This cohort study aims to establish the clinical efficacy of plasma tumor tissue–modified human papillomavirus DNA testing in the diagnosis and surveillance of human papillomavirus–associated oropharyngeal squamous cell carcinoma in a contemporary clinical setting.
Key Points
Question
What is the accuracy of tumor tissue–modified human papillomavirus DNA (TTMV-HPV DNA) liquid biopsy in the diagnosis and surveillance of HPV-associated oropharyngeal squamous cell carcinoma (OPSCC)?
Findings
In this cohort study of 399 patients in a contemporary clinical setting, TTMV-HPV DNA testing at the time of OPSCC diagnosis demonstrated excellent sensitivity and specificity of 91.5% and 100%, respectively. With respect to surveillance, the TTMV-HPV DNA test had a sensitivity of 88.4% and specificity of 100% for detecting a recurrence.
Meaning
These findings support the use of TTMV-HPV DNA testing as an adjunct biomarker for diagnosis and surveillance of HPV-associated OPSCC.
Abstract
Importance
There is growing interest in the use of circulating plasma tumor human papillomavirus (HPV) DNA for diagnosis and surveillance of patients with HPV-associated oropharyngeal squamous cell carcinoma (OPSCC). Recent advances in the assays, combining the identification of circulating HPV tumor DNA and tumor DNA fragment analysis (tumor tissue–modified viral [TTMV]-HPV DNA), have been shown to be highly accurate. However, use of these newer techniques has been limited to small cohort studies and clinical trials.
Objective
To establish the clinical efficacy of plasma TTMV-HPV DNA testing in the diagnosis and surveillance of HPV-associated OPSCC in a contemporary clinical setting.
Design, Setting, and Participants
This retrospective observational cohort study included patients with OPSCC who underwent TTMV-HPV DNA testing between April 2020 and September 2022 during the course of routine clinical care. For the diagnosis cohort, patients with at least 1 TTMV-HPV DNA measurement prior to initiation of primary therapy were included. Patients were included in the surveillance cohort if they had at least 1 TTMV-HPV DNA test performed after completion of definitive or salvage therapy.
Main Outcomes and Measures
Per-test performance metrics, including sensitivity, specificity, positive predictive value, and negative predictive value, for TTMV-HPV DNA testing.
Results
Of 399 patients included in the analysis, 163 were in the diagnostic cohort (median [IQR] age, 63 [56-68.5] years; 142 [87.1%] male), and 290 were in the surveillance cohort (median [IQR] age, 63 [57-70] years; 237 [81.7%] male). Of the 163 patients in the diagnostic cohort, 152 (93.3%) had HPV-associated OPSCC while 11 (6.7%) had HPV-negative OPSCC. The TTMV-HPV DNA sensitivity in pretreatment diagnosis was 91.5% (95% CI, 85.8%-95.4% [139 of 152 tests]), and the specificity was 100% (95% CI, 71.5%-100% [11 of 11 tests]). In the surveillance cohort, 591 tests conducted in 290 patients were evaluated. A total of 23 patients had molecularly confirmed pathologic recurrences. The TTMV-HPV DNA test demonstrated sensitivity of 88.4% (95% CI, 74.9%-96.1% [38 of 43 tests]) and specificity of 100% (95% CI, 99.3%-100% [548 of 548 tests]) in detecting the recurrences. Positive predictive value was 100% (95% CI, 90.7%-100% [38 of 38 tests]), and negative predictive value was 99.1% (95% CI, 97.9%-99.7% [548 of 553 tests]). The median (range) lead time from positive TTMV-HPV DNA test to pathologic confirmation was 47 (0-507) days.
Conclusions and Relevance
This cohort study demonstrated that when evaluated in a clinical setting, the TTMV-HPV DNA assay demonstrated 100% specificity in both diagnosis and surveillance. However, the sensitivity was 91.5% for the diagnosis cohort and 88.4% for the surveillance cohort, signifying that nearly 1 in 10 negative tests among patients with HPV-associated OPSCC was a false negative. Additional research is required to validate the assay’s performance and, if validated, then further research into the implementation of this assay into standard clinical practice guidelines will be required.
Introduction
The rising incidence of oropharyngeal squamous cell carcinoma (OPSCC) is being driven by an increase in the rate of high-risk human papillomavirus (HPV)-associated disease.1,2 In fact, 80% to 90% of all newly diagnosed oropharyngeal cancers are HPV associated.3 It is now well established that this virally mediated cancer represents a distinct clinical entity with an improved prognosis and treatment response profile when compared with its non-HPV–associated cancer counterparts.4,5 These unique tumor characteristics have led to several changes in practice patterns and multiple treatment protocols to de-intensify therapy.6,7 However, in spite of the favorable prognosis, 15% to 25% of patients will relapse with locoregional or distant metastatic disease after standard therapy.3,4,5
While the epidemiological landscape and treatment paradigms are changing, the methods for the diagnosis of primary and recurrent HPV-associated OPSCC remains relatively stagnant. Currently, the diagnosis and treatment of HPV-associated OPSCC is reliant on a tissue-based diagnosis via primary-site biopsy or fine needle aspiration (FNA)/biopsy of a regional lymph node.8,9 The biopsies are informed by clinical examinations, including fiberoptic laryngoscopy and imaging such as computed tomography or positron emission tomographic (PET) scan. However, these methods of detection are not without limitations, including diagnostic accuracy, patient comfort, and most importantly, time.10
Within the past several years, there has been growing interest in the possibility of using circulating cell-free tumor DNA (ctDNA), also known as a liquid biopsy, in the diagnosis and surveillance of HPV-associated OPSCC.11 While there are several HPV biomarker assays actively being studied, a commercially available blood test evaluating tumor tissue–modified viral (TTMV)-HPV DNA has shown considerable promise. This assay uses a distinct method that identifies and quantifies a tumor-associated or tumor-modified pattern of DNA fragments that greatly increases assay specificity for identifying an HPV-associated malignant tumor. However, evaluation of this assay has been limited to small cohort studies and clinical trials.12,13 The aim of this cohort study was to characterize the diagnostic test characteristics of plasma TTMV-HPV DNA testing for the diagnosis and surveillance of HPV-associated OPSCC in a contemporary clinical setting.
Methods
Patient Cohorts
In this retrospective cohort study, we identified patients with a pathologically confirmed diagnosis of OPSCC who underwent TTMV-HPV DNA testing between April 2020 and September 2022 in the Mount Sinai Health System in New York City. Patients were subclassified into 3 cohorts based on their phase of treatment when TTMV-HPV DNA testing was performed. The diagnosis cohort was defined as any patient who underwent TTMV-HPV DNA testing prior to the initiation of therapy. The in-treatment cohort included patients who underwent TTMV-HPV DNA testing during definitive or salvage treatment or within 3 months of the conclusion of therapy. Finally, the surveillance cohort included patients whose TTMV-HPV DNA testing was performed at least 3 months following completion of definitive or salvage therapy, as well as those with pathologically confirmed recurrence who were awaiting initiation of salvage therapy (Figure). For the purposes of this study, we excluded tests performed during the in-treatment phase of care and focused on tests performed during diagnosis and surveillance. Additionally, we excluded individuals who had clinical suggestion of recurrence who were either lost to follow-up or were awaiting confirmatory tissue biopsy. Patients who completed primary therapy or salvage therapy with evidence of persistent disease were also excluded from the cohort because they were considered to be in treatment.
Figure. Cohort Allocation Schematic.
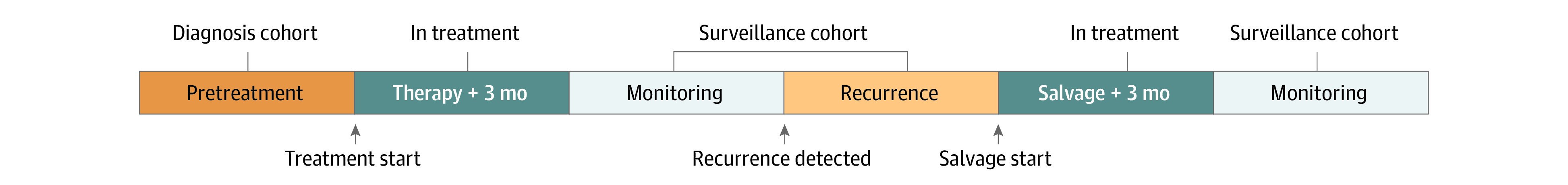
This research was approved by the institutional review board of the Icahn School of Medicine at Mount Sinai, which also granted a waiver of authorization owing to the use of retrospective, deidentified data. The study is reported in accordance with the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) reporting guidelines.
TTMV-HPV Testing
The TTMV-HPV DNA testing was performed using NavDx (Naveris) during the course of routine clinical care at the discretion of the treating health care professional (members of the head and neck surgical oncology, radiation oncology, or medical oncology teams). The TTMV-HPV DNA testing typically coincided with the timing of clinical surveillance examinations. Peripheral blood was collected according to manufacturer guidelines and sent to the Naveris clinical laboratory for sample processing. Using this commercially available blood test, TTMV-HPV DNA can be measured from 5 high-risk HPV subtypes (16, 18, 31, 33, and 35), with excellent limits of detection as previously described.14 The Naveris laboratory uses proprietary methods to distinguish tumor-modified HPV DNA from HPV DNA resulting from an infection.
Study Variables and Outcomes
Patient demographics, tumor clinical/pathologic information, and treatment information were manually extracted from patient medical records (Table 1). The primary outcomes of interest for this study were the per-test performance metrics, including sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV), for TTMV-HPV DNA testing. Secondary outcomes included per-patient performance metrics, as well as lead time detection of recurrence.
Table 1. Patient and Tumor Characteristics for Both Diagnostic and Surveillance Cohorts.
| Characteristic | No. (%) | |
|---|---|---|
| Diagnosis cohort (n = 163) | Surveillance cohort (n = 290) | |
| Age, median (IQR), y | 63 (56-68.5) | 63 (57-70) |
| Sex | ||
| Female | 21 (12.9) | 53 (18.3) |
| Male | 142 (87.1) | 237 (81.7) |
| Follow-up, median (IQR), mo | NA | 40.5 (17-67.5) |
| HPV-associated OPSCC | 152 (93.3) | 290 (100) |
| p16 positive | 150 (98.7) | 277 (95.5) |
| HPV PCR positive | 114 (75) | 246 (84.8) |
| HPV ISH positive | 16 (10.5) | 14 (4.8) |
| AJCC 8th edition tumor classification | ||
| T0 | 12 (7.5) | 14 (4.8) |
| T1 | 37 (23.1) | 107 (36.9) |
| T2 | 72 (44.4) | 121 (41.7) |
| T3 | 25 (15.4) | 27 (9.3) |
| T4 | 15 (9.3) | 21 (7.2) |
| AJCC 8th edition node classification | ||
| N0 | 19 (11.6) | 39 (13.4) |
| N1 | 104 (63.8) | 196 (67.5) |
| N2 | 35 (21.4) | 51 (17.6) |
| N3 | 4 (2.5) | 4 (1.4) |
| Treatment | ||
| Surgery alone | NA | 71 (24.5) |
| Surgery plus adjuvant | NA | 109 (37.6) |
| Radiation alone | NA | 7 (2.4) |
| Chemoradiation | NA | 54 (18.6) |
| Induction chemotherapy plus chemoradiation | NA | 49 (16.9) |
Abbreviations: AJCC, American Joint Committee on Cancer; HPV, human papillomavirus; ISH, in situ hybridization; NA, not applicable; OPSCC, oropharyngeal squamous cell carcinoma; PCR, polymerase chain reaction.
Statistical Analysis
Patient demographic and tumor-specific characteristics were reported descriptively with median (IQR) for continuous variables and numbers (%) for categorical variables. Test metrics were calculated by comparing TTMV-HPV DNA testing with a gold standard of tissue biopsy with confirmatory HPV testing. The TTMV-HPV DNA tests were considered to be positive if the TTMV-HPV DNA score was greater than 5. Previous literature has indicated that TTMV-HPV DNA can be detected in plasma several years before diagnosis; therefore, we defined a true positive test as any positive TTMV-HPV DNA test leading up to a pathologically confirmed recurrence.15 We considered a negative TTMV-HPV DNA test measured within 3 months of a pathologically confirmed recurrence to be a false negative test. The 3-month threshold was selected to coincide with the shortest surveillance interval recommended by the National Comprehensive Cancer Network Guidelines.16 Recurrence lead time was calculated as the time between the first positive TTMV-HPV DNA test and the date of pathologically confirmed recurrence. In the primary analysis for the diagnosis and surveillance cohorts, test metrics were calculated on a per-test basis. For the surveillance cohort, 2 subset analyses were also performed: (1) per-patient performance metrics and (2) per-test performance metrics for patients with known pretreatment positive TTMV-HPV DNA testing. In the per-patient analysis, patients with at least 1 positive test during surveillance were considered to have positive results. We performed all analyses using R, version 4.2.2 (R Foundation for Statistical Computing).
Results
Diagnosis Cohort
The diagnosis cohort consisted of 163 patients presenting with a new diagnosis of OPSCC. The median age was 63 years, and 142 patients (87.1%) in the cohort were male. The majority of patients had oropharyngeal cancers that were HPV associated (n = 152 [93.3%]); however, 11 (6.7%) were not associated with HPV. Among the 152 patients with HPV-associated OPSCC, HPV status was confirmed by p16 staining in 150 patients (98.7%), HPV polymerase chain reaction (PCR) molecular testing in 114 (75%), HPV in situ hybridization in 16 (10.5%), or a combination of methods (Table 1). Of the 152 patients with positive test results for HPV-associated OPSCC, 130 (85.5% [114 by HPV PCR test and 16 by HPV in situ hybridization]) had molecularly confirmed HPV-associated OPSCC.
The per-test sensitivity and specificity for the diagnosis cohort was 91.5% (95% CI, 85.8%-95.4% [139 of 152 tests]) and 100% (95% CI, 71.5%-100% [11 of 11 tests]), respectively (Table 2). The PPV and NPV are not reported for this cohort because the sample has a higher prevalence of HPV-associated OPSCC than the rates cited in the literature, which can bias the true diagnostic PPV and NPV.
Table 2. Per-Test Performance Metrics for the Diagnosis and Surveillance Cohorts.
| TTMV-HPV DNA test result | Diagnosis cohort (n = 163) | Surveillance cohort (n = 591) | Predictive value (95% CI), % | ||
|---|---|---|---|---|---|
| HPV-associated OPSCC | HPV-negative OPSCC | Recurrence | Disease free | ||
| Positive, No. | 139 | 0 | 38 | 0 | 100 (90.7-100)a |
| Negative, No. | 13 | 11 | 5 | 548 | 99.1 (97.9-99.7)b |
| Sensitivity (95% CI), % | 91.5 (85.8-95.4) | NA | 88.4 (74.9-96.1) | NA | NA |
| Specificity (95% CI), % | NA | 100 (71.5-100) | NA | 100 (99.3-100) | NA |
Abbreviations: NA, not applicable; HPV, human papillomavirus; OPSCC, oropharyngeal squamous cell carcinoma; TTMV-HPV DNA, tumor tissue–modified human papillomavirus DNA.
Positive predictive value.
Negative predictive value.
Surveillance Cohort
The surveillance cohort contained 591 tests performed in 290 patients with HPV-associated OPSCC. The median age was 63 years, and the cohort was predominantly male (n = 237 [81.7%]). Median (IQR) follow-up for this cohort was 40.5 (17-67.5) months (Table 1). Of the 290 patients, 162 (55.9%) had 2 or more tests performed during various points in the surveillance period (eFigure 1 in Supplement 1). There were 23 pathologically confirmed recurrences detected in this patient cohort.
On a per-test basis, TTMV-HPV DNA testing demonstrated a sensitivity of 88.4% (95% CI, 74.9%-96.1% [38 of 43 tests]), specificity of 100% (95% CI, 99.3%-100% [548 of 548 tests]), PPV of 100% (95% CI, 90.7%-100% [38 of 38 tests]), and NPV of 99.1% (95% CI, 97.9%-99.7% [548 of 553 tests]) (Table 2). A subset analysis was then performed looking at performance metrics at the patient level and noted sensitivity, specificity, PPV, and NPV of 81.8% (95% CI, 59.7%-94.8% [18 of 22 tests]), 100% (95% CI, 98.6%-100% [268 of 268 tests]), 100% (95% CI, 81.5%-100% [18 of 18 tests]), and 98.5% (95% CI, 96.3%-99.6% [268 to 272 tests]), respectively. Finally, another subset analysis was performed that included the subgroup of patients in the surveillance cohort who had known positive pretreatment TTMV-HPV DNA tests. This corresponded to 51 patients with 115 TTMV-HPV DNA tests. The per-test sensitivity, specificity, PPV, and NPV for this subcohort was 84.6% (95% CI, 54.6%-98.1% [11 of 13 tests]), 100% (95% CI, 96.4%-100% [102 of 102 tests]), 100% (95% CI, 71.5%-100% [11 of 11 tests]), and 98.1% (95% CI, 93.2%-99.8% [102 of 104 tests]), respectively.
For the 13 patients in whom TTMV-HPV DNA testing was the first marker of an eventual pathologically confirmed recurrence, the median (range) lead time was 47 (0-507) days. One patient with a 507-day lead time initially demonstrated avid lung lesions on PET scan and detectable TTMV-HPV DNA approximately 1 year after completion of definitive therapy. Two separate biopsies performed around that time were indeterminate/nondiagnostic due to cellular necrosis. The patient was monitored with serial imaging studies that continued to demonstrate avid lesions on PET scan and a progressive rise in TTMV-HPV DNA levels. A repeat biopsy performed more than 1.5 years after the patient’s initial biopsy confirmed recurrence in the lungs (eFigure 2 in Supplement 1).
Discussion
In this cohort study, we report the diagnostic test characteristics of a commercially available plasma-based TTMV-HPV DNA assay in the detection of HPV-associated OPSCC in the diagnostic setting. Additionally, we report on the clinical utility of this test for surveillance of recurrence using a cohort from a high-volume tertiary care center. We observed that the circulating tumor biomarker demonstrates excellent sensitivity of 91.5% (95% CI, 85.8%-95.4% [139 of 152 tests]) and specificity of 100% (95% CI, 71.5%-100% [11 of 11 tests]) during the diagnostic phase of testing. Additionally, TTMV-HPV DNA testing exhibited a sensitivity of 88.4% (95% CI, 74.9%-96.1% [38 of 43 tests]), specificity of 100% (95% CI, 99.3%-100% [548 of 548 tests]), PPV of 100% (95% CI, 90.7%-100% [38 of 38 tests]), and NPV of 99.1% (95% CI, 97.9%-99.7% [548 of 553 tests]) in the surveillance cohort. If this were confirmed in a prospective clinical validation study, it would suggest that a positive test is confirmatory of HPV-associated OPSCC. The present results demonstrate a median lead time detection of 47 days, with a maximum lead time detection of 507 days. Because testing was performed in a nonprotocolized fashion during routine clinical practice, we expect that the lead times determined in this cohort may be shorter than expected with testing at regular intervals. Furthermore, the time may be more prolonged in slowly growing tumors with low metabolic activity on PET imaging or small-volume disease that is not apparent on clinical examination. Regardless, the lead time provided by positive assay results may allow a window of opportunity for salvage treatment or for the application of adjuvant systemic therapy.
Liquid biopsies have demonstrated considerable promise in several areas of oncology. In the head and neck, virally mediated cancers, like HPV-associated OPSCC, are uniquely suited for detection via liquid biomarkers. Given that the viral and human genomes are distinct and HPV’s oncogenes are well conserved in tumor tissue, they represent optimal targets for ctDNA assays.17 This unique tumor biology coupled with advances in molecular diagnostic techniques has led to the development of multiple assays to detect HPV ctDNA. The multitude of assays, ranging from institutional assays to proprietary commercial assays, vary in substrate used for analysis (ie, saliva, plasma/blood, urine) and molecular detection technique used (ie, quantitative PCR, droplet digital PCR, next generation sequencing).18,19 Combining these different methods, a recent systematic review and meta-analysis found that in 10 studies, including 457 patients, ctDNA-based assays had a pooled sensitivity of 65% and a specificity of 99% in detecting disease.20 While the specificity is quite good across assays, the sensitivity is more highly variable. As this technology continues to evolve with improved levels of sensitivity and specificity, the applications for diagnosis, measuring treatment response, and surveillance will also expand.11
In the context of diagnosis, the field of head and neck oncology generally relies on tissue biopsy and subsequent staining or molecular testing to confirm a diagnosis of squamous cell carcinoma. Often, the most convenient method for diagnosis is an FNA of a neck lymph node; HPV-associated OPSCC often presents with cystic or necrotic nodes, which pose a technical challenge. In this context, FNA only has a 70% to 80% success rate in differentiating a malignant tumor.8,9,21 Nearly 40% of those diagnostic specimens will then go on to have indeterminate p16 staining.9,21 Furthermore, while p16 is a relatively sensitive and specific surrogate marker for high-risk HPV, it does not always adequately predict response to treatment.22 Specimens obtained from FNA are often unable to undergo molecular testing and, therefore, unable to differentiate HPV genotypes that may be eligible for de-escalated treatment.22,23 This often necessitates further open biopsies via an examination under anesthesia, which in addition to their invasiveness, can also lead to delays in diagnosis.24 Operative biopsy, too, is fraught with potential false negatives in the settings of carcinoma of the unknown primary or submucosal lesions that are not readily apparent on endoscopic examination.25,26 Given the limitations of these techniques, ctDNA-based assays have the potential to aid in securing a molecular diagnosis. In another recent meta-analysis evaluating the use of ctDNA at first diagnosis, the authors identified a pooled sensitivity of 81% (95% CI, 78%-84%) and specificity of 98% (95% CI, 96%-99%).27 Subgroup analysis of testing performed using droplet digital PCR, the technique used in the assay examined in this study, demonstrated a sensitivity of 92% (95% CI, 88%-95%) and specificity of 98% (95% CI, 95%-100%). Additionally, in a more recent prospective case-control study using an institutionally developed ctDNA assay, authors demonstrated 98.4% sensitivity and 98.6% specificity for diagnosing HPV-associated OPSCC.21 Using a commercially available assay, the present results are consistent with these previous studies. While this retrospective cohort study demonstrates excellent sensitivity and specificity for TTMV-HPV DNA testing in the context of diagnosis, further prospective work is needed to help elucidate the potential for TTMV-HPV DNA testing as a stand-alone diagnostic test or adjunctive means of obtaining a genotype-specific diagnosis.
The current rate of recurrence for HPV-associated OPSCC is in the range of 15% to 25%.3,28 These recurrences tend to have a different pattern when compared with environmentally associated oropharyngeal cancers.3 There are more frequent cases of oligometastatic disease, metastases to sites outside of the lung, as well as recurrences at time intervals beyond 2 and even 5 years.3,29 The current National Comprehensive Cancer Network Guidelines treat HPV-associated OPSCC surveillance in an identical fashion as HPV-negative OPSCC, with a PET scan at 3 months posttreatment and subsequent clinical examinations with or without imaging depending on the physician’s clinical suspicion.16 This surveillance regimen has limitations, as the currently accepted methods for surveillance are imperfect. For example, PET scans are known for having an excellent NPV in excess of 90%; however, the PPVs are notoriously lower on the order of 62% to 77% and vary with time from treatment.30,31 The frequent false positives can lead to potentially unnecessary biopsies and surgeries.17,32 The NPV of PET scans, while excellent, also has limitations, particularly in cases of small-volume disease or in tumors with low metabolic activity.33,34 Despite the adequate performance metrics for PET scans, it is only considered standard of care to perform a PET scan at 3 months posttreatment. Beyond that time, clinical head and neck examination with a fiberoptic laryngoscope is largely the mainstay when monitoring for recurrence. Unfortunately, these examinations are typically insufficient for detecting asymptomatic recurrences, especially in irradiated tissues. In fact, in asymptomatic patients, current routine surveillance regimens found recurrences 0.3% to 2% of the time.35,36
In this study, a single positive test appeared to confirm the presence of recurrence because the per-test analysis did not identify any falsely positive results. In contrast, a previous study by Chera et al recommended conducting 2 tests to confirm a recurrence because several patients in the study cohort had transiently positive tests that resolved without intervention.12 Discrepancies in these results may be accounted for by differences in treatment modalities, timing of surveillance testing, and possibly refinements in testing techniques. Patients in the study by Chera et al received de-escalated chemoradiotherapy, whereas a majority of the present patient population underwent surgery as part of their definitive management. Additionally, testing in the study by Chera et al was performed as part of a clinical trial protocol with regularly scheduled testing intervals, whereas blood draws for this study were performed during routine clinical surveillance. Finally, alterations to the TTMV-HPV DNA assay that have refined the limits of detection over the last several years may also account for improvements in the specificity of a single test over time.14,37
At present, there are many uncertainties surrounding the appropriate ways to counsel and manage patients using the results of liquid biopsies. Surveillance testing for TTMV-HPV DNA may be useful in resolving cases of clinically or radiographically indeterminate results. However, with careful consideration and appropriate integration into clinical practice guidelines, TTMV-HPV DNA testing may augment aspects of current diagnostic and surveillance algorithms. There are already several efforts in place to determine if liquid biopsy can be used during treatment to de-escalate therapy safely; however, efforts to determine if it can improve oncologic outcomes through efficiencies in diagnosis of new tumors or treatment of recurrences should also be pursued.
Limitations
While we believe the results of this study are informative for many clinicians who are considering integrating TTMV-HPV DNA into their practices, the findings need to be considered in light of several limitations. First, the diagnostic cohort is subject to considerable ascertainment bias in that the testing was applied to patients with known or suspected OPSCC. While the sensitivity is very good (91.5%), this study is not powered or designed to assess use as a screening tool in the general population. Next, with respect to surveillance, this is a retrospective cohort study wherein the majority of patients do not have a pretreatment TTMV-HPV DNA measurement, and as such, we cannot exclude the possibility that a false negative may be the result of the test being negative at baseline as opposed to below the level of detection. Third, our definition of a false negative recurrence using a 3-month time frame may be overly conservative and may artificially lower the calculated sensitivity of this test by classifying a patient who is truly disease free at that point in time as a false negative. As such, we believe that prospective work with scheduled TTMV-HPV DNA testing alongside standard-of-care imaging/clinical examinations will be critical for further elucidating the true sensitivity and appropriate testing interval for this test. Next, there was no standardized protocol for incorporating testing into the surveillance. Therefore, it is hard to make accurate conclusions about lead time detection, as TTMV-HPV testing may have been prompted in some cases by clinical or radiographic findings. Finally, while it is presumed that an earlier detection of disease may lead to improved outcomes, it is beyond the scope of this study to make conclusions about how incorporation of TTMV-HPV DNA testing into practice may affect the overall survival as well as the financial and psychological health of patients.
Conclusions
In this cohort study, we evaluated the performance of TTMV-HPV DNA testing for both diagnosis and surveillance of HPV-associated OPSCC in a contemporary cohort. The TTMV-HPV DNA test demonstrated 100% specificity in both clinical settings. However, the sensitivity was lower for diagnosis (91.5%) and surveillance (88.4%). A positive result appears to be confirmatory of the presence of disease; however, approximately 1 in 10 results in patients with HPV-associated OPSCC may be a false negative; therefore, further workup may be necessary when clinical suggestion of diagnosis or recurrence is high. The present results support the use of TTMV-HPV DNA testing as an adjunct test for diagnosis and surveillance; however, future prospective studies are needed to inform the optimal timing of testing and determine how results should inform treatment decisions.
eFigure 1. TTMV-HPV DNA tests performed per patient (A) and timing (B) of surveillance testing for Surveillance cohort
eFigure 2. Clinical course of a patient with positive TTMV-HPV DNA score preceding a positive biopsy
Data Sharing Statement
References
- 1.Lechner M, Liu J, Masterson L, Fenton TR. HPV-associated oropharyngeal cancer: epidemiology, molecular biology and clinical management. Nat Rev Clin Oncol. 2022;19(5):306-327. doi: 10.1038/s41571-022-00603-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chaturvedi AK, Engels EA, Pfeiffer RM, et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol. 2011;29(32):4294-4301. doi: 10.1200/JCO.2011.36.4596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo T, Kang SY, Cohen EEW. Current perspectives on recurrent HPV-mediated oropharyngeal cancer. Front Oncol. 2022;12:966899. doi: 10.3389/fonc.2022.966899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fakhry C, Westra WH, Li S, et al. Improved survival of patients with human papillomavirus–positive head and neck squamous cell carcinoma in a prospective clinical trial. J Natl Cancer Inst. 2008;100(4):261-269. doi: 10.1093/jnci/djn011 [DOI] [PubMed] [Google Scholar]
- 5.Ang KK, Harris J, Wheeler R, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363(1):24-35. doi: 10.1056/NEJMoa0912217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mensour EA, Alam S, Mawani S, et al. What is the future of treatment de-escalation for HPV-positive oropharyngeal cancer? a review of ongoing clinical trials. Front Oncol. 2022;12:1067321. doi: 10.3389/fonc.2022.1067321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenberg AJ, Vokes EE. Optimizing treatment de-escalation in head and neck cancer: current and future perspectives. Oncologist. 2021;26(1):40-48. doi: 10.1634/theoncologist.2020-0303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rollo F, Dona’ MG, Pellini R, et al. Cytology and direct human papillomavirus testing on fine needle aspirates from cervical lymph node metastases of patients with oropharyngeal squamous cell carcinoma or occult primary. Cytopathology. 2018;29(5):449-454. doi: 10.1111/cyt.12581 [DOI] [PubMed] [Google Scholar]
- 9.Xu B, Ghossein R, Lane J, Lin O, Katabi N. The utility of p16 immunostaining in fine needle aspiration in p16-positive head and neck squamous cell carcinoma. Hum Pathol. 2016;54:193-200. doi: 10.1016/j.humpath.2016.04.002 [DOI] [PubMed] [Google Scholar]
- 10.El-Salem F, Mansour M, Gitman M, et al. Real-time PCR HPV genotyping in fine needle aspirations of metastatic head and neck squamous cell carcinoma: exposing the limitations of conventional p16 immunostaining. Oral Oncol. 2019;90:74-79. doi: 10.1016/j.oraloncology.2019.02.006 [DOI] [PubMed] [Google Scholar]
- 11.Haring CT, Dermody SM, Yalamanchi P, et al. The future of circulating tumor DNA as a biomarker in HPV related oropharyngeal squamous cell carcinoma. Oral Oncol. 2022;126:105776. doi: 10.1016/j.oraloncology.2022.105776 [DOI] [PubMed] [Google Scholar]
- 12.Chera BS, Kumar S, Shen C, et al. Plasma circulating tumor HPV DNA for the surveillance of cancer recurrence in HPV-associated oropharyngeal cancer. J Clin Oncol. 2020;38(10):1050-1058. doi: 10.1200/JCO.19.02444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chera BS, Kumar S, Beaty BT, et al. Rapid clearance profile of plasma circulating tumor HPV type 16 DNA during chemoradiotherapy correlates with disease control in HPV-associated oropharyngeal cancer. Clin Cancer Res. 2019;25(15):4682-4690. doi: 10.1158/1078-0432.CCR-19-0211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gunning A, Kumar S, Williams CK, et al. Analytical validation of NavDx, a cfDNA-based fragmentomic profiling assay for HPV-driven cancers. Diagnostics (Basel). 2023;13(4):725. doi: 10.3390/diagnostics13040725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rettig EM, Faden DL, Sandhu S, et al. Detection of circulating tumor human papillomavirus DNA before diagnosis of HPV-positive head and neck cancer. Int J Cancer. 2022;151(7):1081-1085. doi: 10.1002/ijc.33996 [DOI] [PubMed] [Google Scholar]
- 16.Pfister DG, Spencer S, Adelstein D, et al. Head and Neck Cancers, Version 2.2020, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2020;18(7):873-898. doi: 10.6004/jnccn.2020.0031 [DOI] [PubMed] [Google Scholar]
- 17.Rutkowski TW, Mazurek AM, Śnietura M, et al. Circulating HPV16 DNA may complement imaging assessment of early treatment efficacy in patients with HPV-positive oropharyngeal cancer. J Transl Med. 2020;18(1):167. doi: 10.1186/s12967-020-02330-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie DX, Kut C, Quon H, Seiwert TY, D’Souza G, Fakhry C. Clinical uncertainties of circulating tumor DNA in human papillomavirus–related oropharyngeal squamous cell carcinoma in the absence of National Comprehensive Cancer Network Guidelines. J Clin Oncol. 2023;41(14):2483-2487. doi: 10.1200/JCO.22.00264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adilbay D, Lele S, Pang J, Asarkar A, Calligas J, Nathan CA. Circulating human papillomavirus DNA in head and neck squamous cell carcinoma: possible applications and future directions. Cancers (Basel). 2022;14(23):5946. doi: 10.3390/cancers14235946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Campo F, Zocchi J, Moretto S, et al. Cell-free human papillomavirus-DNA for monitoring treatment response of head and neck squamous cell carcinoma: systematic review and meta-analysis. Laryngoscope. 2022;132(3):560-568. doi: 10.1002/lary.29739 [DOI] [PubMed] [Google Scholar]
- 21.Siravegna G, O’Boyle CJ, Varmeh S, et al. Cell-free HPV DNA provides an accurate and rapid diagnosis of HPV-associated head and neck cancer. Clin Cancer Res. 2022;28(4):719-727. doi: 10.1158/1078-0432.CCR-21-3151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mehanna H, Taberna M, von Buchwald C, et al. ; HNCIG-EPIC group . Prognostic implications of p16 and HPV discordance in oropharyngeal cancer (HNCIG-EPIC-OPC): a multicentre, multinational, individual patient data analysis. Lancet Oncol. 2023;24(3):239-251. doi: 10.1016/S1470-2045(23)00013-X [DOI] [PubMed] [Google Scholar]
- 23.Jalaly JB, Hosseini SM, Shafique K, Baloch ZW. Current status of p16 immunohistochemistry and HPV testing in fine needle aspiration specimens of the head and neck. Acta Cytol. 2020;64(1-2):30-39. doi: 10.1159/000496158 [DOI] [PubMed] [Google Scholar]
- 24.Faden DL. Liquid biopsy for the diagnosis of HPV-associated head and neck cancer. Cancer Cytopathol. 2022;130(1):12-15. doi: 10.1002/cncy.22497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mehta V, Johnson P, Tassler A, et al. A new paradigm for the diagnosis and management of unknown primary tumors of the head and neck: a role for transoral robotic surgery. Laryngoscope. 2013;123(1):146-151. doi: 10.1002/lary.23562 [DOI] [PubMed] [Google Scholar]
- 26.Maghami E, Ismaila N, Alvarez A, et al. Diagnosis and management of squamous cell carcinoma of unknown primary in the head and neck: ASCO guideline. J Clin Oncol. 2020;38(22):2570-2596. doi: 10.1200/JCO.20.00275 [DOI] [PubMed] [Google Scholar]
- 27.Wuerdemann N, Jain R, Adams A, et al. Cell-free HPV-DNA as a biomarker for oropharyngeal squamous cell carcinoma—a step towards personalized medicine? Cancers (Basel). 2020;12(10):2997. doi: 10.3390/cancers12102997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin BM, Wang H, D’Souza G, et al. Long-term prognosis and risk factors among patients with HPV-associated oropharyngeal squamous cell carcinoma. Cancer. 2013;119(19):3462-3471. doi: 10.1002/cncr.28250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang L, Freeman T, Sun S, et al. The use of plasma circulating tumor DNA for early detection of oligometastatic disease in HPV positive oropharyngeal squamous cell carcinoma. Oral Oncol. 2023;139:106357. doi: 10.1016/j.oraloncology.2023.106357 [DOI] [PubMed] [Google Scholar]
- 30.Abgral R, Querellou S, Potard G, et al. Does 18F-FDG PET/CT improve the detection of posttreatment recurrence of head and neck squamous cell carcinoma in patients negative for disease on clinical follow-up? J Nucl Med. 2009;50(1):24-29. doi: 10.2967/jnumed.108.055806 [DOI] [PubMed] [Google Scholar]
- 31.Risør LM, Loft A, Berthelsen AK, et al. FDG-PET/CT in the surveillance of head and neck cancer following radiotherapy. Eur Arch Otorhinolaryngol. 2020;277(2):539-547. doi: 10.1007/s00405-019-05684-2 [DOI] [PubMed] [Google Scholar]
- 32.Lee JY, Garcia-Murillas I, Cutts RJ, et al. Predicting response to radical (chemo)radiotherapy with circulating HPV DNA in locally advanced head and neck squamous carcinoma. Br J Cancer. 2017;117(6):876-883. doi: 10.1038/bjc.2017.258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Erdi YE. Limits of tumor detectability in nuclear medicine and PET. Mol Imaging Radionucl Ther. 2012;21(1):23-28. doi: 10.4274/Mirt.138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eyassu E, Young M. Nuclear Medicine PET/CT Head and Neck Cancer Assessment, Protocols, And Interpretation. StatPearls Publishing LLC; 2023. [PubMed] [Google Scholar]
- 35.Boysen M, Lövdal O, Tausjö J, Winther F. The value of follow-up in patients treated for squamous cell carcinoma of the head and neck. Eur J Cancer. 1992;28(2-3):426-430. doi: 10.1016/S0959-8049(05)80068-1 [DOI] [PubMed] [Google Scholar]
- 36.Kothari P, Trinidade A, Hewitt RJD, Singh A, O’Flynn P. The follow-up of patients with head and neck cancer: an analysis of 1,039 patients. Eur Arch Otorhinolaryngol. 2011;268(8):1191-1200. doi: 10.1007/s00405-010-1461-2 [DOI] [PubMed] [Google Scholar]
- 37.Berger BM, Hanna GJ, Posner MR, et al. Detection of occult recurrence using circulating tumor tissue modified viral HPV DNA among patients treated for HPV-driven oropharyngeal carcinoma. Clin Cancer Res. 2022;28(19):4292-4301. doi: 10.1158/1078-0432.CCR-22-0562 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eFigure 1. TTMV-HPV DNA tests performed per patient (A) and timing (B) of surveillance testing for Surveillance cohort
eFigure 2. Clinical course of a patient with positive TTMV-HPV DNA score preceding a positive biopsy
Data Sharing Statement


