Abstract
Background:
In the current age of diagnostic approaches in cancer, countless efforts have been allocated to identify novel and efficient biomarkers to detect cancer in its early stages. We focused on evaluating the correlation between the progression of gastrointestinal cancer, a leading cause of cancer death worldwide, and human endogenous retrovirus (HERV).
Materials and Methods:
In this study, we conducted a study on the peripheral blood mononuclear cells (PBMC) gathered from gastric and colon cancer patients. We focused on HERV-K rec, np9, gag expression analysis by quantitative real-time PCR, after extraction of RNA and synthesizing cDNA.
Results:
Unlike np9 whose expression increased significantly in the colon and gastric cancers, the mRNA level of the rec gene declined in both cancers. Moreover, our data illustrated that the over-expression of the gag gene was only observed in colon cancerous cells rather than gastric malignancy.
Conclusions:
Overall, given the correlation between the expression level of HERV-associated genes and gastrointestinal cancer, our study suggests that these genes could be considered beneficial markers for cancer diagnosis. However, researchers should conduct studies in future articles on whether these genes can be employed as biomarkers in gastrointestinal cancer.
Keywords: Colon cancer, gastric cancer, HERV-K, real-time PCR
INTRODUCTION
In a wide range of malignancies which are categorized into the big family of gastrointestinal cancers, colon and gastric cancers are the third and second leading cause of cancer death around the world, respectively.[1,2,3] In a study conducted in 2012, it has been reported that 952,000 new cases were diagnosed with gastric cancer,[4] of which only 25% of these patients survived and reached complete remission status.[5,6] Moreover, the results of a survey in Canada revealed that 21500 new cases of colon cancer were diagnosed in 2008, of which among them 8900 patients lost their lives.[7] Numerous risk factors have been enumerated to be linked with the incidence of gastrointestinal cancer, including red meat, alcohol, obesity, age, smoking, sex, and even infectious diseases like helicobacter pillory, and viral diseases.[6,7,8,9,10,11,12,13]
One of the viral infections that can deteriorate the patient's condition in cancer diseases could be human endogenous retrovirus (HERV).[14] The possession of reverse transcriptase (RT) enzyme gives this virus the ability to convert viral RNA into DNA, which in turn integrates into the host germline genome. HERV genes have been merged into the ancestor germ line of ancient infections between 30 and 40 million years ago the first time, however, most HERV-associated genes remained inactivated due to the mutations.[15,16,17] In the structure of HERV genes, some regulatory factors transmit virtually to the next generation.[15] These regulatory factors, which are located between two long terminal repeats (LTRs) could regulate the expression of gag, rec, and np9.[18,19,20] In the longitude list of HERV family members,[18] the highest biological activity of HERV-K makes this virus participate in different human diseases, especially numerous malignancies.[21,22,23] The association between HERV-k and different human cancers, such as bladder cancer, ovarian cancer, melanoma, breast cancer, kidney cancer, and lymphoma has been well-established in several studies; however, its correlation with gastrointestinal cancer has not been studied, thus far. The present study was designed to check the HERV-K rec, np9, gag expression by quantitative real time PCR, after extraction of RNA and synthesizing cDNA.
MATERIALS AND METHODS
Sample
Approval to conduct this study was obtained from the Shahid Beheshti University of Medical Sciences” IR.SBMU.MSP.REC.1401.164 (Grant No 32848). In this study, 25 and 23 patients diagnosed with gastric and colon cancer at Emam Hossein hospital in Tehran were selected, respectively. The expert pathologists tested the existence of gastric and colon cancer and the information on patients is written in Table 1. We also collected 7 ml blood samples from 25 patients with gastric polyp and 23 patients with colon polyp as control groups. Patients who were treated either by corticosteroids, non-steroidal anti-inflammatory drugs, or immunosuppressive drugs were excluded from this study. The consent that was obtained from study participants was verbal in collaboration with a physician.
Table 1.
The information of patients, suffering from gastrointestinal cancer
| Gastric cancer PBMC | Gastric polyp PBMC | Colon cancer PBMC | Colon polyp PBMC | |
|---|---|---|---|---|
| Male | 12 | 11 | 6 | 15 |
| Female | 13 | 14 | 17 | 8 |
| Mean age | 66.7 | 63.5 | 58.2 | 66 |
| <60 | 10 | 7 | 13 | 9 |
| >60 | 15 | 18 | 10 | 14 |
| Well differentiated adenocarcinoma and Moderate | 12 | 15 | ||
| poor differentiate adenocarcinoma | 13 | 8 | ||
| H.Pylori positive | 4 | 5 | Unknown |
Blood collecting
All blood samples were collected in sterile falcon tubes, containing 100 U/ml of heparin, and after that, we diluted the whole blood with PBS. Afterward, peripheral blood mononuclear cells (PBMC) were gathered from 14 ml of blood by using Ficoll. Noteworthy, the PBMCs of all samples were washed with PBS three times.
RNA extraction
The mixture of RNX-plus solution (Cinnagen, Tehran, Iran) and PBMCs was exposed at 37°C for 5 min before adding chloroform. The extracted RNA was purified by using propanol and 70% alcohol. Nanodrop (Eppendorf, Humburg, Germany) examined the number of RNA samples at the wavelength of 260 nm. We confirmed the quality of extracted RNA by running in 1% agarose gel electrophoresis (5S, 18S, and 28S bands were observed).
cDNA synthesis
cDNA kit (Daejeon, South Korea) synthesized the cDNA by reverse transcription reaction. We provided a 20 μl reaction containing 9 ul master mixes, 10 ul of RNA samples, and 1 ul random hexamer. We put this mixture in Bio Intellectica PCR at 50°C for 40 min and at 95°C for 10 min. The sterile water was used to dilute two times these synthesized cDNAs.
Quantitative real-time PCR
The combination of 10 ul BIOFACT™ 2× real-time PCR master mix (for SYBR Green I; BIOFACT, South Korea), 1 ul forward 10 pmol, 1 ul reverse primer 10 pmol, 6 ul sterile water, 2 ul cDNA in a final 20 ul volume was employed which was incubated in one cycle for 10 min at 95°C, 40 cycles for 30s at 95°C; for 30s at 55°C, for 30s at 72°C in Rotor-gene 6000 (Corbett life sciences, Sydney, Australia) in 36-well Gene Discs. The melt curve started at 60°C and finished at 95°C. We used GAPDH housekeeping as an internal control. We summarized primers in Table 2.[24,25]
Table 2.
Nucleotide sequences of primers used for real-time RT-PCR
| Gene | Forward primer (5’- 3’) | Reverse primer (5’- 3’) |
|---|---|---|
| GAPDH | ATGTTCGTCATGGGTGTGAA | GGTGCTAAGCAGTTGGTGGT |
| rec | ATCGAGCACCGTTGACTCACAAGA | GGTACACCTGCAGACACCATTGAT |
| np9 | AGATGTCTGCAGGTGTACCCA | CTCTTGCTTTTCCCCACATTTC |
| gag | AGCAGGTCAGGTGACCGTAAC | GGTGCCATAGCATTGTCTCCT |
Statistical analysis
In this study, we employed Graph-PadPrism software and ANOVA tests to evaluate experimental data and statistical significance, respectively. Indeed, the mean ± standard deviation of three independent assays was used to express experimental data, and a P value less than (P < 0.05) was used for the differences.
RESULTS
Sample
For evaluating whether there is a link between rec, np9, gag genes expression, and gastrointestinal cancer, the blood samples from 25 gastric cancer (12 male, 13 female with an average age of 66.7 years old) and 23 colon cancer (6 male, 17 female with the average age of 58.2 years old) were collected. It's worth mentioning that we also collected the blood sample from 25 patients with gastric polyp (11 male, 14 female; mean age: 63.5) and 23 colon polyp (15 male, 8 female; mean age: 66) as a control group. The expert pathologists tested the existence of gastric and colon cancer and 12 patients with gastric cancer and 15 patients with colon cancer who possessed a tumor stage considered as moderate or well-differentiated adenocarcinoma.
HERV-K rec, np9, gag expression analysis in isolated PBMC of gastric and colon cancer
We checked whether there is a link between the alteration in the expression of HERV-associated genes and gastric and colon cancer, we collected the blood sample from cancerous patients, and then the expression level of np9, gag, and rec was evaluated using RQ-PCR analysis. Although the expression of np9 increased in both gastric and colon cancer as compared to patients who had polyp, however, this increase was not statistically significant [Figure 1]. The data of RQ-PCR analysis revealed that the mRNA level of rec remarkably decreased in both cancers [Figure 2a and b]. Moreover, while the transcription level of gag increased in PBMCs harvested from colon cancer patients, the expression level of this gene remained unchanged in gastric cancer (*P value <0.05) [Figure 3].
Figure 1.
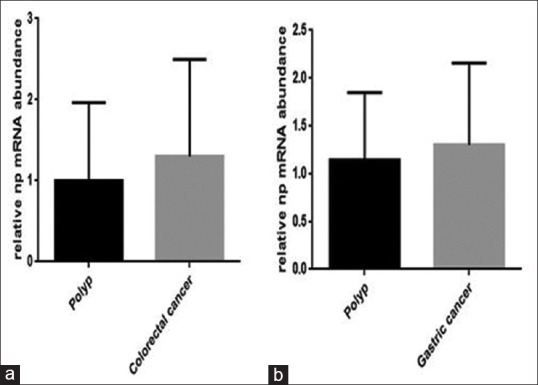
The evaluation of HERV-K np9 expression level in colorectal (a) and gastric cancer- (b) derived PBMC compared to PBMC harvested from polyp patients. Although there is a slight increment in the expression of np9, this elevation in gene expression was not statistically significant
Figure 2.
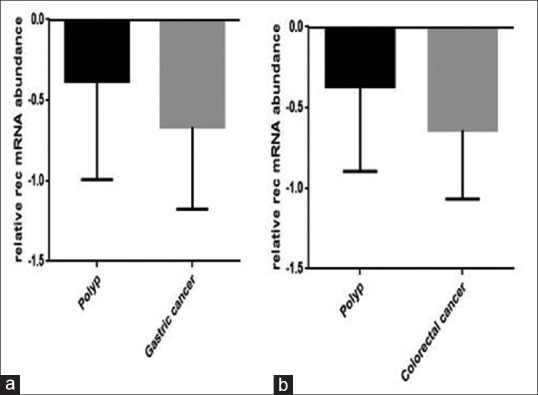
The results of RQ-PCR analysis revealed that the expression of HERV-K rec decreased in PBMC obtained from both gastric (a) and colorectal (b) cancer patients. Values are given as mean ± standard deviation of three independent experiments. *P ≤ 0.05 represents significant changes from untreated control
Figure 3.
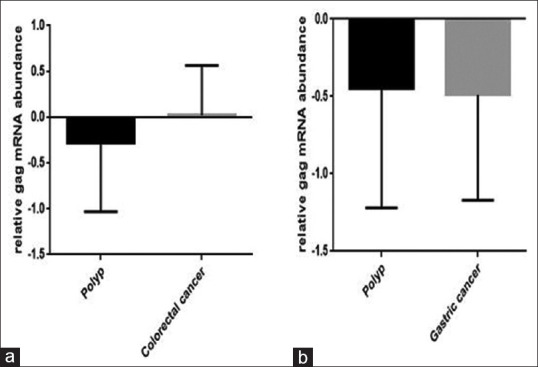
HERV-k gag mRNA expression in the gastric and colorectal cancer. While the expression level of gag increased significantly in colorectal cancer (a), the expression of this gene remained unchanged in the PBMC obtained from gastric cancer patients (b). Values are given as mean ± standard deviation of three independent experiments. *P ≤ 0.05 represents significant changes from untreated control
DISCUSSION
In the era of novel genetic and laboratory experiments, the discovery of the role of viral infection in the initiation of different cancers has revolutionized the current understanding of the pathogenesis of human cancers. The involvement of HERV, a well-known retrovirus, in human malignancies is emphasized by recent studies, introducing the associated genes of this virus, such as gag, rec, and np9 as potent oncogenes.[26] Scientists have classified HERV as a single-stranded positive-sense RNA that can convert their RNA to DNA by RT enzyme.[27] Studies have declared that the expression of HERV-associated genes is specific to some tissues; as placenta, testis cells, and other steroid hormone-regulated tissues displayed over-expressed rec, np9, and rec genes.[15] Moreover, the over-expression of HERV-associated genes has also been detected in different human cancers, such as melanomas, germ cell tumors (GCT), and ovarian cancers.[28] To the best of our knowledge, the correlation between HERV and gastrointestinal cancers has not been evaluated, thus far. Moreover, evidence has declared that the early detection of gastrointestinal cancer could improve the prognosis of the disease and reinforce the response of patients to conventional chemotherapeutic drugs. Given these and to identify a promising novel biomarker of gastrointestinal cancer, this study was designed to search whether the expression level of HERV-related genes, such as rec, gag, and np9 is altered in this cancer.
The data of the current study clearly showed that unlike np9 and rec mRNA, whose expression alteration was not statistically significant, the mRNA gag surged in colon cancer patients as compared to patients who had colon polyp. Noteworthy, we failed to find any links between the expression of this gene and the progression of gastric cancer. The oncogenic effects of HERV-related genes are well-described in a wide array of cancers.[29] In a recent study, the expression of the gag gene was reported to be elevated in the peripheral blood of prostate cancer patients, suggestive of the diagnostic value of this gene in the detection of prostate cancer.[30] Another study also showed that the HERV-K env gene is over-expressed in the PBMC of patients suffering from ovarian cancer, proposing that probably inhibiting the expression of this gene could be used as a therapeutic approach for this type of cancer.[28] Depil et al.[31] reported that HERV-K gag is expressed in normal PBMC lower than that expressed in malignant leukemic cells, suggesting that the increase in this gene could also participate in leukemogenesis. The results of other studies also revealed that elevated HERV-K np9 mRNA probably not only contributes to the development of Kaposi's Sarcoma in HIV+ patients but also is accounted for the invasion of cells in this malignancy.[32]
CONCLUSIONS
There is a significant link between the expression level of HERV-related genes and the development of both gastric and colon cancer. Based on our results and given the expression of these genes in the PBMCs, it is reasonable to assume that the mRNA level of HERV rec, np9, and gag could be exploited as tumor biomarkers for these patients.
Ethics approval and consent to participate
Ethics Approval and Consent to Participate: This study has been conducted in the Department of the School of Medicine Shahid Beheshti University of Medical.
Declaration of patient consent
The authors certify that they have obtained all appropriate patient consent forms. In the form, the patient(s) has/have given his/her/their consent for his/her/their images and other clinical information to be reported in the journal. The patients understand that their names and initials will not be published and due efforts will be made to conceal their identity, but anonymity cannot be guaranteed.
Financial support and sponsorship
Approval to conduct this study was obtained from the Shahid Beheshti University of Medical Sciences” IR.SBMU.MSP.REC.1401.164 (Grant No 32848).
Conflicts of interest
There are no conflicts of interest.
REFERENCES
- 1.Kobayashi H, Enomoto A, Woods SL, Burt AD, Takahashi M, Worthley DL. Cancer-associated fibroblasts in gastrointestinal cancer. Nat Rev Gastroenterol Hepatol. 2019;16:282–95. doi: 10.1038/s41575-019-0115-0. [DOI] [PubMed] [Google Scholar]
- 2.Bagheri M, Pormohammad A, Fardsanei F, Yadegari A, Arshadi M, Deihim B, et al. Diagnostic accuracy of pyrazinamide susceptibility testing in mycobacterium tuberculosis: A systematic review with meta-analysis. Microb Drug Resist. 2022;28:87–98. doi: 10.1089/mdr.2021.0048. [DOI] [PubMed] [Google Scholar]
- 3.Wang Q, Geng W, Guo H, Wang Z, Xu K, Chen C, et al. Emerging role of RNA methyltransferase METTL3 in gastrointestinal cancer. J Hematol Oncol. 2020;19(13):57. doi: 10.1186/s13045-020-00895-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 5.Anderson LA, Tavilla A, Brenner H, Luttmann S, Navarro C, Gavin AT, et al. EUROCARE-5 Working Group. Survival for oesophageal, stomach and small intestine cancers in Europe 1999-2007: Results from EUROCARE-5. Eur J Cancer. 2015;51:2144–57. doi: 10.1016/j.ejca.2015.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meng C, Bai C, Brown TD, Hood LE, Tian Q. Human gut microbiota and gastrointestinal cancer. Genomics Proteomics Bioinformatics. 2018;16:33–49. doi: 10.1016/j.gpb.2017.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Riahi Rad Z, Riahi Rad Z, Goudarzi H, Goudarzi M, Mahmoudi M, Yasbolaghi Sharahi J, et al. MicroRNAs in the interaction between host-bacterial pathogens: A new perspective. J Cell Physiol. 2021;236:6249–70. doi: 10.1002/jcp.30333. [DOI] [PubMed] [Google Scholar]
- 8.Lu L, Mullins CS, Schafmayer C, Zeißig S, Linnebacher M. A global assessment of recent trends in gastrointestinal cancer and lifestyle-associated risk factors. Cancer Commun (Lond) 2021;41:1137–51. doi: 10.1002/cac2.12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haghroosta A, Goudarzi H, Faghihloo E, Ghalavand Z, Ranjbar MM, Langroudi RP. In silico analysis of a chimeric fusion protein as a new vaccine candidate against Clostridium perfringens type A and Clostridium septicum alpha toxins. Comp Clin Path. 2020;29:981–9. doi: 10.1007/s00580-020-03136-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen K, Qiu JL, Zhang Y, Zhao YW. Meta analysis of risk factors for colorectal cancer. World J Gastroenterol. 2003;9:1598–600. doi: 10.3748/wjg.v9.i7.1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nikkhahi F, Robatjazi S, Niazadeh M, Javadi A, Shahbazi GH, Aris P, et al. First detection of mobilized colistin resistance mcr-1 gene in Escherichia coli isolated from livestock and sewage in Iran. New Microbes New Infect. 2021;18(41):100862. doi: 10.1016/j.nmni.2021.100862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hajikhani B, Goudarzi M, Kakavandi S, Amini S, Zamani S, van Belkum A, et al. The global prevalence of fusidic acid resistance in clinical isolates of Staphylococcus aureus: A systematic review and meta-analysis. Antimicrob Resist Infect Control. 2021;1(10):75. doi: 10.1186/s13756-021-00943-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dadashi M, Hajikhani B, Faghihloo E, Owlia P, Yaslianifard S, Goudarzi M, et al. Proliferative effect of FadA recombinant protein from Fusobacterium nucleatum on SW480 colorectal cancer cell line. Infect Disord Drug Targets. 2021;21:623–8. doi: 10.2174/1871526520666200720113004. [DOI] [PubMed] [Google Scholar]
- 14.Dolci M, Favero C, Tarantini L, Villani S, Bregni M, Signorini L, et al. Human endogenous retroviruses env gene expression and long terminal repeat methylation in colorectal cancer patients. Med Microbiol Immunol. 2020;209:189–99. doi: 10.1007/s00430-020-00662-6. [DOI] [PubMed] [Google Scholar]
- 15.Lima-Junior DS, Krishnamurthy SR, Bouladoux N, Collins N, Han SJ, Chen EY, et al. Endogenous retroviruses promote homeostatic and inflammatory responses to the microbiota. Cell. 2021;184:3794–3811.e19. doi: 10.1016/j.cell.2021.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khalfallah Y, Genge A. HERV-K inactive or potential pathogens from within? J Neurol Sci. 2021;423:117359. doi: 10.1016/j.jns.2021.117359. [DOI] [PubMed] [Google Scholar]
- 17.Aliakbar Ahovan Z, Khosravimelal S, Eftekhari BS, Mehrabi S, Hashemi A, Eftekhari S, et al. Thermo-responsive chitosan hydrogel for healing of full-thickness wounds infected with XDR bacteria isolated from burn patients: In vitro and in vivo animal model. Int J Biol Macromol. 2020;164:4475–86. doi: 10.1016/j.ijbiomac.2020.08.239. [DOI] [PubMed] [Google Scholar]
- 18.Padmanabhan Nair V, Liu H, Ciceri G, Jungverdorben J, Frishman G, Tchieu J, et al. Activation of HERV-K (HML-2) disrupts cortical patterning and neuronal differentiation by increasing NTRK3. Cell Stem Cell. 2021;28:1566–81.e8. doi: 10.1016/j.stem.2021.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo Y, Yang C, Liu Y, Li T, Li H, Han J, et al. High Expression of HERV-K (HML-2) might stimulate interferon in COVID-19 patients. Viruses. 2022;7(14):996. doi: 10.3390/v14050996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang C, Guo X, Li J, Han J, Jia L, Wen HL, et al. Significant upregulation of HERV-K (HML-2) transcription levels in human lung cancer and cancer cells. Front Microbiol. 2022;10(13):850444. doi: 10.3389/fmicb.2022.850444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu CH, Grandi N, Palanivelu L, Tramontano E, Lin LT. Contribution of human retroviruses to disease development-a focus on the HIV- and HERV-cancer relationships and treatment strategies. Viruses. 2020;4(12):852. doi: 10.3390/v12080852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niazadeh M, Nikkhahi F, Robatjazi S, Javadi A, Farzam SA, Babaei S, et al. Evaluation of mechanisms of colistin resistance in Klebsiella pneumoniae strains isolated from patients with urinary tract infection in ICU. Iran J Microbiol. 2022;14:31–7. doi: 10.18502/ijm.v14i1.8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tugnet N, Rylance P, Roden D, Trela M, Nelson P. Human endogenous retroviruses (HERVs) and autoimmune rheumatic disease: Is there a link? Open Rheumatol J. 2013;7:13–21. doi: 10.2174/1874312901307010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Golrokh Mofrad M, Samie S, Lak R, Faghihloo E. Detection of major human herpesviruses in Iranian patients with suspected encephalitis using multiplex polymerase chain reaction. Eur Neurol. 2022;85:142–7. doi: 10.1159/000519127. [DOI] [PubMed] [Google Scholar]
- 25.Golrokh Mofrad M, Sadigh ZA, Ainechi S, Faghihloo E. Detection of human papillomavirus genotypes, herpes simplex, varicella zoster and cytomegalovirus in breast cancer patients. Virol J. 2021;18:25. doi: 10.1186/s12985-021-01498-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dewannieux M, Blaise S, Heidmann T. Identification of a functional envelope protein from the HERV-K family of human endogenous retroviruses. J Virol. 2005;79:15573–7. doi: 10.1128/JVI.79.24.15573-15577.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berkhout B, Jebbink M, Zsíros J. Identification of an active reverse transcriptase enzyme encoded by a human endogenous HERV-K retrovirus. J Virol. 1999;73:2365–75. doi: 10.1128/jvi.73.3.2365-2375.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rycaj K, Plummer JB, Yin B, Li M, Garza J, Radvanyi L, et al. Cytotoxicity of human endogenous retrovirus K-specific T cells toward autologous ovarian cancer cells. Clin Cancer Res. 2015;21:471–83. doi: 10.1158/1078-0432.CCR-14-0388. [DOI] [PubMed] [Google Scholar]
- 29.Wallace TA, Downey RF, Seufert CJ, Schetter A, Dorsey TH, Johnson CA, et al. Elevated HERV-K mRNA expression in PBMC is associated with a prostate cancer diagnosis particularly in older men and smokers. Carcinogenesis. 2014;35:2074–83. doi: 10.1093/carcin/bgu114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suntsova M, Garazha A, Ivanova A, Kaminsky D, Zhavoronkov A, Buzdin A. Molecular functions of human endogenous retroviruses in health and disease. Cell Mol Life Sci. 2015;72:3653–75. doi: 10.1007/s00018-015-1947-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Depil S, Roche C, Dussart P, Prin L. Expression of a human endogenous retrovirus, HERV-K, in the blood cells of leukemia patients. Leukemia. 2002;16:254–9. doi: 10.1038/sj.leu.2402355. [DOI] [PubMed] [Google Scholar]
- 32.Dai L, Del Valle L, Miley W, Whitby D, Ochoa AC, Flemington EK, et al. Transactivation of human endogenous retrovirus K (HERV-K) by KSHV promotes Kaposi's sarcoma development. Oncogene. 2018;37:4534–45. doi: 10.1038/s41388-018-0282-4. [DOI] [PMC free article] [PubMed] [Google Scholar]


