Summary
Interferon-induced transmembrane proteins (IFITMs) are restriction factors that block many viruses from entering cells. High levels of type I interferon (IFN) are associated with adverse pregnancy outcomes, and IFITMs have been shown to impair the formation of syncytiotrophoblast. Here, we examine whether IFITMs affect another critical step of placental development, extravillous cytotrophoblast (EVCT) invasion. We conducted experiments using in vitro/ex vivo models of EVCT, mice treated in vivo with the IFN-inducer poly (I:C), and human pathological placental sections. Cells treated with IFN-β demonstrated upregulation of IFITMs and reduced invasive abilities. Transduction experiments confirmed that IFITM1 contributed to the decreased cell invasion. Similarly, migration of trophoblast giant cells, the mouse equivalent of human EVCTs, was significantly reduced in poly (I:C)-treated mice. Finally, analysis of CMV- and bacterial-infected human placentas revealed upregulated IFITM1 expression. These data demonstrate that high levels of IFITM1 impair trophoblast invasion and could explain the placental dysfunctions associated with IFN-mediated disorders.
Subject areas: Biological sciences, Virology, Cell biology
Graphical abstract
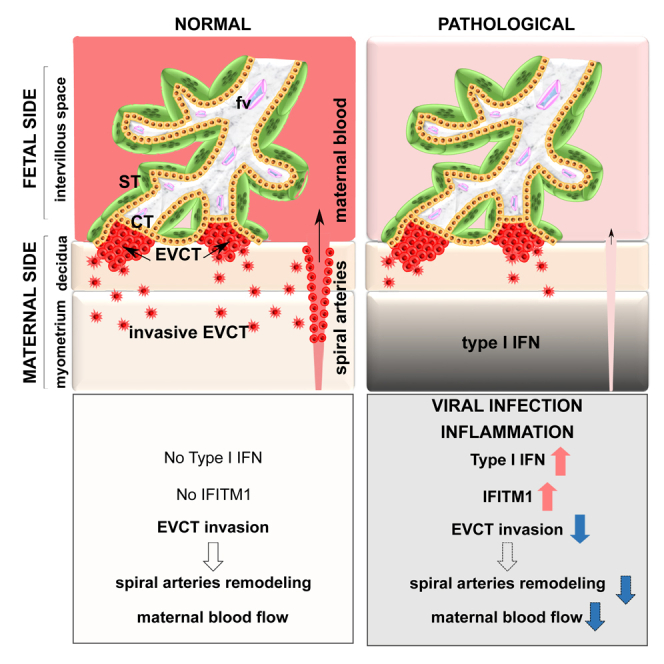
Highlights
-
•
IFITMs expression is induced in IFN-treated trophoblast cells
-
•
IFITM1 overexpression significantly inhibits human trophoblast invasion
-
•
Trophoblast invasion is inhibited in IFN-induced mouse model
-
•
IFITM1 expression is induced in IFN-mediated pregnancy diseases
Biological sciences; Virology; Cell biology
Introduction
The placenta is a transitory organ that arises after implantation and is essential for the development and growth of the fetus, as well as for the maintenance of pregnancy. It plays, therefore, a critical role in pregnancy outcome. The structural and functional unit of the human placenta is the chorionic villus, which is bathed by maternal blood in the intervillous space. The chorionic villus is characterized by a mesenchymal axis containing immune cells and fetal vessels that is covered by a monolayer of epithelial cells (villous cytotrophoblasts) that fuse with the overlying syncytiotrophoblast to renew it during pregnancy. This trophoblast bilayer acts as a placental “barrier” that facilitates exchange between maternal and fetal blood and is also the site of the production of pregnancy hormones.
The chorionic villus is anchored into the uterine wall (decidua basalis) by extravillous cytotrophoblasts (EVCTs) that invade the uterus up to the upper third of the myometrium.1 EVCTs interact with uterine immune cells to permit the immune tolerance of the conceptus and participate in the remodeling of the uterine spiral arteries. Trophoblast invasion thus represents an essential step in the development and function of the placenta, and defective trophoblastic invasion is associated with miscarriage, placental dysfunction, fetal growth restriction (FGR), and preeclampsia.2
Many problems in pregnancy arise as the result of infections, which have been linked to low birth weight, congenital birth defects, complications (intra-uterine growth restriction, preeclampsia, prematurity), and miscarriage.3 These outcomes appear to be mediated by increased levels of inflammatory factors, such as interferons, TNF-α, and interleukins into the maternal circulatory system.4,5 In humans, the response to viral infections is characterized by the upregulation of several interferon-inducible transmembrane proteins (IFITMs), a protein family composed of five genes (IFITM1, 2, 3, 5, and 10). IFITM1, 2, 3, and 5 are located in the same gene cluster in a 26-kb region of the short arm of chromosome 11, while IFITM10 is located 1.4 kb from the centromere of chromosome 11 and its function remains unknown. IFITM2 and IFITM3 share strong sequence homology (91%) which makes it difficult to distinguish between them, especially through the use of antibody-based tests. All IFITMs are membrane proteins. IFITM1 is enriched at the plasma membrane, while IFITM2 and 3 predominate in the intracellular compartment (endosomes), suggesting possible specialization in the roles of these three proteins.6 Instead, IFITM5 is not interferon-responsive and is involved in bone mineralization.7 IFITMs1, 2, and 3 have been described as viral restriction factors with varying degrees of effectiveness (review in8). Specifically, these IFITMs, and more particularly IFITM3, inhibit the cell-cell fusion induced by viral infection by modifying the physico-chemical properties of cell membranes, notably by reducing membrane fluidity.9 Work by our group as well as others has shown that IFITMs also inhibit the fusion of villous cytotrophoblasts.10,11 Moreover, high expression of IFITM1 and IFITM3 has been observed in cancer cells, with effects on proliferation and invasion.12,13,14,15
To date, the role of IFITMs in the invasion of EVCTs has not been studied. Here, we examined the impact of IFITMs (1, 2, and 3) on trophoblast invasion using approaches based on IFN induction and IFITM transduction in the HTR8/SVneo cell line, primary EVCTs, and first-trimester placental explants. We also assessed the effect of the viral mimic poly (I:C) in a mouse model, and performed immunostaining of CMV-/bacterial-infected human placental tissue sections to visualize the expression of IFITMs.
Results
IFN-β induces expression of IFITMs and inhibits trophoblast invasion
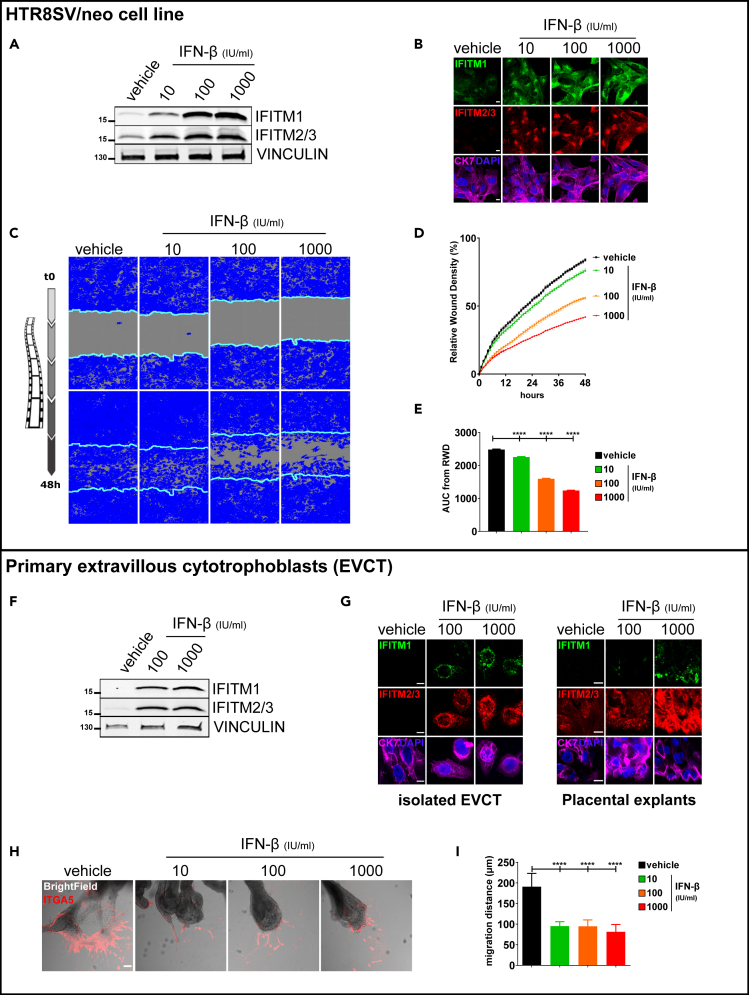
We initiated our study by investigating the effect of IFN-β on trophoblastic expression of IFITMs and on the invasion of human trophoblasts using HTR8/SVneo cells (Figures 1A–1E). Western blot and immunofluorescence (Figures 1A and 1B) revealed induction of IFITM1, 2, or 3 expression after 48 h of IFN-β treatment regardless of the concentration used (10, 100, and 1000 IU/mL). Invasion of IFN-β-treated HTR8/SVneo cells was evaluated using a wound healing assay with Matrigel and monitored for 48 h in a live-imaging device (Figures 1C–1E). Compared to the control treatment (vehicle), IFN-β significantly inhibited cell invasion in a dose-dependent manner (76 ± 2.9%, 56 ± 2.4%, and 42 ± 1.6% versus 84 ± 3.2%, respectively, for 10, 100, and 1000 IU/mL of IFN-β versus vehicle).
Figure 1.
Upregulation of IFITMs in HTR8/SVneo cells following IFN-β treatment was evaluated by western blot and immunofluorescence
IFN-β inhibits invasion of HTR8/SVneo cells (A–E) and primary human EVCTs (F–I). HTR8/SVneo cells and EVCTs were incubated for 48 h with IFN-β (10, 100, or 1000 IU/mL).
(A) Cell lysates (40 μg) were loaded in 4–15% SDS-PAGE gradient gels and blots were probed for IFITM1, IFITM2/3, and vinculin expression. One representative experiment (out of 3) is shown.
(B) Representative image of HTR8/SVneo cells immunostained for cytokeratin 7 (marker of trophoblast, magenta) and IFITM1 (green) or IFITM2/3 (red), and then counterstained with DAPI (blue). Scale bars: 10 μm. Inhibition of HTR8/SVneo cell invasion following IFN-β treatment, quantified using IncuCyte technology (C–E). HTR8/SVneo cells were cultured in a monolayer in 96-well plates. Cells were treated for 48 h, then a wound was made using a 96-well wound maker; this was covered by Matrigel and cells were imaged with Incucyte every 30 min for 48 h.
(C) Representative images of HTR8/SVneo cell invasion at 48 h. The blue region denotes the area of the initial wound (light blue line) covered by advancing cells.
(D) Time course of wound closure expressed as relative wound density (%). Values are reported as mean ± SEM.
(E) Comparison between vehicle and IFN-β-treated cells was performed using an analysis of the area under the curve of the replicates represented in (D). Values are reported as mean + SEM (n = 6 wells/condition, n = 3 experiments). Statistical analysis was performed with one-way ANOVA to compare treatment to vehicle controls. ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Upregulation of IFITMs in EVCTs following IFN-β treatment was evaluated by western blot (F) and immunofluorescence (G).
(F) Cell lysates (40 μg) from isolated EVCTs were loaded in 4–15% SDS-PAGE gradient gels and blots were probed for IFITM1, IFITM2/3, and vinculin expression. One representative experiment (out of 3) is shown.
(G) Representative image of isolated EVCTs and placental explants immunostained for cytokeratin 7 (marker of trophoblast, magenta) and IFITM1 (green) or IFITM2/3 (red), and then counterstained with DAPI (blue). Scale bars: 10 μm. Inhibition of EVCT invasion following IFN-β treatment using villous explant culture model (H–I).
(H) Representative image of extravillous explants cultured on Matrigel and immunostained for integrin α5 (ITGA5, marker of invasion, red). Scale bars: 100 μm.
(I) Statistical analysis of the invasion distance of ITGA5 + EVCTs, as representatively shown in H. Values are presented as mean + SD (n = 3 explants per condition, n = 3 placentas). One-way ANOVA was performed to compare treatment to vehicle control; ∗∗∗∗p < 0.0001.
We next evaluated the effect of IFN-β in primary EVCTs. For this purpose, purified primary EVCTs and first-trimester placental explants were cultured on Matrigel and treated for 48 h with 100 and 1000 IU/mL of IFN-β. Western blot and immunofluorescence revealed that IFN-β treatment resulted in the induction of IFITM1, 2, and 3 (Figures 1F and 1G). The invasion of IFN-β treated-EVCTs was then evaluated by immunofluorescence using integrin α5 (ITGA5) as a marker of the invasion front (Figures 1H and 1I). Compared to control (vehicle), treatment with 10, 100, or 1000 IU/mL of IFN-β significantly inhibited the distance of cell migration (respectively, in μm, 95 ± 11, 95 ± 15, and 82 ± 18 versus 191 ± 32, p < 0.0001).
Overexpression of IFITM1 inhibits trophoblast invasion
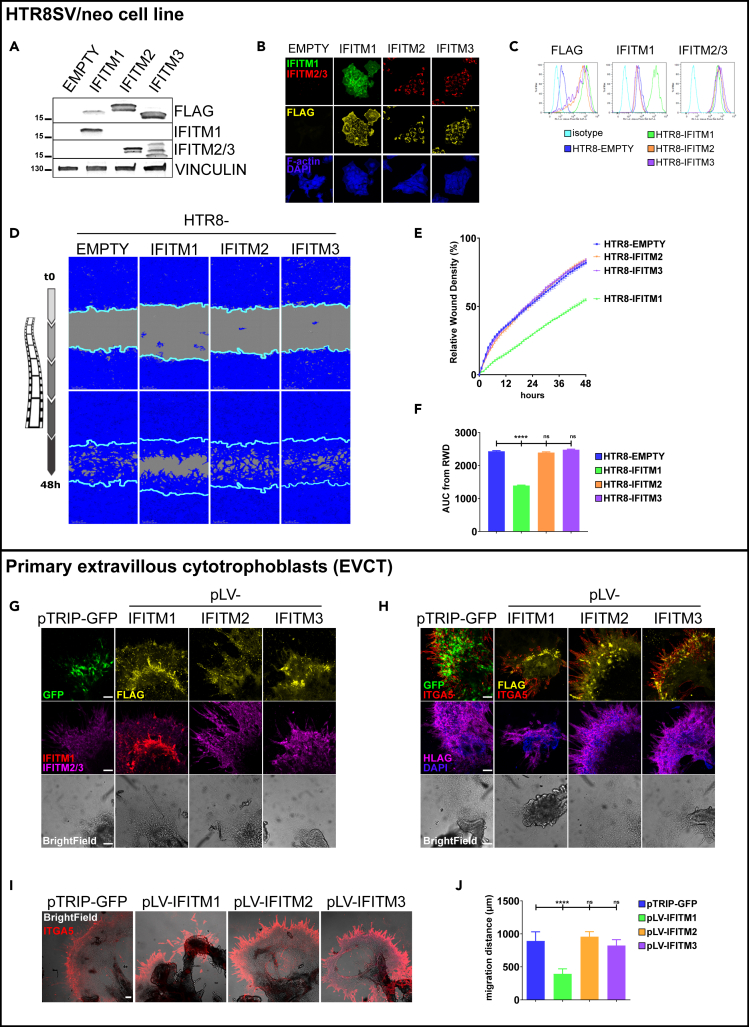
To directly study the role of IFITMs in the invasion of human trophoblasts, we first transduced the HTR8/SVneo cells with lentiviral vectors encoding IFITM1, IFITM2, and IFITM3, with an empty vector (EMPTY) as control. These vectors carry the gene coding for puromycin resistance, so successfully transduced cells were selected with this antibiotic after treatment. Expression of IFITMs was confirmed in transduced cells by western blot, immunofluorescence, and flow cytometry (Figures 2A–2C). In cells transduced with the empty vector (EMPTY cells), no expression of the FLAG tag or IFITM1 was observed, and only low endogenous expression of IFITM2/3 was detected. Instead, the cells transduced with IFITM1, 2, and 3 all demonstrated expression of the FLAG tag. Only IFITM1-transduced cells showed IFITM1 expression and the IFITM2- and IFITM3-transduced cells showed overexpression of IFITM2/3. Interestingly, in the confocal images (Figures 2B and S1B), we observed clear differences in the localization of IFITMs that were consistent with those described in the literature, with IFITM1 mainly located at the plasma membrane and IFITM2 and IFITM3 at the plasma membrane and within endosomal compartments. Transduction had no apparent effect on either cell viability or proliferation (Figure S1).
Figure 2.
Ectopic expression of FLAG and IFITMs in HTR8/SVneo cells transduced with IFITM1–3 was verified by western blot, immunofluorescence, and flow cytometry
IFITM1 inhibits invasion of HTR8/SVneo cells (A–E) and primary human EVCTs.
(A) Cell lysates (40 μg) were loaded in 4–15% SDS-PAGE gradient gels and blots were probed for IFITM1, IFITM2/3, and vinculin expression. A representative experiment is shown.
(B) Representative image of transduced HTR8/SVneo cells immunostained for FLAG (yellow) and IFITM1 (green) or IFITM2/3 (red), and then counterstained with DAPI (blue). Scale bar: 10 μm.
(C) HTR8/SVneo cells transduced with a vector containing IFITM1 (green), IFITM2 (orange), IFITM3 (magenta), or a control empty vector (dark blue) were stained with antibodies for IFITM1, IFITM2/3, and FLAG, as well as corresponding isotypes. Cells were then analyzed by flow cytometry. Inhibition of IFITM1-transduced HTR8/SVneo cell invasion, quantified using IncuCyte technology (D–F). HTR8/SVneo cells transduced with IFITM1–3 were cultured in a monolayer in 96-well plates and a wound was made using a 96-well wound maker; this was covered by Matrigel and cells were imaged with Incucyte every 30 min for 48 h.
(D) Representative images of transduced HTR8/SVneo cell invasion at 48 h. The blue region denotes the area of the initial wound (light blue line) covered by advancing cells.
(E) Time course of wound closure expressed as relative wound density (%). Values are reported as mean ± SEM of the percentage.
(F) Comparison between cells transduced with IFITM1–3 and the empty vector was performed using an analysis of the area under the curve of the replicates represented in (E). Values are represented as mean + SEM (n = 6 wells/condition, n = 3 experiments). Statistical analysis was performed with one-way ANOVA to compare treatment to vehicle controls. ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Ectopic expression of GFP, FLAG, and IFITMs in EVCT explants transduced with IFITM1–3 was verified by immunofluorescence (G–J).
(G) Representative image of placental explants transduced with GFP or IFITM1–3, cultured on Matrigel, and immunostained for FLAG (yellow), GFP (green), and IFITM1 (red) or IFITM2/3 (magenta), and then counterstained with DAPI (blue). Scale bars: 100 μm.
(H) Representative image of EVCT explants transduced with GFP or IFITM1–3, cultured on Matrigel, and immunostained for FLAG (yellow), GFP (green), and ITGA5 (red) or HLAG (magenta), and then counterstained with DAPI (blue). Scale bars: 100 μm. Inhibition of IFITM1-transduced EVCT invasion using villous explant culture model (I–J).
(I) Representative image of transduced extravillous explants cultured on Matrigel and immunostained for integrin α5 (ITGA5, marker of invasion, red). Scale bars: 100 μm.
(J) Statistical analysis of the invasion distance of ITGA5 + EVCTs, as representatively shown in I. Values are presented as mean + SD (n = 3 explants per condition, n = 3 placentas). One-way ANOVA was performed to compare treatment to pTRIP-GFP control; ∗∗∗∗p < 0.0001.
Next, we evaluated the invasive abilities of the IFITM-transduced HTR8/SVneo cells using a wound healing assay with Matrigel that was monitored for 48 h in a live-imaging device (Figures 2D–2F). Compared to control (EMPTY) transduced cells, IFITM1-transduced cells exhibited significantly inhibited cell invasion (55 ± 3.0% versus 82 ± 4.4%, p < 0.0001); instead, no difference was evident in IFITM2- or IFITM3-transduced cells (respectively, 84 ± 3.5% and 84 ± 3.2% versus 82 ± 4.4%). We also tested a treatment with amphotericin B, an antifungal agent capable of counteracting the fusion restriction of IFITM3,16 but no difference was observed (Figure S2).
We next evaluated the effect of IFITM overexpression in primary EVCTs. For this purpose, first-trimester placental explants were grown on Matrigel and transduced with lentiviral vectors encoding IFITM1, IFITM2, or IFITM3, as well as the empty vector (pLV-EMPTY) and a positive vector (pTRIP-GFP) as controls (Figures 2G–2J). We first validated explant transduction with IFITM and FLAG tag expression by immunofluorescence (Figure 2G). In cells transduced with the empty vector (EMPTY) or GFP, no expression of the FLAG tag or IFITM1 was detected, and only low endogenous IFITM2/3 expression was observed. Instead, FLAG tag expression was found in cells transduced with IFITM1, IFITM2, and IFITM3. Only IFITM1-transduced cells showed IFITM1 expression, and IFITM2- and IFITM3-transduced cells showed overexpression of IFITM2/3.
Then, we used immunofluorescence to evaluate the invasion of primary EVCTs transduced with IFITMs, using the FLAG tag or GFP to visualize the transduced cells, integrin α5 (ITGA5) and HLAG as hallmark markers of invasive EVCTs (Figure 2H). All transduced cells (FLAG+ or GFP+) were invasive EVCTs (ITGA5+ HLAG+). Finally, the migration distances of IFITM-transduced EVCTs were evaluated by immunofluorescence using ITGA5 as a marker of the invasion front (Figures 2I and 2J). Compared to GFP-transduced control cells, the only cells that demonstrated significantly inhibited invasion were those that overexpressed IFITM1 (365 ± 83 μm versus 918 ± 151 μm, p < 0.0001); invasion of IFITM2- and IFITM3-transduced cells was similar to controls (respectively, 920 ± 109 μm, 901 ± 135 μm versus 918 ± 151 μm).
Type I IFN induction inhibits invasion of trophoblast giant cells (TGC) in vivo in mice
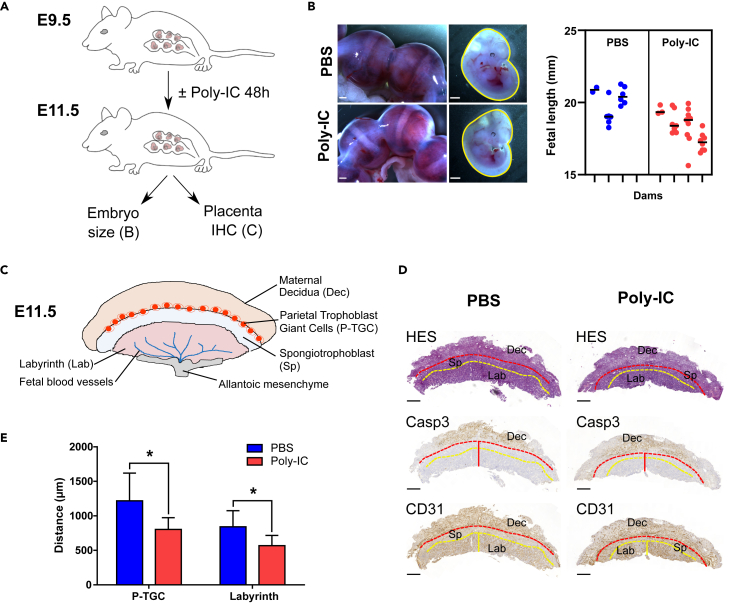
To assess the effect of IFN induction on trophoblast invasion in vivo, we evaluated the migration distances of parietal trophoblast giant cells (P-TGC) and labyrinth thickness in the placentas of mice treated with poly (I:C) (Figure 3). P-TGC arise from the polar trophectoderm from around 7-7.5pc onward. They are intrinsically invasive and phagocytic, and first mediate the invasion of the implanted conceptus into the maternal decidua. They lie between the maternal decidua and the spongiotrophoblast layer. The labyrinth is the site of exchanges between the fetal vasculature and maternal blood spaces, which occurs through syncytialized trophoblast layers (reviewed in17). It forms by days 9.5–10 pc, after the allantois has fused with the chorionic plate. A dose of 60 μg of poly (I:C) was injected intraperitoneally in gestating mice at embryonic day E9.5 (Figure 3A). A 60-μg dose was chosen as this does not induce fetal resorption, which can occur with 200-μg doses in WT mice.10,18 At E11.5, there is a trend for reduced fetal size (p = 0.0579) in dams treated with poly I:C (Figure 3B). Next, P-TGC cell invasion was evaluated by histological analyses of E11.5 placentas using two markers to identify the layers: cleaved caspase 3 (CASP3), which is a marker of apoptosis that is mainly observed in the remodeling decidua, and CD31, a marker of vascular endothelium that is present in the labyrinth and in the decidua but not in the spongiotrophoblast (Figures 3C and 3D). P-TGCs are also directly observed by HES staining, based on their very recognizable phenotype (large, highly polyploids cells). Compared to control (PBS treated) mice, poly (I:C) treatment significantly inhibited TGC invasion (803 ± 168 μm vs. 1218 ± 400 μm, p < 0.05) and significantly reduced labyrinth thickness (567 ± 149 μm vs. 840 ± 233 μm, p < 0.05) (Figure 3E).
Figure 3.
IFN induction inhibits P-TGC invasion and fetal growth in mice
(A) Gestating C57BL/6JRj dams mated with C57BL/6JRj males were injected intraperitoneally with 60 μg poly (I:C) or phosphate-buffered saline (PBS) at E9.5. No fetal resorption was observed in the poly (I:C) injection condition. The fetal lengths were assessed at E11.5.
(B) Representative images of E11.5 uterus and corresponding fetuses. Scale bars: 1 mm. Right panel corresponds to the measurement of fetal length (mm), performed manually using Image J. Numbers of litters were as follows: PBS n = 3 and poly (I:C) n = 4. Mixed model analysis with dam as subject, and fetuses as repeated measures was performed using a nested t-test.
(C) Schematic depiction of the anatomy of the mouse placenta.
(D) Representative images of immunohistochemistry of placental sections at E11.5 for CASP3 and CD31 in both PBS and poly (I:C) treatment groups. The red dashed line represents the P-TGC layer at the decidua (Casp3+)/spongiotrophoblast junction area, the red vertical line represents the distance of P-TGC invasion measured from the chorion to the P-TGC layer, the yellow dashed line depicts the labyrinth/spongiotrophoblast junction area, and the yellow vertical line represents the thickness of the labyrinth (CD31+).
(E) Measurement of the distance of P-TGC invasion and labyrinth thickness in both PBS (n = 3) and poly (I:C) (n = 4) treatment groups. Values are presented as mean + SD. Unpaired t-test was performed to compare treatment to PBS control; ∗p < 0.05, ∗∗∗∗p < 0.0001.
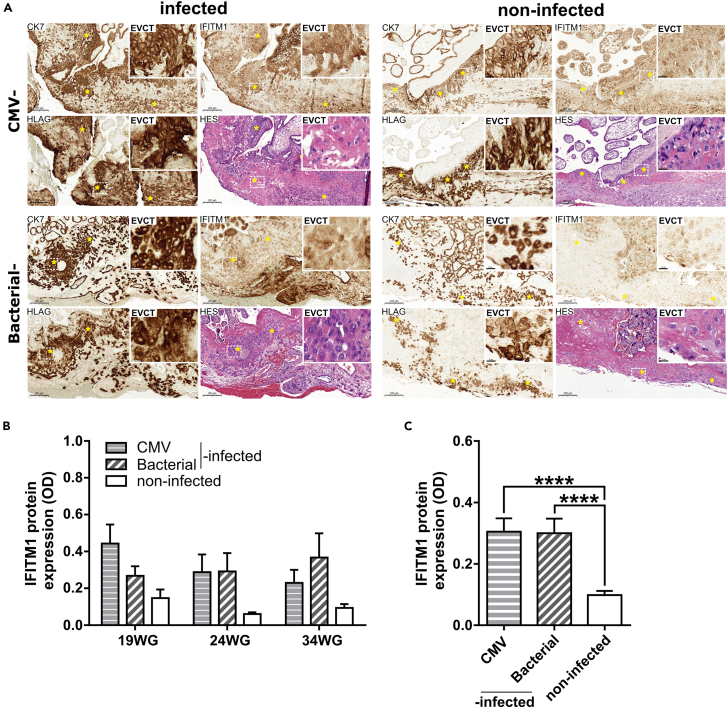
IFITM1 expression is induced in human placental tissues from IFN-mediated pregnancy diseases (TORCH)
After validation of IFITM1 labeling on paraffin section of IFN-β-treated placental explants (Figure S3), IFITM1 expression was then assayed by immunohistochemistry in placental tissue characterized by IFN-induced pathologies of pregnancy (Figure 4A). IFITM1 was detected in villous cytotrophoblasts, syncytiotrophoblast, mesenchyme core, and EVCTs (yellow asterisks). Quantification of the labeling intensity in EVCTs (CK7+/HLAG+) revealed that IFITM1 expression was significantly higher in placental tissue from pregnancies complicated by CMV or bacterial infection compared to that from non-infected pregnancies at all available gestational ages (Figure 4B) or in the entire sample combined (0.305 ± 0.04 and 0.299 ± 0.05 versus 0.098 ± 0.01, p < 0.0001, Figure 4C).
Figure 4.
Induction of IFITM1 expression in placental tissues from IFN-induced pathologies of pregnancy
(A) Representative images of CK7, IFITM1, and HLAG immunostaining and HES staining in placental tissues from CMV- and bacterial-infected patients (left panel) and non-infected controls (right panel). Yellow asterisks represent EVCTs (CK7+, HLGA+). Scale bars: 200 μm. The insets depict a higher magnification of EVCT labeling. Scale bar: 20 μm.
(B) Quantification of IFITM1 protein expression in EVCTs from CMV- and bacterial-infected and non-infected control pregnancies at three different weeks of gestation (19, 24, and 34 WG, n = 1 per condition, 25 non-overlapping ROIs per placenta).
(C) Statistical analyses of IFITM1 protein expression of data shown in B combined by all gestational ages per (n = 3 placentas (19, 24, and 34 WG) per condition (CMV- or bacterial-infected and non-infected), 75 non-overlapping ROIs per condition). Values are presented as mean + SD. One-way ANOVA was performed to compare infected to non-infected controls; ∗∗∗∗p < 0.0001.
Discussion
In earlier work,10 we reported a previously unknown impact of IFITMs on placental development and pregnancy outcome. We described how IFITMs impaired the formation of the multinucleated syncytiotrophoblast by inhibiting the syncytin-mediated cell fusion process, as observed in vivo in mice and in vitro in human trophoblastic cells. Since the syncytiotrophoblast is involved in placental exchanges and endocrine function all throughout gestation, we proposed that overexpression of IFITMs could explain the placental dysfunctions associated with IFN-mediated disorders. In the present study, we aimed to determine whether IFITMs also impair human trophoblast invasion/migration, another critical step of placental development.
During early gestation, trophoblast invasion of the uterine wall is essential for implantation and immune-tolerance of the conceptus, for anchoring the placental villi in the maternal uterus, and for the remodeling of the uterine arteries, which results in low-resistance blood vessels that provide optimal maternal-fetal exchanges. The physiological process of trophoblastic cell invasion is tightly controlled during the first trimester and is required for proper placental development and normal pregnancy outcome.19,20 Indeed, impaired trophoblast invasion has been implicated in gestational pathologies, such as FGR and preeclampsia.21,22
We used different models to study the role of IFITMs on trophoblastic cell invasion in a variety of contexts. Using human primary EVCTs, HTR8/SVneo trophoblast cells, and first-trimester placental explants, we observed that, as we have reported in primary villous trophoblasts,10 IFN-β strongly upregulated IFITM1, IFITM2, and IFITM3.
In vitro, treatment with IFN-β decreased trophoblastic cell invasion in each of these three human placental cell models. Ectopic transduction demonstrated that only IFITM1 decreased significantly the invasion process. IFITM2/3 expression did inhibit trophoblastic fusion.10 However, it is not excluded that endogenous expressions of IFITM2/3 participate to the IFN-induced inhibition of the cell invasion process. Further studies using gene knock-out will need to be performed to further dissect the role of IFITM1 in trophoblast invasion and to rule out the role of endogenous IFITM2/3 in this process.
Trophoblast invasion is often considered as a pseudo-tumoral process because of the similarity of the mechanisms. In the literature, IFITM1 has been reported to be highly expressed in various tumor tissues but, in contrast to our results obtained in trophoblasts, its overexpression has been found to promote, rather than restrict, tumor cell proliferation, invasion, and metastasis.13,23,24,25 Moreover, IFITM2 was also reported to promote proliferation, migration, invasion, and the epithelial-to-mesenchymal transition in gastric cancer cells in vitro.26 Interestingly, IFITM overexpression may be viewed as a biomarker in certain tumors. To note, EVCT differentiation also involved an epithelial-to-mesenchymal transition process. IFN-IFITM signaling appears, thus to be different between cancer cells and pseudotumoral trophoblastic cells. It is interesting to note that using HTR8SVneo, we observed that IFITMs did not modify trophoblast proliferation, another discrepancy with tumor cells. The increase in tumor cell invasion mostly involved EGFR pathway, and interaction with IFITM trough caveolin 1 is considered.27 In trophoblasts, EGFR and caveolin 1 also promote invasion.28,29 Furthermore, IFITM1 has been also shown to interact with CD81 and to promote the growth and metastasis of various cancers.27 In trophoblasts, it was shown that in contrast, CD81 inhibits cell invasion.30 This might represent an interesting signaling to investigate the role of IFITM1-regulated cell invasion between cancer and trophoblastic cells.
We also investigated the expression of IFITM1 in situ using sections of human placental tissue obtained from pregnant patients with increased IFN-associated infections. We observed the overexpression of IFITM1 in all trophoblast subtypes, i.e. villous trophoblasts and syncytiotrophoblasts, as well as invasive extravillous trophoblasts. IFITM1 overexpression was also reported in placentas collected from patients with severe preeclampsia associated with defective trophoblast invasion.31 These findings provide a molecular explanation for the placental dysfunctions and pregnancy complications associated with IFN-mediated diseases such as preeclampsia and “TORCH” infections (toxoplasmosis, other, rubella, cytomegalovirus, and herpes).3 Genetic and autoimmune interferonopathies such as systemic lupus erythematosus are also associated with pregnancy complications,18 and elevated levels of IFN can serve as a serum marker of poor pregnancy outcome in these cases as well.32
To further substantiate these results, we evaluated the effect of IFN induction in vivo in mouse placentas, which share homologous cell types with human placentas. For example, parietal trophoblast giant cells (P-TGCs) in mice and EVCTs in human which are located at the cytotrophoblast-uterine interface. Although the P-TGCs performing intrauterine invasion in mice are not as invasive as EVCT in human that invade up to the one-third of the myometrium, they share some analogous functions like anchoring the placenta to the uterus. In an earlier study, we have reported that the IFN inducer poly (I:C), which mimics a viral infection, induced placental IFITM expressions and promoted fetal resorption and placental abnormalities in wild-type mice.10 In the current study, we observed that poly (I:C) inhibited P-TGC invasion in vivo. As expected, labyrinth development, formed by the fusion of trophoblast layers, is also reduced. Both phenotypes likely contribute to type I IFN-associated placental dysfunction and fetal demise in mice.
Taken together with our previous work,10 this study demonstrates that high levels of IFN-β during gestation induce IFITM expression, that is in turn implicated in adversely affecting the ability of EVCTs to invade the maternal decidua and alter the formation of the syncytiotrophoblast. These IFITM-dependent impairments in trophoblast differentiation can then result in placental dysfunction and may lead to complications during pregnancy. It seems that IFITMs are an interesting example of the biological imperative in pregnancy to achieve balance between protection against viral infection and the potential for deleterious effects on placental development. It will be worth determining how these two opposite functions of IFITM1 shaped their evolution and their expression in placental tissues.
Limitations of the study
Although, we used a variety of experimental systems, including models of human trophoblasts (primary extravillous cytotrophoblasts, HTR8/SVneo cell line, and placental explants [7–12 WG]), an IFN-induced mouse model, and histological sections of human placentas associated with IFN-mediated pregnancy diseases to demonstrate that IFITM1 prevents trophoblast invasion in both humans and mice, we are aware that this study has some limitations. Indeed, further studies using IFITM1 null mice will need to be performed to confirm that IFITM1 contributes to this complex process in vivo.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-CD31 antibody | Abcam | Cat# ab28364; RRID:AB_726362 |
| Rabbit anti-Cleaved Caspase-3 (Asp175) Antibody | Cell Signaling Technology | Cat# 9661; RRID:AB_2341188 |
| Mouse anti-Cytokeratin 7 (clone OV-TL 12/30) | Agilent | Cat# M7018; RRID:AB_2134589 |
| Mouse anti-Cytokeratin 18 CytoDEATH Monoclonal Antibody (clone M30) | Roche | Cat# 12140349001; RRID:AB_1613874 |
| Mouse monoclonal anti-FLAG antibody (clone M2) | Sigma-Aldrich | Cat# F1804; RRID:AB_262044 |
| Mouse monoclonal anti-HLAG (clone MEM-G/9), Biotin | Invitrogen | Cat# MA1-19513; RRID:AB_1076721 |
| Mouse monoclonal anti-HLAG (clone 4H84) | Santa Cruz Biotechnology | Cat# sc-21799; RRID:AB_627938 |
| Mouse IFITM1-Specific Monoclonal antibody (clone 5B5E2) | Proteintech | Cat# 60074-1-Ig; RRID:AB_2233405 |
| Rabbit anti-IFITM1 polyclonal antibody | Sigma-Aldrich | Cat# HPA004810; RRID:AB_1851419 |
| Rabbit anti-Fragilis [IFITM2/3] Monoclonal antibody (clone EPR5242) | Abcam | Cat# ab109429; RRID:AB_10865792 |
| Mouse monoclonal anti-ITGA5/CD49e (clone SAM1) | Immunotech | Cat# IM0770 |
| Mouse anti-Vinculin Monoclonal Antibody (Clone hVIN-1) | Sigma-Aldrich | Cat# V9131; RRID:AB_477629 |
| Goat anti-Mouse IgG2a Cross-Adsorbed Secondary Antibody, Alexa Fluor™ 488 | Invitrogen | Cat# A-21131; RRID:AB_2535771 |
| Goat anti-Mouse IgG2b Cross-Adsorbed Secondary Antibody, Alexa Fluor™ 555 | Invitrogen | Cat# A-21147; RRID:AB_2535783 |
| Goat anti-Mouse IgG1 Cross-Adsorbed Secondary Antibody, Alexa Fluor™ 647 | Invitrogen | Cat# A-21240; RRID:AB_2535809 |
| Goat anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor™ 488 | Invitrogen | Cat# A-11034; RRID:AB_2576217 |
| Goat anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor™ 555 | Invitrogen | Cat# A-21429; RRID:AB_2535850 |
| Donkey anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor™ 680 | Invitrogen | Cat# A10038; RRID:AB_2534014 |
| Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor™ 790 | Invitrogen | Cat# A11374; RRID:AB_2534145 |
| Chemicals, peptides, and recombinant proteins | ||
| Accumax | Invitrogen | Cat# 00-4666-56 |
| DMEM/F-12 | Gibco | Cat# 11320033 |
| Fetal Calf Serum (FCS) | Eurobio | Cat# CVFSVF00-01 |
| Recombinant Human IFN-beta Protein | R&D Systems | Cat# 8499-IF |
| Matrigel® Growth Factor Reduced (GFR) | Corning | Cat# 354230 |
| Paraformaldehyde 16% solution | Electron Microscopy Sciences | Cat# 15710 |
| Phalloidin-iFluor 405 Reagent | Abcam | Cat# ab176752 |
| Phosphatase inhibitors | Calbiochem | Cat# 539131 |
| Poly(I:C) | InvivoGen | Cat# vac-pic |
| Protease inhibitors | Calbiochem | Cat# 539131 |
| Puromycin | InvivoGen | Cat# ant-pr-1 |
| RIPA buffer | Thermo Scientific | Cat# 89901 |
| RPMI 1640 Medium | Gibco | Cat# 21875034 |
| TrypLE Express | Gibco | Cat# 12604021 |
| Critical commercial assays | ||
| Bond Polymer Refine Detection Kit | Leica Biosystems | Cat# DS9800 |
| EasySep™ Release Human Biotin Positive Selection Kit | StemCell Technologies | Cat# 17653 |
| Incucyte® ImageLock 96-well plates | Sartorius | Cat# 4379 |
| micro BCA™ Protein Assay Kit | Thermo Scientific | Cat# 23135 |
| Novolink™ Polymer Detection Systems | Leica Biosystems | Cat# RE7200-CE |
| PowerVision Poly-HRP anti-Rabbit IgG | Leica Biosystems | Cat# PV6119 |
| Experimental models: Cell lines | ||
| HTR-8/SVneo | ATCC | Cat# CRL-3271 |
| 293T | ATCC | Cat# CRL-3216 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6JRj | Janvier Labs | RRID:MGI:2670020 |
| Recombinant DNA | ||
| pLV-Empty (pLV-EF1a-IRES-Puro) | Meyer et al.32 | Addgene Plasmid #85132 |
| pLV-IFITM1-N-FLAG | Buchrieser et al.10 | N/A |
| pLV-IFITM2-N-FLAG | Buchrieser et al.10 | N/A |
| pLV-IFITM3-N-FLAG | Buchrieser et al.10 | N/A |
| pTRIP-GFP | Zennou et al.33 | N/A |
| Software and algorithms | ||
| ImageJ v1.52i | NIH | https://imagej.nih.gov/ij/ |
| Image Studio Lite (version 5.2) | Li-Cor Biosciences | https://www.licor.com/bio/ |
| Incucyte Zoom software | Sartorius | https://www.essenbioscience.com/en/resources/incucyte-zoom-resources-support/ |
| Prism 9.5.1 | GraphPad | https://www.graphpad.com/features |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Séverine A Degrelle (severine.degrelle@inserm.fr; severine.degrelle@inovarion.com).
Materials availability
This study did not generate new unique reagents.
Experimental model and study participant details
Human placental samples & ethical statement
Placental tissues from patients who voluntarily and legally chose to terminate pregnancy during the first trimester were obtained from the Cochin Port-Royal maternity unit (Paris, France). The cytomegalovirus- (CMV) / bacterial- infected and gestational age-matched control placentas (19, 24 and 34 WG) were obtained from the department of anatomical pathology at Necker hospital. These biological samples were obtained following informed written consent from patients and approval from our local ethics committee (CPP 2015-mai-13909).
Isolation of extravillous cytotrophoblasts
EVCT cells were isolated from first-term placentas as described in a previous report.33 Briefly, placental villi were cut into small pieces and enzymatically digested three times in a solution containing equal amounts of TrypLE (Gibco, Fisher Scientific) and Accumax (Invitrogen, Fisher Scientific) for 20 min at 37°C under slight agitation (50 rpm). Pooled cell suspensions were filtered through a 70-mm mesh filter. EVCTs were immunomagnetically purified using a biotin-conjugated anti-HLAG antibody (Invitrogen #MA1-19513, Fisher Scientific) and the EasySep Biotin Selection Kit (StemCell Technologies). The resulting cell pellet was suspended in DMEM/F12 supplemented with 10% FCS and the cells were counted using a TC20™ Automated Cell Counter (Biorad).
Generation of stable IFITM-transduced HTR8/SVneo cell lines
HTR8/SVneo (2 × 104) cells were resuspended in 150 μl of RPMI 1640 medium supplemented with 5% FCS containing 5–25 μl of ultracentrifuged lentiviral vectors. Cells were agitated 30 seconds every 5 minutes for 2.5 h at 37°C in a Thermomixer. Then, cells were continuously selected using 1 μg/ml puromycin (InvivoGen). All cell lines were routinely tested for mycoplasma and found to be negative.
Mouse ethics & poly(I:C) challenge
All procedures with animals were approved by the Animal Ethics Committee on March 9, 2016 (approval 2016-0018), authorized by the French Ministry of Research (#6466), and performed in compliance with European regulation 2010/63 EU. C57BL/6JRj mice were purchased from Janvier Labs (France) and bred at Institut Pasteur.
C57BL/6JRj female mice 7 to 10 weeks of age were used for timed mating with C57BL/6JRj males and checked for the presence of a vaginal plug, indicating a gestational age of E0.5. Pregnant mice were injected intraperitoneally at E9.5 with 60 μg of HMW VacciGrade Poly(I:C) (InvivoGen) (30 μl of 2 mg/ml poly(I:C) in PBS), dose equivalent of approximatively 2,7 mg/kg. Control mice were injected with 30 μl of PBS. Mice were sacrificed at E11.5 and fetuses and placentas were harvested and examined. Images of fetuses were acquired using a ZEISS SteREO Discovery V20 microscope and fetal length was calculated using ImageJ. Placentas were fixed overnight at 4°C in 4% PFA for immunohistochemical staining.
Method details
Cell cultures & IFN-β treatment
EVCTs were cultured in a 35-mm dish (5 × 105 cells, TPP) or a 12-well removable chamber slide (3 × 104 cells/well, Ibidi #81201) coated with growth factor–reduced (GFR) Matrigel (5 mg/ml). HTR8SV/neo cells (ATCC #CRL3271) were directly cultured in a 35-mm dish (5 × 105 cells, TPP) or a 12-well removable chamber slide (3 × 104 cells/well, Ibidi #81201). Two hours after seeding, HTR8/SVneo and primary EVCTs were treated with IFN-β (10, 100, or 1000 IU/ml) in culture medium supplemented with FCS. After 48 h of culture, cells were preserved in one of two ways: 1) fixed with 4% PFA for 20 min at room temperature, washed three times in PBS, then stored in PBS at 4°C until immunostaining was performed, or 2) snap-frozen. Cells to be frozen were washed two times with cold PBS, scraped into 1 ml of cold PBS, and transferred into a 1.5-m tube. After centrifugation (a 10-s pulse at 14.000 rpm), the PBS supernatants were discarded and the cell pellets were snap-frozen in liquid nitrogen and stored at −80°C until western blot.
Cell viability assay
EVCT viability was quantified via immunoblotting and immunostaining of cleaved cytokeratin 18 (cCK18), an early marker for apoptosis. The manufacturers’ protocols were followed as described in previous publications.10,34,35
Human extravillous explant culture, IFN-β treatment, and lentiviral infection
Human first-trimester placental villi (n = 9) were collected and the anchoring villi with attached trophoblast cell columns were dissected under microscope. The explants were cultured in triplicate and placed in a 24-well black plate (Ibidi # 82406) coated with GFR Matrigel (5 mg/ml). For IFN-β treatment, villi were cultured in 500 μl of DMEM/F12 supplemented with 10% FCS with or without IFN-β (10, 100, or 1000 IU/ml). After 48 h of culture, explants were fixed with 4% PFA overnight at 4°C, then washed three times and stored in 1X PBS at 4°C until use. For IFITM transduction/lentiviral infection: After 2 h of incubation, lentiviruses were added in the culture medium for 48 h. Then, explants were fixed with 4% PFA overnight at 4°C, washed 3 times, and stored in 1X PBS at 4°C until use.
Plasmids and lentiviral vectors
The pLV-Empty (pLV-EF1a-IRES-Puro) control plasmid was a gift from Tobias Meyer (Addgene plasmid #8513236). N-terminal FLAG-tagged IFITM1, IFITM2, and IFITM3 were cloned into pLV-Empty using the BamHI and EcoRI restriction enzymes to generate the plasmids pLV-IFITM1-N-FLAG, pLV-IFITM2-N-FLAG, and pLV-IFITM3-N-FLAG. The construction of pTRIP-GFP is described in previous work.37
For lentiviral production, 293T cells (ATCC #CRL-3216) were co-transfected with pLV- and pTRIP-derived plasmids, packaging plasmid R8-2, and the VSV-G plasmid as described in a previous publication.38 Lentiviral particles were collected at 36 h, 48 h, and 72 h post-transfection, ultracentrifuged for 1 h at 4°C at 22000 g, resuspended in PBS, and stored at −80°C until use.
Invasion assay
HTR8/SVneo cells were seeded at 5 × 104 cells/well in a fibronectin-coated 96-well ImageLock Plate (Essen Bioscience). As well described, no chemical treatment such as mitomycin-C to arrest HTR8/SVneo proliferation was needed before the migration/invasion assay.39,40,41,42 So, after overnight incubation, cells were confluent, and scratch wounds (∼200 μm) were made using the accompanying WoundMaker device (Essen Bioscience). For the invasion assay, the culture medium was aspirated, and 50 μl of GFR Matrigel (5 mg/ml) was added. After this solidified, 100 μl of culture medium was added and plates were placed into an automated live-cell imager (IncuCyte Zoom; Essen Bioscience). Images were recorded every hour for 48 h. Experiments were conducted at 37°C with 5% CO2. IncuCyte Zoom software was used to analyze the images and determine the rate of cell invasion, graphed as the percentage of Relative Wound Density (RWD); the area under the curve (AUC) was calculated for each condition from mean RWD values and used to compare the treatment groups.
Immunostaining and image analysis
PFA-fixed cells or explants were incubated with primary antibodies diluted in PBS/1% BSA IgG free/0.05% saponin. After overnight incubation at 4°C, cells were rinsed three times with 0.1% Tween-20 in PBS (PBST), and staining was revealed with an appropriate secondary antibody and/or phalloidin for 1 h, in the dark at room temperature. After three washes in PBST, cells were counterstained with DAPI for 10 min at room temperature. Finally, slides were mounted with Fluorescent Mounting Medium (#S3023, Dako) and stored at 4°C. Confocal microscopy images were obtained with a Leica SP8 inverted microscope equipped with Plan Apo x40/1.3 and x60/1.4 oil objectives and processed with ImageJ Version 1.52i (National Institutes of Health, https://imagej.nih.gov/ij/).
The primary antibodies used were: IFITM1 (#60074-1-Ig, Proteintech) 3 μg/ml, IFITM2/3 (#ab109429, Abcam) 1 μg/ml, HLAG (#MA1-19513, Invitrogen) 1 μg/ml, cytokeratin 7 (CK7, OV-TL 12/30, Dako) 1 μg/ml, FLAG-Tag DYKDDDDK (#F1804, Sigma-Aldrich) 1 μg/ml, and integrin-α5 (ITGA5/CD49e, #IM0770, Immunotech) 2 μg/ml. The secondary antibodies were Highly Cross-Adsorbed Goat anti-Mouse or anti-Rabbit IgG (H+L) coupled with Alexa Fluor 488, 555, or 647 (Invitrogen).
Western blot
Protein extracts were prepared from snap-frozen cell pellets using RIPA Buffer (5 mM Tris-HCl (pH 7.6), 150 mM NaCl, 1% NP-40, 1% Sodium desoxycholate, and 0.1% SDS, containing freshly added protease inhibitors (#539131, Calbiochem) and phosphatase inhibitors (#524629, Calbiochem)), and sonicated. Lysates were centrifuged at 14,000 g for 10 min at 4°C, and supernatants were transferred into a new tube. Protein concentrations were determined using the micro BCA™ Protein Assay Kit (#23135, Thermo Scientific). Equal amounts of proteins (40 μg) were separated on 4–15% SDS-PAGE mini-PROTEAN® TGX™ precast proteunder reducing conditions (DTT) and transferred onto a 0.2-μm nitrocellulose membrane using a Trans-blot® Turbo™ Transfer System (Bio-rad). After blocking with 5% non-fat milk in TBS at 4°C overnight, membranes were incubated with primary antibodies in 1% BSA TBS at room temperature for 3 h. The membranes were then washed and incubated with secondary antibodies at room temperature for 1 h. Membranes were washed and scanned with an Odyssey® Imaging System (Li-Cor). The primary antibodies were: anti-IFITM1 (0.75 μg/ml (1:2000), 60074-1, Proteintech), anti IFITM2/3 (0.15 μg/ml (1:2000), ab109429, Abcam), and anti-vinculin (0.1 μg/ml (1:10000), #V9131, Sigma-Aldrich). The secondary antibodies were: Alexa Fluor® 680-conjugated donkey anti-mouse IgG (H+L), highly cross-adsorbed (1:20000, #A10038, Invitrogen); and Alexa Fluor® 790-conjugated donkey anti-rabbit IgG (H+L), highly cross-adsorbed (1:20000, #A11374, Invitrogen). Signal intensity was quantified using Image Studio Lite (version 5.2) software. The densities were normalized to vinculin.
Histological analyses
For immunostaining on murine samples, freshly PFA-fixed placentas (n = 3 PBS, n = 8 Poly(I:C) were embedded in paraffin, and serial sections (4 μm) were stained with HES. For cleaved caspase 3 and CD31 staining, we used a Bond Leica automated immunostainer instrument and paraffin sections were processed for heat-induced antigen retrieval (ER2 corresponding EDTA buffer, pH 9). For detection of CD31, slides were incubated with CD31 primary antibody (#ab28364, 1:50) for 60 min at room temperature and analyzed with the Bond Polymer Refine Detection Kit (#DS9800 Leica Biosystems). The signal was revealed with DAB and counterstained with hematoxylin. For detection of cleaved caspase 3, slides were incubated with cleaved caspase 3 primary antibody (Cell Signaling #9661, 1:100) for 60 min at room temperature. Then, the slides were incubated with PowerVision Poly-HRP anti-Rabbit IgG (#PV6119, Leica Biosystems). The signal was revealed with DAB. Slides were scanned using a Lamina multilabel slide scanner (Perkin Elmer) using brightfield imaging at a magnification of x20. Measurements of the depth of invasion and labyrinth thickness were performed using ImageJ 1.52 software. Depth of invasion and labyrinth thickness were calculated, respectively, as the migration distance of trophoblast giant cells (delimitated by cCasp3 labeling) or the distance of the CD31 labeling area, delimited by the spongiotrophoblastic junction area.
For immunostaining on human samples, freshly PFA-fixed CMV/bacterial-infected and non-infected placentas (collected at three different weeks of gestation: 19, 24, and 34 WG, n = 1 per condition) were embedded in paraffin, and serial sections (5 μm) were stained with HES. CMV and bacterial infections were detected by the department of anatomical pathology at Necker hospital. CMV infection was detected by PCR and/or by CMV immunostaining showing several intra-nuclear inclusions clustered in villi. Bacterial infection was detected by the observation of lesions of acute chorioamniotitis with polynuclear leukocytes which indicates a bacterial infection. For IFITM1, CK7, and HLAG immunohistochemistry, paraffin sections were processed for heat-induced antigen retrieval and incubated with rabbit anti-IFITM1 antibody diluted 1:250 (200 μg/ml; Sigma-Aldrich HPA004810), mouse anti-cytokeratin 7 (CK7) antibody diluted 1:250 (200 μg/ml; Dako OV-TL 12/30), or mouse anti-HLAG antibody diluted 1:400 (200 μg/ml; Santa Cruz, sc-21799). Staining was visualized using Novolink™ Polymer Detection Systems (Leica Biosystems). Slides were scanned using a Lamina multilabel slide scanner (Perkin Elmer) using brightfield imaging at a magnification of x20. Quantification of IFITM1 protein expression was performed on 25 non-overlapping ROIs (24 × 24 pixels) per sample selected on the basis of EVCT labeling from serial sections (HLAG- & CK7-positive labeling) and this all along the placenta section using the ImageJ 1.52 software thresholding analysis as previously described.43,44,45
Quantification and statistical analysis
All measurements were performed at least in three independent experiments. Data are expressed as the mean +SD or sem. Statistical comparison of each group versus control was performed using GraphPad Prism 9.5.1 software. Results were considered significant if the p value was <0.05 (∗), <0.01 (∗∗), <0.001 (∗∗∗), or <0.0001 (∗∗∗∗).
Acknowledgments
The authors wish to thank the consenting patients and the clinical staff midwives of Cochin Port-Royal, Antony and Montsouris Hospitals for providing placental tissues, the Cellular and Molecular Imaging facility (UMS3612 CNRS, US25 Inserm, Faculté de Pharmacie de Paris, Université Paris Cité, Paris, France), the CB-UTechS facilities (Institut Pasteur, France), Olivia Bawa (AMMICa, Gustave Roussy, Villejuif) for her contribution to the histological analyses, and Lindsay Higgins for English editing (http://www.englishservicesforscientists.com). Work in OS lab is funded by Institut Pasteur, Fondation pour la Recherche Médicale (FRM), ANRS, the Vaccine Research Institute (ANR-10-LABX-77), Labex IBEID (ANR-10-LABX-62-IBEID), ANR/FRM Flash Covid PROTEO-SARS-CoV-2, ANR Coronamito, and IDISCOVR.
Author contributions
S.A.D. conceived and designed the work, analyzed and interpreted the results, and wrote the manuscript; J.B., A.D., and F.P. acquired data, and interpreted the results; L.L. provided resources; O.S. and T.F. provided administrative, technical support, expert advice, and edited the manuscript. All authors agreed on the final version.
Declaration of interests
The authors declare no competing interests.
Published: June 15, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107147.
Supplemental information
Data and code availability
Data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyse the data reported in this paper is available from the lead contact upon request.
References
- 1.Pijnenborg R., Dixon G., Robertson W.B., Brosens I. Trophoblastic invasion of human decidua from 8 to 18 weeks of pregnancy. Placenta. 1980;1:3–19. doi: 10.1016/s0143-4004(80)80012-9. [DOI] [PubMed] [Google Scholar]
- 2.Warner J.A., Zwezdaryk K.J., Day B., Sullivan D.E., Pridjian G., Morris C.A. Human cytomegalovirus infection inhibits CXCL12- mediated migration and invasion of human extravillous cytotrophoblasts. Virol. J. 2012;9:255. doi: 10.1186/1743-422X-9-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coyne C.B., Lazear H.M. Zika virus - reigniting the TORCH. Nat. Rev. Microbiol. 2016;14:707–715. doi: 10.1038/nrmicro.2016.125. [DOI] [PubMed] [Google Scholar]
- 4.Lau S.Y., Guild S.J., Barrett C.J., Chen Q., McCowan L., Jordan V., Chamley L.W. Tumor necrosis factor-alpha, interleukin-6, and interleukin-10 levels are altered in preeclampsia: a systematic review and meta-analysis. Am. J. Reprod. Immunol. 2013;70:412–427. doi: 10.1111/aji.12138. [DOI] [PubMed] [Google Scholar]
- 5.Sharma K., Singh R., Kumar M., Gupta U., Rohil V., Bhattacharjee J. First-Trimester Inflammatory Markers for Risk Evaluation of Pregnancy Hypertension. J. Obstet. Gynaecol. India. 2018;68:27–32. doi: 10.1007/s13224-017-0988-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weston S., Czieso S., White I.J., Smith S.E., Kellam P., Marsh M. A membrane topology model for human interferon inducible transmembrane protein 1. PLoS One. 2014;9 doi: 10.1371/journal.pone.0104341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moffatt P., Gaumond M.H., Salois P., Sellin K., Bessette M.C., Godin E., de Oliveira P.T., Atkins G.J., Nanci A., Thomas G. Bril: a novel bone-specific modulator of mineralization. J. Bone Miner. Res. 2008;23:1497–1508. doi: 10.1359/jbmr.080412. [DOI] [PubMed] [Google Scholar]
- 8.Shi G., Schwartz O., Compton A.A. More than meets the I: the diverse antiviral and cellular functions of interferon-induced transmembrane proteins. Retrovirology. 2017;14:53. doi: 10.1186/s12977-017-0377-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li K., Markosyan R.M., Zheng Y.M., Golfetto O., Bungart B., Li M., Ding S., He Y., Liang C., Lee J.C., et al. IFITM proteins restrict viral membrane hemifusion. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buchrieser J., Degrelle S.A., Couderc T., Nevers Q., Disson O., Manet C., Donahue D.A., Porrot F., Hillion K.H., Perthame E., et al. IFITM proteins inhibit placental syncytiotrophoblast formation and promote fetal demise. Science. 2019;365:176–180. doi: 10.1126/science.aaw7733. [DOI] [PubMed] [Google Scholar]
- 11.Zani A., Zhang L., McMichael T.M., Kenney A.D., Chemudupati M., Kwiek J.J., Liu S.L., Yount J.S. Interferon-induced transmembrane proteins inhibit cell fusion mediated by trophoblast syncytins. J. Biol. Chem. 2019;294:19844–19851. doi: 10.1074/jbc.AC119.010611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borg D., Hedner C., Gaber A., Nodin B., Fristedt R., Jirström K., Eberhard J., Johnsson A. Expression of IFITM1 as a prognostic biomarker in resected gastric and esophageal adenocarcinoma. Biomark. Res. 2016;4:10. doi: 10.1186/s40364-016-0064-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hatano H., Kudo Y., Ogawa I., Tsunematsu T., Kikuchi A., Abiko Y., Takata T. IFN-induced transmembrane protein 1 promotes invasion at early stage of head and neck cancer progression. Clin. Cancer Res. 2008;14:6097–6105. doi: 10.1158/1078-0432.CCR-07-4761. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y., Lu R., Cui W., Pang Y., Liu C., Cui L., Qian T., Quan L., Dai Y., Jiao Y., et al. High IFITM3 expression predicts adverse prognosis in acute myeloid leukemia. Cancer Gene Ther. 2020;27:38–44. doi: 10.1038/s41417-019-0093-y. [DOI] [PubMed] [Google Scholar]
- 15.Yang M., Gao H., Chen P., Jia J., Wu S. Knockdown of interferon-induced transmembrane protein 3 expression suppresses breast cancer cell growth and colony formation and affects the cell cycle. Oncol. Rep. 2013;30:171–178. doi: 10.3892/or.2013.2428. [DOI] [PubMed] [Google Scholar]
- 16.Lin T.Y., Chin C.R., Everitt A.R., Clare S., Perreira J.M., Savidis G., Aker A.M., John S.P., Sarlah D., Carreira E.M., et al. Amphotericin B increases influenza A virus infection by preventing IFITM3-mediated restriction. Cell Rep. 2013;5:895–908. doi: 10.1016/j.celrep.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watson E.D., Cross J.C. Development of structures and transport functions in the mouse placenta. Physiology. 2005;20:180–193. doi: 10.1152/physiol.00001.2005. [DOI] [PubMed] [Google Scholar]
- 18.Yockey L.J., Iwasaki A. Interferons and Proinflammatory Cytokines in Pregnancy and Fetal Development. Immunity. 2018;49:397–412. doi: 10.1016/j.immuni.2018.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aplin J.D. Implantation, trophoblast differentiation and haemochorial placentation: mechanistic evidence in vivo and in vitro. J. Cell Sci. 1991;99(Pt 4):681–692. doi: 10.1242/jcs.99.4.681. [DOI] [PubMed] [Google Scholar]
- 20.Kliman H.J. Uteroplacental blood flow. The story of decidualization, menstruation, and trophoblast invasion. Am. J. Pathol. 2000;157:1759–1768. doi: 10.1016/S0002-9440(10)64813-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huppertz B. Placental origins of preeclampsia: challenging the current hypothesis. Hypertension. 2008;51:970–975. doi: 10.1161/HYPERTENSIONAHA.107.107607. [DOI] [PubMed] [Google Scholar]
- 22.Roberts J.M., Cooper D.W. Pathogenesis and genetics of pre-eclampsia. Lancet. 2001;357:53–56. doi: 10.1016/s0140-6736(00)03577-7. [DOI] [PubMed] [Google Scholar]
- 23.Liang R., Li X., Zhu X. Deciphering the Roles of IFITM1 in Tumors. Mol. Diagn. Ther. 2020;24:433–441. doi: 10.1007/s40291-020-00469-4. [DOI] [PubMed] [Google Scholar]
- 24.Yan J., Jiang Y., Lu J., Wu J., Zhang M. Inhibiting of Proliferation, Migration, and Invasion in Lung Cancer Induced by Silencing Interferon-Induced Transmembrane Protein 1 (IFITM1) BioMed Res. Int. 2019;2019 doi: 10.1155/2019/9085435. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Yu F., Xie D., Ng S.S., Lum C.T., Cai M.Y., Cheung W.K., Kung H.F., Lin G., Wang X., Lin M.C. IFITM1 promotes the metastasis of human colorectal cancer via CAV-1. Cancer Lett. 2015;368:135–143. doi: 10.1016/j.canlet.2015.07.034. [DOI] [PubMed] [Google Scholar]
- 26.Xu L., Zhou R., Yuan L., Wang S., Li X., Ma H., Zhou M., Pan C., Zhang J., Huang N., et al. IGF1/IGF1R/STAT3 signaling-inducible IFITM2 promotes gastric cancer growth and metastasis. Cancer Lett. 2017;393:76–85. doi: 10.1016/j.canlet.2017.02.014. [DOI] [PubMed] [Google Scholar]
- 27.Friedlová N., Zavadil Kokáš F., Hupp T.R., Vojtěšek B., Nekulová M. IFITM protein regulation and functions: Far beyond the fight against viruses. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.1042368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dai Z., Sheng F., Sun N., Ji Y., Liao Q., Sun S., Yang F., Li W. Caveolin-1 promotes trophoblast cell invasion through the focal adhesion kinase (FAK) signalling pathway during early human placental development. Reprod. Fertil. Dev. 2019;31:1057–1067. doi: 10.1071/RD18296. [DOI] [PubMed] [Google Scholar]
- 29.Massimiani M., Vecchione L., Piccirilli D., Spitalieri P., Amati F., Salvi S., Ferrazzani S., Stuhlmann H., Campagnolo L. Epidermal growth factor-like domain 7 promotes migration and invasion of human trophoblast cells through activation of MAPK, PI3K and NOTCH signaling pathways. Mol. Hum. Reprod. 2015;21:435–451. doi: 10.1093/molehr/gav006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen L., Diao Z., Sun H.X., Yan G.J., Wang Z., Li R.T., Dai Y., Wang J., Li J., Ding H., et al. Up-regulation of CD81 inhibits cytotrophoblast invasion and mediates maternal endothelial cell dysfunction in preeclampsia. Proc. Natl. Acad. Sci. USA. 2017;114:1940–1945. doi: 10.1073/pnas.1617601114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gormley M., Ona K., Kapidzic M., Garrido-Gomez T., Zdravkovic T., Fisher S.J. Preeclampsia: novel insights from global RNA profiling of trophoblast subpopulations. Am. J. Obstet. Gynecol. 2017;217:200.e1–200.e17. doi: 10.1016/j.ajog.2017.03.017. [DOI] [PubMed] [Google Scholar]
- 32.Andrade D., Kim M., Blanco L.P., Karumanchi S.A., Koo G.C., Redecha P., Kirou K., Alvarez A.M., Mulla M.J., Crow M.K., et al. Interferon-alpha and angiogenic dysregulation in pregnant lupus patients who develop preeclampsia. Arthritis Rheumatol. 2015;67:977–987. doi: 10.1002/art.39029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okae H., Toh H., Sato T., Hiura H., Takahashi S., Shirane K., Kabayama Y., Suyama M., Sasaki H., Arima T. Derivation of Human Trophoblast Stem Cells. Cell Stem Cell. 2018;22:50–63.e6. doi: 10.1016/j.stem.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 34.Longtine M.S., Barton A., Chen B., Nelson D.M. Live-cell imaging shows apoptosis initiates locally and propagates as a wave throughout syncytiotrophoblasts in primary cultures of human placental villous trophoblasts. Placenta. 2012;33:971–976. doi: 10.1016/j.placenta.2012.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shoaito H., Petit J., Chissey A., Auzeil N., Guibourdenche J., Gil S., Laprévote O., Fournier T., Degrelle S.A. The Role of Peroxisome Proliferator-Activated Receptor Gamma (PPARgamma) in Mono(2-ethylhexyl) Phthalate (MEHP)-Mediated Cytotrophoblast Differentiation. Environ. Health Perspect. 2019;127 doi: 10.1289/EHP3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hayer A., Shao L., Chung M., Joubert L.M., Yang H.W., Tsai F.C., Bisaria A., Betzig E., Meyer T. Engulfed cadherin fingers are polarized junctional structures between collectively migrating endothelial cells. Nat. Cell Biol. 2016;18:1311–1323. doi: 10.1038/ncb3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zennou V., Petit C., Guetard D., Nerhbass U., Montagnier L., Charneau P. HIV-1 genome nuclear import is mediated by a central DNA flap. Cell. 2000;101:173–185. doi: 10.1016/S0092-8674(00)80828-4. [DOI] [PubMed] [Google Scholar]
- 38.Compton A.A., Bruel T., Porrot F., Mallet A., Sachse M., Euvrard M., Liang C., Casartelli N., Schwartz O. IFITM proteins incorporated into HIV-1 virions impair viral fusion and spread. Cell Host Microbe. 2014;16:736–747. doi: 10.1016/j.chom.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Albaghdadi A.J.H., Coyle K., Kan F.W.K. Low-Dose Tacrolimus Promotes the Migration and Invasion and Nitric Oxide Production in the Human-Derived First Trimester Extravillous Trophoblast Cells In Vitro. Int. J. Mol. Sci. 2022;23 doi: 10.3390/ijms23158426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lye P., Bloise E., Nadeem L., Peng C., Gibb W., Ortiga-Carvalho T.M., Lye S.J., Matthews S.G. Breast Cancer Resistance Protein (BCRP/ABCG2) Inhibits Extra Villous Trophoblast Migration: The Impact of Bacterial and Viral Infection. Cells. 2019;8 doi: 10.3390/cells8101150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salomon C., Kobayashi M., Ashman K., Sobrevia L., Mitchell M.D., Rice G.E. Hypoxia-induced changes in the bioactivity of cytotrophoblast-derived exosomes. PLoS One. 2013;8 doi: 10.1371/journal.pone.0079636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshida T., Takada K., Komine-Aizawa S., Kamei Y., Ishihara O., Hayakawa S. Lactobacillus crispatus promotes invasion of the HTR-8/SVneo trophoblast cell line. Placenta. 2021;111:76–81. doi: 10.1016/j.placenta.2021.06.006. [DOI] [PubMed] [Google Scholar]
- 43.Degrelle S.A., Chissey A., Stepanian A., Fournier T., Guibourdenche J., Mandelbrot L., Tsatsaris V. Placental Overexpression of Soluble CORIN in Preeclampsia. Am. J. Pathol. 2020;190:970–976. doi: 10.1016/j.ajpath.2019.12.012. [DOI] [PubMed] [Google Scholar]
- 44.Jensen E.C. Quantitative analysis of histological staining and fluorescence using ImageJ. Anat. Rec. 2013;296:378–381. doi: 10.1002/ar.22641. [DOI] [PubMed] [Google Scholar]
- 45.Deval G., Nedder M., Degrelle S., Rogozarski J., Vignaud M.L., Chissey A., Colzin S., Laguillier-Morizot C., Coumoul X., Boland S., et al. Benzo(a)pyrene and Cerium Dioxide Nanoparticles in Co-Exposure Impair Human Trophoblast Cell Stress Signaling. Int. J. Mol. Sci. 2023;24 doi: 10.3390/ijms24065439. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyse the data reported in this paper is available from the lead contact upon request.