Abstract
Advances in genome sequencing technologies have unlocked new possibilities in identifying disease-associated and causative genetic markers, which may in turn enhance disease diagnosis and improve prognostication and management strategies. With the capability of examining genetic variations ranging from single-nucleotide mutations to large structural variants, whole-genome sequencing (WGS) is an increasingly adopted approach to dissect the complex genetic architecture of neurologic diseases. There is emerging evidence for different structural variants and their roles in major neurologic and neurodevelopmental diseases. This review first describes different structural variants and their implicated roles in major neurologic and neurodevelopmental diseases, and then discusses the clinical relevance of WGS applications in neurology. Notably, WGS-based detection of structural variants has shown promising potential in enhancing diagnostic power of genetic tests in clinical settings. Ongoing WGS-based research in structural variations and quantifying mutational constraints can also yield clinical benefits by improving variant interpretation and disease diagnosis, while supporting biomarker discovery and therapeutic development. As a result, wider integration of WGS technologies into health care will likely increase diagnostic yields in difficult-to-diagnose conditions and define potential therapeutic targets or intervention points for genome-editing strategies.
Neurologic diseases pose an ever-growing health and health economic burdens worldwide; in 2016, neurologic disorders were estimated to be the leading cause of disability-adjusted life-years lost (∼276 million years of healthy life lost due to premature death and disability) and the second leading cause of deaths; a staggering 9.0 million deaths worldwide.1 To dissect the distinct genetic profiles of these diseases, population-scale genome-wide association studies have set the stage by empowering our understanding of single-nucleotide variants in disease pathogenesis. Though an important knowledge gap remains in the missing heritability for these complex diseases. It is possible that genetic variations in less characterised (and thus under-examined) genomic regions contribute to large proportions of unexplained variance in disease risk.
To tackle this problem, newly developed techniques have ushered in a new era of genomic medicine, exemplified by the utilisation of next generation sequencing (NGS) technologies to discover novel disease-causing mutational signatures.2
Whole-genome sequencing (WGS) involves fragmenting and sequencing the entire genome of interest without pre-selecting specific DNA sequences, in contrast to whole-exome sequencing (WES) that focuses on sequencing either isolated or enriched coding portions of the genome.2 At an increasingly competitive cost, WGS-based methods are powerful tools in analysing genetic variations outside of coding regions given their wide sequencing coverage, higher sensitivity, and uniform coverage depth compared to exome sequencing.2 In recent years, advanced WGS technologies have enabled researchers to investigate structural variants (SVs) that explain significant portions of the missing heritability in complex neurologic and neurodevelopmental diseases.3,4
SVs are large alterations of more than 50 base-pairs in the genome, which are classified based on their mode of rearrangement.5 Some SV do not result in copy number changes (e.g., inversion and translocation), while others may involve copy number gains or losses from either insertion or deletion events.5 Somatic mosaic SVs (mSVs) are acquired SVs present in only subsets of cells of the affected individual,6 which behave differently to inherited germline SVs that are present from fertilisation and are present in cells throughout the body. Different models exist to explain how these SVs affect disease risk, one of which suggests that SVs are individually rare variations that collectively contribute to significant disease burdens.7
For diagnosing rare neurodevelopmental disorders, chromosomal microarrays and exome sequencing are commonly used strategies in clinical genetic laboratories, with some now starting to implement WGS to assess cases not diagnosed by microarrays and exome sequencing.8,9 Rexach et al.2 previously highlighted the potential of applying NGS technologies in clinical neurology. It is not until recently that the values of SV characterisation, as well as integrating RNA sequencing (RNA-seq) in the diagnostic setting have been recognised. This review will first describe SVs in the context of neurodegenerative and neurodevelopmental diseases, followed by discussing the clinical applications of WGS-based strategies and current international efforts on WGS integration at the health system level.
Structural Variations in Neurologic and Neurodevelopmental Diseases
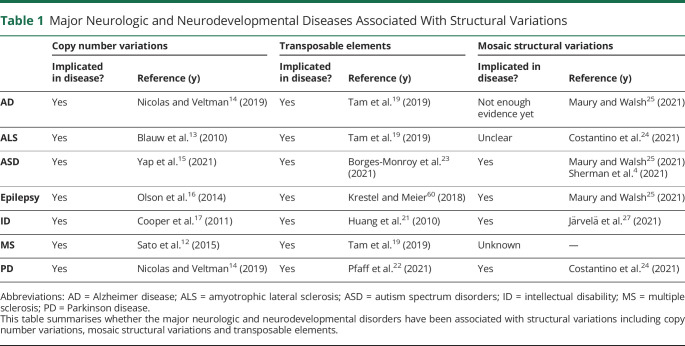
As shown in Table 1, there are several major neurologic and neurodevelopmental disorders that have been associated with different forms of SVs, including inherited germline SVs and mSVs. The following sections will describe germline CNVs and transposable elements, as well as mSVs (e.g., mosaic CNVs) that are implicated in these diseases.
Table 1.
Major Neurologic and Neurodevelopmental Diseases Associated With Structural Variations
Copy Number Variations
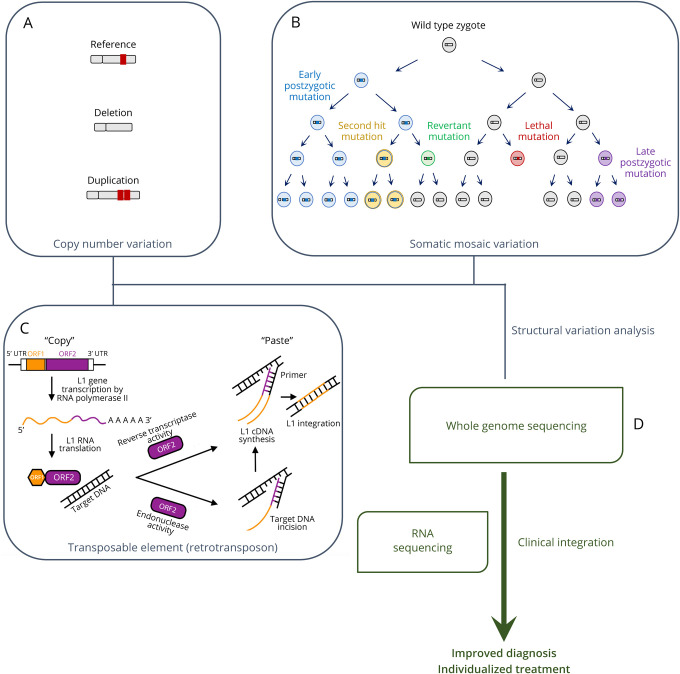
With a size of at least 50 base-pairs, copy number variations (CNVs) are defined as copy-number-varying structural changes in the genome.5 Different modes of variations can generate CNVs, including deletions and duplications (illustrated in Figure, A). A classic example of this is the well documented duplication of the PMP22 gene in Charcot Marie Tooth disease type 1A causing a demyelinating peripheral neuropathy, while the deletion of the same segment of DNA result in Tomaculous neuropathy (hereditary neuropathy with liability to pressure palsies).10 It has been hypothesised that some of these events are the results of non-allelic homologous recombination, where a duplicated DNA segment was mistakenly recognised as the homologue during homologous recombination processes and subsequently led to unequal cross-over. Another potential mechanism underlying CNV generation is the non-homologous end-joining process involving an error-prone DNA repair attempt with little to no homology. Individually rare intragenic CNVs were found to be prevalent (35%) among pathogenic variants for neurologic and neuromuscular disorders,11 while specific CNV types have been associated with several neurodegenerative diseases. For instance, Sato et al.,12 in a Japanese cohort study found that deletion-type CNVs at the T-cell receptor alpha and gamma loci were associated with multiple sclerosis (MS). In people with amyotrophic lateral sclerosis (ALS), rare CNVs in NIPA1 were associated with higher disease susceptibility.13 Further, de novo CNVs have been characterised in adult-onset neurodegenerative diseases, including duplications of the APP locus at chromosome 21 and a partial deletion of intron 1 of the BACE2 gene in early-onset Alzheimer disease (AD), and copy number gains (duplications or triplications) of the SNCA gene in families with Parkinson disease (PD).14 Recent studies have also implicated CNVs in neurodevelopmental diseases such as autism spectrum disorder (ASD),15 epilepsy16 and intellectual disability.17 Overall, there is a growing body of evidence that suggests a role for CNVs in these diseases, and established framework integrating multiple WGS-based detection methods has helped reveal small CNVs that were previously undetected in conditions such as ASD.18 Larger studies of this type would help elucidate the mechanisms by which candidate CNVs may influence disease susceptibility.
Figure. Whole-Genome-Sequencing-Based Structural Variation Analysis Can Yield Clinical Benefits.
(A) Copy number variations generated through deletion and duplication; (B) different types of somatic mosaic variations can occur at different time points with varying consequences; (C) LINE-1 (L1) retrotransposons can integrate into target DNA through a “copy-and-paste” mechanism; (D) complementing by RNA sequencing technologies, whole-genome DNA-sequencing-based methods can be used to detect and analyze disease-associated structural variations, which may in turn enhance diagnostic power and enable individualised treatments.
Transposable Elements
Transposable elements are highly relevant in neurodegenerative diseases,19 they are DNA sequences characterised by their ability to move between different genomic positions. Occupying more than half of the human genome, transposable elements are broadly grouped into 2 classes based on the mechanisms by which they mobilise across the human genome. The Class I elements, commonly termed retrotransposons, utilise RNA intermediates to mobilise in a “copy-and-paste” fashion with target-primed reverse transcriptase as the primary driver20 (see example illustrated in Figure, C). These can be further classified based on the presence of long terminal repeats (LTR). LTR retrotransposons make up approximately 8% of the human genome, and one example of this subclass are the human endogenous retroviruses associated with neurodegenerative diseases including AD, ALS, and MS.19 Next, different forms of non-LTR retrotransposons (∼33% of human genome) have also been implicated in neurologic and developmental diseases. For instance, insertions of long-interspersed element 1 (L1) were observed in X-linked intellectual disability,21 while a WGS study22 suggested a link between the SINE-R/VNTR/Alu elements and progression of PD. In addition, another recent WGS analysis also detected different types of de novo retrotransposon insertions in people with ASD, including the L1 and SINE-R/VNTR/Alu elements.23 Given the emerging links between transposable elements and these diseases, it is important to investigate how these mutational events contribute to disease pathophysiology.
Mosaic Structural Variations
Mosaicism describes a scenario where at least 2 genotypes are present in different cells of the same individual, caused by somatic mutations that can occur post-zygotically at varying timepoints (illustrated in Figure, B). The early post-zygotic variation affects many cells due to the many following proliferation steps, while later-occurring post-zygotic variation affects fewer cells as there are fewer following proliferative steps. The mutational events may also have different consequences: lethal mutations can terminate cell proliferation, whereas revertant mosaicism can revert the mutational signature back to wild type. mSVs are large-scale chromosomal aberrations in some but not all cells of the body. Depending on the underlying processes, mSVs can be either copy-neutral, loss of heterozygosity or in the form of mosaic CNVs. Unlike germline de novo CNVs, mosaic CNVs are associated with aging and mostly present in a subset of cells because the responsible mutational events typically occur post-fertilisation.4 Highly correlated with age, somatic mosaic CNVs have been implicated in neurodegenerative diseases such as PD,24 where somatic SNCA gain of function mosaic CNVs in dopaminergic neurons were differentially increased in cases with PD compared to controls and may contribute to the risk of sporadic synucleinopathies. Several somatic mutations were identified in sporadic early-onset AD, though it remains unclear whether somatic aneuploidy (full chromosomal copy variations) and CNVs influence AD risk.25 For neurodevelopmental diseases, a large study of children with developmental disorders26 revealed a 0.9% burden and 40-fold enrichment of somatic CNVs in cases compared to controls. A more recent study4 described the contribution of large mosaic CNVs (>4 Mb) to the previously unexplained portion of ASD risk. Despite their occurrence at a relatively low rate (∼0.2% compared to ∼5% for germline CNVs), they conferred significant ASD risk where increasing size of the mosaic CNVs was strongly correlated with increased ASD severity.4 In addition, mSVs resulted from maternal uniparental disomy (both copies of chromosome came from the mother) were identified in cases with intellectual disability in another study.27 Partly due to the rarity of known disease-causing mSVs, understanding the relationships between mSVs and disease susceptibility in neurology is a work in progress.
Since SVs may confer significant disease risk even in a modest proportion of the population, as was demonstrated by Sherman et al.4 in the case of mosaic CNVs in ASD genetic susceptibility, it highlights the importance of SV detection and analyses in population-scale studies.
Structural Variation Detection Using Whole-Genome Sequencing
Types of Whole-Genome Sequencing
To investigate SVs at population-scale, most studies to date rely on high-throughput short-read WGS technologies, which produce large quantities of reads that are ∼25–400 bp in length. One example is the Illumina technology that involves generating copies of adapter-tagged DNA fragments and subsequent fluorescent labelling of the newly generated DNA forward strands.28 Compared to short-read WGS, third generation long-read sequencing technologies can produce reads of more than 10 kb in length (and up to 4 Mb). Based on different template topologies and techniques to characterise the DNA sequences, PacBio single-molecule real-time sequencing and Oxford Nanopore Technologies sequencing are both commonly used and widely tested technologies in long-read sequencing.28 By design, short-read WGS has inherent limitations for variant phasing and SV characterisation especially in highly repetitive regions, while long-read WGS is generally better for characterising these genomic regions.
Bioinformatic Toolkits to Analyze Structural Variations
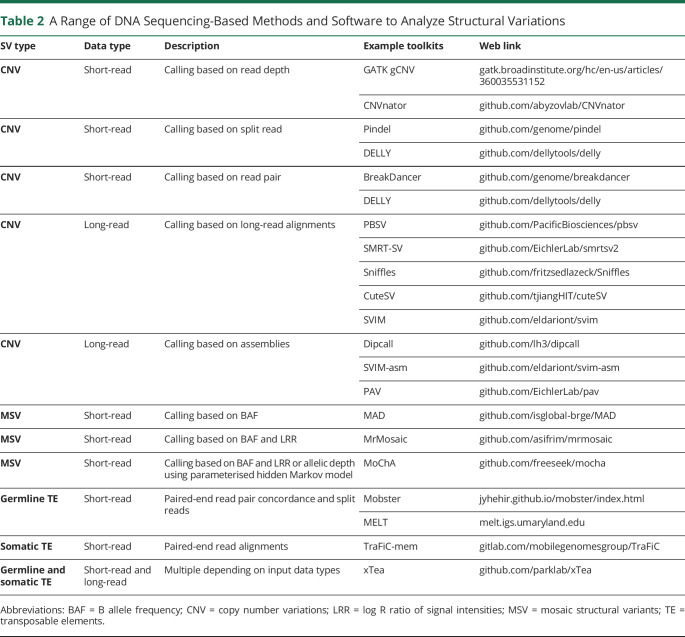
There are different bioinformatic toolkits available to call SVs in short-read and long-read sequencing data (Table 2). For instance, read-pair-based methods such as BreakDancer are especially sensitive in detecting deletion-type CNVs.29 To detect transposable elements, MELT is useful for identifying polymorphic inherited insertions, while TraFiC-mem is commonly used for detecting somatic insertions among case-control pairs. More recently, xTea is a new bioinformatic tool useful for analysing transposable element insertions in both short-read and long-read data. Applying xTea to analyze WGS data from 2,288 ASD families, Borges-Monroy et al.30 detected 86,154 retrotransposon insertions (more than 60% of which were previously undetected) and found higher-than-expected de novo retrotransposon insertions in disease risk genes including Alu, L1 and SINE-R/VNTR/Alu elements. For mosaic CNVs, MoChA is a recently developed software which was employed by Sherman et al.4 in their study of ASD. Further, a study by Ebert et al.31 found approximately 77% concordance between SV calls from short-read and long-read assemblies, and that the main advantage of long-read-based approach is increased sensitivity in detecting smaller SVs (∼83% of <250 bp SVs were not identified in short-read). Recently, Ohori et al.32 utilised long-read WGS and detected a partial MBD5 deletion in an WES-negative case with neurodevelopmental disorder, highlighting the clinical utility of long-read WGS as a useful technique to discover pathogenic SVs.
Table 2.
A Range of DNA Sequencing-Based Methods and Software to Analyze Structural Variations
Whole-Genome Sequencing in Clinical Neurology
Early diagnoses can facilitate disease management strategies and lead to better health outcomes.33 Consequently, genetic tests that can capture disease-causing mutational signatures among pre-symptomatic individuals are invaluable tools in clinical settings. For instance, the most common mutational signature in ALS and frontotemporal dementia (FTD) is the “GGGGCC” hexanucleotide repeat expansion in intron 1 of the C9orf72 gene, where nearly one-third of ALS and FTD cases carry a pathogenic C9orf72 repeat expansion.34 Along with the “CAG” trinucleotide repeat expansion in ATXN2 contributing to a smaller fraction of disease risk (∼0.01%–0.2% depending on number of repeats),35 these short tandem repeat (STR) sites are established genetic risk factors for ALS that can be efficiently analyzed using WGS-based methods. Recently, Roeck et al.36 developed a novel algorithm based on long-read WGS and demonstrated its capability in obtaining accurate lengths and sequences of an ABCA7 tandem repeat that was associated with greater than 4-fold increased risk of AD. Recognising those at risk and therefore potential earlier intervention may improve health outcomes, and these variants are clearly a potential site for gene correction/genomic-editing strategies. Further, early detection or even pre-symptomatic detection may be critical to reduce the disease burden, as often significant neuronal loss has occurred before clinical signs or symptoms develop.
As discussed, different forms of SVs are implicated in neurodegenerative and neurodevelopmental conditions. In the following sections, we will focus on WGS as a tool for detecting complex pathogenic mutations to enhance diagnostic power of genetic tests and facilitate the identification of disease-causative mutations as promising drug targets.
Detecting Structural Variations to Enhance Genetic Diagnosis
Currently, genetic testing is the standard of care for several neurodevelopmental disorders including ASD and intellectual disability, which can involve screening for pathogenic and likely pathogenic CNVs using either WES or chromosomal microarray technologies.9 The overall diagnostic yields of microarray-based tests (∼15%–20%) are generally lower than that of exome-sequencing-based tests (∼31%) for neurodevelopmental disorders.9 As genetic tests continue to evolve, the integration of other modalities such as omics technologies, advanced imaging and WGS technologies into testing may be useful in identifying and assessing pre-symptomatic individuals with genetic disorders.37 Given the emerging roles of SVs in several neurodevelopmental and neurologic disorders, WGS-based detection of SVs could become an essential part of genetic testing.
Despite the modest sample sizes of existing studies on WGS integration in clinical settings, there is mounting evidence that supports the diagnostic superiority of tests integrating WGS when compared to other genetic testing methods. For example, in a nested cohort of 50 people with severe intellectual disability (but without an established genetic diagnosis after extensive genomic microarray analysis and WES), Gilissen et al.8 reported a higher diagnostic yield of WGS-based tests (42%) in comparison to those based on genomic microarrays (12%) and WES technology (27%). Similarly, a more recent study by Lindstrand et al.38 also demonstrated the usefulness of WGS in clinical diagnosis amongst 100 people with severe intellectual disability. An important observation was that WGS performed well at calling a wide range of SV sizes, including those that could not be detected using microarrays due to probe coverage. The resulting diagnostic yield was higher in WGS-based tests (27%) compared to that of microarray-based tests (12%).38 In another study of 103 patients recruited from paediatric subspecialty clinics, Lionel et al.39 also found that WGS performed better than routine clinical tests based on microarrays and targeted sequencing. The resulting diagnostic yield of WGS-based tests was 41% (42 of 103 patients; 17 patients had their diagnoses made by WGS only), a significant improvement (p = 0.01) compared to a 24% yield of routine genetic tests.39
However, some issues faced by WES remain challenging in short-read WGS, such as the detection of small SVs involving highly repetitive regions. A recent study by Sone et al.40 demonstrated that long-read WGS can detect SVs in clinically relevant disease-risk genes that were previously missed with short-read sequencing technologies. Based on the risk genes identified in the study,40 a genetic diagnosis for neuronal intranuclear inclusion disease is now possible, whereas previously diagnoses could only depend on histopathology. Further, recent reports also suggested benefits of combining multiple sequencing technologies compared to relying on one sequencing method. For instance, Chaisson et al.41 found a 7-fold increase in SV discovery utilising multiple state-of-the-art sequencing technologies (e.g., PacBio long-read WGS, Bionano Genomics optical mapping, etc) compared to Illumina short-read sequencing alone. Additionally, a study by Zhao et al.42 on 3 matched trio families from the 1,000 Genomes Project also highlighted the added value of long-read WGS in characterising new catalogues of insertions and transposable elements, though a “blind spot” for long-read WGS was discovered where 88.2% of large CNVs identifiable in 30X short-read WGS depth-based methods were not detected using the long-read assembly.
RNA-seq technology allows researchers to examine a wide range of RNA species and aberrant transcription in human diseases. Complementing DNA sequencing, RNA-seq is an emerging approach to facilitate prioritisation of SVs that alter gene structures in transcribed genomic regions with potential functional implications.43 There is increasing evidence that SVs such as transposable elements are associated with gene expression and splicing alterations,44 though current clinical applications of RNA-seq have mostly focused on single-nucleotide variants to improve variant interpretation and resolve cases with inconclusive diagnoses from standard DNA testing. For instance, Cummings et al.45 were able to apply RNA-seq and perform expression-based annotation of variants to identify positions with similar read counts but markedly different expression levels across tissues such as brain cortex and liver. For haploinsufficient disease genes in the gnomAD database, the expression-based variant annotation reduced annotation error by 22.8% while retaining most (>96%) of the high-confidence pathogenic variants.45 Further, by systematically analysing aberrant gene expression and slicing events in 182 previously undiagnosed individuals, Murdock et al.46 found that the transcriptome-guided approach helped diagnose cases with rare Mendelian conditions (e.g., NSD2-and CLTC-related intellectual disability) that were unresolved by routine genetic tests based on WES or chromosomal microarrays. In a recent case study of lissencephaly, Qashqari et al.47 also demonstrated that RNA-seq was useful in clinical settings where standard DNA tests could not reach conclusive diagnosis. Still in its infancy, the clinical application of RNA-seq is already showing promising potential in streamlining variant detection via WGS-based DNA tests and improving variant interpretation and prioritisation (especially non-coding variants), which in turn yielded functional evidence to facilitate genetic diagnosis for diseases including hypomyelinating leukodystrophy and severe Leighs syndrome.48 As the knowledge base for SVs in human diseases continues to grow, integration of RNA-seq and WGS in future population-scale SV studies may provide new insights to further enhance genetic diagnosis of SV-related diseases.
Towards Wider Implementation of Whole-Genome Sequencing
Factors Limiting Wide-Spread WGS Integration in Clinical Neurology
Despite its significant diagnostic power, a genetic test involving WGS still has limited applications in routine clinical care. At this stage, the utility of offering WGS-based tests remains bounded by the significant knowledge gaps in disease-causing genes and variants to be tested for most neurologic conditions. That is, we cannot realise the full potential of WGS technologies in clinical practice until we develop a better understanding of the genetic architecture of neurologic diseases. A scattergun approach to applying WGS technologies will no doubt generate a vast quantity of data, but without the ability to analyze and interpret the outputs WGS may only complicates diagnostic processes. A potential solution to this issue is the emerging approach of combining RNA-seq with WGS as discussed. Furthermore, there is a gap in preparing frontline clinicians to support the implementation of WGS in routine clinical care.49 Offering WGS-based clinical tests will create a huge extra burden in understanding the output from WGS testing, along with the considerable time and financial burden of analysing and interpreting high-volume WGS outputs. Clearly, for WGS to be implemented as a useful diagnostic tool, a close relationship between clinicians, clinical geneticists, and computational biologists needs to be developed. At the same time, standardised pipelines to generate, analyze, and interpret test results are critical to a wider implementation of WGS in health care.
Finally, the cost of undertaking WGS is only a small component of the overall cost of implementing the test into clinical practice, as it is a complex process starting from analysis and interpretation of the outputs to implementation of the findings, which requires a dedicated and coordinated team approach. Further adding to the complexity of this testing approach is that finding a deleterious variant or SV will have significant implications not only to the person being tested but to their family members. That said, the enhanced ability to provide a formal diagnosis is an important first step in formulating personalised treatment plans, while providing significant closure for a family with an affected individual.
Current Efforts to Integrate Genome Sequencing Into Health Care System
Reducing costs, in addition to advancements in WGS technologies, have enabled generation of WGS data at the national health system level. Through WGS of 660 individuals with rare neurologic and developmental disorders, a UK Biobank study50 provided a diagnostic report implicating likely pathogenic or pathogenic variants to 33% of the cases. The study also identified known and novel mutational signatures (including one deletion-inversion-duplication SV) in the expert-curated list of 1,423 diagnostic-grade genes that have an established causal role in neurologic and developmental disorders. In addition, the higher number of genes (30 genes) required to cover 50% of the diagnostic reports for neurologic and developmental disorders compared to other rare diseases (ranged from 5 to 11 genes) further highlighted the complexity of the genetic landscape in neurologic and neurodevelopmental disorders. As the results indicated, WGS had a much wider coverage of genetic variants in the diagnostic-grade genes than WES.50 More recently, a preprint by Halldorsson et al.51 analyzed WGS data from a larger number of individuals in the UK Biobank study (n = 150,119) identified 895,055 SVs and 2,536,688 STRs, some of which were highly relevant for conditions affecting the brain such as epilepsy and familial hemiplegic migraine. Another study in Swedish health care setting52 also demonstrated the value of using WGS to analyze more complex pathogenic variations such as CNVs and balanced SVs in genetic tests. At later stages of the study, the addition of screening for STRs and genome-wide CNV analysis for relevant indications using WGS resulted in a 7.5% increased diagnostic yield among 285 cases with rare diseases, which included a range of developmental disorders, neurodegenerative, neuromuscular and ataxia diseases.
Ongoing international and collaborative efforts are being made to integrate genomic technologies into health care, including the China Precision Medicine Initiative, Genomics England, the French Plan for Genomic Medicine 2025, Australian Genomics Health Alliance, the All of Us research program in the United States, and the Global Genomic Medicine Collaborative.53 Stark et al.53 recognised 2 key priorities in breaking down existing barriers to genomic implementation. First, there is a need for building a robust evidence-base for clinical applications of genomic technologies through evaluation of its long-term health and health economic impact, which will facilitate the development of standardised evaluation criteria specific to diseases, funding contexts, and health care systems. To explore WGS implementation in health care and to optimise the implementation process (selecting patients, obtaining consent, interpreting WGS test results and providing feedback), the 100,000 Genomes Project is a population-scale research project in the UK that involved WGS of more than 100,000 genomes from approximately 97,000 individuals and their families affected by rare diseases or cancers.54 With far-reaching impact, the project to date has yielded actionable findings for ∼25% of rare disease patients, as well as identifying potential for a therapy or a clinical trial in approximately 50% of cancer patients.
Next, there is also an increasing need to encourage genomic data sharing and accelerate the development of new population genomic databases, specifically in secure data storage and transfer. For both the rigor as well as ease of interpretation and clinical use, it is critical for computational biologists to develop standardised data inputs and outputs in easy-to-understand and interpret formats. To achieve these, there are many technical and regulatory challenges at an international scale. The Global Alliance for Genomics and Health (GA4GH) is an international not-for-profit alliance that focuses its efforts on formulating standardised strategies for secure storage and transfer of genomic data, such as open-access toolkits for data security as well as regulatory and ethical guidance. There are ongoing GA4GH initiatives to identify and address current needs of international genomic communities and subsequently guide framework development, which can in turn promote more effective use of genomic data and motivate international collaborations to accelerate progress in genomic research.
Ongoing WGS-Based Research and Future Directions
Measuring Mutational Constraint to Improve Variant Interpretation and Disease Diagnosis
Enhancing our ability to identify disease-associated genes and improving disease variant interpretation are both crucial to further improve genetic diagnostic yield. Disease gene discovery can gain power from classifying genes by their tolerance to inactivation. Predicted loss-of-function (pLoF) variants are variants that are predicted to render the corresponding genes non-functional. To quantify the extent to which natural selection removes pLoF variants from the population, a constraint score can be calculated by obtaining the ratio of observed pLoF variant counts and expected pLoF variant counts based on DNA mutation rates. Despite current limitations in deriving these metrics from SVs at WGS resolution, Collins et al.5 found strong concordance between depletion of rare pLoF SVs and the existing metrics derived from single-nucleotide variants, indicating a similar pattern of selection against highly pLoF SVs. These metrics for measuring mutational constraint can be useful in improving variant interpretation. For cases with intellectual disability or other neurodevelopmental conditions, there is evidence of higher rates of pLoF de novo variations in genes located within the more constrained deciles of the genome compared to controls, while similar pLoF enrichment patterns were also observed in ASD.55 Compared to conventional methods such as genetic linkage analysis, high-throughput sequencing methods including WGS were better at identifying disease-associated genes under extreme constraint against pLoF variations.55 Consequently, the application of WGS will likely increase the power to identify this form of disease-associated genetic variation. Further, significant enrichment of pLoF variants was observed in highly expressed exonic regions in ASD, where the high-expressed pLoF variants had larger effect sizes compared to the low-expressed variants.45 Through RNA-seq integration, transcript-aware annotation of pLoF variants was demonstrated to selectively remove annotation errors by existing toolkits, while improving the interpretation of the functional impacts posed by rare variants in disease settings.45 Together, these findings suggest that implementation of transcript expression-aware pLoF annotation could enhance rare disease diagnostic strategies. Overall, the curation and characterisation of pLoF variants have been demonstrated to enhance disease variant discovery and interpretation, with clinical relevance in improving rare variant burden testing and disease diagnosis.
WGS-Based Research to Enhance Biomarker Discovery and Therapeutic Development
With an incomplete picture of the genetic landscapes in many neurologic diseases, there is a subsequent scarcity of reliable disease biomarkers for these diseases. As discussed, the incomplete knowledge of genetic variants in these diseases also limits the wide-spread implementation of WGS in clinical diagnosis and care. Consequently, this highlights the need to address this knowledge gap and identify more robust and easily assessable disease biomarkers, which could provide the basis for earlier disease detection in clinical settings, as well as personalised medicine enabling the use of current and future therapies with more precision. Human genetics studies are increasingly used as a tool to aid biomarker discovery, and given the advances in WGS technologies, researchers are now much better equipped in the search for new disease associated variants. For instance, Wilfert et al.56 recently used WGS to study ultra-rare, likely gene-disruptive variants in 3,474 ASD families. They found much of the variant burden (95%) was outside of known risk genes for ASD, and that children with these newly identified rare mutational events were 2.7 times more likely to have ASD. Incorporating methods such as transcript expression-aware pLoF annotation, with larger and more comprehensive WGS studies will likely uncover more candidate genes conferring a greater proportion of unexplained disease risk, as well as novel targets for existing and novel therapeutic interventions.
Further, population-scale WGS could power investigations of drug target evaluation and safety profiling. Minikel et al.57 utilised pLoF variants from sequencing data to quantify constraint and found that on average, targets of approved drugs were slightly more constrained compared to protein-coding genes overall. Complementing preclinical studies that involve animal knockout models, candidate pLoF variants serve as natural in vivo models of human gene inactivation and can thus be valuable in assessing the potential toxicity of drugs under development. For example, LRRK2 inhibition had been suggested as a promising treatment strategy for PD, though its long-term safety profile in humans remained unclear.58 To investigate potential toxicity of LRRK2 inhibition in human, Whiffin et al.58 analyzed 3 large sequencing data sets (gnomAD, UK Biobank, and 23andMe) and identified high-confidence candidate LRRK2 pLoF variants in 1,455 individuals. Based on validated pLoF variants, Whiffin et al.58 confirmed reduction of the LRRK2 protein levels in 82.5% of the cohort, but this was not strongly associated with any disease state or specific health-related phenotype. Consequently, ongoing research in identifying and interpreting pLoF variants (including SVs) from large sequencing cohorts could provide a roadmap for future studies of natural human knockout models and facilitate evaluation of druggable targets during early stages of the therapeutic development process.
Soon, WGS-enhanced tests will likely enable appropriate genetic counselling, preimplantation diagnostics, and initiation of established treatments at an earlier time point. They may also provide points of intervention for personalised genetic medicine where specific treatments aimed at correcting or mitigating the effects of a genetic variant could be developed and applied at the individual level. These clinical benefits were recently showcased by Zou et al.59 in a study of 320 Chinese children with epilepsy, where genome sequencing tests had a 75.0% yield in those with earlier seizure onset and the identified genetic causes for 42 cases (13.1%) were treatable using targeted therapies.
Conclusion
Driven by WGS-based research in neurologic and neurodevelopmental disorders, current understanding of disease-causing mutations is rapidly evolving. Emerging but not yet conclusive evidence suggests a critical role of SVs as significant contributors to the global neurologic disease burden, with particular emphasis on ASD, intellectual disability, epilepsy, AD, MS, and PD. WGS technologies are on the cusp of becoming mainstream diagnostic tools in neurologic practice, with increasing clinical relevance in improving diagnostic power and evaluating genes as potential therapeutic targets. As a result, wider implementation of WGS in health care will likely hold the key to higher diagnostic yields, novel therapeutic targets, and cost-effective preventive measures. Complementing the DNA sequencing technologies, a transcriptome-guided approach involving RNA-seq has emerged as a useful strategy to help realise the full potential of genomic medicine.
Glossary
- AD
Alzheimer disease
- ALS
amyotrophic lateral sclerosis
- ASD
autism spectrum disorder
- CNV
copy number variations
- FTD
frontotemporal dementia
- GA4GH
the Global Alliance for Genomics and Health
- L1
long-interspersed element 1/ LINE-1
- LTR
long terminal repeats
- MS
multiple sclerosis
- mSVs
mosaic SVs
- PD
Parkinson disease
- pLoF
predicted loss-of-function
- RNA-seq
RNA sequencing
- STR
short tandem repeats
- SV
structural variants
- WES
whole-exome sequencing
- WGS
whole-genome sequencing
Appendix. Authors
Contributor Information
Xin Lin, Email: xin.lin@utas.edu.au.
Yuanhao Yang, Email: yuanhao.yang@mater.uq.edu.au.
Phillip E. Melton, Email: phillip.melton@utas.edu.au.
Vikrant Singh, Email: vikrant.singh@utas.edu.au.
Steve Simpson-Yap, Email: steve.simpsonyap@unimelb.edu.au.
Kathryn P. Burdon, Email: kathryn.burdon@utas.edu.au.
Bruce V. Taylor, Email: bruce.taylor@utas.edu.au.
Study Funding
This work was supported by funding from the MS Research Australia (B.V.T.: Macquarie Foundation/MSRA paired clinical fellowship; Y.Z.: postdoctoral fellowship), The Medical Research Future Fund (B.V.T.), and the Australian National Health and Medical Research Council (B.V.T.; Y.Z.: GNT1173155). Y.Y. was supported by the Mater Foundation.
Disclosure
B.V.T. has received compensation for consulting, talks, and advisory/steering board activities for Merck, Novartis, Biogen, and Roche. He receives research funding support from MS Research Australia, Medical Research Future Fund Australia, and the National Health & Medical Research Council Australia. Go to Neurology.org/NG for full disclosure.
References
- 1.Feigin VL, Emma N, Alam T, et al. Global, regional, and national burden of neurological disorders, 1990-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol 2019;18(5):459-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rexach J, Lee H, Martinez-Agosto JA, Németh AH, Fogel BL. Clinical application of next-generation sequencing to the practice of neurology. Lancet Neurol 2019;18(5):492-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Logsdon GA, Vollger MR, Eichler EE. Long-read human genome sequencing and its applications. Nat Rev Genet 2020;21(10):597-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sherman MA, Rodin RE, Genovese G, et al. Large mosaic copy number variations confer autism risk. Nat Neurosci 2021;24(2):197-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins RL, Brand H, Karczewski KJ, et al. A structural variation reference for medical and population genetics. Nature 2020;581(7809):581444-581451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poduri A, Evrony GD, Cai X, Walsh CA. Somatic mutation, genomic variation, and neurological disease. Science 2013;341(6141):1237758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li YR, Glessner JT, Coe BP, et al. Rare copy number variants in over 100,000 European ancestry subjects reveal multiple disease associations. Nat Commun 2020;11(1):255-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilissen C, Hehir-Kwa JY, Thung DT, et al. Genome sequencing identifies major causes of severe intellectual disability. Nature 2014;511:344-347. [DOI] [PubMed] [Google Scholar]
- 9.Savatt JM, Myers SM. Genetic testing in neurodevelopmental disorders. Front Pediatr 2021;9:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu X, Duan X, Zhang Y, Fan D. Clinical and genetic diversity of PMP22 mutations in a large cohort of Chinese patients with charcot-marie-tooth disease. Front Neurol 2020;11:630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Truty R, Paul J, Kennemer M, et al. Prevalence and properties of intragenic copy-number variation in Mendelian disease genes. Genet Med 2019;21(1):114-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sato S, Yamamoto K, Matsushita T, et al. Copy number variations in multiple sclerosis and neuromyelitis optica. Ann Neurol 2015;78(5):762-774. [DOI] [PubMed] [Google Scholar]
- 13.Blauw HM, Al-Chalabi A, Andersen PM, et al. A large genome scan for rare CNVs in amyotrophic lateral sclerosis. Hum Mol Genet 2010;19(20):4091-4099. [DOI] [PubMed] [Google Scholar]
- 14.Nicolas G, Veltman JA. The role of de novo mutations in adult-onset neurodegenerative disorders. Acta Neuropathol 2019;137(22):183-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yap CX, Alvares GA, Henders AK, et al. Analysis of common genetic variation and rare CNVs in the Australian Autism Biobank. Mol Autism 2021;12(1):1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olson H, Shen Y, Avallone J, et al. Copy number variation plays an important role in clinical epilepsy. Ann Neurol 2014;75(6):943-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cooper GM, Coe BP, Girirajan S, et al. A copy number variation morbidity map of developmental delay. Nat Genet 2011;43(9):838-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Werling DM, Brand H, An JY, et al. An analytical framework for whole-genome sequence association studies and its implications for autism spectrum disorder. Nat Genet 2018;50(5):727-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tam OH, Ostrow LW, Gale Hammell M. Diseases of the nERVous system: retrotransposon activity in neurodegenerative disease. Mob DNA 2019;10(1):32-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elbarbary RA, Lucas BA, Maquat LE. Retrotransposons as regulators of gene expression. Science 2016;351(6274):aac7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang CR, Schneider AM, Lu Y, et al. Mobile interspersed repeats are major structural variants in the human genome. Cell 2010;141(7):1171-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfaff AL, Bubb VJ, Quinn JP, Koks S. Reference SVA insertion polymorphisms are associated with Parkinson's Disease progression and differential gene expression. NPJ Parkinson's Dis 2021;7(1):1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borges-Monroy RC, Chu C, Dias C, et al. Whole-genome analysis of de novo and polymorphic retrotransposon insertions in autism spectrum disorder. Mob DNA 2021;12(1):28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Costantino I, Nicodemus J, Chun J. Genomic mosaicism formed by somatic variation in the aging and diseased brain. Genes (Basel) 2021;12(7):1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maury EA, Walsh CA. Somatic copy number variants in neuropsychiatric disorders. Curr Opin Genet Dev 2021;68:9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.King DA, Jones WD, Crow YJ, et al. Mosaic structural variation in children with developmental disorders. Hum Mol Genet 2015;24(10):2733-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Järvelä I, Määttä T, Acharya A, et al. Exome sequencing reveals predominantly de novo variants in disorders with intellectual disability (ID) in the founder population of Finland. Hum Genet 2021;140(7):1011-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Logsdon GA, Vollger MR, Eichler EE. Long-read human genome sequencing and its applications. Nat Rev Genet 2020;21(10):597-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whitford W, Lehnert K, Snell RG, Jacobsen JC. Evaluation of the performance of copy number variant prediction tools for the detection of deletions from whole genome sequencing data. J Biomed Inform 2019;94:103174. [DOI] [PubMed] [Google Scholar]
- 30.Borges-Monroy R, Chu C, Dias C, et al. Whole-genome analysis reveals the contribution of non-coding de novo transposon insertions to autism spectrum disorder. Mobile DNA 2021;12(1):28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ebert P, Audano PA, Zhu Q, et al. Haplotype-resolved diverse human genomes and integrated analysis of structural variation. Science 2021;372(6537):eabf7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohori S, Tsuburaya RS, Kinoshita M, et al. Long-read whole-genome sequencing identified a partial MBD5 deletion in an exome-negative patient with neurodevelopmental disorder. J Hum Genet 2021;66(7):697-705. [DOI] [PubMed] [Google Scholar]
- 33.Owen MJ, Niemi AK, Dimmock DP, et al. Rapid sequencing-based diagnosis of thiamine metabolism dysfunction syndrome. N Engl J Med 2021;384(22):2159-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turner MR, Al-Chalabi A, Chio A, et al. Genetic screening in sporadic ALS and FTD. J Neurol Neurosurg Psychiatry 2017;88(12):1042-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neuenschwander AG, Thai KK, Figueroa KP, Pulst SM. Amyotrophic lateral sclerosis risk for spinocerebellar ataxia type 2 ATXN2 CAG repeat alleles: a meta-analysis. JAMA Neurol 2014;71(12):1529-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Roeck A, De Coster W, Bossaerts L, et al. NanoSatellite: accurate characterization of expanded tandem repeat length and sequence through whole genome long-read sequencing on PromethION. Genome Biol 2019;20(1):239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hou YC, Yu HC, Martin R, et al. Precision medicine integrating whole-genome sequencing, comprehensive metabolomics, and advanced imaging. Proc Natl Acad Sci U S A 2020;117(6):3053-3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lindstrand A, Eisfeldt J, Pettersson M, et al. From cytogenetics to cytogenomics: whole-genome sequencing as a first-line test comprehensively captures the diverse spectrum of disease-causing genetic variation underlying intellectual disability. Genome Med 2019;11(1):68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lionel AC, Costain G, Monfared N, et al. Improved diagnostic yield compared with targeted gene sequencing panels suggests a role for whole-genome sequencing as a first-tier genetic test. Genet Med 2018;20(4):435-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sone J, Mitsuhashi S, Fujita A, et al. Long-read sequencing identifies GGC repeat expansions in NOTCH2NLC associated with neuronal intranuclear inclusion disease. Nat Genet 2019;51(8):1215-1221. [DOI] [PubMed] [Google Scholar]
- 41.Chaisson MJP, Sanders AD, Zhao X, et al. Multi-platform discovery of haplotype-resolved structural variation in human genomes. Nat Commun 2019;10(1):1784-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao X, Collins RL, Lee WP, et al. Expectations and blind spots for structural variation detection from long-read assemblies and short-read genome sequencing technologies. Am J Hum Genet 2021;108(5):919-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mahmoud M, Gobet N, Cruz-Dávalos DI, Mounier N, Dessimoz C, Sedlazeck FJ. Structural variant calling: the long and the short of it. Genome Biol 2019;20(1):246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cao X, Zhang Y, Payer LM, et al. Polymorphic mobile element insertions contribute to gene expression and alternative splicing in human tissues. Genome Biol 2020;21(1):185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cummings BB, Karczewski KJ, Kosmicki JA, et al. Transcript expression-aware annotation improves rare variant interpretation. Nature 2020;581(7809):452-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murdock DR, Dai H, Burrage LC, et al. Transcriptome-directed analysis for Mendelian disease diagnosis overcomes limitations of conventional genomic testing. J Clin Invest 2021;131(1):e141500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qashqari H, Ramani A, Gonorazky H, et al. Child neurology: RNA sequencing for the diagnosis of lissencephaly. Neurology 2021;97(12):e1253–e1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yépez VA, Gusic M, Kopajtich R, et al. Clinical implementation of RNA sequencing for Mendelian disease diagnostics. Genome Med 2022;14(1):38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jaitovich Groisman I, Hurlimann T, Shoham A, Godard B. Practices and views of neurologists regarding the use of whole-genome sequencing in clinical settings: a web-based survey. Eur J Hum Genet 2017;25(7):801-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Turro E, Astle WJ, Megy K, et al. Whole-genome sequencing of patients with rare diseases in a national health system. Nature 2020;583(7814):96-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Halldorsson BV, Eggertsson HP, Moore KHS, et al. The sequences of 150,119 genomes in the UK biobank. bioRxiv. Preprint posted online March 01, 2022. doi: 10.1101/2021.11.16.468246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stranneheim HL, Lagerstedt-Robinson K, Magnusson M, et al. Integration of whole genome sequencing into a healthcare setting: high diagnostic rates across multiple clinical entities in 3219 rare disease patients. Genome Med 2021;13(1):1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stark Z, Dolman L, Manolio TA, et al. Integrating genomics into healthcare: a global responsibility. Am J Hum Genet 2019;104(1):13-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.The 100000 genomes project. Genomics England. Accessed March 26, 2022. genomicsengland.co.uk/about-genomics-england/the-100000-genomes-project/. [Google Scholar]
- 55.Karczewski KJ, Francioli LC, Tiao G, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020;581(7809):434-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilfert AB, Turner TN, Murali SC, et al. Recent ultra-rare inherited variants implicate new autism candidate risk genes. Nat Genet 2021;53:1125-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Minikel EV, Karczewski KJ, Martin HC, et al. Evaluating drug targets through human loss-of-function genetic variation. Nature 2020;581(7809):459-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whiffin N, Armean IM, Kleinman A, et al. The effect of LRRK2 loss-of-function variants in humans. Nat Med 2020;26(6):869-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zou D, Wang L, Liao J, et al. Genome sequencing of 320 Chinese children with epilepsy: a clinical and molecular study. Brain 2021;144(12):3623-3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Krestel H, Meier JC. RNA editing and retrotransposons in neurology. Front Mol Neurosci 2018;11:163. [DOI] [PMC free article] [PubMed] [Google Scholar]