Abstract
Liver fibrosis is the excessive accumulation of extracellular matrix proteins. Due to the lack of an accurate test for an early diagnosis of liver fibrosis and the invasiveness of the liver biopsy procedure, there is an urgent need for effective non-invasive biomarkers for screening the patients. we aimed to evaluate the diagnostic performance of circulating miRNAs (miR-146b, −194, −214) and their related mechanisms in the pathogenesis of liver fibrosis. The expression levels of miR-146b, −194, and −214 were quantified in whole blood samples from NAFLD patients using real-time PCR. The competing endogenous RNA (ceRNA) network was constructed and a gene set enrichment analysis (GSEA) was performed for HSC activation-related genes. Also, the transcription factor (TF)-miR co-regulatory network and the survival plot for three miRNAs and core genes were illustrated. The qPCR results showed that the relative expression of miR-146b and miR-214 significantly increased in NAFLD patients, while miR-194 showed significant down-regulation. The ceRNA network analysis implicated NEAT1 and XIST as sponge candidates for these miRNAs. The GSEA results identified 15 core genes involved in HSC activation, primarily enriched in NF-κB activation and autophagy pathways. STAT3, TCF3, RELA, and RUNX1 were considered potential transcription factors connected to miRNAs in the TF-miR network. Our study elucidated three candidate circulating miRNAs differentially expressed in NAFLD that could serve as a promising non-invasive diagnostic tool for early detection strategies. Also, NF-κB activation, autophagy, and negative regulation of the apoptotic process are the main potential underlying mechanisms regulated by these miRNAs in liver fibrosis pathogenesis.
Keywords: Circulating miRNA, Non-alcoholic fatty liver disease, Liver fibrosis, Hepatic stellate cell, Non-invasive diagnosis
1. Introduction
Nonalcoholic fatty liver disease (NAFLD) is the most common chronic liver disorder, ranging from steatosis to nonalcoholic steatohepatitis (NASH) with an increased risk of fibrosis, cirrhosis, and hepatocellular carcinoma (HCC) [1,2]. Progressive steatosis may lead to hepatocyte ballooning and inflammation, where pro-inflammatory cytokines increase [3]. Subsequently, the excessive accumulation of extracellular matrix proteins (ECM), mainly collagen fibers, leads to the development of liver fibrosis. Hepatic stellate cells (HSCs) have a crucial role in fibrogenesis, where activated HSCs (aHSCs) constitute a significant source of collagen in the liver and can abundantly secrete ECM proteins and cytokines [4].
microRNAs (miRNAs) are small non-coding RNAs that regulate post-transcriptional gene expression, playing an essential role in various biological processes such as cell proliferation, apoptosis, and differentiation [5,6]. Several studies have shown that miRNAs play a role in NAFLD progression and exhibit dysregulation at different stages of the disease [[7], [8], [9], [10]]. Circulating miRNAs can be detected in the body fluids including blood, plasma, and urine, bound to extracellular vesicles (EVs) or RNA-binding proteins. Due to the stability of circulating miRNAs, they could serve as diagnostic biomarkers for liver diseases [11].
Serum biomarkers, especially ALT (alanine aminotransferase) and AST (aspartate aminotransferase), in addition to serum scoring systems such as NAFLD fibrosis score, Fibrosis-4 (Fib-4) score, and AST-to-platelet ratio index (APRI), have also been used widely for non-invasive screening of liver fibrosis [12]. Nevertheless, they aren't tissue-specific and have limited sensitivity and specificity to diagnose the early stages of fibrosis [[12], [13], [14], [15], [16]]. Although liver biopsy is still the gold standard for NAFLD diagnosis, it has some drawbacks, including sampling error, invasive and expensive procedure with the complications such as pain and bleeding [1,17]. Therefore, liver biopsy prescription is limited because of these disadvantages, and alternative non-invasive methods are needed for disease screening.
In the present study, the association of miR-146b, miR-194, and miR-214 expression levels was investigated with NAFLD severity to use them as reliable non-invasive biomarkers for early diagnosis of patients. Also, we identified the competing endogenous RNA (ceRNA) network by integrating the lncRNA-miRNA-mRNA interactions and performed the functional enrichment analyses for genes related to HSCs activation as core genes. Moreover, the upstream regulatory network of core genes was constructed to determine the potential related mechanisms for regulating gene expression during HSCs activation. Finally, survival analysis of core genes was executed to identify their prognostic value for disease.
2. Patients and methods
2.1. Study groups and subjects
70 NAFLD patients were included in the Gastroenterology and Liver Diseases Clinic, Taleghani General Hospital. Using ultrasound, the level of liver steatosis (defined as >5% of hepatocytes) was determined, and fibroscan was performed to measure liver stiffness. The routine laboratory test was conducted for aspartate transaminase (AST), alanine transaminase (ALT), high-density lipoprotein cholesterol (HDL), low-density lipoprotein cholesterol (LDL), total cholesterol level, and serum triglyceride level (TG) alkaline phosphatase (ALP), fasting blood sugar (FBS), and platelet. Inclusion criteria were set as follows: (1) age over 18 years; (2) no significant alcohol consumption (more than 70 g/week); (3) no evidence of autoimmune disease or viral hepatitis. A subset of patients (N = 34) agreed and was divided into three groups based on disease severity: NASH (fibrosis stage 0; ALT >40, N = 7), Fibrosis (fibrosis stages 1-3, N = 19), and Cirrhosis (fibrosis stage 4, N = 8).
2.2. miRNA expression level measurement
A total of 3 ml whole blood from participants was collected into ethylenediaminetetraacetic acid (EDTA) tubes. Immediately, RNA was purified from 200 μl of blood using Super RNA Extraction Kit (AnaCell, PC1010) according to the manufacturer's instruction manual and stored at −80 °C. The RNA quality and concentration were measured by BioTek™ Epoch™ Microplate Nanodrop. Then, 200 ng of total RNA was reverse-transcribed using miRNA-specific primers (AnaCell, CS0025) and stored in −20 °C. The qPCR reaction was performed by RealQ Plus 2x Master Mix Green NO Rox (Ampliqon, A323402) and Rotor-Gene Q (Qiagen) cycler using the following profile: 95 °C for 10 min; and 40 cycles at 95 °C for 15 s, 59 °C for 30 s, and 72 °C for 15 s. The miRBase database was used to design the sequence of primers shown in Table 1. The miRNAs' relative expression was calculated based on 2-ΔΔCt and normalized with U6 as the reference gene.
Table 1.
Primer sequences of miRNAs.
| Gene | miRBase accession | Primer sequence |
|---|---|---|
| hsa-miR-146b-5p | MIMAT0002809 | Fwd: ACACTATAGCTGGGTGAGAACTGAATTCC |
| hsa-miR-194-5p | MIMAT0000460 | Fwd: ACACTATAGCTGGGTGTAACAGCAACTC |
| hsa-miR-214-3p | MIMAT0000271 | Fwd: ACACTATAGCTGGGACAGCAGGCACAGA |
| U6 | - | Fwd: CTCGCTTCGGCAGCACA |
| Universal reverse: TGGTGTCGTGGAGTCGGCAATTCAGTTG | ||
2.3. ceRNA network construction
To discover interactions between miRNA and target genes (MTIs), various databases was utilized, including miRTarBase [18] and TarBase 8 [19], for experimentally validated interactions, as well as TargetScan [20], miRWalk [21], miRDB [22], and DIANA-microT [19] for predicted MTIs. Each database incorporates different features, prediction models, and data sources such as sequence complementarity, conservation across species, site accessibility, thermodynamics, machine learning-based approach, binding energy, and RNA secondary structure to provide a comprehensive collection of miRNA-target interactions. These elements are employed to generate a comprehensive compilation of miRNA-target interactions. To ensure accuracy, genes that appeared in at least three databases among the predicted miRNA-target gene interactions, in addition to the experimentally validated interactions were selected. To narrow down the number of target genes, the STRING plugin in Cytoscape 3.7.2 was employed to construct protein-protein interactions (PPI) for each miRNA-target genes pair. Subsequently, the cytohubba plugin was used to identify and select the top 15 genes based on their degree. Moreover, DIANA-LncBase v3 [23], miRNet [24], and RNA Interactome [25] were employed to retrieve the lncRNA-miRNA interactions. By combining the miRNA-mRNA and lncRNA-miRNA interactions, the ceRNA network was constructed and visualized using Cytoscape 3.7.2 [26].
2.4. Investigation of the candidate miRNAs’ effect on HSCs activation
The MTIs was limited to the genes that participated in the HSCs activation to better understand the role of selected miRNAs during this process. The COREMINE medical database (https://coremine.com/medical) was utilized to obtain the quarry genes, and functional enrichment analysis was performed based on common target genes of miR-146b/194/214, and those involved in HSCs activation. To this aim, the EnrichR database [27] was used to analyze the gene ontology (GO) and pathway enrichment analyses. The cutoff criterion of adjusted p-value <0.05 was considered statistically significant. The venny online tool was used to find the overlapping genes [28].
2.5. Upstream gene regulatory network analysis
The upstream gene regulatory network (GRN) of HSCs activation core genes was constructed by integrating the transcription factor and kinase enrichment analyses to identify the potential regulatory factors involved in controlling the expression of these core genes during HSC activation. Core genes were selected based on common signaling pathways and biological processes of miR-146b, miR-194, and miR-214. The eXpression2Kinases (X2K) web [29] was utilized to get the GRN data and which was illustrated by Cytoscape. The minimum number of articles supporting protein-protein interaction was considered three. Furthermore, TF-miRNA interactions were constructed using TransmiR v2.0 [30] and RegNetwork [31] to determine the co-regulatory relationships between TFs, miRNAs, and genes.
2.6. Survival analysis
Kaplan Meier-plotter online tool was used to determine the association between the expression levels of core genes and the overall survival time of HCC patients [32]. The hazard ratio (HR) and log-rank p-value were calculated automatically. To determine the optimal cutoff for dividing the input data into high and low expression groups, the data were iterated over variable values ranging from the lower quartile to the upper quartile. Cox regression was performed for each setting, and the most statistically significant cutoff value was selected as the best cutoff [33]. Alcohol consumption and hepatitis virus were set to “none,” and log-rank p-value <0.05 was considered the significance threshold.
2.7. Statistical analysis
Clinical data were analyzed using SPSS 26.0 software and GraphPad Prism 9. The Shapiro-Wilk test was employed to investigate the normality of distributions. miRNAs expression levels were compared by student's t-test and Mann-Whitney U. A one-way analysis of variance (ANOVA) was conducted for comparing more than two groups, followed by post-hoc tests. Receiver Operating Characteristic (ROC) curves were plotted by GraphPad Prism 9 using relative gene expression values to evaluate the diagnostic accuracy of miRNAs, ALT, and AST. Data are presented as mean ± standard error of the mean (SEM), and the p-value <0.05 was considered statistically significant.
3. Results
3.1. Study participants
The characteristics of the subjects are summarized in Table 2. Among the total of 34 patients, the mean age was 46.8 ± 1.9, with 61.7% being male. While there were no significant differences observed between ALT and AST in the patient groups, there was a significant difference in total cholesterol levels (p = 0.03). Additionally, platelet counts were found to be significantly different between the NASH group and the other groups (p = 0.01). Interestingly, the mean ages of NASH, Fibrosis, and Cirrhosis patients were 37.33 (range 33–43), 46.16 (range 23–61), and 55.63 (range 36–69), respectively. These findings suggest a positive correlation between age and disease severity within the population. Furthermore, the results of BMI indicated that most of the patients were classified as overweight rather than obese.
Table 2.
Clinical characteristics of healthy controls and NAFLD patients.
| Control (N = 7) | NASH (N = 7) | Fibrosis (N = 19) | Cirrhosis (N = 8) | P-value | |
|---|---|---|---|---|---|
| Gender (males/females) | 3/2 | 5/2 | 10/9 | 6/2 | - |
| Age (years) | 35.3 ± 3.2 | 37.3 ± 1.8 | 46.2 ± 2.4 | 55.6 ± 3.6 | 0.0008* |
| BMI | 22.2 ± 2.1 | 29.1 ± 1.9 | 31.2 ± 1.1 | 28.3 ± 2.1 | 0.01* |
| TG [mg/dl] | 107.6 ± 2.5 | 201.7 ± 32.24 | 167.0 ± 20.79 | 123.3 ± 20.88 | 0.19 |
| LDL [mg/dl] | 98.4 ± 1.2 | 137.1 ± 17.01 | 107.0 ± 9.42 | 84.29 ± 12.46 | 0.07 |
| HDL [mg/dl] | 61.6 ± 1.2 | 85.86 ± 42.21 | 43.06 ± 2.62 | 36.14 ± 8.17 | 0.17 |
| T.Cholesterol [mg/dl] | 177.6 ± 12.5 | 206.7 ± 21.82 | 173.8 ± 9.82 | 138.0 ± 17.85 | 0.03* |
| FBS [mg/dl] | 86.4 ± 2.3 | 87.71 ± 4.67 | 106.6 ± 7.1 | 97.86 ± 3.70 | 0.18 |
| ALT [U/l] | 27.1 ± 6.3 | 92.50 ± 22.78 | 57.06 ± 11.53 | 43.29 ± 7.96 | 0.08 |
| AST [U/l] | 30.6 ± 2.6 | 41.43 ± 9.71 | 35.67 ± 4.66 | 59.29 ± 13.50 | 0.14 |
| Plt | 251.4 ± 12.3 | 253.7 ± 29.72 | 247.2 ± 13.94 | 153.4 ± 34.57 | 0.01* |
ALT: Alanine Aminotransferase; AST: Aspartate Aminotransferase; BMI: Body Mass Index; FBS: Fast Blood Sugar; HDL: High-Density Lipoprotein; LDL: Low-Density Lipoprotein; PLT: Platelet; TG: Triglycerides. *Significant (p < 0.05).
3.2. Expression of candidate miRNAs in NAFLD patients
The relative expression of miR-146b, miR-194, and miR-214 was carried out by qPCR in NAFLD versus healthy control groups. miR-146b and miR-214 were significantly up-regulated in NAFLD patients, while miR-194 expression level was notably decreased (p < 0.05) (Fig. 1, a). miR-146b relative expression in NASH, fibrosis, and cirrhosis was 5.03 ± 1.36, 6.41 ± 1.07, and 5.68 ± 2.47, respectively (Fig. 1, b). For miR-194, the expression level in NASH, fibrosis, and cirrhosis was 0.29 ± 0.1, 0.37 ± 0.053, and 0.33 ± 0.06 (Fig. 1, c). The relative expression of miR-214 was 63.73 ± 16.69, 106.7 ± 13, and 57.79 ± 4.6 in NASH, fibrosis, and cirrhosis (Fig. 1, d). Moreover, miR-214 expression decreased significantly during progression from fibrosis to cirrhosis (p < 0.05).
Fig. 1.
The expression levels of circulating miRNAs in NAFLD patients. a) The heatmap shows the up-regulation of miR (−146b, −214) and down-regulation of miR-194 in NAFLD patients. b) circulating miR-146b expression level significantly increased in NASH and Fibrosis patients. c-d) circulating miR-194 significantly down-regulated in NASH, Fibrosis, and Cirrhosis patients, while the expression of miR-214 was significantly higher in patients' groups. * p-value <0.05, ** p-value <0.01, *** p-value <0.001, **** p-value <0.0001.
ROC curve analysis was performed for miRNAs relative gene expression, ALT and AST values to assess the diagnostic performance. miR-146b, miR-194, and miR-214 exhibited the highest area under the curves (AUCs) when comparing NAFLD stages to healthy controls. Furthermore, miR-214 demonstrated significant discrimination between Fibrosis and Cirrhosis, with an AUC of 0.88 (Supplementary Fig. 1, a-c). ALT showed significant discrimination between NASH and healthy controls, with an AUC of 0.85, while AST displayed significant distinction between Cirrhosis patients and healthy controls, with an AUC of 0.78 (Supplementary Fig. 1, d-e). However, neither ALT nor AST exhibited significant differences among the different stages of NAFLD in patients.
3.3. ceRNA network for miR-146b, miR-194, and miR-214
The ceRNA network was constructed based on the interactions between lncRNA-miRNA and miRNA-mRNA to determine the potential mechanisms of ceRNAs in NAFLD. Various databases were utilized to obtain the interactions, and Cytoscape was used to visualize the network. Based on the presence in at least two databases, we selected 17, 30, and 15 lncRNAs for miR-146b, miR-194, and miR-214, respectively. Also, we chose 78 MTIs for miR-146b, 229 MTIs for miR-194, and 664 ones for miR-214 (Fig. 2). NEAT1 and XIST were common in all networks and acted as sponges for targeting miRNAs. Moreover, SNHG16 and MALAT1 were in common between miR-146b/214 and miR-146b/194, respectively, whereas MIR17HG and PVT1 overlapped between miR-194/214.
Fig. 2.
The ceRNA networks of lncRNA-miRNA-mRNA interactions. MiR-146b (a), miR-194 (b), and miR-214 (c) are connected to 17, 30, and 15 lncRNAs, respectively. Additionally, 78, 229, and 664 mRNAs were chosen as target genes for miR-146b, miR-194, and miR-214, respectively. The top 15 target genes for each miRNA were selected using the cytohubba plugin. The degree of each gene is represented by the length of the connection line.
3.4. miR-146b, miR-194, and miR-214 potential roles in fibrogenesis
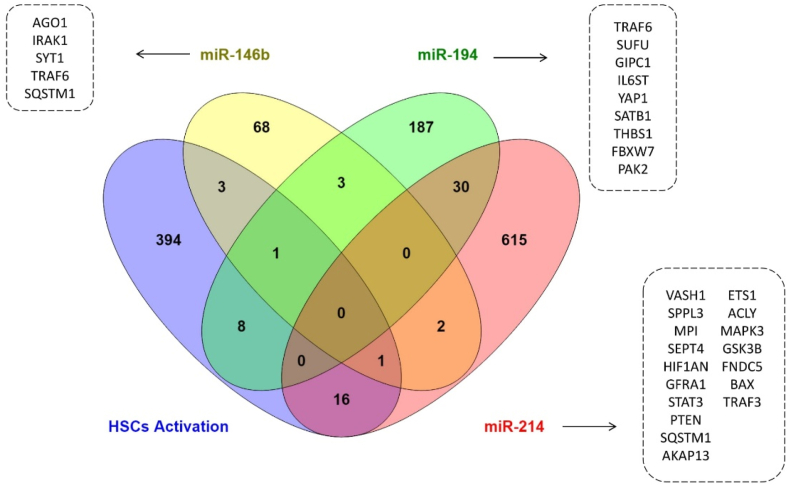
To understand the potential role of these miRNAs in fibrogenesis, functional and pathway enrichment analyses were performed for common genes between MTIs and those related to HSCs activation (Fig. 3). The results revealed that NF-kappaB activation, signaling by interleukins, cell death signaling by NRIF, immune system, senescence, and autophagy signaling pathways are common signaling pathways involved in the activation of HSCs by miR-146b, miR-194, and miR-214 (Fig. 4, Supplementary Table 1). The GO analysis showed that regulation of programmed cell death and apoptotic process, cytokine-mediated signaling pathway, regulation of intracellular signal transduction, mitochondrion organization, and protein ubiquitination are the most important biological processes during fibrogenesis (Fig. 4, Supplementary Table 1). Based on the common signaling pathways and biological processes, GSK3B, FBXW7, STAT3, PTEN, THBS1, TRAF3, SEPT4, AKAP13, PAK2, IRAK1, TRAF6, MAPK3, BAX, IL6ST, and SQSTM1 are core genes which targeted by three miRNAs and play a vital role in HSCs activation.
Fig. 3.
The venn diagram of three miRNAs' target genes and HSC activation-related genes. 5 genes are in common for miR-146b, 9 genes for miR-194, and 17 genes for miR-214.
Fig. 4.
The gene ontology (GO) and pathway enrichment analyses for common MTIs and HSC activation-related genes.
3.5. Upstream regulatory network for HSC activation core genes
To understand the potential regulatory mechanisms besides miRNAs, the regulatory network analysis was conducted based on TF-intermediate gene-kinase interactions. We found that STAT3, TCF3, RELA, RCOR1, PBX3, UBTF, SPI1, FOS, RUNX1, and SOX2 are the top ten TFs that reg + ulate the core genes. Moreover, MAPK1, MAPK3, AKT1, ERK2, ERK1, MAPK8, MAPK14, CK2ALPHA, JNK1, and CSNK2A1 were the top ten kinases connected to the network through 18 intermediate proteins (Fig. 5, a). The TF-miRNA-gene co-expression network was generated based on the integrated data (Fig. 5, b). It shows that STAT3 and TCF3 regulate miR-146b, miR-194, and miR-214, whereas RELA interacts with miR-194 and miR-214. miR-214 regulated RUNX1 and STAT3, considered the hub miRNA with 12 interactions. Among the TFs, STAT3 and TCF3 had the most interactions with miRNAs and core genes by the degree of 11 and 8, respectively. STAT3 are connected to other TFs, including RUNX1, SOX2, SPL1, RCOR1, PBX, and FOS.
Fig. 5.
The upstream regulatory networks for core genes. a) The regulatory network of top ten TFs, kinases, and intermediate proteins' interactions. b) The TF-miRNA co-regulatory network. STAT3 is common between core genes and TFs.
3.6. Overall survival analysis
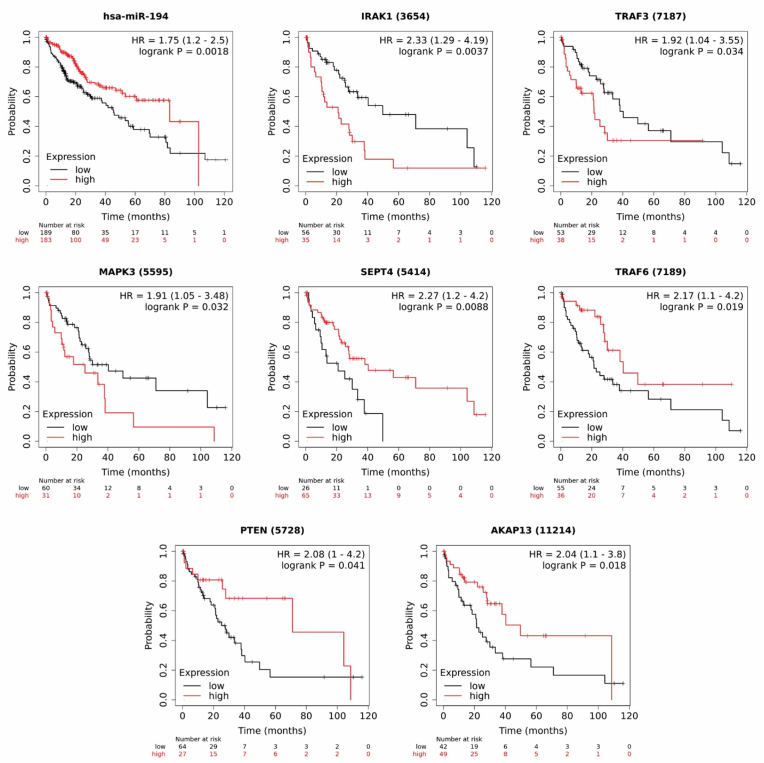
Since NAFLD can progress to HCC, we analyzed the association of core genes and miRNAs with the overall survival time of HCC patients to determine their prognosis value using Kaplan Meier (KM)-plotter online tool. For plotting the overall survival time of HCC patients based on mRNA expression, liver cancer RNA-seq data from a total of 364 patients was utilized. After excluding cases related to alcohol consumption and hepatitis virus, there remained a cohort of 91 patients [34]. Regarding miRNA expression, liver cancer miRNA data was used to plot the overall survival time of HCC patients [35]. The KM survival plots showed that down-regulation of miR-194 increased the hazard ratio (HR) significantly to 1.75 with a 95% confidence interval (CI: 1.2–2.5) in HCC patients (Fig. 6). miR-146b and miR-214 were not found to be statistically significant. Additionally, down-regulation of AKAP13, SEPT4, TRAF6, and PTEN decreased the survival time significantly with the HR of 2.04 (95% CI: 1.1–3.8), 2.27 (95% CI: 1.2–4.2), 2.17 (95% CI: 1.1–4.2), and 2.08 (95% CI: 1–4.2), respectively. Conversely, high expression levels of TRAF3 (HR = 1.92, 95% CI: 1.1–3.8), IRAK1 (HR = 2.33,95% CI: 1.29–4.19), and MAPK3 (HR = 1.91,95% CI: 1.05–3.48) are significantly associated with increasing the risk of mortality (Fig. 6).
Fig. 6.
Overall survival analysis for candidate miRNAs and core genes in HCC patients. Low expression of miR-194 increased the mortality risk to 1.75. Up-regulation of IRAK1, TRAF3, and MAPK3 decreased the survival probability. Conversely, down-regulation of SEPT4, TRAF6, PTEN, and AKAP13 were associated with poor survival outcome. The number of patients for each gene, indicated as "number at risk". HR= Hazard Ratio. logrank p < 0.05 considered significance threshold.
4. Discussion
NAFLD is the most common chronic liver disease worldwide, triggered by fat accumulation in liver cells [2]. One of the most critical problems for managing NAFLD is that patients remain asymptomatic in the early stages until they progress to cirrhosis. To date, liver biopsy has been used as the gold standard to definitive diagnose NAFLD. However, its aggressive procedure increases the need for non-invasive tests to diagnose the disease [1,3]. miRNAs, which are involved in post-transcriptional regulation, can release from cells, circulate in body fluids, and become stable by binding to argonate proteins or vesicular membranes [5,36]. This feature led to many studies on the circulating miRNAs in the development of biomarkers for several diseases.
In line with our analysis of the NAFLD patients’ characteristics, the results indicated that most patients are categorized in overweight (44.11%) and obese (41.17%) groups. However, BMI is not in correlation with disease progression. It has been suggested that central obesity is more important than general obesity and BMI, explaining NAFLD incidence in patients with normal BMI but increased waist circumference [37]. We found that the disease severity increased with the age of patients. Accordingly, the mean age of the NASH stage is 37.33 ± 1.78, versus 55.63 ± 3.58 in the Cirrhosis group (p < 0.05). It shows the importance of early diagnosis to impede the progression of NAFLD. Conversely, the ALT enzyme, used as a diagnostic marker in NAFLD, decreased gently from NASH to Cirrhosis. Altogether, finding a reliable biomarker for the early detection of NAFLD is substantial.
Recent studies have reported the roles of miR-146b, miR-194, and miR-214 in controlling the HSCs activation. Previous research has indicated that miR-146b plays a critical role in regulating inflammatory responses and immune signaling pathways, which are key factors in the development and progression of NAFLD [38]. Studies have highlighted the dysregulation of miR-194 in liver fibrosis. Its altered expression has been associated with the activation of hepatic stellate cells, the main contributors to the excessive deposition of extracellular matrix in fibrotic liver tissue [39,40]. miR-214 has been implicated in fibrogenesis and fibrosis-related processes in the liver. It is involved in the regulation of hepatic stellate cell activation, extracellular matrix remodeling, and pro-fibrotic signaling pathways [41,42]. Recently, it has been reported that the expression of miR-146b and miR-214 is up-regulated in liver fibrosis, while miR-194 was down-regulated. The same expression changes of these miRNAs were observed during HSCs activation [39,40,[42], [43], [44], [45], [46], [47]]. However, the expression levels of circulating miRNAs-146b, −194, and −214 have not been studied in the NAFLD. Here, we investigated the expression level of these miRNAs in the whole blood of NAFLD patients. Our results revealed that miR-146b significantly up-regulated in patients' blood. Although the expression of miR-146b is significantly increased in NASH and Fibrosis (p < 0.05), its increment is not significant in Cirrhosis patients, necessitating further investigations at this stage. Despite the absence of a significant change in miR-146b expression in Cirrhosis patients, its expression levels still provide valuable insights into the pathogenesis of liver disease. It is important to note that miR-146b plays a crucial role in regulating inflammatory responses and immune signaling pathways. Therefore, even without a significant up-regulation, miR-146b may still have functional relevance in cirrhosis. The lack of significant change in miR-146b expression may be attributed to the complex interplay of various factors within the liver microenvironment during cirrhosis. Moreover, additional blood samples are required to investigate the precise alteration of miR-146b during Cirrhosis. Ge et al. identified the up-regulation of miR-146b in hepatic fibrosis rats and its correlation with fibrosis-related genes, including TGF-β, TIMP1, and COL-1α [45]. Further analysis showed that the miR-146b expression level increased in aHSCs and participated in HSC activation by targeting the KLF4 3′UTR [47].
Comparing the circulating miR-194 expression level between NAFLD patients and healthy controls revealed a significant down-regulation, according to the previous studies. RecentIn vitro investigations on HSCs and a high-fat diet (HFD) induced NAFLD model indicated that miR-194 expression level was decreased [39,44,48]. Wu et al. examined the miR-194 expression in human fibrotic liver tissues, aHSCs, and CCL4-induced mice models. They reported that the expression of miR-194 was notably decreased during liver fibrosis [40].
miR-214 has been known as a fibrotic regulator in various tissues, including the liver. Its increased expression level has been reported in NAFLD mice models and aHSCs [43]. The investigation of miR-214 hepatic expression during liver cirrhosis progression in rats demonstrated the high up-regulation in the fibrotic area [41]. Ma et al. identified the increased level of miR-214 in activated rat HSCs, CCL4-treated and HFD-induced rats and mice, and human cirrhosis samples [42]. In agreement with the previous reports, the up-regulation of circulating miR-214 in patients’ blood was showed in our study. Besides, a significant decrease was detected in fibrosis to cirrhosis progression. It might be related to the inhibitory effect of miR-214 on myofibroblast differentiation, which is disclosed in fibroblastic MT-9 cells [42].
By integrating the interactions between miR-146, miR-194, miR-214, and related target genes or lncRNAs, we established the ceRNAs network to illustrate their potential regulatory roles in NAFLD. Based on the ceRNA hypothesis, RNAs can communicate with miRNA response elements (MREs) and act as molecular sponges for miRNAs [49]. The lncRNA-miRNA-mRNA interactions illustrated that NEAT1 and XIST sponged all three miRNAs, suggesting that these lncRNAs may have an important function in fibrogenesis. Many studies reported the up-regulation of NEAT1 in NAFLD and liver fibrosis, leading to increased lipid deposition, inflammation response, and fibrosis through targeting mTOR/S6K1, miR-146a/ROCK1, AMPK/SREBP1, and miR-506/GLI3 [50]. Yu et al. disclosed the NEAT1/miR-122-/KLF6 signaling cascade in liver fibrosis, where NEAT1 overexpression affects HSC activation by sponging miR-122 [51]. A recently published study suggested that miR-139 sponged by NEAT1, regulated liver fibrosis by targeting the β-catenin/SOX9/TGF-β1 pathway [52]. Some investigations showed the up-regulation of lncRNA XIST in liver fibrosis and HCC [53,54], while other studies declared its down-regulation in NAFLD mice and HCC [[55], [56], [57]]. Xie et al. demonstrated that XIST enhanced the ethanol-activated HSCs autophagy and activation by miR-29b/HMGB1 pathway [53]. These results indicate that more investigations are needed to clarify the exact role of lncRNA XIST in NAFLD and fibrogenesis.
We performed a gene set enrichment analysis to explore the potential roles of miR-146b, miR-194, and miR-214 in HSC activation and fibrogenesis. Pathway analysis revealed that NF-κB activation and p75 neurotrophin receptor signaling are the most significant pathways, function by IRAK1, TRAF6, and SQSTM1. It has been proved that p75NTR is expressed in aHSCs and promotes hepatocytes proliferation [58]. Several studies have shown the role of NF-κB in liver disorders, including fibrosis, by regulating the inflammatory responses. Inhibition of NF-κB pathway prevents HSC activation, decreases inflammatory cytokines levels, and ameliorates liver fibrosis [[59], [60], [61]]. IRAK1/TRAF6 is a well-known axis, which functions as s signal transducer in the NF-κB pathway. It is demonstrated that inhibition of IRAK1 and TRAF6 by miR-146a/b suppresses the NF-κB activation and pro-inflammatory cytokine production [[62], [63], [64]]. Autophagy is an essential function during HSC activation, which controls the loss of cytoplasmic lipid droplets to provide cellular energy [65,66]. The results demonstrated that our candidate miRNAs are contributed to autophagy via targeting SQSTM1, IL6ST, THBS1, GSK3B, and PTEN. These mRNAs, along with IRAK1, TRAF6, PAK2, and STAT3, are enriched in negative regulation of apoptosis, based on the biological process analysis. Also, it has been reported that NF-κB has an anti-apoptosis effect in HSCs [67].
Next, the potential miRNA-TF co-regulatory network was analyzed in liver fibrogenesis. We performed the upstream regulatory network analysis for HSC activation core genes. Fig. 5 illustrates the top ten TFs, protein kinases, and intermediate proteins, regulating the core genes. The co-regulatory network demonstrated that STAT3, TCF3, RELA, and RUNX1 are connected to the miRNAs. It has been indicated that TGF-β and IL-6 promote HSC activation, mediated by the STAT3 signaling pathway [[68], [69], [70]]. Previous researches have shown that suppressing the STAT3 activity leads to inhibition of HSC activation [[70], [71], [72]]. Recently, Wang et al. highlighted the role of RUNX1 as a master regulator responsible for the HSCs activation, which was up-regulated in aHSCs [73]. In addition, it has been found that the expression of RUNX1 is up-regulated in NAFLD and correlates with NASH severity, controlling the angiogenic and inflammatory activities [74,75]. In line with our results, the miRNA-TF co-regulatory network suggests that miR-214/RUNX1 regulates the activation of HSCs by targeting BAX, PTEN, and IL6ST.
5. Conclusion
In conclusion, we provided a potential panel of three miRNAs- (146b, 194, 214) which can significantly distinguish NAFLD patients and healthy individuals, improving the early non-invasive diagnosis. Except for miR-146b, which could not significantly discriminate the Cirrhosis group, miR-194 and miR-214 accurately diagnose the patients from onset of NAFLD until its end-stage. Also, miR-214 can differentiate Cirrhosis from Fibrosis. Furthermore, our results suggested the important role of lncRNAs NEAT1 and XIST, in addition to NF-κB activation and autophagy. These miRNAs can affect HSC activation through interaction with STAT3 and RUNX1 transcription factors. Our findings shed light on the new potentially diagnostic panel and related molecular mechanisms for NAFLD pathogenesis.
Ethics approval and consent to participate
The Ethics Committee approved all experimental procedures and protocols of this study at the Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran. The written informed consent was signed by all participants. All experiments were performed in accordance with the relevant guidelines and regulations.
Availability of data and materials
The data generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
CRediT authorship contribution statement
Taha Aghajanzadeh: Investigation, Formal analysis, Data curation, Writing – original draft, Visualization, Writing – review & editing, All authors reviewed and approved the final manuscript. Mahmood Talkhabi: Methodology, Validation, Writing – review & editing, All authors reviewed and approved the final manuscript. Mohammad Reza Zali: Writing – review & editing, All authors reviewed and approved the final manuscript. Behzad Hatami: Resources, All authors reviewed and approved the final manuscript. Kaveh Baghaei: Conceptualization, Methodology, Validation, Writing – review & editing, Supervision, Project administration, All authors reviewed and approved the final manuscript.
Declaration of competing interest
The authors declare that they have no competing interests.
Acknowledgments
Not applicable.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ncrna.2023.06.004.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Huang T., Behary J., Zekry A. Non‐alcoholic fatty liver disease (NAFLD): a review of epidemiology, risk factors, diagnosis and management. Intern. Med. J. 2019;50(9):1038–1047. doi: 10.1111/imj.14709. [DOI] [PubMed] [Google Scholar]
- 2.Michelotti G.A., Machado M.V., Diehl A.M. NAFLD, NASH and liver cancer. Nat. Rev. Gastroenterol. Hepatol. 2013;10(11):656–665. doi: 10.1038/nrgastro.2013.183. [DOI] [PubMed] [Google Scholar]
- 3.Farrell G.C., McCullough A.J., Day C.P. John Wiley & Sons; 2013. Non-alcoholic Fatty Liver Disease: A Practical Guide. [Google Scholar]
- 4.Zhang C.-Y., et al. Liver fibrosis and hepatic stellate cells: etiology, pathological hallmarks and therapeutic targets. World J. Gastroenterol. 2016;22(48) doi: 10.3748/wjg.v22.i48.10512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eulalio A., Huntzinger E., Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132(1):9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 6.Miyaaki H., et al. Significance of serum and hepatic micro RNA‐122 levels in patients with non‐alcoholic fatty liver disease. Liver Int. 2014;34(7):e302–e307. doi: 10.1111/liv.12429. [DOI] [PubMed] [Google Scholar]
- 7.Khalifa O., et al. Disease markers; 2020. Noncoding RNAs in Nonalcoholic Fatty Liver Disease: Potential Diagnosis and Prognosis Biomarkers; p. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Su Q., et al. MicroRNAs in the pathogenesis and treatment of progressive liver injury in NAFLD and liver fibrosis. Adv. Drug Deliv. Rev. 2018;129:54–63. doi: 10.1016/j.addr.2018.01.009. [DOI] [PubMed] [Google Scholar]
- 9.Baffy G. MicroRNAs in nonalcoholic fatty liver disease. J. Clin. Med. 2015;4(12):1977–1988. doi: 10.3390/jcm4121953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gjorgjieva M., et al. miRNAs and NAFLD: from pathophysiology to therapy. Gut. 2019;68(11):2065–2079. doi: 10.1136/gutjnl-2018-318146. [DOI] [PubMed] [Google Scholar]
- 11.Musaddaq G., et al. Circulating liver-specific microRNAs as noninvasive diagnostic biomarkers of hepatic diseases in human. Biomarkers. 2019;24(2):103–109. doi: 10.1080/1354750X.2018.1528631. [DOI] [PubMed] [Google Scholar]
- 12.Nallagangula K.S., et al. Liver fibrosis: a compilation on the biomarkers status and their significance during disease progression. Future Sci OA. 2018;4(1):Fso250. doi: 10.4155/fsoa-2017-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bataller R., Brenner D.A. Liver fibrosis. J. Clin. Investig. 2005;115(2):209–218. doi: 10.1172/JCI24282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soresi M., et al. Non invasive tools for the diagnosis of liver cirrhosis. World J. Gastroenterol. 2014;20(48):18131–18150. doi: 10.3748/wjg.v20.i48.18131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stål P. Liver fibrosis in non-alcoholic fatty liver disease-diagnostic challenge with prognostic significance. World J. Gastroenterol.: WJG. 2015;21(39) doi: 10.3748/wjg.v21.i39.11077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lambrecht J., et al. The miRFIB-Score: a serological miRNA-based scoring algorithm for the diagnosis of significant liver fibrosis. Cells. 2019;8(9):1003. doi: 10.3390/cells8091003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Piazzolla V.A., Mangia A. Noninvasive diagnosis of NAFLD and NASH. Cells. 2020;9(4):1005. doi: 10.3390/cells9041005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang H.-Y., et al. miRTarBase Updates to the experimentally validated microRNA–target interaction database. Nucleic Acids Res. 2020;48(D1):D148–D154. doi: 10.1093/nar/gkz896. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vlachos I.S., Hatzigeorgiou A.G. Drug Target miRNA. Springer; 2017. Functional analysis of miRNAs using the DIANA tools online suite; pp. 25–50. [DOI] [PubMed] [Google Scholar]
- 20.Agarwal V., et al. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4 doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sticht C., et al. miRWalk: an online resource for prediction of microRNA binding sites. PLoS One. 2018;13(10) doi: 10.1371/journal.pone.0206239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Y., Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;48(D1):D127–D131. doi: 10.1093/nar/gkz757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karagkouni D., et al. DIANA-LncBase v3: indexing experimentally supported miRNA targets on non-coding transcripts. Nucleic Acids Res. 2020;48(D1):D101–D110. doi: 10.1093/nar/gkz1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang L., et al. miRNet 2.0: network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res. 2020;48(W1):W244–W251. doi: 10.1093/nar/gkaa467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin Y., et al. RNAInter in 2020: RNA interactome repository with increased coverage and annotation. Nucleic Acids Res. 2020;48(D1):D189–D197. doi: 10.1093/nar/gkz804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shannon P., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie Z., et al. Gene set knowledge discovery with Enrichr. Current protocols. 2021;1(3):e90. doi: 10.1002/cpz1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oliveros J.C. 2007. VENNY. An Interactive Tool for Comparing Lists with Venn Diagrams.http://bioinfogp.cnb.csic.es/tools/venny/index.html [Google Scholar]
- 29.Clarke D.J.B., et al. eXpression2Kinases (X2K) Web: linking expression signatures to upstream cell signaling networks. Nucleic Acids Res. 2018;46(W1):W171–W179. doi: 10.1093/nar/gky458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tong Z., et al. TransmiR v2. 0: an updated transcription factor-microRNA regulation database. Nucleic Acids Res. 2019;47(D1):D253–D258. doi: 10.1093/nar/gky1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Z.-P., et al. RegNetwork: an integrated database of transcriptional and post-transcriptional regulatory networks in human and mouse. Database. 2015;2015 doi: 10.1093/database/bav095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Győrffy B. Discovery and ranking of the most robust prognostic biomarkers in serous ovarian cancer. Geroscience. 2023:1–10. doi: 10.1007/s11357-023-00742-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lánczky A., Győrffy B. Web-based survival analysis tool tailored for medical research (KMplot): development and implementation. J. Med. Internet Res. 2021;23(7) doi: 10.2196/27633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Menyhárt O., Nagy Á., Győrffy B. Determining consistent prognostic biomarkers of overall survival and vascular invasion in hepatocellular carcinoma. R. Soc. Open Sci. 2018;5(12) doi: 10.1098/rsos.181006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagy Á., et al. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci. Rep. 2018;8(1):9227. doi: 10.1038/s41598-018-27521-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang K., et al. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc. Natl. Acad. Sci. USA. 2009;106(11):4402–4407. doi: 10.1073/pnas.0813371106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pang Q., et al. Central obesity and nonalcoholic fatty liver disease risk after adjusting for body mass index. World J. Gastroenterol.: WJG. 2015;21(5):1650. doi: 10.3748/wjg.v21.i5.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang W., et al. MiR‐146b attenuates high‐fat diet‐induced non‐alcoholic steatohepatitis in mice. J. Gastroenterol. Hepatol. 2015;30(5):933–943. doi: 10.1111/jgh.12878. [DOI] [PubMed] [Google Scholar]
- 39.Venugopal S.K., et al. Liver fibrosis causes downregulation of miRNA-150 and miRNA-194 in hepatic stellate cells, and their overexpression causes decreased stellate cell activation. Am. J. Physiol. Gastrointest. Liver Physiol. 2010;298(1):G101–G106. doi: 10.1152/ajpgi.00220.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu J.-C., et al. MicroRNA-194 inactivates hepatic stellate cells and alleviates liver fibrosis by inhibiting AKT2. World J. Gastroenterol. 2019;25(31):4468. doi: 10.3748/wjg.v25.i31.4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Izawa T., et al. Anti-fibrotic role of miR-214 in thioacetamide-induced liver cirrhosis in rats. Toxicol. Pathol. 2015;43(6):844–851. doi: 10.1177/0192623315573587. [DOI] [PubMed] [Google Scholar]
- 42.Ma L., et al. MicroRNA-214 promotes hepatic stellate cell activation and liver fibrosis by suppressing Sufu expression. Cell Death Dis. 2018;9(7):1–13. doi: 10.1038/s41419-018-0752-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ceccarelli S., et al. Dual role of microRNAs in NAFLD. Int. J. Mol. Sci. 2013;14(4):8437–8455. doi: 10.3390/ijms14048437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davoodian P., et al. Effect of TGF-β/smad signaling pathway blocking on expression profiles of miR-335, miR-150, miR-194, miR-27a, and miR-199a of hepatic stellate cells (HSCs) Gastroenterol Hepatol Bed Bench. 2017;10(2):112–117. [PMC free article] [PubMed] [Google Scholar]
- 45.Ge S., et al. Deep sequencing analysis of microRNA expression in porcine serum-induced hepatic fibrosis rats. Ann. Hepatol. 2014;13(4):439–449. [PubMed] [Google Scholar]
- 46.Ge S., et al. HMGB1 inhibits HNF1A to modulate liver fibrogenesis via p65/miR-146b signaling. DNA Cell Biol. 2020;39(9):1711–1722. doi: 10.1089/dna.2019.5330. [DOI] [PubMed] [Google Scholar]
- 47.Ge S., et al. MicroRNA-146b regulates hepatic stellate cell activation via targeting of KLF4. Ann. Hepatol. 2017;15(6):918–928. doi: 10.5604/16652681.1222111. [DOI] [PubMed] [Google Scholar]
- 48.Torres L., Cogliati B., Otton R. Green tea prevents NAFLD by modulation of miR-34a and miR-194 expression in a high-fat diet mouse model. Oxid. Med. Cell. Longev. 2019;2019 doi: 10.1155/2019/4168380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Y., et al. Competitive endogenous RNA is an intrinsic component of EMT regulatory circuits and modulates EMT. Nat. Commun. 2019;10(1):1–12. doi: 10.1038/s41467-019-09649-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bu F.t., et al. LncRNA NEAT1: shedding light on mechanisms and opportunities in liver diseases. Liver Int. 2020;40(11):2612–2626. doi: 10.1111/liv.14629. [DOI] [PubMed] [Google Scholar]
- 51.Yu F., et al. NEAT1 accelerates the progression of liver fibrosis via regulation of microRNA-122 and Kruppel-like factor 6. J. Mol. Med. 2017;95(11):1191–1202. doi: 10.1007/s00109-017-1586-5. [DOI] [PubMed] [Google Scholar]
- 52.Wang Q., et al. miR-139-5p sponged by LncRNA NEAT1 regulates liver fibrosis via targeting β-catenin/SOX9/TGF-β1 pathway. Cell Death Discovery. 2021;7(1):1–15. doi: 10.1038/s41420-021-00632-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xie Z.Y., et al. Long noncoding RNA XIST enhances ethanol‐induced hepatic stellate cells autophagy and activation via miR‐29b/HMGB1 axis. IUBMB Life. 2019;71(12):1962–1972. doi: 10.1002/iub.2140. [DOI] [PubMed] [Google Scholar]
- 54.Liu L., et al. LncRNA XIST promotes liver cancer progression by acting as a molecular sponge of miR-200b-3p to regulate ZEB1/2 expression. J. Int. Med. Res. 2021;49(5) doi: 10.1177/03000605211016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang C., et al. Impact of high-fat diet on liver genes expression profiles in mice model of nonalcoholic fatty liver disease. Environ. Toxicol. Pharmacol. 2016;45:52–62. doi: 10.1016/j.etap.2016.05.014. [DOI] [PubMed] [Google Scholar]
- 56.Zhang Y., et al. lncRNA XIST regulates proliferation and migration of hepatocellular carcinoma cells by acting as miR-497-5p molecular sponge and targeting PDCD4. Cancer Cell Int. 2019;19(1):1–13. doi: 10.1186/s12935-019-0909-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chang S., et al. Long non-coding RNA XIST regulates PTEN expression by sponging miR-181a and promotes hepatocellular carcinoma progression. BMC Cancer. 2017;17(1):1–14. doi: 10.1186/s12885-017-3216-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Passino M.A., et al. Regulation of hepatic stellate cell differentiation by the neurotrophin receptor p75NTR. Science. 2007;315(5820):1853–1856. doi: 10.1126/science.1137603. [DOI] [PubMed] [Google Scholar]
- 59.Son G., et al. Selective inactivation of NF-κB in the liver using NF-κB decoy suppresses CCl4-induced liver injury and fibrosis. Am. J. Physiol. Gastrointest. Liver Physiol. 2007;293(3):G631–G639. doi: 10.1152/ajpgi.00185.2007. [DOI] [PubMed] [Google Scholar]
- 60.Bai F., et al. Trolline ameliorates liver fibrosis by inhibiting the NF-κB pathway, promoting HSC apoptosis and suppressing autophagy. Cell. Physiol. Biochem. 2017;44(2):436–446. doi: 10.1159/000485009. [DOI] [PubMed] [Google Scholar]
- 61.Liao X., et al. MicroRNA‐326 attenuates hepatic stellate cell activation and liver fibrosis by inhibiting TLR4 signaling. J. Cell. Biochem. 2020;121(8–9):3794–3803. doi: 10.1002/jcb.29520. [DOI] [PubMed] [Google Scholar]
- 62.Yang G., Zhao Y. Overexpression of miR-146b-5p ameliorates neonatal hypoxic ischemic encephalopathy by inhibiting IRAK1/TRAF6/TAK1/NF-αB signaling. Yonsei Med. J. 2020;61(8):660. doi: 10.3349/ymj.2020.61.8.660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou C., et al. MicroRNA-146a inhibits NF-κB activation and pro-inflammatory cytokine production by regulating IRAK1 expression in THP-1 cells. Exp. Ther. Med. 2019;18(4):3078–3084. doi: 10.3892/etm.2019.7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen Y., et al. MicroRNA-146a-5p negatively regulates pro-inflammatory cytokine secretion and cell activation in lipopolysaccharide stimulated human hepatic stellate cells through inhibition of toll-like receptor 4 signaling pathways. Int. J. Mol. Sci. 2016;17(7):1076. doi: 10.3390/ijms17071076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hernández–Gea V., et al. Autophagy releases lipid that promotes fibrogenesis by activated hepatic stellate cells in mice and in human tissues. Gastroenterology. 2012;142(4):938–946. doi: 10.1053/j.gastro.2011.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thoen L.F., et al. A role for autophagy during hepatic stellate cell activation. J. Hepatol. 2011;55(6):1353–1360. doi: 10.1016/j.jhep.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 67.Sun B., Karin M. NF-κB signaling, liver disease and hepatoprotective agents. Oncogene. 2008;27(48):6228–6244. doi: 10.1038/onc.2008.300. [DOI] [PubMed] [Google Scholar]
- 68.Kagan P., et al. Both MAPK and STAT3 signal transduction pathways are necessary for IL-6-dependent hepatic stellate cells activation. PLoS One. 2017;12(5) doi: 10.1371/journal.pone.0176173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xiang D.-M., et al. The HLF/IL-6/STAT3 feedforward circuit drives hepatic stellate cell activation to promote liver fibrosis. Gut. 2018;67(9):1704–1715. doi: 10.1136/gutjnl-2016-313392. [DOI] [PubMed] [Google Scholar]
- 70.Choi S., et al. A novel STAT3 inhibitor, STX-0119, attenuates liver fibrosis by inactivating hepatic stellate cells in mice. Biochem. Biophys. Res. Commun. 2019;513(1):49–55. doi: 10.1016/j.bbrc.2019.03.156. [DOI] [PubMed] [Google Scholar]
- 71.Cummins C.B., et al. Luteolin-mediated inhibition of hepatic stellate cell activation via suppression of the STAT3 pathway. Int. J. Mol. Sci. 2018;19(6):1567. doi: 10.3390/ijms19061567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Park Y.J., et al. (–)-Catechin-7-O-β-d-Apiofuranoside inhibits hepatic stellate cell activation by suppressing the STAT3 signaling pathway. Cells. 2020;9(1):30. doi: 10.3390/cells9010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang Z.-Y., et al. Single-cell and bulk transcriptomics of the liver reveals potential targets of NASH with fibrosis. Sci. Rep. 2021;11(1):1–15. doi: 10.1038/s41598-021-98806-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kaur S., et al. Increased expression of RUNX1 in liver correlates with NASH activity score in patients with non-alcoholic steatohepatitis (NASH) Cells. 2019;8(10):1277. doi: 10.3390/cells8101277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huang J., et al. RUNX1 regulates SMAD1 by transcriptionally activating the expression of USP9X, regulating the activation of hepatic stellate cells and liver fibrosis. Eur. J. Pharmacol. 2021;903 doi: 10.1016/j.ejphar.2021.174137. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.