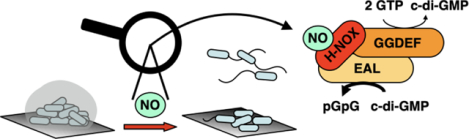
Abstract
Transitions between motile and biofilm lifestyles are highly regulated and fundamental to microbial pathogenesis. H-NOX (heme-nitric oxide/oxygen-binding domain) is a key regulator of bacterial communal behaviors, such as biofilm formation. A predicted bi-functional cyclic di-GMP metabolizing enzyme, composed of diguanylate cyclase (DGC) and phosphodiesterase (PDE) domains (avi_3097), is annotated downstream of an hnoX gene in Agrobacterium vitis S4. Here, we demonstrate that avH-NOX is a nitric oxide (NO)-binding hemoprotein that binds to and regulates the activity of avi_3097 (avHaCE; H-NOX-associated cyclic di-GMP processing enzyme). Kinetic analysis of avHaCE indicates a ~4-fold increase in PDE activity in the presence of NO-bound avH-NOX. Biofilm analysis with crystal violet staining reveals that low concentrations of NO reduce biofilm growth in the wild-type A. vitis S4 strain, but the mutant ΔhnoX strain has no NO phenotype, suggesting that H-NOX is responsible for the NO biofilm phenotype in A. vitis. Together these data indicate that avH-NOX enhances cyclic di-GMP degradation to reduce biofilm formation in response to NO in A. vitis.
Keywords: nitric oxide, NO, cyclic di-GMP, c-di-CMP, biofilm, H-NOX, HaCE, Agrobacterium vitis
ACCESSION IDS: avHaCE: avi_3097, avH-NOX: avi_3098
Graphical Abstract
INTRODUCTION
The transition between a planktonic, motile lifestyle and a biofilm, sessile lifestyle is a highly regulated process that is fundamental to microbial pathogenesis in both animal and plant hosts (1). Bacterial biofilms are bacterial communities encased in a self-secreted extracellular polymeric matrix and usually attached to a surface. They are estimated to be 100- to 1000-fold harder to kill with antibiotics than planktonic cells and responsible for up to 80% of chronic infections (2). Biofilm dispersal results in motile cells that can move on to colonize another host or surface, but also renders the motile cells more susceptible to antibiotics (3). Thus, the drivers of biofilm regulation are relevant targets for prevention and alleviation of biofilm-derived infections and diseases.
A well-studied regulatory protein, H-NOX (heme-nitric oxide/oxygen binding domain) has been shown to have an important role in regulating bacterial communal behaviors, such as biofilm formation and motility, symbiosis, and quorum sensing, by controlling the activity of stand-alone bacterial signaling proteins in response to nitric oxide (NO) (4, 5). NO is a diatomic gaseous signaling molecule in both eukaryotic and prokaryotic organisms (4, 6, 7). There is evidence for NO-mediated cyclic di-GMP regulation of bacterial communal behaviors (8–17), but the molecular basis of these signaling pathways is poorly understood.
The majority of predicted bacterial H-NOX proteins are associated with histidine kinases in two-component signaling pathways, but some are associated with diguanylate cyclases (DGC) and/or cyclic di-GMP phosphodiesterases (PDE). We have named H-NOX-associated cyclic di-GMP synthesis/hydrolysis enzymes HaCEs (8, 12). DGCs synthesize, and PDEs hydrolyze, the bacterial 2nd messenger bis-(3’−5’)-cyclic dimeric guanosine monophosphate (cyclic di-GMP, c-di-GMP). Cyclic di-GMP is correlated with the motility-to-sessility transition in various bacterial species (18), including Agrobacterium tumefaciens (19, 20). At high intracellular levels, cyclic di-GMP promotes biofilm formation and low intracellular cyclic di-GMP levels contribute to biofilm dispersal. It affects motility by targeting pathways involved in the biosynthesis of flagella, pili, extracellular DNA, and polysaccharides (21).
NO/H-NOX regulation of HaCE domains and biofilm formation has been characterized in Shewanella woodyi (12) and Legionella pneumophila (10). In L. pneumophila, lpgHaCE is composed of an enzymatically active DGC domain and an inactive PDE domain; DGC activity is suppressed in response to NO-binding to H-NOX (10). In contrast, in S. woodyi, swHaCE contains enzymatically active DGC and PDE domains, and in response to NO, DGC activity is suppressed and PDE activity activated (12). Although the details of the mechanisms vary, both systems involve NO/H-NOX driven decreases in intracellular cyclic di-GMP concentration and biofilm formation. These observations are consistent with the NO-induced biofilm dispersal phenotype that has been exhibited in many bacterial species (8, 16, 17).
Thus, NO has previously been shown to affect biofilm development and dispersal in numerous species (8, 16, 17, 22) and H-NOX is a well-characterized NO sensing protein (4, 10–13, 15, 23–38), but NO/H-NOX/HaCE systems are not well understood. Here, to gain a deeper understanding of NO/H-NOX/HaCE signaling, we characterize a third system from Agrobacterium vitis, a plant pathogen responsible for the production of crown gall in grapes. We have previously shown that avi_3097 from Agrobacterium vitis strain S4 is a HaCE containing enzymatically active PDE and DGC domains (39). In this study, we use biochemical and genetic characterization to show that avi_3098 is an H-NOX protein that acts as a NO sensor and is responsible for the NO biofilm phenotype in A. vitis. Further, we found that H-NOX and HaCE directly interact and that NO-bound H-NOX upregulates the PDE activity of HaCE in A. vitis. The data presented here reveal a common molecular mechanism for NO signaling pathways involving H-NOX/HaCEs.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and material.
All reagents were purchased in their highest available purity.
All strains and plasmid used in this study are listed in Table 1. Escherichia coli DH5α and BL21(DE3)pLysS strains were routinely used for plasmid amplification and protein purification, and were routinely grown with appropriate antibiotics in Luria broth (LB) at 37°C and 250 rpm or on LB agar plates (10 g/L LB and 10 g/L bacto agar) at 37°C. Antibiotics were used at the following concentrations unless noted, 100 μg/ml ampicillin and 35 μg/mL chloramphenicol.
Table 1.
Bacterial strains, plasmid, and primers used in this study
| Name | Description | Reference or source |
|---|---|---|
| Agrobacterium vitis - Bacterial Strain | ||
| S4 | Wild type | T. J. Burra |
| ΔhnoX | S4 avi_3098::pVIK165, KmR | T. J. Burra |
| ΔhnoX/phnoX | ΔhnoX mutant containing pPZP201-avi_3098, KmR, SpR | T. J. Burra |
| Escherichia coli - Bacterial Strain | ||
| BL21(DE3) pLysS | Expression strain, CmR | |
| DH5 | Plasmid amplification | |
| S17–1/pir | Host Strain for biparental mating | (40) |
| Plasmid | ||
| pET20b | Cloning vector, AmpR | Novagen |
| pET20b-avi_3097 WT | pET20b carrying avi_3097 WT, AmpR | This work |
| pET20b -avi_3098 WT | pET20b carrying avi_3098 WT, AmpR | This work |
| pET20b-avi_3097 GGAAF | pET20b carrying avi_3097 GGAAF, AmpR | This work |
| pET20b-avi_3097 AAL | pET20b carrying avi_3097 AAL, AmpR | (39) |
| pVIK165 | Suicide vector, KmR | (41) |
| pPZP201 | Broad-host range cloning vector, SpR | (42) |
| pPZP201-avi_3098 (phnoX) | pPZP201 carrying avi_3098, SPR | This work |
| Complementation - Primers | ||
| avi_3098 FWD comp | TAGAGGATCCATGAAAGGGATGGTTTTTACGGAAATG | This work |
| avi_3098 REV comp | AGATCTGCAGTCACGATGCGCTCTCGGCGATC | This work |
| Amplification of avH-NOX and avHaCE WT from Agrobacterium vitis S4 genomic DNA - Primers | ||
| avi_3097 WT FWD | GGGCCCGCGCATATGGTAAACCAGCCATTTGAG | (39) |
| avi_3097 WT REV | CCCGGGCGCCTCGAGAGGGCTCTTTCCAGCTGCAAGGCT | (39) |
| avi_3098 WT FWD | GGGCCCGCGCATATGAAAGGGATGGTTTTTACG | This work |
| avi_3098 WT REV | CCCGGGCGCCTCGAGCGATGCGCTCTCGGCGATCAC | This work |
| Site-directed mutagenesis - Primers | ||
| avi_3097 GGAAF FWD | GTGGCACGGCTGGGTGGTGCCGCCTTTGCATTGATTCTCGAC | This work |
| avi_3097 GGAAF REV | GTCGAGAATCAATGCAAAGGCGGCACCACCCAGCCGTGCCAC | This work |
| avi_3097 AAL FWD | CGGATACTGGGGGTGGCCGCCCTGGCACGCTGGCAGCAT | (39) |
| avi_3097 AAL REV | ATGCTGCCAGCGTGCCAGGGCGGCCACCCCAGTATCCG | (39) |
Gift from Thomas Burr at Cornell University, Geneva, NY, USA
Agrobacterium vitis strains were routinely grown for 36 h at 25°C with agitation at 250 rpm in mannitol-glutamic acid: Luria-Bertani (MG/L) medium (2.5 g yeast extract, 5.0 g tryptone, 5.0 g NaCl, 5.0 g/L mannitol, 1.15 g L-glutamic acid, 250 mg KH2PO4, 100 mg MgSO4 7H2O per liter, pH 7.0) supplemented with 1 mg/mL thiamine. For growth kinetics and biofilm experiments, subcultures in MG/L after 36 h were diluted 1:100 in modified AB minimal media (300 mg MgSO4 7H2O, 150 mg KCl, 10 mg CaCl2 2H2O, 3g K2HPO4, 1g KH2PO4, and 5g/L glucose per liter) supplemented with 10% mannitol. FeSO4 7 H2O (2.5 mg/L) was not included in the AB minimal medium used in our studies. For mutant (ΔhnoX) and mutant complemented (ΔhnoX/phnoX) A. vitis S4 strains, liquid media or plates were supplemented with kanamycin (50 μg/mL) for ΔhnoX and kanamycin (50 μg/mL) and spectinomycin (100 μg/mL for liquid cultures or 400 μg/mL for plates) for ΔhnoX/phnoX. Cultures of A. vitis were grown on either LB or AB agar plates at 25C.
Construction of expression vectors for A. vitis HaCE and H-NOX.
Expression vector for HaCE WT (avi_3097) and H-NOX WT (avi_3098) in A. vitis was constructed for protein expression. The expression vectors for HaCE was constructed using a previously published method (39). In detail, A. vitis S4 genomic DNA was purified from cells using the Wizard® SV Genomic DNA Purification System (Promega). Polymerase chain reaction (PCR) was used to amplify HaCE and H-NOX from purified A. vitis S4 genomic DNA using phusion polymerase and the primers containing appropriate restriction sites (listed on Table 1). The PCR products were digested, cloned between the NdeI and XhoI sites of pet20b to generate a C-terminal or N-terminal hexahistidine (His6)-tagged for avi_3097 or avi_3098, respectively, and then transformed into DH5α cells and plated on LB agar plates containing ampicillin (50 μg/mL). The plasmid was isolated using a DNA extraction kit (Qiagen), and then sequenced at the Stony Brook DNA Sequencing Facility to confirm the wild type avi_3097 and avi_3098 genes in pet20b. For HaCE variants, QuikChange PCR-based site-directed mutagenesis was used to generate the double mutants, GGAAF variant (D158A and E159A) and AAL variant (E284A and V285A), from wild type avi_3097 plasmid in pET20b. Extracted DNA from DH5α cells was sequenced at Stony Brook DNA Sequencing Facility.
Expression and purification of His6-tagged avHaCE proteins.
Plasmid DNA harboring the gene encoding avi_3097 WT or variants were transformed into BL21(DE3)pLysS competent cells for protein over-expression, and purified using previously published methods (39). In brief, cultures were grown in 2XYT medium (16 g tryptone, 10 g yeast extract, 5 g NaCl per liter) at 37C to an OD600 of ~ 1.2. Then, protein expression was induced with isopropyl -D-thiogalactopyranoside (IPTG, 10 μM), and then the induced cells were incubated at 18C under agitation (250 rpm) for 16 h. Cells were harvested by centrifugation and then lysed by sonication in lysis buffer containing 50 mM Tris-HCl pH 7.4, 50 mM arginine, 50 mM glutamic acid, 200 mM NaCl, 500 μM EDTA, 5 mM -mercaptoethanol, 10% glycerol and 1 mM phenylmethylsulfonyl fluoride (PMSF). Cellular debris was removed by centrifugation (18,500 rpm for 1 h at 4 C), and then the cleared lysate was loaded onto a Ni-NTA column equilibrated in buffer (50 mM Tris-HCl, pH 7.4, 50 mM arginine, 50 mM glutamic acid, 200 mM NaCl, 500 μM EDTA, 5 mM -mercaptoethanol, 10% glycerol). The column was washed with 10 column volumes of the same buffer, and then with buffer containing increasing amounts of imidazole. After washing, the protein was eluted with 250 mM imidazole, and the eluted protein was desalted using a PD-10 column (GE Healthcare Life Sciences) and stored at −80C.
Expression and purification of GST-tagged WT avHaCE.
WT HaCE (avi_3097) containing GST at the N-terminus, was generated by QuikChange site-directed mutagenesis using a GST-tagged avGGAAF construct in vector pGEX-4T-2 (GE Healthcare) as the template. GST-tagged avHaCE was grown in Lennox L Broth (10 grams tryptone, 5 grams sodium chloride, 5 grams yeast extract per liter) to an OD600 of ~0.6, after which expression was induced by the addition of 250 μM IPTG and allowed to proceed for 16 hours at 18 °C under agitation (250 rpm). Cells were harvested by centrifugation and then lysed in phosphate buffered saline (PBS, 8 g NaCl, 2 g KCl, 1.44 g Na2HPO4, 0.24 g KH2PO4 per liter, pH 7.4) containing 1% Triton X-100 and 1 mM PMSF. Cellular debris were removed as described above and the cleared lysate was loaded onto Glutathione Sepharose 4B resin and washed with 10 column volumes of PBS. GST-tagged avHaCE was eluted from the column using 50 mM Tris-HCl (pH 8.0) containing 20 mM reduced L-glutathione. Fractions eluted from the column were analyzed by SDS-PAGE. The fractions containing the fused protein were pooled and concentrated, and then desalted, to remove glutathione, into 50 mM Tris-HCl (pH 8.0) buffer using a PD-10 column.
Protein concentration determination.
Protein concentrations were determined using the method of Bradford with bovine serum albumin (BSA) as the standard (43). Protein purity was assessed using SDS-PAGE with a 12.5% gel.
Pull-down assay to determine HaCE and H-NOX protein interactions.
Interactions between HaCE and H-NOX constructs were monitored using pull-down assays as described previously (12). In brief, His6-tagged H-NOX (2.5 μM) was incubated with GST-tagged HaCE (or isolated HaCE domain) immobilized onto Glutathione Sepharose 4B beads overnight at 4 °C. The beads were thoroughly washed to remove unbound H-NOX and then subjected to SDS-PAGE and followed by Western analysis using polyclonal 6X anti-His tag antibody (Abcam). Each experiment was repeated at least three times.
Formation and UV-visible analysis of H-NOX complexes.
Purified H-NOX was incubated with 20 mM potassium ferricyanide at room temperature for 20 min to generate oxidized Fe(III)-heme. The protein was desalted using a PD-10 column equilibrated in 50 mM HEPES (pH 7.5) and 200 mM NaCl to remove excess potassium ferricyanide. The Fe(III)-H-NOX was exposed to an anaerobic environment using a COY Anaerobic Chamber (Coy Laboratory Products) to deoxygenate the protein. Oxygen-free Fe(III)-H-NOX was treated with 60 mM sodium dithionite (Na2S2O4) at room temperature for 20 min to generate Fe(II) bound heme protein. The protein was again desalted as previously described, but under anaerobic conditions. NO-bound Fe(II)-H-NOX was formed by incubating the protein with the NO donor diethylammonium (Z)-1-(N,N-diethylamino)diazen-1-ium-1,2-diolate (DEA-NONOate, Cayman Chemicals) dissolved in 10 mM sodium hydroxide and followed by desalting. Carbon monoxide(CO)-bound Fe(II)-H-NOX was generated by bubbling CO gas into the protein for 15 min in an airtight vial. Electronic absorption spectra of all complexes were measured on a Cary 100 UV-Vis spectrophotometer equipped with a constant temperature bath set to 25°C.
NO dissociation kinetics.
Electronic absorption spectra were measured on a Cary 100 UV-Vis spectrophotometer equipped with a constant temperature bath set to 20°C. H-NOX complexes were prepared in an anaerobic glove bag as described above. NO dissociation rate was measured as previously published (44). In detail, the absorption spectrum of NO bound Fe(II)-H-NOX was measured at various times in the presence of saturating CO and 60 mM Na2S2O4. The absorbance change (ΔAbs = Abst – Abs0) was plotted versus wavelength (Fig. S1A in supplemental material). The difference between the maximum and minimum (ΔΔAbs) was than plotted versus time (Fig. S1B in supplemental material), and NO dissociation rate (koff) was determined by a nonlinear fit of the data using two phase exponential association in GraphPad Prism version 4.03 for Windows (Graph Pad Software, San Diego, CA USA, www.graphpad.com). Standard deviation was determined from at least three independent trials.
Cyclase and PDE activity of avHaCE proteins in the absence of H-NOX.
Steady-state kinetics of cyclase activity were carried out using a modified Invitrogen EnzChek™ pyrophosphate assay kit; the method is described in the procedures below for avHaCE activity in the presence of H-NOX as well as the supplemental materials and in Liu et al. (12). For analysis of avHaCE cyclase activity in the absence of H-NOX, initial velocities of 250 nM AAL were measured with various concentrations of GTP (0–100 μM) at 25 °C and pH 7.5 (50 mM TRIS-HCl buffer containing 5 mM MgCl2 and 2 mM sodium azide). The steady-state kinetic parameters of the AAL variant for GTP were determined from fits of data to the Hill equation (Equation 1), where K’ is the concentration of substrate ([S]) required to give half the maximal velocity (Vmax) and n represents the number of substrate binding sites per molecule of enzyme.
| (1) |
Steady-state kinetics of PDE activity in the absence of H-NOX were determined by monitoring reactions of 15 nM GGAAF variant with various concentrations of cyclic di-GMP (0–20 μM). In detail, reactions were conducted at 25 °C at pH 7.5 in buffer containing 50 mM Tris-HCl and 5 mM MgCl2. Aliquots were removed at various times points, between 0–50 min, and quenched by the addition of 10 μL of 0.1 M CaCl2 (10 mM final concentration). Thereafter, the quenched reactions were treated with 10 μL of calf-intestinal phosphatase (CIP from New England Biolabs, 10 units/mL) to convert with a 1:1 ratio, product pGpG into phosphate. Standards containing 0 to 30 μM of inorganic phosphate were also prepared in 50 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 10 mM CaCl2 and 1 unit of CIP. The CIP treated reactions and phosphate standards were transferred to Falcon 96-well plates and incubated with 20 μL of SensoLyte® malachite green phosphate assay (AnaSpec) for 10 min at room temperature. Then the absorbance was read at 620 nm using a VICTOR™ X multilabel plate reader (Perkin Elmer, Inc.). The absorbance readings were corrected by subtracting out absorbance background from protein only and substrate only controls. For the reactions, the pGpG product concentration was calculated from the linear plot of absorbance at 620 nM versus phosphate standard concentrations (Fig. S2). Thereafter, initial velocities (v0) were determined by plotting the product concentration versus time, and then fitting with a linear regression equation (Equation 2).
| (2) |
The kinetic parameters of GGAAF for cyclic di-GMP were obtained from fits of data to Equation 3, the Michalis-Menten equation. All fits were generated using Origin software (OriginLab).
| (3) |
Cyclase and PDE activity of avHaCE proteins in the presence of H-NOX.
Cyclase and PDE activity assays were conducted on avHaCE variants in the presence of unligated or NO-bound Fe(II)-H-NOX. The AAL or GGAAF variant was pre-incubated with 10X H-NOX anaerobically for 10 min. Reactions were initiated by the addition of either GTP or cyclic di-GMP, and then initial velocities were determined using Invitrogen EnzChek™ assay kit with either pyrophosphatase or CIP to detect phosphate produced from the cleavage of pyrophosphate in the cyclase (AAL) reactions or cleavage of terminal phosphate from pGpG in the PDE (GGAAF) reactions. Phosphate from the resultant products (pGpG or pyrophosphate) was monitored at 360 nm as a function of time on a Cary 100 spectrophotometer. More details on this assay is described in the supplemental materials. The resulting initial velocities for cyclase and PDE reactions in the presence of H-NOX was normalized to the initial velocity of the corresponding enzyme only (without H-NOX) reactions.
Construction of H-NOX mutant strain.
To determine the effects of H-NOX on biofilm formation and bacterial growth, an H-NOX mutant (ΔhnoX) was generated using a modified published procedure (45). In brief, avi_3098 gene was amplified using primers containing SacI and XbaI restriction sites and A. vitis S4 genomic DNA. The PCR product was purified, digested with restriction enzymes SacI and XbaI, and then cloned into suicide vector pVIK165. The plasmid was then transformed into E. coli S17–1/ by electroporation and plated on LB agar plates (50 μg/mL kanamycin). Thereafter, biparental mating was performed between E. coli S17–1/pir::pVIK165-avi_3098 gene and A vitis S4 and then plated on potato dextrose agar plates supplemented with kanamycin (50 μg/mL). Mutation was confirmed using PCR.
Construction and complementation of plasmid.
For H-NOX complementation into ΔhnoX A. vitis S4 strain (ΔhnoX/phnoX), avi_3098 was amplified from A. vitis S4 genomic DNA using primers containing PstI and BamHI restriction sites (avi_3098 FWD comp and avi_3098 REV comp in Table 1). The PCR products were then ligated to pPZP201 vector using Taq DNA Ligase (New England Biolabs) and transformed into DH5 cells, and then plated on LB agar plates containing spectinomycin (100 μg/mL). The positive colonies were isolated using a DNA extraction kit (Qiagen). The plasmid pPZP201-avi_3098 was confirmed by restriction digest and sequencing (Stony Brook DNA Sequencing Facility). For complementation, the plasmid pPZP201-avi_3098 was introduced into ΔhnoX strain by electroporation. Transformants were selected on AB minimal medium plates containing kanamycin (100 μg/mL) and spectinomycin (400 μg/mL) and were verified by PCR.
Sessile biofilm and planktonic growth analysis of A. vitis strains.
Media used for biofilm studies and planktonic growth kinetics were supplemented with the following antibiotics, kanamycin (50 μg/mL) for ΔhnoX and kanamycin (50 μg/mL) and spectinomycin (100 μg/mL) for ΔhnoX/phnoX. Bacterial liquid cultures of A. vitis strains were grown in MG/L media for 36 h at 25C. The subcultures were diluted 1:100 in AB media (10% mannitol; 2 mL for biofilms or 50 mL for planktonic growth) supplemented with and without 20 μM DETA NONOate. For planktonic growth analysis, the subcultures were grown at 25C under agitation (250 rpm) and OD at 600 nm was recorded periodically for up to 72 h.
Crystal violet-binding assay was used for biofilm studies and performed using modified published procedures (46, 47). For biofilm quantitation, diluted A. vitis subcultures (200 μL) were transferred into a 96-well polystyrene plate and incubated for 48 h at 25C. After 48 h, 100 μL was transferred to a 96-well plate to measure the OD at 600 nm using a VICTOR™ X multilabel plate reader. The incubated plates were then rinsed three times with doubly distilled water and allowed to dry for ~ 2 h. The sessile bacteria were stained by the addition of 225 μL of crystal violet to the wells and then incubated for 15 min at room temperature. After 15 min, crystal violet was discarded, and the stained biofilms were solubilized in 225 μL of 30% acetic acid and incubated for 15 min at room temperature under light agitation. Thereafter, the solubilized solutions (100 μL) were transferred to a 96-well plate and the absorbance was measured at 570 nm. The data are reported as crystal violet-stain biofilms at 570 nm divided by the OD of planktonic cells at 600 nm (CV/OD).
RESULTS AND DISCUSSION
We have previously reported that avi_3097 in A. vitis strain S4 encodes an enzyme with catalytically active DGC and PDE domains, i.e. avHaCE (39). Directly upstream of avi_3097, an hnoX gene is encoded by avi_3098. It thus seemed reasonable to hypothesize that, as in S. woodyi (12) and L. pneumphila (10), NO binds to avH-NOX and regulates the cyclic di-GMP output of avHaCE and thus biofilm formation.
avH-NOX and avHaCE form a complex in vitro.
To help establish our hypothesis, we first performed a pull-down assay to establish if the co-cistronic proteins avi_3097 (avHaCE) and avi_3098 (avH-NOX) are binding partners (Fig. S3). Briefly, in the presence of glutathione-S-transferase (GST)-tagged avHaCE, hexahistidine (His6)-tagged avH-NOX was pulled down, whereas GST alone was unable to pull down His6-tagged avH-NOX. These data confirmed that avH-NOX and avHaCE are binding partners.
Ligand binding properties of H-NOX: avH-NOX is a NO sensor.
A sequence alignment comparing avH-NOX to other characterized H-NOX proteins from facultative anaerobes, as well as the H-NOX domains from mammalian soluble guanylyl cyclases (sGCs), shows a high degree of sequence homology (Fig. S4) and suggests that avi_3098 is an H-NOX protein containing a histidine-ligated protoporphyrin IX hemoprotein that may detect and bind gaseous signaling molecules at the ferrous iron center. Therefore, since we propose that avH-NOX regulates the enzymatic activity of avHaCE via sensing NO, our first step was to confirm that it is a NO sensor. Therefore, the gene encoding the putative H-NOX from A. vitis S4 was cloned, and then purified as a mixture of Fe(II)- and Fe(III)-bound heme. We then pursued characterization of the purified hemoprotein. Both thermodynamic (Fig. 1) and kinetic (Fig. S1) ligand binding properties of avH-NOX were evaluated with UV-vis spectroscopy. A comparison of the ligand binding properties of avH-NOX with other bacterial H-NOX proteins and sGC from bovine lung is summarized in Table 2.
FIGURE 1.
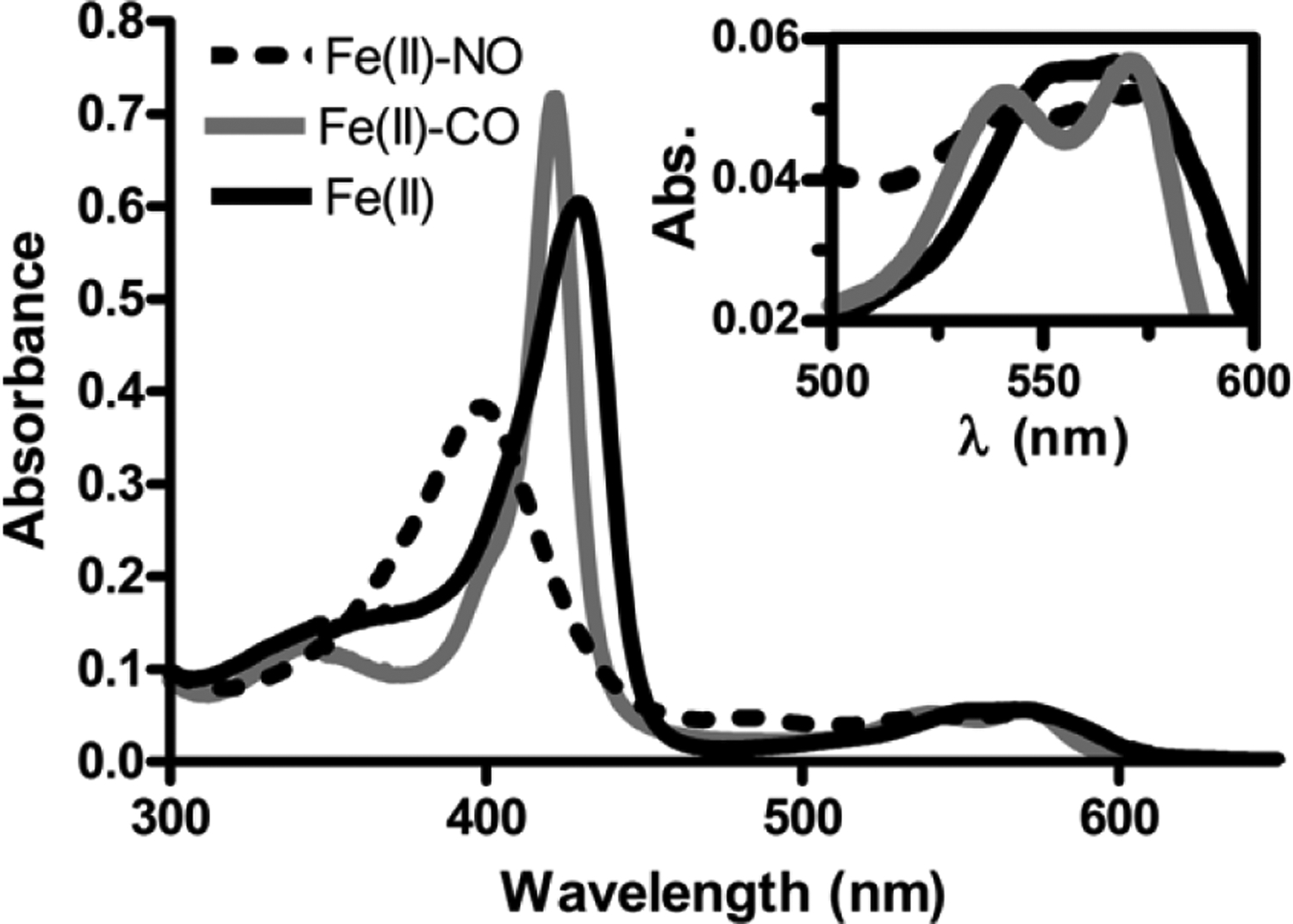
Electronic absorption spectra of avH-NOX as the unligated Fe(II) complex (solid black line), the Fe(II)-CO complex (solid gray line), and the Fe(II)-NO complex (dashed black line) in 50 mM HEPES pH 7.5 buffer and 200 mM NaCl at room temperature. Insert shows the region at wavelength range 500 – 600 nm.
TABLE 2.
Ligand binding properties of H-NOX proteins from multiple bacterial species: electronic absorption maxima and NO dissociation rate constants
To characterize the thermodynamic ligand-binding properties, the as-purified mixture of Fe(III)/Fe(II) avH-NOX was first oxidized with potassium ferricyanide to fully oxidize the sample, and then the sample was reduced with dithionite to form the 5-coordinate Fe(II)-unligated complex; the electronic spectrum of this complex has a characteristic Soret band at 429 nm. Incubation of Fe(II)-unligated avH-NOX with the NO donor diethylamine (DEA) NONOate results in a complex with a Soret band at 398 nm. This blue shift has been observed in many well-characterized 5-coordinate Fe(II)-NO hemoproteins (48, 50–52), including H-NOX proteins (12, 15, 35, 44, 48, 53), in which the proximal histidine heme ligand dissociates upon binding of NO. Likewise, exposure of the Fe(II)-unligated protein to CO gas results in formation of the 6-coordinate Fe(II)-CO complex with a characteristic Soret band at 422 nm. Like the other H-NOX family members, H-NOX from A. vitis does not appreciably bind molecular oxygen; we observe no shift in the absorption maxima of the Fe(II)-unligated H-NOX complex upon exposure to air (Fig. S5).
Evaluation of the NO dissociation rate constant revealed a koff(NO) of 8.2 ± 3 × 10−4 s−1 at 20°C (Fig. S1), which is very similar to the dissociation rate constants measured for other members of the H-NOX family (Table 2) (12, 15, 44, 49). Assuming a nearly diffusion-limited kon of ~ 108 M−1 s−1 at 20°C, as has been observed for other H-NOX proteins, the KD(NO) for avH-NOX is in the picomolar range (12). From both experiments, we can conclude that avH-NOX has ligand binding properties consistent with a sensitive and selective NO sensor.
Steady-state kinetics of HaCE: avHaCE is phosphodiesterase dominant.
As mentioned above, avi_3097 (avHaCE), annotated just downstream of avH-NOX, was previously characterized as a bi-functional enzyme containing enzymatically active DGC (containing a GGDEF motif) and PDE (containing a EVL motif) domains (39). The DGC domain converts two molecules of guanosine 5’-triphosphate (GTP) to cyclic di-GMP and pyrophosphate, and the PDE domain, converts cyclic di-GMP to 5’-phosphoguanylyl-3’,5’-guanosine (pGpG).
To determine if avHaCE activity is modulated by avH-NOX, we expressed and purified avHaCE wild-type and variants and analyzed the enzymatic activity of these enzymes with and without avH-NOX. To simplify our studies, we generated two avHaCE active site variants by site-directed mutagenesis to disrupt either DGC activity, to study PDE activity alone (the GGAAF variant is D158A/E159A avHaCE), or to disrupt PDE activity to study cyclase activity alone (the AAL variant is E284A/V285A avHaCE). We have previously shown that the avHaCE AAL variant has DGC activity when GTP is used as the substrate but does not catalyze the hydrolysis of cyclic di-GMP to form pGpG (39). Likewise, the avHaCE GGAAF variant lacks the ability to convert GTP to cyclic di-GMP, but catalyzes the linearization of cyclic di-GMP to pGpG.
Table 3 shows the steady-state kinetic parameters of the AAL and GGAAF variants of avHaCE. To assess PDE activity, we used a malachite green colorimetric assay to quantify inorganic phosphate released upon treatment of product 5’-pGpG with calf-intestinal phosphatase (CIP) (54). For the cyclase assay, we used a previously described modified Invitrogen EnzChek™ assay to quantify inorganic phosphate released upon treatment of the cyclase reaction product pyrophosphate with inorganic pyrophosphatase (12). Please refer to the Materials and Methods and supplemental materials for more details on the enzyme assays used in this study.
TABLE 3.
Steady-state kinetic parameters for AAL (DGC activity) and GGAAF (PDE activity) variants of AvHaCE
| Enzyme | kcat (s−1) | KM (μM) | |
|---|---|---|---|
| AAL | 0.031 ± 0.001 | 17.0 ± 0.72 | 0.018 ± 0.001 |
| GGAAF | 0.036 ± 0.002 | 5.2 ± 0.9 | 0.068 ± 0.001 |
The initial velocity versus GTP concentration plot for the AAL variant is a sigmoidal curve, indicating cooperative behavior in the diguanylate cyclase domain (Fig. 2A), whereas the GGAAF variant obeys Michalis-Menten kinetics (Fig. 2B and Fig. S6). The KM of the AAL variant for GTP was determined to be 17.0 ± 0.72 μM with a kcat/KM of (0.018 ± 0.001) 10−1 μM−1 s−1 (Table 3). The value of n was determined to be 2.28 ± 0.117, indicating that there are at least two GTP binding sites in the diguanylate cyclase domain of the dimeric HaCE. The KM of the GGAAF variant for GTP was determined to be 5.2 ± 0.9 μM with a kcat/KM of (0.068 ± 0.001) 10−1 μM−1 sec−1 (Table 3). In comparison to other diguanylate cyclases, the catalytic efficiency of avHaCE is ~400-fold lower than WspR (kcat/KM = 0.75 s−1 μM−1, KM = 5.97 ± 0.80 μM, kcat = 4.50 ± 0.12 s−1) and ~8-fold lower than swHaCE (kcat/KM = 0.014 ± 0.002 sec−1, KM = 7.74 ± 0.90 μM, and kcat = 0.105 ± 0.005 s−1) (12, 55).
FIGURE 2.
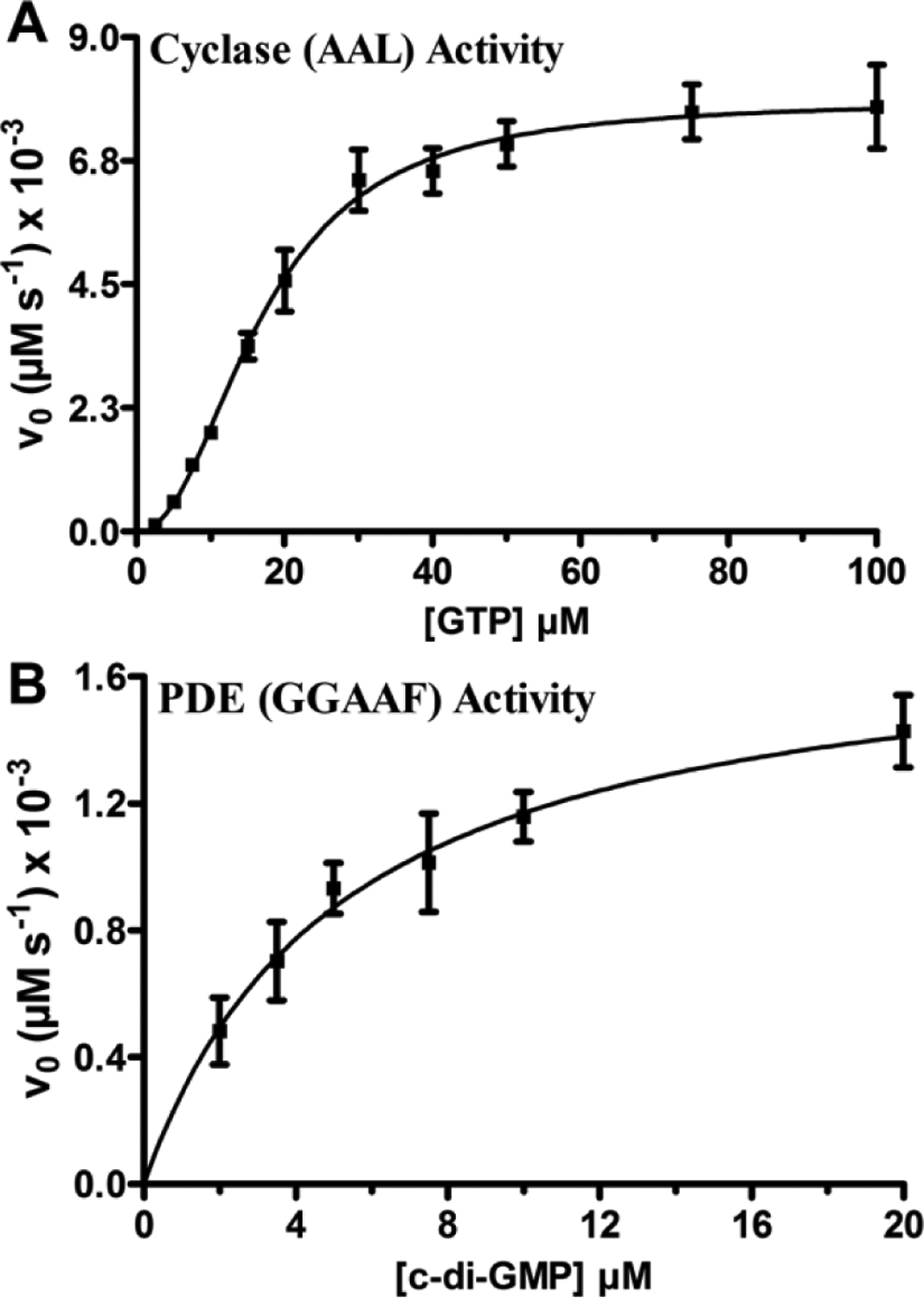
Steady-state kinetics of cyclase and phosphodiesterase activities of the AAL and GGAAF variants of avHaCE, respectively. Initial velocity measurements of the diguanylate cyclase activity of the AAL variant (250 nM) as a function of GTP concentration (0 μM - 100 μM) in 50 mM Tris-HCl pH 7.5 buffer containing 5 mM MgCl2 at 25C (A) and phosphodiesterase activity of the GGAAF variant (15 nM) as a function of cyclic di-GMP concentration (0 μM – 20 μM) in 50 mM Tris-HCl pH 7.5 buffer containing 5 mM MgCl2 at 25C (B). The data were fit to the Hill equation (plot A) or Michaelis−Menten equation (plot B), respectively. Error bars represent standard deviation from the mean determined from at least three independent trials.
In comparison to other phosphodiesterases, the catalytic efficiency of avHaCE is ~30-fold lower than that of RocR (kcat/KM = 0.21 ± 0.02 μM−1 s−1, KM = 3.2 ± 0.3 μM, and kcat = 0.67 ± 0.03 s−1) and ~200-fold lower than swHaCE (kcat/KM = 1.16 ± 0.20 μM−1 s−1, KM = 1.31 ± 0.22 μM, and kcat = 1.52 ± 0.05 s−1) (12, 56). Within avHaCE, kcats of the cyclase and phosphodiesterase activities are similar, but the KM for GTP binding to the cyclase domain is 3-fold greater than cyclic di-GMP binding to the phosphodiesterase domain. Thus, the comparison of catalytic efficiency values indicates that avHaCE is a PDE-dominant bifunctional enzyme with a ~4-fold more active phosphodiesterase domain compared to its cyclase domain in the absence of avH-NOX. This was also the case for the HaCE studied from S. woodyi, but in swHaCE, the phosphodiesterase domain is overwhelmingly more active, by ~90-fold, in comparison to the cyclase domain (12). In contrast, in L. pneumophila, HaCE is cyclase dominant due to an inactive phosphodiesterase domain (10).
HaCE activity in the presence of H-NOX: NO-bound avH-NOX increases phosphodiesterase activity.
To understand how the H-NOX/HaCE system in A. vitis responds to NO to regulate cyclic di-GMP metabolism in vivo, we next evaluated how avHaCE activity is modulated by avH-NOX in vitro by measuring the cyclase and phosphodiesterase activities in the presence of Fe(II)-unligated or NO-bound avH-NOX. In both assays, the initial velocities are reported as the change in absorbance at 360 nm per minute (ΔAbs360nm min−1) [the absorption maxima (360 nm) of the enzymatic conversion of 2-amino-6-mercapto-7-methylpurine ribonucleoside (MESG) to ribose 1-phosphate and 2-amino6-mercapto-7-methylpurine by purine ribonucleoside phosphorylase (PNP)] using a modified Invitrogen EnzChek™ assay kit as described by Liu et al. (12) and in more detail in the supplemental materials.
We found that in the presence of 10X Fe(II)-unligated avH-NOX, both initial velocities for phosphodiesterase and cyclase activities were reduced by ~50% compared to the corresponding activities in the absence of H-NOX (Fig. 3). In the presence of 10X Fe(II)-NO-bound avH-NOX, however, the cyclase activity was unchanged relative to the cyclase activity in the presence of Fe(II)-unligated avH-NOX, while the phosphodiesterase activity increased 4-fold upon addition of NO (Fig. 3).
FIGURE 3.
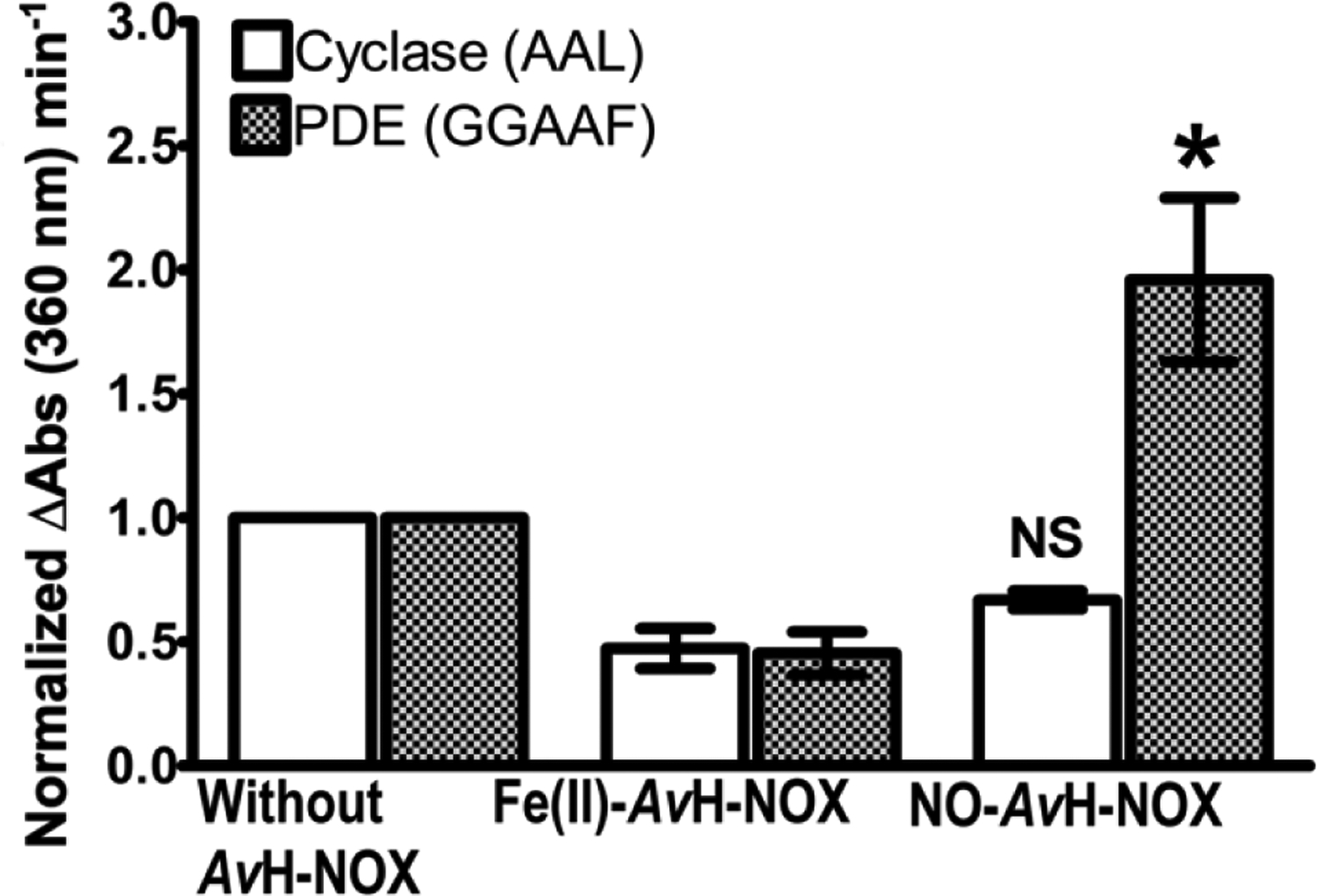
Initial velocities (ΔAbs360nm min−1) determined using a modified Invitrogen EnzChek™ assay were plotted as a function of enzyme activity without and with avH-NOX as the Fe(II)-unligated (Fe(II)-avH-NOX) or Fe(II)-NO (NO-avH-NOX) complex. The initial velocity of the cyclase activity of the AAL variant (250 nM) with and without 2.5 μM Fe(II)-avH-NOX or NO-avH-NOX is reported relative to the initial velocity of the cyclase activity of the AAL variant in the absence of avH- NOX (white bars). The initial velocity of the phosphodiesterase activity of the GGAAF variant (50 nM) with and without 0.50 μM Fe(II)-avH-NOX or NO-avH-NOX, is reported relative to the initial velocity of the phosphodiesterase activity of the GGAAF variant in the absence of avH- NOX (shaded bars). Errors bars represent the standard deviation from the mean of at least three independent trials. NS = not significant and * = p < 0.05 are for the comparison of AAL (white bars) or GGAAF (patterned bars) activities in the presence of NO-avH-NOX to those activities in the presence of Fe(II)-avH-NOX.
The details of the effect of H-NOX on cyclic di-GMP cyclase and phosphodiesterase activities in other H-NOX/HaCE systems has varied, but NO-bound H-NOX always results in lower cyclic di-GMP accumulations. In L. pneumophila, lpgHaCE cyclase activity is lower in the presence of Fe(II)-NO-bound H-NOX than in the presence of Fe(II)-unligated H-NOX, while the phosphodiesterase domain is incapable of turning over cyclic di-GMP, and thus insensitive to the ligation state of H-NOX (10). In S. woodyi, both cyclic di-GMP cyclase and phosphodiesterase activities are regulated by NO/H-NOX; the cyclase activity of swHaCE is markedly enhanced in the presence of Fe(II)-H-NOX in comparison to the presence of Fe(II)-NO-bound H-NOX, while the phosphodiesterase activity of swHaCE is markedly enhanced in the presence of Fe(II)-NO-bound H-NOX in comparison to Fe(II)-unligated H-NOX (12). Finally, in A. vitis, NO-bound H-NOX results in no significant change in cyclase activity, but an increase in phosphodiesterase activity.
Possible structural basis for differential regulation of HaCE activities by H-NOX.
It is possible that a PAS domain, present in swHaCE, but not avHaCE, explains the differential regulation of HaCE cyclase activity by H-NOX. Although the mechanistic details of H-NOX regulation of HaCE are not well understood, there are some structural details available to guide our thinking. H-NOX has been shown to bind partner proteins (26, 36) via an N-terminal -helical region (26, 31). NO binding induces H-NOX conformational changes including rotation of the H-NOX “signaling helix” (26, 28, 30, 33, 36l), leading to increased binding interaction with its partner (26, 35, 36l). A study focused on sGC, which is composed of a fused N-terminus H-NOX domain followed by a PAS domain and C-terminus cyclase, showed that the sGC PAS domain participates in the signal transfer mechanism by forming additional contacts with the H-NOX domain in response to NO binding, leading to increased C-terminal cyclase domain activity (57). Therefore, in S. woodyi, it is possible that the swHaCE PAS domain could introduce additional binding interactions within the H-NOX/HaCE regulatory complex, facilitating regulation of both the cyclic di-GMP cyclase and phosphodiesterase domains (see schematic in Fig. 4). Perhaps H-NOX generally regulates cyclase activity through PAS/cyclase domain contacts and regulates phosphodiesterase activity through direct interaction with the phosphodiesterase domain. It would follow, therefore, that avHaCE lacks a PAS domain, and thus also lacks H-NOX regulation of its cyclase domain.
FIGURE 4.
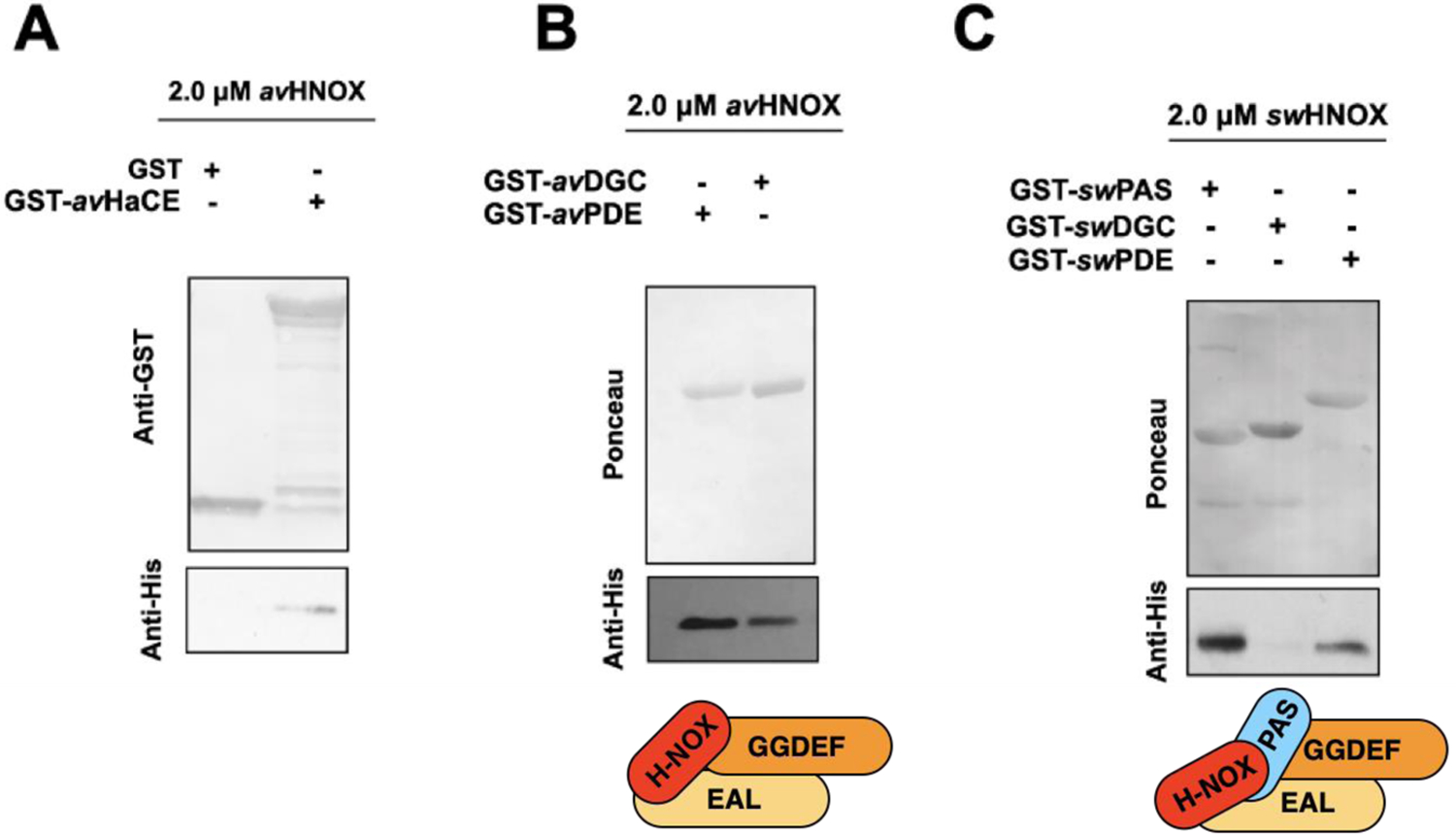
Interaction of H-NOX with HaCE domains. The top panel in A, B, and C illustrates the protein loading of the bait protein (GST or GST-tagged HaCE or HaCE domains) present in the pull-down assay, detected by either anti-GST immunoblotting or ponceau protein staining. The bottom panel illustrates the amount of H-NOX pulled down by the GST-tagged bait protein and detected by anti-His immunoblotting. (A) avH-NOX interacts with the full-length GST tagged avHaCE (lane 2). (B) avH-NOX interacts with avPDE (lane 1) and avDGC (lane 2); domain interactions shown schematically in the bottom panel. (C) swH-NOX interacts with swPAS (lane 1) and with swPDE (lane 3) but not with swDGC (lane 2); domain interactions shown schematically in the bottom panel.
To support our hypothesis that H-NOX interacts with PAS domains to regulate DGC activity in bacterial H-NOX/HaCE complexes, we performed H-NOX pull-down experiments using the isolated domains of partner HaCE proteins with both the A. vitis and S. woodyi homologs. avHaCE, as well as the isolated DGC and PDE domains from A. vitis (avDGC and avPDE) were each cloned into a vector that expresses GST at the N-terminus of the HaCE construct, generating GST-tagged avHaCE constructs. The GST constructs were used to pull down his-tagged avH-NOX (Fig. 4). This experiment shows that avH-NOX interacts with the full length avHaCE and both avDGC and the avPDE domains (Fig. 4A and 4B). Therefore avH-NOX makes molecular contacts with both the DGC and PDE domains of avHaCE. Qualitatively, it is also notable that the interaction of avH-NOX with the avPDE domain is consistently stronger than with the avDGC domain, which is consistent with the trend we see for activity (Fig. 3).
We have previously shown that swH-NOX interacts with full length swHaCE (12). In that study we determined that swH-NOX binds full length swHaCE as a 2+2 dimer in both the Fe(II) and NO complexes, indicating NO does not trigger protein association or dissociation, although the Kds of the interaction likely vary. In this study, to better understand the structural basis underlying the differences in regulation of HaCE by H-NOX, we sought to understand which domains of swHaCE interact with swH-NOX. Thus, we performed His-tagged H-NOX pull-down experiments with the GST-tagged individual domains of swHaCE, i.e., swPAS, swDGC and swPDE. We found that swH-NOX interacts with swPAS domain and swPDE, but not swDGC (Fig. 4C). Again, qualitatively, the interaction of swH-NOX with the swPAS domain is stronger than with the swPAS domain.
More research is required to fully understand the regulation of multiple enzyme active sites by H-NOX domains, but the data presented here suggest our hypothesis may be correct. Taken together, these results suggest that H-NOX regulation of cyclase domains is mediated through interaction with a PAS domain, in both bacterial DGCs and mammalian sGCs, while H-NOX regulation of PDEs is mediated by a direct H-NOX/PDE binding interface. This study provides some structural insight into how domain arrangement in HaCE influences the signal transfer mechanism in these NO regulatory systems.
Bacterial growth and biofilm formation: avH-NOX is responsible for NO regulation of biofilm formation in A. vitis.
We have shown that NO-bound avH-NOX increases the activity of the PDE domain in vitro, which should lead to decreased intracellular cyclic di-GMP levels and reduced biofilms in vivo. Thus, we propose that NO could act as a switch for the sessile-to-motile transition in A. vitis through NO/H-NOX/HaCE regulation of cyclic di-GMP concentration. To test this hypothesis in A. vitis, we constructed an hnoX deletion strain of A. vitis S4 using standard methods. Thereafter, static biofilm assay and planktonic growth curves were employed to elucidate the role of H-NOX in biofilm (48 h at 25C) and planktonic growth (up to 72 h at 25C) assays in the presence and absence of a NO donor. First, the concentration dependence of the NO donor diethylenetriamine (DETA) NONOate on A. vitis growth was determined at room temperature in AB minimal medium. We determined that 20 μM or less NONOate is nontoxic to the A. vitis wild-type strain under the tested conditions (Fig. S7). The NO concentration in solution was estimated to be ~1000-fold less than the NONOate concentration. This estimate is based on a previous study that measured NO release from 200 μM DETA NONOate, which had a relatively steady NO dissociation rate as the NO concentration decreased from 80 nM to 60 nM after 24 h at 25C in marine media broth (12). DETA NONOate from Cayman Chemicals has a half-life (t1/2) of 56 h at 22–25C.
As illustrated in figure 5, we employed a static biofilm assay using crystal violet staining to compare biofilm formation of wild-type and ΔhnoX strains of A. vitis S4 after 48 h of growth in AB media in the presence and absence of 20 μM DETA NONOate (~20 nM NO). The CV-stained biofilm cells were normalized to the OD600 readings of replicate cultures to account for differences in planktonic growth in the presence of NO (Fig. S8). The wild-type strain displayed a significant decrease in biofilm growth in the presence of NO, as expected, and consistent with the well-documented role of NO on biofilm formation in bacteria. Furthermore, similar to the S. woodyi H-NOX/HaCE system (12), hnoX deletion resulted in less biofilm than the wild-type strain, and no NO phenotype. Plasmid complementation of hnoX in the mutant background (ΔhnoX/phnoX), however, restored wild-type biofilm growth and the NO response. The reduction in biofilm for the ΔhnoX strain (Fig. 5) can be correlated to the phosphodiesterase-dominant HaCE activity we observed in the absence of H-NOX in our in vitro kinetic studies (Table 3 and Fig. 2). The results in A. vitis and S. woodyi are in contrast to L. pneumophila where the hnoX deletion strain displayed a hyperbiofilm phenotype, which was rationalized by an unregulated cyclase dominant lpgHaCE in the absence of H-NOX (10).
FIGURE 5.
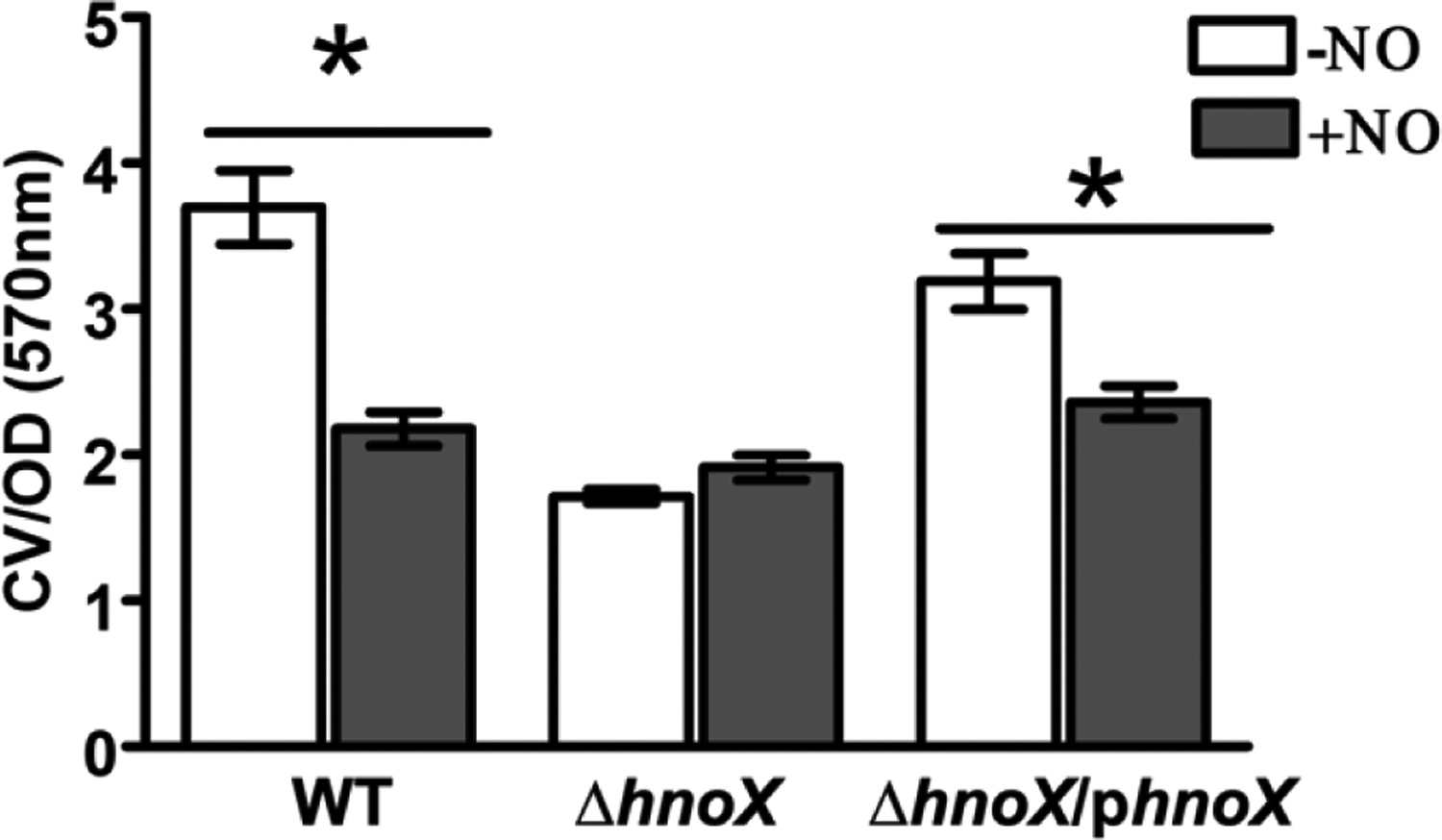
Biofilms of wild-type, ΔhnoX, and ΔhnoX/phnoX strains of A. vitis S4 grown in polystyrene plates for 48 h in AB media in the presence and absence of NO donor (20 μM DETA NONOate) and quantified with crystal violet (CV) staining. CV measurements at 570 nm were normalized to OD readings of the cultures before staining with CV. Error bars represent the standard deviation from the mean of at least three independent trials, and within each trial, each biofilm condition was run a minimum of 6 times. * = p < 0.05.
As illustrated in figure 6, we also monitored the planktonic growth of A. vitis S4 in AB media at 25C in the presence and absence of NO. Results showed that ΔhnoX growth was similar to WT growth, demonstrating that the absent hnoX gene does not present a delay in cell growth. Wild-type growth in the presence ~20 nM NO is like its growth in the absence of NO. However, for the ΔhnoX strain, there is a marked impairment of cell growth when grown in the presence of NO, but complementation restores wild-type growth in the presence of NO.
FIGURE 6.
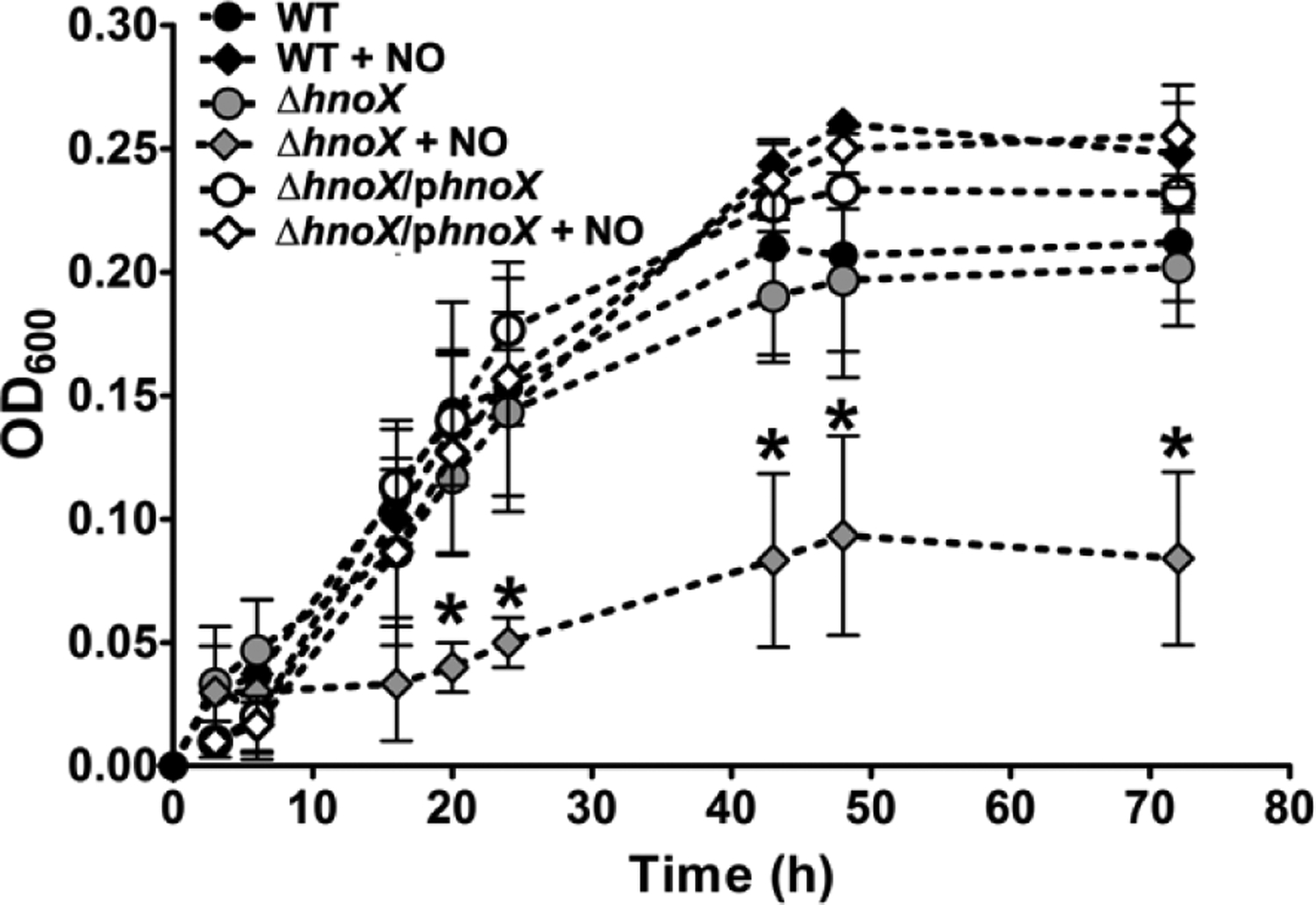
Planktonic growth curves of wild-type, ΔhnoX, and ΔhnoX/phnoX strains of A. vitis S4 in AB minimal medium supplemented with or without 20 μM DETA NONOate at 25C. The OD600 was measured up to 72 h. Error bars represent the standard deviation from the mean of at least three independent trials. * = p < 0.05 in comparison to ΔhnoX/phnoX in the absence of NO.
These results implicate H-NOX in both biofilm and planktonic growth responses to the presence of NO at low concentrations. It does not seem likely that reactive nitrogen species are being generated at this NO concentration, even though these studies were carried out under aerobic conditions; indeed, we observe no impact on wild-type growth in the presence of 20 μM DETA NONOate, and H-NOX has never before been implicated in a protective role. Nonetheless, it should be noted that A. vitis apparently lacks several common NO detoxifying enzymes, so it is possible that H-NOX plays a protective role in this bacterium. This is an observation that will require further study.
PERSPECTIVES AND CONCLUSIONS
Biofilm formation is a complex regulatory process that enhances plant pathogen infectivity (47, 58, 59). A. vitis S4 is a tumorigenic plant pathogen and the causative agent for crown gall disease in grapevines (60). As shown in a related tumorigenic pathogen, Agrobacterium tumefaciens, these pathogens first infect plants by forming biofilms on the plant roots (61), then planktonic cells can travel through the xylem to form crown gall tumors at wound sites above ground (60). This transition between motile and biofilm growth modes is, in part, regulated by alterations in intracellular cyclic di-GMP levels (18, 21).
As demonstrated here, cellular cyclic di-GMP levels may be modulated by NO; we have presented data that indicate low levels of NO can reduce biofilm formation by regulating the activity of an H-NOX/HaCE system in A. vitis. Thus, NO could perhaps be one of the mechanisms that control A. vitis infection process in crown gall disease. Indeed, NO has been implicated in virulence and enhanced survival of pathogens in plant hosts (62). Our model for the NO/H-NOX/HaCE system in A. vitis is illustrated in figure 7. Upon NO binding, avH-NOX upregulates the PDE domain of avHaCE (Fig. 3), which subsequently lowers intracellular cyclic di-GMP levels and leads to a reduction in biofilms (Fig. 4).
FIGURE 7.
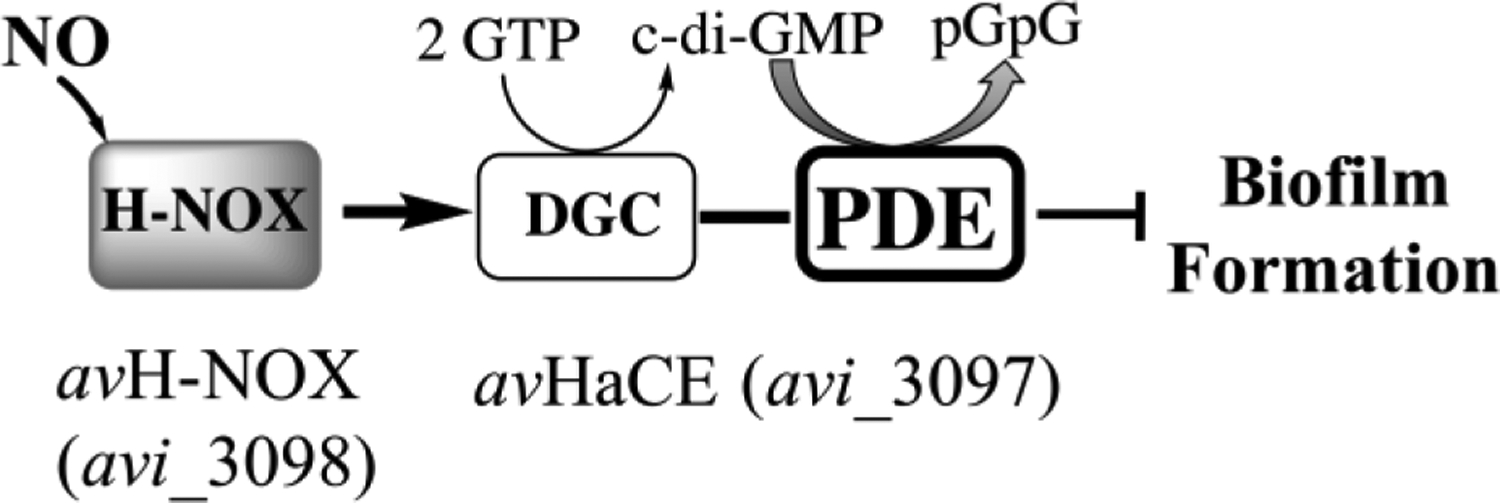
Schematic model for the H-NOX/HaCE system in A. vitis. H-NOX enhances the PDE activity of HaCE in response to NO leading to increased cyclic di-GMP degradation and reduced biofilm formation in A. vitis.
A. vitis could encounter exogenous NO numerous ways in its natural environment. Both denitrifying and nitrifying bacteria are present in the soil that produce NO as an intermediate through the reduction of nitrate to N2 gas by denitrification or the oxidation of ammonium to nitrate by nitrification (63). The dismutation of nitrite at low pH is another process that may result in NO exposure (63). Finally, A. vitis may also experience exogenous NO produced by the host plant. It has been demonstrated that grapevines produce NO as a defense response against invading pathogens (62, 64–66).
In conclusion, our results show that H-NOX is important for biofilm formation and may play a role in protection against NO toxicity in A. vitis. Importantly, we characterized the third H-NOX/HaCE bacterial system that can sense NO to regulate cyclic di-GMP metabolism. Thus far, the reoccurring theme in these NO/H-NOX/HaCE regulatory pathways is NO-induced reduction in cyclic di-GMP output from HaCE, either due to the upregulation of phosphodiesterase activity and/or the downregulation of cyclase activity upon detection of NO-bound H-NOX. Future studies will be required to examine a possible protective role of H-NOX in A. vitis.
Supplementary Material
ACKNOWLEDGMENTS
The A. vitis strains were constructed and gifted by the Burr Laboratory at Cornell University prior to Dr. Burr’s retirement in 2017. We thank Desen Zheng and Supaporn Kaewnum for help with these strains. This work was supported by NIGMS, NIH IRACDA NY-CAPS K12GM102778 (D.E.W).
Footnotes
SUPPLEMENTAL MATERIALS
Supplemental material for this article may be found at https://doi.org/xxxxx. avH-NOX NO dissociation rate constant data and analysis (Figure S1); inorganic phosphate standard curve (Figure S2); H-NOX/HaCE co-immunoprecipitation (Figure S3); H-NOX multiple sequence alignment (Figure S4); avHaCE does not bind molecular oxygen (Figure S5); PDE activity analysis of avHaCE (Figure S6); A. vitis growth curve in the presence and absence of NO (Figure S7); static biofilm assays (Figure S8); modified Invitrogen EnzChek™ pyrophosphate assay method.
REFERENCES:
- ([1.).Romling U, and Balsalobre C (2012) Biofilm infections, their resilience to therapy and innovative treatment strategies, J Intern Med 272, 541–561. [DOI] [PubMed] [Google Scholar]
- ([2.).Ceri H, Olson ME, Stremick C, Read RR, Morck D, and Buret A (1999) The Calgary Biofilm Device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms, Journal of clinical microbiology 37, 1771–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ([3.).Stewart PS, and Costerton JW (2001) Antibiotic resistance of bacteria in biofilms, Lancet 358, 135–138. [DOI] [PubMed] [Google Scholar]
- ([4.).Cary SP, Winger JA, Derbyshire ER, and Marletta MA (2006) Nitric oxide signaling: no longer simply on or off, Trends Biochem Sci 31, 231–239. [DOI] [PubMed] [Google Scholar]
- ([5.).Nisbett LM, and Boon EM (2016) Nitric Oxide Regulation of H-NOX Signaling Pathways in Bacteria, Biochemistry 55, 4873–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ([6.).Moncada S, Palmer RM, and Higgs EA (1991) Nitric oxide: physiology, pathophysiology, and pharmacology, Pharmacological Reviews 43, 109–142. [PubMed] [Google Scholar]
- ([7.).Shimizu T, Huang D, Yan F, Stranava M, Bartosova M, Fojtikova V, and Martinkova M (2015) Gaseous O2, NO, and CO in signal transduction: structure and function relationships of heme-based gas sensors and heme-redox sensors, Chem Rev 115, 6491–6533. [DOI] [PubMed] [Google Scholar]
- ([8.).Arora DP, Hossain S, Xu Y, and Boon EM (2015) Nitric oxide regulation of bacterial biofilms, Biochemistry 54, 3717–3728. [DOI] [PubMed] [Google Scholar]
- ([9.).Barraud N, Schleheck D, Klebensberger J, Webb JS, Hassett DJ, Rice SA, and Kjelleberg S (2009) Nitric oxide signaling in Pseudomonas aeruginosa biofilms mediates phosphodiesterase activity, decreased cyclic diguanosine-5’-monophosphate levels and enhanced dispersal, J Bacteriol 191, 7333–7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ([10.).Carlson HK, Vance RE, and Marletta MA (2010) H-NOX regulation of c-di-GMP metabolism and biofilm formation in Legionella pneumophila, Molecular Microbiology 77, 930–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ([11.).Henares BM, Higgins KE, and Boon EM (2012) Discovery of a Nitric Oxide Responsive Quorum Sensing Circuit in Vibrio harveyi, Acs Chem Biol 7, 1331–1336. [DOI] [PubMed] [Google Scholar]
- ([12.).Liu N, Xu Y, Hossain S, Huang N, Coursolle D, Gralnick JA, and Boon EM (2012) Nitric oxide regulation of cyclic di-GMP synthesis and hydrolysis in Shewanella woodyi, Biochemistry 51, 2087–2099. [DOI] [PubMed] [Google Scholar]
- ([13.).Plate L, and Marletta MA (2012) Nitric Oxide Modulates Bacterial Biofilm Formation through a Multicomponent Cyclic-di-GMP Signaling Network, Molecular Cell 46, 449–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ([14.).Plate L, and Marletta MA (2013) Nitric oxide-sensing H-NOX proteins govern bacterial communal behavior, Trends Biochem Sci 38, 566–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ([15.).Wang Y, Dufour YS, Carlson HK, Donohue TJ, Marletta MA, and Ruby EG (2010) H-NOX-mediated nitric oxide sensing modulates symbiotic colonization by Vibrio fischeri, Proceedings of the National Academy of Sciences of the United States of America 107, 8375–8380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ([16.).Barraud N, Hassett DJ, Hwang S-H, Rice SA, Kjelleberg S, and Webb JS (2006) Involvement of Nitric Oxide in Biofilm Dispersal of Pseudomonas aeruginosa, J. Bacteriol 188, 7344–7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ([17.).Barraud N, Storey MV, Moore ZP, Webb JS, Rice SA, and Kjelleberg S (2009) Nitric oxide-mediated dispersal in single- and multi-species biofilms of clinically and industrially relevant microorganisms, Microb Biotechnol 2, 370–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ([18.).Romling U, Galperin MY, and Gomelsky M (2013) Cyclic di-GMP: the first 25 years of a universal bacterial second messenger, Microbiology and molecular biology reviews : MMBR 77, 1–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ([19.).Xu J, Kim J, Koestler BJ, Choi JH, Waters CM, and Fuqua C (2013) Genetic analysis of Agrobacterium tumefaciens unipolar polysaccharide production reveals complex integrated control of the motile-to-sessile switch, Mol Microbiol 89, 929–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ([20.).Feirer N, Xu J, Allen KD, Koestler BJ, Bruger EL, Waters CM, White RH, and Fuqua C (2015) A Pterin-Dependent Signaling Pathway Regulates a Dual-Function Diguanylate Cyclase-Phosphodiesterase Controlling Surface Attachment in Agrobacterium tumefaciens, mBio 6, e00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ([21.).Hengge R (2009) Principles of c-di-GMP signalling in bacteria, Nat Rev Micro 7, 263–273. [DOI] [PubMed] [Google Scholar]
- ([22.).Hossain S, Nisbett LM, and Boon EM (2017) Discovery of Two Bacterial Nitric Oxide-Responsive Proteins and Their Roles in Bacterial Biofilm Regulation, Accounts of chemical research. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ([23.).Boon EM, Huang SH, and Marletta MA (2005) A molecular basis for NO selectivity in soluble guanylate cyclase, Nature chemical biology 1, 53–59. [DOI] [PubMed] [Google Scholar]
- ([24.).Boon EM, and Marletta MA (2005) Ligand specificity of H-NOX domains: from sGC to bacterial NO sensors, J Inorg Biochem 99, 892–902. [DOI] [PubMed] [Google Scholar]
- ([25.).Guo Y, Cooper MM, Bromberg R, and Marletta MA (2018) A Dual-H-NOX Signaling System in Saccharophagus degradans, Biochemistry 57, 6570–6580. [DOI] [PubMed] [Google Scholar]
- ([26.).Guo Y, Iavarone AT, Cooper MM, and Marletta MA (2018) Mapping the H-NOX/HK Binding Interface in Vibrio cholerae by Hydrogen/Deuterium Exchange Mass Spectrometry, Biochemistry 57, 1779–1789. [DOI] [PubMed] [Google Scholar]
- ([27.).Guo Y, and Marletta MA (2019) Structural Insight into H-NOX Gas Sensing and Cognate Signaling Protein Regulation, Chembiochem 20, 7–19. [DOI] [PubMed] [Google Scholar]
- ([28.).Herzik MA Jr., Jonnalagadda R, Kuriyan J, and Marletta MA (2014) Structural insights into the role of iron-histidine bond cleavage in nitric oxide-induced activation of H-NOX gas sensor proteins, Proceedings of the National Academy of Sciences of the United States of America 111, E4156–4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ([29.).Hespen CW, Bruegger JJ, Guo Y, and Marletta MA (2018) Native Alanine Substitution in the Glycine Hinge Modulates Conformational Flexibility of Heme Nitric Oxide/Oxygen (H-NOX) Sensing Proteins, Acs Chem Biol 13, 1631–1639. [DOI] [PubMed] [Google Scholar]
- ([30.).Hespen CW, Bruegger JJ, Phillips-Piro CM, and Marletta MA (2016) Structural and Functional Evidence Indicates Selective Oxygen Signaling in Caldanaerobacter subterraneus H-NOX, Acs Chem Biol 11, 2337–2346. [DOI] [PubMed] [Google Scholar]
- ([31.).Lahiri T, Luan B, Raleigh DP, and Boon EM (2014) A structural basis for the regulation of an H-NOX-associated cyclic-di-GMP synthase/phosphodiesterase enzyme by nitric oxide-bound H-NOX, Biochemistry 53, 2126–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ([32.).Mukhopadyay R, Sudasinghe N, Schaub T, and Yukl ET (2016) Heme-independent Redox Sensing by the Heme-Nitric Oxide/Oxygen-binding Protein (H-NOX) from Vibrio cholerae, The Journal of biological chemistry 291, 17547–17556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ([33.).Muralidharan S, and Boon EM (2012) Heme flattening is sufficient for signal transduction in the H-NOX family, Journal of the American Chemical Society 134, 2044–2046. [DOI] [PubMed] [Google Scholar]
- ([34.).Olea C, Boon EM, Pellicena P, Kuriyan J, and Marletta MA (2008) Probing the Function of Heme Distortion in the H-NOX Family, Acs Chem Biol 3, 703–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ([35.).Price MS, Chao LY, and Marletta MA (2007) Shewanella oneidensis MR-1 H-NOX Regulation of a Histidine Kinase by Nitric Oxide†, Biochemistry 46, 13677–13683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ([36.).Rao M, Herzik MA Jr., Iavarone AT, and Marletta MA (2017) Nitric Oxide-Induced Conformational Changes Govern H-NOX and Histidine Kinase Interaction and Regulation in Shewanella oneidensis, Biochemistry 56, 1274–1284. [DOI] [PubMed] [Google Scholar]
- ([37.).Wu G, Liu W, Berka V, and Tsai AL (2013) The selectivity of Vibrio cholerae H-NOX for gaseous ligands follows the “sliding scale rule” hypothesis. Ligand interactions with both ferrous and ferric Vc H-NOX, Biochemistry 52, 9432–9446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ([38.).Wu G, Liu W, Berka V, and Tsai AL (2017) Gaseous ligand selectivity of the H-NOX sensor protein from Shewanella oneidensis and comparison to those of other bacterial H-NOXs and soluble guanylyl cyclase, Biochimie 140, 82–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ([39.).Nesbitt NM, Arora DP, Johnson RA, and Boon EM (2015) Modification of a bi-functional diguanylate cyclase-phosphodiesterase to efficiently produce cyclic diguanylate monophosphate, Biotechnol Rep (Amst) 7, 30–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ([40.).Simon R, Priefer U, and Pühler A (1983) A Broad Host Range Mobilization System for In Vivo Genetic Engineering: Transposon Mutagenesis in Gram Negative Bacteria, Bio/Technology 1, 784–791. [Google Scholar]
- ([41.).Kalogeraki VS, and Winans SC (1997) Suicide plasmids containing promoterless reporter genes can simultaneously disrupt and create fusions to target genes of diverse bacteria, Gene 188, 69–75. [DOI] [PubMed] [Google Scholar]
- ([42.).Hajdukiewicz P, Svab Z, and Maliga P (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation., Plant Mol Biol. 25, 989–994. [DOI] [PubMed] [Google Scholar]
- ([43.).Bradford M (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding., Anal Biochem 7, 248–254. [DOI] [PubMed] [Google Scholar]
- ([44.).Boon EM, Davis JH, Tran R, Karow DS, Huang SH, Pan D, Miazgowicz MM, Mathies RA, and Marletta MA (2006) Nitric oxide binding to prokaryotic homologs of the soluble guanylate cyclase beta1 H-NOX domain, The Journal of biological chemistry 281, 21892–21902. [DOI] [PubMed] [Google Scholar]
- ([45.).Hao G, and Burr TJ (2006) Regulation of long-chain N-acyl-homoserine lactones in Agrobacterium vitis, J Bacteriol 188, 2173–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ([46.).O’Toole GA (2011) Microtiter dish biofilm formation assay, Journal of visualized experiments : JoVE, e2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ([47.).Abarca-Grau AM, Penyalver R, López MM, and Marco-Noales E (2011) Pathogenic and non-pathogenic Agrobacterium tumefaciens, A. rhizogenes and A. vitis strains form biofilms on abiotic as well as on root surfaces., Plant Pathology 60, 416–425. [Google Scholar]
- ([48.).Stone JR, and Marletta MA (1994) Soluble guanylate cyclase from bovine lung: activation with nitric oxide and carbon monoxide and spectral characterization of the ferrous and ferric states, Biochemistry 33, 5636–5640. [DOI] [PubMed] [Google Scholar]
- ([49.).Kharitonov VG, Sharma VS, Magde D, and Koesling D (1997) Kinetics of nitric oxide dissociation from five- and six-coordinate nitrosyl hemes and heme proteins, including soluble guanylate cyclase, Biochemistry 36, 6814–6818. [DOI] [PubMed] [Google Scholar]
- ([50.).Gerzer R, Bohme E, Hofmann F, and Schultz G (1981) Soluble guanylate cyclase purified from bovine lung contains heme and copper, FEBS letters 132, 71–74. [DOI] [PubMed] [Google Scholar]
- ([51.).Reynolds MF, Parks RB, Burstyn JN, Shelver D, Thorsteinsson MV, Kerby RL, Roberts GP, Vogel KM, and Spiro TG (2000) Electronic absorption, EPR, and resonance raman spectroscopy of CooA, a CO-sensing transcription activator from R. rubrum, reveals a five-coordinate NO-heme, Biochemistry 39, 388–396. [DOI] [PubMed] [Google Scholar]
- ([52.).Stone JR, Sands RH, Dunham WR, and Marletta MA (1995) Electron paramagnetic resonance spectral evidence for the formation of a pentacoordinate nitrosyl-heme complex on soluble guanylate cyclase, Biochemical and biophysical research communications 207, 572–577. [DOI] [PubMed] [Google Scholar]
- ([53.).Karow DS, Pan D, Tran R, Pellicena P, Presley A, Mathies RA, and Marletta MA (2004) Spectroscopic characterization of the soluble guanylate cyclase-like heme domains from Vibrio cholerae and Thermoanaerobacter tengcongensis, Biochemistry 43, 10203–10211. [DOI] [PubMed] [Google Scholar]
- ([54.).Zhu S, Gan Z, Li Z, Liu Y, Yang X, Deng P, Xie Y, Yu M, Liao H, Zhao Y, Zhao L, and Liao F (2009) The measurement of cyclic nucleotide phosphodiesterase 4 activities via the quantification of inorganic phosphate with malachite green, Anal Chim Acta 636, 105–110. [DOI] [PubMed] [Google Scholar]
- ([55.).De N, Navarro MV, Raghavan RV, and Sondermann H (2009) Determinants for the activation and autoinhibition of the diguanylate cyclase response regulator WspR, Journal of molecular biology 393, 619–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ([56.).Rao F, Qi Y, Chong HS, Kotaka M, Li B, Li J, Lescar J, Tang K, and Liang ZX (2009) The functional role of a conserved loop in EAL domain-based cyclic di-GMP-specific phosphodiesterase, J Bacteriol 191, 4722–4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ([57.).Underbakke ES, Iavarone AT, and Marletta MA (2013) Higher-order interactions bridge the nitric oxide receptor and catalytic domains of soluble guanylate cyclase, Proceedings of the National Academy of Sciences of the United States of America 110, 6777–6782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ([58.).Danhorn T, and Fuqua C (2007) Biofilm formation by plant-associated bacteria, Annu Rev Microbiol 61, 401–422. [DOI] [PubMed] [Google Scholar]
- ([59.).Morris CE, and Monier JM (2003) The ecological significance of biofilm formation by plant-associated bacteria, Annu Rev Phytopathol 41, 429–453. [DOI] [PubMed] [Google Scholar]
- ([60.).Burr TJ, Bazzi C, Sule S, and Otten L (1998) Crown Gall of Grape: Biology of Agrobacterium vitis and the Development of Disease Control Strategies, Plant Dis 82, 1288–1297. [DOI] [PubMed] [Google Scholar]
- ([61.).Matthysse AG, Marry M, Krall L, Kaye M, Ramey BE, Fuqua C, and White AR (2005) The effect of cellulose overproduction on binding and biofilm formation on roots by Agrobacterium tumefaciens, Mol Plant Microbe Interact 18, 1002–1010. [DOI] [PubMed] [Google Scholar]
- ([62.).Arasimowicz-Jelonek M, and Floryszak-Wieczorek J (2014) Nitric oxide: an effective weapon of the plant or the pathogen?, Molecular plant pathology 15, 406–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ([63.).Pilegaard K (2013) Processes regulating nitric oxide emissions from soils, Philos Trans R Soc Lond B Biol Sci 368, 20130126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ([64.).Vandelle E, Poinssot B, Wendehenne D, Bentejac M, and Alain P (2006) Integrated signaling network involving calcium, nitric oxide, and active oxygen species but not mitogen-activated protein kinases in BcPG1-elicited grapevine defenses, Mol Plant Microbe Interact 19, 429–440. [DOI] [PubMed] [Google Scholar]
- ([65.).Santolini J, Andre F, Jeandroz S, and Wendehenne D (2017) Nitric oxide synthase in plants: Where do we stand?, Nitric Oxide 63, 30–38. [DOI] [PubMed] [Google Scholar]
- ([66.).Bellin D, Asai S, Delledonne M, and Yoshioka H (2013) Nitric oxide as a mediator for defense responses, Mol Plant Microbe Interact 26, 271–277. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


