Abstract
Background and Aims
Studying the relationship between phenotypic and genetic variation in populations distributed across environmental gradients can help us to understand the ecological and evolutionary processes involved in population divergence. We investigated the patterns of genetic and phenotypic diversity in the European crabapple, Malus sylvestris, a wild relative of the cultivated apple (Malus domestica) that occurs naturally across Europe in areas subjected to different climatic conditions, to test for divergence among populations.
Methods
Growth rates and traits related to carbon uptake in seedlings collected across Europe were measured in controlled conditions and associated with the genetic status of the seedlings, which was assessed using 13 microsatellite loci and the Bayesian clustering method. Isolation-by-distance, isolation-by-climate and isolation-by-adaptation patterns, which can explain genetic and phenotypic differentiation among M. sylvestris populations, were also tested.
Key Results
A total of 11.6 % of seedlings were introgressed by M. domestica, indicating that crop–wild gene flow is ongoing in Europe. The remaining seedlings (88.4 %) belonged to seven M. sylvestris populations. Significant phenotypic trait variation among M. sylvestris populations was observed. We did not observe significant isolation by adaptation; however, the significant association between genetic variation and the climate during the Last Glacial Maximum suggests that there has been local adaptation of M. sylvestris to past climates.
Conclusions
This study provides insight into the phenotypic and genetic differentiation among populations of a wild relative of the cultivated apple. This might help us to make better use of its diversity and provide options for mitigating the impact of climate change on the cultivated apple through breeding.
Keywords: Population structure, isolation by distance, isolation by ecology, local adaptation, climate change, apple tree, crop wild relatives
INTRODUCTION
Knowledge of the spatial phenotypic and genetic variation among populations is essential for understanding the ecological (biotic and abiotic factors) and evolutionary (gene flow, selection, drift and mutation) processes involved in population divergence and adaptation (Savolainen et al., 2013; Sork, 2018). Plant species distributed across climatic gradients typically experience spatial variation in selection, genetic drift and gene flow, which drives genetic and phenotypic divergences among populations (Svenning et al., 2015). Climate influences demographic processes such as population expansion and contraction, the extent of gene flow among populations and, ultimately, the degree of genetic divergence among populations (Edwards et al., 2022). For instance, changes in the climate since the Last Glacial Maximum (LGM) 20 000 years ago have driven the genetic composition of the European crabapple and many other tree species (Comes and Kadereit, 1998; Kremer et al., 2002; Pyhäjärvi et al., 2008; Cornille et al., 2013a; Riordan et al., 2016; Lander et al., 2021; Parisod, 2021; Yamada et al., 2021). Climate can also shape phenotypic variation among populations. Populations occurring in the same climate can share physiological tolerances to climatic conditions, including plant carbon uptake via photosynthesis. Carbon uptake traits condition plant size and growth, reproduction and survival in different climatic conditions (Hutyra et al., 2007; Nicotra et al., 2010; Pasho et al., 2012; Way and Montgomery, 2015; Mercado et al., 2018; Hartmann et al., 2020; Kühn et al., 2021). Variation in plant carbon uptake in response to climatic conditions can result from phenotypic plasticity, i.e. the ability of individual genotypes to produce different phenotypes when exposed to other environmental conditions, in this case, climate (Dusenge et al., 2019). In some cases, local climate can impose divergent selection on carbon uptake traits, which leads to reproductive isolation among populations; loci under adaptive divergence act as a local barrier to gene flow (Keller et al., 2011; Franks et al., 2014; Aitken and Bemmels, 2015; Ramírez-Valiente et al., 2017; Alexandre et al., 2020). Climate can therefore lead to a long-term reduction in gene flow and local adaptation. Whether the phenotypic variation observed in species distributed across large climatic ranges results from their demographic and/or adaptive histories remains an intense topic of investigation (Li et al., 2012; Tiffin and Ross-Ibarra, 2014; Briscoe Runquist et al., 2020). Investigating this question can help us to predict how plants might respond to climate change and how species adapt to their environment.
There are multiple ways to investigate whether genetic and phenotypic variation among populations distributed across climatic gradients results from selection, genetic drift and/or gene flow. A first step is to use a common garden experiment to investigate the genetic basis of phenotypic variation among populations. Different populations occurring across a climatic gradient might display clinal variation, i.e. differences in a trait that might result from phenotypic plasticity or local genetic adaptation (Savolainen et al., 2013; de Villemereuil et al., 2016). Measuring candidate traits for adaptation to climate [e.g. phenology (Brachi et al., 2013) or traits related to plant carbon uptake (Savolainen et al., 2013; de Villemereuil et al., 2016)] in individuals from different populations in the same environmental conditions can help us to elucidate the genetic basis of phenotypic variation across populations without the confounding effects of the environment. Ideally, common garden trials should include the main genetic groups across the species distribution (de Villemereuil et al., 2020). The association of neutral genotypic variation with phenotypic variation can also be used as evidence of adaptive divergence among populations (Shafer and Wolf, 2013; Wang and Bradburd, 2014). The correlation between neutral genetic differentiation and environmental or phenotypic divergence among populations, independent of geographical distance, referred to as isolation by ecology (IBE), is an extension of the isolation-by-distance (IBD) model (Wright, 1943). Patterns of IBE have been increasingly used as an indicator of adaptive divergence between populations (Shafer and Wolf, 2013). In the IBE model, natural selection, which results from several factors, including climate, can indirectly increase neutral genetic and phenotypic differentiation between populations by promoting general barriers to gene flow (Nosil et al., 2009; Orsini et al., 2013; Shafer and Wolf, 2013; Wang and Bradburd, 2014). Although it can be challenging to map the processes underlying IBE patterns, testing for it is valuable to understand better how natural selection shapes neutral genetic and phenotypic variation. Therefore, evidence from common garden experiments and IBE patterns can contribute to understanding how genotypes, phenotypes and the environment interact to influence population divergence and, potentially, local adaptation.
Fruit trees are a significant component of terrestrial ecosystems (Petit and Hampe, 2011). They are grown in managed plantations and orchards to provide a variety of economically important products (Boyd et al., 2013). Recent breeding efforts have involved the repeated use of a limited number of commercial cultivars, leading to a reduction in genetic diversity that can lead to the loss of valuable alleles of genes that are not directly targeted by human selection (Myles et al., 2011; Warschefsky and von Wettberg, 2019; Migicovsky et al., 2021). Wild relatives of crop fruit trees [crop wild relatives (CWRs)] harbour phenotypic and genetic diversity that is potentially of high valuable for future breeding programmes in the context of climate change (Zhang et al., 2017; Hoban et al., 2018; Hübner and Kantar, 2021). However, phenotypic variation in CWRs in relationship to climate variation has rarely been studied in fruit trees (Kremer and Hipp, 2019). Key traits to study in this context are related to plant carbon uptake. Climate impacts plant carbon uptake (Aubin et al., 2016), which impacts fruit quality characteristics and production (Demestihas et al., 2017). These are topical issues because native CWRs can be threatened by crop-to-wild gene flow from nearby domesticated trees (Delplancke et al., 2011; Cornille et al., 2015; Diez et al., 2015; Feurtey et al., 2017; Flowers et al., 2019; Liu et al., 2019). Therefore, studying the genetic and phenotypic variation among CWR fruit tree populations is timely to guide future breeding programmes. It might also contribute to our understanding of the evolutionary and ecological drivers of population divergence, such as climate.
The European crabapple, Malus sylvestris (L.) Mill., is a CWR of the cultivated apple, Malus domestica (Cornille et al., 2012, 2014, 2019; Peace et al., 2019). Substantial crop-to-wild gene flow has been observed across M. sylvestris populations in Europe [≤23.1 % of naturally occurring individuals are introgressed by M. domestica (Cornille et al., 2015)]. Crop–wild hybrids sampled in a forest in France and grown in controlled conditions showed higher growth rates than wild seedlings (Feurtey et al., 2017). Population genetic analyses also identified five pure (i.e. not introgressed by M. domestica) populations in Scandinavia, western France, eastern France, Eastern Europe and Italy (Cornille et al., 2015). These five populations resulted from past contractions and expansions associated with the LGM (Cornille et al., 2013a; Cornille et al., 2015). It is not known whether these five populations distributed across a large area with different climatic conditions show a phenotypic variation that could result from local adaptation to past and/or present climates.
We investigated the spatial phenotypic and genetic variation among populations of a wild contributor to the cultivated apple genome, M. sylvestris, to test for adaptive divergence, particularly to climate. Plant growth and traits related to carbon uptake were measured in 584 M. sylvestris seedlings grown in controlled conditions and genotyped using 13 microsatellite markers. We first assessed the genetic status of each seedling (pure vs. crop–wild hybrid). Then, we compared growth traits and traits related to plant carbon uptake among seedlings from different European genetic groups. We also formally tested the impact of geography (IBD) and ecology (IBE tested with phenotypic traits and climate) on genetic variation observed from 13 microsatellite markers. We investigated the following questions. First, do growth rates and carbon uptake traits vary between populations of the European crabapple, and are those traits heritable and therefore good candidates for the responses to selection? Second, is there any association between phenotypic variation and genetic variation, taking into account geographical distance, which would suggest adaptive divergence? Third, do we detect a pattern of isolation by climate in the European crabapple that could suggest local adaptive divergence to climate?
MATERIALS AND METHODS
Plant material, experimental design and trait measurements
A total of 584 seeds were collected from 90 M. sylvestris mother trees (between 3 and 15 seeds per mother tree; Supplementary data Table S1) from 22 geographical sites in Europe representing the five main genetic groups previously detected by Cornille et al. (2015): Austria (n = 89, two sites), Denmark (n = 91, three sites), Spain (n = 39, one site), France (n = 220, eight sites), Italy (n = 32, one site) and Romania (n = 117, seven sites) (Supplementary data Table S1). The length, width and weight of 30 seeds per mother tree were measured with an Opto-Agri (Optomachine, Riom, France).
In mid-April 2019, 584 seeds were washed, sterilized (in 0.5 % chlorine for 20 min), and stratified for 3 months at 4 °C in the dark in a mix of damp sand and vermiculite. Then, seeds were sown in jiffy pellets, and each pellet was placed randomly in a 20-hole array. Seeds were grown in controlled conditions for 2 months (from mid-July to mid-September 2019: 22 ± 1 °C, 60 ± 5 % relative humidity, a 16 h:8 h (light:dark) photoperiod, and a light level of 40–60 µmol m−2 s−1). Each 20-hole array was rotated daily in the growth chamber to avoid any micro-environmental variation in plant response, and plants were watered weekly.
During the 2-month experiment, the number of leaves and height of each seedling was recorded. Owing to the low germination rate, some accessions could not be evaluated (i.e. plant height and number of leaves could not be recorded for 19 of 584 seedlings, resulting in n = 565; Table 1). Seedlings were measured every 2–3 days, starting from days 7–11 after commencing the experiment.
Table 1.
Number of Malus sylvestris seedlings used in this study for population genetic analyses inferred with STRUCTURE for K = 8, with 13 microsatellite markers and phenotyping (growth and carbon uptake-related traits).
| Clusters | N pure | N ww | N cw | N dom | N no cluster | Total measured for phenotypic traits | Population name |
|---|---|---|---|---|---|---|---|
| Q1 (light green) | 32 | 46 | 7 | 0 | 10 | 92 | FR-W |
| Q2 (yellow) | 0 | 52 | 5 | 0 | 4 | 57 | FR-E |
| Q3 (pink) | 28 | 1 | 0 | 0 | 28 | FR-Lor | |
| Q4 (blue) | 61 | 21 | 1 | 0 | 6 | 85 | DA |
| Q5 (purple) | 23 | 4 | 3 | 0 | 4 | 34 | IT |
| Q6 (dark green) | 66 | 17 | 1 | 0 | 1 | 83 | AUT |
| Q7 (red) | 77 | 34 | 5 | 0 | 0 | 113 | RO |
| Q8 (black; M. domestica) | 40 | 0 | 46 | 21 | 8 | 73 | DOM |
| Total | 287 | 175 | 68 | 21 | 33 | 551 (584) | |
| Total measured for height and number of leaves | 282 | 167 | 63 | 21 | 32 | 533 (565) | |
| Total measured for leaf chlorophyll and flavonol contents, NBI | 129 | 82 | 22 | 6 | 18 | 239 (257) | |
Abbreviations: NBI, nitrogen balance index; Npure, the number of seedlings assigned to a wild gene pool with a membership coefficient > 0.9; Nww, the number of wild–wild hybrids (i.e. seedlings with a membership coefficient > 0.1 to a wild gene pool other than its own wild gene pool and a membership coefficient < 0.1 to the Malus domestica gene pool); Ncw, the number of crop–wild hybrids (i.e. seedlings assigned to the M. domestica gene pool with a membership coefficient > 0.1); Nno cluster, seedlings that could not be assigned to any defined gene pool. The total measured for phenotypic traits is the number of individuals measured for each phenotypic trait and included in the statistical analyses; the number in parentheses represents the initial sample size before data were filtered for statistical analyses. ‘Wild population name’ specifies populations defined with STRUCTURE for K = 8 excluding crop–wild hybrids and seedlings from misidentified mother trees (i.e. including only wild pure and wild–wild hybrids). Populations are as follows: Austria (AUT), Denmark (DA), Eastern France (FR-E), Lorraine in France (FR-Lor), Western France (FR-W), Italy (IT), Romania (RO) and M. domestica (DOM).
In the last week of the experiment, the superficial flavonol and chlorophyll contents were measured, and the nitrogen balance index (NBI) was calculated for three leaves per seedling. The superficial chlorophyll content is the chlorophyll concentration in the leaf epidermis (in micrograms per square centimetre), and the superficial flavonol content is an index of the flavonoid concentration (in micrograms per square centimetre) in this upper layer and is related to phenol accumulation and ultraviolet protection. The NBI assesses the nitrogen status of the leaf by calculating the ratio of chlorophyll to flavonols (related to nitrogen/carbon allocation) and is currently used as a proxy to estimate the foliar nitrogen content (Demotes-Mainard et al., 2008). Leaf chlorophyll, flavonol content and NBI are parameters correlated with plant carbon uptake via photosynthesis. Flavonol is a phenolic compound that contributes to plant vigour, acclimatization and adaptation to environmental constraints through various mechanisms, including its antioxidant activity. These traits were measured using a portable Dualex® device (Force-A, Orsay, France), which uses a combination of fluorescence signals at various excitation bands to quantify pigments and chemical compounds and has already been calibrated on the apple tree (Hamann et al., 2018). Given that carbon uptake-related traits must be measured on the same day, a subsample of 257 of the 565 seedlings (Table 1; numbers in parentheses) was measured because of time limitations. Seedlings measured for carbon uptake-related traits were selected such that at least one seedling per mother tree and three seedlings per geographical site were sampled.
DNA extraction, microsatellite genotyping and genetic ancestry of the seedlings
At the end of the experiment, the leaves of each seedling were sampled for microsatellite genotyping. Genomic DNA was extracted using the NucleoSpin Plant DNA Extraction Kit II (Macherey & Nagel, Düren, Germany), according to the manufacturer’s instructions. Microsatellites were amplified by multiplex PCR with the Multiplex PCR Kit (QIAGEN, Inc.). We used 13 microsatellite markers (Ch01f02, Ch01f03, Ch01h01, Ch01h10, Ch02c06, Ch02c09, Ch02c11, Ch02d08, Ch03d07, Ch04c07, Ch05f06, GD12 and Hi02c07) in four multiplexes [MP01–MP04 (Cornille et al., 2012)].
Polymerase chain reaction was performed in a final reaction volume of 15 μL (7.5 μL of QIAGEN Multiplex Master Mix, 10–20 μm of each primer, with the forward primer labelled with a fluorescent dye, and 10 ng of template DNA). We used a touch-down PCR program (initial annealing temperature of 60 °C, decreasing by 1 °C per cycle to 55 °C). Genotyping was performed at the GENTYANE platform (INRAE Clermont-Ferrand) on an ABI PRISM X3730XL, with 2 mL of GS500LIZ size standard (Applied Biosystems). Alleles were scored with the GENEMAPPER v.4.0 software (Applied Biosystems). We retained only multilocus genotypes presenting <10 % of missing data.
Clones or closely related individuals can bias inferences about the population structure. We estimated the kinship coefficient between pairs of individuals (Fij) with SPAGeDI 1.5d (Loiselle et al., 1995; Hardy and Vekemans, 2002) and removed individuals with high genetic relatedness with Fij > 0.5.
The individual-based Bayesian clustering method implemented in STRUCTURE v.2.3.3 (Pritchard et al., 2000) was used to estimate the admixture between M. domestica and M. sylvestris, in addition to the population genetic structure of M. sylvestris. STRUCTURE uses Markov chain Monte Carlo simulations to infer the proportion of ancestry of genotypes from K distinct clusters. The underlying algorithm minimizes deviations from Hardy–Weinberg, and linkage disequilibria. The value of K ranged from one to ten. Ten independent runs were carried out for each value of K, and 500 000 Markov chain Monte Carlo (iterations after a burn-in of 50 000 steps were used. Clumpak (greedy algorithm) (Kopelman et al., 2015) was used to identify distinct modes in the ten replicated runs for each value of K. STRUCTURE analyses were run for the entire dataset (n = 584), plus 40 M. domestica genotypes from Western Europe (Supplementary data Table S2) included as a reference for the cultivated apple gene pool (Cornille et al., 2013b). The R package pophelper v.2.3.0 was used (Francis, 2016) to visualize bar plots. The amount of additional information explained by increasing K was determined using the ΔK statistic (Evanno et al., 2005), as implemented in STRUCTURE HARVESTER (Earl and vonHoldt, 2012). However, ΔK provides statistical support for the strongest but not the finest population structure (Puechmaille, 2016). Natural populations can display a hierarchical genetic structure, with a fine-scale population structure. Visual inspection of the bar plots was used to identify the value of K for which all clusters have well-assigned individuals and where additional clusters at higher values of K do not have well-assigned individuals. Therefore, the K value was the finest one, which can be higher than the K value of the strongest population structure identified by ΔK.
We used the membership coefficients at the best K value (inferred with STRUCTURE) for the crop and wild seedlings and defined the genetic status of each seedling. Initially, we separated cultivated apples (seedlings from mother trees that were misidentified in the field, i.e. with a membership proportion to the M. domestica gene pool, Pdom, > 0.9, referred to as ‘dom’ hereafter), crop–wild hybrids (seedlings with 0.1 > Pdom > 0.9, referred to as ‘cw’ hereafter) and pure M. sylvestris (seedlings with a membership coefficient to a given wild apple cluster > 0.9 and with Pdom < 0.1, referred to as ‘pure’ hereafter). We then separated ‘pure’ wild seedlings (i.e. seedlings with membership coefficients to a given wild gene pool > 0.9) from wild–wild admixed seedlings (i.e. seedlings with a membership coefficient to a given wild apple cluster < 0.9, referred to as ‘ww’ hereafter). Two effects were then tested using the statistical models described below: the genetic status effect (i.e. dom, cw, ww and pure) and the wild apple population effect (i.e. corresponding to the ‘pure’ populations detected with STRUCTURE). A seedling that could not be assigned to any cluster (with a membership coefficient to any cluster < 0.5) was removed from further analysis.
We computed descriptive population genetic estimates for each population (i.e. each cluster inferred with STRUCTURE, excluding admixed individuals with a membership coefficient < 0.9). Heterozygosity (expected and observed), Weir and Cockerham F-statistics and deviations from Hardy–Weinberg equilibrium were calculated with genepop v.4.2 (Raymond and Rousset, 1995; Rousset, 2008).
Phenotypic traits and correlations
Height, leaf growth rate and carbon-related traits were estimated for each seedling. The absolute height and leaf growth rates [AGR (Radford, 1967)], relative growth rates [RGR (Briggs et al., 1920)] and whole AGRs were estimated as follows:
| (1) |
| (2) |
| (3) |
Note that for whole AGRs, the beginning of the experiment corresponded to days 7 and 11 for leaf and height measurements, respectively, and the last measurement was performed on day 60. The internode ratio, which represents the ratio between the number of leaves and the height of the seedling at day 60, was also considered as a fitness trait, because this value plays an essential role in apple tree architecture (Ripetti et al., 2008).
Seven phenotypic traits were therefore calculated for the entire dataset (565 seedlings; Table 1): height_AGR, height_RGR, whole_height_AGR, leaf_AGR, leaf_RGR, whole_leaf_AGR and internode. In addition, chlorophyll (Chl) content, flavonol (Flav) content and NBI were measured in a subsample of 257 seedlings (Table 1).
A preliminary exploration of the variation among phenotypic traits was performed. We used the cor R function to assess the correlation between phenotypic traits and seed sizes (length, width and weight) of the mother trees, and between phenotypic traits. The corrplot R package was used to visualize the correlations (Wei and Simko, 2017). Principal component analyses of the phenotypic traits of the seedlings were conducted with the FactoMineR R package (Lê et al., 2008).
Statistical analyses of phenotypic variation
A previous study demonstrated that crop-to-wild gene flow impacts early-stage growth rate (Feurtey et al., 2017). The effect of the genetic status of seedlings (dom, cw, ww or pure; Table 1) on fitness variation among seedlings was therefore tested. A linear mixed model was fitted to the data as follows:
| (4) |
where Yijk is the phenotypic trait (height or leaf growth rate or a carbon uptake-related trait) of a seedling from mother k from population i with the genetic status j, wild population of origin is the fixed effect of the population of origin i of the seedling inferred with STRUCTURE, genetic status is the fixed effect of the genetic status of the seedling (i.e. pure, dom, ww or cw), the interaction between the two fixed effects, mother tree is a normally distributed random effect with its own mean and variance parameters and eijkl is the residual. The mother tree of each seedling was used as a random factor to avoid pseudo-replication owing to the presence of multiple half-siblings (i.e. from the same mother tree). We ran the model (4), but replaced the genetic status effect with the Pdom fixed effect. We gradually removed interactions and effects depending on their significance. In addition, we evaluated the differences in the effect on trait variation using a contrast analysis. We fitted the data to the model using the lme4 R package (Bates et al., 2015). The statistical significance of an effect was assessed with a type II Wald χ2 test.
For phenotypic traits estimated from the number of leaves (leaf_AGR, leaf_RGR and whole_leaf_AGR), a log-link function was used, and the residual distribution was fitted to a negative binomial distribution (function glm.nb in R package lme4). For phenotypic traits defined from the height of the seedling (height_AGR, height_RGR and whole_height_AGR) and for chlorophyll content, flavonol content and NBI, a similar linear mixed model was run, but with a residual term that was assumed to be normally distributed.
Heritability estimates
The level of heritability estimate provides an indication of the potential extent of the response to the selection of a trait. Therefore, computing heritability can allow us to determine whether the traits measured in this study could be good candidates to respond to selection. Heritability estimates were calculated using only pure and wild–wild M. sylvestris seedlings (Table 1). We fitted each fitness proxy with a linear mixed model as follows:
| (5) |
where Yijk is the fitness proxy (growth rate or carbon uptake-related trait) of the kth seedling from mother tree i, member of the jth genetic cluster, μ the overall fixed mean of the population, Fi the random effect of the ith mother tree, Cj is the fixed effect of the jth genetic cluster and eijk the random error term. The model was fitted using REML (restricted maximum likelihood). Calculations were performed by the lme function of the R library nlme (Pinheiro et al., 2018). The output of lme provides estimates for the variance components, the corresponding s.d., and the best unbiased linear predictors for random effects. Genetic parameters were then calculated as follows:
the additive genetic variance: , with representing the between-mother tree variance;
the corresponding coefficient of variation: ;
the phenotypic variation: , with representing the residual variance;
the corresponding coefficient of variation: ;
Narrow-sense heritability: ; and
Dickerson’s approximation of s.d: .
Test for isolation by ecology
Only pure and wild–wild hybrid M. sylvestris seedlings were sampled for the IBE analysis (n = 449, 21 sites; Table 1). The IBE pattern, i.e. the contribution of climatic distance (isolation by climate) and phenotypic distance [isolation by adaptation (IBA)] to the genetic structure, taking into account geographical distance, was evaluated using distance-based redundancy analysis (db-RDA), which can be used when the response variable is a distance matrix [here, it is a genetic distance matrix (FST) across 21 sampled sites] and the explanatory variables are in vector form as follows. The geographical distance between sampled sites, estimated with SPAGeDI 1.5d (Hardy and Vekemans, 2002), which underlies an IBD process, was represented by vectors with positive eigenvalues of a principal coordinate of a neighbour matrix (PCNM) (Borcard and Legendre, 2002). A total of 19 bioclimatic variables, downloaded from the Worldclim2 database (30″ resolution; https://www.worldclim.org/data/worldclim21.html) and representing annual and seasonal trends and extremes averaged over the years 1970–2000 and averaged for the Pleistocene period (20 000 years ago) (Gamisch, 2019), were used to test for an isolation-by-climate pattern. Growth rate (n = 551; Table 1) and carbon uptake-related traits (n = 239; Table 1) averaged per site were used to test for an IBA pattern.
To identify the variables that explained the genetic structure of M. sylvestris, a db-RDA using the ‘capscale’ function (Oksanen et al., 2014) was run on a complete model that included all investigated variables (i.e. PCNM components, growth rates, carbon uptake-related traits and 19 bioclimatic variables). The best variables were selected for an optimum model with the function ‘step’ based on the Akaike information criterion (AIC). Given that db-RDA does not provide information on the relative contribution of each variable of the model, a variance partitioning analysis was run using the ‘varpart’ function from the R package ‘vegan’ (Peres-Neto et al., 2006).
RESULTS
Genetic status of seedlings
No clones or closely related individuals were detected (Supplementary data Fig. S1); therefore, 584 seedlings were included in the STRUCTURE analyses.
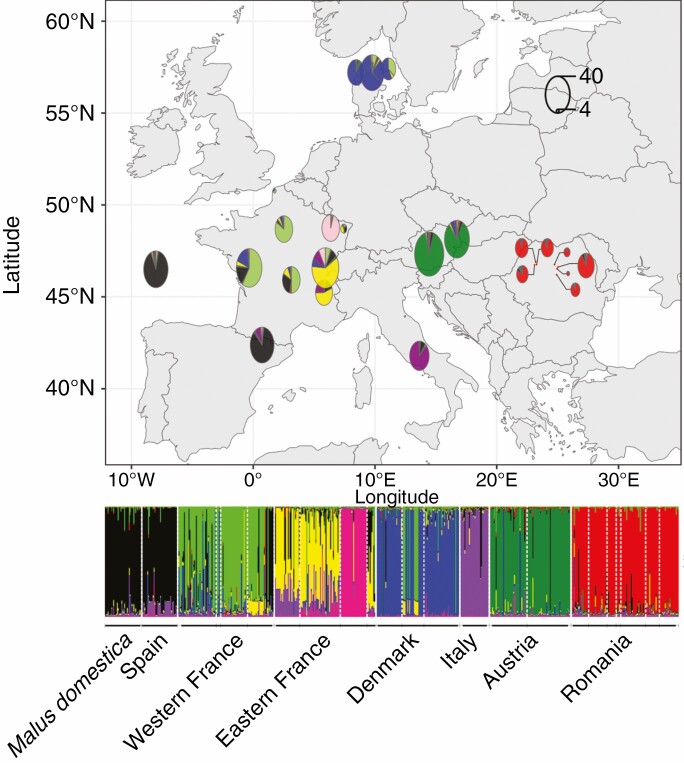
STRUCTURE revealed a clear spatial population genetic structure of M. sylvestris in Europe and crop–wild admixture (Fig. 1; Supplementary data Fig. S2). From K = 2 to K = 8, several clusters appeared. When K > 8, STRUCTURE did not reveal any further substructures but only additional clusters with highly admixed individuals (Supplementary data Fig. S2). Therefore, although the ΔK indicated that the most likely value of K was five (Supplementary data Fig. S3), K = 8 was the finest population structure and was retained in subsequent analyses. For K = 8, we found that the M. domestica reference varieties were admixed with Italian and Western French M. sylvestris. Note that cultivars used as references originated from Western Europe (Supplementary data Table S2). Conversely, we detected 68 M. sylvestris seedlings with Pdom > 0.1 (considered to be cw hybrids), corresponding to 11.6 % of the seedlings (n = 584; Fig. 1; Table 1; Supplementary data Figs S4–S6). We also found 21 seedlings with a membership coefficient to the M. domestica gene pool > 0.9, corresponding to 4 % of the seedlings. Nearly all Spanish seedlings were assigned to the M. domestica gene pool with membership coefficients > 0.1 (i.e. 26 cw hybrids and 13 individuals assigned to the M. domestica gene pool) and showed admixture only with the wild Italian purple gene pool (Supplementary data Figs S4 and S6). A total of 33 individuals could not be assigned to any cluster (i.e. individuals with a membership coefficient to any cluster < 0.5).
Fig. 1.
Bayesian clustering of Malus sylvestris seedlings sampled in this study (n = 584) and reference samples of Malus domestica (n = 40) inferred with STRUCTURE at K = 8 and its associated map of mean membership per sampled site. Each individual is represented by a vertical bar partitioned into clusters. Visualization was improved by sorting genotypes by country. Countries are separated by a white line. The M. domestica reference samples are shown on the far left of the map in the Atlantic. Circle size is proportional to the number of individuals within the cluster (scale shown in the top right-hand corner).
We, therefore, identified 68 cw, 21 dom, 167 ww and 282 pure seedlings (Table 1; n = 551). After removal of cw hybrids (n = 68), seedlings sampled from misidentified mother trees (n = 21), the M. domestica reference samples (n = 40) and individuals with a membership coefficient to any cluster < 0.5, the following seven wild apple populations (i.e. groups of seedlings with a membership coefficient > 0.5 to a wild apple cluster) were defined: French Western (FR-W; n = 77), French Eastern (FR-E; n = 50), French Lorraine (FR-Lor; n = 28), Danish (DA; n = 78), Italian (IT; n = 27), Austrian (AUT; n = 81) and Romanian (RO; n = 108) (Supplementary data Fig. S6). Each M. sylvestris population exhibited a high level of genetic variation (Supplementary data Table S3). The Romanian population was the most genetically differentiated and was close to the Austrian population; the Danish and French Western populations were the closest genetically (Supplementary data Fig. S7), which is congruent with previous results from 26 microsatellite markers (Cornille et al., 2015).
No effect of seedling genetic status on phenotypic variation
Variations and correlations among phenotypic traits are presented in the Supplementary data (Figs S8–S12). Significant correlations between seed length and weight of the mother tree and height of the seedling were observed (P < 0.01). Leaf growth rates (leaf_RGR, leaf_AGR and leaf_whole AGR) were significantly correlated, as were height growth rates (height_RGR, height_AGR and height_whole_AGR). Chlorophyll content was positively correlated with NBI, whereas flavonol content was negatively correlated with NBI. Heritability estimates were moderate to high for all traits except growth rates based on leaf number (AGR_leaf, RGR_leaf and whole_AGR_leaf; Supplementary data Table S4). Given the limited sample size, these estimates must be taken cautiously, as reflected by the large values of s.d.
We did not find any significant effect of the genetic status of the seedlings (i.e. pure, ww, cw or dom; Supplementary data Table S5; Fig. S13) or Pdom (Supplementary data Table S6; Fig. S14) on phenotypic traits, except for a marginally significant effect (P = 0.04; Supplementary data Table S6) of Pdom on the whole height AGR: seedlings with higher levels of introgression by M. domestica had higher whole height AGRs. We, therefore, removed the seedling genetic status and Pdom effects from the model (4), in addition to dom and cw individuals. Thus, the final model included only wild apple seedlings (i.e. pure and ww, n = 449) and focused on the wild population of origin effect (Table 2).
Table 2.
Final model depicting the effects of the Malus sylvestris population to which each seedling belonged (i.e. cluster inferred with STRUCTURE for K = 8) on phenotypic traits (i.e. height, number of leaves, internode, chlorophyll and flavonol contents and nitrogen balance index) measured on 533 individuals.
| Explanatory variable | Population | Mother tree | Model | Mother tree | |||||
|---|---|---|---|---|---|---|---|---|---|
| Fitness | χ2 | P-value | d.f. | REML | s.d. | AIC | R 2 | Corrected R2 | R 2 |
| Height_AGR | 17.863 | 0.007*** | 6 | 1229 | 0.204 | 1248 | 0.047 | 0.091 | 0.044 |
| Height_RGR | 12.846 | 0.045* | 6 | −2264 | 0.006 | −2245 | 0.041 | 0.147 | 0.106 |
| Leaf_AGR | 4.302 | 0.6359 | 6 | 134 | 0.036 | 152 | 0.01 | 0.028 | 0.018 |
| Leaf_RGR | 4.302 | 0.6359 | 6 | 134 | 0.036 | 152 | 0.01 | 0.028 | 0.018 |
| Whole height AGR | 22.243 | 1.00e-03*** | 6 | 630 | 0.175 | 650 | 0.074 | 0.192 | 0.118 |
| Whole leaf AGR | 36.326 | 2.38e-06*** | 6 | −1277 | 0.009 | −1258 | 0.09 | 0.118 | 0.028 |
| Height | 31.623 | 1.93e-05*** | 6 | 4113 | 11.69 | 4,131 | 0.119 | 0.301 | 0.182 |
| Number of leaves | 22.285 | 0.001*** | 6 | – | 0.084 | 2,659 | 0.052 | 0.064 | 0.012 |
| Chlorophyll | 14.418 | 0.025* | 6 | 1181 | 1.352 | 1199 | 0.074 | 0.171 | 0.097 |
| Flavonol | 6.7752 | 0.342 | 6 | −124 | 0.091 | −105 | 0.043 | 0.299 | 0.256 |
| NBI | 1.838 | 0.934 | 6 | 1735 | 7.6617 | 1754 | 0.011 | 0.224 | 0.213 |
| Internode (number of leaves/height) | 17.768 | 0.007*** | 6 | −1328 | 0.009 | −1309 | 0.044 | 0.073 | 0.029 |
Abbreviations: AIC, Akaike information criterion; NBI, nitrogen balance index; –, model without any significant effect. ***P < 0.001; **0.01 < P < 0.001; *0.05 < P < 0.01.
Significant variation in growth rates and chlorophyll content among populations
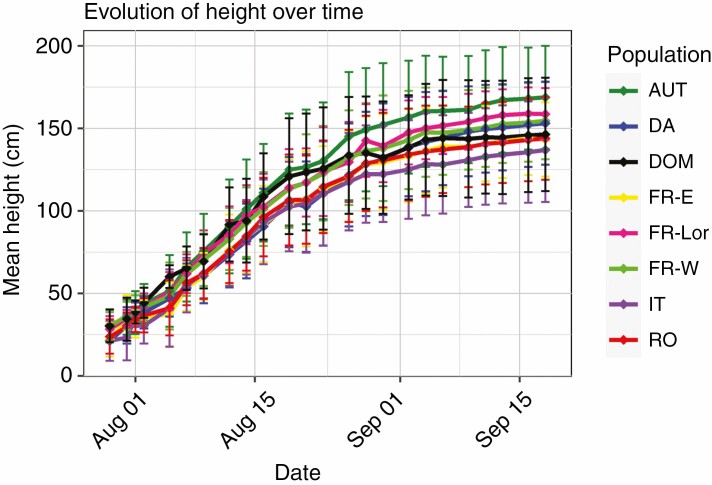
The mean height variation during the experiment among seedlings from different populations is shown in Figure 2. There was significant variation among seedlings from other populations in certain growth-related traits (Table 2). On average, seedlings belonging to the Austrian population were taller (+11 cm, P = 0.047), whereas Romanian (−14.9 cm, P = 0.008) and Italian (−18.7 cm, P = 0.044) seedlings were shorter (Fig. 2; Supplementary data Fig. S15) than seedlings from other populations. Seedlings from other populations did not show a significant difference in height. In addition, the number of leaves and height traits were negatively correlated (r = −0.3, P < 0.001). The Austrian population presented the lowest number of leaves (average = 5, s.d. = 4), whereas seedlings belonging to the Romanian population had the highest number of leaves (average = 8, s.d. = 7; Supplementary data Figure S16). The Romanian population also had the largest internode (+0.02 leaves cm−1, P = 0.024).
Fig. 2.
Cumulative height growth over time in apple seedlings in controlled conditions. Seedlings measured were pure and wild–wild hybrid Malus sylvestris seedlings (n = 449) and seedlings assigned to the Malus domestica gene pool (n = 21), as detected with STRUCTURE for K = 8. The 40 reference M. domestica individuals were not measured in controlled conditions and are therefore not shown here. Vertical lines represent the s.d. Populations: Austria (AUT, n = 81), Denmark (DA, n = 78), M. domestica (DOM, n = 21, which includes 13 Spanish genotypes and seedlings from other countries), Eastern France (FR-E, n = 50), Lorraine in France (FR-Lor, n = 28), Western France (FR-W, n = 77), Italy (IT, n = 27) and Romania (RO, n = 108).
Chlorophyll content differed among populations, with seedlings from the Italian population producing, on average, more chlorophyll (+4.14 µg cm−2, P = 0.039; Supplementary data Fig. S17) than seedlings from other populations. Flavonol content and NBI did not differ significantly among populations.
Significant IBD and IBC
Correlation plots between bioclimatic variables are provided in Supplementary data Figs S18 and S19; however, all variables were included in the analysis, because db-RDA can cope with correlated variables. The optimal model was chosen according to its best AIC value. The optimal model explained ≤25.9 % of the genetic structure (adjusted R2 = 69.9 %, P = 0.001) and contained seven variables (four geographical and three bioclimatic variables) (Table 3): the geographical distance is represented by the first, second, third and sixth axes of the PCNM analysis and three past climatic variables (Bio3, isothermality; Bio6, minimum temperature of the coldest month; and Bio 9, mean temperature of the driest quarter). In total, IBD explained 47 % of the variance of the wild apple tree population genetic structure, whereas IBC explained 22 % (Supplementary data Fig. S20). Taking geographical distance into account, we did not find a pattern of IBA, i.e. covariation between phenotype and genetic divergences.
Table 3.
Contribution of geography and climate to the genetic variation observed among Malus sylvestris seedlings. Distance-based redundancy analyses tested the effects of geography, climate and phenotype on the genetic differentiation (from 13 microsatellites) among 21 sites in the European crabapple. Only the significant variables are listed.
| db-RDA | ||||
|---|---|---|---|---|
| Percentage of variance explained | d.f. | P-value | Adjusted R2 | |
| Global analysis | 25.9 | 7 | 0.001 | 69.9 |
| Residuals | 11.3 | 13 | – | |
| Geography (IBD, PCNM 1-2-3-6) | 14.9 | 4 | <0.015 | |
| Environment (IBC_LGM, BIO3_LGM, BIO6_LGM, BIO9_LGM) | 11.04 | 3 | <0.015 | |
| Residuals | 11.3 % | – | – | |
Abbreviations: BIO3_LGM, isothermality (BIO2/BIO7) (×100); BIO6_LGM, minimum temperature of the coldest month; BIO9_LGM, mean temperature of the driest quarter; IBD, isolation by distance; IBC_LGM, isolation by climate during the Last Glacial Maximum.
DISCUSSION
This study assessed the relationship between phenotypic and genetic variation among populations of a wild contributor to the cultivated apple genome (Cornille et al., 2012), M. sylvestris, a CWR species that occurs naturally along a climatic gradient in Europe. Bayesian clustering revealed a substantial number of seedlings introgressed by M. domestica. With the hybrids removed, seven populations of M. sylvestris distributed across Europe were detected and showed phenotypic variation in growth and chlorophyll content. Based on the IBA pattern estimated from the phenotypic traits measured in this study, this phenotypic variation was not adaptive. However, the significant association between population genetic variation and the LGM climate suggests that the European crabapple might be locally adapted to the past climate conditions of the LGM. The results of this study, therefore, indicate the occurrence of adaptive divergence related to climate in a wild contributor to the cultivated apple genome. This might help us to use its diversity better, providing options for mitigating the impact of climate change on the cultivated apple through breeding (Warschefsky et al., 2014; Prohens et al., 2017; Satori et al., 2022).
Ongoing crop-to-wild gene flow in the European crabapple
We showed substantial gene flow from M. domestica to the European crabapple gene pool, with 11.6 % of seedlings, mainly from Western Europe, introgressed by M. domestica. Introgression rates were lower compared with previous studies [i.e. 37 % in the study by Cornille et al. (2013b) and 23.1 % in the study by Cornille et al. (2015)]. However, these studies genotyped more mother trees (i.e. n = 756 and n = 1889, respectively), which could explain the difference in estimates of crop-to-wild gene flow. Crop-to-wild gene flow is therefore still ongoing in the European crabapple. The higher number of seedlings from Western Europe that are introgressed by M. domestica can be explained by the use of reference Western European cultivated apple varieties. Alleles specific to Eastern and Northern cultivated apple varieties could be missed and might decrease the probability of detecting crop-to-wild introgression events in non-Western wild populations (from Eastern and Northern Europe). Note that the Spanish seedlings sampled in this study were the progeny of trees growing in a location known to have high levels of introgression by M. domestica (G. Alins, IRTA, Lleda, Spain, pers. comm.). It is even possible that the mother trees of these seedlings were M. domestica and not M. sylvestris.
The consequences of crop-to-wild introgression on phenotypic variation between crop and wild individuals are poorly understood in perennial fruit trees. One study has shown that crop–wild hybrid apple seedlings have higher growth rates and germinate earlier than wild apple seedlings (Feurtey et al., 2017). We did not detect any effect of the genetic status of a seedling (pure, ww, cw or dom) or the level of introgression (Pdom) on growth and carbon uptake-related fitness proxies. This could be attributable to the low number of samples from the cw and dom categories.
Signs of local adaptation to past climate in the European crabapple
In controlled conditions, seedlings from different populations exhibited significantly different growth and physiology. Seedlings belonging to the Austrian population were the tallest, with the highest absolute growth rate and the lowest number of leaves; in contrast, Romanian seedlings were the shortest and had the lowest absolute growth rate and the highest number of leaves. Italian seedlings had the highest chlorophyll content. We therefore tested whether this phenotypic variation was adaptive. For instance, the seedlings from the Austrian population might be fitter in the climatic conditions simulated in this controlled condition experiment. However, considering the geographical distance, we found no significant covariation between genetic and phenotypic variation. This suggests a lack of divergent selection on carbon uptake or growth traits, which are often associated with plant responses to climate (Bussotti et al., 2015). Therefore, the phenotypic variation we observed among populations in controlled conditions might result from genetic drift alone and not from divergent selection. Alternatively, although the specific traits we selected are among the traits that are generally considered to be related to responses to climate (Kühn et al., 2021), they might not be perfect candidates for investigating divergent selection by climate. Another explanation for the lack of observation of divergent selection on carbon uptake or growth traits could be that we did not phenotype enough seedlings from each genetic group. We observed a high variation in each phenotypic trait and their heritability estimates, suggesting that the traits we studied might be relevant but that a larger number of seedlings could be phenotyped and analysed. However, some studies have found that even with large sample sizes, the s.e. of heritability estimates can still be large and vary significantly between experimental designs (Visscher and Goddard, 2015). The reasonably high heritability estimates for most traits considered here could be consistent with relatively weak within-population selection, enabling the maintenance of ample additive genetic variation (Wheelwright et al., 2014). Furthermore, high variation in seedling traits combined with high heritability estimates could suggest a substantial amount of genetic variation for adaptation to work on. Altogether, the effect of the population of origin on phenotypic traits suggests a genetic basis for this variation. However, we need to increase the sample size and measure new traits to draw more precise conclusions on the occurrence of adaptive phenotypic variation in the European crabapple.
Although the association between phenotypic and genetic variation was not significant we observed a significant association between LGM climate and genetic variation in the European crabapple, suggesting local adaptation. Given that M. sylvestris is distributed across various climatic conditions, we indeed investigated the role of climate in shaping the genetic variation among populations of the European crabapple without considering phenotypic trait variation. We tested for an IBE pattern, where the pattern of neutral genetic variation covaries with ecological variables (here, climate). There was no combined effect of geographical and climatic distance (IBD ∩ IBC), which allowed us to assess the contribution of these processes separately (Wang and Bradburd, 2014). We showed that IBD and IBC played a significant role (adjusted R2 = 47 % and adjusted R2 = 22 %, respectively) in the genetic differentiation of European crabapple populations. Weak but significant IBD has been identified previously in wild apple relatives of the cultivated apple (i.e. M. sylvestris, M. orientalis and M. sieversii; Cornille et al., 2013a, b; Cornille et al., 2015). Here, we used 13 of the 26 microsatellite markers used in previous studies; the lack of resolution of the 13 simple sequence repeats might explain the lack of IBD. However, this is unlikely, because the Bayesian inferences method was previously able to detect genetic clusters in wild apples (Cornille et al., 2013a, b; Cornille et al., 2015). A weak IBD pattern suggests that M. sylvestris has high dispersal capacities (Coart et al., 2006; Larsen et al., 2006; Cornille et al., 2013a; Cornille et al., 2015; Feurtey et al., 2017; Reim et al., 2017). A weak IBD is explained by a self-incompatibility system that prevents self-fertilization (Brown, 1992), pollen dispersal by bees, beetles and flies, and endozoochorous seed dispersal by large mammals, such as ungulates, wild boars, brown bears or humans (Larsen et al., 2006).
We showed that in addition to IBD, IBC persisted after considering the geographical distance. Climate can impose divergent selection pressures on different locations and thus reduce gene flow between populations. For instance, divergent selection imposed by climate can limit the reproductive success of individuals moving between different climates for which they are adapted, such that IBC contributes to genetic differentiation among populations (Wang and Bradburd, 2014). The main variables explaining genetic differentiation in the European crabapple were related to temperature during the LGM. This suggests that the European crabapple might be adapted to its past climate but not to its current one. In wind-dispersed trees, local adaptation to the current climate has been demonstrated (Savolainen et al., 2013; Kremer and Hipp, 2019; Pyhäjärvi et al., 2020), but to our knowledge, no study has shown local adaptation to past climate conditions in an insect-pollinated tree species.
Factors other than climate can also shape the adaptive divergence between populations. Local adaptation to biotic factors, such as the presence of other species, is possible. Malus sylvestris is a species that needs high levels of light and is not very competitive. Some of the observed variations of the seedlings in growth rate, number of leaves and chlorophyll content might derive from adaptations that would be advantageous within the local niche. In Romania, the populations sampled were in forest edges, middle succession woodlands (with hawthorn and wild pear) or grasslands; no wild apples were found in mature forests. There, M. sylvestris does not compete with other woody species but with grasses and shrubs in the seedling phase; this might explain the slower growth in height and the larger number of leaves (leading to a larger leaf area and more shading of the competing grasses). In France, the trees are present at the edge of mature forests, and in Austria, in mature forests. It is hard to draw clear conclusions; further investigations of the ecology of the wild populations in situ are needed. The rhizosphere composition among seedlings from different populations can impact the phenotypic variation and, potentially, plant fitness in response to climate (Trivedi et al., 2022). Here, seeds were cleaned with chlorine to avoid the effect of local micro-organism community and seed growth. Still, investigations of phenotypic variation among cleaned and uncleaned seeds can also help to assess the role of the rhizosphere in the divergence of the wild apple population. Local adaptation of fruit trees to biotic factors, including parasites (Olvera-Vazquez et al., 2021), also deserves further investigation. Besides selection, the role of phenotypic plasticity in enabling growth and optimal fitness in changing environments also needs to be evaluated carefully (Benito Garzón et al., 2011).
Further investigations are needed on local adaptation and phenotypic plasticity in response to climate in the European crabapple
Our study raises questions regarding the response of wild apple populations to climate change. More data are needed to draw clear conclusions. The adaptation of tree species to climate is complex (Bussotti et al., 2015). For instance, in one study, Eucalyptus camaldulensis, variation in leaf traits and performance was unrelated to the climate of genotype provenance (Asao et al., 2020), whereas in another part of Australia, the same species displays variation in several photosynthetic traits that are related to the climate of genotype provenance (Dillon et al., 2018). In contrast, collective differences in leaf morphology and photosynthetic physiology associated with the length of the growing season, temperature and the level of insolation in several Populus species might be adaptive (Keller et al., 2011; Kaluthota et al., 2015). Further investigations on local adaptation and phenotypic plasticity in response to climate in the European crabapple are needed. Genomic data will help in determining the relative influence of adaptive and neutral processes on climate-driven divergence by scanning the genomes of trees from different populations in Europe. Comparing the fitness of seedlings from different populations in reciprocal transplants (in controlled or natural conditions) will also allow us to investigate local adaptation and phenotypic plasticity. Further studies using additional genetic markers (single nucleotide polymorphism) and measuring phenotypic traits in different climatic conditions are therefore needed. This study is, nevertheless, a starting point for future breeding and conservation programmes of a CWR of an emblematic temperate fruit tree, because it characterized the phenotypic and genetic variation of seedlings in the wild that can be used as ex situ sources to enrich the crop gene pool. Some of the seedlings included have sequenced genomes and have been planted in several orchards in France (https://www.ideev.universite-paris-saclay.fr/en/the-orchard) and are measured each year for several phenotypic traits. Information and samples from these orchards can be requested from the corresponding author. Genetic variation of trees in these ex situ orchards can help to enrich the cultivated apple gene pool. Certain traits related to adaptation to climate change can be introduced into future apple varieties (Warschefsky et al., 2014; Prohens et al., 2017; Satori et al., 2022).
SUPPLEMENTARY DATA
Supplementary data are available at Annals of Botany online and consist of the following.
Table S1: details of Malus sylvestris material used in this study. Table S2: Malus domestica reference samples used in this study. Table S3: genetic diversity estimates for the seven populations of Malus sylvestris inferred at K = 8 with STRUCTURE. Table S4: variance and heritability of the traits measured in 533 individuals. Table S5: effects of status of the apple seedling and of the population to which they belong on phenotypic traits measured on 533 individuals. Table S6: effects of admixture proportion of a wild genotype assigned to the Malus domestica gene pool and of the population to which they belong on phenotypic traits measured on 533 individuals. Figure S1: distribution of kinship between pairs of Malus sylvestris and Malus domestica individuals. Figure S2: Bayesian clustering for Malus sylvestris and Malus domestica, inferred from K = 2 to K = 9 with STRUCTURE v.2.3.3. Figure S3: ΔK estimated for each K cluster inferred with STRUCTURE v.2.3.3 for Malus sylvestris and Malus domestica. Figure S4: distribution of crop–wild hybrids for the apple tree in Europe inferred with STRUCTURE v.2.3.3 at K = 8. Figure S5: distribution of the proportion of admixture of seedlings used in this experiment with the Malus domestica gene pool, Pdom, inferred with STRUCTURE at K = 8. Figure S6: distribution of the genetic status of the seedlings inferred at K = 8 with STRUCTURE, summed up per geographical site. Figure S7: pairwise genetic differentiation between populations of pure wild Malus sylvestris seedlings inferred at K = 8 with STRUCTURE. Figure S8: Pearson correlations between seed size of the mother trees and plant height and the number of leaves of the seedlings in Malus sylvestris. Figure S9: principal component analysis representing variation among phenotypic traits. Figure S10: correlation plot among phenotypic traits measured on the apple seedlings in controlled conditions. Figure S11: principal component analysis among calculated growth rate variables. Figure S12: correlation plot among growth rates obtained using cor() R-function for seedlings used in this study. Figure S13: evolution of mean height of seedling over time in growth chamber depending on the status defined with STRUCTURE for K = 8. Figure S14: the mean absolute growth rate of seedlings plotted against their level of Malus domestica ancestry. Figure S15: variation in height among the eight populations detected with STRUCTURE at K = 8 for seedlings of Malus sylvestris grown in controlled conditions. Figure S16: variation in the number of leaves among the eight populations detected with STRUCTURE at K = 8 for seedlings of Malus sylvestris grown in controlled conditions. Figure S17: variation in chlorophyll content among the eight populations detected with STRUCTURE at K = 8 for the seedling of Malus sylvestris grown in controlled conditions. Figure S18: correlation plot among the 19 bioclimatic variables averaged over 1970–2000 used to test for isolation by climate in Malus sylvestris. Figure S19: correlation plot among the 19 bioclimatic variables averaged for the Pleistocene period used to test for isolation by climate in Malus sylvestris. Figure S20: variance partitioning analysis of db-RDA results obtained for Malus sylvestris.
ACKNOWLEDGEMENTS
We are grateful to the INRAE MIGALE bioinformatics platform (http://migale.jouy.inra.fr) for providing help and support, particularly Véronique Martin, Eric Montaubon and Valentin Loux. We also thank Adrien Falce, Olivier Langella and Benoit Johannet for help and support on the INRAE Génétique Quantitative et Evolution – Le Moulon lab cluster and the Plateforme de Génotypage GENTYANE INRAE UMR 1095. We thank Einar Flensted-Jensen, Michel Bonnessee, Alberto Dominici, Jean-Luc Ricaud, Eric Wolff, Alain Guinet and Line Caminade, Richard Le Menn and Bernard Gambier for providing seeds. USDA is an equal opportunity provider, employer and lender. Mention of trade names or commercial products is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture. A.C., G.M.V., A.R., S.B. and T.M.U. conceived and designed the experiments; A.C., G.M.V., A.F., T.M.U. and T.K. obtained the funding; A.C., G.M.V., A.F., T.M.U., K.A., S.O.-V., R.R., X.C., T.K. and C.R. sampled the material; X.C., C.R., A.V., A.R., G.L., K.A.O., R.R., M.L.G., H.B., V.C., H.C., S.O.-V. and M.F. performed the molecular biology analyses; A.C., A.F., K.A., and X.C. analysed the data. The manuscript was written by A.C., K.A., and A.F., with essential input from other co-authors.
Contributor Information
X Chen, Université Paris Saclay, INRAE, CNRS, AgroParisTech, GQE – Le Moulon, 91190 Gif-sur-Yvette, France.
K Avia, Université de Strasbourg, INRAE, SVQV UMR-A 1131, F-68000 Colmar, France.
A Forler, Université Paris Saclay, INRAE, CNRS, AgroParisTech, GQE – Le Moulon, 91190 Gif-sur-Yvette, France.
C Remoué, Université Paris Saclay, INRAE, CNRS, AgroParisTech, GQE – Le Moulon, 91190 Gif-sur-Yvette, France.
A Venon, Université Paris Saclay, INRAE, CNRS, AgroParisTech, GQE – Le Moulon, 91190 Gif-sur-Yvette, France.
A Rousselet, Université Paris Saclay, INRAE, CNRS, AgroParisTech, GQE – Le Moulon, 91190 Gif-sur-Yvette, France.
G Lucas, Université Paris-Saclay, CEA, CNRS, Institute for Integrative Biology of the Cell (I2BC), Gif-sur-Yvette 91198, France.
A O Kwarteng, Université Paris Saclay, INRAE, CNRS, AgroParisTech, GQE – Le Moulon, 91190 Gif-sur-Yvette, France.
R Rover, Université Paris Saclay, INRAE, CNRS, AgroParisTech, GQE – Le Moulon, 91190 Gif-sur-Yvette, France.
M Le Guilloux, Université Paris Saclay, INRAE, CNRS, AgroParisTech, GQE – Le Moulon, 91190 Gif-sur-Yvette, France.
H Belcram, Université Paris Saclay, INRAE, CNRS, AgroParisTech, GQE – Le Moulon, 91190 Gif-sur-Yvette, France.
V Combes, Université Paris Saclay, INRAE, CNRS, AgroParisTech, GQE – Le Moulon, 91190 Gif-sur-Yvette, France.
H Corti, Université Paris Saclay, INRAE, CNRS, AgroParisTech, GQE – Le Moulon, 91190 Gif-sur-Yvette, France.
S Olverà-Vazquez, Université Paris Saclay, INRAE, CNRS, AgroParisTech, GQE – Le Moulon, 91190 Gif-sur-Yvette, France.
M Falque, Université Paris Saclay, INRAE, CNRS, AgroParisTech, GQE – Le Moulon, 91190 Gif-sur-Yvette, France.
G Alins, Institut de Recerca i Tecnologia Agroalimentàries, IRTA-Fruit Production, PCiTAL, Parc 21 de Gardeny, edifici Fruitcentre, 25003 Lleida, Spain.
T Kirisits, Institute of Forest Entomology, Forest Pathology and Forest Protection (IFFF), Department of Forest and Soil Sciences, University of Natural Resources and Life Sciences, Vienna (BOKU), Peter-Jordan-Straße 82 (Franz Schwackhöfer-Haus), A-1190 Vienna, Austria.
T M Ursu, NIRDBS, Institute of Biological Research Cluj-Napoca, 48 Republicii St., Cluj-Napoca, Romania.
A Roman, NIRDBS, Institute of Biological Research Cluj-Napoca, 48 Republicii St., Cluj-Napoca, Romania.
G M Volk, USDA-ARS National Laboratory for Genetic Resources Preservation, 1111 South Mason Street, Fort Collins, CO 80521, USA.
S Bazot, Ecologie Systématique et Evolution, CNRS, AgroParisTech, Ecologie Systématique Evolution, Université Paris‐Saclay, Orsay, France.
A Cornille, Université Paris Saclay, INRAE, CNRS, AgroParisTech, GQE – Le Moulon, 91190 Gif-sur-Yvette, France.
FUNDING
A.R. and T.M.U. were funded, in part. through project PN2019-2022/19270201—BIODIVERS 3. We thank the Laboratoire d’Excellence BASC (Biodiversité, Agroécosystèmes, Société, Climat), the Institut Diversité Écologie et Évolution du Vivant (IDEEV), the European LEADER program and ATIP-Avenir CNRS Inserm for funding.
LITERATURE CITED
- Aitken SN, Bemmels JB.. 2015. Time to get moving: assisted gene flow of forest trees. Evolutionary Applications 9: 271–290. doi: 10.1111/eva.12293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandre H, Truffaut L, Klein E, et al. 2020. How does contemporary selection shape oak phenotypes? Evolutionary Applications 13: 2772–2790. doi: 10.1111/eva.13082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asao S, Hayes L, Aspinwall MJ, et al. 2020. Leaf trait variation is similar among genotypes of Eucalyptus camaldulensis from differing climates and arises in plastic responses to the seasons rather than water availability. New Phytologist 227(3): 780–793. [DOI] [PubMed] [Google Scholar]
- Aubin I, Munson AD, Cardou F, et al. 2016. Traits to stay, traits to move: a review of functional traits to assess sensitivity and adaptive capacity of temperate and boreal trees to climate change. Environmental Reviews 24: 164–186. doi: 10.1139/er-2015-0072. [DOI] [Google Scholar]
- Bates D, Mächler M, Bolker B, Walker S.. 2015. Fitting linear mixed-effects models using lme4. Journal of Statistical Software, Articles 67: 1–48. [Google Scholar]
- Benito Garzón M, Alía R, Robson TM, Zavala MA.. 2011. Intra-specific variability and plasticity influence potential tree species distributions under climate change. Global Ecology and Biogeography 20: 766–778. doi: 10.1111/j.1466-8238.2010.00646.x. [DOI] [Google Scholar]
- Borcard D, Legendre P.. 2002. All-scale spatial analysis of ecological data by means of principal coordinates of neighbour matrices. Ecological Modelling 153: 51–68. doi: 10.1016/s0304-3800(01)00501-4. [DOI] [Google Scholar]
- Boyd IL, Freer-Smith PH, Gilligan CA, Godfray HCJ.. 2013. The consequence of tree pests and diseases for ecosystem services. Science 342: 1235733. doi: 10.1126/science.1235733. [DOI] [PubMed] [Google Scholar]
- Brachi B, Villoutreix R, Faure N, et al. 2013. Investigation of the geographical scale of adaptive phenological variation and its underlying genetics in Arabidopsis thaliana. Molecular Ecology 22: 4222–4240. doi: 10.1111/mec.12396. [DOI] [PubMed] [Google Scholar]
- Briggs AW, Kidd F, West C.. 1920. A quantitative analyses of plant growth: part II. Annals of Applied Biology 7: 202–223. [Google Scholar]
- Briscoe Runquist RD, Gorton AJ, Yoder JB, et al. 2020. Context dependence of local adaptation to abiotic and biotic environments: a quantitative and qualitative synthesis. The American Naturalist 195: 412–431. doi: 10.1086/707322. [DOI] [PubMed] [Google Scholar]
- Brown SK. 1992. Genetics of apple. In: Janick J. ed. Plant breeding reviews, vol. 9. John Wiley & Sons; New York, 333–366. [Google Scholar]
- Bussotti F, Pollastrini M, Holland V, Brüggemann W.. 2015. Functional traits and adaptive capacity of European forests to climate change. Environmental and Experimental Botany 111: 91–113. doi: 10.1016/j.envexpbot.2014.11.006. [DOI] [Google Scholar]
- Coart E, Van Glabeke S, De Loose M, Larsen AS, Roldán-Ruiz I.. 2006. Chloroplast diversity in the genus Malus: new insights into the relationship between the European wild apple (Malus sylvestris (L.) Mill.) and the domesticated apple (Malus domestica Borkh.). Molecular Ecology 15: 2171–2182. doi: 10.1111/j.1365-294X.2006.02924.x. [DOI] [PubMed] [Google Scholar]
- Comes HP, Kadereit JW.. 1998. The effect of Quaternary climatic changes on plant distribution and evolution. Trends in Plant Science 3: 432–438. doi: 10.1016/s1360-1385(98)01327-2. [DOI] [Google Scholar]
- Cornille A, Antolín F, Garcia E, et al. 2019. A multifaceted overview of apple tree domestication. Trends in Plant Science 24: 770–782. doi: 10.1016/j.tplants.2019.05.007. [DOI] [PubMed] [Google Scholar]
- Cornille A, Feurtey A, Gélin U, et al. 2015. Anthropogenic and natural drivers of gene flow in a temperate wild fruit tree: a basis for conservation and breeding programs in apples. Evolutionary Applications 8: 373–384. doi: 10.1111/eva.12250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornille A, Giraud T, Bellard C, et al. 2013a. Post-glacial recolonization history of the European crabapple (Malus sylvestris Mill.), a wild contributor to the domesticated apple. Molecular Ecology 22: 2249–2263. doi: 10.1111/mec.12231. [DOI] [PubMed] [Google Scholar]
- Cornille A, Giraud T, Smulders MJM, Roldán-Ruiz I, Gladieux P.. 2014. The domestication and evolutionary ecology of apples. Trends in Genetics 30: 57–65. [DOI] [PubMed] [Google Scholar]
- Cornille A, Gladieux P, Giraud T.. 2013b. Crop-to-wild gene flow and spatial genetic structure in the wild closest relatives of the cultivated apple. Evolutionary Applications 6: 737–748. doi: 10.1111/eva.12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornille A, Gladieux P, Smulders MJM, et al. 2012. New insight into the history of domesticated apple: secondary contribution of the European wild apple to the genome of cultivated varieties. PLoS Genetics 8: e1002703. doi: 10.1371/journal.pgen.1002703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delplancke M, Alvarez N, Espíndola A, et al. 2011. Gene flow among wild and domesticated almond species: insights from chloroplast and nuclear markers. Evolutionary Applications 5: 317–329. doi: 10.1111/j.1752-4571.2011.00223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demestihas C, Plénet D, Génard M, Raynal C, Lescourret F.. 2017. Ecosystem services in orchards. A review. Agronomy for Sustainable Development 37: 12. [Google Scholar]
- Demotes-Mainard S, Boumaza R, Meyer S, Cerovic ZG.. 2008. Indicators of nitrogen status for ornamental woody plants based on optical measurements of leaf epidermal polyphenol and chlorophyll contents. Scientia Horticulturae 115: 377–385. doi: 10.1016/j.scienta.2007.10.006. [DOI] [Google Scholar]
- Diez CM, Trujillo I, Martinez-Urdiroz N, et al. 2015. Olive domestication and diversification in the Mediterranean Basin. New Phytologist 206: 436–447. [DOI] [PubMed] [Google Scholar]
- Dillon S, Quentin A, Ivković M, Furbank RT, Pinkard E.. 2018. Photosynthetic variation and responsiveness to CO2 in a widespread riparian tree. PLoS One 13: e0189635. doi: 10.1371/journal.pone.0189635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dusenge ME, Duarte AG, Way DA.. 2019. Plant carbon metabolism and climate change: elevated CO2 and temperature impacts on photosynthesis, photorespiration and respiration. New Phytologist 221: 32–49. [DOI] [PubMed] [Google Scholar]
- Earl DA, vonHoldt BM.. 2012. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources 4: 359–361. [Google Scholar]
- Edwards SV, Robin VV, Ferrand N, Moritz C.. 2022. The evolution of comparative phylogeography: putting the geography (and more) into comparative population genomics. Genome Biology and Evolution 14: evab176. doi: 10.1093/gbe/evab176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J.. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology 14: 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Feurtey A, Cornille A, Shykoff JA, Snirc A, Giraud T.. 2017. Crop‐to‐wild gene flow and its fitness consequences for a wild fruit tree: towards a comprehensive conservation strategy of the wild apple in Europe. Evolutionary Applications 10: 180–188. doi: 10.1111/eva.12441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers JM, Hazzouri KM, Gros-Balthazard M, et al. 2019. Cross-species hybridization and the origin of North African date palms. Proceedings of the National Academy of Sciences of the United States of America 116: 1651–1658. doi: 10.1073/pnas.1817453116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis R. 2016. POPHELPER: an R package and web app to analyse and visualise population structure. Molecular Ecology Resources 17: n/a–n/a. [DOI] [PubMed] [Google Scholar]
- Franks SJ, Weber JJ, Aitken SN.. 2014. Evolutionary and plastic responses to climate change in terrestrial plant populations. Evolutionary Applications 7: 123–139. doi: 10.1111/eva.12112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamisch A. 2019. Oscillayers: a dataset for the study of climatic oscillations over Plio-Pleistocene time-scales at high spatial-temporal resolution. Global Ecology and Biogeography 28: 1552–1560. doi: 10.1111/geb.12979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann FA, Czaja S, Hunsche M, Noga G, Fiebig A.. 2018. Monitoring physiological and biochemical responses of two apple cultivars to water supply regimes with non-destructive fluorescence sensors. Scientia Horticulturae 242: 51–61. doi: 10.1016/j.scienta.2018.07.008. [DOI] [Google Scholar]
- Hardy OJ, Vekemans X.. 2002. SPAGeDI: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Molecular Ecology Notes 2: 618–620. doi: 10.1046/j.1471-8286.2002.00305.x. [DOI] [Google Scholar]
- Hartmann H, Bahn M, Carbone M, Richardson AD.. 2020. Plant carbon allocation in a changing world – challenges and progress: introduction to a Virtual Issue on carbon allocation. New Phytologist 227: 981–988. doi: 10.1111/nph.16757. [DOI] [PubMed] [Google Scholar]
- Hoban S, Volk G, Routson KJ, Walters C, Richards C.. 2018. Sampling wild species to conserve genetic diversity. In: Greene SL, Williams KA, Khoury CK, Kantar MB, Marek LF, eds. North American Crop Wild Relatives, Vol. 1, Conservation Strategies. Cham: Springer International, 209–228. [Google Scholar]
- Hübner S, Kantar MB.. 2021. Tapping diversity from the wild: from sampling to implementation. Frontiers in Plant Science 12: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutyra LR, Munger JW, Saleska SR, et al. 2007. Seasonal controls on the exchange of carbon and water in an Amazonian rain forest. Journal of Geophysical Research, Biogeosciences 112: 1–16. doi: 10.1029/2006jg000365. [DOI] [Google Scholar]
- Kaluthota S, Pearce DW, Evans LM, Letts MG, Whitham TG, Rood SB.. 2015. Higher photosynthetic capacity from higher latitude: foliar characteristics and gas exchange of southern, central and northern populations of Populus angustifolia. Tree Physiology 35: 936–948. doi: 10.1093/treephys/tpv069. [DOI] [PubMed] [Google Scholar]
- Keller SR, Soolanayakanahally RY, Guy RD, Silim SN, Olson MS, Tiffin P.. 2011. Climate-driven local adaptation of ecophysiology and phenology in balsam poplar, Populus balsamifera L. (Salicaceae). American Journal of Botany 98: 99–108. doi: 10.3732/ajb.1000317. [DOI] [PubMed] [Google Scholar]
- Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I.. 2015. Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Molecular Ecology Resources 15: 1179–1191. doi: 10.1111/1755-0998.12387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer A, Hipp AL.. 2019. Oaks: an evolutionary success story. New Phytologist 226: 987–1011. doi: 10.1111/nph.16274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer A, Kleinschmit J, Cottrell J, et al. 2002. Is there a correlation between chloroplastic and nuclear divergence, or what are the roles of history and selection on genetic diversity in European oaks? Forest Ecology and Management 156: 75–87. doi: 10.1016/S0378-1127(01)00635-1. [DOI] [Google Scholar]
- Kühn N, Tovar C, Carretero J, Vandvik V, Enquist BJ, Willis KJ.. 2021. Globally important plant functional traits for coping with climate change. Frontiers of Biogeography 13(4): 1–18. [Google Scholar]
- Lander TA, Klein EK, Roig A, Oddou-Muratorio S.. 2021. Weak founder effects but significant spatial genetic imprint of recent contraction and expansion of European beech populations. Heredity 126: 491–504. doi: 10.1038/s41437-020-00387-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen A, Asmussen C, Coart E, Olrik D, Kjær E.. 2006. Hybridization and genetic variation in Danish populations of European crab apple (Malus sylvestris). Tree Genetics & Genomes 2: 86–97. [Google Scholar]
- Lê S, Josse J, Husson F.. 2008. FactoMineR: an r package for multivariate analysis. Journal of Statistical Software 25: 1–18. [Google Scholar]
- Li J, Li H, Jakobsson M, Li SEN, Sjödin PER, Lascoux M.. 2012. Joint analysis of demography and selection in population genetics: where do we stand and where could we go? Molecular Ecology 21: 28–44. doi: 10.1111/j.1365-294X.2011.05308.x. [DOI] [PubMed] [Google Scholar]
- Liu S, Cornille A, Decroocq S, et al. 2019. The complex evolutionary history of apricots: species divergence, gene flow and multiple domestication events. Molecular Ecology 28: 5299–5314. doi: 10.1111/mec.15296. [DOI] [PubMed] [Google Scholar]
- Loiselle BA, Sork VL, Nason J, Graham C.. 1995. Spatial genetic structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae). American Journal of Botany 82: 1420–1425. doi: 10.1002/j.1537-2197.1995.tb12679.x. [DOI] [Google Scholar]
- Mercado LM, Medlyn BE, Huntingford C, et al. 2018. Large sensitivity in land carbon storage due to geographical and temporal variation in the thermal response of photosynthetic capacity. New Phytologist 218: 1462–1477. doi: 10.1111/nph.15100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migicovsky Z, Gardner KM, Richards C, et al. 2021. Genomic consequences of apple improvement. Horticulture Research 8: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myles S, Boyko AR, Owens CL, et al. 2011. Genetic structure and domestication history of the grape. Proceedings of the National Academy of Sciences of the United States of America 108: 3530–3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicotra AB, Atkin OK, Bonser SP, et al. 2010. Plant phenotypic plasticity in a changing climate. Trends in Plant Science 15: 684–692. doi: 10.1016/j.tplants.2010.09.008. [DOI] [PubMed] [Google Scholar]
- Nosil P, Funk DJ, Ortiz-Barrientos D.. 2009. Divergent selection and heterogeneous genomic divergence. Molecular Ecology 18: 375–402. doi: 10.1111/j.1365-294X.2008.03946.x. [DOI] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, et al. 2014. Vegan: community ecology package (vol. 1, pp. 1–190).
- Olvera-Vazquez SG, Alhmedi A, Miñarro M, et al. 2021. Experimental test for local adaptation of the rosy apple aphid (Dysaphis plantaginea) to its host (Malus domestica) and to its climate in Europe. PCI Ecology Pre-registration version. https://forgemia.inra.fr/amandine.cornille/local_adaptation_dp [Google Scholar]
- Orsini L, Vanoverbeke J, Swillen I, Mergeay J, De Meester L.. 2013. Drivers of population genetic differentiation in the wild: isolation by dispersal limitation, isolation by adaptation and isolation by colonization. Molecular Ecology 22: 5983–5999. doi: 10.1111/mec.12561. [DOI] [PubMed] [Google Scholar]
- Parisod C. 2021. Plant speciation in the face of recurrent climate changes in the Alps. Alpine Botany 132(1): 21–28. Springer Science and Business Media LLC. doi: 10.1007/s00035-021-00259-6. [DOI] [Google Scholar]
- Pasho E, Julio Camarero J, Vicente-Serrano SM.. 2012. Climatic impacts and drought control of radial growth and seasonal wood formation in Pinus halepensis. Trees 26: 1875–1886. doi: 10.1007/s00468-012-0756-x. [DOI] [Google Scholar]
- Peace CP, Bianco L, Troggio M, et al. 2019. Apple whole genome sequences: recent advances and new prospects. Horticulture Research 6: 59. doi: 10.1038/s41438-019-0141-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peres-Neto PR, Legendre P, Dray S, Borcard D.. 2006. Variation partitioning of species data matrices: estimation and comparison of fractions. Ecology 87: 2614–2625. doi: 10.1890/0012-9658(2006)87[2614:vposdm]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Petit RJ, Hampe A.. 2011. Some evolutionary consequences of being a tree. Annual Review of Ecology, Evolution, and Systematics 37: 187–214. [Google Scholar]
- Pinheiro J, Bates D, DebRoy S, Sarkar D, R Core Team. 2018. nlme: linear and nonlinear mixed effects models. R Package Version 3.1-137. http://CRAN.R-project.org/package=nlme.
- Pritchard JK, Stephens M, Donnelly P.. 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prohens J, Gramazio P, Plazas M, et al. 2017. Introgressiomics: a new approach for using crop wild relatives in breeding for adaptation to climate change. Euphytica 213: 158. [Google Scholar]
- Puechmaille SJ. 2016. The program structure does not reliably recover the correct population structure when sampling is uneven: subsampling and new estimators alleviate the problem. Molecular Ecology Resources 16: 608–627. doi: 10.1111/1755-0998.12512. [DOI] [PubMed] [Google Scholar]
- Pyhäjärvi T, Kujala ST, Savolainen O.. 2020. 275 years of forestry meets genomics in Pinus sylvestris. Evolutionary Applications 13: 11–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyhäjärvi T, Salmela MJ, Savolainen O.. 2008. Colonization routes of Pinus sylvestris inferred from distribution of mitochondrial DNA variation. Tree Genetics & Genomes 4: 247–254. [Google Scholar]
- Radford PJ. 1967. Growth analysis formulae – their use and abuse. Crop Science 7: 171–175. doi: 10.2135/cropsci1967.0011183x000700030001x. [DOI] [Google Scholar]
- Ramírez-Valiente JA, Center A, Sparks JP, et al. 2017. Population-level differentiation in growth rates and leaf traits in seedlings of the Neotropical live oak Quercus oleoides grown under natural and manipulated precipitation regimes. Frontiers in Plant Science 8: 585. doi: 10.3389/fpls.2017.00585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond M, Rousset F.. 1995. GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. Journal of Heredity 86: 248–249. doi: 10.1093/oxfordjournals.jhered.a111573. [DOI] [Google Scholar]
- Reim S, Proft A, Heinz S, et al. 2017. Pollen movement in a Malus sylvestris population and conclusions for conservation measures. Plant Genetic Resources 15: 12–20. [Google Scholar]
- Riordan EC, Gugger PF, Ortego J, et al. 2016. Association of genetic and phenotypic variability with geography and climate in three southern California oaks. American Journal of Botany 103: 73–85. doi: 10.3732/ajb.1500135. [DOI] [PubMed] [Google Scholar]
- Ripetti V, Escoute J, Verdeil JL, Costes E.. 2008. Shaping the shoot: the relative contribution of cell number and cell shape to variations in internode length between parent and hybrid apple trees. Journal of Experimental Botany 59: 1399–1407. doi: 10.1093/jxb/ern049. [DOI] [PubMed] [Google Scholar]
- Rousset F. 2008. genepop’007: a complete re-implementation of the genepop software for Windows and Linux. Molecular Ecology Resources 8: 103–106. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
- Satori D, Tovar C, Faruk A, et al. 2022. Prioritising crop wild relatives to enhance agricultural resilience in sub-Saharan Africa under climate change. Plants, People, Planet 4: 269–282. [Google Scholar]
- Savolainen O, Lascoux M, Merila J.. 2013. Ecological genomics of local adaptation. Nature Reviews Genetics 14: 807–820. doi: 10.1038/nrg3522. [DOI] [PubMed] [Google Scholar]
- Shafer ABA, Wolf JBW.. 2013. Widespread evidence for incipient ecological speciation: a meta-analysis of isolation-by-ecology. Ecology Letters 16: 940–950. doi: 10.1111/ele.12120. [DOI] [PubMed] [Google Scholar]
- Sork VL. 2018. Genomic studies of local adaptation in natural plant populations. Journal of Heredity 109: 3–15. [DOI] [PubMed] [Google Scholar]
- Svenning J-C, Eiserhardt WL, Normand S, Ordonez A, Sandel B.. 2015. The influence of paleoclimate on present-day patterns in biodiversity and ecosystems. Annual Review of Ecology, Evolution, and Systematics 46: 551–572. doi: 10.1146/annurev-ecolsys-112414-054314. [DOI] [Google Scholar]
- Tiffin P, Ross-Ibarra J.. 2014. Advances and limits of using population genetics to understand local adaptation. Trends in Ecology & Evolution 29: 673–680. doi: 10.1016/j.tree.2014.10.004. [DOI] [PubMed] [Google Scholar]
- Trivedi P, Batista BD, Bazany KE, Singh BK.. 2022. Plant–microbiome interactions under a changing world: responses, consequences and perspectives. New Phytologist 234: 1951–1959. doi: 10.1111/nph.18016. [DOI] [PubMed] [Google Scholar]
- de Villemereuil P, Gaggiotti OE, Goudet J.. 2020. Common garden experiments to study local adaptation need to account for population structure. Journal of Ecology 110: 1005–1009. doi: 10.1111/1365-2745.13528. [DOI] [Google Scholar]
- de Villemereuil P, Gaggiotti OE, Mouterde M, Till-Bottraud I.. 2016. Common garden experiments in the genomic era: new perspectives and opportunities. Heredity 116: 249–254. doi: 10.1038/hdy.2015.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher PM, Goddard ME.. 2015. A general unified framework to assess the sampling variance of heritability estimates using pedigree or marker-based relationships. Genetics 199: 223–232. doi: 10.1534/genetics.114.171017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang IJ, Bradburd GS.. 2014. Isolation by environment. Molecular Ecology 23: 5649–5662. doi: 10.1111/mec.12938. [DOI] [PubMed] [Google Scholar]
- Warschefsky E, Penmetsa RV, Cook DR, von Wettberg EJB.. 2014. Back to the wilds: tapping evolutionary adaptations for resilient crops through systematic hybridization with crop wild relatives. American Journal of Botany 101: 1791–1800. doi: 10.3732/ajb.1400116. [DOI] [PubMed] [Google Scholar]
- Warschefsky EJ, von Wettberg EJB.. 2019. Population genomic analysis of mango (Mangifera indica) suggests a complex history of domestication. New Phytologist 222: 2023–2037. [DOI] [PubMed] [Google Scholar]
- Way DA, Montgomery RA.. 2015. Photoperiod constraints on tree phenology, performance and migration in a warming world. Plant, Cell & Environment 38: 1725–1736. doi: 10.1111/pce.12431. [DOI] [PubMed] [Google Scholar]
- Wei T, Simko V.. 2017. R package “corrplot”: visualization of a correlation matrix (Version 0.84). https://github.com/taiyun/corrplot.
- Wheelwright NT, Keller LF, Postma E.. 2014. The effect of trait type and strength of selection on heritability and evolvability in an island bird population. Evolution 68: 3325–3336. doi: 10.1111/evo.12499. [DOI] [PubMed] [Google Scholar]
- Wright S. 1943. Isolation by distance. Genetics 28: 114–138. doi: 10.1093/genetics/28.2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Kokubugata G, Fujii S, et al. 2021. Refugia during the last glacial period and the origin of the disjunct distribution of an insular plant. Journal of Biogeography 48: 1460–1474. doi: 10.1111/jbi.14090. [DOI] [Google Scholar]
- Zhang H, Mittal N, Leamy LJ, Barazani O, Song B-H.. 2017. Back into the wild—apply untapped genetic diversity of wild relatives for crop improvement. Evolutionary Applications 10: 5–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.