Abstract
The goal of this study is to develop a general strategy for bacterial engineering using an integrated synthetic biology and machine learning (ML) approach. This strategy was developed in the context of increasing L-threonine production in Escherichia coli ATCC 21277. A set of 16 genes was initially selected based on metabolic pathway relevance to threonine biosynthesis and used for combinatorial cloning to construct a set of 385 strains to generate training data (i.e., a range of L-threonine titers linked to each of the specific gene combinations). Hybrid (regression/classification) deep learning (DL) models were developed and used to predict additional gene combinations in subsequent rounds of combinatorial cloning for increased L-threonine production based on the training data. As a result, E. coli strains built after just three rounds of iterative combinatorial cloning and model prediction generated higher L-threonine titers (from 2.7 g/L to 8.4 g/L) than those of patented L-threonine strains being used as controls (4–5 g/L). Interesting combinations of genes in L-threonine production included deletions of the tdh, metL, dapA, and dhaM genes as well as overexpression of the pntAB, ppc, and aspC genes. Mechanistic analysis of the metabolic system constraints for the best performing constructs offers ways to improve the models by adjusting weights for specific gene combinations. Graph theory analysis of pairwise gene modifications and corresponding levels of L-threonine production also suggests additional rules that can be incorporated into future ML models.
Keywords: Strain engineering, Threonine, ML, Hybrid-machine learning, E. coli, AI-Driven
Highlights
-
•
Increased L-Threonine production in bacteria.
-
•
Machine learning for metabolic engineering.
-
•
Hybrid deep learning models used to predict additional gene combinations.
-
•
Metabolic engineering based on multiple gene modifications.
-
•
Metabolic engineering of E. coli for optimization of increased threonine production.
1. Introduction
SynBio is a field of science that involves engineering organisms for useful purposes by redesigning them to have new properties. Such organism redesign and engineering open unexplored routs to study fundamental principles of organization and behavior of complex biological systems. Besides its impact on basic research, SynBio has untapped potential to produce improved industrial organisms which are used in many areas of human activity, from agriculture (Zhang et al., 2017) to chemical production (Clomburg et al., 2017) and pharmacology (Guo et al., 2017). Microbial production of amino acids (AA), which reached $26 billion in 2021 (https://www.grandviewresearch.com/industry-analysis/amino-acids-market) is one of such areas. We chose AA bioproduction to demonstrate real-world utility of synthetic biology guided by machine learning for effective organism engineering.
In the 1970s, strains of Corynebacterium glutamicum, Brevibacterium flavum, and Escherichia coli were constructed to produce threonine with titers measured in tens of grams per liter (Kase and Nakayama 1972) (Hirakawa et al., 1973). Gradual improvements of AA production driven by genetic selection and gene cloning described in (Wittmann and Becker 2007), and continued since, drove costs of these products down to less than $1.5 per kilogram (bulk price at Alibaba, 2022). A comprehensive review of newer strategies employed for strain engineering in AA bioproduction (with the emphasis on metabolic engineering, flux analysis and comparative genomics) is presented in (Becker and Wittmann 2012). Exhaustive summary of AA strain development tools and approaches, starting with classical mutagenesis and system metabolic engineering and including such modern instruments as optogenetic gene regulation, AA sensors, riboswitches and so on, can be found in (Lee and Wendisch 2017) and in (Ma et al., 2017). An example of such approach is the utilization of a DNA scaffold system, in which a zinc finger protein served as an adapter for binding enzymes of the threonine biosynthetic pathway which increase the proximity of enzymes and local concentrations of metabolites, and led to improved threonine production rates (Lee et al., 2013). The same time, machine learning was missing in the list of impactful strategies in reviews cited above.
An early example of effective incorporation of system biology and metabolic analysis for threonine production in E. coli can be found in (Lee et al., 2007). Rational reengineering of metabolic fluxes in a wild type C. glutamicum resulted in high levels of lysine production, as described in (Becker et al., 2011). Similar approach was later applied for bioproduction of diaminopentane, a nylon building block (Kind et al., 2014). Comparative-genomics provided an additional tool to choose impactful genes and combine mutations found in genomes of strains selected for high levels of lysine production which generated new improved C. glutamicum strains (Ohnishi et al., 2002) (Ikeda et al., 2006). In the last paper, 16 genes chosen by metabolic analysis as potentially affecting lysine production were a source of beneficial mutations which were pooled together to improve production. A similar approach to strain engineering was applied to threonine production of E. coli (Zhu et al., 2019) (Zhao et al., 2020), but in these studies, instead of transferring mutations, authors up- or down-regulated key genes selected by flux analysis. Rather complicated metabolic analysis allowed them to cherry-pick additional genes that would positively-affect threonine production in E. coli including elimination of proP or ProVWX transporters (Wang et al., 2021) and overexpression of the phaCAB gene cluster (Wang et al., 2019). Reduction of the genome size (Lee et al., 2009) and use of photosynthesis as a carbon source (Korosh et al., 2017) were also applied to improve microbial production. Lab evolution aimed to increase sugar utilization was applied in (Papapetridis et al., 2018).
Multiple OMICs datatypes are being used to guide engineering of industrial microorganisms. For example, key genes affecting 5-methyltetrahydrofolate biosynthesis were identified combining modular gene engineering and transcriptomics. (Yang et al., 2022). Quantitative kinetic analysis of microbial metabolism and Bayesian inference were applied to modeling the central carbon metabolism and lysine production in (St John et al., 2019). Dynamic metabolomics applied in guided Design Build Test Learn (DBTL) cycling was reviewed in (Vavricka et al., 2020). Meta-analysis of adaptive laboratory evolution (ALE) aggregated data from 63 ALE experiments in Escherichia coli K-12 MG1655 revealed global trends that underlie ALE-derived strain design principles. (Phaneuf et al., 2020). Technical approaches described in this paper present a successful case of information extraction from independently selected high-producing industrial organisms.
RNA-seq data may contain clues to additional genes (besides more obvious candidates derived from flux analysis) which can affect bioproduction. However, the multidimensional nature of metabolic and regulatory interaction complicates the process of finding such genes. A possible solution to this problem can be found in gene expression clustering described in (Sastry et al., 2019), in which unsupervised machine learning was applied to a compendium of 250 E. coli RNA-seq datasets. Ninety-two statistically independent signals which modulate the expression of specific gene sets–iModulons–were identified in this work. The current release of iModulonDB covers three organisms (Escherichia coli, Staphylococcus aureus and Bacillus subtilis) with 204 iModulons, which can be expanded to additional organisms (Rychel et al., 2021). Near-saturated targeted mutagenesis was applied to four primary routes that affect lysine flux (Bassalo et al., 2018). 16,300 mutations were incorporated in 19 genes involved in lysine biosynthesis, lysine degradation, lysine transport, and expression regulation to test their effect on lysine production. Reaction stoichiometry, thermodynamics, and mass action kinetics that form modeling frameworks used to describe how organisms allocate resources towards both growth and bioproduction are reviewed in (Suthers et al., 2021). This review focuses on the latest algorithmic advancements that have integrated these principles into a quantitative framework.
Numerous high-yielding production strains were engineered using methods described above. With all their success, there is an intrinsic problem of knowledge gaps, which must be bridged in such knowledge-driven strain engineering. Finding optimal combinations of modifications of genes represents another, even bigger problem. If one wants to construct a production strain which contains 7–10 “improved” genes from a list of 20–40 candidates, they must find the best variant in 107 gene combinations.
Machine learning (ML) algorithms make predictions by extracting patterns directly from experiments. That is why, unlike metabolic modeling (MM), which is based on balances derived from reconstructed metabolic networks, ML is much less sensitive to knowledge gaps. Moreover, ML can be applied to navigate vast combinatorial spaces with models built with much smaller training sets, potentially solving the second problem outlined above. At the same time, ML has its own limitations: its models may not generalize and extrapolate well (as noted in (Oyetunde et al., 2018)), although DBTL cycling allows models to iteratively validate and correct themselves. ML also depends on mechanistic inputs to build initial training sets. So, integration of MM and ML should produce mechanism-guided machine learning frameworks for prediction and characterization of the function of complex biological systems, something that is required for reliable organism engineering.
Several important papers describe ML applications for organism engineering, although we do not know examples of such microorganisms used in the existing industrial processes. Microbial production data curated from ∼100 papers together with additional features derived from the genome-scale metabolic model simulations were used to predict productivity by data augmentation and ensemble learning (e.g., support vector machines, gradient boosted trees, and neural networks in a stacked regressor model (Oyetunde et al., 2019). High-dimensional support vector machine (SVM) models were successfully applied to predict enzymes that mediate alternative branches in plant alkaloid biosynthesis from homologous candidate sequences which were successfully validated in complex pathway engineering experiments (Moliner et al., 2019). SVM was also applied to predicting microorganism growth temperatures and enzyme catalytic optima with confirmations by meta-analysis with Pearson correlation coefficients from 0.75 to 0.96 (Li et al., 2019). ML studies of Pseudomonas putida KT2440 transcriptomes reveals its transcriptional regulatory network (Lim et al., 2022). Integration of knowledge mining, genome-scale modeling (GSM), and ML were applied to predict Yarrowia lipolytica titers achievable in bioproduction of organic acids and terpenoids. Pathway fluxes of central metabolism were estimated using GSMs and flux balance analysis to provide metabolic features used to train ML ensemble models, which were used to predict strain production titers with R2 up to 0.87 (Czajka et al., 2021). Synthetic biology tools for metabolic control including ML-based metabolic modeling, and CRISPR-derived methods for transcription inhibition and activation are reviewed in (Lv et al., 2022). Most of these papers although informative, do not provide a direct guidance for strain engineering.
A different paper was published by (Zhang et al., 2020), in which a biosensor that detects tryptophan titer was used to sample data from constructed strains which provided a high-quality training set for ML which was used to optimize the metabolic pathway of tryptophan production. Constraint-based modeling for predicting single gene targets retrieved 192 impactful genes, covering 259 biochemical reactions. A 7776-member combinatorial library (which was assembled from different preconstructed parts consisting of five genes selected from GSM simulations, each controlled by six different promoters selected from transcriptomics data mining) was analyzed to build predictive models for tryptophan biosynthesis rate in yeast. More than 500 of the possible genetic designs from the library were constructed and generated 124,000 experimental data points used for training of a model, which suggested specific improved engineering combinations.
Flux analysis, comparisons of mutations in the independent strains, and RNA-seq data can generate testable lists of gene candidates for combinatorial organism engineering. However, finding the most productive combinations of such genes requires going through a vast space of possible combinations. Our work contains strong evidence that ML can be a reliable guiding tool in this quest. We chose threonine production as a test case because 50 years of strain development by large research groups has produced efficient industrial strains that can serve as a benchmark for our study. Additionally, the impact of many metabolic genes on threonine production is well understood, which facilitates mechanistic interpretation of the effects observed in our study.
2. Methods
2.1. Strains
All strains are derived from Escherichia coli K-12. Constructed strains represent combinations of (1) individual or combined deletions of one to three out of 8 E. coli genes; (2) two host strains; (3) several modifications of Thr operon and the asd gene (inserted in chromosome or cloned in plasmid); and (4) 7 cloned and overexpressed “supplementary’ genes. These modification, and other factors varied in collected samples are listed in Supplementary Table 1 and are reflected in sample names, as shown in Supplementary Table 2.
Axygen 1.1 mL 96-deep-well plates were used to grow the cultures using QuickSeal breathable membrane at 37 C, with 80% humidity at 1000 RPM in 200 μL of minimal seed media (KH2PO4 1 g/L, Bis-Tris 40 g/L, (NH4)2SO4 10 g/L, Glucose, 7.5 g/L, MgSO4.7H2O 0.3 g/L, pH 7.0) containing proline (300 mg/mL), isoleucine (100 mg/mL), methionine (200 mg/mL), lysine (100 mg/mL), diaminopimelate (100 mg/mL), and thiamine (1 mg/mL) with appropriate antibiotics. After about 24 h, 20 μL was transferred to 220 μL of minimal fermentation media (KH2PO4, 1 g/L, Bis-Tris, 40 g/L, (NH4)2SO4, 30 g/L, Glucose, 30 g/L, MgSO4.7H2O,1.2 g/L, Na3Citrate, 1.0 g/L, MnSO4.H2O, 0.02 g/L, FeSO4, 0.03 g/L, pH 7.0) containing the same amino acids and thiamine as the seed media but without antibiotics. IPTG was added at 5 h at a final concentration of 1 mM, to induce the threonine operon.
2.2. Cloning and chromosomal manipulations
2.2.1. Construction of plasmids
Plasmids were made by Gibson assembly using the NEB Gibson Assembly cloning kit or the NEBuilder HiFi DNA Assembly kit. Q5 DNA polymerase from NEB was used for PCR reactions, which were treated with DpnI to cut residual plasmid or chromosomal DNA. PCR fragments were treated with the NEB Monarch PCR clean up kit. DNA concentrations were determined using a nanodrop.
Three core plasmids were constructed containing the threonine pathway genes, thrABC and asd genes. The thrABC and the asd genes were cloned into modified vector pSR58.6 (Schmidl et al., 2014) which resulted in a plasmid containing colE1 origin, the chloramphenicol resistance gene, with the thrABC-asd-gfp operon controlled by tac promoter, and lacIq gene. The resulting three constructs have the following content Plasmids pfb6.4.2 and pfb6.4.3 and contain the feedback resistant thrA gene G433R from ATCC21277. Plasmid pfb6.4.3 contains two copies of the lacIq gene, which increased the growth rate of plasmid-carrying strains when compared with pfb6.4.2.
Several supplemental plasmids were constructed for overexpression of enzymes by cloning the following genes: E. coli genes, rhtA, zwf, aspC, ppc, aceBA, pntAB and a E. coli codon-optimized, Rizobium etli pyc gene (Gokarn et al., 1999). They were cloned in a modified vector pSR43.6 (Schmidl et al., 2014), resulting in plasmids containing the p15A origin, spectinomycin resistance gene, and the gene of interest controlled by a constitutive promoter J23108 (Moore et al., 2016). Combinations of aspC, ppc, and pntAB were made in the same vector under the control of the same constitutive promoter. These plasmids contained pntAB-aspC, pntAB-ppc, aspC-ppc, and pntAB-aspC-ppc).
2.2.2. Chromosomal deletions
Eight individual genes, eleven combinations of two genes, and three combinations of three genes (all genes used are shown in Table 1) were deleted using lambda red gene-replacement system described in (Datsenko and Wanner 2000). Chromosomal deletions were made in strains MG1655 and ATCC 21277 using the Gene Bridges Quick & Easy E. coli Gene Deletion Kit. All the deletions were made to remove the entire coding region from the start codon to the stop codon and the deleted genes replaced by Km resistance marker. They were confirmed by PCR. The deletions were moved to different strains and combined with other deletions by P1 transduction (Thomason et al., 2007). Kan genes were flipped out using the flanking frt sites according to the Gene Bridges protocol.
Table 1.
Genes used in combinatorial cloning and their metabolic functions. Mod column shows which gene was deleted “- “, or upregulated “+”, Modifications of genes with indicated ‘-’ mode appear in the text with ‘D’ prefix (Dtdh).
| Gene name | Enzyme Name | Role | Expected effect | Mod | Reference |
|---|---|---|---|---|---|
| thrABC | Bifunctional aspartokinase/homoserine dehydrogenase 1, homoserine kinase, and threonine synthetase | Thr/Lys/Met biosynthesis | Amplification should activate threonine overproduction | + | (Kozlov Iu et al., 1980) |
| asd | Aspartate semialdehyde dehydrogenase | Thr/Met biosynthesis | Amplification should activate threonine overproduction | + | (Debabov 2003) |
| lysC | Lysine-sensitive aspartokinase 3 | Thr/Lys/Met biosynthesis | Increased expression of lysine feedback resistant allele is expected to improve production | – | (Ogawa-Miyata et al., 2001) |
| metL | Bifunctional aspartokinase/homoserine dehydrogenase 2 | Thr/Lys/Met biosynthesis | Deletion/Increased expression are expected to affect methionine and SAM production | – | (Neidhardt and Curtiss 1996) |
| rhtA | Threonine/homoserine exporter | Threonine export | Increased expression is expected to improve threonine production | + or - | (Livshits et al., 2003) |
| aceBA | Malate synthase A and Isocitrate lyase | Carbon backbone | Increased expression of the glyoxylate shunt is expected to improve threonine production | + | (Liu et al., 2019) |
| ppc | Phosphoenolpyruvate carboxylase | Carbon backbone | Increased expression of ppc is expected to improve threonine production (more precursor available) | + | (Lee et al., 2007) |
| pyc | Pyruvate carboxylase | Carbon backbone | Increased expression facilitates the flow of carbon to oxaloacetate and aspartate in C. glutamicum, expected to cause the same effect in E.coli | + | (Peters-Wendisch et al., 2001) |
| ptsG | PTS system glucose-specific EIICB component | Carbon backbone | Deletion of the PTS system for glucose uptake is expected to make more precursor available | – | (Zhu et al., 2019) |
| dhaM | PEP-dependent dihydroxyacetone kinase, phosphoryl donor subunit | Carbon backbone | Deletion is expected to disrupt putative allosteric regulation of dihydroxyacetone kinase (encoded by dhaK) and negative regulation of dha operon | – | (Gutknecht et al., 2001) (Bachler et al., 2005) |
| zwf | Glucose-6-phosphate 1-dehydrogenase | Carbon backbone, Co-factor biosynthesis |
Increased expression is expected to increase availability of threonine precursors (glucose-6-p and NADPH) | + | (Becker et al., 2007) |
| pntAB | NAD(P) transhydrogenase (membrane-bound) | Co-factor biosynthesis | Increased expression expected to compensate for NADPH depletion | + | (Liu et al., 2019) |
| aspC | Aspartate aminotranferase | Threonine biosynthesis/Competing pathways | Increased expression is expected to increase precursor pool | + | (Zhao et al., 2020) |
| lysA | Diaminopimelate decarboxylase | Competing pathways | Deletion is expected to make more precursors available | – | (Lee et al., 2007) |
| dapA | Dihydrodipicolinate synthase | Competing pathways | Deletion is expected to affect availability of all threonine precursors (aspartate, glutamate, NADPH) | – | |
| tdh | L-threonine 3-dehydrogenase | Threonine catabolism | Deletion is expected to diminish threonine degradation | – | (Lee et al., 2007) |
2.2.3. Genome insertions
The tac promoter was inserted into the genome by of MG1655 and ATCC21277 as described above replacing threonine leader peptide (thrL) and native thr promoter and upstream of asd geneomoter replacing its pr. Finally, the kan marker gene was flipped out. Two version of tac-controlled thrABC operon, one with a wild type thrA gene and another which was feedback-resistant to threonine (thrA*), were inserted in both host strains replacing wild type copy with its regulatory region.
2.2.4. P1 transduction
P1 transduction was done with P1vir using protocols described in (Thomason et al., 2007).
2.3. Measurements
2.3.1 Threonine was measured using the BioVision PicoProbe Threonine Assay Kit (Fluorometric). The protocol was modified for use in 384 well plates (Corning Assay Plate 384 well low Volume Black with Clear Bottom #3540) and scaled down to 20 μL instead of 100 μL. The threonine samples were diluted 100 to 1000-fold to be in the linear range of the assay, and standard curves were run on each 384 well plate. The fluorescence measurements were made on the Hidex Sense Plate Reader.
2.3.2 Glucose was measured using the Sigma-Aldrich glucose assay kit (GAGO20) scaled down for use in 96 well assay plates (Corning Clear Flat Bottom Assay Plate #9017). Each well contained 70 μL assay mix 5 μL sample (100-fold dilution). Each plate had a glucose standard curve for calculating glucose concentration.
2.4. ML models used to predict optimal strain design from production data and strain composition
We used a Deep Neural Network to predict threonine production from combinations of strain engineering elements shown in Table 1 and Supplementary Table 2. The feature vector consisted of indicators for individual strain modifications, with multi-valued modifications such as the core threonine operon specification one-hot encoded, for a total of 33 input dimensions and one output dimension (threonine titer). When multiple experiments were performed on identical samples, the trimean of all results was used as the target.
DeepLearning4J was used for model training, prediction, and hyperparameter tuning. For most models, we used a batch normalization layer followed by 2–9 feed-forward layers with no gradient normalization and an output layer loss function of L2 (squared error). To minimize the effect of outliers, the model search was tuned to optimize the mean absolute error rather than the squared error. The activation functions tested were hard hyperbolic tangent, rectified linear unit, Gaussian error linear unit, and the normalized exponential function Softmax. The same function was used for all inner layers, but a second function was sometimes used for the input layer. The model was validated using 9-fold cross-validation. The mean error during cross-validation for the models constructed was 3.1% of the total output range, with an IQR less than 1.0% of the total output range. (The output range varied from 1.2 g/L for the smallest model to 6.4 for the largest.)
One significant concern was the performance of the model beyond the range of the data used for training and testing the model. The upper range of the production used to train the first model was 2.6 g/L. We were interested in values over 2.0 g/L, but only 18 samples of a total 1963 were in this range. We therefore decided to use 1.2 g/L as a cutoff, giving us 50 samples in range. We built a second model that was trained and tested only on the 1913 samples at 1.2 or below. This second model was then run against the full set to determine what predictions it would make for the samples producing above 1.2 g/L.
The model predictions were compared against the actual values from the experimental run using a simple utility to classify predictions and actual results by whether or not they were above 0.8. There were 241 samples not used to train the model, including the original 50 with production levels over 1.2 g/L and 191 holdouts used as a testing set. Of 50 samples with actual production values over 1.2, 20 were predicted over 0.8 by the model, a success rate of 40%. No samples with production values less than 0.8 were predicted to have output over 0.8, which indicates a low rate of false positives.
2.5. Graph reconstruction
Cytoscape 3.9.1 (Otasek et al., 2019) was used to reconstruct the graphs from the data presented in Supplementary Table 3 and for the topological analysis.
3. Theory
3.1. Overall strategy
Our approach, which we call “agnostic strain engineering”, relies on general computational tools, flux analysis and comparative genomics. It does not depend on extensive process-specific information (hence the term “agnostic”), and, in principle, can be applied to a broad set of strain-construction projects. The approach consists of the following steps: (1) flux-analysis-based selection of genes whose inactivation or overexpression can affect bioproduction of a specific chemical; (2) combinatorial cloning aimed to construct an initial training set of strains for ML analysis; (3) ML modeling to predict optimal combinations of modified genes; and (4) DBTL cycling, in which subsequent sets of strains suggested by ML are constructed achieving a gradual strain improvement (Fig. 1). Building successful ML models, which predict better combinations of modified genes and guide strain engineering, is the key element of the approach.
Fig. 1.
AI-driven “agnostic” strain engineering HTP-High Throughput.
3.2. Gene selection
Sixteen genes and operons were selected to be incorporated in the construction of initial combinatorial training set based on their positions in the network of metabolic reactions of E. coli iJO1366 (Orth et al., 2011) metabolic model (Fig. 2). The focus was made on reactions in a proximity to the biosynthetic pathway and responsible for a supply of precursors for the biosynthesis of threonine, threonine efflux, as well as pathways of threonine degradation. The enzymes in the threonine biosynthetic pathway: aspartate kinase, homoserine dehydrogenase, homoserine kinase, and threonine synthase are in an operon whose expression is controlled by threonine, also aspartate kinase and homoserine dehydrogenase are allosteric regulated by threonine. These regulatory mechanisms must be altered to ensure the consistent increased flow of the metabolites. The flux through the pathway was found to be strongly dependent on aspartate concentrations that we decided to improve via modulation of the upstream functions of anaplerotic pathways, glyoxylate shunt and diminishing of phosphoenolpyruvate loss to the PTS metabolite transport.
Fig. 2.
Genes chosen for modification and their positions in metabolic fluxes. Selected pathways (dashed line) and reactions (solid line) are highlighted as follows: (green) carbon backbone biosynthesis; (blue) threonine biosynthesis; (pink) competing pathways and threonine catabolism; (orange) threonine efflux, and (red) NADPH regeneration. Genes subjected for deletion are indicated in red and genes subjected for induction are indicated in purple. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Besides its role in threonine biosynthesis, aspartate is a precursor for biosynthesis of methionine and lysine. The impact of a crosstalk in this metabolic system is hard to quantitate using the flow models as the fluxes through these pathways are subjected to additional regulation and the kinetics of the enzymes may be nonlinearly affected by high, non-physiological, concentrations of the precursor(s). Lysine biosynthesis, however, also shares two other precursors with biosynthesis of threonine (NADPH and glutamate) and was considered as the impactful explicit competitor. The anaplerotic reactions chosen for modulation (phosphoenolpyruvate carboxylase and pyruvate carboxylase) were shown to impact lysine biosynthesis in C. glutamicum (Peters-Wendisch et al., 2001) and our model predicted their differential impact on threonine production in the engineered strains with and without inhibition of lysine biosynthesis.
NADPH restoration is a noted bottleneck in the biosynthesis of threonine. A strong impact of membrane NADPH transhydrogenase (encoded by pntAB) on NADPH restoration has been demonstrated earlier (Sauer et al., 2004) and this gene became one of the first candidates for a modulation. We also chose glucose-6-phosphate 1-dehydrogenase, which has a double function affecting carbon and NADPH availability. To reduce experimental complexities of combinatorial approach, we limited the number of seed functions for the first round of ML and did not include other potentially impactful metabolic sources of NADPH, functions supporting a provision of glutamate, and other threonine degradative functions, in this round of ML. We also did not include the housekeeping pathways competing for aspartate, such as pyrimidine and asparagine biosynthesis.
Table 1 shows genes selected for combinatorial cloning which encode enzymes involved in: (1) glycolytic biosynthesis of carbon backbone (aspartate) via phosphoenolpyruvate (ppc, pyc, zwf), (2) glyoxylate shunt (aceAB), (3) biosynthesis of threonine from aspartate (asd, aspC, threABC, lysC, metL), (4) restoration of NADPH from NADP+ (zwf) or NADP+ and NADH (pntAB), (5) pathways competing for phosphoenolpyruvate (ptsG, dhaM) and the precursors for biosynthesis of threonine from aspartate (dapA, lysA), (6) threonine catabolism (tdh), and (7) threonine efflux (rhtA).
These genes were selected based on generic information on the roles of the proteins they code in E. coli pathways. The impact of most of these proteins on threonine production was already demonstrated (see references in Table 1), which makes threonine test case seemingly less “agnostic”, but having such prior information allowed us to find mechanistic explanations and additional validation of phenomena observed in high-producing ML-designed variants. Additionally, even with all the information available for the impacts of individual genes, there was no systematic understanding of the effects of their combinations, a major challenge which we address in our study.
4. Results
4.1. Initial combinatorial cloning
Strains constructed in the study consisted of 4 major variable elements: (1) the thrABCasd core operon; (2) a chromosomal knockout of a “negative” gene or a group of such genes expected to decrease threonine production; (3) overexpressed “positive” genes or operons expected to increase production; (4) a bacterial host.
The following construction parts were used for combinatorial cloning:
-
1.
“Positive” genes rhtA, zwf, aspC, ppc, aceBA, pntAB and a codon-optimized pyc gene from Rizobium etli (Gokarn et al., 1999) cloned using a modified vector pSR43.6 (Schmidl et al., 2014), resulting in plasmids containing the p15A origin, the spectinomycin resistance gene, and the gene of interest controlled by the constitutive promoter J23108 (Moore et al., 2016).
-
2.
The thrABC and the asd genes cloned into modified vector pSR58.6 (Schmidl et al., 2014) which resulted in a plasmid containing colE1 origin, the chloramphenicol resistance gene, with the thrABC-asd-gfp operon controlled by tac promoter, and lacIq gene (as described in Methods). Plasmids pfb6.4.2 and pfb6.4.3 contain the feedback resistant thrA gene G433R from ATCC21277. Plasmid pfb6.4.3 contains two copies of the lacIq gene, which increased the growth rate of plasmid-carrying strains when compared with pfb6.4.2.
-
3.
Eight individual “negative” genes (metL, lysA, ptsG, lysC, dapA, rhtA, tdh, dhaM), and six combinations of two genes (dapA-tdh,lysA-tdh, lysC-tdh, metL-tdh, ptsG-tdh, rhtA-tdh) were deleted using lambda red gene-replacement system described in (Datsenko and Wanner 2000). After individual deletions were constructed, they were moved between strains using P1 transduction (Thomason et al., 2007). In the text and the figures, modifications of “negative” genes are indicated by ‘D’ (deleted) prefix (as Dtdh).
-
4.
These gene combinations were introduced into two host strains, MG1655 (wild type) and ATCC21277 (one of the early production strains).
Seven plasmids carrying “positive” genes were individually transformed into each of 70 strains carrying chromosomal deletions of “negative” genes and different versions of a core threonine operon, all of it incorporated into two E. coli hosts, which resulted in 385 combinatorial clones. (Their composition, presence, or absence of modified or deleted genes, was verified using PCR). 1998 samples, representing different growth time points and results of IPTG induction were grown in 96-deep-well plates in synthetic media. Threonine was measured in the media using the BioVision PicoProbe. Threonine Assay Kit and glucose was measured using the Sigma-Aldrich glucose assay kit as described in Methods. Table 1, Supplement, contain the data on threonine titer, yield, and OD at 24 h. Strain modifications and growth conditions are reflected in sample names as shown in Supplementary Table 2.
In the initial run, the best engineered strain produced 2.6 g/L, while the industrial control strains produced 3.1–3.8 g/L (These industrial strains represent variants constructed more than a decade ago. No published information is available on most recent ones, but based on personal communications, their titer may be increased by 20–30%.) By the end of the project, after three DBTL cycles, 64 of the engineered strains had produced more than 4 g/L of threonine, the best making 8.4 g/l. Each sample was tested multiple times and the trimean of all tests was used as the computed production level for the sample. If one test's production differed from the others by more than 1.2 g/L it was discarded as erroneous outlier. Over the course of the project, 959 samples were tested 3 times, 1095 twice, 502 only once, and 943 four or more times. The mean threonine production levels ranged from 0.0 to 8.4 g/L, heavily weighted toward 0.0. The average error in a test (comparing the observed titer to the trimean value used in the models) was 0.22 g/L.
In general, strains carrying an upregulated feedback resistant threonine operon and a pair of “positive” modifications of gene expression produced up to 2.6 g/L of threonine when grown in minimal media in microtiter-plates. Modifications found to contribute to high threonine production most were: (1) chromosomal location of the IPTG-induced threonine operon; (2) addition of overexpressed ppc, pntAB, or aspC; and (3) deletions of tdh, dapA and metL.
4.2. Optimization of strain design by deep learning
A set of strains carrying pairwise combinations of genes selected by flux analysis was used to train a deep learning (DL) model to predict threonine production in strains carrying more complex combinations of engineering elements (gene parts). We expect that a single improved strain may carry up to 8 different modified genes (knockouts or overexpressed), which creates a virtual space of over 107 variant strains to be analyzed by DL. The key question—whether the effects of pairwise gene combinations observed in the initial set of strains would be representative of far more complex constructs in the much larger virtual space—was answered by experimental validation of DL models.
4.2.1. Building DL models
Of the 1998 samples described in the previous section, 35 were derived from various industrial control strains. The remaining 1963 were used to train a deep learning (DL) model to predict production from combinations of strain-engineering elements used as descriptive attributes (features). Feature vectors were constructed with indicators for individual strain modifications, with multi-valued modifications (such as the core threonine operon specification) one-hot encoded for a total of 26 input dimensions and one output dimension (threonine titer). Hyperparameter searches were optimized for lowest mean absolute error.
When validated with holdout (testing) samples, our initial model (shown in a Fig. 3A) had mean absolute error of 0.06 g/l and Pearson coefficient of 0.85, which was highly encouraging. However, when it was applied to the 1,376,256-variant virtual space of possible gene combinations, no strain was predicted to produce more than 3 g/L. This could have been either a result of poor initial selection of genes used for combinatorial design, or a limitation of DL models, which simply cannot predict production outcomes exceeding values observed in the training set.
Fig. 3.
Validation of predictions of threonine concentration generated by a deep learning model. A. Predicted versus actual for 1724 training samples (blue) and 191 holdout testing samples (red). B. Predicted versus actual for a model trained on samples producing less than 1.2 g/L of threonine. Red dots to the right of 1.2 g/L show poor quantitative performance of the model outside of its training range. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
To test the second hypothesis (and better understand the model's predictive performance in the tail of distribution of threonine production values), we trained a DL model with samples that only produced less than 1.2 g/L (a cutoff chosen to insure at least 50 samples were considered “high”). We evaluated such model with the holdout set of 241 samples, 50 of which had production levels greater than or equal to 1.2 g/L (Fig. 3B). High-producing samples in the holdout set showed very little correlation between the predicted and actual numeric values of threonine production (Pearson's coefficient of 0.41). This meant that regression analysis could not generate accurate numeric predictions. Nonetheless, there was a significant clustering of samples that produced over 1.2 g/L and that were predicted to produce over 1.2 g/L. In our first test, in which no machine learning model was used, only 1% of samples produced over 1.2 g/L. The above exercise tells us that we can expect 40% of the samples chosen by the model to produce at that high level—a clear improvement. Thus, using the model enables us to choose better samples for each subsequent test run, even though the predictions of the model have a low accuracy by most metrics.
The question being addressed - “Which combinatorial designs would produce high quantities of threonine?” - can be treated as a classification problem, which can be solved by putting strains in groups exceeding or falling below of certain production cutoffs. The classification approach is, however, complicated by the fact that the target class of interest, a highly productive set of strains, represents only a small percentage of the initial training set (0.7%). Thus, a model that classified all strains as low producing variants would have over 99% accuracy. Our solution to this problem was to use neural network regression to predict the production level first. Then, the resulting model would assign numeric weights to modifications and combinations thereof that are used to compute predicted production levels even if those levels are well below the value selected for “high” production. In addition, when selecting strains to test for the next round, the numerical production value provided a natural way to sort the most-likely candidates from the rest.
Once the numeric regression model is generated, its results are classified depending on whether or not they are above a certain threshold, and this is used to generate classification metrics for the model, a hybrid regression/classification approach. Using 1.2 g/L as a cutoff between high-producing and low-producing variants, the confusion matrix for the testing set in the first model is shown in Table 2 below.
Table 2.
Confusion matrix for initial model using 1.2 g/L cutoff for first model.
| Predicted ≥ 1.2 | Predicted <1.2 | |
|---|---|---|
| Actual ≥ 1.2 | 2 | 1 |
| Actual <1.2 | 1 | 192 |
Like the full dataset, the testing set is heavily biased toward negative results, so the value of the standard accuracy measure is limited, and an MCC (Mathews Correlation Coefficient) score is more useful. The MCC score is 0.66, indicating a good correlation.
4.2.2. DBTL cycling
4.2.2.1. Overview
The initial DL model was used to predict improved strain designs (defined as producing over 1.2 g/L). 71 strains predicted to be high-producing, plus 176 additional strains representing slight variations of these or carrying gene combinations that were insufficiently explored in the first round were constructed, tested and included in the training set used for the second round.
Table 3 shows the results of three testing runs. The first run was unique in that there was no ML data. So, the lower rows indicate numbers computed from the previous run and used to select samples to test for the next run.
Table 3.
The results of three testing runs.
| statistic | Run 1 | Run 2 | Run 3 | Detailed description |
|---|---|---|---|---|
| Tot size | 1998 | 948 | 1131 | Number of samples run. |
| New size | 1998 | 786 | 862 | Number of samples new to this run. |
| Max prod all | 5.49 | 5.83 | 8.39 | Maximum production output. |
| Max prod constructed | 2.90 | 5.83 | 8.39 | Maximum production output for a constructed strain. |
| Max prod control | 5.49 | 5.49 | Maximum production output for a control strain. | |
| Tot strains | 393 | 286 | 583 | Number of strains run. |
| New strains | 393 | 247 | 431 | Number of strains new to this run. |
| Tot constructed | 385 | 280 | 583 | Number of constructed strains in this run. |
| New constructed | 385 | 247 | 431 | Number of constructed strains new to this run. |
| Predictions computed | 1376256 | 57986 | Total number of predictions computed in virtual space to build run. | |
| High predictions computed | 72900 | 1933 | Total number of predictions in virtual space ≥ 1.20. | |
| Tot predictions | 2057 | 3643 | Number of samples with predicted values from the model used to create the run. | |
| New predictions | 412 | 726 | Number of samples with predicted values new to this run. | |
| Max prediction | 2.63 | 8.30 | Maximum prediction from the model used to create the run. | |
| AUC | 0.65 | 0.86 | Area-Under-Curve for classification by production level of samples new to the run. |
Tot size is the total number of samples run at that time. Note that when we build the model later, all samples, even those from previous runs, are used. New size is the number of samples that had never been run before. The rest are being re-tested for verification. Max prod all is the maximum production output. There is a clear increase here, which is why we felt we were improving. Tot strains and new strains identify unique strains. For a given strain, we might run it at different time intervals, but we almost always run it both induced and uninduced. Tot constructed and new constructed indicate how many of the strains were built by us, rather than being controls. Note that in the last run all of them were constructed. The other rows are self-explanatory. The increase in AUC indicates that the predictions are getting better, but the max prod constructed is the real measure of our success.
The DL model from the second round was the final model produced by the project. It used a batch normalization layer followed by seven feed-forward layers. The inner layer widths were 22, 19, 16, 13, 10, 7, and 4. The activation function chosen was rectified linear unit (RELU). The mean error during cross-validation was 2.5% of the total output range (here 0.0 to 8.4), with an inter-quartile range equal to 0.4% of the total output range. Because of the scarcity of training data with yields of 1.2 g/L or greater, mean absolute error increased at higher output, 0.22 for strains yielding less than 1.2 g/L, and 1.3 for high-yielding strains. This model's predictions on the training and testing sets are shown in Fig. 4 below.
Fig. 4A.
Second ML model; Validation with holdout samples. 4B Predicted versus actual for a model trained on the second DBLT round. Only samples introduced in the third experiment run for which predictions were made (726 samples) are shown. The 1.2 g/L cutoff is shown in red, the 4.0 g/L cutoff in green. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Once again using 1.2 as the cutoff between low-producing and high-producing, the confusion matrix for the second model is shown in Table 4. The MCC for this model is 0.80, indicating a strong correlation.
Table 4.
Confusion matrix for initial model using 1.2 g/L cutoff.
| Predicted ≥ 1.2 | Predicted <1.2 | |
|---|---|---|
| Actual ≥ 1.2 | 13 | 5 |
| Actual <1.2 | 3 | 284 |
4.2.2.2. ML1 round
1998 samples derived from 393 strains engineered in the MM-guided construction round were used to train the first DL model. In this model, an exhaustive set of all possible combinations of features was generated and used as input to the trained model to predict threonine production. This set of feature combinations (referred to subsequently as samples) was processed to eliminate samples that were logically impossible (for example, knocking out and promoting the same gene), leaving 1,376,256 total samples. Of these strains, 72,900 had predicted threonine yield greater than 1.2 g/L (roughly 5.3%). The strains with the highest predicted threonine production were chosen, and these were further filtered to insure they were no more than two construction steps from existing strains. A total of 71 strains together with another group of 176 designed “around” these strains were built. Of the 71 predicted strains, 45 (63%) were confirmed in the experiment, a 24-fold enrichment over their overall 2.6% fraction in the first run, which was designed using metabolic modeling alone. Of these 71 AI-directed variants, 12 produced more threonine compared with the best industrial control strain NRRL B-21593.
4.2.2.3. ML2 round
278 new strains constructed in ML1 round were added to the training set. Adding these strains shifted a ratio of “high-producing” (higher than 1.2 g/L) variants from 2.6% (MM-round) to 7.8% (MM + ML1 rounds) and moved highest measured threonine production from 2.6 to 5.8 g/L. As expected, a new ML model used to suggest engineering designs for the ML2 round was able to significantly extend prediction range (from 3 to 8.3 g/L). Out of 321 strains were predicted to produce more than 1.2 g/L of threonine using the hybrid regression/classification approach as discussed above, 50% were experimentally confirmed (64 of these strains produced more than 4 g/L). For 104 strains predicted to produce more than 4 g/l, 19 (18%) were experimentally confirmed. The best strain from among those predicted over 4 g/L produced 8.15 g/L as average of three measurements. We observed significantly increased production characteristics of the strains constructed in the second round, but inability to rate strain candidates within a class obviously forces us to construct more strains to catch the best variants. A similarly low Pearson's coefficient was observed for experimentally tested strains in the ML-generated designs of the first DBLT round, which was then explained by demonstrated inability of ML to extrapolate (as seen in Fig. 3B). However, this explanation cannot be applied to the second round, in which training-set production values were increased to nearly 6 g/l. A reason for ML's inability to generate accurate numeric predictions in this case can be due to increase of combinatorial complexity in subsequent DBTL cycles and due to underrepresentation of specific gene combinations in the training sets.
4.2.2.4. Summary of DBTL cycling
Using 1.2 g/L as a cutoff for high performance, our first model had a predictive accuracy of 99%, true positive rate of 66%, and a fallout rate of 0.5% in validation with holdout samples when applied to all strains constructed in the first round of engineering and carrying pairs of modifications. This means that within the tested range of the model (in this case, up to 2.6 g/L) we expect to catch 66% of the high-output strains and to only pick up 2.5% of the low-producing variants by accident. When expanded to a virtual space in which more complex modifications were allowed, the model performed slightly worse, with only 63% of the predicted high-producing strains being confirmed in the actual experiments (Table 5). Our second model had a predictive accuracy of 97%, true positive rate of 73%, and a fallout rate of 1.0%. The model predicted that 321 of the tested strains would perform over 1.2 g/L. In actual experiments, 50% of these samples were confirmed. Similarly, 104 strains were predicted to perform over 4.0 g/L and 19 of them exceeded that production level. Using the 1.2 cutoff, the second model had an F1-score of 0.76 and an MCC of 0.79.
Table 5.
Summary of 3 DBTL rounds.
| Rounds | Max production, average for 3 measurements | Success, 1.2 g/L | predicted, 1.2 g/L | success, 4.0 g/L | predicted, 4.0 g/L |
|---|---|---|---|---|---|
| MM | 2.7 | NA | NA | NA | NA |
| ML 1 | 6.2 | 63% | 71 | NA | 0 |
| ML 2 | 8.4 | 50% | 321 | 18% | 104 |
The theoretical yield of threonine conversion from glucose is 81% g/g (122% mole/mole (Lee et al., 2007)). (Together with the production rate of a target molecule, yield is the most important property of an industrial strain). In the conditions tested, yield of the control industrial strains vary from 22 to 50%. Several of the most productive strains constructed in the first DBLT round had glucose utilization yields of 24–28%, the best strain from the second round has yield of ∼43%.
To summarize, we found that: (1) deep learning models demonstrate 91–98% accuracy in validation with holdout samples when labels (production values) of such samples stay within training set values; (2) regression, but not classification accuracy, deteriorates for samples outside of the training range of production values; (3) up to 63% of the strains predicted to produce more threonine than a cut-off (chosen based on distribution of productivity values in the training set) were confirmed experimentally, and (4) 64 best deep learning-designed strains produced more threonine than the industrial strains used as a controls.
4.3. Graph analysis of pairwise combinations of gene modifications used for ML-training
Though an individual impact of each gene in strains with triplet or pairwise combinations of modifications is not explicitly obvious, it can be assessed by AI from a training set and guide strain engineering (as demonstrated above). In order to illustrate how an AI model may lead to predictive conclusion, we analyze graphs topology to infer relations between the gene modifications. We define that a pair of gene modifications is in a relation if it is combined in one engineered strain. If gene modifications are presented as graph vertices, and their paired combinations are represented by the adjacent edges, we can use the corresponding values of threonine production (Supplementary Table 3) as edge weights. In this weighted graph (Fig. 5) each node can be characterized by; (1) a number of adjacent edges (pairwise gene modification combinations in threonine producing strains); (2) weights of the adjacent edges (an impact of the paired modifications on threonine production); and (3) a position in respect to other nodes/gene modifications (that would reflect a gene's network outreach and indicate its impactful potential). Multiple highly weighted edges adjacent to one node suggest a strong impactful potential of the corresponding gene modification on threonine production. Strong interconnectivity of the impactful nodes and particular topological characteristics of this interconnectivity (such as closeness of a graph, when all the nodes have highly weighted connections to at least two other nodes in a group) could indicate a potentially high synergistic value of a combination of the corresponding gene modifications in one strain.
Fig. 5.
A Cytoscape 3.9.1 graph reconstruction from the data on threonine production by strains with paired gene modifications (Supplementary Table 3). A. The Degree Sorted Circular Layout with genes placed anticlockwise in accordance to a number of their adjacent edges. B, C. Hierarchical layout for modifications that appear in pairwise-modified strains with > 1.2 g/l threonine production. Each gene modification is presented as a node and each edge presents a combination of the adjacent gene modifications in one engineered strain. The color of an edge corresponds to a threonine production value at 24h growth point with induced thrABC/asd operon (according to the scale shown). The edges of closed graphs defined by gene modifications with the most positive impact on threonine production are highlighted in C. Abbreviations for gene modifications are as explained in Table 1. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The graphs have been constructed using Cytoscape 3.9.1 (Otasek et al., 2019). The Degree Sorted Circular Layout of the graph (Fig. 5A) places genes in accordance to a number of the adjacent edges in anticlockwise direction starting from Dtdh, which has a highest connectivity degree. The edge weights (yellow-green edges as <1.2 g/l and blue ones as >1.2 g/l associations) are distributed unevenly, which indicates different individual impact of different gene modifications. Notably, one gene modification (Dtdh) is adjacent to only and few (DdapA, pntAB) to almost only >1.2 g/L threonine production levels, where the others (zwf, ptsG, lysC) to only <1.2 g/l. Two main hubs, Dtdh and DdapA, are the most impactful in terms of their highly weighted network outreach (characterized by the values of Degree and the Average Shortest Path-the number of links to reach any other node in the graph) and their high Betweenness Centrality (a measure based on shortest paths and a way of detecting the amount of influence a node has over the flow of information in a graph) (Supplementary Table 4).
Fig. 5 B shows a graph for gene modifications that are associated with threonine production which is higher than 1.2 g/L. The hierarchical layout places nodes in accordance with their rank from the top to the bottom of the graph, the highest having more linear connections. Overall, Dtdh, and DdapA appear as invariable, required modifications, and we can suggest potential effects for other gene modifications, shown on this graph as subordinated, when they are paired with Dtdh, or DdapA. We use a term ‘driver’ to highlight a primary role of these modifications in increased threonine production. DdhaM, which is also characterized by a strong Connectivity and Betweenness Centrality is, however, present in strains with <1.2 g/l of threonine production and though might have a lesser ‘driving’ significance. DrhtA demonstrates strong connectivity characteristics, but moderate associated levels of threonine production that are never the highest for any of the paired modifications. It suggested rather a neutral role of this genomic intervention, turned into negative in strains with more complex constructs (Supplementary Table 1).
Among the engineered strains of the ML-led rounds, the pntAB, Dtdh,DdapA, DdhaM,DMetL combination of gene modifications showed the strongest impact on threonine production (Supplementary Table 1). Connectivity of the nodes corresponding to these gene modifications has unique topological characteristics in the reconstructed graph, and this group of nodes can be graphically distinguished (Fig. 5 C, highlighted by pink color). All pairwise combinations of pntAB, Dtdh and DdapA led to the highest levels of threonine production, that is reflected in the structure, where the corresponding highly weighted edges form a closed graph (a structure where each node is connected to at least two other nodes). This graph could be explicitly extended to DdhaM and DmetL nodes. Though the edge connecting DhaM and DmetL is associated with only 0.94 g/l threonine production value, adjacent Dtdh-DmetL and Ddham-DdapA edges are strongly weighted. All these considerations can be incorporated in the AI algorithm in favor of a line combination Dtdh-DmetL-DdhaM-DdapA in a ML model.
The power of ML-guided design goes beyond our mechanistic understanding of systems. A graph analysis of predictions made by ML increases our confidence in them by providing sanity checks. Besides this, graph analysis, which is based on the independent, purely algebraic approach can be integrated in ML models making them more selective. A predictive ML algorithm ranks likewise associations between paired gene modifications favoring consistently larger weights (threonine production levels) withing a group of gene modifications paired in different combinations, and any data associated with gene pairs, including a prior knowledge of the effects of their combinations and omics data (gene proximity in metabolic network, imodulones or genomic neighborhoods, co-expression, and etc.) and other strain characteristics (growth rate, substrate consumption, etc), can be integrated via adjustment of the weights of the pairwise connections. The dataset used in our example was effective in revealing of a driving role of Dtdh and DdapA. Our analysis also pointed to gene combinations (pntAB, Dtdh, DdapA), and (pntAB, Dtdh, DdapA, DdhaM, DdmetL), which should be found in strains with improved threonine production, as it is also suggested by ML. There are still some other combinations that look promising from the connectivity point of view. A larger dataset size would be required to apply other topological analysis algorithms that can improve an overall accuracy of ML predictions.
4.4. Mechanistic analysis of the results of ML-driven design
1038 clones were constructed in three engineering rounds. 64 produced more than 4 g/L of threonine, with the best making 8.4 g/L (average for 3 tests) (Supplementary Table 1). ML-guidance was used to suggest engineering designs and up to 63% of such designs were confirmed experimentally. However, the approach also has certain shortcomings, which are discussed above, making ML-guided strain engineering less precise. Mechanistic analysis of observed effects of gene combinations used for strain construction has a potential to help with it.
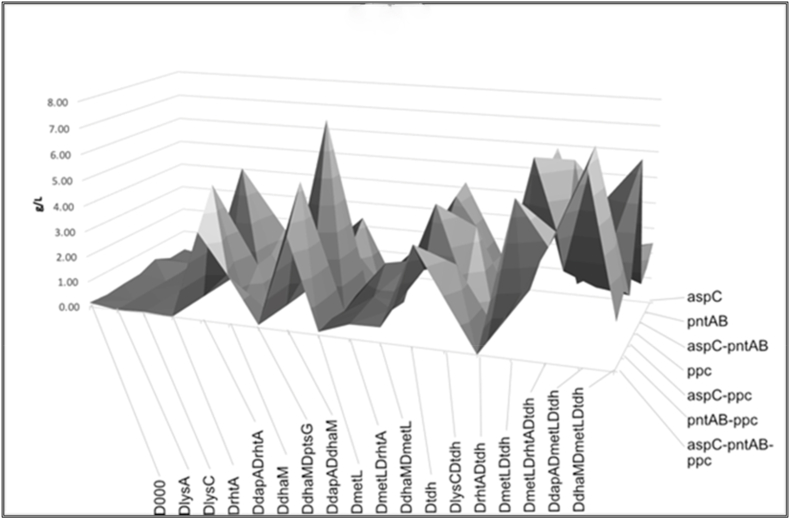
Threonine production as a function of gene modifications is presented in Fig. 6, which shows both conservation of gene patterns among high-producing variants and seemingly unpredictable abrupt effects of relatively small differences in gene composition.
Fig. 6.
Patterns in the effects of individual genes and their combinations on threonine production in the engineered strains, based on data presented inSupplementary Table 1. Deletions of tdh, dapA genes and overexpression of pntAB, aspC and ppc genes look most effective in different combinations.
ATCC21277 parental strain occurred to be the most promising host. and, as expected, overexpression of asd and thrABC was required to achieve highest levels of production. Though strains bearing Ddap and Dtdh produce high levels of threonine even without thr operon induction, its induction increases threonine production further. Effects of Dtdh and Ddap were confirmed in multiple complex constructs, where almost all strains bearing these modifications produced comparably high levels of threonine. PntAB induction was not equally effective in all combinations that is probably due to differential TCA and, consequently, NADH production rate modulations in different strains, and potential confronting action of differentially expressed soluble transhydrogenase encoded by sthA. Upregulated ppc (with or without upregulated aspC in a place of pntAB) had a slightly lesser but also beneficial effect on threonine production. We do not see any positive effect of overexpression of zwf (that would, however, correspond to its modest effect in NADPH production in E. coli (Lindner et al., 2018) (Olavarria et al., 2014). So far, the best-producing strains combine upregulated pntAB, and deletions of tdh, dapA, metL, dhaM. A loss or a substitution of any gene from the pntAB, tdh, dapA, metL, dhaM combination in the best producing strains is associated with a drop in a threonine production level (Fig. 7).
Fig. 7.
Combinations of genes targeted in the best constructed strains. Each color represents one complex construct/one strain: numbers on the right are the levels of threonine production. The most impactful genes are highlighted (red outline, white font). Blue arrows point to the interpretable functional connections between the modifications likely to be responsible for the observed outcomes. The genes tdh, dapA, dhaM and metL were deleted and the rest of the genes were overexpressed. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
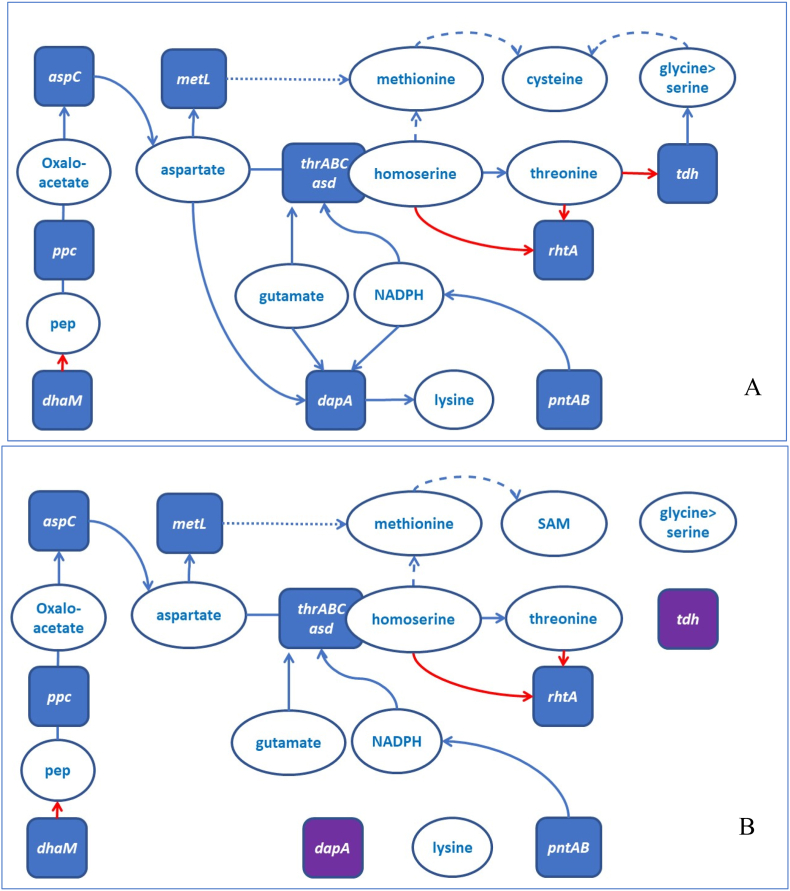
The outcomes of the strain engineering highlight a complexity of the interactions between the gene modifications favored by ML predictions (represented by blue arrows on Fig. 7). Although a prediction of impactful combinations using only prior knowledge of bacterial metabolism remains a challenge, it is possible to explain why a particular combination of modifications is effective in concrete cases predicted by ML. Moreover, as shown below, analysis of the results of ML predictions can be used to improve the algorithms and to increase predictability via rule-guided ML models. Metabolic shifts in a proximity of the threonine biosynthetic pathways, which may explain observed results, are shown in Fig. 8.
Fig. 8.
A simplified diagram of metabolic connections and metabolic shifts in a proximity of the modulated threonine biosynthetic pathways. A. Strains without gene deletions, B. Strains with dapA and tdh being deleted. Red arrows suggest metabolite depletion. Dashed lines-general directions of metabolic effects or metabolite flows. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
As it is summarized in the diagram, a deletion of dapA, the first gene in lysine biosynthetic pathway, affects flows of aspartate, glutamate, NADPH, pyruvate and succinate, that would impact threonine and methionine biosynthesis, respiration and biosynthesis of the cofactors originated from the TCA intermediates. Consequently, it has much stronger effect than a deletion of lysA, which encodes a terminal enzyme in the pathway. Deletion of dhaM accompanying deletion of dapA can be especially beneficial to compensate for PEP depletion (Gutknecht et al., 2001). Similarly, tdh deletion not only stops catabolic degradation of threonine but likely shifts equilibrium in serine-methionine-cysteine metabolic system and SAM-dependent regulation, due to a decreased l-threonine-3-dehydrogenase-dependent production of glycine (Weissbach and Brot 1991) (Lee et al., 2007). Though an exact role of metL deletion is not clear, in a complement to Dtdh it may affect regulation of methionine biosynthesis at the DNA level (affecting the downstream operon) or protein level (suggesting a regulatory or metabolite channeling role of MetL). PntAB, which encodes for the main supplier of NADPH (Lindner et al., 2018) in most engineered strains, can support two reactions in threonine biosynthesis and one reaction in biosynthesis of glutamate, making its induction one of the most impactful modifications. Another impactful modification, induction of rhtA, have a role in both threonine and l-homoserine efflux, where the latter, if accumulated, may affect a diversion of the pathway towards methionine and SAM (Wang et al., 2005) and even suppress biosynthesis of glutamate (Kotre et al., 1973).
From these examples, we see that topology of metabolic network can be utilized as a prior knowledge in weighting of particular gene associations in a ML model. A significance of Dmetl and DdhaM, for instance, was underestimated in ML predictions for combined constructs with two or more modifications of genes from pntAB, Dtdh or DdapA group, which could be compensated by weighting derived from mechanistic analysis. We can also find additional constrains for ML models analyzing cases in which complex constructs have performed worse than predicted. In these cases, the main overestimation can be explained by a diminishing value of sequential functions in linear pathways (functions encoded by ppc, aspC, for instance), simultaneous activation of which would not necessarily lead to the ML-predicted synergism. Lowering weights of the consequent functions in a linear metabolic pathway will be considered in future ML models.
5. Discussion
Finding an optimal combination of genes with altered levels of expression to achieve maximal production of targeted metabolite in the engineered bacterial strain, was a key problem addressed in this study. ML models, which connect threonine production and gene modifications (taken as features in the training sets of engineered strains), provided a solution to this problem guiding engineering designs with sufficient accuracy. With 1034 clones constructed in three engineering rounds, we were able to increase threonine production from 2.6 g/l in the first round to 8.6 g/L in the third one, having inference accuracy (defined as accuracy of prediction for a strain to belong to a high-producing class) up to 63%. In microtiter plates, 64 of these strains produced more threonine than several industrial strains used for comparison (strains were grown in synthetic buffered high-glucose media in extensively aerated deep-well plates reaching an optical density of 3.0–5.0 in 24 h). This cell density is 10–20 times lower than observed in industrial fermentation conditions in which threonine titer may reach 130 g/L in 48–72 h.
To apply DL to strain design, two problems had to be addressed: (1) inability of regressor models to extrapolate to unobserved threonine yields, and (2) heavy bias towards low-producing strains in the training data, which cause a classifier that simply classified everything as low-producing to be 98% accurate. Second problem was resolved via a hybrid DL approach in which, at first, a regression model was constructed, so that information about the effects of the engineering changes would be represented in the form of equation coefficients, and then, interpretation of the continuous output variable of the model as a classifier predicted low-producing and high-producing variants. Prediction of actual production levels was imprecise at the higher ranges of threonine production values, but classification was sufficiently accurate to guide experiments.
The first problem—inability to extrapolate beyond unobserved experimental data—was partially eliminated by the hybrid regression-classification approach described above, and even more, by expansion of training sets, when higher producing variants were constructed in the subsequent round of strain engineering and added to the training. We expected these factors to eliminate problems of regression analysis, but it was not the case (as seen in Fig. 4B). Although production of the constructed strains was increased in subsequent rounds, the inability to rate strain candidates within a class forces us to construct more strains and potentially lose some of the best variants. A likely reason for ML's inability to generate accurate numeric predictions may be the increase in combinatorial complexity in subsequent DBTL cycles, and underrepresentation of specific gene combinations in the training sets.
To deal with this and to increase the ML model predictability one can introduce additional restraints, for instance, increase weights of specific modifications and their pairwise combinations, based on a prior knowledge, or analysis of the results of the initial DBTL cycles, as explained in our graph analysis example. Genes can be ranked in accordance with their metabolic network-based interference with metabolites in the biosynthetic pathway, with ones sharing the highest number of metabolites (as dapA or pntAB in our set) being attributed a larger weight. In a wider set, genes can be also weighted in accordance with their proximity and topology of connectivity to the biosynthetic pathway. For example, our mechanistic analysis suggests that weights of functions in a linear pathway could be decreased due to their potential partial redundancy. The weighting of gene combinations may be expanded to the data on imodulones or genomic neighborhoods, co-expression, and any other indicator of potential synergism of these genes or their deletions in the engineered strains. Utilizing other characteristics of strain performance, such as growth rate and a substrate consumption rate, weighting each feature via integrated values of potentially leading parameters may also increase accuracy of predictions.
Mechanistic analysis of the most efficient combinations of gene modifications revealed their complex interplay underlying non-linear effects of their integration into one producing strain (Fig. 8). Some of the relationships separating ‘drivers’, such as Dtdh and DdapA, from secondary modifications, can be discovered by in graph theory approach. We have applied the graph analysis to characterize the effect of the modifications that in combination with the weights of the corresponding edges show how each modification can affect threonine biosynthesis and modulate outcomes of other introduced genomic changes. Graphical analysis of potential hierarchy of modifications presents a clear, though still hypothetical, structure of the functional subordination in the engineered strains, where modification of tdh, dapA and pntAB genes would be the most essential after modulation of thrABCasd to ensure high threonine production. Pairwise combinations of Dtdh, DdapA, pntAB are reflected in a closed graph of the highly weighted edges preceding the core of the most successful combinations of engineered gene modifications. A graph analysis is an independent purely algebraic approach connecting topology of metabolic networks and productivity of engineered strains, and as such, may be integrated in ML model increasing its accuracy.
Our results demonstrate that engineering of industrial strains based on general computational tools and “general” metabolic and comparative genomic information can produce high-yielding microbial strains in a predictable and accelerated manner. The following sequence of elements, a majority of which were tested in our study, represent a general algorithm for such organism engineering: (1) flux-based gene selection of the most impactful genes for strain engineering; (2) combinatorial cloning producing initial training set of gene combinations associated with production titers; (3) building and validating of ML models predicting improved strain design; (4) performing subsequent DBTL rounds to improve models and model-designed strains.
Two important factors which can further improve this strategy are: (1) solving the problem with the accuracy of regression modeling, (2) expansion, refining and weighting of gene repertoire used for combinatorial cloning. To address the first problem, besides pure computational approaches, both metabolic and statistical analysis can be applied to find mechanical connections or conserved patterns separating groups of strains by their “predictability”. If successful, it should lead to a much more productive design of training sets and add important precision to numeric predictions generated by ML models. Set of genes used for combinatorial cloning in our study, although not including all possible impactful genes, was sufficient to assemble highly productive variants. In many less studied cases, we, however, envision a necessity of “fishing expeditions” going beyond flux analysis, which will be needed to find additional gens driving processes of interest. Three possible directions for such gene-finding efforts are: (1) metabolic analysis of RNA-Seq samples; (3) whole genome comparison of producing strains; (3) genome scanning in which ordered genomic libraries of knockouts or overexpressed variants are tested in production strains.
Historically, organism construction for bioproduction started by using random mutagenesis and genetic selection. Knowledge-driven strategies, like genomics and flux analysis, further empower our engineering toolbox and became a new driving force in strain development. Limitations highlighted in the introduction, however, constrain the utility of these strategies. ML offered an orthogonal approach which allows to explore vast combinatorial spaces by discovering correlations between engineering designs and their properties which we often cannot explain. Integration of ML capabilities and various knowledge-driven techniques is required to maximize effectiveness of organism engineering, and our project is a step in this direction. Our accelerated “agnostic” engineering strategy can significantly expand the list of chemicals produced in biomanufacturing by eliminating its important bottleneck, slow and poorly predictable process of strain engineering.
6. Conclusions
Functional (metabolic) models form a foundation of knowledge-driven organism engineering. However, gaps in the underlying stoichiometric matrices and lack of integration of metabolic effects of regulatory networks limit power of such models and make unreachable many important applications of synthetic biology. Going from individual gene modifications to an effective combination of target genes is a major challenge even with all existing prior knowledge on metabolic systems. ML can provide a “universal glue” capable to fill these gaps and guide organisms engineering based on patterns extracted from the engineered strains performance and gene expression data. We see no limitations in applying such strategy to numerous fundamental and applied engineering projects.
Credit author statement
Paul Hanke: Conceptualization, Methodology, Investigation, Validation, Writing-Original Draft, Writing – review & editing.
Bruce Parrello: Methodology, Software, Validation, Formal analysis, Data Curation, Writing-Original Draft, Writing-Review & editing, Visualization.
Olga Vasieva: Conceptualization, Methodology, Software, Formal analysis, Writing-Original Draft, Writing-Review & editing, Visualization.
Chase Akins: Investigation, Validation.
Philippe Chlenski: Data Curation, Formal analysis.
Gyorgy Babnigg: Supervision, Investigation, Validation.
Chris Henry: Supervision, Formal analysis.
Fatima Foflonker: Formal analysis.
Thomas Brettin Supervision, Project administration.
Dionysios Antonopoulos Supervision, Writing-Original Draft, Project administration, Funding acquisition.
Rick Stevens: Supervision.
Michael Fonstein: Conceptualization, Methodology, Investigation, Writing – Original Draft, Supervision, Project administration, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This material is based upon work supported by Laboratory Directed Research and Development (LDRD) funding from Argonne National Laboratory (ANL), provided by the Director, Office of Science, of the U.S. Department of Energy (DOE) under Contract No. DE-AC02-06CH11357. A portion of this work was also supported by the U.S. DOE, Office of Science, Biological and Environmental Research program's Secure Biosystems Design project entitled, “Rapid Design and Engineering of Smart and Secure Microbiological Systems”, at ANL. Argonne is a U.S. DOE national laboratory managed by UChicago Argonne, LLC.
We would like to thank Ross Overbeek, Marie-Francoise Gros, and Philippe Noirot for many productive discussions. Additionally, we thank members of the Environmental Sample Preparation and Sequencing Facility at ANL, specifically Stephanie M. Greenwald and Sarah M. Owens, for their assistance with sequence data generation.
Handling Editor: Mattheos Koffas
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mec.2023.e00225.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Links are included to all data in the supplementary Information.
References
- Bachler C., Schneider P., Bahler P., Lustig A., Erni B. Escherichia coli dihydroxyacetone kinase controls gene expression by binding to transcription factor DhaR. EMBO J. 2005;24(2):283–293. doi: 10.1038/sj.emboj.7600517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassalo M.C., Garst A.D., Choudhury A., Grau W.C., Oh E.J., Spindler E., Lipscomb T., Gill R.T. Deep scanning lysine metabolism in Escherichia coli. Mol. Syst. Biol. 2018;14(11) doi: 10.15252/msb.20188371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J., Klopprogge C., Herold A., Zelder O., Bolten C.J., Wittmann C. Metabolic flux engineering of L-lysine production in Corynebacterium glutamicum--over expression and modification of G6P dehydrogenase. J. Biotechnol. 2007;132(2):99–109. doi: 10.1016/j.jbiotec.2007.05.026. [DOI] [PubMed] [Google Scholar]
- Becker J., Wittmann C. Systems and synthetic metabolic engineering for amino acid production - the heartbeat of industrial strain development. Curr. Opin. Biotechnol. 2012;23(5):718–726. doi: 10.1016/j.copbio.2011.12.025. [DOI] [PubMed] [Google Scholar]
- Becker J., Zelder O., Hafner S., Schroder H., Wittmann C. From zero to hero--design-based systems metabolic engineering of Corynebacterium glutamicum for L-lysine production. Metab. Eng. 2011;13(2):159–168. doi: 10.1016/j.ymben.2011.01.003. [DOI] [PubMed] [Google Scholar]
- Clomburg J.M., Crumbley A.M., Gonzalez R. Industrial biomanufacturing: the future of chemical production. Science. 2017;355(6320) doi: 10.1126/science.aag0804. [DOI] [PubMed] [Google Scholar]
- Czajka J.J., Oyetunde T., Tang Y.J. Integrated knowledge mining, genome-scale modeling, and machine learning for predicting Yarrowia lipolytica bioproduction. Metab. Eng. 2021;67:227–236. doi: 10.1016/j.ymben.2021.07.003. [DOI] [PubMed] [Google Scholar]
- Datsenko K.A., Wanner B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debabov V.G. In: The Threonine Story. Microbial Production of L-Amino Acids. Faurie R., Thommel J., Bathe B., et al., editors. Springer Berlin Heidelberg; Berlin, Heidelberg: 2003. pp. 113–136. [Google Scholar]
- Gokarn R., Eiteman M.A., Altman E. Pyruvate Carboxylase overexpression for enhanced production of oxaloacetate-derived biochemicals in microbial cells. PCT WO 99/53035. 1999 [Google Scholar]
- Guo W., Sheng J., Feng X. Mini-review: in vitro metabolic engineering for biomanufacturing of high-value products. Comput. Struct. Biotechnol. J. 2017;15:161–167. doi: 10.1016/j.csbj.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutknecht R., Beutler R., Garcia-Alles L.F., Baumann U., Erni B. The dihydroxyacetone kinase of Escherichia coli utilizes a phosphoprotein instead of ATP as phosphoryl donor. EMBO J. 2001;20(10):2480–2486. doi: 10.1093/emboj/20.10.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa T., Tanaka T., Watanabe K. l-Threonine production by Auxotrophs of E. coli. Agric. Biol. Chem. 1973;37(1):123–130. [Google Scholar]
- Ikeda M., Ohnishi J., Hayashi M., Mitsuhashi S. A genome-based approach to create a minimally mutated Corynebacterium glutamicum strain for efficient L-lysine production. J. Ind. Microbiol. Biotechnol. 2006;33(7):610–615. doi: 10.1007/s10295-006-0104-5. [DOI] [PubMed] [Google Scholar]
- Kase H., Nakayama K. Production of l-threonine by analog-resistant Mutants. Agric. Biol. Chem. 1972;36(9):1611–1621. [Google Scholar]
- Kind S., Neubauer S., Becker J., Yamamoto M., Volkert M., Abendroth G., Zelder O., Wittmann C. From zero to hero - production of bio-based nylon from renewable resources using engineered Corynebacterium glutamicum. Metab. Eng. 2014;25:113–123. doi: 10.1016/j.ymben.2014.05.007. [DOI] [PubMed] [Google Scholar]
- Korosh T.C., Markley A.L., Clark R.L., McGinley L.L., McMahon K.D., Pfleger B.F. Engineering photosynthetic production of L-lysine. Metab. Eng. 2017;44:273–283. doi: 10.1016/j.ymben.2017.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov Iu I., Kochetova L.P., Livshits V.A., Mashko S.V., Moshentseva V.N. [Cloning of threonine operon genes in Escherichia coli cells] Genetika. 1980;16(1):66–77. [PubMed] [Google Scholar]
- Lee J.H., Jung S.-C., Bui L.M., Kang K.H., Song J.-J., Kim S.C. Improved Production of <span class="sc">l</span>-Threonine in <span class="named-content genus-species" id="named-content-1">Escherichia coli</span> by Use of a DNA Scaffold System. Appl. Environ. Microbiol. 2013;79(3):774–782. doi: 10.1128/AEM.02578-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Sung B.H., Kim M.S., Blattner F.R., Yoon B.H., Kim J.H., Kim S.C. Metabolic engineering of a reduced-genome strain of Escherichia coli for L-threonine production. Microb. Cell Factories. 2009;8:2. doi: 10.1186/1475-2859-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Wendisch V.F. Production of amino acids - genetic and metabolic engineering approaches. Bioresour. Technol. 2017;245(Pt B):1575–1587. doi: 10.1016/j.biortech.2017.05.065. [DOI] [PubMed] [Google Scholar]
- Lee K.H., Park J.H., Kim T.Y., Kim H.U., Lee S.Y. Systems metabolic engineering of Escherichia coli for L-threonine production. Mol. Syst. Biol. 2007;3:149. doi: 10.1038/msb4100196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Rabe K.S., Nielsen J., Engqvist M.K.M. Machine learning applied to predicting microorganism growth temperatures and enzyme catalytic optima. ACS Synth. Biol. 2019;8(6):1411–1420. doi: 10.1021/acssynbio.9b00099. [DOI] [PubMed] [Google Scholar]
- Lim H.G., Rychel K., Sastry A.V., Bentley G.J., Mueller J., Schindel H.S., Larsen P.E., Laible P.D., Guss A.M., Niu W., Johnson C.W., Beckham G.T., Feist A.M., Palsson B.O. Machine-learning from Pseudomonas putida KT2440 transcriptomes reveals its transcriptional regulatory network. Metab. Eng. 2022;72:297–310. doi: 10.1016/j.ymben.2022.04.004. [DOI] [PubMed] [Google Scholar]
- Lindner S.N., Ramirez L.C., Krusemann J.L., Yishai O., Belkhelfa S., He H., Bouzon M., Doring V., Bar-Even A. NADPH-auxotrophic E. coli: a sensor strain for testing in vivo regeneration of NADPH. ACS Synth. Biol. 2018;7(12):2742–2749. doi: 10.1021/acssynbio.8b00313. [DOI] [PubMed] [Google Scholar]
- Liu J., Li H., Xiong H., Xie X., Chen N., Zhao G., Caiyin Q., Zhu H., Qiao J. Two-stage carbon distribution and cofactor generation for improving l-threonine production of Escherichia coli. Biotechnol. Bioeng. 2019;116(1):110–120. doi: 10.1002/bit.26844. [DOI] [PubMed] [Google Scholar]
- Livshits V.A., Zakataeva N.P., Aleshin V.V., Vitushkina M.V. Identification and characterization of the new gene rhtA involved in threonine and homoserine efflux in Escherichia coli. Res. Microbiol. 2003;154(2):123–135. doi: 10.1016/S0923-2508(03)00036-6. [DOI] [PubMed] [Google Scholar]
- Lv X., Hueso-Gil A., Bi X., Wu Y., Liu Y., Liu L., Ledesma-Amaro R. New synthetic biology tools for metabolic control. Curr. Opin. Biotechnol. 2022;76 doi: 10.1016/j.copbio.2022.102724. [DOI] [PubMed] [Google Scholar]
- Ma Q., Zhang Q., Xu Q., Zhang C., Li Y., Fan X., Xie X., Chen N. Systems metabolic engineering strategies for the production of amino acids. Synth Syst Biotechnol. 2017;2(2):87–96. doi: 10.1016/j.synbio.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moliner M., Roman-Leshkov Y., Corma A. Machine learning applied to zeolite synthesis: the missing link for realizing high-throughput discovery. Acc. Chem. Res. 2019;52(10):2971–2980. doi: 10.1021/acs.accounts.9b00399. [DOI] [PubMed] [Google Scholar]
- Moore S.J., Lai H.E., Kelwick R.J., Chee S.M., Bell D.J., Polizzi K.M., Freemont P.S. EcoFlex: a Multifunctional MoClo kit for E. coli synthetic biology. ACS Synth. Biol. 2016;5(10):1059–1069. doi: 10.1021/acssynbio.6b00031. [DOI] [PubMed] [Google Scholar]
- Neidhardt F.C., Curtiss R. ASM Press; 1996. Escherichia Coli and Salmonella: Cellular and Molecular Biology. [Google Scholar]
- Ogawa-Miyata Y., Kojima H., Sano K. Mutation analysis of the feedback inhibition site of aspartokinase III of Escherichia coli K-12 and its use in L-threonine production. Biosci. Biotechnol. Biochem. 2001;65(5):1149–1154. doi: 10.1271/bbb.65.1149. [DOI] [PubMed] [Google Scholar]
- Ohnishi J., Mitsuhashi S., Hayashi M., Ando S., Yokoi H., Ochiai K., Ikeda M. A novel methodology employing Corynebacterium glutamicum genome information to generate a new L-lysine-producing mutant. Appl. Microbiol. Biotechnol. 2002;58(2):217–223. doi: 10.1007/s00253-001-0883-6. [DOI] [PubMed] [Google Scholar]
- Olavarria K., De Ingeniis J., Zielinski D.C., Fuentealba M., Munoz R., McCloskey D., Feist A.M., Cabrera R. Metabolic impact of an NADH-producing glucose-6-phosphate dehydrogenase in Escherichia coli. Microbiology (Read.) 2014;160(Pt 12):2780–2793. doi: 10.1099/mic.0.082180-0. [DOI] [PubMed] [Google Scholar]
- Orth J., Conrad T.M., Na J., Lerman J.A., Nam H., Adam M Feist A.M., Pallson B.Ø. A comprehensive genome-scale reconstruction of Escherichia coli metabolism--2011. Mol. Syst. Biol. 2011;7(1):535. doi: 10.1038/msb.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otasek D., Morris J.H., Boucas J., Pico A.R., Demchak B. Cytoscape Automation: empowering workflow-based network analysis. Genome Biol. 2019;20(1):185. doi: 10.1186/s13059-019-1758-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyetunde T., Bao F.S., Chen J.W., Martin H.G., Tang Y.J. Leveraging knowledge engineering and machine learning for microbial bio-manufacturing. Biotechnol. Adv. 2018;36(4):1308–1315. doi: 10.1016/j.biotechadv.2018.04.008. [DOI] [PubMed] [Google Scholar]
- Oyetunde T., Liu D., Martin H.G., Tang Y.J. Machine learning framework for assessment of microbial factory performance. PLoS One. 2019;14(1) doi: 10.1371/journal.pone.0210558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papapetridis I., Verhoeven M.D., Wiersma S.J., Goudriaan M., van Maris A.J.A., Pronk J.T. Laboratory evolution for forced glucose-xylose co-consumption enables identification of mutations that improve mixed-sugar fermentation by xylose-fermenting Saccharomyces cerevisiae. FEMS Yeast Res. 2018;18(6) doi: 10.1093/femsyr/foy056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters-Wendisch P.G., Schiel B., Wendisch V.F., Katsoulidis E., Möckel B., Sahm H., Eikmanns B.J. Pyruvate carboxylase is a major bottleneck for glutamate and lysine production by Corynebacterium glutamicum. J. Mol. Microbiol. Biotechnol. 2001;3(2):295–300. [PubMed] [Google Scholar]
- Phaneuf P.V., Yurkovich J.T., Heckmann D., Wu M., Sandberg T.E., King Z.A., Tan J., Palsson B.O., Feist A.M. Causal mutations from adaptive laboratory evolution are outlined by multiple scales of genome annotations and condition-specificity. BMC Genom. 2020;21(1):514. doi: 10.1186/s12864-020-06920-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rychel K., Decker K., Sastry A.V., Phaneuf P.V., Poudel S., Palsson B.O. iModulonDB: a knowledgebase of microbial transcriptional regulation derived from machine learning. Nucleic Acids Res. 2021;49(D1):D112–D120. doi: 10.1093/nar/gkaa810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sastry A.V., Gao Y., Szubin R., Hefner Y., Xu S., Kim D., Choudhary K.S., Yang L., King Z.A., Palsson B.O. The Escherichia coli transcriptome mostly consists of independently regulated modules. Nat. Commun. 2019;10(1):5536. doi: 10.1038/s41467-019-13483-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer U., Canonaco F., Heri S., Perrenoud A., Fischer E. The soluble and membrane-bound transhydrogenases UdhA and PntAB have divergent functions in NADPH metabolism of Escherichia coli. J. Biol. Chem. 2004;279(8):6613–6619. doi: 10.1074/jbc.M311657200. [DOI] [PubMed] [Google Scholar]
- Schmidl S.R., Sheth R.U., Wu A., Tabor J.J. Refactoring and optimization of light-switchable Escherichia coli two-component systems. ACS Synth. Biol. 2014;3(11):820–831. doi: 10.1021/sb500273n. [DOI] [PubMed] [Google Scholar]
- St John P.C., Strutz J., Broadbelt L.J., Tyo K.E.J., Bomble Y.J. Bayesian inference of metabolic kinetics from genome-scale multiomics data. PLoS Comput. Biol. 2019;15(11) doi: 10.1371/journal.pcbi.1007424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suthers P.F., Foster C.J., Sarkar D., Wang L., Maranas C.D. Recent advances in constraint and machine learning-based metabolic modeling by leveraging stoichiometric balances, thermodynamic feasibility and kinetic law formalisms. Metab. Eng. 2021;63:13–33. doi: 10.1016/j.ymben.2020.11.013. [DOI] [PubMed] [Google Scholar]
- Thomason L.C., Costantino N., Court D.L. E. coli genome manipulation by P1 transduction. Curr Protoc Mol Biol Chapter. 2007;1 doi: 10.1002/0471142727.mb0117s79. Unit 1.17. [DOI] [PubMed] [Google Scholar]
- Vavricka C.J., Hasunuma T., Kondo A. Dynamic metabolomics for engineering biology: accelerating learning cycles for bioproduction. Trends Biotechnol. 2020;38(1):68–82. doi: 10.1016/j.tibtech.2019.07.009. [DOI] [PubMed] [Google Scholar]
- Wang J., Ma W., Fang Y., Yang J., Zhan J., Chen S., Wang X. Increasing L-threonine production in Escherichia coli by overexpressing the gene cluster phaCAB. J. Ind. Microbiol. Biotechnol. 2019;46(11):1557–1568. doi: 10.1007/s10295-019-02215-0. [DOI] [PubMed] [Google Scholar]
- Wang L., Li J., March J.C., Valdes J.J., Bentley W.E. luxS-dependent gene regulation in Escherichia coli K-12 revealed by genomic expression profiling. J. Bacteriol. 2005;187(24):8350–8360. doi: 10.1128/JB.187.24.8350-8360.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Fang Y., Wang Z., Zhang S., Wang L., Guo Y., Wang X. Improving L-threonine production in Escherichia coli by elimination of transporters ProP and ProVWX. Microb. Cell Factories. 2021;20(1):58. doi: 10.1186/s12934-021-01546-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissbach H., Brot N. Regulation of methionine synthesis in Escherichia coli. Mol. Microbiol. 1991;5(7):1593–1597. doi: 10.1111/j.1365-2958.1991.tb01905.x. [DOI] [PubMed] [Google Scholar]
- Wittmann C., Becker J. In: The L-Lysine Story: from Metabolic Pathways to Industrial Production. Amino Acid Biosynthesis ∼ Pathways, Regulation and Metabolic Engineering. Wendisch V.F., editor. Springer Berlin Heidelberg; Berlin, Heidelberg: 2007. pp. 39–70. [Google Scholar]
- Yang H., Yang J., Liu C., Lv X., Liu L., Li J., Du G., Chen J., Liu Y. High-level 5-methyltetrahydrofolate bioproduction in Bacillus subtilis by combining modular engineering and transcriptomics-guided global metabolic regulation. J. Agric. Food Chem. 2022;70(19):5849–5859. doi: 10.1021/acs.jafc.2c01252. [DOI] [PubMed] [Google Scholar]
- Zhang J., Petersen S.D., Radivojevic T., Ramirez A., Perez-Manriquez A., Abeliuk E., Sanchez B.J., Costello Z., Chen Y., Fero M.J., Martin H.G., Nielsen J., Keasling J.D., Jensen M.K. Combining mechanistic and machine learning models for predictive engineering and optimization of tryptophan metabolism. Nat. Commun. 2020;11(1):4880. doi: 10.1038/s41467-020-17910-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.P., Sun J., Ma Y. Biomanufacturing: history and perspective. J. Ind. Microbiol. Biotechnol. 2017;44(4–5):773–784. doi: 10.1007/s10295-016-1863-2. [DOI] [PubMed] [Google Scholar]
- Zhao L., Lu Y., Yang J., Fang Y., Zhu L., Ding Z., Wang C., Ma W., Hu X., Wang X. Expression regulation of multiple key genes to improve L-threonine in Escherichia coli. Microb. Cell Factories. 2020;19(1):46. doi: 10.1186/s12934-020-01312-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Fang Y., Ding Z., Zhang S., Wang X. Developing an l-threonine-producing strain from wild-type Escherichia coli by modifying the glucose uptake, glyoxylate shunt, and l-threonine biosynthetic pathway. Biotechnol. Appl. Biochem. 2019;66(6):962–976. doi: 10.1002/bab.1813. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Links are included to all data in the supplementary Information.