Abstract
Objective: Long non-coding RNAs (lncRNAs) may have a significant regulatory effect on the progression of hepatocellular carcinoma (HCC), according to recent data. This study aims to investigate how SNHG20, a small nucleolar RNA host gene, contributes to the development of HCC. Methods: LncRNA SNHG20, miR-5095, and MBD1 gene levels were determined using reverse transcription qPCR (RT-qPCR). Huh-7 and HepG2 cell bioactivities were evaluated using the CCK-8 kit, EdU, flow cytometry, and wound-healing migration tests. To assess the metastasis of Huh-7 and HepG2 cells, a transwell assay was used. The amounts of invasion- and proliferation-associated proteins were determined using western blot. Using the miRDB (www.mirdb.org) software, the possible target genes of lncRNA and miRNA were predicted, and this prediction was then verified by a twofold luciferase reporter test. To determine the pathologic alteration and Ki67 level in tumor tissues, H&E staining and IHC were employed. TUNEL was conducted to assess the presence of apoptotic bodies in the tumor tissues. Results: lncRNA SNHG20 exhibited a high expression in HCC cells (P<0.01). LncRNA SNHG20 knockdown inhibited HCC cell metastasis (P<0.01) and accelerated apoptosis (P<0.01). LncRNA SNHG20 acted as a sponge of miR-5095 in HCC. In addition, miR-5095 overexpression inhibited HCC cell metastasis (P<0.01) and accelerated apoptosis (P<0.01); and miR-5095 negatively targeted MBD1. Furthermore, LncRNA SNHG20 regulated HCC progression through the miR-5095/MBD1 axis, and LncRNA SNHG20 knockdown inhibited HCC growth. Conclusion: LncRNA SNHG20 accelerates HCC progression by the miR-5095/MBD1 axis, indicating lncRNA SNHG20 can be used as a biomarker for patients with HCC.
Keywords: LncRNA SNHG20, hepatocellular carcinoma, MiR-5095, MBD1
Introduction
Hepatocellular carcinoma (HCC) is one of the most prevalent malignant tumors worldwide and a frequent cause of cancer-related deaths [1]. Even though screening and therapy have made significant advancements, the morbidity and mortality of HCC is unfortunately increasing [2].
Since non-coding genes account for most of the total DNA, and carcinogenic mutations occur mostly in non-coding genomes, non-coding genes are the main inducers of tumorigenesis. Long non-coding RNAs (lncRNAs) may be crucial in the formation of tumors, according to previous research [3,4]. Therefore, the regulation of non-standard non-coding transcription may affect the tumor phenotype [5]. LncRNAs, as intracellular regulatory factors, have functional activities in tumor proliferation and metastasis [6]. Many biologic processes are affected by lncRNAs because they are involved in miRNA-mediated gene regulation, acting as miRNA sponges [7,8]. To date, the functions of a variety of lncRNAs have been elucidated in HCC. Huang et al. reported that as a growth regulator, lncRNA taurine up-regulated 1 (TUG1) regulated the progression of HCC through epigenetic silencing of KLF transcription factor 2 (KLF2) [9]. Furthermore, according to Liang et al., the release of HCC exosomes was encouraged by the lncRNA HOX transcript antisense RNA (HOTAIR) [10]. However, the fundamental mechanism of lncRNA short nucleolar RNA host gene 20 (SNHG20) in regulating HCC is unknown, despite reports of its involvement in laryngeal squamous cell carcinoma and gastric cancer [11].
A non-coding short RNA with an evolutionary length of 18 to 20 bp is known as a microRNA (miRNA) [12]. miRNAs control post-transcriptional translation, has and have been linked to the development of several illnesses [13,14]. According to studies, miR-5095 participates in the regulation of the bioactivity of esophageal cancer cells by acting as a sponge for circRAD23B [15]. According to Zhao et al., miR-5095 affects how responsive cervical cancer cells are to radiation by modulating the pliable nature of lncRNA LINC00958 [16]. Through suppressing the expression of miR-5095, LINC01296’s ectopic expression encourages the growth and migration of non-small cell lung cancer cells [17]. Based on previous research, we speculate that miR-5095 is regulated by the sponging of LncRNA SNHG20. However, a correlation between miR-5095 and LncRNA SNHG20 has not been investigated.
DNA methylation is a crucial method of controlling cancer epigenetics, and DNA methylation is strongly tied to MBD1 (methyl-CpG binding domain protein 1) [18]. At this point, several miRNAs can regulate the function of MBD. According to Liu et al., miR-184 targets MBD1 to regulate the bioactivity of neural stem cells [19]. Furthermore, in cervical cancer, MBD1 is targeted by miR-4429 to accelerate cell proliferation [20]. We speculate that MBD1 may be regulated by miR-5095 to play a role in the HCC development based on a prior study. Namely, MBD1 may be regulated by miR-5095 and involved in HCC progression. However, this has not yet been investigated.
Therefore, further exploring the pathogenesis of HCC will be helpful to the demand for novel treatment strategies, which has important practical significance. This investigation attempts to determine whether the miR-5095/MBD1 axis is involved in HCC development, aiming to find a marker for the evaluation and outlook of HCC.
Materials and methods
Cell lines and culture
HCC cells, including Huh-7, MHCC97-H, HepG2, and SMMC-7721 cells were provided by Procell Life Technologies Co., Ltd., Wuhan, China. Dulbecco’s modified Eagle’s medium (Roche, Basel, Switzerland, Roche), 10% fetal bovine serum (Basel, Switzerland, Roche), and 1% penicillin-streptomycin solution (Beijing, China, Solarbio) were employed to cultivate cells in a humid culture tank with 5% carbon dioxide at 37°C.
Hepatoblastoma, eventually discovered to be the source of the HepG2 cell line, was once believed to be a HCC cell line. The HepG2 cell line has been the subject of various investigations, despite being thought of as hepatoblastoma, according to the most recent study [21,22]. The source of the cell was verified using STR, and the STR certificate is included in the supplemental information.
Cell transfection
The miR-5095 mimics (5’-UUACAGGCGUGAACCACCGCG-3’), miR-5095 inhibitor (5’-CGCGGTGGTTCACGCCTGTAA-3’), pGL3-LncRNA SNHG20 (sh-SNHG20, 5’-GCCACUCACAAGAGUGUAUTT-3’), pGL3-MBD1 (sh-MBD1, 5’-CCGGGAACAGAGAATGTTTAA-3’), and its negative controls NC mimics (5’-ACUCUAUCUGCACGCUGACUU-3’), NC inhibitor (5’-CAGUACUUUUGUGUAGUACAA-3’) and pGL3-NC (sh-NC, 5’-GGATACGGAGTACTATAGC-3’) were synthesized by Biotechnology Co., Ltd., Beijing, China. The transfection doses were 2 μg/well for Huh-7 and HepG2 cells in 6-well plates. All transfections were carried out using the transfection agent LiposomeTM 3000 (Kusatsu Takayama, Japan). Huh-7 and HepG2 cells that had been transfected 48 hours earlier were employed for the following tests.
Reverse transcription qPCR (RT-qPCR)
Total RNA was extracted from HCC cell lines using TRIZOL reagents (Takara, Kusatsu, Japan), and RNA concentrations were calculated at a wavelength of 260 nm. The cDNA was produced using the M-MLV Reverse Transcriptase (RNase H) Kit (TaKaRa, Ohtsu, Japan). RT-qPCR kit (Thermo Fisher Scientific) was used to specifically measure the levels of the MBD1 and SNHG20 LncRNA genes [23]. The expression of miR-5095 was determined using the Taqman microRNA assay kit (Applied Biosystems; Thermo Fisher Scientific, Inc.). GAPDH was used as the endogenous control for LncRNA SNHG20 and MBD1, while U6 served as the internal reference for miR-5095 in the data analysis. The 2-ΔΔCt method was employed to identify fold changes. The entire process was performed in triplicate. Table 1 lists the primers used.
Table 1.
Primer sequences
| Primer name | Primer sequences |
|---|---|
| F-LncRNA SNHG20 | 5’-ATGGCTATAAATAGATACACGC-3’ |
| R-LncRNA SNHG20 | 5’-GGTACAAACAGGGAGGGA-3’ |
| F-miR-5095 | 5’-AACGAGACGACGACAGAC-3’ |
| R-miR-5095 | 5’-TACAGGCGTGAAC-CACC-3’ |
| F-MBD1 | 5’-CTGCATCTGCGTCTTCACAT-3’ |
| R-MBD1 | 5’-CACACCCCACAGTCCTCTTT-3’ |
| F-U6 | 5’-GCTCGCTTCGGCAGCACA-3’ |
| R-U6 | 5’-GAGGTATTCGCACCAGAGGA-3’ |
| F-GAPDH | 5’-GAGTCAACGGATTTGGTCGT-3’ |
| R-GAPDH | 5’-TTGATTTTGGAGGGATCTCG-3’ |
Noted: MBD1: Methyl-CpG-Binding Domain Protein 1. LncRNA: long non-coding RNA. GAPDH: Glyceraldehyde-3-phosphate dehydrogenase.
Western blot
Using a cell lysis solution that comprises 25 mM Tris-HCl, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, and 0.1% sodium dodecyl sulfate, the total protein was extracted from samples of HCC cells (Beyotime, Nanjing, China). All of the antibodies adopted in this work were from Abcam (Cambridge, UK, 1:1000), and they were utilized to measure the proliferative proteins PCNA (ab29) and Ki-67 (ab270650), and the apoptosis-related proteins Bax (ab32503), Bcl-2 (ab32124), cleaved-caspase-3 (ab32042), cleaved-caspase-9 (ab2324), with β-actin (ab8226) as internal reference. Band intensities acquired by ImageJ software (Inc., California, USA) for western blot quantifications were adjusted to β-actin [24].
Cell counting kit-8 (CCK-8) assay
The Beyotime Cell Count Kit was used to assess the viability of Huh-7 and HepG2 cells (Beyotime, Nanjing, China). Huh-7 and HepG2 cells were specifically grown on a 96-well plate. At 0 h, 24 h, 48 h and 72 h, a solution of 10 μL CCK-8 was introduced to each well. Subsequently, a fresh 96-well plate was filled with the medium and the combined CCK-8 solution. A fluorescent microplate reader (Sigma, St. Louis, Missouri, USA) was used to measure the absorbance at 450 nm.
5-ethynyl-20-deoxyuridine (EdU) assay
Huh-7 and HepG2 cells were transfected with the respective plasmids and injected into a 24-well plate after 48 hours. Thereafter, the prepared EDU medium (Abcam, Cambridge, UK, soluble in water to 10 mM and in DMSO to 100 mM) was added to the plate. After two hours of incubation, the culture was removed, and the cells were trypsinized and washed twice with 1×PBS (pH=7.4). Huh-7 and HepG2 cells were fixed with formaldehyde for 30 minutes, after which they were dyed with glycine, washed twice with 1×PBS, left in 0.5% Triton X-100 for 10 minutes, and again washed twice with 1×PBS. EdU staining was performed according to the operating instructions of the Cell Light TMEDU Cell Proliferation Assay (Sigma, St. Louis, Missouri, USA). The stained samples were visualized using an inverted fluorescence microscope (Olympus, Tokyo, Japan).
Flow cytometry analysis
Cell apoptosis was analyzed using flow cytometry. Initially, matching plasmids were transfected into 1×105 Huh-7 or HepG2 cells, and the cells were then grown in a 12-well plate with serum-free DMEM for 24 hours. After that, Huh-7 or HepG2 (5 L/well) were treated with Annexin V-FITC and Propylene Iodide (PI), and incubated at 37°C for 2 hours. With BD-FACSAriaTM Fusion, the number of apoptotic cells was counted (BD Biosciences, New York, USA). The apoptosis rate was calculated as Q4 (early apoptosis) + Q2 (late apoptosis). To analyze the data, ModiFit software 5.0 (Olympus, Tokyo, Japan) was used.
Wound-healing migration assay
The wound-healing assay was conducted in both Huh-7 and HepG2 cells. A 24-well culture plate received Huh-7 and HepG2 cells, respectively. The relevant plasmid was transfected once the cell population had expanded to 80% of its original size. After 24 hours of transfection, a wound gap was created by gently scratching a single layer of cells was crossed with the tip of a 200 μL. The migratory potential of Huh-7 and HepG2 cells was evaluated by observing the cells on culture plates (0-48 h) under an inverted microscope (Olympus, Tokyo, Japan).
Transwell assay
Cell invasion and migration were assessed in the Transwell chamber with 8 μm diameter membrane holes. The Transwell chamber of migration does not require matrigel coating, in contrast to the invasion experiment. In the top compartment, Huh-7 or HepG2 cells were implanted in a medium devoid of serum. A culture containing 20% fetal bovine serum was put into the bottom chamber at the same time. After 24 hours of culture, the cells remaining on the top chamber were removed with a cotton brush, and the cells in the bottom chamber were fixed with 4% polyformaldehyde and 0.1% crystalline amethyst staining. The stained cells were counted in five randomly selected fields under an inverted microscope to assess the invasion ability of the cells.
Bioinformatic and luciferase reporter assay
The miRDB database (www.mirdb.org) was used to forecast potential target genes. Fluorinase vectors (pGL3 Luciferase Reporter Vectors), obtained from Sangon Biotech, included wild-type or mutant miR-5095 binding regions of wild-type or mutant lncRNA SNHG20 and MBD1 (Shanghai, China). On a 6-well plate, 500 ng of wild-type or mutant LncRNA SNHG20 and MBD1 plasmids were injected into each well. According to the directions, liposomes TM3000 were applied to transfect Huh-7 or HepG2 cells with miR-5095 mimics (100 nM) and NC mimics (100 nM) (Takara, Kusatsu, Japan). A twofold photochemical luminescence reporting gene analysis system (Foster City Applied Biosystics, USA) was employed to measure luciferase activity and normalize it using Renira luciferase activity.
Tumorigenesis assays
We purchased four-week-old female BALB/c mice from Zhejiang Vital River Laboratory Animal Technology. All animal studies were carried out following the protocol that has been authorized by the Animal Laboratory Center’s Ethics Committee at Hangzhou Medical College (YANSHEN 2020-220). For the subcutaneously implanted tumor experiment, groups of BALB/c nude mice (7 mice per group) were subcutaneously injected with 1×106 Huh-7 cells transfected with either sh-LncRNA SNHG20 or mock vectors obtained from Gene Pharmaceutical Co., Ltd., (Shanghai, China). The cells were suspended in 100 μL PBS before injection. The nude mice were then acquired and fed in the Zhejiang University Animal Center. Once every week, the tumor’s development was observed. After the mice were euthanized by intraperitoneal injection of sodium pentobarbital overdose (260 mg/kg body weight) after 21 days, the tumors were promptly excised, and tumor weight and volume were examined. Finally, the tumors were preserved and stained for histologic studies using H&E staining and immunohistochemistry (IHC) [25,26]. According to the guidelines, H&E staining and IHC were carried out to assess the pathologic alterations and the levels of Ki67 in tumor samples.
TUNEL assay
Beyotime (Shanghai, China) provided colorimetric TUNEL apoptosis assay kits, which were utilized to assess the presence of apoptotic small bodies in HCC tissues [27]. The dewaxed tissue slices from the xenograft were subjected to incubation with 20 μg/mL proteinase K without DNase at room temperature for 15 min. After three washes with PBS, the samples were blocked with a blocking solution (Beyotime, Shanghai, China) at room temperature for 20 min. Following three washes with PBS, 50 μL of biotin labeling solution was added. Then the sample was incubated at room temperature for 1 h in the dark. The stained sections were then incubated with 50 μL of streptavidin HRP working solution and stop reaction solution for 10 min at room temperature. A light microscope (Olympus Microscope) was utilized to obtain the images, and the percentage of TUNEL-positive cells (brown) was computed.
Statistical analysis
The results from three independent experiments were statistically evaluated using GraphPad Prism version 5.0 software (GraphPad Software, Inc.). The data were presented as mean ± standard deviation. Two independent groups were compared using an unpaired t-test. One-way repeated measures ANOVA or two-way repeated measures ANOVA with Tukey’s post-test was employed to examine data among several groups or time points. A difference was statistically significant when P<0.05.
Results
LncRNA SNHG20 expressed highly in HCC
LncRNA SNHG20 has been found to be present in various HCC cell lines, including Huh-7, MHCC97-H, HepG2, and SMMC-7721. It was revealed that lncRNA SNHG20 was strongly expressed in HCC cells (P<0.01) in contrast with the control, particularly in Huh-7 and HepG2 cells (Figure 1). As a result, Huh-7 and HepG2 cells were chosen for further functional research.
Figure 1.
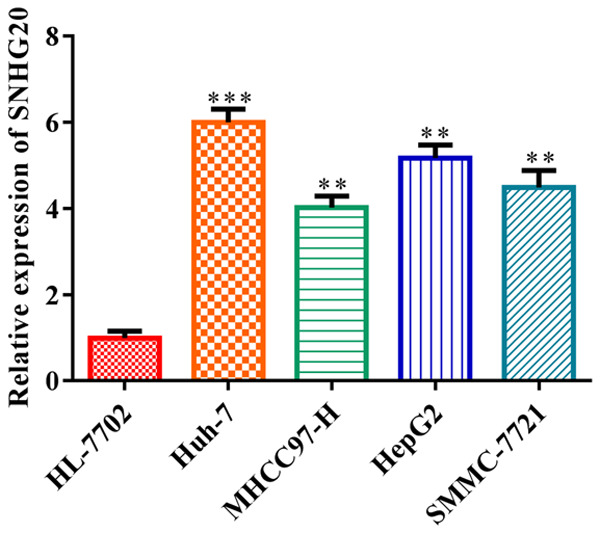
LncRNA SNHG20 was highly expressed in HCC cells. RT-qPCR was conducted to evaluate the expressions of LncRNA SNHG20 in HCC cell lines, including Huh-7, MHCC97-H, HepG2 and SMMC-7721. Data indicate the mean of three independent experiments. Error bars represent SD. In contrast to the HL-7702 group, **P<0.01, ***P<0.001. LncRNA: long non-coding RNA.
LncRNA SNHG20 knockdown inhibited HCC cell proliferation but accelerated apoptosis
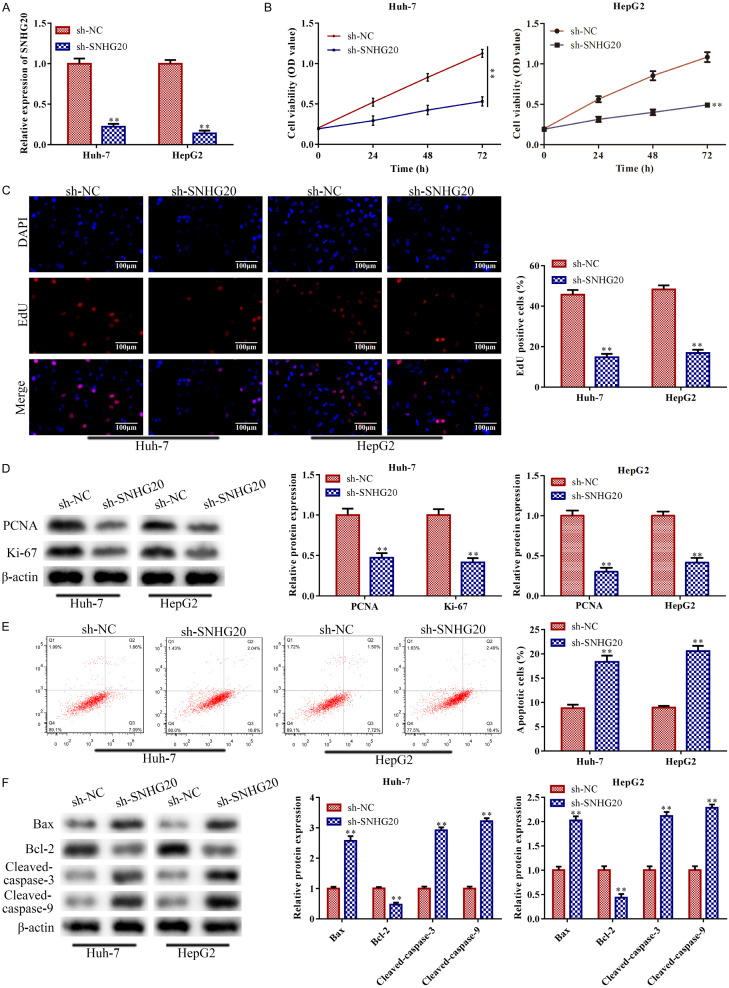
The expression of LncRNA SNHG20 was suppressed in Huh-7 and HepG2 cells in comparison to the control, according to RT-qPCR analysis (P<0.01; Figure 2A). Cell viability of Huh-7 and HepG2 was decreased by LncRNA SNHG20 knockdown, according to CCK-8 assay analysis (P<0.01; Figure 2B). LncRNA SNHG20 knockdown consistently suppressed the levels of proteins related to cell proliferation, such as PCNA and Ki-67 in the EdU experiment (P<0.01; Figure 2C). The results were also supported by western blot. This was the case for Huh-7 and HepG2 cell proliferation (P<0.01; Figure 2D). Subsequently, LncRNA SNHG20 knockdown accelerated Huh-7 and HepG2 cell death in comparison to the control (P<0.01; Figure 2E). This was also supported by western blot. LncRNA SNHG20 knockdown boosted pro-apoptotic protein expression and lowered Bcl-2 levels, indicating that it accelerated apoptosis (P<0.01; Figure 2F).
Figure 2.
LncRNA SNHG20 knockdown inhibited HCC cell proliferation while accelerating apoptosis. Huh-7 and HepG2 cells were transfected with sh-LncRNA SNHG20, with sh-NC acting as the negative control, in order to knock down LncRNA SNHG20. A: To measure the knockdown efficiency of LncRNA SNHG20, RT-qPCR was adopted. B: Cell viability of Huh-7 and HepG2 cells was determined using CCK-8 assay. C, D: EdU assay (magnification, ×200) and western blot were conducted to measure the proliferation ability of Huh-7 and HepG2 cells. E, F: FCMs assay and western blot were conducted to evaluate the apoptosis of Huh-7 and HepG2 cells. Data are the average of three independent experiments. The error bars show SD. In contrast with the sh-NC group, **P<0.01. sh-NC was used as the negative control. LncRNA: long non-coding RNA.
LncRNA SNHG20 knockdown inhibited HCC cell metastasis
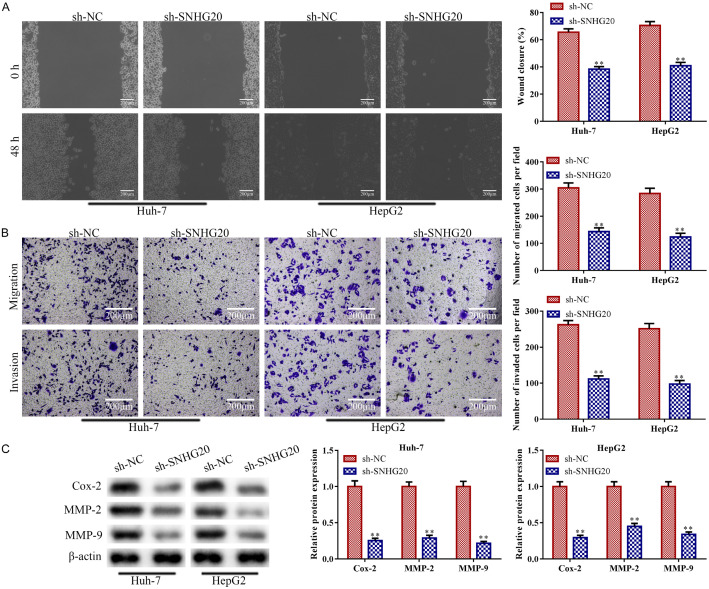
The migration and invasion ability of Huh-7 and HepG2 cells were decreased by LncRNA SNHG20 knockdown in contrast with those in the control group (P<0.01; Figure 3A and 3B). In addition, western blot analysis revealed that LncRNA SNHG20 knockdown significantly reduced the expressions of proteins related to the invasion ability (Cox-2, MMP2, and MMP9) (P<0.01; Figure 3C), which are essential for angiogenesis and tumor spread [28].
Figure 3.
LncRNA SNHG20 knockdown prevented HCC cells from migrating and invading. Huh-7 and HepG2 cells were transfected with sh-LncRNA SNHG20, with sh-NC acting as the negative control, to knock down lncRNA SNHG20. A: Huh-7 and HepG2 cell movement was evaluated using a wound-healing migration assay (magnification, ×200); B: Transwell assay was implemented to quantify the invasion and migration of Huh-7 and HepG2 cells (magnification, ×200); C: Western blotting was applied to ascertain the concentrations of proteins related to the invasion ability, such as Cox-2, MMP-2, and MMP-9. Data are the average of three independent experiments. The error bars show SD. In contrast with the sh-NC group, **P<0.01. sh-NC was used as the negative control. lncRNA: long non-coding RNA.
LncRNA SNHG20 functions as a sponge of miR-5095 in HCC cells
Bioinformatic research revealed that LncRNA SNHG20 and miR-5095 may have a common binding site (Figure 4A). When Huh-7 and HepG2 cells co-transfected with wild-type plasmid of LncRNA SNHG20 and miR-5059 mimics, luciferase activity was reduced (P<0.01), but the luciferase activity did not change (P>0.05) in Huh-7 and HepG2 cells transfected with the mutant LncRNA SNHG20-containing plasmid. The double luciferase reporter assay confirmed the prediction (Figure 4B). Moreover, RT-qPCR analysis revealed that HCC cell lines in general (P<0.05), and Huh-7 and HepG2 cells in particular (P<0.01), showed lower levels of miR-5059 than the control HL-7702 cell lines (Figure 4C). Also, the analyses of RT-qPCR suggested that, in Huh-7 and HepG2 cells, sh-LncRNA SNHG20 up-regulated the level of miR-5059 in contrast with the absence of sh-NC (P<0.01; Figure 4D).
Figure 4.
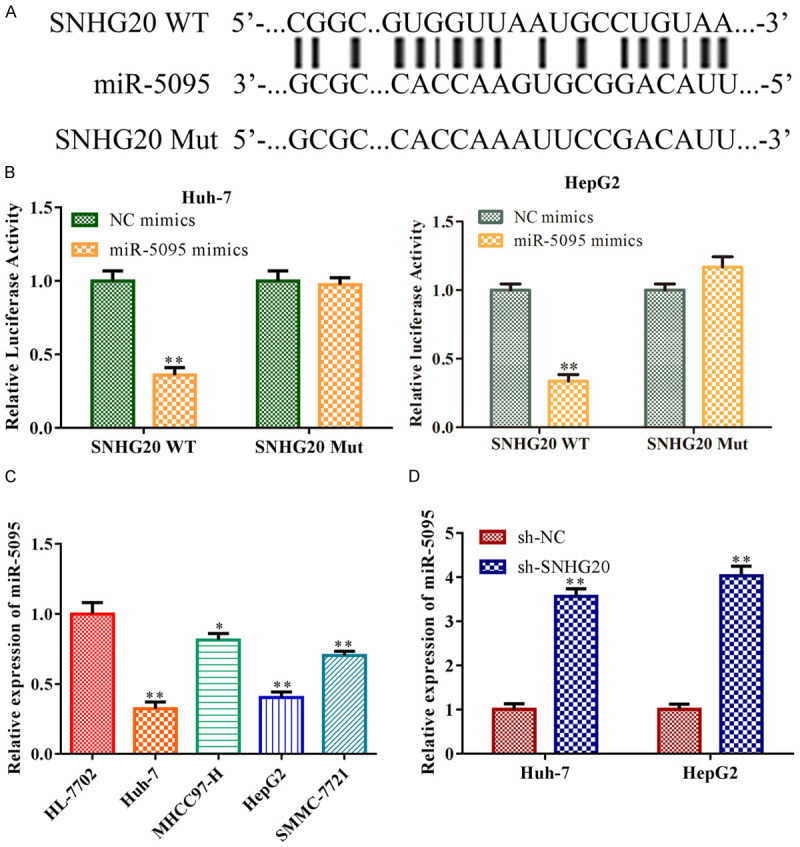
LncRNA SNHG20 functions as a sponge of miR-5095 in HCC cells. A: miRDB program identified the location of the probable binding site between LncRNA SNHG20 and miR-5059. To overexpress miR-5059, Huh-7, and HepG2 cells were transfected with miR-5059 mimics. NC mimics acted as the negative control of miR-5059 mimics. The direct binding connection between LncRNA SNHG20 and miR-5059 was verified. B: The dual luciferase reporter gene experiment; C: The levels of miR-5095 in HCC cell lines such as Huh-7, MHCC97-H, HepG2, and SMMC-7721 were measured by RT-qPCR. Huh-7 and HepG2 cells were transfected with sh-LncRNA SNHG20, with sh-NC acting as the negative control, to knock down LncRNA SNHG20; D: RT-qPCR measured level of miR-5095 in Huh-7 and HepG2 cells. Data are the average of three independent experiments. The error bars show SD. In contrast with the NC group, **P<0.01. sh-NC was used as the negative control. WT: wild type; MUT: mutated. LncRNA: long non-coding RNA.
miR-5095 overexpression inhibited HCC cell metastasis but accelerated apoptosis
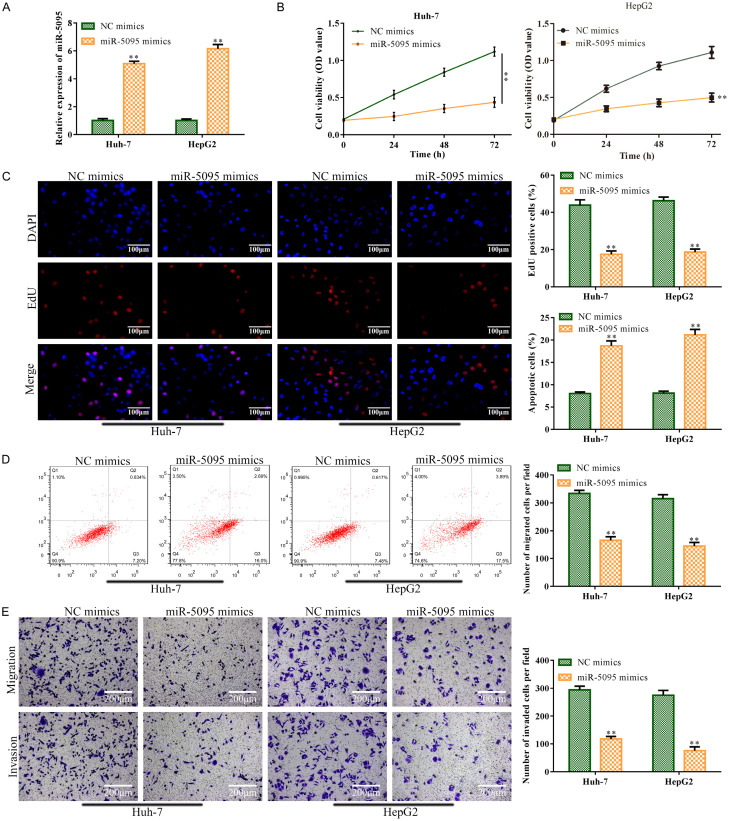
After miR-5095 mimic transfection, RT-qPCR analysis revealed that, in comparison to the control, miR-5095 was substantially expressed in Huh-7 and HepG2 cells (P<0.01; Figure 5A). After that, CCK-8 and EdU assay results revealed that miR-5095 overexpression reduced Huh-7 and HepG2 cell viability and proliferation when compared to the control (P<0.01; Figure 5B and 5C). In contrast, FCMs analysis revealed that miR-5095 overexpression sped up Huh-7 and HepG2 cell death in comparison to the control (P<0.01; Figure 5D). Transwell assay results showed that miR-5095 overexpression reduced the invasion and metastasis of Huh-7 and HepG2 cells when compared to the control (P<0.01; Figure 5E).
Figure 5.
miR-5095 overexpression prevented HCC cells from proliferating, migrating, and invading, while accelerating the apoptosis. Huh-7 and HepG2 cells were infected with miR-5095 mimics to overexpress miR-5095. The NC mimics served as the negative control for miR-5095 mimics. A: miR-5059 concentration was identified using RT-qPCR. B: Cell viability of Huh-7 and HepG2 cells was determined using CCK-8 assay. C: EdU test was carried out to analyze the Huh-7 and HepG2 cell proliferation (magnification, ×200); D: Huh-7 and HepG2 cell apoptosis was tested using the FCMs assay. E: Huh-7 and HepG2 cell migration and invasion were analyzed using Transwell assay (magnification, ×200). Data are the average of three independent experiments. The error bars show SD. In contrast to the NC mimics group, **P<0.01. NC mimics was used as the negative control. WT: wild type; MUT: mutated.
miR-5095 negatively targeted MBD1
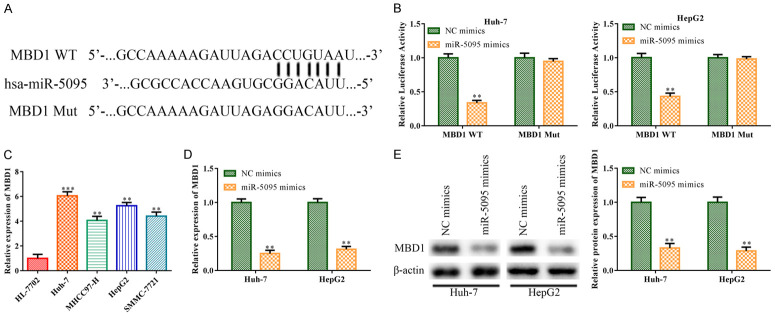
According to a bioinformatic study, there may be a binding site between miR-5095 and MBD1 (Figure 6A), suggesting that the miR-5095 binding sequence was found in the 3’-UTR of MBD1. The physical interaction between MBD1 and miR-5095 was then demonstrated by the luciferase reporter assay. Co-transfection of wild-type MBD1 and miR-5095 mimic plasmid resulted in a significant reduction in luciferase activity in Huh-7 and HepG2 cells (P<0.01). However, this reduction in luciferase activity was not observed (P>0.05) when the putative binding site mutation occurred (Figure 6B). In addition, RT-qPCR findings revealed that MBD1 was substantially expressed in HCC cells, including Huh-7 (P<0.001), MHCC97-H (P<0.01), HepG2 (P<0.01) and SMMC-7721 (P<0.01), particularly in Huh-7 and HepG2 cells (Figure 6C). Besides, RT-qPCR and western blot analyses showed that the expression level of MBD1 was decreased in Huh-7 and HepG2 cells upon transfection with miR-5095 mimics, compared to that in the control NC mimics group (P<0.01; Figure 6D and 6E).
Figure 6.
miR-5095 negatively targeted MBD1. A: miRDB program was able to determine the probable binding location between miR-5095 and MBD1. B: Huh-7 and HepG2 cells were transfected with miR-5095 mimics, with NC mimics acting as a negative control. To validate the direct combining interaction between miR-5095 and MBD1, a dual luciferase reporter gene test was carried out; C: Huh-7, MHCC97-H, HepG2, and SMMC-7721 cells were used as HCC cell lines in RT-qPCR to identify the MBD1 expression, and HL-7702 cells were used as the negative control; D: MBD1 expressions in Huh-7 and HepG2 cells were measured using RT-qPCR in response to miR-5095 overexpression. E: Huh-7 and HepG2 cells that had miR-5095 overexpressed were used to determine the level of MBD1 protein. The data represent the average of three independent experiments. The error bars indicate SD. In contrast to the NC mimics group, **P<0.05, ***P<0.001. NC mimics was used as the negative control. MBD1: Methyl-CpG-Binding Domain Protein 1.
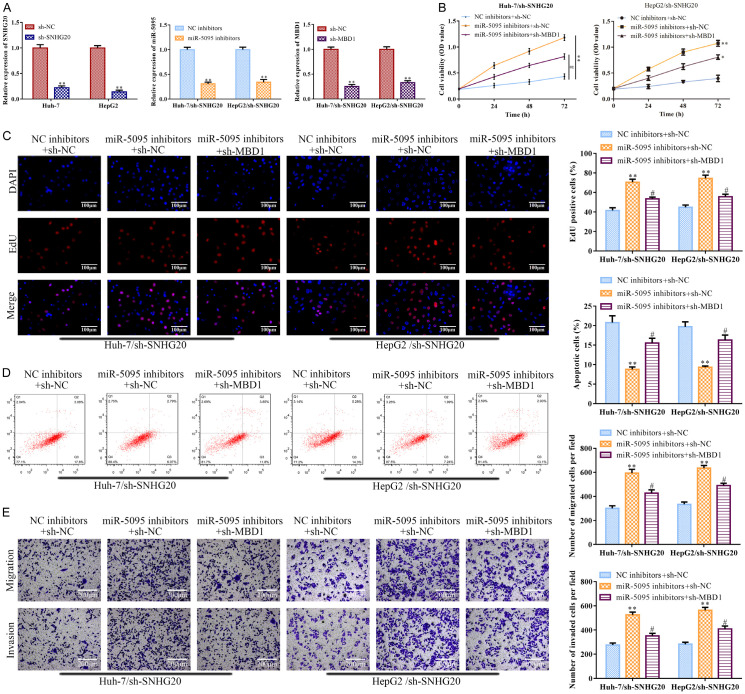
LncRNA SNHG20 regulated HCC progression through the miR-5095/MBD1 axis
LncRNA SNHG20 knockdown increased miR-5095 levels (Figure 4D), while miR-5095 inhibitor antagonized the enhanced effect, according to the RT-qPCR analysis (P<0.01; Figure 7A left). LncRNA SNHG20 knockdown consistently reduced the expression of MBD1, but MBD1 knockdown counteracted this effect (P<0.01; Figure 7A right). CCK-8 and EdU assays revealed that the effects of miR-5095 inhibitors were subsequently attenuated by MBD1 depletion (P<0.05), whereas the impacts of LncRNA SNHG20 knockdown encouraged the decrease in Huh-7 and HepG2 cell viability, proliferation, migration, and invasion (P<0.01; Figure 7B, 7C and 7E). According to the flow cytometry, miR-5095 inhibitor inhibited the apoptosis induced by LncRNA SNHG20 knockdown in Huh-7 and HepG2 cells (P<0.01), but MBD1 knockdown reversed the effects of the miR-5095 inhibitor on LncRNA SNHG20 knockdown in Huh-7 and HepG2 cells (P<0.05; Figure 7D).
Figure 7.
LncRNA SNHG20 regulated HCC progression through the miR-5095/MBD1 axis. In Huh-7 and HepG2 cells, miR-5095 inhibitor or sh-MBD1 was transfected to suppress the LncRNA SNHG20 for 24 hours; the negative control for the miR-5095 inhibitor was an NC inhibitor, while the negative control for sh-MBD1 was sh-NC. A: MBD1 or miR-5095 levels were determined using RT-qPCR; B: Huh-7 and HepG2 cell viability was assessed with CCK-8. C: EdU test was conducted (magnification, ×200) to evaluate the proliferation of Huh-7 and HepG2 cells. D: Huh-7 and HepG2 cell apoptosis was assessed using the FCMs assay. E: Transwell experiment was conducted to evaluate the invasion and migration of Huh-7 and HepG2 cells (magnification, ×200). Data are the average of three independent experiments. The error bars reflect SE. In contrast to the NC inhibitors group, *P<0.05, **P<0.001; in contrast to the miR-505 inhibition group, #P<0.05. sh-NC, NC inhibitors or NC inhibitors + sh-NC was used as the negative control. WT: wild type; MUT: mutated. LncRNA: long non-coding RNA. MBD1: Methyl-CpG-Binding Domain Protein 1.
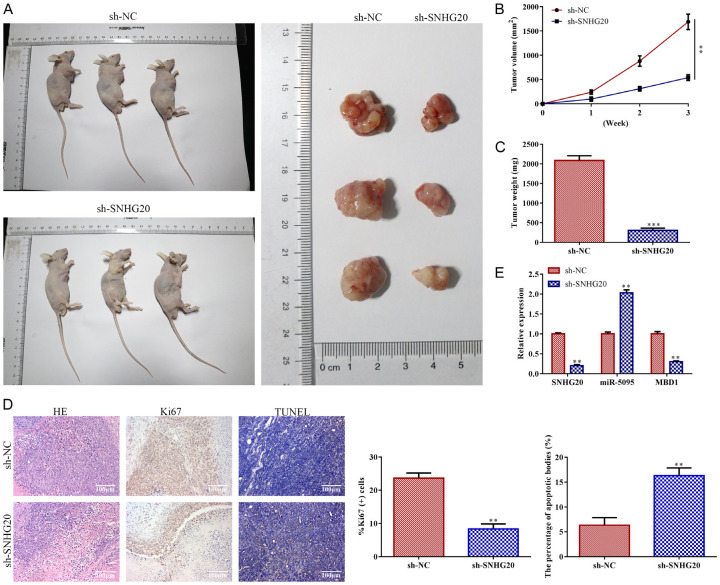
LncRNA SNHG20 knockdown inhibited tumor growth
Primarily, BALB/C nude mice were injected with Huh-7 cells, in which LncRNA SNHG20 was suppressed, for tumor information. According to tumor phenotypic statistics between 0-21 days, LncRNA SNHG20 knockdown reduced the volume of the tumors in comparison to the control (P<0.01; Figure 8A and 8B). The tumor weight in the LncRNA SNHG20 knockdown group was less than that of the control group (P<0.001; Figure 8C). Additionally, LncRNA SNHG20 knockdown dramatically reduced the cell density and level of Ki67 protein expression in tumor tissue. TUNEL analysis revealed that, as compared to controls, LncRNA SNHG20 knockdown expedited the development of apoptotic bodies (P<0.01; Figure 8D). Moreover, RT-qPCR analysis revealed that LncRNA SNHG20 knockdown decreased the levels of LncRNA SNHG20 and MBD1 (P<0.01) and increased the expression level of miR-5095 (P<0.01) when compared to control (P<0.01; Figure 8E).
Figure 8.
LncRNA SNHG20 knockdown inhibited tumor growth. For LncRNA SNHG20 knockdown, Huh-7 cells were transfected with sh-LncRNA SNHG20, with sh-NC acting as the negative control. After that, BALB/C nude mice were injected subcutaneously with LncRNA SNHG20 knockdown-Huh-7 cells to collect tumor information (n=7, 0-21 days). A: Tumor phenotypic statistics (21 days); B: Tumor volume (0-21 days); C: Tumor weight (21 days). D: H&E staining (magnification, ×200), IHC (magnification, ×200), and TUNEL test were carried out to analyze the development of apoptotic bodies as well as the pathologic alterations and levels of Ki67 in tumor tissues (21 days); E: LncRNA SNHG20, miR-5095, and MBD1 levels were measured by RT-qPCR in HCC tumor tissues (magnification, ×200). Data are the average of three independent experiments. The error bars show SE. **P<0.01, ***P<0.001 in contrast to the si-NC group. sh-NC was used as the negative control. H&E: hematoxylin and eosin. LncRNA: long non-coding RNA.
Discussion
IncRNA, a novel class of non-coding RNAs that has a length greater than 200 bp, is involved in molecular processes [29]. Through regulating target gene expression by sponging miRNA, it has been demonstrated that lncRNA plays a significant role in the pathogenesis of various disorders [30]. Concerning cancer types, lncRNA SNHG20 often exhibits aberrant expression in HCC, colorectal cancer, and bladder cancer [31]. Functional research in vitro showed that LncRNA SNHG20 knockdown prevented HCC cell metastasis while speeding up the apoptosis. These results suggested that lncRNA SNHG20 might serve as a therapeutic target in HCC.
According to previous research, RNA interference-based LncRNA SNHG20 knockdown prevented GC cells from proliferation and invasion by suppressing the production of miR-495-3p [11]. Similar to how LncRNA SNHG20 acted as a sponge for miR-5095, RT-qPCR revealed that the level of miR-5095 was suppressed in HCC cells; however, the inhibitory impact was reduced by LncRNA SNHG20 knockdown. Further analysis found that miR-5095 overexpression accelerated the mortality but restrained the expansion of HCC cells. Our findings demonstrated that LncRNA SNHG20 accelerated the growth of HCC by sponging miR-5095.
Research has revealed a strong correlation between MBD1 and the formation and growth of several cancers. According to Liu et al., MBD1 played a role in the epithelial-mesenchymal transition of gallbladder cancer and was substantially expressed in gallbladder tumors [32]. Our findings indicated that MBD1 was substantially expressed in HCC, indicating a promoting function of MBD1 in the advancement of HCC. Moreover, miRNAs target the gene MBD1 to contribute to play a role in regulating tumor growth. By targeting MBD1, for instance, miR-4429 has been shown to accelerate the development of cervical cancer [20]. The findings in this paper demonstrated that miR-5095 negatively targeted MBD1, and additional investigations have shown that the miR-5095/MBD1 axis, facilitated by LncRNA SNHG20, promotes HCC metastasis and inhibits apoptosis.
However, our current understanding of the functional mechanisms of lncRNA SNHG20 in HCC remains limited. This study also has certain limitations that should be acknowledged. For instance, it is unclear whetherlncRNA SNHG20 would affect HCC through common downstream signaling pathways. Another intriguing aspect that remains to be explored is whether lncRNA SNHG20 plays a role in modulating the immunological microenvironment of HCC.
Our study uncovered several key findings regarding lncRNA SNHG20 in the context of HCC. First, we observed that lncRNA SNHG20 was highly expressed in HCC. Additionally, when lncRNA SNHG20 was suppressed, it led to a reduction in metastasis and an acceleration of apoptosis in HCC cells. Conversely, overexpression of lncRNA SNHG20 resulted in decreased metastasis but enhanced apoptosis. Furthermore, we identified lncRNA SNHG20 as a miR-5095 sponge in HCC cells. Our findings also indicated that lncRNA SNHG20 negatively targeted MBD1. Moreover, the miR-5095/MBD1 axis regulated the development of HCC through lncRNA SNHG20. In conclusion, the miR-5095/MBD1 axis plays a role in promoting the development of HCC through lncRNA SNHG20. Our findings suggest a viable HCC prognostic and therapeutic target.
Acknowledgements
All authors would like to give our sincere gratitude to the reviewers for their constructive comments. This work was supported by the Natural Science Foundation of Zhejiang Province of China (LY19H160049).
Disclosure of conflict of interest
None.
References
- 1.Yu J, Yang M, Zhou B, Luo J, Zhang Z, Zhang W, Yan Z. CircRNA-104718 acts as competing endogenous Rna and promotes hepatocellular carcinoma progression through microRNA-218-5p/TXNDC5 signaling pathway. Clin Sci (Lond) 2019;133:1487–1503. doi: 10.1042/CS20190394. [DOI] [PubMed] [Google Scholar]
- 2.Giordano S, Columbano A. MicroRNAs: new tools for diagnosis, prognosis, and therapy in hepatocellular carcinoma? Hepatology. 2013;57:840–847. doi: 10.1002/hep.26095. [DOI] [PubMed] [Google Scholar]
- 3.Kondo Y, Shinjo K, Katsushima K. Long non-coding RNAs as an epigenetic regulator in human cancers. Cancer Sci. 2017;108:1927–1933. doi: 10.1111/cas.13342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wei JW, Huang K, Yang C, Kang CS. Non-coding RNAs as regulators in epigenetics (review) Oncol Rep. 2017;37:3–9. doi: 10.3892/or.2016.5236. [DOI] [PubMed] [Google Scholar]
- 5.Fang Y, Fullwood MJ. Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genomics Proteomics Bioinformatics. 2016;14:42–54. doi: 10.1016/j.gpb.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339:159–166. doi: 10.1016/j.canlet.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 7.Li Y, Xu J, Guo YN, Yang BB. LncRNA SNHG20 promotes the development of laryngeal squamous cell carcinoma by regulating miR-140. Eur Rev Med Pharmacol Sci. 2019;23:3401–3409. doi: 10.26355/eurrev_201904_17704. [DOI] [PubMed] [Google Scholar]
- 8.Wang XY, Wang L, Xu PC, Huang FJ, Jian X, Wei ZC, Chen YQ. LINC01605 promotes the proliferation of laryngeal squamous cell carcinoma through targeting miR-493-3p. Eur Rev Med Pharmacol Sci. 2019;23:10379–10386. doi: 10.26355/eurrev_201912_19677. [DOI] [PubMed] [Google Scholar]
- 9.Huang MD, Chen WM, Qi FZ, Sun M, Xu TP, Ma P, Shu YQ. Long non-coding RNA TUG1 is up-regulated in hepatocellular carcinoma and promotes cell growth and apoptosis by epigenetically silencing of KLF2. Mol Cancer. 2015;14:165. doi: 10.1186/s12943-015-0431-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang L, Peng X, Li Y, Zhang X, Ma Y, Wu C, Fan Q, Wei S, Li H, Liu J. Long non-coding RNA HOTAIR promotes exosome secretion by regulating RAB35 and SNAP23 in hepatocellular carcinoma. Mol Cancer. 2019;18:78. doi: 10.1186/s12943-019-0990-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cui N, Liu J, Xia H, Xu D. LncRNA SNHG20 contributes to cell proliferation and invasion by upregulating ZFX expression sponging miR-495-3p in gastric cancer. J Cell Biochem. 2019;120:3114–3123. doi: 10.1002/jcb.27539. [DOI] [PubMed] [Google Scholar]
- 12.Shi Y, Fang N, Li Y, Guo Z, Jiang W, He Y, Ma Z, Chen Y. Circular RNA LPAR3 sponges microRNA-198 to facilitate esophageal cancer migration, invasion, and metastasis. Cancer Sci. 2020;111:2824–2836. doi: 10.1111/cas.14511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bandiera S, Pfeffer S, Baumert TF, Zeisel MB. miR-122--a key factor and therapeutic target in liver disease. J Hepatol. 2015;62:448–457. doi: 10.1016/j.jhep.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 14.Elton TS, Selemon H, Elton SM, Parinandi NL. Regulation of the MIR155 host gene in physiological and pathological processes. Gene. 2013;532:1–12. doi: 10.1016/j.gene.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 15.Lan X, Liu X, Sun J, Yuan Q, Li J. CircRAD23B facilitates proliferation and invasion of esophageal cancer cells by sponging miR-5095. Biochem Biophys Res Commun. 2019;516:357–364. doi: 10.1016/j.bbrc.2019.06.044. [DOI] [PubMed] [Google Scholar]
- 16.Zhao H, Zheng GH, Li GC, Xin L, Wang YS, Chen Y, Zheng XM. Long noncoding RNA LINC00958 regulates cell sensitivity to radiotherapy through RRM2 by binding to microRNA-5095 in cervical cancer. J Cell Physiol. 2019;234:23349–23359. doi: 10.1002/jcp.28902. [DOI] [PubMed] [Google Scholar]
- 17.Hu X, Duan L, Liu H, Zhang L. Long noncoding RNA LINC01296 induces non-small cell lung cancer growth and progression through sponging miR-5095. Am J Transl Res. 2019;11:895–903. [PMC free article] [PubMed] [Google Scholar]
- 18.Li L, Chen BF, Chan WY. An epigenetic regulator: methyl-CpG-binding domain protein 1 (MBD1) Int J Mol Sci. 2015;16:5125–5140. doi: 10.3390/ijms16035125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu C, Teng ZQ, Santistevan NJ, Szulwach KE, Guo W, Jin P, Zhao X. Epigenetic regulation of miR-184 by MBD1 governs neural stem cell proliferation and differentiation. Cell Stem Cell. 2010;6:433–444. doi: 10.1016/j.stem.2010.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu D, Huang K, Wang T, Zhang X, Liu W, Yue X, Wu J. NR2F2-AS1 accelerates cell proliferation through regulating miR-4429/MBD1 axis in cervical cancer. Biosci Rep. 2020;40:BSR20194282. doi: 10.1042/BSR20194282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Tian Y, Zhong W, Wang N, Wang Y, Zhang Y, Zhang Z, Li J, Ma F, Zhao Z, Peng Y. Artemisia argyi essential oil inhibits hepatocellular carcinoma metastasis via suppression of DEPDC1 dependent Wnt/beta-catenin signaling pathway. Front Cell Dev Biol. 2021;9:664791. doi: 10.3389/fcell.2021.664791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ren Y, Wang Y, Hao S, Yang Y, Xiong W, Qiu L, Tao J, Tang A. NFE2L3 promotes malignant behavior and EMT of human hepatocellular carcinoma (HepG2) cells via Wnt/Beta-catenin pathway. J Cancer. 2020;11:6939–6949. doi: 10.7150/jca.48100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong SM, Cui JH, Zhang W, Zhang XW, Kou TC, Cai QC, Xu S, You S, Yu DS, Ding L, Lai JH, Li M, Luo KJ. Inhibition of translation initiation factor eIF4A is required for apoptosis mediated by microplitis bicoloratus bracovirus. Arch Insect Biochem Physiol. 2017;96 doi: 10.1002/arch.21423. [DOI] [PubMed] [Google Scholar]
- 24.Cui JH, Dong SM, Chen CX, Xiao W, Cai QC, Zhang LD, He HJ, Zhang W, Zhang XW, Liu T, Ding L, Yang Y, Lai JH, Zhu QS, Luo KJ. Microplitis bicoloratus bracovirus modulates innate immune suppression through the eIF4E-eIF4A axis in the insect spodoptera litura. Dev Comp Immunol. 2019;95:101–107. doi: 10.1016/j.dci.2019.02.010. [DOI] [PubMed] [Google Scholar]
- 25.Ding Y, Wang H, Shen H, Li Z, Geng J, Han H, Cai J, Li X, Kang W, Weng D, Lu Y, Wu D, He L, Yao K. The clinical pathology of severe acute respiratory syndrome (SARS): a report from China. J Pathol. 2003;200:282–289. doi: 10.1002/path.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen X, Du J, Jiang R, Li L. MicroRNA-214 inhibits the proliferation and invasion of lung carcinoma cells by targeting JAK1. Am J Transl Res. 2018;10:1164–1171. [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Yuan J, Guo M, Xiang R, Xie T, Zhuang X, Dai W, Li Q, Lai Q. Upregulation of long intergenic non-coding RNA LINC00326 inhibits non-small cell lung carcinoma progression by blocking Wnt/Beta-catenin pathway through modulating the miR-657/dickkopf WNT signaling pathway inhibitor 2 axis. Biol Direct. 2023;18:3. doi: 10.1186/s13062-023-00359-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu Y, Shi C, Zeng L, Liu G, Jiang W, Zhang X, Chen S, Guo J, Jian X, Ouyang J, Xia J, Kuang C, Fan S, Wu X, Wu Y, Zhou W, Guan Y. High COX-2 expression in cancer-associated fibiroblasts contributes to poor survival and promotes migration and invasiveness in nasopharyngeal carcinoma. Mol Carcinog. 2020;59:265–280. doi: 10.1002/mc.23150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rathinasamy B, Velmurugan BK. Role of lncRNAs in the cancer development and progression and their regulation by various phytochemicals. Biomed Pharmacother. 2018;102:242–248. doi: 10.1016/j.biopha.2018.03.077. [DOI] [PubMed] [Google Scholar]
- 30.Lai XN, Li J, Tang LB, Chen WT, Zhang L, Xiong LX. MiRNAs and LncRNAs: dual roles in TGF-β signaling-regulated metastasis in lung cancer. Int J Mol Sci. 2020;21:1193. doi: 10.3390/ijms21041193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao W, Ma X, Liu L, Chen Q, Liu Z, Zhang Z, Ma S, Wang Z, Li H, Wang Z, Wu J. SNHG20: a vital lncRNA in multiple human cancers. J Cell Physiol. 2019;234:14519–14525. doi: 10.1002/jcp.28143. [DOI] [PubMed] [Google Scholar]
- 32.Wensheng L, Bo Z, Qiangsheng H, Wenyan X, Shunrong J, Jin X, Quanxing N, Xianjun Y, Xiaowu X. MBD1 promotes the malignant behavior of gallbladder cancer cells and induces chemotherapeutic resistance to gemcitabine. Cancer Cell Int. 2019;19:232. doi: 10.1186/s12935-019-0948-1. [DOI] [PMC free article] [PubMed] [Google Scholar]