Abstract
Background:
Nanotechnology has introduced valuable carriers for vaccine delivery. The success of vaccination depends on many factors, such as the intact and safe presentation of vaccine candidates to immune cells. We have conjugated branched PEI-2k and oleic acid (OL) as the building block of the cationic micelle. We aimed to introduce a novel carrier for vaccine candidates.
Materials and Methods:
We conjugated polyethyleneimine and OL (POA) to synthesize the building blocks of cationic micelles. The critical micelle concentration (CMC), size and zeta potential of micelles, and their stability in 60 days were determined. Loading, encapsulation efficiency, and in vitro release study were assessed using bovine serum albumin (BSA) as a protein model. Furthermore, the cytotoxicity and hemocompatibility of developed nanosized micelles were evaluated to ascertain the biocompatibility of fabricated micelles. Cell uptake of cationic micelles in the macrophage cell line was also followed up.
Results:
The conjugation of two polymer parts was confirmed by Fourier transform infrared spectroscopy and 1H nuclear magnetic resonance techniques. The CMC of the developed micelles was around 5.62 × 10−8 mg/ ml, whereas the loading and encapsulation efficiencies were 16.5% and 70%, respectively. The size and zeta potential of the cationic micelles were 96.53 ± 18.53 nm and 68.3 mV, respectively. The release of BSA from POA micelles after 8 and 72 hours was 8.5% and 82%, respectively. Finally, fluorescence microscopy showed that the prepared micelles were successfully and effectively taken up by RAW264.7 cells.
Conclusion:
These results may provide a cutting-edge vaccine delivery solution and open up a new avenue for future vaccine research.
Keywords: Bovine serum albumin, drug delivery systems, micelle, nanoparticles, oleic acid, polyethyleneimine
INTRODUCTION
Recent advances in nanomedicine have significantly impacted the development of smart and multifunctional nanoplatforms to deliver bioactive agents to cells. Among them, nanoparticle-based vaccine delivery platforms have been gradually attracting more attention due to their unique features such as their intrinsic adjuvanticity, ease of surface modification, ability to efficiently deliver antigens, and co-delivery of adjuvants along with antigens, and more importantly, their nanoscale size.[1,2] Besides these features, surface charge, shape, and targeting moieties can affect the efficiency of a nanovaccine.[3] Vaccination has supplied the possibility of controlling and directing the immune system's response to infectious and some noninfectious acute diseases. Over the past years, several approaches have been applied for designing vaccine formulation, including live attenuated, inactivated, protein subunit, peptide, toxoid, and DNA/RNA vaccines.[4] In the formulation of classical vaccines, the whole organism, which contains several proteins, is used to induce immunity, whereas most of these proteins have no role in immunogenicity, and in some cases, these additional proteins can lead to the formation of some unwanted side effects. Using protein, peptide, or DNA/RNA as a subunit vaccine seems more effective,[5] but these types of vaccines are prone to degradation and have a short half-life, as well as their low immunogenicity nature has created another challenge in vaccine design strategies.
Antigen-presenting cells’ (APC) successful uptake, processing, and localization of antigens are necessary to induce specific adaptive immunity and strong memory formation. The administration of adjuvant carriers in vaccine formulation can enhance antigen delivery quality and has a crucial role in provoking immunity.[6]
An adjuvant induces a moderate inflammatory reaction that results in the recruitment of immune cells and a much more efficient adaptive immunity formation.[7] Adjuvants can act as immunomodulatory agents by activating inflammasomes in the cytoplasm and producing cytokines. The stability of nanocarriers with adjuvant potency is also essential for successful APC uptake and presentation without the induction of chronic inflammation.[6]
Several nanoparticles have been recognized as ideal carriers to deliver antigens with intrinsic adjuvant effects, liposomes, virus-like particles, metal and nonmetal inorganic nanoparticles, polymeric nanoparticles, and micelles.[4]
Micelles have two individual parts: a hydrophobic core and a hydrophilic shell or corona. Spontaneous self-assembly of an amphiphilic molecule in the water contributes to forming these unique constructs.[4,5] Several studies have reported the utility of micelles as a drug carrier for the protected transportation of hydrophobic drugs.[5] Some research has demonstrated that micelles have adjuvant properties in addition to antigen encapsulation as well.[4]
Overall, two strategies have been employed to deliver protein/peptide vaccine candidates using micelles: (i) the production of peptide amphiphiles (PAs) by linking a peptide to a hydrophobic tail and (ii) the covalent corporation of an antigen to the hydrophilic shell.[4,5]
The tendency of micelles to self-assemble a structure has provided a great option to control their size during their preparation. Because of their small size (mostly <100 nm), they can deliver antigens to the APCs in the injection site and can transport antigens to the lymph nodes through the lymphatic system.[1,8] The successful transition of an antigen to the lymph node can make an excellent opportunity for exogenous antigen cross-presentation by resident CD8+ dendritic cells. This cross-presentation can help to achieve cellular immunity from vaccination.[5]
Cationic micelle-based delivery systems seem to be promising gene and peptide delivery carriers. Using branched polyethyleneimine 2k (PEI-2k) as a hydrophilic part of the micelle has provided cationic micelles an enormous chance to deliver mRNA and peptides as vaccine candidates in individual studies.[9] In previous studies, PEI was covalently linked to stearic acid and formed an amphiphilic cationic micelle block. Their findings showed successful cellular and humoral immunity induction against their candidate vaccines.[10,11]
In this study, we have conjugated branched PEI-2k and oleic acid (OL) as the building block of the cationic micelle. We aimed to introduce a novel carrier for vaccine candidates by characterizing its physico-chemical properties, examining its encapsulation capacity and release behavior, along with in vitro cytotoxicity. Also, the biocompatibility and hemocompatibility of fabricated POA micelles were investigated in detail. Furthermore, its cellular uptake via a macrophage cell line was exploited to predict APCs’ reactions to this nanoparticle as a vaccine carrier agent. It is interesting to note that the current study is a part of our ongoing research studies in the field of nanoparticle-based vaccine delivery platforms to hasten their translation into the (pre) clinical trials.
MATERIALS AND METHODS
PEI, branched) MW = 1800, purity ≥99%, CAS number: 9002-98-6), was obtained from Alfa Aesar (Heysham, United Kingdom). N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC, CAS number: 25952-53-8), N-hydroxysuccinimide (NHS, CAS number: 6066-82-6), dichloromethane (DCM, CAS number: 75-09-2), and dimethyl sulfoxide (DMSO, CAS number: 67-68-5) were purchased from Merck (Kenilworth, USA). OL (purity ≥99%, CAS number: 112-80-1), thiazolyl blue tetrazolium bromide (MTT, CAS number: 57360-69-7), and curcumin were acquired from Sigma-Aldrich (St. Louis, MO, USA). Dulbecco's modified Eagle's medium (DMEM) was obtained from INOCLON (Tehran, Iran). Fetal bovine serum (FBS, Product number: F7524), penicillin–streptomycin (pen-strep, Product number: P0781), and BSA (CAS number: 9048-46-8) were acquired from Sigma-Aldrich (St. Louis, MO, USA). RAW264.7 cell lines were purchased from the Iranian Biological Resource Center (Tehran, Iran, IBRCC 10072).
Synthesis of PEI-2k-OL (POA)
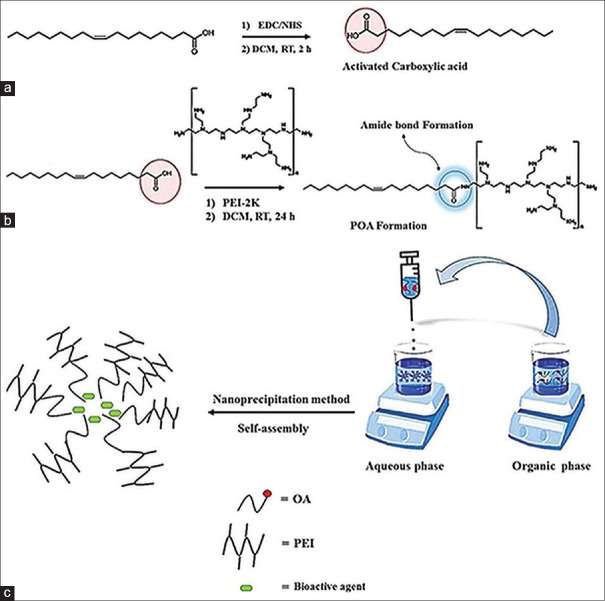
Amphiphiles are synthetic or natural molecules composed of two parts: (i) a hydrophilic part and (ii) a hydrophobic part. Synthetic amphiphiles are generally prepared by linking these two parts through covalent bonds. Due to their hydrophilic and hydrophobic regions, these amphiphilic molecules could undergo self-assembly in an aqueous medium to form a vesicle-like structure.[12] To prepare the amphiphilic POA molecule, PEI was covalently conjugated to OL through an amide bond. In this case, carbodiimide-mediated amide formation remains the most frequently used technique for amine and carboxylic acid conjugation. This technique mainly consists of two steps: (i) activation of carboxylic acids and (ii) conjugation reaction, as shown in Figure 1a and b. Briefly, the carboxylic acid groups of the OLs were first activated by dissolving 72.1 mg of OL in 10 mL of DCM, then NHS (34.5 mg) and EDC (57.5 mg) were added, and the mixture was stirred for 2 h. After the carboxylic acid groups were successfully activated, the conjugation reaction was preceded by the addition of the PEI-2k solution (752.1 mg in DCM) into the reaction mixture, and stirring was continued for 24 h at the same temperature (25°C). The resulting product was then precipitated in cold diethyl ether to obtain the pure POA molecules. Then, it was dialyzed (cut-off 12 kDa) against distilled water for 48 h for further purification of the final products to ensure that the purification process had successfully proceeded. Verification of conjugation was performed by hydrogen-1 nuclear magnetic resonance (1HNMR) in CDCl3 (Bruker DRX-250 AVANCE spectrometer, Billerica, Massachusetts, USA) at 250 MHz and Fourier transform infrared (FTIR) spectroscopy using a (Bruker Tensor 27, Billerica, Massachusetts, USA) FTIR spectrometer.
Figure 1.
Synthesis of amphiphilic POA molecules and their chemical structures. (a) Activation of carboxylic acid groups of OL, (b) conjugation of PEI and OL parts through an amide bond formation. (c) Preparation of cationic POA micelles and their loading process
Synthesis of cationic micelles (POA micelles)
The nanoprecipitation method was used to encapsulate the cargo to investigate the efficacy of the developed POA conjugate as a promising protein delivery system [Figure 1c]. In brief, 4 mg of the POA polymer was dissolved in methanol (organic phase), and then the resulting solution was added dropwise to stirring distilled water (aqueous phase) within half an hour. Then, stirring was continued for at least 4 h to ensure the evaporation of the organic phase and spontaneous formation of nanosized micelles. Finally, the resultant micelles were centrifuged at 20,000 g for 15 min to remove any impurities and washed with distilled water. Then, the precipitated product was resuspended in distilled water and centrifuged again; this process was repeated three times to obtain the impurity-free POA nanosized micelles. Note that for the preparation of POA micelles containing bioactive agents (BSA) in the organic phase besides POA molecules, the cargo molecules (BSA) were also dissolved, and then the same procedures were performed as mentioned in the preparation of blank POA [Figure 1c].
Characterization of POA micelles
Micellization behaviors
The critical micelle concentration (CMC) of POA micelles was established by fluorescence measurement.[13] Pyrene with a final concentration of 6.0 × 10−7 M was used as a fluorescent probe. The different concentrations of POA polymer (1 to 10−9 g/l) were added to volumetric flasks, which already had dried pyrene solution (6.0 × 10−7 M) in acetone. After 24 h, fluorescence intensity was measured at λem = 390 nm and λex = 339 nm using a fluorescence spectrophotometer (Hitachi F-2500 fluorescence spectrophotometer, Corston, United Kingdom).
Size and zeta potential analysis of POA micelles
The average size and zeta potential of the POA micelles were analyzed by the zeta sizer (Zetasizer Nano ZS, Malvern, United Kingdom). Transmission electron microscopy (TEM) was also used to determine POA micelles morphology by the negative staining technique (TEM Philips EM 208S, Hillsboro, USA). The micelles were prepared and stored at 4°C for 2 months to evaluate their physical stability.
Evaluation of loading and encapsulation efficiency
As previously stated, the nanoprecipitation method was applied to encapsulate BSA molecules, and spectrometric techniques were used to measure the BSA loading and encapsulation efficiency. In brief, 4 mg of the POA polymer was dissolved in methanol; after mixing with 1 mg/300 μg of BSA solution, the resulting mixture was added to stirring distilled water at pH 4.7 for 30 min, and the reaction was continued for at least 4 h [Figure 1c]. At the end of the reaction, the pH of the solution was adjusted to 7.4. After ultra-centrifugation, the precipitate was dissolved in methanol, and the BSA loading percent and encapsulation efficiency were evaluated by measuring the optical density (OD) at 280 nm (Thermo Scientific NanoDrop 2000, Waltham, USA). The loading process was carried out at pH 7.4 similarly, and the results of the two reaction states were compared. Protein loading percent and encapsulation efficiency percent (EE%) were calculated by the following formulas.
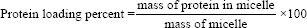

In vitro release assessment
The release study was carried out by the sample and separation method.[14] About 72 mg of freeze-dried loaded micelles was dispersed into 7.5 ml of phosphate-buffered saline (PBS) (pH 7.4). The resulting solution was divided into five microtubes (500 μl) and incubated at 37°C. At defined time intervals (8, 16, 24, 36, and 72 h), each sample was ultra-centrifuged, and the amount of released BSA was assessed by measuring absorbance at 280 nm. All of the release study samples were assessed in triplicate.
In vitro biocompatibility estimation
Hemolysis study
The human blood sample was collected in an ethylenediaminetetraacetic acid-containing tube and then centrifuged at 2000 g for 5 min. After removing the serum and washing it three times with sterile PBS (pH 7.4), the red blood cell (RBC) sample was diluted ten times with PBS. About 0.5 ml of the resulting solution was added to 0.5 ml of different POA micelle concentration samples (PBS and deionized water were used as the negative and positive controls, respectively) and then incubated at 37°C and 100 rpm (Kuhner shaker incubator, Birsfelden (Basel), Switzerland) for 4 h. Samples were centrifuged (10000 g for 15 min), and the absorbance of the supernatants was determined at 540 nm using a spectrophotometer (Biochrom WPA spectrophotometer, Cambridge, United Kingdom). The percentage of hemolysis was calculated based on the following formula.
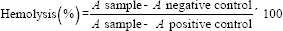
A = Absorbance at 540 nm.
Cell viability assay
The RAW 264.7 cells (a macrophage cell line) were used to evaluate POA micelle cytotoxicity. In brief, 4 × 103 cells/well were cultured (DMEM enriched with 10% FBS and 1% pen-strep antibiotic) and seeded in a 96-well plate. After overnight incubation, the different micelle concentrations (5, 25, 50, 75, and 100 μg/ml) were added to each well. After 24, 48, and 72 h of incubation, 10 μl of MTT solution (5 mg/ml) in 90 μl of DMEM was added to each well. The MTT solution was removed after 4 h, and then formazan crystals were dissolved in 100 μl of dimethyl sulfoxide (DMSO). Finally, the absorbance was measured at 570 nm and a reference wavelength of 690 nm using an enzyme-linked immunosorbent assay (ELISA) plate reader (Infinite M200, Tecan, Mannedorf, Switzerland).
Macrophage cell line uptake
The RAW 264.7 cells were cultured and seeded at a density of 2 × 104 cells/well in a 12-well plate. After 24 h, we treated cells as follows: non-treated cells (as negative control), DMSO as the solvent of curcumin, 25 μg/ml free POA micelle, 20 μM, and 75 μM free curcumin (as positive controls), and 25 μg/ml curcumin-loaded micelles. After 24 h of incubation, cellular uptake was evaluated by fluorescence microscopy (ZEISS Primovert iLED, Oberkochen, Germany).
Statistical analysis
All data are expressed as mean ± standard deviation (SD). The MTT assay result analysis was carried out using the two-way analysis of variance (ANOVA) method in GraphPad prism7 software. All experiments were performed in triplicate.
RESULTS
Confirmation of PEI-OL conjugation by 1H NMR and FTIR Spectroscopy
1HNMR result analysis showed successful conjugation of PEI-OL. As depicted in Figure 2, the 1HNMR spectra of OL show defined proton assignment around 5.3 and 11.1 ppm, which refer to (–CH=CH–) and OH groups, respectively. The PEI 1HNMR spectrum illustrates peaks around 2.5 ppm which belong to protons of the PEI skeleton –CH2–CH2–. The recorded peaks of PEI-OL 1HNMR in CDCL3 were 0.86 (–CH3), 1.25 (CH3 (–CH2)5–), (–CH2CH2CH=CH–), and (–CH=CH–CH2–(CH2)3–); 1.98 (–CH2CH=CHCH2–); and 5.32 ppm (–CH=CH–). The peaks from 2.1 to 3.6 ppm refer to the proton of the PEI backbone –NCH2CH2N– and –CH2CO– from OL. The new peak at 3.39 ppm approves PEI-OL conjugation. In addition, the pure PEI and OL were also characterized by FTIR spectroscopy.[15] The FTIR spectra of OL, PEI, and POA are shown in Figure 3. In the OL FTIR spectrum, the peak at 3040 cm− 1 was attributed to the C–H stretching (C = C–H), and the broad peak at 2500–3500 cm–1 was assigned to the O–H of carboxylic acid groups. Moreover, the peak of carbonyl groups (C = O) was also observed at 1712 cm–1, and the CH2 rocking vibration was also assigned at 720 cm–1 [Figure 3].[16] In the FTIR spectrum of PEI, the peaks at 3300–3500 cm− 1 were attributed to the stretching vibrations of amine groups (N–H), and 2851 and 2934 cm− 1 corresponded to the C–H stretching vibrations, whereas C–N stretching was observed at 1141 cm−1. Potassium bromide (KBr) pellets of PEI-OL conjugates were used to determine whether the conjugation reaction was processed successfully or not. The most distinct peak presented in the FTIR spectrum of PEI-OL is 1650 cm−1 which is attributed to the amide groups and highlights that the conjugation successfully occurred and PEI covalently bonds to the OL[17] [Figure 3]. All other peaks observed in OL and PEI spectra were observed in the PEI-OL spectrum, indicating the PEI-OL conjugation.
Figure 2.
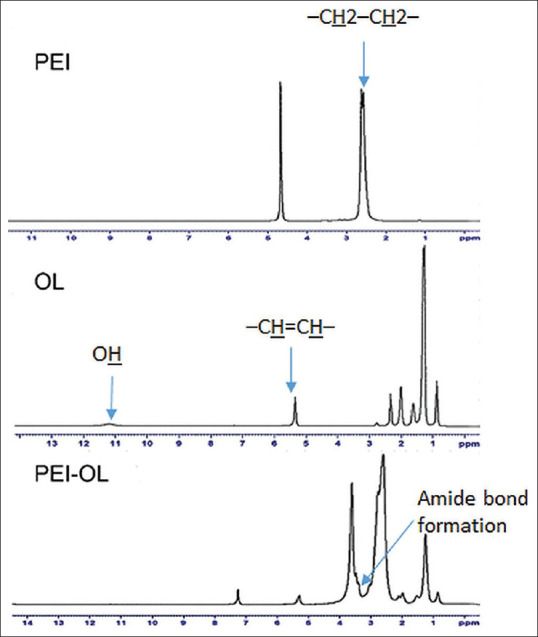
1HNMR results of PEI in D2O, OL, and PEI-OL in CDCl3 as the solvent. The results demonstrate successful PEI-OL synthesis as building blocks of POA micelles
Figure 3.
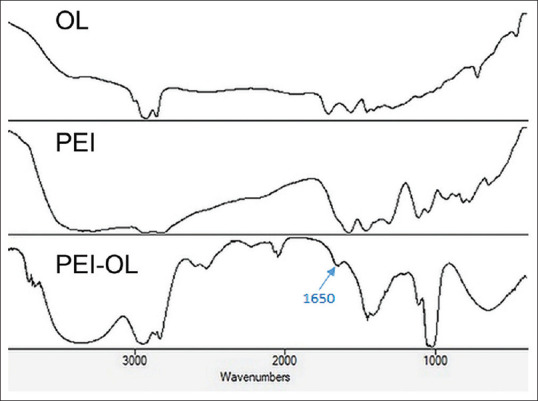
Displays the FTIR result of PEI-OL. The conjugation of two parts of the polymer has been verified
CMC determination
In Figure 4, the Boltzmann-type curve of pyrene fluorescent intensity against polymer concentration indicated that the CMC of fabricated POA micelles is around 5.62 × 10−8 mg/ ml.
Figure 4.
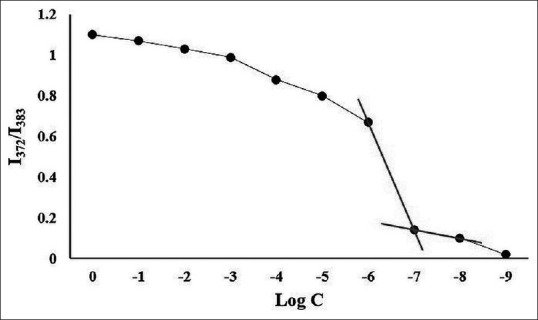
Definition of POA micelles’ CMC. The plot shows the pyrene fluorescent intensity ratio of I372/I383 against the PEI-OL concentration logarithm. In the CMC plot, a significant fluorescent intensity shift in the concentration of 5.62 × 10-8 mg/ml is illustrated, which is determined as the POA micelles’ CMC
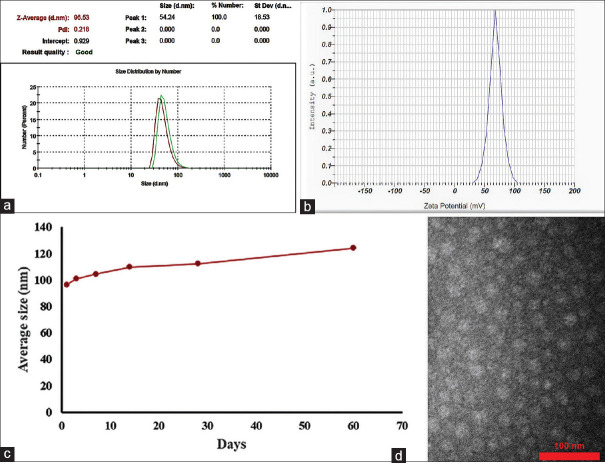
Size and zeta potential
The average hydrodynamic size and the zeta potential of synthesized micelles were approximately 96.53 ± 18.53 nm [Figure 5a] and 68.3 mV, respectively [Figure 5b]. Table 1 and Figure 5c present the data regarding the stability of the POA micelle. Within 2 months, the mean value of size changes was 13.83 nm, whereas the polydispersity index (PDI) was kept below 0.3.[18] Based on these findings, it can be concluded that fabricated POA micelle did not show any noticeable change in the particle size and PDI during 60 days, and these results underline the excellent physical stability. The TEM images present that the prepared POA micelles were spherical in shape with no signs of aggregation [Figure 5d].
Figure 5.
(a and b) Show the POA micelles’ size and zeta potential. (c) The micelles’ size changes within a 60-day period. (d) illustrates the TEM image of POA micelles
Table 1.
Micelle’s size changes in 2 months of tracking
| Days | 1 | 3 | 7 | 14 | 28 | 60 |
|---|---|---|---|---|---|---|
| Average size (nm) | 96.5 | 101 | 104.4 | 109.8 | 112.4 | 124.2 |
Loading and encapsulation efficiency
The loading reaction pH was set at isoelectric point (IEP) of BSA and the POA-loaded micelles were prepared. The loading and encapsulation efficiency was calculated as 16.5% and 70%, respectively.
In vitro release study
As mentioned in the loading section, 1 mg of BSA was used in the loading reaction, and regarding its 70% encapsulation efficiency (700 μg BSA in the final POA-loaded micelle), the release percent of BSA was determined in defined time ranges [Figure 6]. Table 2 also shows the amount (μg) of released BSA at a defined timeline. In our study, the release of BSA from POA micelles after 8 and 72 h incubation in PBS was 8.5% (59.5 μg) and 82% (574 μg), respectively.
Figure 6.
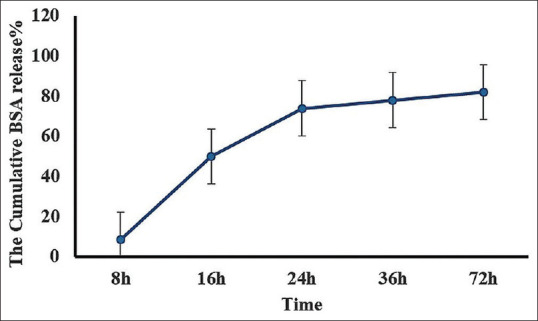
Release of BSA from POA micelles in a 72 h period study. All experiments were performed in triplicate
Table 2.
Shows the amount of released BSA at the defined timeline
| Time (h) | 8 | 16 | 24 | 36 | 72 |
|---|---|---|---|---|---|
| Mean of released BSA (µg) | 59.5 | 348.81 | 516.81 | 546 | 574 |
POA micelles hemocompatiblity
As shown in Figure 7, the hemolysis activity of POA micelles was generally dose-dependent in 4 h of incubation at 37°C. It was also found that POA at concentrations of ≤25 μg/ml is practically safe and non-toxic.
Figure 7.
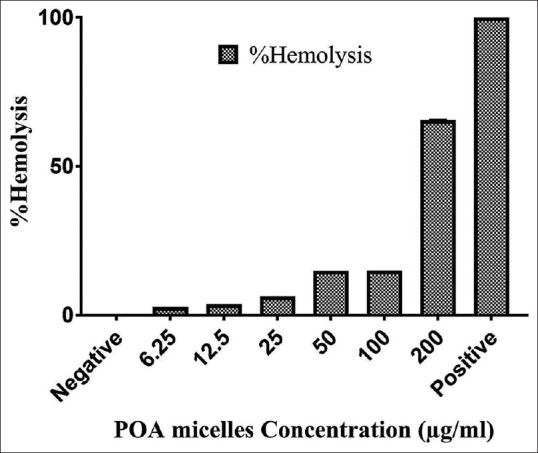
The effect of POA micelles’ concentration on red blood cells. All experiments were performed in triplicate
Cell survival interpretation
Using the MTT assay, we assess metabolic activity in mitochondria that occurred in viable cells.[19] Different concentrations of POA micelles were incubated for 72 h. Our results showed that POA micelles’ toxicity is time- and dose-dependent [Figure 8]. In the case of 24 h of incubation, the concentration of 5 and 25 μg/ml did not show any cytotoxicity within 48 h, but at higher concentrations, it exhibited cell toxicity in the range of 22% (25 μg/ml) to 80.1% (100 μg/ml). However, in 72 h of incubation, all concentrations exhibited toxic effects on cell growth, and cell viability decreased to 20% and 96% for the concentrations of 25 μg/ml and 100 μg/ml, respectively.
Figure 8.
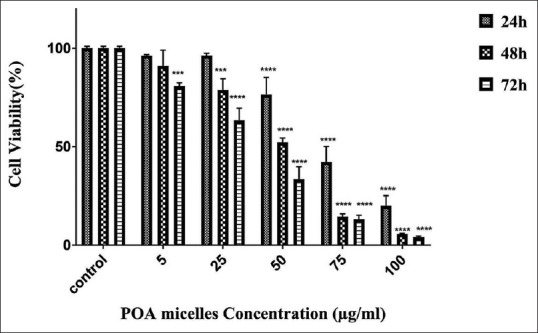
The cell cytotoxicity study. The graph shows the cell viability percent in 72 h period of incubation with POA micelles. All experiments were performed in triplicate. Statistical significance: ***P = 0.0002, ****P < 0.0001
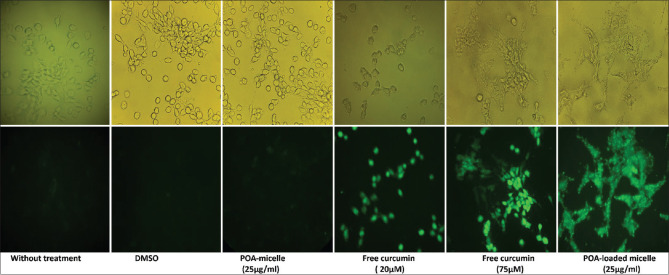
Cellular uptake of curcumin-loaded POA micelles by RAW264.7 cells
As displayed in Figure 9, fluorescence microscopy results showed that RAW 264.7 cells can effectively uptake curcumin-loaded POA micelles.
Figure 9.
The curcumin-loaded POA micelles’ uptake by the RAW264.7 cell line. The light (top row) and fluorescent (bottom row) microscopic images of the different treatments of RAW264.7 cells are shown (40× magnification)
DISCUSSION
Using modern vaccine generations such as subunit vaccines (protein, peptide, and DNA/RNA) seems to be more effective, but they are prone to degradation.[5] Cationic micelles have made an excellent opportunity to protect and deliver vaccine candidates to immune cells.[10,11] In this research, we synthesized and characterized the building blocks of novel cationic micelles as protein delivery vehicles.
According to the 1HNMR and FTIR spectra of PEI-OL [Figures 2 and 3], the emerging peaks attributed to the amide bond formation highlight that the conjugation successfully occurred, and PEI was covalently bonded to the OL as the building block of cationic micelles.[17]
Pyrene as a probe was used to determine the CMC value of developed POA micelles. The CMC is the minimum concentration of the polymer, which contributes to micelle formation in a water-based solvent. A lower CMC can grant more stable micelles in the solution. The ratio of the length of the hydrophobic to the hydrophilic part can affect the value of the CMC.[17] There are five indicative peaks in the pyrene fluorescent spectrum.[18] The polarity of the pyrene microenvironment can influence its peaks relative to fluorescent intensity. Measurement of the fluorescent intensity with the ratio of 372/384 nm is susceptible to the polarity of the pyrene microenvironment. The entrance of the pyrene in the micelle, a less polar microenvironment than water, can remarkably decrease the value of 372/384 nm.[17]
In general, the hydrodynamic size of micelles ranges from 10 to 200 nm and is predominantly spherical. Their size and shape are directly affected by the molecular weight, the molar mass ratio between two parts of the polymer (hydrophobic and hydrophilic parts), and the polymer structure.[19] To monitor the physical stability of developed micelles, both size and PDI of POA micelle were measured for 2 months (3, 7, 14, 28, and 60 days after synthesis).
Micelle stability is affected by many factors, including the character of building blocks and environmental conditions. Both segments of building blocks (hydrophobic and hydrophilic parts) are essential to maintain the micellar structure. Hydrophobicity and interaction of hydrophobic chains together in the core of the micelle can influence the micelle stability, and the more hydrophobic the chain is, the more stable the micelle is. The interaction of polymer blocks (hydrophilic part) with the solvent (water) is also a key factor in the stability of micelles.[19] In a study that Owens and his colleagues conducted, they used polyethyleneglycol (PEG) as the hydrophilic segment in the polymer structure, and they found that by increasing the length of PEG chains and surface density, the dipole–dipole/hydrogen binding of PEG with water improved and prohibition of water molecules’ penetration into the core of the micelle resulted in increased physical stability.[20] Probably, using branched PEI provided a high surface coverage to interact with aqueous molecules and prevented the exposure of the hydrophobic core and micelle destabilization.
The pH of the loading condition was set up at 4.7, which is the IEP of BSA. BSA has the lowest solubility at this pH, which can improve loading and the encapsulation efficiency from 2.4% and 10% (at pH 7.4 in a soluble form) to 16.5% and 70% (at pH 4.7), respectively. Similarly, Shi et al. found that by optimizing the pH of the loading condition to the IEP of recombinant human erythropoietin (pH 3.8), the loading efficiency of protein into poly (ethylene glycol)–poly (d, l-lactide) (PEG–PLA) di-block copolymeric micelles was significantly improved.[21] The protein loading efficiency was highly affected by the interaction of the protein with the hydrophobic segment.[22] BSA has seven segments called fatty acid-binding sites (FA1–FA7) for binding to different mono-/polyunsaturated fatty acids.[20] It is expected that in our study, BSA had hydrophobic interaction with OL in the core of the POA micelle. Saha et al. used BSA to study protein interaction with anionic and cationic surfactants in line with our data. In their study, BSA had hydrophobic interaction with the mixture of anionic sodium dodecyl sulfate (SDS) and cationic dodecyltrimethylammonium bromide (DTAB) and formed connected protein-decorated micelle-like complexes.[23]
As mentioned above, the interaction of BSA with the fatty acid chain can consequently affect both the loading efficiency and release profile of BSA. Some considerations should be noticed regarding the release profiles of a nanocarrier for being used as a vaccine delivery vehicle. Nanovaccines can have different fates once injected by subcutaneous, intramuscular, or intradermal route of administration. The following possibilities are considered for subcutaneous injection of nanovaccines: taking up by resident dendritic cells or monocyte-derived dendritic cells, entrance to the lymphatic system, or blood circulation.[24] It is shown that subcutaneous administration can recruit neutrophils and monocytes after 2 h of injection, and it peaked at 12 and 24 h after administration. Between 2 and 12 h after injection, the migration of dendritic cells from the skin to draining lymph nodes happened.[25] In another observation, macrophage migration began 3 h after injection and reached its maximum value at 6 h.[26] Therefore, conserving the nanocarrier's cargoes at least during the first 6 h after inoculation seems to be crucial.
Once nanoparticles enter the body, they can interact directly with blood components such as red blood cells, leukocytes, platelets, endothelial cells, and plasma proteins, interfering with their function. More specifically, it has been reported that the direct contact of nanoparticles with erythrocytes can cause hemolysis.[27] By the in vitro hemolysis assay, we can have a view of nanoparticles’ effects on the erythrocytes. Generally, 10% to 25% hemolysis is a borderline level, and nanoparticles with hemolysis below 10% are considered safe, whereas hemolysis above 25% is deemed hemolytic for nanoparticles.[27]
It is worth mentioning that nanocarriers as vaccine delivery agents are mainly administrated subcutaneously, intradermally, or intramuscularly. As mentioned above, these nanoparticles can also enter blood circulation after administration. However, nanoparticles of 10 nm in size could enter vascular capillaries.[5] The dynamic light scattering (DLS) result shows that a negligible number of POA micelles with an average size of 96.53 nm exist, which can enter the blood circulation from the administration site.
The cell culture study is the first step for estimating the behavior of nanoparticles under in vivo conditions. This assay is more ethical, easier, and less expensive to perform than the animal study.[28,29]
The particle size, shape, hydrophobicity, zeta potential, surface roughness, and surface biocompatibility affect nanoparticles’ toxicity.[30] Nanoparticles can affect cells’ activity through various mechanisms: inducing oxidation by reactive oxygen species, damaging cell membrane and cytoskeleton, causing DNA damage and disrupting transcription, disturbing the mitochondria, causing apoptosis, etc., For instance, nanoparticles of different sizes can affect cell viability by at least one of the mentioned mechanisms.[31] Zeta potential is another factor that affects nanoparticles’ cytotoxicity. To spell out this effect, cytotoxicity was examined on the L929 mouse fibroblast cell line in a study that employed four different polymeric nanoparticles with the same size but different zeta potentials (−30 ~ +40 mV). The results showed that cytotoxicity is highly affected by the zeta potential, concentration, and time trends. It can also be deduced from the findings that nanoparticles with negative zeta potential had less cytotoxicity than positive ones. The MTT results revealed that nanoparticles with a positive surface charge could reduce the cell survival rate and causes severe damage to the cell membrane.[32] Empty POA micelles had a high positive charge that led to cytotoxicity. It is notable that once BSA was formulated in the nanoparticle, the positive charge of cationic micelle was dramatically reduced from 68.3 mV to 3.6 mV (data not shown). In other words, the positive charge is reduced by loading protein into the micelles and consequently exerts low cell toxicity.
Cellular uptake analysis of nanoparticles is vital when they are supposed to be used in an in vivo condition and is considered a major cargo transporter route.[33] Monocyte-derived macrophage cells are resident in many mammals’ tissues. When a vaccine delivery agent is injected subcutaneously, intramuscularly, or intradermally, resident macrophages can uptake them.[26] We applied the RAW264.7 macrophage cell line to assess whether the POA micelles were efficiently taken up or not. Curcumin has autofluorescent properties, and its green fluorescence emission is visible under a 535–600 nm filter.[33] We used curcumin-loaded POA micelles to follow up on their uptake capability by RAW264.7 cells. In this case, as control groups, 20 and 75 μM of curcumin were used as the low and high concentrations of free (unloaded) curcumin, respectively. Size and surface charge can affect the cellular uptake of nanoparticles. Small size and positive charge are considered favorable properties for a carrier in cellular uptake.[5] POA micelles’ positive charge and small size (<100 nm) facilitate their uptake by RAW 264.7 cells.
CONCLUSION
In this project, the successful conjugation of branched PEI-2k to OL was carried out to build the blocks of a cationic micelle for protein delivery. Both size (<100 nm) and positive zeta potential of developed micelles highlight their potential capability to deliver cargoes to the resident immune cells; they can also enter the lymphatic system and convey loaded protein to draining lymph-node APCs. Successful uptake of POA micelles by RAW264.7 cells confirms their internalization into the cell, and the utility of this nanoplatform is thus underlined. MTT and hemolysis assays approved POA micelle's biocompatibility and hemocompatibility, respectively. The loading results indicated that optimizing the pH is the crucial parameter in the entrapment of the bioactive agents. Altogether, these results offer compelling evidence for the potential capability of this nanoplatform as a promising strategy for protein delivery.
Ethics approval and consent to participate
The research was approved by the Ethics Committee, Zanjan University of Medical Sciences (approval number: IR.ZUMS.REC.1398.017).
Financial support and sponsorship
This work was supported by the Student Research Committee [grant number: A-12-(873-9], Cancer Gene Therapy Research Center [grant number: A-12-(873-10] at the Zanjan University of Medical Sciences and the Iran National Science Foundation (INSF) [grant number: 98002828].
Conflicts of interest
There are no conflicts of interest.
Acknowledgements
We would like to thank our great colleagues at the Medical Biotechnology department at the Zanjan University of Medical Sciences. We convey our special thanks to the ZUMS Cancer Gene therapy Research Center, Student Research Committee (Zanjan University of Medical Sciences), and Iran National Science Foundation (INSF) for their financial support.
REFERENCES
- 1.Facciola A, Visalli G, Lagana P, La Fauci V, Squeri R, Pellicano GF, et al. The new era of vaccines: The “nanovaccinology”. Eur Rev Med Pharmacol Sci. 2019;23:7163–82. doi: 10.26355/eurrev_201908_18763. [DOI] [PubMed] [Google Scholar]
- 2.Sharma J, Carson CS, Douglas T, Wilson JT, Joyce S. Nano-Particulate Platforms for Vaccine Delivery to Enhance Antigen-Specific CD8+ T-Cell Response. Vaccine Design: Springer Humana New York USA. 2022:367–98. doi: 10.1007/978-1-0716-1892-9_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang H, Wang Q, Sun X. Lymph node targeting strategies to improve vaccination efficacy. J Control Release. 2017;267:47–56. doi: 10.1016/j.jconrel.2017.08.009. [DOI] [PubMed] [Google Scholar]
- 4.Guo S, Fu D, Utupova A, Sun D, Zhou M, Jin Z, et al. Applications of polymer-based nanoparticles in vaccine field. Nanotechnol Rev. 2019;8:143–55. [Google Scholar]
- 5.Trimaille T, Verrier B. Micelle-based adjuvants for subunit vaccine delivery. Vaccines (Basel) 2015;3:803–13. doi: 10.3390/vaccines3040803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boraschi D, Italiani P. From antigen delivery system to adjuvanticy: The board application of nanoparticles in vaccinology. Vaccines (Basel) 2015;3:930–9. doi: 10.3390/vaccines3040930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ott G, Van Nest G. Development of Vaccine Adjuvants: A Historical Perspective. Vaccine Adjuvants and Delivery Systems: John Wiley & Sons Hoboken NJ USA. 2007:1–31. [Google Scholar]
- 8.Luo Z, Shi S, Jin L, Xu L, Yu J, Chen H, et al. Cationic micelle based vaccine induced potent humoral immune response through enhancing antigen uptake and formation of germinal center. Colloids Surf B Biointerfaces. 2015;135:556–64. doi: 10.1016/j.colsurfb.2015.07.079. [DOI] [PubMed] [Google Scholar]
- 9.Alfagih IM, Aldosari B, AlQuadeib B, Almurshedi A, Alfagih MM. Nanoparticles as Adjuvants and Nanodelivery Systems for mRNA-Based Vaccines. Pharmaceutics. 2021;13:45. doi: 10.3390/pharmaceutics13010045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeng Q, Jiang H, Wang T, Zhang Z, Gong T, Sun X. Cationic micelle delivery of Trp2 peptide for efficient lymphatic draining and enhanced cytotoxic T-lymphocyte responses. J Control Release. 2015;200:1–12. doi: 10.1016/j.jconrel.2014.12.024. [DOI] [PubMed] [Google Scholar]
- 11.Zhao M, Li M, Zhang Z, Gong T, Sun X. Induction of HIV-1 gag specific immune responses by cationic micelles mediated delivery of gag mRNA. Drug Deliv. 2016;23:2596–607. doi: 10.3109/10717544.2015.1038856. [DOI] [PubMed] [Google Scholar]
- 12.Rezaei SJT, Sarijloo E, Rashidzadeh H, Zamani S, Ramazani A, Hesami A, et al. pH-triggered prodrug micelles for cisplatin delivery: Preparation and in vitro/vivo evaluation. React Funct Polym. 2020;146:104399. [Google Scholar]
- 13.Wilhelm M, Zhao CL, Wang Y, Xu R, Winnik MA, Mura JL, et al. Poly (styrene-ethylene oxide) block copolymer micelle formation in water: A fluorescence probe study. Macromolecules. 1991;24:1033–40. [Google Scholar]
- 14.Amatya S, Park EJ, Park JH, Kim JS, Seol E, Lee H, et al. Drug release testing methods of polymeric particulate drug formulations. J Pharm Investig. 2013;43:259–66. [Google Scholar]
- 15.Iijima M, Kawaharada Y, Tatami J. Effect of fatty acids complexed with polyethyleneimine on the flow curves of TiO2 nanoparticle/toluene suspensions. J Asian Ceram Soc. 2016;4:277–81. [Google Scholar]
- 16.Premaratne W, Priyadarshana W, Gunawardena S, De Alwis A. Synthesis of nanosilica from paddy husk ash and their surface functionalization. J Sci Univ Kelaniya. 2013;8:33–48. [Google Scholar]
- 17.Wang L, Zhang X, Cui Y, Guo X, Chen S, Sun H, et al. Polyethyleneimine-oleic acid micelle-stabilized gold nanoparticles for reduction of 4-nitrophenol with enhanced performance. Transit Met Chem. 2020;45:31–9. [Google Scholar]
- 18.Pal A, Chaudhary S. Ionic liquid induced alterations in the physicochemical properties of aqueous solutions of sodium dodecylsulfate (SDS) Colloids Surf A Physicochem Eng Asp. 2013;430:58–64. [Google Scholar]
- 19.Owen SC, Chan DP, Shoichet MS. Polymeric micelle stability. Nano Today. 2012;7:53–65. [Google Scholar]
- 20.Fatima S, Sen P, Sneha P, Priyadoss CG. Hydrophobic interaction between Domain I of Albumin and B Chain of detemir may support Myristate-dependent Detemir-Albumin binding. Appl Biochem Biotechnol. 2017;182:82–96. doi: 10.1007/s12010-016-2312-4. [DOI] [PubMed] [Google Scholar]
- 21.Shi Y, Huang W, Liang R, Sun K, Zhang F, Liu W, et al. Improvement of in vivo efficacy of recombinant human erythropoietin by encapsulation in PEG–PLA micelle. Int J Nanomedicine. 2013;8:1–11. doi: 10.2147/IJN.S38648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Owens DE, 3rd, Peppas NA. Opsonization, biodistribution, and pharmacokinetics of polymeric nanoparticles. Int J Pharm. 2006;307:93–102. doi: 10.1016/j.ijpharm.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 23.Saha D, Ray D, Kumar S, Kohlbrecher J, Aswal VK. Interaction of a bovine serum albumin (BSA) protein with mixed anionic–cationic surfactants and the resultant structure. Soft Matter. 2021;17:6972–84. doi: 10.1039/d1sm00264c. [DOI] [PubMed] [Google Scholar]
- 24.Jarvi NL, Balu-Iyer SV. immunogenicity challenges associated with subcutaneous delivery of therapeutic proteins. BioDrugs. 2021;35:125–46. doi: 10.1007/s40259-020-00465-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schetters STT, Kruijssen LJW, Crommentuijn MHW, Kalay H, den Haan JMM, van Kooyk Y. Immunological dynamics after subcutaneous immunization with a squalene-based oil-in-water adjuvant. FASEB J. 2020;34:12406–18. doi: 10.1096/fj.202000848R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee SB, Lee HW, Singh TD, Li Y, Kim SK, Cho SJ, et al. Visualization of macrophage recruitment to inflammation lesions using highly sensitive and stable radionuclide-embedded gold nanoparticles as a nuclear bio-imaging platform. Theranostics. 2017;7:926–34. doi: 10.7150/thno.17131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de la Harpe KM, Kondiah PPD, Choonara YE, Marimuthu T, du Toit LC, Pillay V. The hemocompatibility of nanoparticles: A review of cell–nanoparticle interactions and hemostasis. Cells. 2019;8:1209. doi: 10.3390/cells8101209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lewinski N, Colvin V, Drezek R. Cytotoxicity of nanoparticles. Small. 2008;4:26–49. doi: 10.1002/smll.200700595. [DOI] [PubMed] [Google Scholar]
- 29.Patravale V, Dandekar P, Jain R. Nanotoxicology: Evaluating toxicity potential of drug-nanoparticles. Nanoparticulate Drug Delivery. Elsevier Amsterdam Netherlands. 2012:123–55. [Google Scholar]
- 30.Sharifi S, Behzadi S, Laurent S, Forrest ML, Stroeve P, Mahmoudi M. Toxicity of nanomaterials. Chem Soc Rev. 2012;41:2323–43. doi: 10.1039/c1cs15188f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sukhanova A, Bozrova S, Sokolov P, Berestovoy M, Karaulov A, Nabiev I. Dependence of nanoparticle toxicity on their physical and chemical properties. Nanoscale Res Lett. 2018;13:44. doi: 10.1186/s11671-018-2457-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shao XR, Wei XQ, Song X, Hao LY, Cai XX, Zhang ZR, et al. Independent effect of polymeric nanoparticle zeta potential/surface charge, on their cytotoxicity and affinity to cells. Cell prolif. 2015;48:465–74. doi: 10.1111/cpr.12192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chuah LH, Roberts CJ, Billa N, Abdullah S, Rosli R. Cellular uptake and anticancer effects of mucoadhesive curcumin-containing chitosan nanoparticles. Colloids Surf B Biointerfaces. 2014;116:228–36. doi: 10.1016/j.colsurfb.2014.01.007. [DOI] [PubMed] [Google Scholar]