this study was obtained from the UniProt database, and amino acid similarity was judged based on the BLOSUM 62 matrix. Peptides were determined to be regions of interest if they were present in collagen V but not the other types. Multiple alignment of each collagen type was done using COBALT to further validate the peptides identified. Three dimensional models of the collagen subunits obtained from the Swiss-Model Repository were also used to predict locations of the peptide of interest on the proteins.
Stem Cell culture
Human iPSC line IMR90 from WiCell Research Institute were routinely cultured in the mTeSR1 medium (StemCell Technologies) on 1:100 diluted growth factor reduced Matrigel (Corning) coated dishes in a humidified 5% CO2 37°C incubator. Cells were passaged every 2 or 3 days at ratios of 1:3 to 1:4 using Dispase (StemCell Technologies), as described in our previous study. 18
Stepwise differentiation
A 5-stage serum-free differentiation protocol based on our previous study was used to induce iPSC differentiation into pancreatic lineage with slight modification shown in Figure 5(a).3,9 Briefly, iPSCs were treated with Accutase (Stemcell Technologies) and seeded to Matrigel-coated plates at 0.5 million cells/ml. In Stage 1 (definitive endoderm, 5 days): cells were cultured in RPMI 1640 (Corning) supplemented with 50 ng/ml activin A (PeproTech), 1 mM sodium butyrate (NaB, Sigma-Aldrich), and B27 (Gibco) for 24 h. The concentration of NaB was reduced to 0.5 mM for the next 4 days. In Stage 2 (posterior foregut, 5 days): cells were cultured in RPMI 1640 supplemented with 50 ng/ml keratinocyte growth factor (KGF, PeproTech), 100 ng/ml Noggin (PeproTech), 250 µM ascorbic acid (Vc, Sigma-Aldrich), 1 µM retinoic acid (RA, Sigma-Aldrich), 300 nM (-)-indolactam V (ILV, AdipoGen), 100 nM LDN193189 (LDN, Sigma-Aldrich), and B27 for 5 days. In Stage 3 (pancreatic progenitor, 5 days): cells were cultured in DMEM/F12 (Gibco) supplemented with 1 µM RA, 300 nM ILV, 200 nM LDN, 10 µg/ml heparin (HP, Sigma-Aldrich), 1 µM 3,3′,5-Triiodo-L-thyronine sodium salt (T3, Sigma-Aldrich), 10 µM RepSox (Rep, MedChemExpress), B27, and 20 mM glucose (Sigma) for 5 days. In Stage 4 (endocrine lineage, 7 days): cells were cultured in RPMI 1640 supplemented with 10 µg/ml HP, 1 µM T3, 10 µM Rep, 1 mM N-acetyl cysteine (N-Cys, Sigma-Aldrich), 0.5 µM R428 (SelleckChem), 10 μM trolox (Enzo Life Sciences), 100 nM γ-secretase inhibitor XX (SiXX, Millipore), 10 mM nicotinamide (Nic, Sigma-Aldrich), B27, and 20 mM glucose for 7 days. At end of stage 4, cells were detached with Dispase, and transferred to 24-well ultra-low attachment plate for 3D culture. In Stage 5 (mature endocrine cells, 9–13 days): cells were cultured in CMRL supplement (Corning) containing 2% bovine serum albumin (BSA, Sigma-Aldrich), 1 µM T3, 10 µM Rep, 100 nM SiXX, and 10 mM Nic for 9–13 days. Angiopoietin-2 (Ang2, Peprotech) was applied to differentiation media at 20 ng/ml from Stage 4 until the end of Stage 5. 9 The differentiation media were exchanged every other day. For 3D culture, half of the medium was changed every other day. The peptide, synthesized by Biomatik (Canada) was added to the differentiation medium of Stage 2 (posterior foregut) at 40 ng/ml.
Figure 5.
The peptide promotes the generation of insulin secreting cells during iPSC-pancreatic endocrine differentiation with enhanced glucose responsive insulin secretion of i-islets. (a) A schematic diagram of a five-stage islet development protocol (DE: definitive endoderm; PF: posterior foregut; EP: endocrine progenitor; EL: endocrine lineage; EC: mature endocrine cells). iPSCs were cultured and induced to differentiate to islet organoids on Matrigel-coated plates. The peptide (P1) was added to the differentiation medium during Stage 2. At the end of differentiation, the i-islets were immunofluorescently labeled for (b) C-peptide (CP, red) and glucagon (GCG, green); (c) pancreatic polypeptide (PPY, red) and somatostatin (SST, green); (d) CP (red) and MAFA (green). (e) Representative images for GCG (red) and MAFB (green). Cells were counterstained with DAPI (blue or gray). Scale bars: 50 μm. (f–i) Semi-quantitative analysis of cellularity of the i-islets corresponding to individual representative images. Image analysis was performed using ImageJ software (n = 9–12 images for each condition). Results are shown as mean ± SD. ***: p < 0.001 compared to the control group. (j) The insulin stimulation index from i-islets generated in the control and P1 groups by glucose-stimulated insulin secretion analysis (n = 5 for each group). Results are shown as mean ± SD. *: p < 0.05.
TaqMan quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from cells using RNeasy Mini Kit (Qiagen). RNA was quantified using a Synergy H1 Microplate Reader. qRT-PCR was performed using QuantiTect Multiplex PCR Kit (Qiagen) on a CFX Connect Real-Time PCR system (Bio-Rad) as described in our previous work. 5 Primer-probe sets used were listed in Supplemental Table S1.
RNA-sequencing
RNA samples were sent to LC Sciences (Houston, TX) for library preparation, mRNA-sequencing, and analysis. It was performed on an Illumina Novaseq 6000 sequencing system. Sequencing libraries were constructed using a TruSeq stranded mRNA library kit from Illumina. Approximately 40 million reads were obtained for each sample. FASTQ files were aligned to the GRCh38 human reference. Differentially expressed genes were analyzed using EdgeR algorithm. Genes in the peptide treated group that are up- or down-regulated more than 1.5-fold from the control group with a p-value of <0.05 were considered significant. Gene set enrichment analysis (GSEA) was carried out at https://www.gsea-msigdb.org/gsea/index.jsp, heatmaps were generated at http://www.heatmapper.ca/expression/, and the BLAST was used to help identify genes of interest.
Flow cytometry
Cells were washed twice with PBS and digested using trypsin-EDTA. The cells were fixed and permeabilized using Foxp3/Transcription Factor Fixation/Permeabilization solution (Thermo Fisher Scientific). The cells were stained with primary and secondary antibody or isotype antibodies (Supplemental Table S2), and resuspended in PBS containing 2% bovine serum albumin (Gibco), followed by analyzing on ZE5 Cell Analyzer (Bio-Rad). Statistics and graphing were performed using FlowJo (BD Biosciences), and gating was determined using differentiated cells stained with secondary antibodies and isotype antibodies.
Immunofluorescence staining and microscopy
Cells were fixed and permeabilized using the Foxp3/Transcription Factor Fixation/Permeabilization solution as described in our previous work. 9 After subsequent staining and washing with primary and secondary antibodies, the cell nuclei were counterstained with a mounting medium containing DAPI (Vector Laboratories). The cells were examined under a Zeiss 880 multiphoton laser scanning microscope. Samples processed with secondary antibodies and isotype antibodies were used as negative controls. The antibodies used in immunostaining were listed in Supplemental Table S2.
Cryosectioning for immunofluorescence microscopy
The i-islets were rinsed with PBS, fixed with 4% paraformaldehyde at room temperature for 1–2 h, and washed with PBS. They were incubated in 30% sucrose (w/v) at 4°C overnight, embedded in Optimal Cutting Temperature compound (OCT) (Thermo Fisher Scientific) and stored at 4°C overnight, after which the samples were snap frozen in liquid nitrogen. Frozen samples were cryosectioned at 10 µm and the sections were mounted on TruBond adhesive slides (Electron Microscopy Sciences) for staining with primary and secondary antibodies, as described elsewhere. 3 The cell nuclei were counterstained with ProLong Diamond Antifade Mountant with DAPI (ThermoFisher Scientific) for imaging using a Zeiss 880 multiphoton laser scanning microscope. For calculating the percentage of different cell types in the organoids, the images (n = 9–12) of aggregates were quantified using the ImageJ software (Version 1.53t). Samples processed with secondary antibodies were used as negative controls. The antibodies used were listed in Supplemental Table S2.
Glucose stimulated insulin secretion analysis
At the end of differentiation, organoids were washed and incubated in Krebs–Ringer buffer solution (KRB, Boston BioProducts) containing 1 mM glucose for 4 h at 37°C in a water bath. After rinsing with KRB, aggregates were stimulated with KRB containing 2 mM glucose, 20 mM glucose, and 30 mM KCl, respectively, at 37°C in the water bath for 30 min. The respective supernatants were collected. The number of organoids from each sample were counted using an Olympus phase contrast microscope and used for normalization. Glucose stimulated insulin secretion was performed using an insulin enzyme-link immunosorbent assay (ELISA) kit (Mercodia). 3 The aggregates around 100–300 µm were counted as organoids. Insulin stimulation index was calculated as the ratio of insulin secreted in high (20 mM) to low glucose (2 mM).
Immunostaining of iPSC-derived cells treated with FITC-conjugated peptide and cell fractionation
The peptide WWASKS was synthesized and fluorescently labeled with FITC by Biomatik. The IMR90 cells were seeded on Matrigel-coated Nunc Lab-Tek 8-well chambered cover glass (Thermo Fisher Scientific) and cultured in mTeSR1 for 24 h. The cells were differentiated to definitive endoderm, and then treated with 60 µg/ml fluorescently labeled peptide (P1-FITC) in a posterior foregut differentiation medium for 24 h. The cells were then fixed with 4% paraformaldehyde for 10 min and washed with PBS three times. The cell membrane was stained using CellMask Deep Red plasma membrane stain (Thermo Fisher Scientific) and incubated at 37°C for 10 min. After rinsing with PBS, the cells were incubated with a mounting medium containing DAPI. The samples were imaged using a Zeiss 880 multiphoton laser scanning microscope. Differential interference contrast (DIC) images were used as confocal transmitted scan. Samples without P1-FITC treatment and CellMask Deep Red plasma membrane staining were used as negative controls. Cell fractionation was performed by isolating cytosol, membrane, and nucleus from the FITC-peptide treated group using a cell compartment kit from QIAGEN. The FITC-peptide untreated group served as a control. The presence of the peptide in these cellular fractions was determined by detecting FITC-peptide using a Synergy H1 microplate reader. The experiments were carried out in quadruplicates.
Western blot analysis
Immunoblotting was performed as detailed in our early study. 5 Briefly, total proteins were extracted using Pierce RIPA cell lysis buffer (Thermo Fisher Scientific). Protein concentration was quantified by BCA assay. The protein samples was loaded to the 4–20% Precast Protein Gels (Bio-Rad) and the electrophoresis was performed under 70 V for 20 min, followed by 200 V for 25 min. The proteins were then blotted to PVDF membranes. The membranes were incubated in 5% nonfat milk in Tris-buffered saline with 0.1% Tween (TBST) containing 150 mM NaCl, 25 mM Tris-HCl, and 0.1% Tween 20 (pH 7.0), for 1 h to prevent nonspecific binding. Primary antibody was added to the membrane containing buffer and incubated at 4°C overnight. After washing using TBST, the membranes were further incubated with HRP-conjugated secondary antibody at room temperature for 1 h, followed by washing with TBST. The SuperSignal West Pico Plus Chemiluminescent Substrate (Fisher Scientific) was used to detect protein expressions on the blot. Cytoplasmic and nuclear proteins were extracted using a kit from Thermo Fisher Scientific. HRP-conjugated β-actin antibody (Sigma) served as a loading control for the assay. Proteins were visualized with chemiluminescent substrate reagents (Fisher Scientific). The antibodies used in Western blot were listed in Supplemental Table S3.
Statistical analysis
Statistical significance was calculated by unpaired two-tailed Student’s t-test and p < 0.05 was recognized as statistically significant. Data analysis was performed using Prism 7.0 (GraphPad Software Inc.). Numeric data were shown as means ± standard deviation (SD) if not otherwise indicated and were derived from at least three independent experiments.
Results and discussion
Identify dpECM-derived peptide motifs that endorse pancreatic lineage specification
In our previous study, we have discovered a dpECM protein COL5 capable of inducing islet organogenesis and morphogenesis during iPSC pancreatic differentiation. 3 To identify peptide motifs of COL5 that possess developmental activities to induce endocrine lineage specification during iPSC pancreatic differentiation, we performed BLAST and COBALT alignments between each subunit of COL5 and other collagen types shown in Figure 1(a). The probability of a substitution between two amino acids with a positive score on the BLOSUM 62 matrix is greater than that of random chance. 19 This is indicative of biochemical similarity between the two amino acids. From these alignments, we discovered a short peptide motif, WWASKS that is unique to COL5. It is present in the α2 subunit of COL5. We used 3D models of the collagen subunits obtained from the Swiss-Model Repository to predict the locations of the WWASKS peptide on COL5 (Figure 1(b) and (c)).
Figure 1.
Enhanced generation of endocrine progenitors in WWASKS stimulated iPSC pancreatic differentiation. (a) Alignment of collagen subtypes with region of interest highlighted. (b) Zoomed-in tertiary structure of peptide sequence WWASKS obtained from the Swiss-Model Repository. Arrows indicate the first and last amino acids of the peptide. (c) Full view of type V collagen with red box indicating the peptide location. (d) A schematic diagram of a three-stage endocrine progenitor development protocol (DE: definitive endoderm; PF: posterior foregut; EP: endocrine progenitor). iPSCs were differentiated into pancreatic progenitor cells. The peptide (P1) was added to the differentiation medium during posterior foregut differentiation stage. (e) NKX6.1+ PDX1+ cell subsets detected in the peptide and control groups. (f) Average percentages of NKX6.1 and PDX1 and co-expressing cells in iPSC-derived pancreatic progenitors. Results are shown as mean ± SD (n = 3). *: p < 0.05 compared to the control group. (g and h) NKX6.1 and PDX1 expressing cells in iPSC-derived pancreatic progenitors that were immunofluorescently labeled for NKX6.1 (red) and PDX1 (green). Cells were counterstained with DAPI (blue). Scale bar: 200 µm.
The dpECM protein-derived peptide endorses endocrine progenitor lineage specification from iPSCs
To evaluate the developmental activity of the candidate peptide, we differentiated iPSCs into pancreatic progenitors in the presence of the WWASKS during the stage of posterior foregut formation (Figure 1(d)). At the end of the endocrine progenitor stage, we performed a dual-color flow cytometric analysis of NKX6.1 and PDX1 dual-positive cells in the WWASKS group as well as in the control group (Figure 1(e)), as mounting evidence shows that the concurrent expression of PDX1 and NKX6.1 during endocrine progenitor cell maturation is associated with enhanced differentiation of iPSCs into mature endocrine cells.20,21 We found that the percentage of NKX6.1+ PDX1+ cells in the peptide group was considerably higher than that in the control group, with values of 65% versus 53% (Figure 1(e)). Additionally, the percentage of NKX6.1− PDX1− cells was noticeably lower in the peptide group. It was 22.3% in the peptide treated group as compared to 31.4% in the control group (Figure 1(f)). Next, we confirmed the upregulated expression and co-localization of NKX6.1 and PDX1 in the peptide group through immunofluorescence microscopy (Figure 1(g) and (h)). These imaging results were in agreement with quantitative measurements detected by flow cytometry (Figure 1(e) and (f)).
Assessment of the effect of the identified peptide on iPSC pancreatic lineage differentiation by global transcriptome profiling
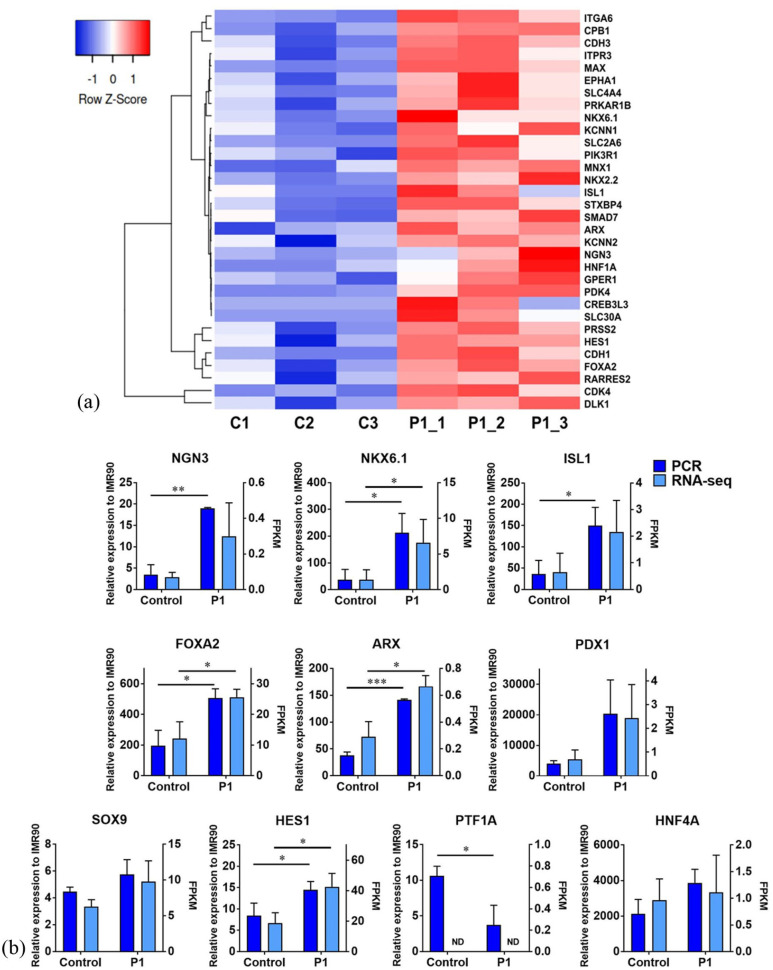
To ascertain the effect of the WWASKS peptide on pancreatic lineage differentiation, we interrogated global transcriptome profiles of the peptide treated and the control groups by RNA-seq. We observed upregulation of 732 genes and downregulation of 678 genes in the peptide group. Approximately 59,201 genes were not affected by the WWASKS stimulation (Supplemental Figure S1). A total of 30 genes associated with pancreatic development and function were identified to be significantly upregulated in the peptide treated group (Figure 2(a)). Generally, these genes correspond to Gene Ontology (GO) terms relating to the pancreas, β cell development, and regulation of insulin secretion in GO terms. These genes include FOXA2, ARX, NKX2.2, NKX6.1, E-cadherin (CDH1), HNF1α, ISL1, etc. For instance, there was a considerable enhancement of NKX6.1 expression under the WWASKS stimulation. NKX6.1 is a key transcription factor that is important for β cell development, function, and proliferation. It plays a key role in insulin production and secretion. 22 Studies showed that the inactivation of NKX6.1 in mice leads to the development of diabetes. 22 Furthermore, the knockout of NKX6.1 causes β cells to lose their identity and to behave similarly to δ cells. 22 ARX is another upregulated gene in the peptide group known to play a critical role in the development of pancreatic α cells and pancreatic polypeptide cells. It is a key transcription factor for α cells,23,24 necessary for ɑ cell differentiation and is expressed by both islet and acinar cells.25,26 When ARX gene is knocked out from endocrine cells, there are decreased numbers of lineages positive for pancreatic polypeptide, glucagon, and insulin, 24 suggesting the importance of ARX in endocrine function. NKX2.2 is a homeodomain transcription factor that plays an essential role in pancreatic endocrine cell specification and differentiation in mouse development. It is also significantly upregulated in the peptide group, with a 6.64-fold increased expression compared to the control group. Even after development, this transcription factor plays an important role in β cell function. 27 NKX2.2-repressor transgenic mice exhibit both a loss of β cell function and islet architecture. 27 Another gene, CDH1, was upregulated in the peptide group. CDH1, an islet gene, plays an important role in islet cell-cell communications, islet function, and β cell proliferation.28,29 Therefore, upregulation of CDH1 is indicative of the efficient differentiation of iPSCs into endocrine progenitors. In addition, the differential expression of genes associated with pancreatic dysfunction were significantly downregulated in the peptide group compared to the control (Supplemental Figure S3). These genes are associated with GO terms such as “endocrine resistance,” “insulin resistance,” and “type II diabetes mellitus.” Additionally, we found that an additional 49 pancreatic signature genes were also upregulated considerably in the peptide group (Supplemental Figure S2). The increased expression of these genes under WWASKS stimulation suggested the enhanced iPSC differentiation toward endocrine progenitor cells.
Figure 2.
Enhanced gene expression of endocrine progenitor signatures in WWASKS stimulated iPSC pancreatic differentiation. iPSCs were differentiated into pancreatic progenitor cells in the absence (Control) or presence of the WWASKS (P1) and differential gene expression analysis was carried out. (a) The peptide increases the expression of genes associated with pancreatic function. C1 ~ C3 and P1_1~P1_3 represent three batches of RNA-seq results from control and peptide group (n = 3). (b) The y-axis at left side represents the expression levels quantified and normalized to iPSCs by qRT-PCR. The y-axis at right side represents the gene expressions quantified by RNA-seq. Results are from three independent experiments and shown as mean ± SD. *: p < 0.05; and **: p < 0.01 compared to the control group. ND: not detected.
To cross-check RNA-seq results, we performed TaqMan qRT-PCR to examine the expression of key endocrine progenitor marker genes and islet genes, including NGN3 and PDX1, pancreatic islet markers NKX6.1, ISL1, ARX, and FOXA2; pancreatic ductal progenitor maker SOX9; pancreatic acinar progenitor markers HES1 and PTF1A; and hepatocyte marker HNF4A (Figure 2(b)). Differentiation in the absence of the WWASKS served as a control. We observed a remarkable elevation of NKX6.1, NGN3, ISL1, FOXA2, and ARX expressions in the peptide group. The expression levels of NKX6.1, NGN3, ISL1, FOXA2, and ARX were 5.7, 5.4, 4.1, 2.6, and 3.7-fold upregulated, respectively, in the WWASKS group (Figure 2(b)). These increases align with those analyzed by RNA-seq (Figure 2(a) and (b)). NGN3 is a master regulator of endocrine cell development necessary for islet cell differentiation. 30 It plays a crucial role in the establishment of endocrine function in the pancreas. 31 The upregulation of this gene in the WWASKS group is indicative of enhanced development of endocrine progenitors under the peptide stimulation. Interestingly, we observed a significant upregulation of ISL1 expression in the peptide group (Figure 2(a) and (b)). ISL1 is a pancreatic marker that plays an imperative role in the proliferation and differentiation of pancreatic endocrine progenitors. 32 ISL1 knockout in murine β cells leads to insufficient insulin secretion and decreased glucose tolerance. 32
To further examine the effect of the WWASKS on the formation of pancreatic acinar and duct progenitors, we characterized the expressions of pancreatic acinar and duct progenitor markers PDX1, PTF1α, SOX9, and HES1 by both qRT-PCR and RNA-seq. Our data suggested that the expressions of PDX1 and SOX9 were not affected by the WWASKS stimulation (Figure 2(b)). Interestingly, PTF1α was found to be downregulated by ~3.3-fold and HES1 showed ~2-fold increase in the peptide group. PTF1α is necessary for acinar cell maintenance. 33 PTF1α and SOX9 are typically downregulated in endocrine cells.34,35 These studies suggested that a decreased expression of PTF1α results in a shift in cell fate toward endocrine cells. 34 Besides, acinar cell differentiation is accelerated by HES1 inactivation. 35 It seems that the acinar cell lineage specification is synergistically suppressed due to the upregulated expression of HES1 by WSSASKS. Overall, a decreased expression of PTF1α and an increased expression of both NGN3 and ISL1 under the peptide stimulation endorse the generation of endocrine progenitor cells from iPSCs. Furthermore, no significant difference in the expression level of HNF4A, a hepatocyte marker, was observed between the peptide and control groups. PDX1 is a marker for both acinar/duct progenitors and endocrine progenitors, and SOX9 is a pancreatic ductal progenitor marker. There were no significant differences in gene expression of these two markers, suggesting that the peptide favors the pancreatic endocrine progenitor lineage specification from iPSCs. The comparison of a panel of key gene expressions unveiled a general tendency of consistency between qRT-PCR and RNA-seq analyses (Figure 2(b)). However, qRT-PCR demonstrated more profound sensitivity and specificity due to the nature of highly sensitive and targeted detection of gene expression by TaqMan RT-PCR. 36
Remarkably, pancreatic tip progenitor cell marker genes, including FOXF1, GLI3, TBX3, and FGFR1, were not differentially expressed between the control and peptide groups (Supplemental Figure S4). Thus, our results indicate that the cells stimulated by the peptide are more committed toward pancreatic endocrine lineages. Overall, these results revealed that the peptide is able to substantially promote the differentiation of iPSCs into pancreatic progenitors.
The peptide facilitates iPSC lineage commitment by downregulating hypoxia-associated genes
A hypoxic niche is beneficial to maintaining undifferentiated state of pluripotent stem cells.37,38 iPSCs heavily rely on glycolysis. 39 Interestingly, we found considerable enrichment of hypoxia-associated genes in the control group (p-value = 0.000) when performing GSEA to compare the peptide with the control group (Figure 3(a)). Figure 3(b) illustrates the expression results for genes typically upregulated under hypoxic conditions. Of the 12 differentially expressed genes, 10 were significantly downregulated in the peptide group. One example of a hypoxia-associated gene showing remarkably reduced expression in the peptide group is DNA damage inducible transcript 4 (DDIT4) (Figure 3(b)), a marker for both glycolysis and hypoxia. 40 We confirmed DDIT4 protein expression by Western blot analysis (Figure 3(d) and (e)). There was approximately 61% reduction in DDIT4 protein expression in the peptide group. Another example of a hypoxia-associated gene with significantly reduced expression in the peptide group is polo-like kinase 2 (PLK2). Its expression is downregulated under hypoxia. 41 We detected a significant reduction in PLK2 expression in the peptide group (Figure 3(d) and (e)). In addition, placental growth factor (PGF) is known to be involved in angiogenesis 42 and is released under hypoxic conditions. 42 Prolyl 4-hydroxylase subunit alpha 1 (P4HA1) which functions to stabilize hypoxia inducible factor 1 alpha (HIF1A) 43 was also remarkably downregulated in the peptide group (Figure 3(b)). These results revealed that the WWASKS is able to promote iPSC differentiation into endocrine progenitors along with reduced necrosis. The downregulation of the hypoxia pathway in pancreatic endocrine lineages during iPSC differentiation is consistent with a previous study reported by Nair et al. 44 It is expected that pluripotent stem cells express high levels of hypoxia-related genes, as a low oxygen tension is preferred in maintaining the undifferentiated status of stem cells. 45 Our RNA-seq confirmed this. Moreover, iPSCs rely on glycolysis as their main source of energy, regardless of the availability of oxygen. 45 As cells differentiate, their metabolism shifts toward oxidative phosphorylation. 45 This is the case during the differentiation into pancreatic progenitors and endocrine progenitors as well as mature islets. Mature pancreatic islets have a high demand for oxygen. In fact, even mild hypoxia negatively affects their function, as shown by the decrease in glucose responsive insulin secretions resulting from hypoxic culturing conditions. 46 Therefore, the decreased expression of hypoxia-related genes in the WWASKS treated group indicates that they are more genotypically similar to endocrine cells than iPSCs.
Figure 3.
The peptide promotes iPSC lineage commitment by downregulating hypoxia-associated genes. (a) Gene set enrichment analysis showing hypoxia genes were enriched significantly in iPSC-derived pancreatic progenitor cells in the control group (Con) that lacks the peptide (P1) stimulation. (b) Heatmap signifying significantly altered expression of genes involved in hypoxia in peptide treated(P1_1~P1_3) and the control (C1~C3) group. (c) Genes involved in maintenance of pluripotency were downregulated in the iPSC-derived pancreatic progenitors under the peptide stimulation. (n = 3). (d) Key molecular expressions associated with hypoxia and pluripotency were downregulated by Western blot analysis. (e) Semi-quantitative analysis of relative protein expressions associated with hypoxia and pluripotency. The protein expressions in the peptide group were normalized to the loading control β-actin. Results are from three independent experiments and shown as mean ± SD. *: p < 0.05. ***: p < 0.001.
Additionally, to examine the extent to which the iPSCs had differentiated toward endocrine progenitors, we compared the expression of genes associated with the maintenance of pluripotency between the peptide-stimulated and the control groups. We identified genes responsible for maintaining pluripotency versus those facilitating the loss of pluripotency based on the GO and Kyoto Encyclopedia of Genes and Genomes (KEGG) terms “pluripotency” and “stem cell population maintenance.” We found that several genes involved in maintaining pluripotency were remarkably downregulated in the peptide group. Eleven significantly downregulated genes were identified (Figure 3(c)). For example, the gene MYC was significantly downregulated in the peptide group with a fold change of 0.37. MYC plays an important role in the establishment and maintenance of pluripotency in stem cells by regulating the transcription of thousands of genes. 47 The downregulation of this protein expression was confirmed by Western blot (Figure 3(d) and (e)). Several genes associated with the maintenance of pluripotency were found to be downregulated significantly in the WWASKS treated group. These results suggest that the cells treated with the peptide are differentiated more effectively into endocrine progenitor cells compared to the control group.
The peptide suppresses in vitro development of non-pancreatic organs
We identified genes that play a role in the development of organs besides the pancreas using GO terms. Several genes associated with the development of other organs were downregulated significantly in the peptide group. Genes involved in the development of the nervous system are shown in Figure 4(a). A total of 35 considerably downregulated genes associated with nervous development were identified. One such gene TRPC5, or short transient receptor potential channel 5, is important in nervous system development as well as the function of mature neurons. 48 This gene is downregulated significantly in the WWASKS group, with a fold change of 0.10. Similarly, the transcription factor FEZF2 is downregulated significantly in the peptide group, with a fold change of 0.29. FEZF2 is a key regulator in the differentiation of neural stem cells shown to direct their fate into glutamatergic subcerebral projecting neurons. 49 We selected a number of molecules to assess their protein expression by Western blot to confirm the downregulation of these molecules. Both TRPC5 and FEZF2 proteins showed approximately 2-fold decrease in the WWASKS group (Figure 4(e) and (f)).
Figure 4.
The suppression of expression of genes associated with other organ development by the peptide during iPSC pancreatic differentiation. (a–d) Genes involved in (a) nervous system, (b) heart (c) cartilage, (d) and skeletal system development were analyzed. (e) Detection of representative protein expressions by Western blot analysis. iPSC-derived pancreatic progenitor cells were harvested and total proteins were extracted for Western blot analysis. β-actin was used as a loading control for normalization. (f) Semi-quantitative analysis of relative protein expressions in the peptide group compared to the control. Results are from three independent experiments and shown as mean ± SD. *: p < 0.05. **: p < 0.01.
Besides, we found 14 genes associated with heart development, including NTRK3, GJA5, and MSX2, were downregulated in the differentiated cells under the peptide stimulation (Figure 4(b)). NTRK3, GJA5, and MSX2 are essential for heart development in animal models.50–52 GJA5 plays an important role in the arteriogenesis during acute ischemic cardiovascular disease. 53 We validated these protein expressions and found that expression of NTRK3, GJA5, and MSX2 reduced significantly to approximately 2.5-, 2.7-, and 2-fold in peptide treated group (Figure 4(f)). Furthermore, ten genes involved in the development of cartilage were also found to be downregulated (Figure 4(c)). MSX2 was downregulated in the peptide group, and it is important in the development of the frontal bone. 54 Moreover, ten genes involved in the skeletal system were significantly downregulated in the peptide group (Figure 4(d)). For instance, GJA5 was significantly downregulated in the peptide treated group. This gene plays a role in skeletal development. 55 As mentioned above, the expression levels of MSX2 and GJA5 proteins reduced significantly in the peptide-stimulated group (Figure 4(e) and (f)).
The peptide promotes the generation of insulin secreting cells from iPSCs with enhanced glucose responsive insulin secretion of i-islets
To further assess the effect of the peptide on pancreatic islet organoid development from iPSCs, we induced iPSC differentiation into pancreatic islet organoids using a five-stage differentiation protocol developed in our previous study with slight modification by temporal stimulation of cells with WWASKS, as shown in Figure 5(a).3,9 The morphology and architecture of the i-islets were characterized by visualizing the expression of the hormones and key transcription factors in i-islets through immunofluorescence microscopy. These markers, include C-peptide (CP), glucagon (GCG), somatostatin (SST), pancreatic polypeptide (PPY), mature β cell transcription factor MAFA, and mature α cell transcription factor MAFB (Figure 5(b)–(e)). To determine the cell composition of the organoids, the images of each experimental condition (n = 9–12) were analyzed by ImageJ and the percentage of each subtype of the organoids were calculated. The results revealed that the peptide stimulation increased the synthesis of the insulin-secreting β cells. The percentage of CP+ GCG− population was remarkably elevated in the WWASKS group from 28.5% to 41.1% as compared to the control group (Figure 5(f)). The populations of other islet cell types, GCG+ α, SST+ δ, and PPY+ γ subsets, didn’t show significant difference between the control and peptide group (Figure 5(g)–(i)), indicating that peptide stimulation promotes the generation of more β cells. Notably, the insulin stimulation index, which is defined as a ratio of insulin released from organoids at high glucose (20 mM) to that at low glucose (2 mM), increased significantly in the peptide treated group (4.8 ± 2.3) as compared with the control group (1.9 ± 0.6) (p = 0.045) (Figure 5(j)). Importantly, previous study showed that the insulin stimulation index from freshly isolated islets after 6 h culture was 6.21 ± 0.80 and dropped to 2.64 ± 0.25 after 48 h of culture. 56 Our experimental results revealed that WWASKS stimulation improves the i-islets’ function with an enhanced glucose-responsive insulin secretion capacity. The insulin stimulation index in peptide group was close to that from freshly isolated islets. 56
The peptide activates canonical Wnt signaling pathway during iPSC differentiation
To unveil mechanisms underlying the enhanced pancreatic endocrine lineage specification by WWASKS, we added FITC-conjugated WWASKS to the differentiation medium during posterior foregut differentiation stage. The localization of the peptide was visualized at 24 h post peptide treatment. We observed that the peptide localized to the plasma membrane of the iPSC-derived cells (Figure 6(a)). The peptide that we identified belongs to category of ultrashort peptide, which is precisely defined as peptides consisting of up to seven amino acids. 57 The advantages of ultrashort peptides include easier synthesis, less cost, higher mechanical stability, better bio- and cytocompatibility, and less immunogenicity, compared with their longer analogs. 57 Some peptides have the ability to penetrate cells, thus also known as cell penetrating peptides (CPPs), which are 6–30 amino acids long. They may carry a variety of cargoes across the cellular membranes. 58 Ma et al. reported that short peptides, P13 and M6, which support the self-renewal of hESCs, bound to the plasma membrane without penetrating into cytoplasm. 59 To examine whether WWASKS penetrates into the cytosol, we carried out a cell fractionation experiment. The FITC-peptide untreated group served as a control for the experiment. As shown in Figure 6(e), presence of the peptide was detected in both cytosol and plasma membrane, but not in the cell nucleus, suggesting the penetration of the peptide into the cytosol. We speculate that this penetration occurred due to the extra small size of the peptide because it is well known that cells can engulf small molecules such as short peptides. It appears that the peptide was diluted in the cytosol due to the relatively large space of the cytosol compared to membrane space, which hindered detection by imaging. The membrane-bound peptides are highly concentrated locally, making them visualizable under a confocal fluorescence microscope.
Figure 6.
The peptide activates canonical Wnt signaling pathway. (a) The binding of the peptide to the plasma membrane in iPSC-derived cells during differentiation detected by confocal microscopy. The cells were immunofluorescently labeled for cell plasma membrane (red). FITC-conjugated peptide (green) was added to the culture on day 1 of posterior foregut differentiation. Cells were counterstained with DAPI (blue). White scale bars: 50 μm. Blue scale bars: 20 μm. (b) Transcription factors that activate or are target of Wnt signaling pathway were significantly upregulated in the iPSC-derived pancreatic progenitors under the peptide stimulation compared to the control group (n = 3). (c) β-catenin translocation from the cytoplasm to the nucleus in the peptide (P1) and the control (Con) group. β-actin served as loading control. (d) Relative β-catenin expression level after normalization to β-actin (n = 4). Results are from four independent experiments and shown as mean ± SD. *: p < 0.05; ns: not significant. (e) Localization of the peptide in iPSC-derived cells detected through cell fractionation assay (n = 4).
In addition, according to the RNA-seq analysis, at least 11 transcription factors involved in the Wnt signaling activation were upregulated substantially in the WWASKS group (Figure 6(b)). These factors include DLX5, CBFB, TCF25, SLC9A3R1, TLE2, CDH3, LBH, GLI1, TLE6, MITF, and FOLR1.60–66 The gene SFRP2, which is an inhibitor of canonical Wnt signaling, was significantly downregulated (Figure 6(b)).67,68 To interpret the involvement of Wnt signaling in the WWASKS stimulated iPSC-pancreatic progenitor differentiation, we performed cellular fractionation analysis. We constantly observed strong translocation of β-catenin to the nucleus in the peptide group by Western blot analysis (Figure 6(c)). There was approximately 90% more β-catenin expression in the nucleus in the peptide group than that in the control, while the expression levels of cytoplasmic β-catenin in both control and peptide groups were similar (Figure 6(c) and (d)). Hence, our experimental results discovered that the peptide identified in this study activates canonical Wnt signaling pathway, leading to augmentation of pancreatic endocrine lineage and islet organoid differentiation. Previous studies reported that temporal activation of canonical Wnt signaling is required to promote pancreatic progenitor differentiation from hPSCs and further insulin-expressing endocrine cells. 69 Our previous study also uncovered that an activated Wnt signaling offered by porous membrane substrates facilitates hESC definitive endoderm (DE) differentiation. 5 It has also been found that the degree of Wnt signaling activity regulates DE formation from hPSC. 70 Therefore, the WWASKS treatment during the formation of posterior foregut resulted in temporal activation of canonical Wnt signaling and may contribute to subsequent endocrine progenitor specification, which is consistent with others’ and our previous findings.
Conclusion
Short peptides play important roles in processing biological information, regulating transcription, and facilitating stem cell differentiation.10,12 On the other hand, the generation of islet organoids containing all the major endocrine cell types from stem cells remains a challenge. 17 In this study, we identified, for the first time, a dpECM-derived developmental short peptide WWASKS. It is capable of directing iPSC differentiation toward endocrine progenitors and subsequently islet organoids. Of interest, the peptide endorses the generation of NKX6.1+ PDX1+ cell population, pivotal to the development of pancreatic endocrine tissues. The upregulation of NKX6.1+ PDX1+ cell population under the peptide stimulation suggests that the peptide is critical to iPSC differentiation into endocrine progenitor cells. Remarkably, the peptide is able to significantly enhance the insulin stimulation index with more mature β cells during iPSC islet organoid differentiation. In the iPSC-derived cells at the endocrine progenitor stage under the peptide stimulation, the efficiency of pancreatic endocrine cell lineage specification was considerably increased while suppressing other organ lineage specification. Another interesting finding is that the short peptide discovered by this study is able to suppress several genes involved in the differentiation of the nervous system, heart, cartilage and skeletal system development. This indicated that the peptide directed iPSC differentiation away from other lineages when committing to pancreatic endocrine lineages. Furthermore, our experimental results discovered that genes that are typically upregulated in hypoxic conditions were significantly enriched in the control group. This suggested that the iPSCs under WWASKS stimulation are differentiated more effectively. Future work is necessary to further elucidate the mechanisms underlying the enhancements of in vitro endocrine cell development by the peptide stimulation. This includes identification of the interaction between the peptide and cellular surface molecules. Taken together, we demonstrated a unique dpECM-derived peptide possessing capability for the promotion of iPSC pancreatic endoderm progenitor differentiation and subsequent generation of glucose responsive insulin secreting islet organoids.
Supplemental Material
Supplemental material, sj-docx-1-tej-10.1177_20417314231185858 for Extracellular matrix-derived peptide stimulates the generation of endocrine progenitors and islet organoids from iPSCs by Emma S Heaton, Ming Hu, Tianzheng Liu, Huang Hui, Yinfei Tan, Kaiming Ye and Sha Jin in Journal of Tissue Engineering
Footnotes
Author contributions: SJ and KY conceived and designed the experiments; MH, EH, TL, and HH performed the experiments; MH, EH, YT, and SJ analyzed and interpreted the data; EH and SJ wrote the manuscript; SJ, KY, and MH contributed to revision of the manuscript. All of the authors read and approved the final manuscript.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was partially supported by National Science Foundation CBET1928855, CBET1919830, and National Institute of Health EB027391-01.
ORCID iD: Sha Jin  https://orcid.org/0000-0002-8033-8110
https://orcid.org/0000-0002-8033-8110
Supplemental material: Supplemental material for this article is available online.
References
- 1.Sim EZ, Shiraki N, Kume S.Recent progress in pancreatic islet cell therapy. Inflamm Regen 2021; 41(1): 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bi H, Karanth SS, Ye K, et al. Decellularized tissue matrix enhances self-assembly of islet organoids from pluripotent stem cell differentiation. ACS Biomater Sci Eng 2020; 6: 4155–4165. [DOI] [PubMed] [Google Scholar]
- 3.Bi H, Ye K, Jin S.Proteomic analysis of decellularized pancreatic matrix identifies collagen V as a critical regulator for islet organogenesis from human pluripotent stem cells. Biomater 2020; 233: 119673. [DOI] [PubMed] [Google Scholar]
- 4.Chmielowiec J, Szlachcic WJ, Yang D, et al. Human pancreatic microenvironment promotes β-cell differentiation via non-canonical WNT5A/JNK and BMP signaling. Nat Commun 2022; 13: 1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jin S, Yao H, Krisanarungson P, et al. Porous membrane substrates offer better niches to enhance the Wnt signaling and promote human embryonic stem cell growth and differentiation. Tissue Eng Part A 2012; 18: 1419–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rezania A, Bruin JE, Arora P, et al. Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nat Biotechnol 2014; 32: 1121–1133. [DOI] [PubMed] [Google Scholar]
- 7.Hai N, Shin DW, Bi H, et al. Mechanistic analysis of physicochemical cues in promoting human pluripotent stem cell self-renewal and metabolism. Int J Mol Sci 2018; 19: 3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang W, Jin S, Ye K.Development of islet organoids from H9 human embryonic stem cells in biomimetic 3D scaffolds. Stem Cells Dev 2017; 26: 394–404. [DOI] [PubMed] [Google Scholar]
- 9.Karanth SS, Sun S, Bi H, et al. Angiopoietins stimulate pancreatic islet development from stem cells. Sci Rep 2021; 11: 13558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caputi S, Trubiani O, Sinjari B, et al. Effect of short peptides on neuronal differentiation of stem cells. Int J Immunopathol Pharmacol 2019; 33: 2058738419828613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khavinson VKH, Tendler SM, Vanyushin BF, et al. Peptide regulation of gene expression and protein synthesis in bronchial epithelium. Lung 2014; 192: 781–791. [DOI] [PubMed] [Google Scholar]
- 12.Khavinson V, Linkova N, Diatlova A, et al. Peptide regulation of cell differentiation. Stem Cell Rev Rep 2020; 16: 118–125. [DOI] [PubMed] [Google Scholar]
- 13.Khavinson VKH, Durnova AO, Polyakova VO, et al. Effects of pancragen on the differentiation of pancreatic cells during their ageing. Bull Exp Biol Med 2013; 154: 501–504. [DOI] [PubMed] [Google Scholar]
- 14.Zhou J, Wang X, Pineyro MA, et al. Glucagon-like peptide 1 and exendin-4 convert pancreatic AR42J cells into glucagon- and insulin-producing cells. Diabetes 1999; 48: 2358–2366. [DOI] [PubMed] [Google Scholar]
- 15.Drucker DJ.Glucagon-like peptides: regulators of cell proliferation, differentiation, and apoptosis. Mol Endocrinol 2003; 17: 161–171. [DOI] [PubMed] [Google Scholar]
- 16.Llacua A, de Haan BJ, Smink SA, et al. Extracellular matrix components supporting human islet function in alginate-based immunoprotective microcapsules for treatment of diabetes. J Biomed Mater Res A 2016; 104: 1788–1796. [DOI] [PubMed] [Google Scholar]
- 17.Heaton ES, Jin S.Importance of multiple endocrine cell types in islet organoids for type 1 diabetes treatment. Transl Res 2022; 250: 68–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin S, Yao H, Weber JL, et al. A synthetic, xeno-free peptide surface for expansion and directed differentiation of human induced pluripotent stem cells. PLoS One 2012; 7: e50880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eddy SR.Where did the BLOSUM62 alignment score matrix come from? Nat Biotechnol 2004; 22: 1035–1036. [DOI] [PubMed] [Google Scholar]
- 20.Hogrebe NJ, Maxwell KG, Augsornworawat P, et al. Generation of insulin-producing pancreatic β cells from multiple human stem cell lines. Nat Protoc 2021; 16: 4109–4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walczak MP, Drozd AM, Stoczynska-Fidelus E, et al. Directed differentiation of human iPSC into insulin producing cells is improved by induced expression of PDX1 and NKX6.1 factors in IPC progenitors. J Transl Med 2016; 14: 341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor BL, Liu FF, Sander M.Nkx6.1 is essential for maintaining the functional state of pancreatic beta cells. Cell Rep 2013; 4: 1262–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lam CJ, Chatterjee A, Shen E, et al. Low-level insulin content within abundant non-β islet endocrine cells in long-standing type 1 diabetes. Diabetes 2019; 68: 598–608. [DOI] [PubMed] [Google Scholar]
- 24.Gage BK, Asadi A, Baker RK, et al. The role of ARX in human pancreatic endocrine specification. PLoS One 2015; 10: e0144100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee CS, Sund NJ, Vatamaniuk MZ, et al. Foxa2 controls Pdx1 gene expression in pancreatic beta-cells in vivo. Diabetes 2002; 51: 2546–2551. [DOI] [PubMed] [Google Scholar]
- 26.Lee CS, Sund NJ, Behr R, et al. Foxa2 is required for the differentiation of pancreatic alpha-cells. Dev Biol 2005; 278: 484–495. [DOI] [PubMed] [Google Scholar]
- 27.Doyle MJ, Sussel L.Nkx2.2 regulates beta-cell function in the mature islet. Diabetes 2007; 56: 1999–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carvell MJ, Marsh PJ, Persaud SJ, et al. E-cadherin interactions regulate beta-cell proliferation in islet-like structures. Cell Physiol Biochem 2007; 20: 617–626. [DOI] [PubMed] [Google Scholar]
- 29.Serrill JD, Sander M, Shih HP.Pancreatic exocrine tissue architecture and integrity are maintained by E-cadherin during postnatal development. Sci Rep 2018; 8: 13451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rukstalis JM, Habener JF.Neurogenin3: a master regulator of pancreatic islet differentiation and regeneration. Islets 2009; 1: 177–184. [DOI] [PubMed] [Google Scholar]
- 31.Lee JC, Smith SB, Watada H, et al. Regulation of the pancreatic pro-endocrine gene neurogenin3. Diabetes 2001; 50: 928–936. [DOI] [PubMed] [Google Scholar]
- 32.Ediger BN, Du A, Liu J, et al. Islet-1 is essential for pancreatic β-cell function. Diabetes 2014; 63: 4206–4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masui T, Swift GH, Hale MA, et al. Transcriptional autoregulation controls pancreatic ptf1a expression during development and adulthood. Mol Cell Biol 2008; 28: 5458–5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dong PD, Provost E, Leach SD, et al. Graded levels of ptf1a differentially regulate endocrine and exocrine fates in the developing pancreas. Genes Dev 2008; 22: 1445–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hosokawa S, Furuyama K, Horiguchi M, et al. Impact of sox9 dosage and Hes1-mediated notch signaling in controlling the plasticity of adult pancreatic duct cells in mice. Sci Rep 2015; 5: 8518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin S, Zhang B, Weisz OA, et al. Receptor-mediated entry by equine infectious anemia virus utilizes a pH-dependent endocytic pathway. J Virol 2005; 79: 14489–14497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ezashi T, Das P, Roberts RM.Low O2 tensions and the prevention of differentiation of hES cells. Proc Natl Acad Sci USA 2005; 102: 4783–4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mohyeldin A, Garzón-Muvdi T, Quiñones-Hinojosa A.Oxygen in stem cell biology: a critical component of the stem cell niche. Cell Stem Cell 2010; 7: 150–161. [DOI] [PubMed] [Google Scholar]
- 39.Ito K, Suda T.Metabolic requirements for the maintenance of self-renewing stem cells. Nat Rev Mol Cell Biol 2014; 15: 243–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brugarolas J, Lei K, Hurley RL, et al. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev 2004; 18: 2893–2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Künzel SR, Sekeres K, Kämmerer S, et al. Hypoxia-induced epigenetic silencing of polo-like kinase 2 promotes fibrosis in atrial fibrillation. bioRxiv 2018; 445098. [Google Scholar]
- 42.Arnould T, Thibaut-Vercruyssen R, Bouaziz N, et al. PGF(2alpha), a prostanoid released by endothelial cells activated by hypoxia, is a chemoattractant candidate for neutrophil recruitment. Am J Pathol 2001; 159: 345–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiong G, Stewart RL, Chen J, et al. Collagen prolyl 4-hydroxylase 1 is essential for HIF-1α stabilization and TNBC chemoresistance. Nat Commun 2018; 9: 4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nair GG, Liu JS, Russ HA, et al. Recapitulating endocrine cell clustering in culture promotes maturation of human stem-cell-derived β cells. Nat Cell Biol 2019; 21: 263–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang X, Trinh T, Aljoufi A, et al. Hypoxia signaling pathway in stem cell regulation: Good and Evil. Curr Stem Cell Rep 2018; 4: 149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Myasnikova D, Osaki T, Onishi K, et al. Synergic effects of oxygen supply and antioxidants on pancreatic β-cell spheroids. Sci Rep 2019; 9: 1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chappell J, Dalton S.Roles for MYC in the establishment and maintenance of pluripotency. Cold Spring Harb Perspect Med 2013; 3: a014381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Puram SV, Riccio A, Koirala S, et al. A TRPC5-regulated calcium signaling pathway controls dendrite patterning in the mammalian brain. Genes Dev 2011; 25: 2659–2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zuccotti A, Le Magueresse C, Chen M, et al. The transcription factor Fezf2 directs the differentiation of neural stem cells in the subventricular zone toward a cortical phenotype. Proc Natl Acad Sci USA 2014; 111: 10726–10731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Werner P, Paluru P, Simpson AM, et al. Mutations in NTRK3 suggest a novel signaling pathway in human congenital heart disease. Hum Mutat 2014; 35: 1459–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guida V, Ferese R, Rocchetti M, et al. A variant in the carboxyl-terminus of connexin 40 alters GAP junctions and increases risk for tetralogy of Fallot. Eur J Hum Genet 2013; 21: 69–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kwang SJ, Brugger SM, Lazik A, et al. Msx2 is an immediate downstream effector of Pax3 in the development of the murine cardiac neural crest. Dev 2002; 129: 527–538. [DOI] [PubMed] [Google Scholar]
- 53.Lu XJ, Wang HT.Reduced Gja5 expression in arterial endothelial cells impairs arteriogenesis during acute ischemic cardiovascular disease. Exp Ther Med 2017; 14: 4339–4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Han J, Ishii M, Bringas P, Jr, et al. Concerted action of Msx1 and Msx2 in regulating cranial neural crest cell differentiation during frontal bone development. Mech Dev 2007; 124: 729–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pizard A, Burgon PG, Paul DL, et al. Connexin 40, a target of transcription factor Tbx5, patterns wrist, digits, and sternum. Mol Cell Biol 2005; 25: 5073–5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Noguchi H, Naziruddin B, Jackson A, et al. Fresh islets are more effective for islet transplantation than cultured islets. Cell Transplant 2012; 21: 517–523. [DOI] [PubMed] [Google Scholar]
- 57.Apostolopoulos V, Bojarska J, Chai TT, et al. A global review on short peptides: frontiers and perspectives. Mol 2021; 26: 430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taylor RE, Zahid M.Cell penetrating peptides, novel vectors for gene therapy. Pharm 2020; 12: 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ma R, Ren Z, Li B, et al. Novel venom-based peptides (P13 and its derivative-M6) to maintain self-renewal of human embryonic stem cells by activating FGF and TGFβ signaling pathways. Stem Cell Res Ther 2020; 11: 243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xia L, Wu L, Bao J, et al. Circular RNA circ-CBFB promotes proliferation and inhibits apoptosis in chronic lymphocytic leukemia through regulating miR-607/FZD3/Wnt/β-catenin pathway. Biochem Biophys Res Commun 2018; 503: 385–390. [DOI] [PubMed] [Google Scholar]
- 61.Rieger ME, Sims AH, Coats ER, et al. The embryonic transcription cofactor LBH is a direct target of the Wnt signaling pathway in epithelial development and in aggressive basal subtype breast cancers. Mol Cell Biol 2010; 30: 4267–4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu G, Zhu Y, Liu H, et al. LncRNA mir194-2HG promotes cell proliferation and metastasis via regulation of miR-1207-5p/TCF19/Wnt/β-Catenin signaling in liver cancer. OncoTargets Ther 2020; 13: 9887–9899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ploper D, Taelman VF, Robert L, et al. MITF drives endolysosomal biogenesis and potentiates Wnt signaling in melanoma cells. Proc Natl Acad Sci 2015; 112: E420–E429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Paina S, Garzotto D, DeMarchis S, et al. Wnt5a is a transcriptional target of Dlx homeogenes and promotes differentiation of interneuron progenitors in vitro and in vivo. J Neurosci 2011; 31: 2675–2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Noubissi FK, Goswami S, Sanek NA, et al. Wnt Signaling Stimulates Transcriptional Outcome of the Hedgehog Pathway by Stabilizing GLI1 mRNAWnt Signaling Stabilizes GLI1 mRNA. Cancer Res 2009; 69: 8572–8578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen P-C, Kuraguchi M, Velasquez J, et al. Novel roles for MLH3 deficiency and TLE6-like amplification in DNA mismatch repair-deficient gastrointestinal tumorigenesis and progression. PLoS Genet 2008; 4: e1000092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martin V, Agirre X, Jiménez-Velasco A, et al. Methylation status of Wnt signaling pathway genes affects the clinical outcome of Philadelphia-positive acute lymphoblastic leukemia. Cancer Sci 2008; 99: 1865–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ito K, Lim AC, Salto-Tellez M, et al. RUNX3 attenuates beta-catenin/T cell factors in intestinal tumorigenesis. Cancer Cell 2008; 14: 226–237. [DOI] [PubMed] [Google Scholar]
- 69.Nostro MC, Sarangi F, Ogawa S, et al. Stage-specific signaling through TGFβ family members and WNT regulates patterning and pancreatic specification of human pluripotent stem cells. Dev 2011; 138: 861–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jiang Y, Chen C, Randolph LN, et al. Generation of pancreatic progenitors from human pluripotent stem cells by small molecules. Stem Cell Rep 2021; 16: 2395–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-tej-10.1177_20417314231185858 for Extracellular matrix-derived peptide stimulates the generation of endocrine progenitors and islet organoids from iPSCs by Emma S Heaton, Ming Hu, Tianzheng Liu, Huang Hui, Yinfei Tan, Kaiming Ye and Sha Jin in Journal of Tissue Engineering