Abstract
In this proof-of-concept study, we developed a single-cell method that provides genotypes of somatic alterations found in coding regions of messenger RNAs and integrates these transcript-based variants with their matching cell transcriptomes. We used nanopore adaptive sampling on single-cell complementary DNA libraries to validate coding variants in target gene transcripts, and short-read sequencing to characterize cell types harboring the mutations. CRISPR edits for 16 targets were identified using a cancer cell line, and known variants in the cell line were validated using a 352-gene panel. Variants in primary cancer samples were validated using target gene panels ranging from 161 to 529 genes. A gene rearrangement was also identified in one patient, with the rearrangement occurring in two distinct tumor sites.
Graphical Abstract
Graphical Abstract.
INTRODUCTION
Single-cell genomics has proven to be a highly informative method for analyzing cancer and other disease tissues. Single-cell RNA sequencing (scRNA-seq) provides a granular view of an individual cell’s gene expression. One can characterize different cell types, cellular heterogeneity from complex tumor samples and differential gene expression among individual cells. Most scRNA-seq approaches focus on gene expression. However, single-cell genomic approaches can examine other features such as copy number and even somatic mutations. These additional genomic features increase the overall yield of valuable information from single cancer cells. However, identifying cancer mutations based on scRNA-seq is not commonly attempted given specific limitations of the current short-read approach.
Quantitative measurement of single-cell messenger RNA (mRNA), in the form of sequence reads from complementary DNA (cDNA), requires a combination of cell barcode and transcript sequence. Single-cell short-read sequencing methods typically require fragmenting the full-length cDNA into lower molecular weight species for library preparation. The resulting short reads are several hundred bases, starting from either the 5′ or 3′ end of a given transcript. This sequence information allows one to count the number of transcripts expressed within an individual cell. However, these short reads have significant limitations for the analysis of cancer cell transcriptomes due to the fragmentation that eliminates transcript sequence features closer to the nonbarcoded end of the molecule. This loss can include somatic allelic variants present in the internal mRNA coding sequence, chimeric rearrangements and alternative splicing events that alter transcript isoform structure. Overall, short-read sequencing leads to a loss of valuable transcript information such as genetic variants that can only be derived from an intact cDNA molecule.
Increasingly, single-molecule long-read sequencing is being used for genomic studies of gene expression and characterizing cDNAs (1–5). There are two sequencers in this class available from Oxford Nanopore or Pacific Biosciences. Both generate long reads with lengths of 1 kb and higher, all from single DNA molecules. With intact cDNAs from single cells, the library preparation for long-read sequencers does not truncate the molecules. As a result, long reads can readily cover an entire cDNA molecule. This sequence information can be used to identify transcript structure and genetic variants present in exon sequences (5).
For single-cell genomics, targeted sequencing of specific gene transcripts provides an opportunity to identify transcript structure and genetic variant features present among individual cells. Targeted sequencing provides higher coverage and reduces the cost of analyzing single cells. There are a variety of methods used for single-cell sequencing of specific target cDNAs. For example, polymerase chain reaction (PCR) amplification of specific targets from single-cell libraries with amplicon sequencing provides higher coverage of genes (6). Several steps are required for developing PCR assays to amplify gene targets from scRNA-seq libraries. Requirements include identifying specific primer sequences for a given cDNA target and optimizing PCR conditions to reduce artifacts. When one develops assays for multiplexing PCR, amplification artifacts complicate this method and place practical limits on the total number of amplification targets.
Another common method involves bait capture of single-cell cDNAs. These assays use biotinylated oligonucleotide probes that hybridize to a target. This process enriches a specific cDNA molecule of interest from scRNA-seq libraries (2,3). The bait capture approach can be scaled up to enrich many genes. However, the development of these assays requires extensive testing of probes and optimizing amplification steps as part of the capture process. In addition, the experimental workflow has multiple manipulation steps that add to the complexity of the process.
Several studies have demonstrated a new approach for targeted single-molecule sequencing that leverages attributes of the Oxford Nanopore platform (7). Referred to as adaptive sampling, this method involves directly assessing DNA molecules for specific target sequences (8–10). A reference file with a set of target genomic coordinates is provided. The process involves on-the-fly base calling from each DNA molecule per given nanopore, sequence alignment of data, real-time control of the nanopore voltage and selection of those molecules with an extended long read of the target sequence. Once the target sequence is identified, the nanopore instrument proceeds to sequence the remainder of the molecule. This method enables direct sampling and enrichment of specific DNA molecules without prior preparative steps. It does not require library manipulation for selective PCR amplification or bait hybridization enrichment of the target molecule of interest. Importantly, this approach reduces any potential biases in library content by not requiring any pre-amplification step.
We conducted a proof-of-concept study to determine the feasibility of applying nanopore adaptive sampling for targeted scRNA-seq. The objective was to conduct targeted sequencing of specific single-cell gene cDNAs and genotype somatic genetic alterations among individual cells from a cancer line and primary tumors. For this approach, one introduces the single-cell cDNA library into the nanopore sequencer, with the controller evaluating for matching sequences of a given cDNA molecule as it passes through the pore. Then, the target cDNAs are sequenced with long reads, covering the entire length of the cDNA. Variants that are present in the exons, even when they are positioned far from the 5′ and 3′ ends, are detectable. We tested the capability of adaptive sampling by targeting cDNAs from single-cell libraries derived from a cancer cell line. Subsequently, we sequenced a set of different cancers, including metastatic and lymphoid malignancies (Table 1). These tumors had previously undergone diagnostic cancer gene sequencing and the clinical reports of coding cancer mutations were available for each patient (Table 2). From these cell line and patient samples and using the same scRNA-seq library, we integrated the scRNA-seq short- and long-read data. The long-read data were used to identify the prior reported cancer mutations or the induced CRISPR edits among single cells. We determined whether previously reported cancer mutations could be mapped among the single cells from different tumor sites. We observed that mutation genotypes can be identified if there is sufficient expression of the target gene. Overall, our study demonstrated the feasibility of single-cell identification of genetic alterations with adaptive long-read sequencing of cDNAs for genes with sufficiently high native expression.
Table 1.
Cancer samples used for single-cell adaptive sequencing
| Source ID or cell line | Tumor ID | Tumor type | Tumor site or experimental condition | Mutation and somatic variant discovery | Number of gene targets | Number of gene targets detected | Number of coding substitution mutations or CRISPR targets |
|---|---|---|---|---|---|---|---|
| Jurkat | C1 | T-cell leukemia | Cell line | Exome sequencing | 352 | 352 | |
| C2 | Cell line transduced with CRISPR | Amplicon sequencing | 16 | 16 (targets) | |||
| 8605 | T1 | Appendiceal carcinoma | Primary appendix tumor | UCSF cancer sequencing panel | 529 | 498 | 5 |
| T2 | Metastasis in the left ovary | 496 | 5 | ||||
| 8629 | T3 | Appendiceal carcinoma | Metastasis in the omentum | STAMP-FoundationOne sequencing panel | 330 | 312 | 4 |
| T4 | Metastasis in the small intestine | 319 | 4 | ||||
| 6408 | T5 | B-cell lymphoma | Metastasis in the right inguinal lymph node | Heme-STAMP cancer sequencing panel | 161 | 154 | 9 |
| T6 | Metastasis in the right cervical lymph node | 155 | 9 |
Table 2.
Substitution cancer mutations
| ID | Tumor type | Sample | Gene | Coordinates | Mutation | AA change | Clinical significance |
|---|---|---|---|---|---|---|---|
| 8605 | Appendix carcinoma | T1 | APC | chr5:112839667 | C>T | A1358V | Uncertain significance |
| GNAS | chr20:58909365 | C>A | R844S | Pathogenic | |||
| KMT2D | chr12:49031792 | C>T | V4305I | Likely benign | |||
| KRAS | chr12:25245350 | C>A | G12V | Pathogenic | |||
| POLD1 | chr19:50406302 | G>A | V455M | Uncertain significance | |||
| 8629 | Appendix carcinoma | T3, T4 | GNAS | chr20:58909365 | C>T | R844C | Pathogenic |
| KRAS | chr12:25245350 | C>T | G12D | Pathogenic | |||
| SF3B1 | chr2:197402110 | T>C | K700E | Likely pathogenic | |||
| SMAD2 | chr18:47841840 | G>C|T | S464* | Nonsense | |||
| 6408 | Follicular lymphoma | T7a | BCL2 | chr18:63318582 | C>T | E29K | Deleterious |
| BCL2 | chr18:63318653 | C>A | G5V | Benign | |||
| BCL2 | chr18:63318494 | T>C | H58R | Benign | |||
| BCL2 | chr18:63318411 | G>A | L86F | Benign | |||
| BCL2 | chr18:63318320 | G>A | S116F | Benign | |||
| CREBBP | chr16:3736766 | A>G | Y1482H | Deleterious | |||
| DNMT3A | chr2:25300227 | T>G | E30A | Likely benign | |||
| EP300 | chr22:41151887 | A>G | S958G | Benign | |||
| NF1 | chr17:31358550 | A>G | I2681V | Likely benign |
aTargeted cancer sequencing done on right axillary node (not T5 or T6).
MATERIALS AND METHODS
Patient samples and processing
Patients with metastatic appendiceal cancer were consented on IRB protocol 44036 approved by Stanford University. Tumor tissues were obtained from surgery and stored in RPMI medium before dissociation. Single-cell suspensions were obtained from tissue fragments using enzymatic and mechanical dissociation. Cells were washed twice in RPMI + 10% fetal bovine serum (FBS), filtered through 70 μm (Flowmi, Bel-Art SP Scienceware, Wayne, NJ), followed by 30 μm (Miltenyi) or 40 μm filter (Flowmi). Cryofrozen cells were rapidly thawed in a bead bath at 37°C followed by above washing and filtering steps. Live cell counts were obtained on a Bio-Rad TC20 Cell Counter (Bio-Rad, Hercules, CA) or a Countess II FL Automated Cell Counter (Thermo Fisher Scientific) using 1:1 trypan blue dilution. Cells were concentrated between 500 and 1500 live cells/μl for subsequent single-cell library preparation. The patient with follicular lymphoma was consented on IRB protocol 13500 approved by Stanford University. Fine needle aspirate specimens from two spatially separated nodal tumor sites were obtained and prepared as previously described (11).
Cell lines and induction of CRISPR mutations
The Jurkat cell line (ATCC TIB-152) and a Cas9-stable version of Jurkat (SL555, GeneCopoeia, Inc., Rockville, MD) were maintained in RPMI medium supplemented with 10% FBS at 37°C under standard CO2 conditions. We produced an oligonucleotide pool for the guide RNA (gRNA) library (IDT, Coralville, IA). Amplified gRNAs were cloned to lentiGuide-Puro (Addgene plasmid #52963). To transduce Cas9-expressing Jurkat cell for CRISPR editing, we used the spinoculation method. The lentiviral supernatant and 8 μg of polybrene (Sigma–Aldrich, St Louis, MO) were added to 1.0 × 105 Cas9-stable Jurkat. The mixture was centrifuged at 800 × g at 32°C for 30 min. Cell pellets were resuspended to fresh media, and after 72 h, transduced cells were selected by puromycin (Life Technologies, Carlsbad, CA). Additional details about this CRISPR-edited cell line are fully described by Kim et al. (6).
To identify specific CRISPR mutations, we generated single-cell full-length cDNAs from transduced Jurkat cells as previously described (6). One nanogram of single-cell cDNA library was used to amplify transcripts with a set of primers flanking the CRISPR edit site. KAPA HiFi HotStart ReadyMix (Roche, Basel, Switzerland) was used for amplification. Extension time was 60 s. Amplicons were pooled at equimolar concentrations. The libraries were prepared with 900 fmol of pooled amplicon for PromethION Flow Cell FLO-PRO002 (Oxford Nanopore Technologies) using Native Barcoding Expansion and Ligation Sequencing Kit (Oxford Nanopore Technologies) as per the manufacturer’s protocol. Libraries were sequenced on Oxford Nanopore PromethION for 72 h.
Single-cell library preparation and short-read sequencing
Sequencing libraries were prepared using Chromium NextGEM Single Cell 5′ Library & Gel Bead Kit v1.1 or v2 (10X Genomics, Pleasanton, CA) as per the manufacturer’s protocol. gRNA direct capture for Jurkat CRISPR assay has been performed as previously described using 6 pmol of scaffold binding oligonucleotides (6). The cDNA and gene expression libraries were amplified with either 14 or 16 cycles of PCR, depending on the starting amount. The size distribution of gene expression libraries was confirmed via gel electrophoresis (Thermo Fisher Scientific, Waltham, MA). The libraries were quantified using a Qubit fluorescent assay (Invitrogen). Short-read sequencing was performed on Illumina sequencers (Illumina, San Diego, CA).
Nanopore long-read sequencing of single-cell libraries
We amplified the entire single-cell cDNA material using the following primer sequences: partial read 1: CTACACGACGCTCTTCCGATCT and non-poly(dT): AAGCAGTGGTATCAACGCAGAG. KAPA HiFi HotStart 2X ReadyMix (Roche, Basel, Switzerland) was used for PCR amplification with 250 nM of each primer. Following PCR, the amplicons were purified using 1.5× volume equivalents of Ampure XP beads. Libraries were quantified with Qubit (Thermo Fisher Scientific). The library was diluted to a total concentration of 600 fmol and loaded onto a MinION R9.4.1 Flow Cell and sequenced for 72 h as per the manufacturer’s instructions (LSK-110, Oxford Nanopore Technologies). For the PromethION runs, 900 fmol of pooled amplicon was loaded onto a PromethION Flow Cell FLO-PRO002 (Oxford Nanopore Technologies) and sequenced for 72 h.
Our patient samples included hematologic and solid epithelial tumors. Each patient had one of their tumor sites analyzed with one of three different cancer gene panels used for diagnostic tumor sequencing. The number of target genes per gene panel ranged from 130 to 529. For each gene panel, the canonical exon coordinates for each gene were identified and organized into a bed file. For adaptive sequencing of each sample, we uploaded the genomic bed file corresponding to their diagnostic sequencing, into the instrument control software. Live base calling was based on the ‘fast’ model enabled rapid alignment and subsequent enrichment of reads that overlapped the target regions.
Bioinformatic analysis
Short-read processing and cell type assignment
Cell Ranger (10X Genomics) version 3.1.0 ‘mkfastq’ and ‘count’ commands were used with default parameters and alignment to GRCh38 to generate matrix of unique molecular identifier (UMI) counts per gene and associated cell barcode. We constructed Seurat objects from each dataset using Seurat (version 4.0.1) (12,13) to apply quality control filters. Quality controls included removing cells that expressed <200 genes, had >30% mitochondrial genes or had UMI counts >6000 indicating potential doublets. Genes detected in <3 cells were removed. We normalized data using ‘SCTransform’ and used the first 20 principal components with a resolution of 0.8 for clustering. We then removed computationally identified doublets from each dataset using DoubletFinder (version 2.0.2) (14). The ‘pN’ value was set to default value of 0.25 as the proportion of artificial doublets. The ‘pK’ value representing the PC neighborhood size was calculated using 20 principal components. The ‘nExP’ value was set to expected doublet rate according to Chromium Single Cell 3′ v2 Reagent Kit User Guide (10X Genomics). These parameters were used as input to the ‘doubletFinder_v3’ function to identify doublet cells.
For determining cell type, clusters were annotated based on cell type-specific marker genes as indicated below. Among our tumor biopsies, we had appendiceal carcinomas that are epithelial in origin and lymphomas that are B cell derived. For the appendiceal cancers, we identified epithelial cells (EPCAM, TFF3, MUC2), fibroblasts (DCN, COL1A1, LUM), endothelial cells (VWF, PLVAP, PECAM1), T cells (CD3D, IL7R, CD8A), natural killer (NK) cells (NKG7, GNLY), B or plasma cells (MS4A1, CD79A), mast cells (TPSAB1) and macrophages or dendritic cell lineages (CD68, CD14, FCGR3A, HLA-DRA).
For the lymphoma samples, we included MS4A1, CD19, CD79A (B cells), CD3E, CD3D, CD2 (T cells), CD8A, CD8B (CD8+ T cells), CD4 (CD4+ T cells), LEF1, CCR7, NOSIP (naïve T cells), IL7R, SELL (memory T cells), CD4, IL2RA, FOXP3 (T regulatory cells), GZMA, NKG7 (T effector cells), GNLY, NCAM1 (NKT/NK cells), and CD14 and LYZ (myeloid cells). The classification of malignant versus nonmalignant B cells was based on calculating the average expression of each kappa and lambda variable region gene for the different clusters (15). The expression of a clonal light chain provided assignment for the malignant B-cell clusters. In contrast, the normal B-cell cluster expressed heterogeneous BCR light chain variable genes.
Adaptive long-read processing
The adaptive sequencing runs from the Oxford Nanopore and their sequence output were filtered to include just the reads within one of the targeted regions, per the gene panel bed file. This step involved using the log file provided by the sequencer. The log file indicates whether each read was ejected (‘unblock’) or accepted (‘stop_receiving’—enriched). The accepted reads, which contain full-length cDNA, were bioinformatically selected using the fast5_subset command from the ont_fast5_api package. These data were iteratively processed using the ‘super-accuracy’ base calling mode with Guppy (v5.0.16) and were aligned to the reference genome GRCh38 using minimap2 (v2.22) (16). To infer the presence of a full-length cDNA transcript, the first 65 bases of soft-clipped sequence closest to the aligned bases were evaluated for A or T homopolymers (depending on the orientation of the alignment). A homopolymer of length ≥12 was assessed as being a polyA tail, indicative of a full-length transcript.
Integration of short and long reads from single-cell cDNA
As previously described, we developed a method to match the short and long reads from overlapping single cells (6). The Cell Ranger processing of short reads provides a list of cell barcodes. We compared this whitelist of known barcodes to barcode sequences extracted from the soft-clipped sequences in the aligned long reads. The Python pysam module was used to identify soft-clipped portions of aligned reads. The next step was a machine learning approach utilizing a cosine similarity function (CountVectorizer from scikit-learn Python module, with k-mer length of 8) to identify potential barcode matches within the soft-clipped sequences. Using the five highest ranking cosine similarity scores per read, the edit distance between the long-read barcode sequence and the whitelisted barcode was calculated. Barcode matches with the lowest edit distance were selected. Then, the highest cosine similarity score was selected for final evaluation. If the paired barcode edit distance was <3, it was considered a successful match, otherwise the read was not considered a match to any of the barcodes identified in short-read sequencing and was excluded from further integrated analysis. From the resulting file, any exactly matched barcode/UMI combinations were removed as PCR duplicates.
CRISPR genotyping analysis
Using the long-read data from targeted cDNAs, we identified the genotypes of CRISPR mutations from the Jurkat cell line. After aligning each nanopore read and confirming the coordinates of the target, we evaluated a 2-bp sliding window that was tiled across the putative cleavage site. Insertions, deletions or base substitutions were identified among the long reads. We performed this analysis for each gRNA target per given cell and summarized the mutation frequency of the CRISPR target.
CRISPR-induced exon skipping analysis
For each read, we used the exon coordinates for the SRSF5 gene, to determine which exons were present in the long-read transcript (6). Exon coordinates were based on the Ensembl canonical transcripts from the GENCODE version 38 GTF file (17). Transcripts that began at exon 1 and included exon 5 were evaluated and considered to have skipped exon 4 if <12 bases were aligned to that exon.
Single-cell analysis of cancer mutations from tumor biopsies
We had a set of tumor samples originating from patients with metastatic cancer. These patients had one of their tumor sites undergo diagnostic cancer genome sequencing. The clinical sequencing reports provided a list of mutations that led to amino acid changes, frameshifts or premature stops—this information was compiled for our study. For mutations reported in GRCh37 coordinates, we conducted a liftover procedure to convert to GRCh38 coordinates. For mutations reported as amino acid changes, we conducted an analysis with the CADD application to lift these mutations to the GRCh38 reference coordinates (18). We used the pileup command from the Python pysam module to identify the specific nanopore reads that had the reported mutations (19,20). As an additional validation of the tumor mutation calls, we used Longshot to call variants (21). Longshot is designed for germline variant calling of long reads, so some parameters were adjusted to provide more sensitive variant calling appropriate for somatic mutations. Longshot was run with variant phasing disabled, a strand bias P-value cutoff of 0.0001 and variant density filter set to 10:100:50 (10 variants within 100 bp with genotype quality ≥50) to filter out any variants in a very dense cluster. The cell barcode for each long read was identified as described in the ‘Adaptive long-read processing’ section, and using this information the cells in the short-read Seurat object were annotated as having either reference or alternate base values. Standard Seurat functions such as DimPlot and VlnPlot were then used to visualize the differences in cell type distribution and gene expression level, for those cells with the variant versus the wild type.
To determine whether there were rearrangements, we used cuteSV (22). The following parameters were applied: maximum distance to cluster reads together for insertion or deletion: 100; maximum base pair identity to merge breakpoints for insertion or deletion: 0.3.
RESULTS
Overview of the approach
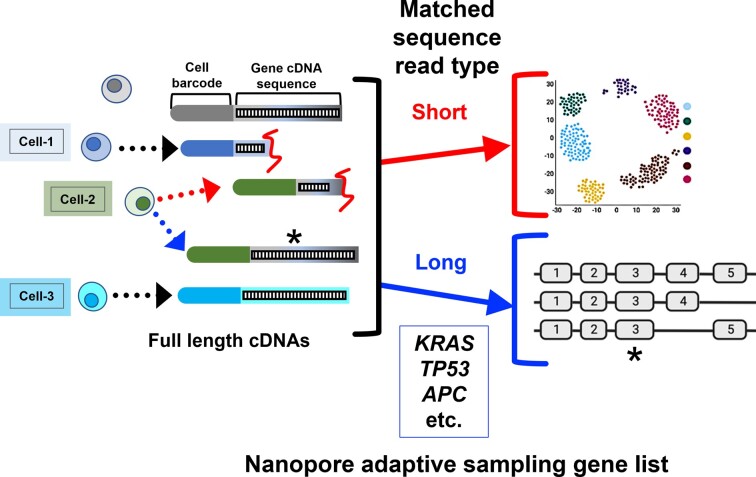
We determined whether one could apply nanopore adaptive sampling for single-cell genotyping of somatic genetic variants and identification of rearrangements (Figure 1A). For this study, we used a cancer cell line and several different tumors across different anatomic sites. These tumors had prior targeted deep sequencing results from diagnostic DNA testing; we then checked for these same results in the long-read cDNA transcripts from single cells.
Figure 1.
An adaptive sampling method for sequencing target cDNAs from scRNA-seq. (A) Overview of single-cell library preparation, long- and short-read sequencing analysis, and integration of results from both modalities. Integrative Genomics Viewer (IGV) screenshot of SRSF5 targeting sites from cells with gRNAs: (B) SRSF5-1 and (C) SRSF5-2. (D) Boxplot showing CRISPR-induced mutation rate for all genes targeted.
This scRNA-seq genotyping approach involved the following steps: (i) We generated single-cell cDNAs (10X Genomics) from the sample (see the ‘Materials and Methods’ section). (ii) A portion of the single-cell cDNA underwent library preparation for conventional Illumina short-read sequencing that requires fragmenting the cDNA. (iii) A proportion of the same single-cell cDNA library, without fragmentation, was prepared for Oxford Nanopore sequencing. (iv) Adaptive sampling was used to target the cDNA of specific genes. (v) The long reads were pre-processed, aligned and evaluated for CRISPR edits, cancer mutations and rearrangements among the individual cells. (vi) The presence of a variant was identified by direct examination of the altered genomic position among the sequence reads and also by variant calling on the long-read data (see the ‘Materials and Methods’ section). (vii) To infer cell type, we integrated the scRNA-seq short- and long-read data by matching the cell barcodes. This step allowed us to assign each mutation to specific cell types, which is an important step for analyzing primary tumor samples. The long-read coding variants matched those identified from deep targeted diagnostic sequencing with the exception of genes with low expression and lacked sufficient sequence coverage.
Single-cell mutation mapping of a cancer cell line
We first analyzed the Jurkat cell line that is derived from a T-cell leukemia. This cell line has undergone prior genome sequencing with reported mutations (23). The cells were grown without any CRISPR genome modifications and then underwent single-cell cDNA preparation and cDNA amplification. As noted above, the same library was split into two aliquots and used for both short- and long-read sequencing.
We evaluated the accuracy of variant calling from long-read sequencing of the targeted gene set by investigating the Jurkat cell line and using the matched short-read sequencing as the ground truth reference. To create a confident ground truth reference, we used only those genes for which there were at least 100 short-read transcripts and called the variant if the short-read had a variant frequency of >1%. Two hundred forty-nine genes had sufficient short-read transcripts (range 107–8052 reads) and a high percentage had consistent mutation calling between long reads and short reads.
We demonstrated an overall accuracy of 95.18% with 95% confidence interval (91.73%, 97.49%) for long-read variant calls if variants are called when the long-read variant frequency is at least 4%. There were 2 false negative calls and 10 false positive calls. The number of long-read transcripts sequenced per gene ranges from 7 to 6679 and the recall rate is 97.98%. Notably, the false negative rate is controlled under 3% when the number of long-read transcripts per gene is between 7 and 1600 (Supplementary Figure S1).
We used the results from a short-read scRNA-seq analysis of the Jurkat cells to identify individual cells and their gene expression levels (see the ‘Materials and Methods’ section). There were 5881 cells detected, with an average of 38 966 reads per cell (Supplementary Table S1). Next, we evaluated the expression levels of 319 genes that had been previously reported to have mutations in this cell line (Supplementary Table S2) (23). These genes covered a range of different expression levels that were corroborated by both the short- and long-read data (Supplementary Figure S2).
For mutation identification, we used the same single-cell cDNA library for nanopore adaptive sampling with an Oxford MinION sequencer, albeit without fragmenting the cDNA. For targeting, the adaptive sampling list covered 319 gene targets (Supplementary Table S2). After alignment and processing, a total of 5881 cells were recovered from the long-read data (Supplementary Table S1). A total of 1.47 million reads aligned to the target genes, with an average read length of 944 bp. There were an average of 188 long reads per cell representing an average of 88.2 genes per cell. Across all cells, each gene had an average of 3733 reads and 1743 cell barcodes. We compared the cell barcodes between the short- and long-read data. Short-read sequencing identified 5881 cell barcodes of which 5873 (>99%) overlapped with the long-read data.
Overall, 292 of the 351 mutations in targeted genes were shown to be present among the Jurkat cells, representing 83% of the gene-based mutations that have been previously reported. The 58 variants not detected were due to either low coverage or variation in mutations across the cell line. Thirty of the 58 had low coverage (<20 long-read transcripts); 26 had adequate coverage and variant was not detected in either short reads or long reads. The remaining two variants were detected in short reads at <1% frequency but were not detected in the long reads. In total, we identified 910 cells with mutations among 1663 cells. As another general metric for the appearance of a mutation, we determined the transcript allele frequency. This value reflects the ratio of reads identified with the mutation over the total number of reads per cell. The detected variant frequency varied among genes; however, limited conclusions can be made since in general only one transcript was sequenced per cell for a given gene. The C466Y mutation in the TOP1MT gene had a mean variant allele frequency (VAF) of 90.7% with 95% confidence interval (84.6%, 96.8%), meaning most cells had this mutation in at least one allele. In contrast, the VAF for the L142L mutation in the ACAT2 gene was 32.7% with 95% confidence interval (31%, 34.5%), which suggests a lower penetrance for this mutation (Supplementary Table S3 and Supplementary Figure S3).
Single-cell genotyping of somatic CRISPR edits in Jurkat cells
Next, we assessed whether adaptive sampling and targeted sequencing could identify de novo CRISPR-introduced edit mutations from single cells. We used CRISPR-edited Jurkat cells that stably expressed Cas9, as previously described (6). We transduced this Jurkat cell line with a multiplexed gRNA library containing 32 gRNAs targeting 16 genes. There were two guides per gene. We also included five control gRNAs (Supplementary Tables S4 and S5). These transduced cells underwent processing to generate a single-cell cDNA library. As part of the short-read analysis, we identified which gRNAs were expressed within a given individual cell (see the ‘Materials and Methods’ section). This assay relies on using a primer to polymerase extend over the gRNA adjacent to a given cell barcode, followed by sequencing in which both gRNA and cell barcode appear in the same read (24). With the paired gRNA and cell barcode sequence, one determines the distribution of expressed gRNAs across individual cells.
The cells expressing a given gRNA were identified and matched with the long-read adaptive sequencing of single-cell cDNAs. With the targeted long reads, we identified the CRISPR-induced edits among the target gene cDNAs among the single cells that also expressed the specific gRNA (Figure 1B–D and Supplementary Figure S4). We have evaluated CRISPR-induced mutations using both Illumina short-read and nanopore long-read sequencing techniques and demonstrated a high degree of concordance between the two methods (25). This proves that nanopore long-read sequencing can reliably characterize CRISPR mutations. The average number of target long reads matching the gene target was 5.32 per cell. As expected, CRISPR mutations were identified at the target gene site among the cells expressing the gRNA. The average target mutation frequency from cells with the guide was 79.0% and significantly higher than the wild-type cells (P = 2.9e−07) (Figure 1D).
Furthermore, we detected CRISPR-induced transcript isoform alterations at single-cell resolution. For example, the gRNA SRSF5-2 introduced skipping of exon 4 in 14.81% of cells with the gRNA (95% confidence interval: 1.16%, 28.5%) (Figure 1B and C). All cells skipping exon 4 had zero bases aligned to that exon. Of the cells without the gRNA, 99.5% (95% confidence interval: 98.7%, 100%) did not skip exon 4 (Supplementary Figure S5). This result confirms that adaptive long reads were informative for identifying CRISPR edits.
For validation, we PCR amplified the cDNA targets with long-read sequencing. We generated amplicons of the gene targets from the same scRNA-seq library. These amplicons underwent long-read sequencing (6). We then identified the matching cell barcodes between the two datasets. Comparing the adaptive versus the amplicon sequencing, all identified mutations overlapped, further validating the adaptive results (Supplementary Figure S6).
Identifying single-cell mutations from tumors
We applied this adaptive nanopore sampling to identify somatic alterations among single cells from tumors. These samples originated from patient biopsies. We analyzed matched pairs of tumors from three patients with advanced cancer present in different anatomic sites. The first and second patients had metastatic appendiceal cancer. The third patient had follicular lymphoma, a B cell-derived tumor, affecting distinct nodal regions throughout the body.
Every patient had their tumor tested with diagnostic cancer gene sequencing, either from a primary site or from a metastatic site (Supplementary Table S2). From the targeted sequencing of each patient’s cancer, we evaluated the reported coding variants (Table 2). The reports provided all nonsynonymous variants, including those classified as benign, pathogenic or of unknown significance. For this study, we are genotyping the base detected at a given genomic coordinate, and therefore excluded any variants that were insertions, deletions or frameshifts since these tend to have less predictable presentation within the aligned sequence and are also more prone to be affected by nanopore sequencing errors. There were only two such variants excluded across all the samples and we inspected the variant positions in the read data with IGV. One variant was a 1-bp deletion not visible in IGV due to the prevalence of homopolymers around the deletion site and the propensity for this type of indel error in nanopore sequencing. The other variant was a 69-bp splice site deletion that is not directly detectable in cDNA. The exons on either side of the deletion were not present in the gene transcripts.
Each tumor sample underwent single-cell library preparation and the same single-cell libraries were used for both short- and long-read sequencing (see the ‘Materials and Methods’ section). Gene expression profiles from the short-read sequencing revealed cell types. For the genes reported to have mutations via diagnostic sequencing, we used the long-read sequencing to identify base calls at the genomic coordinates of these variants. We also performed de novo variant calling for these same genes using Longshot as described in the ‘Materials and Methods’ section. Subsequently, we matched the cell barcodes between the long- and short-read sequences to integrate gene expression, cell type and mutation status.
Across the six tumor samples, the number of single cells identified by short-read sequencing ranged from 7748 to 16 219 and the median number of genes per cell ranged from 468 to 1468 (Supplementary Table S1). The spread in median genes per cell was attributable to differences in the cell types. Lymphomas are composed of B cells that have a significantly higher number of expressed genes per cell compared to epithelial cells such as those originating from appendiceal cancer. This observation is consistent with what has been noted from single-cell studies of lymphocytes and solid tissues (15,26).
The proportion of full-length transcripts was determined by detection of a polyA tail within the soft-clipped portion of the aligned reads. The median percent of full-length transcripts across all genes in the gene panels for each sample was between 58% and 75%. For most of the clinical diagnostic genes, the percent of full-length transcripts was between 60% and 82%, though several genes expressed in the lymphoma samples were lower at between 42% and 51% (Supplementary Table S6).
We determined the number of unique cells in the adaptive long-read data by matching the cell barcode sequences with the list of cell barcodes identified by Cell Ranger in the short-read data. This ranged between 7732 and 15 786 cells per sample (Supplementary Table S1). The median number of transcripts per cell ranged from 10 to 83 and the average number of target genes per cell ranged between 7.6 and 27.0 (Supplementary Table S1). Overall, the yield of reads was higher from the B-cell lymphoma than from the appendiceal epithelial tumors.
Single-cell mutations among appendiceal cancers
We analyzed a set of tumors from two patients (P8605 and P8629) with appendiceal cancer. This cancer originates from the epithelial cells of the appendix, a vestigial organ connected to the right colon. The target gene list covered 529 genes for P8605 and 330 genes for P8629.
8605’s appendiceal cancer and metastasis
Patient 8605 had an appendiceal carcinoma (T1) and a metastatic site (T2) located in the left ovary (Figure 2A). The patient’s primary tumor site underwent diagnostic cancer sequencing. Based on the clinical report, the T1 tumor had five nonsynonymous substitution variants in the cancer driver genes APC, GNAS, KRAS, KMT2D and POLD1, with only the GNAS and KRAS variants being pathogenic per ClinVar (Table 2) (27).
Figure 2.
Single-cell mutations from the T1 and T2 appendiceal cancers. (A) Location of tumor samples for patient 8605, and variants detected from clinical diagnostic sequencing having sufficient long-read depth for analysis. (B) IGV screenshots for T1 alignments, covering the lengths of KRAS and GNAS genes. (C) UMAP clustered plot showing integration of T1 and T2 samples. (D) UMAP clustered plot annotated with cell types and dot plot showing expression of cell type markers. (E) IGV screenshot of T1 and T2 alignments showing GNAS R844S mutation position, UMAP plot highlighting location of cells with GNAS mutation and violin plot showing relative expression of mutated and wild-type GNAS epithelial cells. (F) IGV screenshot of T1 and T2 alignments showing KRAS G12V mutation position, UMAP plot highlighting location of cells with KRAS mutation and violin plot showing relative expression of mutated and wild-type KRAS epithelial cells.
The primary appendiceal cancer and its matched metastasis underwent scRNA-seq with both short and long reads (Figure 2B–D). The short-read sequencing provided single-cell transcriptome information that informed cell identity, and the sequencing metrics are shown in Supplementary Table S1. Based on short-read sequencing, the T1 appendiceal site had a total of 12 127 cells with an average of 889 genes per cell (Figure 2C). The T2 metastatic site had 14 214 cells with an average of 655 genes per cell (Figure 2C). With this scRNA-seq data, we defined the different cell types in each sample, including epithelial, stromal and immune cells. The canonical genes that defined the epithelial cells included MUC2, TFF3 and EPCAM (Figure 2D).
Using the panel of 529 genes, we generated single-cell long-read sequence data from these target cDNAs. After alignment, we identified the long reads of the target genes that matched cell barcodes occurring in the short-read data (see the ‘Materials and Methods’ section). We analyzed the adaptive long-read data for both tumor sites (Figure 2E and F, and Supplementary Table S1). For the T1 tumor, 67% of long-read transcripts were matched to a short-read cell barcode, resulting in identification of 11 914 cells and 498 of the 529 genes targeted. For the T2 metastasis, 69% of long-read transcripts were matched to a short-read cell barcode with 14 077 cells and 496 of the 529 targeted genes identified. Transcripts not matched to a short-read cell barcode are due to either errors in sequencing of the long-read barcode or the cDNA library sampling process (i.e. cells sampled for short-read sequencing but not long reads, or vice versa). The average number of target genes per cell for the T1 tumor was 11.5 and for the T2 metastasis was 12.8.
Variants in all clinically identified genes were found in the T1 and T2 tumors, albeit with very few cells detected expressing the tumor suppressor genes APC, KMT2D or POLD1 (Table 3). In examining the single-cell short-read data for the T1 tumor, the gene expression levels of KMT2D, POLD1 and APC were generally low with transcript counts between 0.01 and 0.04 per cell (Supplementary Figure S7). The low coverage could be due to loss of function in these genes, or due to just low native expression, and is not determinable with cDNA analysis methods. There is insufficient power to form any conclusion on cell type and mutation status for these variants.
Table 3.
Single-cell identification of cancer mutations
| Epithelial cells | Other cell types | Epithelial cells | Other cell types | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Source ID | Tumor type | Gene | Amino acid change | Tumor ID | Cells w/ transcript coverage | Cells w/ variant | Cells w/ transcript coverage | Cells w/ variant | % Mut | Tumor ID | Cells w/ transcript coverage | Cells w/ variant | Cells w/ transcript coverage | Cells w/ variant | % Mut |
| 8605 | Appendiceal cancer | APC | A1358V | T1 | 0 | 0 | 2 | 1 | 50% | T2 | 0 | 0 | 1 | 1 | 100% |
| KRAS | G12V | 48 | 42 | 218 | 2 | 17% | 1 | 0 | 204 | 2 | 1% | ||||
| KMT2D | V4305I | 1 | 1 | 0 | 0 | 100% | 0 | ||||||||
| POLD1 | V455M | 0 | 0 | 2 | 2 | 100% | 0 | 0 | 2 | 2 | 100% | ||||
| GNAS | R844S | 110 | 76 | 416 | 13 | 17% | 11 | 5 | 494 | 3 | 2% | ||||
| 8629 | Appendiceal cancer | SF3B1 | K700E | T3 | 6 | 0 | 15 | 0 | 0% | T4 | 0 | 0 | 9 | 0 | 0% |
| KRAS | G12D | 62 | 27 | 134 | 3 | 15% | 15 | 5 | 221 | 1 | 3% | ||||
| SMAD2 | S464* | 19 | 17 | 14 | 0 | 52% | 2 | 2 | 31 | 0 | 6% | ||||
| GNAS | R844C | 108 | 0 | 214 | 0 | 0% | 17 | 0 | 330 | 1 | 0% | ||||
| GNAS | R844H | 126 | 59 | 256 | 8 | 18% | 23 | 11 | 406 | 8 | 4% | ||||
| B cells | Other cell types | B cells | Other cell types | ||||||||||||
| Source ID | Tumor type | Gene | Amino acid change | Tumor ID | Cells w/ transcript coverage | Cells w/ variant | Cells w/ transcript coverage | Cells w/ variant | % Mut | Tumor ID | Cells w/ transcript coverage | Cells w/ variant | Cells w/ transcript coverage | Cells w/ variant | % Mut |
| 6408 | B-cell lymphoma | DNMT3A | E30A | T5 | 46 | 19 | 11 | 3 | 39% | T6 | 22 | 13 | 56 | 21 | 44% |
| CREBBP | Y1482H | 96 | 37 | 16 | 1 | 34% | 72 | 34 | 64 | 3 | 27% | ||||
| NF1 | I2681V | 23 | 14 | 6 | 4 | 62% | 18 | 11 | 3 | 2 | 62% | ||||
| BCL2 | S116F | 870 | 831 | 14 | 6 | 95% | 563 | 497 | 57 | 20 | 83% | ||||
| BCL2 | L86F | 839 | 796 | 13 | 6 | 94% | 549 | 482 | 54 | 17 | 83% | ||||
| BCL2 | H58R | 745 | 696 | 11 | 5 | 93% | 499 | 475 | 55 | 19 | 89% | ||||
| BCL2 | E29K | 798 | 711 | 11 | 4 | 88% | 520 | 479 | 54 | 19 | 87% | ||||
| BCL2 | G5V | 845 | 1 | 12 | 0 | 0% | 545 | 1 | 59 | 0 | 0% | ||||
| EP300 | S958G | 51 | 14 | 7 | 4 | 31% | 18 | 9 | 22 | 12 | 53% | ||||
We examined the GNAS R844S mutation in the T1 and T2 tumors—for general visualization, we combined the data from both tumors for UMAP and violin plots (Figure 2E). GNAS is a proto-oncogene that represents the Gsα subunit of heterotrimeric G proteins and is involved in production of cyclic AMP-based signal transduction (28). For the T1 tumor, there were 526 cells with long reads of the GNAS transcript, and short-read transcriptome data (Table 3). Among the 110 epithelial cells with GNAS long-read transcripts covering the variant site, 76 cells had an R844S mutation allele. Most cells only had one GNAS transcript sequenced, and since somatic variants are typically heterozygous it is likely that all or most epithelial cells harbored the mutation. Of the remaining 416 cells that were not classified as epithelial cells, 95% of the GNAS alleles were wild type. The nonepithelial cells with mutated alleles likely indicate either a misclassification of cell type from the short-read data or a false positive variant call.
Next, we evaluated the T2 metastasis for this same GNAS mutation (Figure 2E). There were 505 cells that had long reads of the GNAS transcript and matching short-read transcriptome data (Table 3). We identified 11 epithelial cells, and sampling of ∼1 allele per cell identified five mutation transcripts and six wild-type transcripts, consistent with heterogeneous expression of the mutation in each cell. Of the remaining 494 nonepithelial cells, over 99% of the transcript alleles were GNAS wild type.
We then identified cells in T1 and T2 with the KRAS G12V mutation (Table 3 and Figure 2F). This mutation is a hotspot that enables KRAS activity and acts as an oncogenic driver. For T1, 42 of the 48 epithelial cells with KRAS transcripts had a G12V allele, indicating at least 88% of these cells had the mutation. Only 1 of the 205 T2 cells with KRAS transcripts was an epithelial cell, so no conclusions on presence or absence of mutation can be drawn. For the cells not classified as epithelial, <1% of cells in both T1 and T2 had a G12V allele. The low frequency of observed mutations in nonepithelial cells would be consistent with a false positive variant call. Alternatively, the cells with the mutation may be misclassified epithelial cells.
There was no evidence of mutation-related transcript instability in either GNAS R844S or KRAS G12V epithelial cell transcripts in T1 or T2. Transcripts harboring the GNAS or KRAS mutations had stable gene expression compared to their respective wild-type transcripts (Figure 2E and F).
As validation of positive variant calls, we ran the Longshot program (Supplementary Table S7; see the ‘Materials and Methods’ section). The KRAS and GNAS mutations were called for T1 but were not called for T2 given the low VAF for these genes. No mutations were called for APC, KMT2D and POLD1 due to low read depth in both samples (Table 3).
As targeted negative controls for T1 and T2, we genotyped the variant calls from other patient samples. Coverage ranged from 2 to 45 transcripts per gene, and there were no variants detected in T1, or in six of the seven genes for T2 (Supplementary Table S8). There was one variant transcript out of eight total transcripts for the EP300 gene in T2, indicating a false positive sequencing error.
8629’s appendiceal metastasis
For patient 8629, we had biopsies from two metastatic sites (T3 and T4) of an appendiceal cancer (Figure 3A). These implants were located on the omentum (T3), a tissue covering the abdominal viscera and in the small intestine (T4). Based on the diagnostic tumor sequencing, the primary appendiceal tumor had four genes with substitution variants, including GNAS, KRAS, SF3B1 missense variants and a SMAD2 nonsense mutation, with all variants being pathogenic or likely pathogenic (Table 2). These samples underwent both short- and long-read scRNA-seq (Figure 3B and Supplementary Table S1). Based on the short-read sRNA-seq, the T3 and T4 metastatic sites had over 10 000 cells, and over 400 genes per cell on average (Figure 3C). With this single-cell transcriptome data, we defined the different cell types in each sample, including epithelial, stromal and immune cells (Figure 3D). After applying standard quality control filtering (see the ‘Materials and Methods’ section), the T3 site had 8814 cells of which 1760 were epithelial, 2142 were stromal and 4912 were immune cells. The T4 site had 14 511 cells of which 293 were epithelial, 5863 were stromal and 8355 were immune cells.
Figure 3.
Single-cell mutations from the T3 and T4 appendiceal cancers. (A) Location of tumor samples for patient 8629, and variants detected from clinical diagnostic sequencing having sufficient long-read depth for analysis. (B) IGV screenshots for T3 alignments, covering the length of KRAS and GNAS genes. (C) UMAP clustered plot showing integration with T3 and T4 samples. (D) UMAP clustered plot annotated with cell types and dot plot showing expression of cell type markers. (E) IGV screenshot of T3 and T4 alignments showing KRAS G12V mutation position, UMAP plot highlighting location of cells with KRAS mutation and violin plot showing relative expression of mutated and wild-type KRAS epithelial cells. (F) IGV screenshot of T3 and T4 alignments showing GNAS R844C and R844H mutation positions, UMAP plot highlighting location of cells with GNAS R844H mutation and violin plot showing relative expression of mutated and wild-type GNAS R844H epithelial cells.
We analyzed the long-read data for both sites (Supplementary Table S1). The target list consisted of 330 genes (Supplementary Table S2). For the T3 tumor, 59% of the long-read barcodes matched a short-read barcode. These data define a set of 9929 cells with long-read coverage for 312 of the 330 genes targeted. For the T4 site, 70% of long reads matched a short-read barcode resulting in 15 786 cells, with long-read coverage for 319 of the 330 genes targeted. The average number of target genes per cell for T3 tumor was 8.4 and for the T4 metastasis was 7.6.
There were long reads covering the coding mutation sites for GNAS, KRAS, SMAD2 and SF3B (Table 3). For the T3 and T4 metastases, the KRAS G12D mutation was the most prevalent. The combined single-cell data for this mutation are shown in the UMAP and violin plots (Figure 3E). KRAS G12D is a common hotspot mutation found among colon and appendiceal cancers and is a critical oncogenic driver. One hundred ninety-six T3 cells and 236 T4 cells had matching long- and short-read transcriptome data for KRAS (Table 3). The G12D allele was detected in 27 of 62 epithelial cells in T3, and 10 of 15 epithelial cells in T4, suggesting that most or all of the epithelial cells harbored this mutation. Nonepithelial cells in these samples included T cells, macrophages, dendritic cells and fibroblasts. Only four of the 355 nonepithelial cells across T3 and T4 had transcripts with the G12D mutation.
The SMAD2 S464* truncation was also found in the T3 and T4 samples. This gene is an intracellular signal transducer and transcriptional modulator activated by transforming growth factor beta (29). T3 and T4 each had 33 cells with long reads and matching short-read transcriptome data for SMAD2 (Table 3). The SMAD2 S464* nonsense mutation was detected in 17 out of 19 epithelial cells in T3 and both of the epithelial cells in T4. All nonepithelial cells in both samples had the SMAD2 wild-type transcript.
Three hundred twenty-two cells in T3 and 347 cells in T4 were identified with transcripts covering the GNAS R844C variant position (Table 3 and Figure 3F). One hundred eight and 17 cells, respectively, were classified as epithelial cells and no R844C mutation was detected in any of the GNAS transcripts. One of the 330 nonepithelial cells in T4 did have a transcript allele with the R844C transcript, and may be a false positive based on the very low frequency of this observation and the fact that the cell is not an epithelial cell. However, visualization in IGV did show evidence of a GNAS R844H mutation, which after genotyping was detected in 59 of 126 epithelial cell transcripts in T3 and 11 of 23 epithelial cell transcripts in T4 (Figure 3F). This finding was notable since it was consistent between the two independent samples for this patient and suggests a miscall in the original clinical sequencing report. The mutation found in long reads was 1 bp away from the clinical sequencing call, and in the same codon. There were no R844C mutations found in T3, and one found in the TME in T4. In contrast, ∼50% of the epithelial cells in both T3 and T4 had the R844H mutation per long-read sequencing, indicating this is likely to be a true positive call. In nonepithelial cells, the R844H mutation was detected in 3% and 2% of cells in T3 and T4, respectively, which is within the range of expected sequencing error, or may be misclassification of cell type.
SF3B1 transcripts were detected in 21 cells in T3 and 9 cells in T4, and no transcripts had the K700E mutation. A loss-of-function variant in the tumor suppressor gene SF3B1 would not be detectable by this method since no mRNA would be transcribed, and so could account for this finding. Alternatively, this gene may not be highly expressed and therefore there is insufficient power for detection of mutations.
There was no evidence of mutation-related transcript instability in either the GNAS R844H or KRAS G12D epithelial cell transcripts in T3 or T4. Transcripts harboring the GNAS or KRAS mutations had stable gene expression compared to their respective wild-type transcripts (Figure 3E and F).
We applied the Longshot variant caller (see the ‘Materials and Methods’ section) as a means of cross-validating variants (Supplementary Table S7). For the T3 metastasis, the KRAS G12D mutation was not identified, but the SMAD2 S464* truncation was called. Of note, Longshot did not identify the GNAS R844C but did identify the GNAS R844H mutation, consistent with our genotyping analysis. For the T4 metastasis, the mutations were not detected by Longshot due to low gene coverage. The Longshot caller is not designed to identify variants present at low allelic fractions.
As targeted negative controls for T3 and T4, we genotyped the variant calls from other patient samples. Coverage ranged from 0 to 22 transcripts per gene. For T3, there were no long-read transcripts for APC, and no variants detected in any of the other seven genes (Supplementary Table S8). For T4, one of the three APC transcripts had the variant, no transcripts were found for POLD1 and no variants were detected in the other six genes.
Single-cell mutations from a multifocal B-cell lymphoma
For patient 6408, we analyzed follicular lymphoma samples from two distinct nodal tumor sites (T5 and T6). This type of lymphoma is derived from germinal center B cells and affects the lymphatic system, commonly enlarging the affected lymph nodes. Most patients present with multifocal disease involving multiple lymph node regions. The T5 tumor sample came from a right inguinal lymph node in the groin and T6 tumor sample from a right cervical lymph node located in the neck (Figure 4A). The diagnostic sequencing was conducted on a distinct, third lymphoma site from the right axillary lymph node. Coding variants were reported in five genes that included BCL2, CREBBP, DNMT3A, EP300 and NF1 (Table 2). Only the BCL2 E29K and CREBBP Y1482H variants are predicted to be deleterious. BCL2, CREBBP and EP300 are known to be recurrently altered in follicular lymphoma (30).
Figure 4.
Single-cell mutations from the T5 and T6 B-cell lymphomas. (A) Location of tumor samples for patient 6408 and location of biopsy taken for clinical diagnostic sequencing, plus mutations detected from targeted sequencing having sufficient long-read depth for analysis. (B) IGV screenshots for T5 alignments, covering the length of CREBBP and BCL2 genes. (C) UMAP clustered plot showing integration of T5 and T6 samples. (D) UMAP clustered plot annotated with cell types and dot plot showing expression of cell type markers. (E) IGV screenshot of T5 and T6 alignments showing BCL2 mutation positions, UMAP plot highlighting location of cells with BCL2 S116F mutation and violin plot showing relative expression of mutated and wild-type BCL2 S116F B cells, with an asterisk indicating significant difference in expression (adjusted P-value <0.05). (F) IGV screenshot of T5 and T6 alignments showing CREBBP Y1482H mutation position, UMAP plot highlighting location of cells with CREBBP mutation and violin plot showing relative expression of mutated and wild-type CREBBP B cells.
For this analysis, the two lymphoma tumor samples underwent both adaptive long- and short-read scRNA-seq (Figure 4A and B, and Supplementary Table S1). Based on the short-read sequencing, the T5 site had a total of 7748 cells with an average of 1468 genes per cell (Figure 4C). The T6 site had 11 865 cells with an average of 1438 genes per cell (Figure 4C). Tumor B cells were identified by restricted expression of the immunoglobulin chains as well as transcriptional phenotypes, as we have previously published (15). These tumor B cells clustered separately from the macrophages, NK and other T cells (Figure 4D).
For adaptive sampling of the lymphomas, the target list consisted of 161 genes involved in blood-based malignancies (Supplementary Table S2). We analyzed the adaptive long-read data for both tumor sites (Supplementary Table S1). After matching the cell barcodes between the long- and short-read data, the T5 tumor had 7732 cells, while the T6 tumor had 11 835 cells. Among the 161 targeted genes, we identified expression of 154 and 155 genes for T5 and T6, respectively. The average number of target genes per cell for the T5 right inguinal lymph node was 27 and for the T6 right cervical node was 22. Mutations in BCL2, CREBBP, DNMT3A, EP300 and NF1 were detected among single cells of these tumors (Table 3 and Figure 4E and F). The relative number of tumor cells with a mutation in each sample was similar across the different genes and across the two sites.
Mutations in BCL2 were the most prevalent among single cells across both tumor sites (Table 3 and Figure 4E). BCL2 inhibits apoptosis and its overexpression prevents cancer cell death (30). BCL2 is typically overexpressed in follicular lymphoma due to a hallmark t(14;18)(q32;q21) IGH/BCL2 translocation. This translocation involves the BCL2 gene on chromosome 18 to the IGH (immunoglobulin heavy chain gene) on chromosome 14, bringing BCL2 close to the potent enhancer sequences of the IGH gene and driving BCL2 overexpression (30). In the presence of this translocation, BCL2 is also a target of somatic hypermutation. This high mutation rate is a result of activation-induced cytidine deaminase activity that alters cytosine in DNA, resulting in mutation-inducing repair processes. The cluster of BCL2 variants in both T5 and T6 occurred in a hotspot and were all phased, meaning they were ordered in tandem on the same molecule, as observed in another recent study (30). This cluster thus represents a somatic mutation haplotype. Of the five BCL2 variants reported from the targeted sequencing of the third tumor site, our adaptive sampling results confirmed four, with between 87% and 94% of cells harboring each of the four mutations. The fifth BCL2 variant was observed in only 1 of 844 cells spanning that genomic location and is likely a false positive sequencing error.
There were a small number of cells classified as T cells or macrophages in short-read analysis, which expressed BCL2 and had transcripts harboring one or more of the variants (Table 3). This could be the result of cell type misclassification, an error in the barcode matching process, library artifacts or sequencing error. For barcode matching, errors between short and long reads may be possible since an edit distance of 2 between short- and long-read barcodes is allowed due to the nanopore sequencing error rate. Manual examination of the six cells in T5 with S116F variant transcripts showed that one cell was a misclassified B cell, and four of the other cells had long-read transcripts that were library artifacts. The BCL2 transcripts from these four cells presented as a concatenation of transcripts from two different cells but were not detected as doublets by DoubletFinder. No determination could be made regarding the other cell that expressed T-cell canonical markers and could be a false positive sequencing error.
In the T5 tumor, a CREBBP transcript was detected in 112 cells (Table 3 and Figure 4F). The Y1482H variant was found among 37 of 96 tumor B cells and in 1 of 16 cells not assigned to the tumor cell type. The EP300 S958G, DNMT3A E30A and NF1 I2681V variants were found at between 31% and 53% frequency among the tumor B cells (Table 3). As noted, the T6 tumor had a variant pattern like the T5 tumor albeit with fewer cells, and in general a similar or slightly lower percentage of mutation-bearing cells (Table 3).
There was no evidence of mutation-related transcript instability in CREBBP Y1482H B-cell transcripts in T5 or T6. Transcripts harboring the CREBBP variant had stable gene expression compared to the wild-type transcript (Figure 4E). BCL2 gene expression in tumor B cells was moderately higher in transcripts harboring the S116F variant compared to the wild-type transcript, with P-value = 0.011 using the Welch two-sample t-test (Figure 4F).
We used Longshot to cross-validate variants from the long-read data. Four of the five BCL2 variants were called in the T5 lesion, as well as the variants in DNMT3A, CREBBP, NF1 and EP300. Read depth at the fifth BCL2 variant position was high as with the other four variants. However, there was no variant present at this position. Since the diagnostic sequencing was done on a third lesion, this suggests that the third lesion arose later than T5 and T6 and acquired an additional variant in the hypermutated BCL2 region. In the region between the first and fourth BCL2 variants, three other variants were called by Longshot and supported by visual inspection of the reads (Figure 5A). This result is consistent with somatic hypermutation events in BCL2. The positive and negative calls for the T6 diagnostic variants were identical to T5: four of the five BCL2 variants were called, plus the variants in in DNMT3A, CREBBP, NF1 and EP300 (Table 3). In contrast to T5, only two of the three additional BCL2 variants were found. The variant at chr18:63318573 that occurs at ∼50% frequency in T5 was not present in T6, suggesting that the T6 lesion arose prior to T5.
Figure 5.
(A) IGV screenshots of T5 and T6 lymphomas and locations of BCL2 variants called by Longshot. Coding mutations are labeled in gray and additional variants detected by Longshot are labeled in blue. (B) Schematic of translocation detected by cuteSV. An IGV screenshot showing primary alignments to IGH-D2 on chromosome 14 with soft-clipped sequence to the right, plus secondary alignments of the same reads to a region downstream of BCL2 3′ UTR on chromosome 18.
As targeted negative controls for T5 and T6, we genotyped the variant calls from other patient samples. There were no transcripts for APC, KMT2D, POLD1 or SMAD2 in either sample. Across both samples, there were 386 SF3B1 transcripts, with 1 (0.3%) harboring the variant, and 2250 KRAS transcripts, with 5 (0.2%) harboring the variant. GNAS was highly expressed in both samples with variant transcripts occurring at a frequency between 0.9% and 1.4% (Supplementary Table S8), which is consistent with expected false positive rate due to sequencing error.
Identification of a translocation among single cells in lymphoma
Finally, we used the cuteSV program to call structural variants from T5 and T6 (22), since a hallmark IGH/BCL2 translocation is frequently found in follicular lymphoma. An IGH/BCL2 rearrangement was identified in both the T5 and T6 tumors and shared the same breakpoints in both (Figure 5B). The breakpoints were in IGH-D2 and ∼5 kb downstream from BCL2 3′ UTR, both typical for this translocation (31). We determined that multiple long reads supported the rearrangement. Interestingly, the mutated BCL2 allele was more highly expressed than the wild-type allele, which is consistent with the IGH:BCL2 translocation driving overexpression of this gene. This result represents the first demonstration where single-cell sequencing reveals the presence of a rearranged chimeric transcript.
DISCUSSION
This proof-of-concept study demonstrates a new approach for single-cell identification of cancer mutations. This method integrates nanopore adaptive sequencing and scRNA-seq in order to identify cell type-specific somatic mutations. With the adaptive sampling feature of Oxford Nanopore’s sequencer, one selects specific target cDNAs, derived from mRNAs, based on a list of gene coordinates. The most extensive gene list in our study consisted of 529 genes. Adaptive sampling enabled these targets to be sequenced with an enriched number of reads compared to the remainder of the cDNA population. The nanopore long reads cover the entire cDNA molecule, which enables comprehensive determination of coding mutations present in the mRNA sequence. The same single-cell cDNA library is also subjected to conventional short-read sequencing, which provides the transcriptome features of the same cells. By matching cell barcodes, the long- and short-read data are integrated, thus providing both full-length mRNA sequence features and single-cell gene transcriptomes.
We tested this method on single-cell cDNA libraries obtained from a cancer cell line and tumor biopsies. For the Jurkat cell line, our approach enabled direct identification of CRISPR edits, allowing screening for CRISPR genotypes occurring within coding regions, as well as transcript isoform changes within noncoding regions. For tumor biopsies, previous diagnostic sequencing of the tumor samples identified substitution variants in coding regions of cancer-associated genes. Our method successfully detected nearly all of these coding variants in the single cells from the same samples. We also identified a translocation resulting in a chimeric transcript in two tumor sites from a single patient, highlighting the potential of our approach to identify gene chimeras resulting from rearrangements.
One useful aspect of integrating long-read mutation calling with conventional short-read scRNA-seq is the potential improvement in calling cell types. For example, in the appendiceal cancers, we identified somatic mutations among cells that did not fall within the classified epithelial clusters. However, these mutation-bearing cells were likely to be cancer epithelial cells. Therefore, integrating long- and short-read scRNA-seq with the addition of somatic variants may improve classification of certain cell types in cancer.
Targeted scRNA-seq with adaptive sampling offers several advantages over whole transcriptome sequencing. First, it is more cost-effective while providing higher read coverage. Second, the simplified library preparation workflow eliminates the need for prior enrichment steps as the cDNA molecules are selected for sequencing based on their sequence properties. Furthermore, this method does not require cDNA fragmentation since there is no inherent limit on the length of the molecule being sequenced. For CRISPR edits, the direct detection of genotypes is advantageous since the genotype resulting from CRISPR engineering in a single cell can vary: cells may not be edited at all, or there may be several potential genotypes resulting from the edit. In contrast to other methods that rely on the presence of the single-guide RNA sequence in a cell to infer CRISPR-induced variants, this method detects the actual resulting genotype. As CRISPR is increasingly used in various applications, the direct genotyping among single cells offered by our approach may prove valuable.
This study identified specific issues of adaptive sampling for identifying transcript-based mutations with scRNA-seq. Because the sampling depends on the intrinsic expression levels of a given mRNA, transcripts with low expression provide fewer molecules for sequencing. When analyzing single cells, the transcript yield is already low. Therefore, some transcripts with low expression are missed, which reduces their single-cell representation and leads to a loss of sensitivity in detecting mutations in these low-abundance transcripts. One approach to overcome this limitation involves enriching and amplifying the target genes from a single-cell cDNA library. Our future work will involve integrating adaptive nanopore sampling and single cDNA targeted amplification.
Additionally, since this method relies on sequencing mRNA transcripts, it cannot directly detect loss-of-function variants that trigger nonsense-mediated decay (NMD), such as certain frameshift or nonsense variants. Inferring NMD based on expression level is also challenging due to the inherently variable expression level across transcripts. Therefore, short-read analysis comparing the gene expression level of samples with known mutations to those with wild-type transcript sequences will generally be a more effective way to infer the presence of NMD in modified transcripts.
There are many potential applications for this approach. For example, one could identify the specific set of mutations that define the subclonal populations of a tumor. This type of analysis may prove useful in the study of other diseases beyond cancer. For example, clonal hematopoiesis of indeterminate potential involves hematopoietic stem cells that have genetically distinct subpopulations defined by the presence of somatic mutations. This approach provides a way to determine which cell types account for mutations with low allelic fractions that were identified with bulk genomic DNA sequencing. As we demonstrated, this approach can also identify gene fusions and may provide a new way of screening cancers for rearrangements. As we have described in our previous work (6), targeted sequencing of specific cDNAs provides detailed information about transcript isoforms that play a key role in regulating cell terminal differentiation. Thus, one could have integrated long- and short-read analysis to define the associations between alternative isoforms and specific cell types.
In summary, our study has introduced a powerful single-cell sequencing and analysis approach that enables the identification of somatic mutations in mRNA coding regions and their association with specific cell types. Our method employs nanopore adaptive sampling of single-cell cDNA libraries for the detection of CRISPR edits, gene rearrangements and somatic mutations with high accuracy and resolution. By combining the genotype information with single-cell gene expression data, we can pinpoint which cells and cell types harbor these genetic alterations. This method has broad applications particularly in the identification of specific subclonal populations of tumors and in other diseases where clonal subpopulations are involved. Our approach opens new avenues for exploring the complex interplay between genetic alterations and cellular phenotypes at the single-cell level, providing deeper insights into the fundamental mechanisms underlying cellular function and disease pathogenesis.
DATA AVAILABILITY
The sequencing data for the Jurkat cell line have been deposited in the NCBI Sequence Read Archive (SRA) database under the accession number PRJNA708300 (23). Tumor sequencing data is available from the NCBI dbGaP (phs002188.v3 (pending release) and phs001818.v4). Scripts for analysis are publicly available on GitHub (https://github.com/sgtc-stanford/scCRISPR) (24) and Zenodo (https://zenodo.org/badge/latestdoi/365008149) (25) under MIT license.
Supplementary Material
ACKNOWLEDGEMENTS
Authors’ contributions: S.M.G., H.S.K., S.R., X.B., B.T.L. and H.P.J. were involved in conception and design of the study, development of methodology, acquisition of sequencing data, analysis and interpretation of data, and writing of the manuscript. A.F.A-N. and C.I.A. managed the acquisition of appendiceal samples, mutation information and sequencing. S.H., T.S. and R.L. provided lymphoma samples, mutation information and single-cell libraries. S.M.G., H.S.K., S.R. and H.P.J. wrote the manuscript. H.P.J. oversaw all aspects of the study.
Contributor Information
Susan M Grimes, Division of Oncology, Department of Medicine, Stanford University School of Medicine, Stanford, CA 94305, USA.
Heon Seok Kim, Division of Oncology, Department of Medicine, Stanford University School of Medicine, Stanford, CA 94305, USA.
Sharmili Roy, Division of Oncology, Department of Medicine, Stanford University School of Medicine, Stanford, CA 94305, USA.
Anuja Sathe, Division of Oncology, Department of Medicine, Stanford University School of Medicine, Stanford, CA 94305, USA.
Carlos I Ayala, Department of Surgery, Stanford University School of Medicine, Stanford, CA 94305, USA.
Xiangqi Bai, Division of Oncology, Department of Medicine, Stanford University School of Medicine, Stanford, CA 94305, USA.
Alison F Almeda-Notestine, Division of Oncology, Department of Medicine, Stanford University School of Medicine, Stanford, CA 94305, USA.
Sarah Haebe, Division of Oncology, Department of Medicine, Stanford University School of Medicine, Stanford, CA 94305, USA.
Tanaya Shree, Division of Oncology, Department of Medicine, Stanford University School of Medicine, Stanford, CA 94305, USA.
Ronald Levy, Division of Oncology, Department of Medicine, Stanford University School of Medicine, Stanford, CA 94305, USA.
Billy T Lau, Division of Oncology, Department of Medicine, Stanford University School of Medicine, Stanford, CA 94305, USA.
Hanlee P Ji, Division of Oncology, Department of Medicine, Stanford University School of Medicine, Stanford, CA 94305, USA; Department of Engineering, Stanford University, Stanford, CA 94305, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Cancer Online.
FUNDING
National Institutes of Health [R33 CA247700 to H.P.J., H.S.K., B.T.L. and S.R., R01HG006137 to H.P.J. and H.S.K., R35HG011292-01 to B.T.L. and H.S.K., R35 CA197353 to R.L., 5T32HL120824-04 to T.S., K08CA252637 to T.S.]; Clayville Foundation (to H.P.J.); Leukemia & Lymphoma Society [TRP 6539-18 to R.L.]; Hoogland Lymphoma Research Fund(to R.L.); American Cancer Society [PF-17-239-01-LIB to T.S.]; Deutsche Krebshilfe [70113507 to S.H.]; Don and Ruth Seiler Fund (to C.I.A. and H.P.J.).
Conflict of interest statement. None declared.
REFERENCES
- 1. Arzalluz-Luque A., Conesa A. Single-cell RNAseq for the study of isoforms—how is that possible?. Genome Biol. 2018; 19:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gupta I., Collier P.G., Haase B., Mahfouz A., Joglekar A., Floyd T., Koopmans F., Barres B., Smit A.B., Sloan S.A. et al. Single-cell isoform RNA sequencing characterizes isoforms in thousands of cerebellar cells. Nat. Biotechnol. 2018; 36:1197–1202. [DOI] [PubMed] [Google Scholar]
- 3. Singh M., Al-Eryani G., Carswell S., Ferguson J.M., Blackburn J., Barton K., Roden D., Luciani F., Giang Phan T., Junankar S. et al. High-throughput targeted long-read single cell sequencing reveals the clonal and transcriptional landscape of lymphocytes. Nat. Commun. 2019; 10:3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lebrigand K., Magnone V., Barbry P., Waldmann R. High throughput error corrected Nanopore single cell transcriptome sequencing. Nat. Commun. 2020; 11:4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tian L., Jabbari J.S., Thijssen R., Gouil Q., Amarasinghe S.L., Voogd O., Kariyawasam H., Du M.R.M., Schuster J., Wang C. et al. Comprehensive characterization of single-cell full-length isoforms in human and mouse with long-read sequencing. Genome Biol. 2021; 22:310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kim H.S., Grimes S.M., Hooker A.C., Lau B.T., Ji H.P. Single-cell characterization of CRISPR-modified transcript isoforms with nanopore sequencing. Genome Biol. 2021; 22:331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Loose M., Malla S., Stout M. Real-time selective sequencing using nanopore technology. Nat. Methods. 2016; 13:751–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Baslan T., Kovaka S., Sedlazeck F.J., Zhang Y., Wappel R., Tian S., Lowe S.W., Goodwin S., Schatz M.C. High resolution copy number inference in cancer using short-molecule nanopore sequencing. Nucleic Acids Res. 2021; 49:e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Miller D.E., Sulovari A., Wang T., Loucks H., Hoekzema K., Munson K.M., Lewis A.P., Fuerte E.P.A., Paschal C.R., Walsh T. et al. Targeted long-read sequencing identifies missing disease-causing variation. Am. J. Hum. Genet. 2021; 108:1436–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Payne A., Holmes N., Clarke T., Munro R., Debebe B.J., Loose M. Readfish enables targeted nanopore sequencing of gigabase-sized genomes. Nat. Biotechnol. 2021; 39:442–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Haebe S., Shree T., Sathe A., Day G., Czerwinski D.K., Grimes S.M., Lee H., Binkley M.S., Long S.R., Martin B. et al. Single-cell analysis can define distinct evolution of tumor sites in follicular lymphoma. Blood. 2021; 137:2869–2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018; 36:411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hafemeister C., Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019; 20:296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McGinnis C.S., Murrow L.M., Gartner Z.J. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 2019; 8:329–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Andor N., Simonds E.F., Czerwinski D.K., Chen J., Grimes S.M., Wood-Bouwens C., Zheng G.X.Y., Kubit M.A., Greer S., Weiss W.A. et al. Single-cell RNA-seq of follicular lymphoma reveals malignant B-cell types and coexpression of T-cell immune checkpoints. Blood. 2019; 133:1119–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018; 34:3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Frankish A., Diekhans M., Jungreis I., Lagarde J., Loveland J.E., Mudge J.M., Sisu C., Wright J.C., Armstrong J., Barnes I. et al. GENCODE 2021. Nucleic Acids Res. 2021; 49:D916–D923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rentzsch P., Witten D., Cooper G.M., Shendure J., Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019; 47:D886–D894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bonfield J.K., Marshall J., Danecek P., Li H., Ohan V., Whitwham A., Keane T., Davies R.M. HTSlib: C library for reading/writing high-throughput sequencing data. Gigascience. 2021; 10:giab007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. 1000 Genome Project Data Processing Subgroup Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Edge P., Bansal V. Longshot enables accurate variant calling in diploid genomes from single-molecule long read sequencing. Nat. Commun. 2019; 10:4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jiang T., Liu Y., Jiang Y., Li J., Gao Y., Cui Z., Liu Y., Liu B., Wang Y. Long-read-based human genomic structural variation detection with cuteSV. Genome Biol. 2020; 21:189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tate J.G., Bamford S., Jubb H.C., Sondka Z., Beare D.M., Bindal N., Boutselakis H., Cole C.G., Creatore C., Dawson E. et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019; 47:D941–D947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Replogle J.M., Norman T.M., Xu A., Hussmann J.A., Chen J., Cogan J.Z., Meer E.J., Terry J.M., Riordan D.P., Srinivas N. et al. Combinatorial single-cell CRISPR screens by direct guide RNA capture and targeted sequencing. Nat. Biotechnol. 2020; 38:954–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim H.S., Grimes S.M., Sathe A., Lau B.T., Ji H.P. Single cell CRISPR base editor engineering and transcriptional characterization of cancer mutations. 2022; bioRxiv doi:02 November 2022, preprint: not peer reviewed 10.1101/2022.10.31.514258. [DOI]
- 26. Sathe A., Grimes S.M., Lau B.T., Chen J., Suarez C., Huang R.J., Poultsides G., Ji H.P. Single-cell genomic characterization reveals the cellular reprogramming of the gastric tumor microenvironment. Clin. Cancer Res. 2020; 26:2640–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Landrum M.J., Chitipiralla S., Brown G.R., Chen C., Gu B., Hart J., Hoffman D., Jang W., Kaur K., Liu C. et al. ClinVar: improvements to accessing data. Nucleic Acids Res. 2020; 48:D835–D844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Landis C.A., Masters S.B., Spada A., Pace A.M., Bourne H.R., Vallar L. GTPase inhibiting mutations activate the alpha chain of Gs and stimulate adenylyl cyclase in human pituitary tumours. Nature. 1989; 340:692–696. [DOI] [PubMed] [Google Scholar]
- 29. Yang J., Wahdan-Alaswad R., Danielpour D. Critical role of Smad2 in tumor suppression and transforming growth factor-beta-induced apoptosis of prostate epithelial cells. Cancer Res. 2009; 69:2185–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Patel A.A., Smith S.M. Clinical and biological prognostic factors in follicular lymphoma. Hematol. Oncol. Clin. North Am. 2020; 34:647–662. [DOI] [PubMed] [Google Scholar]
- 31. Vaandrager J.W., Schuuring E., Philippo K., Kluin P.M. V(D)J recombinase-mediated transposition of the BCL2 gene to the IGH locus in follicular lymphoma. Blood. 2000; 96:1947–1952. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data for the Jurkat cell line have been deposited in the NCBI Sequence Read Archive (SRA) database under the accession number PRJNA708300 (23). Tumor sequencing data is available from the NCBI dbGaP (phs002188.v3 (pending release) and phs001818.v4). Scripts for analysis are publicly available on GitHub (https://github.com/sgtc-stanford/scCRISPR) (24) and Zenodo (https://zenodo.org/badge/latestdoi/365008149) (25) under MIT license.