Abstract
Background and Aims
The evolution of ecological specialization is favoured under divergent selection imposed by increased environmental heterogeneity, although specialization can limit the geographical range of organisms, thus promoting endemism. The Atlantic Forest (AF) is an ancient montane domain with high plant endemism, containing different environments for plant specialization. Miconia is the most diverse genus of woody flowering plant within the AF domain, including AF-endemic and non-endemic lineages. We hypothesized that Miconia species have faced increased environmental heterogeneity and consequently have been selected towards increased specialization in the AF domain, and this increased specialization has greatly reduced species geographical ranges, ultimately promoting endemism. Hence, we made the following predictions: (1) AF-endemic species should face greater environmental heterogeneity than non-endemic species; (2) AF-endemic species should be more specialized than non-endemic species; (3) specialization should lead to smaller geographical ranges; (4) specialization and small geographical ranges among AF-endemic species should conform to a selection-driven evolutionary scenario rather than to a neutral evolutionary scenario; and (5) small geographical ranges among AF-endemic species should date back to the occupation of the AF domain rather than to more recent time periods.
Methods
We used geographical, environmental and phylogenetic data on a major Miconia clade including AF-endemic and non-endemic species. We calculated Rao’s Q to estimate the environmental heterogeneity faced by species. We used georeferenced occurrences to estimate the geographical ranges of species. We applied environmental niche modelling to infer species niche breadth. We inferred the most likely evolutionary scenario for species geographical range and niche breadth via a model-fitting approach. We used ancestral reconstructions to evaluate species geographical range throughout time.
Key Results
Atlantic Forest-endemic species faced 33–60 % more environmental heterogeneity, with the increase being associated with montane landscapes in the AF. The AF-endemic species were 60 % more specialized overall, specifically over highly variable environmental gradients in AF montane landscapes. Specialization strongly predicted small geographical ranges among AF-endemic species and was a major range-limiting factor among endemic lineages. The AF-endemic species have evolved towards specialization and small geographical ranges under a selection-driven regime, probably imposed by the great environmental heterogeneity in AF montane landscapes. The AF-endemic species underwent a major reduction of geographical range immediately after their evolution, indicating a long-standing effect of selective pressures in the AF domain.
Conclusion
Environmental heterogeneity imposes selective pressures favouring ecological specialization and small geographical ranges among plant lineages in the AF domain. This selection-driven process has probably promoted plant endemism in the AF domain throughout its history.
Keywords: Environmental heterogeneity, habitat specificity, hotspot, landscape complexity, Melastomataceae, Miconia, niche breadth evolution, range size, specialization
INTRODUCTION
Ecological specialization can constrain the geographical range of organisms and, consequently, impact on spatial patterns of biodiversity. Specialization occurs when an organism can explore fewer types of conditions or resources than another organism (Devictor et al., 2010), translating into an n-dimensional niche with narrower breadth (Hutchinson, 1957). Given their narrower niche requirements, specialists have fewer options of suitable areas than generalists (Brown, 1984). Consequently, specialists often have smaller geographical ranges than their generalist relatives (Slatyer et al., 2013). The relationship between specialization and small geographical range becomes evident in plant species. For instance, when comparing monkeyflower species (Mimulus L.) in Western North America, species with narrower thermal tolerances are more geographically restricted than species with broader tolerances (Sheth and Angert, 2014). Nonetheless, small geographical ranges can also result from geographical barriers, historical processes or biotic interactions acting upon plant species and plant dispersal traits (Sheth et al., 2020). Additionally, specialization can still allow for large geographical ranges when suitable areas are widely available (Sheth et al., 2020). Assessing the relative role of range-limiting factors is necessary to understand how species can become spatially concentrated, leading to the uneven distribution of biodiversity across the globe (Kier et al., 2005; Jenkins et al., 2013). Moreover, detecting major factors that constrain geographical range can aid conservation efforts to alleviate small geographical ranges, which forecast greater extinction risk (Staude et al., 2020). Based on that, tracking plant specialization and where it can thrive could explain the origin of megadiverse regions and, ultimately, provide the means to manage them.
Under the heterogeneity–specialization model, specialization is more likely to evolve and persist owing to divergent selective pressures imposed by different but spatially close environments (i.e. environmental heterogeneity) (Brown and Pavlovic, 1992). Each environment presumably imposes a challenge to organisms, thus requiring a specific trait set to maximize fitness (Levins, 1962). Hence, the more contrasting the environments, the more different their fitting trait sets become, such that mixed trait sets would provide reduced fitness in any environment (Levins, 1962). In an environmentally heterogeneous background, organisms are more likely to find contrasting environmental conditions when dispersing themselves or their propagules (Brown and Pavlovic, 1992). Under such a scenario, mixed-trait organisms will have reduced fitness regardless of the environment, whereas organisms with any fitting trait set will have enhanced fitness when dispersing to their respective suitable environment and reduced fitness otherwise (Brown and Pavlovic, 1992). According to these assumptions, fitting-trait organisms would become increasingly frequent in their respective suitable environment, and dispersals would be likely to lead to reduced fitness, thus favouring specialization (Brown and Pavlovic, 1992). This model has found support in different scales of plant biology. Plant ramets develop different carbon-allocation strategies in contrasting soil patches, revealing different suitable strategies across environments (Wijesinghe and Hutchings, 1999). Moreover, plant species have locally adapted populations in adjacent montane habitats, indicating that environmental differences promote selection-driven divergence despite geographical proximity (Hamann et al., 2016). Finally, tree species in topographically complex forests display greater habitat specificity than those in more even forests (Potts et al., 2004), suggesting specialization as the fittest response under greater environmental heterogeneity.
As consequence of specialization, environmental heterogeneity can also promote endemism via the spatial accumulation of species. Endemism occurs when organisms are found only in a given focal area (Anderson, 1994). This pattern has been assigned mainly to climatically stable areas (i.e. refugia) that have sheltered species during past climatic shifts (Haffer, 1969). Under the refugial model, climatic shifts drive populations to extinction outside refugia and, consequently, promote speciation by fragmenting large-ranged ancestral species into small-ranged descendant ones (Harrison and Noss, 2017). This model assumes that species cannot cope with novel climatic ranges (i.e. non-evolving niche breadths), thus undergoing an overall geographical range reduction (Harrison and Noss, 2017). Alternatively, the environmental heterogeneity model has long been hypothesized to underlie species diversity by promoting endemism (MacArthur, 1965; Ricklefs, 1976). Environmental heterogeneity favours specialization to different but spatially close niches (Brown and Pavlovic, 1992), and the resulting specialization impairs geographical expansion (Brown, 1984), ultimately promoting species accumulation in the focal area. Illustrating this process, tropical plant communities over increased topographic variation have greater spatial overlap among tree species than communities over more even terrains (Brown et al., 2013) Likewise, phytogeographical domains with greater climatic heterogeneity harbour greater plant species diversity than their same-latitude but more homogeneous counterparts (Jiménez and Ricklefs, 2014). Following this, high plant endemism has been associated with specialization in environmentally complex regions (Asthon, 1969; Gentry, 1992). For instance, in the tropical Andes, many endemic plant species inhabit specific habitats along the steep montane gradients (Young et al., 2002). Likewise, in South Africa’s Cape Floristic Region, plant endemism is often associated with edaphic specialization across different soil types (Grobler and Cowling, 2021). Based on that, environmental heterogeneity has probably led to high plant endemism in various floras worldwide.
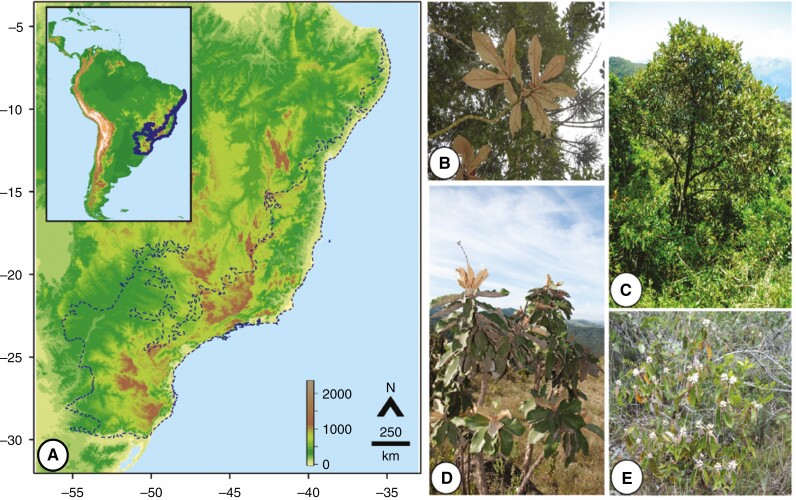
In the Neotropics, the Atlantic Forest (AF) domain is an acknowledged conservation hotspot (Myers et al., 2000), harbouring ~14 900 plant species, of which 9800 (65 %) are endemic (Fig. 1A; The Brazil Flora Group, 2021). Endemism in the AF domain has often been related to forest refugia during the Pleistocene [2.58–0.01 million years ago (Mya); Carnaval and Moritz, 2008]. Proposed forest refugia currently contain greater genetic diversity of different plant species than non-refugial areas, supporting their past role as shelters (e.g. Turchetto-Zolet et al., 2016; Leal et al., 2018; Mäder et al., 2021). However, as with most hypotheses of forest refugia, endemic plant lineages in the AF domains have been assigned to Pleistocene climatic shifts primarily based on ancestral reconstructions of geographical distribution, but without verifying the assumptions of geographical range reduction and speciation (Harrison and Noss, 2017). Although palaeo-projections based on niche modelling have indicated smaller geographical ranges during the Pleistocene (e.g. Bünger et al., 2016), these projections assume species with non-evolving niches a priori (Soberón and Nakamura, 2009). At the same time, plant endemism has also been associated with environmental heterogeneity in the AF domain. Montane landscapes underlie steep environmental gradients and delimit different ecoregions in the AF domain (Neves et al., 2017), thus providing a variable and spatially aggregated niche space. Accordingly, phylogenetic beta-diversity among plant communities is strongly correlated with mountain gradients in the AF domain (Mariano et al., 2020), indicating specialization across montane habitats. Finally, montane regions have the greatest phylogenetic endemism in the AF domain (Brown et al., 2020), suggesting specialization as a major promoter of plant endemism.
Fig. 1.
The Atlantic Forest (AF) domain in the Neotropics and some representative Miconia species from this study. (A) The elevational variation in the Neotropics, and more specifically, in the AF domain. Colour scale at the bottom right indicates elevation (in metres). The AF boundaries are represented by blue lines. (B) Miconia robusta Cogn. under a forest canopy in the AF. (C) Miconia formosa Cogn. on a forest border in the AF. (D) Miconia ferruginata DC. in a campos rupestres. (E) Miconia macuxi Meirelles, Caddah & R. Goldenb. in an Amazonian campinarana.
Miconia Ruiz & Pav. (Melastomataceae) sensuMichelangeli et al. (2019) is the most diverse genus of woody flowering plants in the AF domain, containing different endemic lineages in this domain (Flora do Brasil, 2020). Miconia species have berry fruits dispersed by various vertebrates and some insects, which act as keystone species in different Neotropical ecosystems (Messeder et al., 2021). Miconia species mainly rely on bees for pollination, although fruit and seed production also happen via apomixis, reducing pollination dependence (Renner, 1989). Hence, Miconia species have great dispersal potential, which becomes evident in their invasiveness over island systems (Meyer and Florence, 1996). However, different Miconia lineages display high levels of endemism in the AF domain (Reginato and Michelangeli, 2016; Goldenberg et al., 2018), indicating range-limiting factors associated with this domain. One such lineage is the Miconia supersect. Discolores Caddah & Goldenberg (Fig. 1B–E), a major clade (~77 spp.) whose species occur in the AF and other Neotropical domains (Caddah et al., 2022). In this group, most endemic species (~25 spp.) are restricted to the AF domain, while a few are restricted to other Neotropical domains (Caddah et al., 2022). In turn, non-endemic species (~50 spp.) are spread over the Neotropics, mostly in the AF, Amazonia, Caatinga, Cerrado and Mesoamerican domains in different combinations (Caddah et al., 2022). This pronounced endemism in the AF domain has previously been related to Pleistocene refugia (Caddah et al., 2022), but without an evaluation of species geographical range during this period. Nonetheless, species with different geographical distributions also occupy a different range of habitats, indicating the evolution of different niche breadths. The AF-endemic species inhabit evergreen or semi-deciduous forests at montane elevations (>500 m). In contrast, non-endemic species can occupy riverine forests, white-sand woodlands (campinaranas), Neotropical savannas or rocky grasslands (campos rupestres) (Caddah et al., 2022). Based on that, the Miconia supersect. Discolores allows evaluation of the relative impact of historical processes and ecological specialization on plant endemism in the AF domain.
In the present study, we investigated whether plant endemism in the AF would result from the evolution of specialization owing to greater environmental heterogeneity in this domain, using Miconia supersect. Discolores as a study system. We hypothesized that Miconia species have faced increased environmental heterogeneity and, consequently, have been selected towards increased specialization in the AF domain, and this increased specialization has greatly reduced the geographical range of species, ultimately promoting endemism. Hence, we made the following predictions: (1) AF-endemic species should face greater environmental heterogeneity than non-endemic species; (2) AF-endemic species should be more specialized than non-endemic species; (3) specialization should lead to smaller geographical ranges; (4) specialization and small geographical ranges among AF-endemic species should conform to a selection-driven evolutionary scenario rather than to a neutral evolutionary scenario; and (5) small geographical ranges among AF-endemic species should date back to the occupation of the AF domain rather than to more recent time periods. For the first prediction, we applied a spatial metric to estimate environmental heterogeneity around species distributions. For the second prediction, we inferred environmental specialization of species via niche modelling. For the third prediction, we measured species geographical range and inferred its association with environmental specialization. For the fourth prediction, we applied a model-fitting approach to infer the most likely evolutionary scenario of environmental specialization and geographical range. For the fifth prediction, we applied ancestral reconstructions to assess species geographical ranges over time.
MATERIALS AND METHODS
Unless otherwise stated, we performed data analyses in the R environment v.4.1.0 (R Development Core Team, 2021). The R code and all datasets are available at https://github.com/eknery/specialization_endemism_AF. Throughout, we have cited only the main functions and their respective R packages. We considered a significance level of 0.05 for frequentist statistical tests.
Study area
Our study area was the AF domain in the Neotropics (Fig. 1A). This phytogeographical domain is located within coordinates 02°50ʹ–33°45ʹS and 34°45ʹ–55°15ʹW, encompassing an area of 1 110 182 km2 (IBGE, 2012).The AF largely overlaps the eastern Brazilian highlands, ranging from sea level to 2892 m above (IBGE, 2012). From south to north, three major mountain ranges divide the domain: the Serra Geral, the Serra do Mar and the Serra da Mantiqueira. These mountain ranges have probably resulted from different geological processes from the Early Cretaceous to the Palaeogene (145–23 Mya; e.g. Hiruma et al., 2010). Despite its mainly tropical latitude, the AF domain hosts different climatic zones, from wet tropical to subtropical with dry seasons or frosts (Alvares et al., 2013). Soil types vary the most from east to west, following the elevational gradient, including variation regarding granulation, drainage and organic matter content (Cunha et al., 2019).
Study system
Our study system was the Miconia supersect. Discolores, a plant lineage including shrubs, treelets and trees (Fig. 1B–E). Given that lineage-wise ecological data are not available, we surveyed studies recording variation in Miconia species fitness associated with environmental conditions. We considered studies including any species in the Miconia supersect. Discolores, and we assumed that other species in the group would probably be affected by the same environmental conditions. Those environmental variables affecting fitness components were considered relevant for ecological niche modelling. Species in the Miconia supersect. Discolores are mostly dominant locally in forest habitats (Arellano and Macía, 2014), and their fruit dispersal relies mainly on birds (Allenspach and Dias, 2012; Maruyama et al., 2013; Santos et al., 2017). Their seeds are dormant-like and can withstand mild dry seasons in soil (Silveira et al., 2013; Escobar and Cardoso, 2015a), and they require high light availability for germination (Escobar and Cardoso, 2015b). Moreover, seed germination is reduced under increased temperature variation (Oliveira et al., 2014; Escobar and Cardoso, 2015b). After recruitment, seedling growth is not limited by soil nutrient availability (Denslow et al., 1998), but it decreases sharply in alkaline soils (Haridasan, 1988). Once established, mature plants reach lower photosynthetic rates during dry seasons (Monteiro and Prado, 2006), and they also display reduced flowering in shady conditions (Silva et al., 2016).
Environmental variables
To estimate environmental heterogeneity and model species environmental niches, we selected four variables that reflect environmental conditions that affect the fitness of the study Miconia species, as previously highlighted above. We also favoured a minor set of variables to avoid niche model overfitting (Mod et al., 2016). We downloaded raster layers of the diurnal temperature range (diurnal range/annual range, in degrees Celsius), precipitation seasonality (coefficient of variation, in millimetres), solar radiation (in kilojoules per square metre per day) and soil pH from the WorldClim and WoSIS repositories (Fick and Hijmans, 2017; Batjes et al., 2020). We set the coordinate reference system to WGS84 and raster resolution to 2.5ʹ (~5 km × 5 km grid cells). At this resolution, grid cells can encompass different communities where a species inhabits, hence they represent well the environmental conditions experienced by the whole species across its geographical distribution [i.e. beta niche sensuAckerly et al. (2006); see Environmental niche modelling section for further explanation]. Hereafter, we term the grid cells ‘sites’.
Phylogenetic reconstruction
To enable comparative phylogenetic analyses, we conducted a molecular-based phylogenetic reconstruction of the Miconia supersect. Discolores. To date, the most comprehensive phylogenetic study on the Miconia supersect. Discolores has included 57 species, but sampling has focused mostly on the AF domain (Caddah et al., 2022). Hence, we sought to improve species sampling from other geographical domains by including unpublished but already-curated molecular data from GenBank (Benson et al., 2005). Our phylogenetic sample covered most species (66 spp., 85 %) from the Miconia supersect. Discolores (92 % cover the AF domain, and 82 % cover other domains altogether) (Caddah et al., 2022). To improve inferences on the last common ancestor, we included 16 allied species of the Miconia supersect. Discolores (Goldenberg et al., 2008). To allow divergence time estimates, we also included 30 species representing the major clades of Miconia (Goldenberg et al., 2008). We set Eriocnema fulva, Physeterostemon fiaschii and Physeterostemon thomasii as the outgroup. Based on preliminary analyses, we excluded two species (Miconia amoena and Miconia eriodonta) that reduced the overall support of phylogenetic hypotheses. Our phylogenetic sampling included 115 species.
The molecular dataset included all loci sequenced for more than half of the species in the Miconia supersect. Discolores: the nuclear ribosomal ETS and ITS, and the plastidial intergenic spacers accD-psaI, atpF-atpH, psbK-psbL, trnS-trnG. We retrieved sequences from GenBank (Supplementary data Table S1), and we aligned same-locus sequences with MAFFT, under the GINSi strategy (Katoh and Standley, 2013). The aligned sequences provided 3894 characters. We assessed congruence and substitution patterns of sampled loci by functions of the ‘phangorn’ package (Schliep et al., 2017). To assess congruence among loci, we first generated neighbour-joining bootstrap trees (n = 100) for each locus using the NJ function. Then, we compared trees by a principal coordinate analysis on the Robison and Foulds’ distance (1981) using the RF.dist function. To assess substitution patterns in each locus, we evaluated 18 substitution models by the corrected Akaike information criterion (AICc) using the modelTest function. Loci differed regarding topology, hence we considered each locus as a single partition in phylogenetic reconstruction (Supplementary data Fig. S1).
We applied the Bayesian phylogenetic reconstruction implemented in BEAST 2.0, which uses a Markov chain Monte Carlo (MCMC) method to optimize phylogenetic trees (Drummond and Rambaut, 2007). Instead of assuming substitution models a priori, we applied a reversible-jump MCMC to use different substitution models (~30) over simulations, allowing a more extensive search on the tree space (Bouckaert and Drummond, 2017). We set a relaxed log-normal molecular clock for each locus, using hyperparameters to estimate substitution rates (Drummond et al., 2006). We assumed the calibrated Yule model for tree topology, and we set two secondary calibration points by Gavrutenko (2020): (1) the last common ancestor of Eriocnema+Phyeseterostemon+Miconia (14.71–29.9 Mya); and (2) the Miconia crown [Miconia IV + Miconia V groups in Goldenberg et al. (2008)] (7.05–13.44 Mya). The MCMC lasted 7 × 107 iterations, with sampling every 7 × 104 iterations. We verified the convergence of MCMC and sampling sufficiency (effective sample size, ESS > 200) via Tracer v.1.7 (Rambaut et al., 2018). We generated a mean-valued maximum clade credibility (MCC) after a 20 % burn-in via TreeAnnotator (Drummond and Rambaut, 2007). The MCC tree agreed with previous phylogenetic trees of the Miconia supersect. Discolores (Goldenberg et al., 2008; Caddah, 2013), but with higher statistical support and greater taxonomic representation (Supplementary data Fig. S2).
To consider the inherent uncertainty of phylogenetic reconstruction, we conducted comparative analyses on a sample of phylogenetic trees. We drew a random sample of phylogenetic trees (n = 100) from the burn-in posterior distribution.
Species occurrence dataset
To infer the geographic range and niche breadth of species, we built a taxonomically curated dataset of species occurrences for the Miconia supersect. Discolores. Taxonomically curated datasets have 24 % fewer misidentifications than non-curated datasets (Freitas et al., 2020). Moreover, taxonomically curated datasets provide equally suitable information for environmental niche modelling compared with more comprehensive datasets, despite their reduced sample sizes (Fourcade, 2016).
We gathered species occurrences while considering taxonomic nomenclature, voucher association and identification by experts. First, we researched all synonyms for each species (106 names; Supplementary data List S1; Meirelles, 2015; Caddah et al., 2020; Goldenberg and Bacci, 2020), in order that occurrences could reflect the geographical range of species despite their different naming among countries or research groups. Second, we downloaded occurrences (latitude and longitude) representing herbarium vouchers in the SpeciesLink (splink.org.br) and PBI:Miconiae (sweetgum.nybg.org/melastomataceae) databases. We did not consider GBIF a suitable source of species occurrence for the AF, because three-quarters (75 %) of GBIF entries hold invalid spatial data for this domain (Colli-Silva et al., 2020). Third, we retained only occurrences identified by an expert on Miconia taxonomy (68 experts in Supplementary data List S2). We recognized taxonomic expertise when the researcher had published monographs on Miconia groups, floras of Melastomataceae including Miconia, or descriptions of new Miconia species. We also recognized taxonomic expertise when the researcher had authored unpublished floras of poorly known localities, such as some areas of Amazonia. Our initial effort resulted in 9724 occurrences.
Afterwards, we conducted procedures to improve dataset quality. First, we excluded same-voucher occurrences within species and occurrences with low geographical precision or erroneous placement (e.g. municipality centroids and sea). Second, we assigned geographical coordinates to incomplete occurrences, following a standardized dataset of Neotropical localities (available from the first author upon request). Finally, to avoid spatial pseudo-replication, we conducted spatial thinning over occurrences using the thin function in the ‘spThin’ package (Aiello-Lammens et al., 2015). We excluded same-species occurrences <7 km apart, the greatest distance within a 5 km × 5 km site. For eight species known from few populations (Miconia angelana, M. capixaba, M. dura, M. kollmannii, M. kriegeriana, M. mellina, M. penduliflora and M. suberosa), we considered that same-site occurrences were not pseudo-replicates but rather a rough approximation of abundance. After improvement, our dataset comprised 5521 species occurrences.
Nonetheless, herbarium-based datasets might reflect the sampling bias by taxonomists rather than species distributions, a known pattern in the AF domain (Ostroski et al., 2020). Hence, to detect spatial sampling bias, we applied a normal kernel density estimation to species occurrences with the kde2d function in the ‘MASS’ package (Venables and Ripley, 2002). We found that sampling was five times denser towards the southeastern AF (Supplementary data Fig. S3). To avoid the effect of sampling bias, we applied the background-group strategy to environmental niche modelling (for further explanation, see the Environmental niche modelling section).
Geographical distribution of species
To assess species geographical distributions, we quantified species occurrences over Neotropical domains. We considered eight Neotropical domains: Amazonia, Andes, Atlantic Forest, Caatinga, Cerrado, Chaco, Mesoamerica and Pampa, whose boundaries were based on the shapefile of Olson et al. (2001) (Supplementary data Fig. S4). We overlapped species occurrences in the shapefile to quantify their absolute presence in each domain (Supplementary data Table S2). Most species (45 spp.) were in more than one domain, and the most frequent distributions were as follows: AF + Cerrado (eight spp.); Amazonia + Cerrado (six spp.); AF + Caatinga + Cerrado (four spp.); and AF + Amazonia + Caatinga + Cerrado (four spp.). Another 17 geographical distributions including more than one domain had low frequencies (three or fewer spp.) (Supplementary data Table S2). The remaining species (21 spp.) had single-domain distributions: AF (17 spp.); Amazonia (three spp.); and Cerrado (one sp.).
However, absolute presence is misleading when defining endemism because it is sensitive to erroneous records and does not consider biological processes that can revoke endemism temporarily, such as peripheral shrinking populations (Lima et al., 2020). Hence, we followed Lima et al. (2020) in defining optimal thresholds to detect endemic plant species in the AF domain based on their percentage presence. We assigned species to three geographical distributions based on their percentage presence in the AF domain: ‘AF-endemic’, ≥90 % of occurrences inside the AF domain; ‘outside the AF domain’, ≤10 % of occurrences inside the AF domain; ‘AF and other domains’, between 10 and 90 % of occurrences inside the AF domain. The percentage presence in the AF domain had a bimodal distribution, peaking at 10 and 90 % (Supplementary data Fig. S5), indicating these values as non-arbitrary divisors for discrete states.
We did not use each domain and their combinations as geographical distributions for different reasons. First, our objective was to understand the underlying processes of endemism in the AF, because this domain contains most endemic species in our study system. Hence, geographical distribution coding should reflect the degree of restriction to the focal area (e.g. Vasconcelos et al., 2020). Second, historical biogeographical analyses have already been conducted by Caddah et al. (2022), which indicated an Amazonian origin for our study system. Third, in our study system, non-endemic species can occupy two or more domains in different combinations, rendering 22 types of geographical distribution in ancestral reconstructions. Given that geographical distributions are treated as evolutionary regimes (see the Evolutionary scenario inference section), our phylogenetic sampling would not have the statistical power to infer parameters for many regimes.
Environmental heterogeneity estimates
To estimate the environmental heterogeneity, we applied Rao’s Q index over environmental raster layers using the paRao function in the ‘rasterdiv’ package (Rocchini et al., 2021). The Q index is the average dissimilarity between a focal site and other sites within a given spatial frame (Rocchini et al., 2017). To calculate the Q index, we measured dissimilarity among sites as the Euclidean distance based on the four environmental raster layers. We scaled environmental values to z-scores to avoid overweighting attributable to different scales. We considered a spatial frame including the eight immediate neighbours of each site, hence Q-values would approximate the environmental heterogeneity found during dispersal events, the relevant scale for the heterogeneity–specialization model (Brown and Pavlovic, 1992). After calculating the Q raster (Supplementary data Fig. S6), we used species occurrences to extract their respective Q-values. The Q-value distributions of species often departed from normality (Supplementary data Fig. S7); therefore, we considered the median Q-value as the proxy for the overall environmental heterogeneity faced by a species.
To evaluate whether species with different geographical distributions faced distinct environmental heterogeneity, we applied a phylogenetic generalized least squares (PGLS) model using the gls function in the ‘nlme’ package (Pinheiro et al., 2021). We considered the species Q-value as the response variable and species geographical distribution as a fixed factor. Given that species can inherit occupied sites from ancestors, we sought to estimate phylogenetic inheritance of environmental heterogeneity. We fitted the Brownian motion (BM) and the Ornstein–Uhlenbeck process (OU) models to species median Q-values by using the fitContinuous function in the ‘geiger’ package (Harmon et al., 2008). The OU model had the best fit according to the AICc; therefore, BM-based methods, such as phylogenetic independent contrasts, were considered unsuitable (Díaz-Uriarte and Garland, 1996). Based on the OU model, we calculated the variance–covariance matrix of species Q-values and implemented this matrix into the PGLS models to correct residual variation. We ln-transformed Q-values because the relationship between variables was exponential.
We also applied PGLS modelling to assess whether montane landscapes would influence the environmental heterogeneity faced by species. In this second model, we considered the species Q-value as the response variable and species median elevation as the predictor variable. The variance–covariance matrix calculation and data transformation proceeded as described above.
Environmental niche modelling
Niche modelling is a proxy for the overall environmental conditions occupied by a species. At a finer spatial scale, each species occupies a relative position along each environmental gradient within a given community, the alpha niche, which changes across communities owing to abiotic and biotic factors (Ackerly et al., 2006). At a coarser spatial scale, each species experiences environmental variation across its geographical range, the beta niche, which changes with evolutionary processes acting upon the whole species (Ackerly et al., 2006). Geographically referenced data, such as those applied to niche modelling, can approximate the beta niche scale (Ackerly et al., 2006). Hence, we used occurrences on raster layers to extract environmental values occupied by species, and we considered niche models as a proxy for the environmental niche at a beta scale.
Niche modelling should also indicate unoccupied environmental values, which are provided by absence records. Data derived from scientific collections lack absence records (Ponder et al., 2001); therefore, their application to niche modelling requires strategies that take this limitation into account. The background-group strategy assigns virtual absences of a given focal species based on presences of other spatially related species (Ponder et al., 2001). This strategy assumes that sampling effort is equally effective in detecting the focal species and the spatially related species, hence the presence of the latter without the presence of the former is considered an absence of the focal species (Ponder et al., 2001). Consequently, this strategy incorporates spatial bias from presence records into absence records, avoiding the problem that niche inferences reflect sampling bias (Phillips et al., 2009). We considered this strategy suitable for the Miconia supersect. Discolores because the group has been subject to taxonomic studies that have surveyed all species simultaneously (Caddah et al., 2022). Hence, we defined the background group of each modelled species as the occurrences of all other species, except sites where the modelled species occur.
To infer species environmental niche, we applied the hypervolume modelling method to species environmental values. The hypervolume method applies kernel functions to observed environmental values to infer the probability density function over n environmental variables (Blonder et al., 2014). The inferred probability functions delimit an n-dimensional volume, the hypervolume, whose size is a proxy for the overall niche breadth (Blonder et al., 2014). Hence, we performed environmental niche modelling using functions from the ‘hypervolume’ package (Blonder et al., 2014). We had already scaled environmental values to z-scores to avoid overweighting owing to the measurement scale.
To evaluate the predictive performance of hypervolume models, we used a k-fold cross-validation procedure under different probability thresholds. We considered environmental values from species occurrences as ‘presences’ and environmental values from the respective background group as ‘absences’. We split presences randomly into three sets (k = 3), using two sets (two-thirds) as training and one set (one-third) as testing over three different rounds. For species known from few localities, we validated hypervolume models by using a leave-one-out procedure (k = n). We built species hypervolume models based on the training by the hypervolume_gaussian function, using a normal bandwidth approximation by the estimate_bandwidth function (Blonder et al., 2018). We evaluated the predictive performance of models with the true skill statistics (TSS; sensitivity + specificity − 1) (Allouche et al., 2006), considering different probability thresholds (0.05, 0.25, 0.5, 0.75 and 0.95). Sensitivity was the proportion of the training classified as ‘presence’, and specificity was the proportion of the background group classified as ‘absence’. We averaged same-threshold rounds to evaluate model performance, and we used the maximum TSS value to choose the best threshold to model the species environmental niche.
After model evaluation (Supplementary data Table S3), we applied the hypervolume method to the environmental values of species, using the probability threshold of the maximum TSS value. We retrieved the hypervolume size as a proxy for species environmental niche breadth. Small hypervolumes sizes indicated increased environmental specialization. We calculated the 50 % interquartile range (IQR) over each probability density distribution as a measure of species niche breadth over each environmental gradient.
Nonetheless, specialization inferred from species occurrences can be an artefact of spatial autocorrelation (Cardillo et al., 2018). Under great spatial autocorrelation, distinct environmental values are found only at great distances (Legendre, 1993). Hence, when inferring niche breadth via occurrence-based modelling, specialization can be inferred for small-ranged species only because species occurrences are over regions with great spatial autocorrelation (Cardillo et al., 2018). To assess whether spatial autocorrelation influenced specialization inference, we estimated the local Moran’s I around species occurrences (Anselin, 1995). We calculated the I of environmental variables at a local scale, 3 × 3 sites, using the MoranLocal function in the ‘raster’ package (Hijmans, 2021). We then extracted the I-values from species occurrences, and we considered the median I-values of species as an estimate of the spatial autocorrelation in regions occupied by species.
To evaluate whether species with different geographical distributions had distinct niche breadths, we applied a PGLS model using the gls function in the ‘nlme’ package (Pinheiro et al., 2021). We considered the species hypervolume size as the response variable and species geographical distribution as a fixed factor with three levels. We set species median I-values as a covariate to evaluate the effect of spatial autocorrelation. We fitted the BM and OU models to hypervolume size by using the fitContinuous function in the ‘geiger’ package (Harmon et al., 2008). The OU model had the best fit according to the AICc. Based on the OU model, we calculated the covariance matrix for species hypervolume size and implemented it into the PGLS model to correct residual variation. We applied square-root transformation to conform residual variation to a normal distribution.
We also applied PGLS modelling to evaluate whether species with different geographical distributions differed regarding niche breadth over each environmental gradient. In each model, we considered the species IQR values for an environmental variable as the response variable and species geographical distribution as a fixed factor with three levels. The variance–covariance matrix calculation proceeded as described above, but without transforming the response variables.
Species geographical range
To infer species geographical ranges, we calculated the area within the minimal convex polygon (i.e. convex hull) delimited by species occurrences. Although less accurate for oddly shaped geographical distributions (Burgman and Fox, 2003), the convex hull is an accurate estimator of range size reduction (Darroch and Saupe, 2018). Given that we sought to evaluate geographical range reduction during the Pleistocene, we considered the convex hull area a reliable proxy for species geographical range. We delimited the convex hulls around species occurrences using the st_convex_hull function, and we measured the area (in square kilometres) within the convex hulls using the st_area function, with all functions being from the ‘sf’ package (Pebesma, 2018).
To evaluate whether species geographical range was related to specialization, we applied PGLS models using the gls function in the ‘nlme’ package (Pinheiro et al., 2021). In the first model, we considered the species convex hull area as the response variable and species hypervolume size as the predictor variable. In the second model, we kept the same response variable, but nested species hypervolume size within geographical distribution. We set species median I-values as a covariate in all models. We fitted the BM and OU models to the convex hull area using the fitContinuous function in the ‘geiger’ package (Harmon et al., 2008). The OU model had the best fit according to the AICc. Based on the OU model, we calculated the variance–covariance matrix of species convex hull area and implemented it into the PGLS models. We applied a cubic-root transformation to the convex hull area because the relationship between variables was cubic. We selected the best-fitting model by AICc.
Evolutionary scenario inference
To assess when endemism to the AF domain had evolved, we conducted an ancestral reconstruction of species geographical distribution using the ‘BioGeoBEARS’ package (Matzke, 2013). We applied the dispersal–extinction–cladogenesis (DEC) model, which represents the evolution of species geographical distribution via random dispersal and local extinction, while considering different modes of geographical area inheritance (Ree and Smith, 2008). We considered two geographical areas, AF and other domains, consequently allowing three geographical distributions: ‘AF-endemic’, ‘AF and other domains’ and ‘outside the AF domain’. We did not use time-structured transition rates because the AF domain is much older than our study system. We fitted the DEC model to our sample of phylogenetic trees (n = 100), and we used the maximum marginal likelihood at each node to assign ancestral species to a geographical distribution on each phylogenetic tree.
To infer the most likely evolutionary scenario of niche breadth and geographical range, we applied a model-fitting approach to species hypervolume size and convex hull area, considering species geographical distributions as evolutionary regimes. We used evolutionary models for continuous-scaled phenotypes based on the Brownian motion (BM) and the Ornstein–Uhlenbeck (OU) process. Under the BM models, phenotypic divergence results solely from an evolutionary rate (σ) representing stochastic variation through time (Felsenstein, 1988). In turn, under OU models, phenotypic divergence results not only from a an evolutionary rate (σ) but also from an attraction force (α) towards some optimum value (ϴ) (Hansen, 1997). As a general interpretation, BM models would represent evolution under genetic drift and mutation or fluctuating directional selection, whereas OU models would represent evolution under consistent stabilizing selection or constraints (Beaulieu et al., 2012).
Based on this approach, we contrasted seven evolutionary models with a different number of parameters (i.e. complexity) implemented in the ‘OUwie’ package (Beaulieu et al., 2012). The simplest model (BM1) assumed a single σ value for all evolutionary regimes, and the most complex model (OUMVA) assumed variable σ, ϴ and α values for each evolutionary regime. The other models had an intermediate number of parameters (Table 1). We applied all evolutionary models to species hypervolume size and convex hull area, considering different phylogenetic trees (n = 100). For each phylogenetic tree, we evaluated model fit by the AICc and ranked models by delta AICc (ΔAICc). When the lowest ΔAICc model was more complex than the second lowest ΔAICc model, we chose the more complex model only if it scored more than two over the simpler model.
Table 1.
Description of evolutionary models applied to species hypervolume size and convex hull area. Parameters represent different aspects of evolutionary regimes (σ, evolutionary rates; ϴ, optimum value; and α, attraction force). ‘Single’ indicates that parameter values were the same for evolutionary regimes, whereas ‘variable’ indicates that parameter values could vary among evolutionary regimes
| Model | Parameters | ||
|---|---|---|---|
| σ | ϴ | α | |
| BM1 | Single | – | – |
| BMS | Variable | – | – |
| OU1 | Single | Single | Single |
| OUM | Single | Variable | Single |
| OUMV | Variable | Variable | Single |
| OUMA | Single | Variable | Variable |
| OUMVA | Variable | Variable | Variable |
After model fitting, we assessed whether evolutionary regimes departed from expectations under neutral evolution. We first estimated σ in BM1 for species hypervolume size and convex hull area on each phylogenetic tree by the Ouwie function in the ‘OUwie’ package. Using the estimated σ values, we simulated the evolution of hypervolume size and convex hull area under the BM model by the fastBM function in the ‘phytools’ package. We ran 100 simulations on each phylogenetic tree, randomly setting an observed value as a possible ancestral state (z0) at each simulation. We calculated the mean value of each simulation to produce a null distribution (n = 10 000) for each trait. Finally, we compared the mean values in each evolutionary regime with the null distributions to assess whether evolutionary regimes would be expected under neutral evolution.
Ancestral reconstruction of geographical range
To evaluate the effect of Pleistocene climatic shifts on species geographical range, we reconstructed ancestral convex hull areas using our sample of phylogenetic trees (n = 100). We estimated ancestral values based on the best-fitting evolutionary models using the OUwie.anc function in the ‘OUwie’ package. From each phylogenetic tree, we retrieved the convex hull area, geographical distribution based on DEC, and age for every inner node (i.e. ancestral species). Then we compared mean ancestral values in each geographical distribution across ages, and we evaluated visually whether ancestral values underwent a reduction during the Pleistocene. We acknowledge that OU-based reconstructions have limited interpretation at deep nodes of phylogenetic trees (Royer-Carenzi and Didier, 2016); therefore, we have focused our discussion on relative rather than absolute values of convex hull area.
It is noteworthy that geographical range size is an emergent species-level trait, meaning that it is absent at the organism level, albeit affected by organism-level processes (Grantham, 1995). Species geographical range derives from heritable components, mostly organismal requirements, tolerances and dispersal mode, and non-heritable components, broadly including geographical features and historical processes (Sheth et al., 2020). These heritable components would provide some theoretical basis to infer ancestral geographical ranges based on evolutionary models for heritable phenotypes. Supporting this approach, the fossil records of some taxonomic groups display a positive relationship between ancestral and descendant range sizes (Jablonski, 1987). Moreover, based on in silico simulations, medium to large heritability values can produce the typical distribution of geographical range values observed in most taxonomic groups (Borregaard et al., 2012). Hence, we treated species geographical range as an evolving phenotype, but we also acknowledged effects from non-heritable components of geographical range.
RESULTS
Our sampling included 66 species of the Miconia supersect. Discolores: 23 species were AF-endemic and 33 non-endemic. Among the non-endemic species, 18 were spread in the AF and other domains, and 25 were outside the AF domain. The AF-endemic species had fewer occurrences (41 ± 63, mean ± s.d.) overall than species spread in the AF and other domains (181 ± 276) and species outside the AF domain (53 ± 46).
Environmental heterogeneity: higher Q-values for AF-endemic species and at higher elevations
Following our first prediction, Q-values, a proxy for the environmental heterogeneity faced by species, were 33 and 60 % higher for AF-endemic species (0.16 ± 0.03, mean ± s.d.) than for species spread in the AF and other domains (0.12 ± 0.02) and species outside the AF domain (0.10 ± 0.04), respectively (t = −3.0 and −6.2, P = 0.003 and 0.001, respectively; Fig. 2A). Considering each environmental variable separately, Q-values were also higher for AF-endemic species than for non-endemic species (Supplementary data Fig. S8). Species at high elevations, mostly AF-endemic species, had increased Q-values (slope = 0.53, t = 7.0, P < 0.001), and elevation explained about one-third of Q-value variation (R2 = 0.36; Fig. 2B).
Fig. 2.
Rao’s Q-value, representing environmental heterogeneity faced by species, and its relationship with species geographical distribution and elevation. (A) Species Q-value and its relationship with species geographical distribution. Box-and-whisker plots represent the 50 and 95 % interquartile ranges, and violin plots represent density distributions. *Significant differences from AF-endemic species. (B) Log species Q-value and its relationship with species elevation (in kilometres). The line represents the inferred relationship based on a phylogenetic generalized least squares (PGLS) model. The slope value indicates the inferred effect of species elevation, and the R2 value indicates model fit. *Significant effect of the predictor variable.
Environmental niche modelling: smaller hypervolumes among AF-endemic species
Following our second prediction, the hypervolume size, a proxy for species overall niche breadth, was 60 and 59 % smaller for AF-endemic species (1.84 ± 1.54, mean ± s.d.) than for species spread in the AF and other domains (4.64 ± 3.34) and species outside the AF domain (4.55 ± 4.60), respectively (t = 2.97 and 2.03, P = 0.004 and 0.046, respectively; Fig. 3). The value of Moran’s I, a proxy for the spatial autocorrelation in regions occupied by species, did not affect species hypervolume size (t = 0.88, P = 0.37), indicating that small hypervolume sizes (i.e. specialization) were not associated with highly autocorrelated regions. Interquartile range values calculated over solar radiation and soil pH differed between AF-endemic and non-endemic species, whereas the diurnal temperature range and precipitation seasonality did not differ (Supplementary data Fig. S9). Interquartile range values over solar radiation were 27 % smaller among AF-endemic species (896) than among species spread in the AF and other domains (1200) and species outside the AF domain (1201) (t = 2.8 and 2.7, P = 0.005 and 0.006, respectively). Interquartile range values of soil pH were 28 and 47 % smaller among AF-endemic species (0.31) than among species in the AF and other domains (0.43) and species outside the AF domain (0.59), respectively (t = 2.2 and 4.1, P = 0.003 and 0.001, respectively). Hence, the smaller hypervolume sizes of AF-endemic species reflected a narrow niche breadth over solar radiation and soil pH gradients.
Fig. 3.
Hypervolume size, representing species’ environmental niche breadth, and its relationship with species geographical distribution. Box-and-whisker plots represent the 50 and 95 % interquartile ranges, and violin plots represent density distributions. *Significant differences from AF-endemic species.
Geographical range: smaller areas with smaller hypervolumes, mostly among AF-endemic species
Convex hull area, a proxy for species geographical range, was 14 and 20 times smaller for AF-endemic species (12 ± 15 km2, median ± IQR) than for species spread in the AF and other domains (169 ± 574 km2) and species outside the AF domain (223 ± 324 km2), respectively.
Following our third prediction, convex hull area was positively associated with hypervolume size. When comparing PGLS models on the relationship between convex hull area and hypervolume size, the nested model (AICc = 294, R2 = 0.57) had a better fit and explained a greater part of convex hull area variation than the simpler model (AICc = 308; R2 = 0.38). Under the nested PGLS model, hypervolume size had a great positive effect on convex hull area among AF-endemic species (slope = 0.73, t = 2.6, P = 0.009), but only a moderate effect among species spread in the AF and other domains (slope = 0.44, t = 3.1, P = 0.002) and species outside the AF domain (slope = 0.31, t = 3.6, P = 0.001; Fig. 4).
Fig. 4.
Convex hull area, representing species geographical range, and its relationship with hypervolume size, representing species’ environmental niche breadth. Convex hull area is cubic root-transformed to represent relationships in a linear fashion. Each line represents a relationship between variables according to species geographical distribution based on a nested phylogenetic generalized least squares (PGLS) model. Slope values indicate the inferred effect of species hypervolume size, and the R2 value indicates the overall model fit. *Significant effects of geographical distribution on the inferred relationship.
Evolutionary scenarios: best-fitting OUMV models indicate distinct selective regimes
Phylogenetic reconstruction indicated that the Miconia supersect. Discolores dates to 8.1 Mya (95 % highest posterior density = 5.9–10.8 Mya; Fig. 5). The last common ancestor of the group was outside the AF domain, and most descendant species remained outside. Endemism in the AF domain has evolved independently three times (numbered squares in Fig. 5). First, a vicariant process originated an AF-endemic lineage between 6.2 and 4.4 Mya, which then diversified in the AF domain. Different dispersal processes to other domains originated non-endemic lineages between 2.4 and 0.1 Mya, during the Pleistocene. Second, a vicariant process originated an AF-endemic lineage between 2.0 Mya and the present, during the Pleistocene. Third, a vicariant process originated an AF-endemic lineage between 0.8 Mya and the present, during the Pleistocene.
Fig. 5.
Phylogenetic reconstruction of Miconia supersect. Discolores, inferred ancestral geographical distributions, and their hypervolume sizes and convex hull areas. Phylogenetic relationships are represented by the maximum clade credibility tree. Pie charts at nodes represent the proportional likelihood of ancestral species geographical distributions based on the DEC model, and pie charts at tips indicate geographical distributions of extant species. Numbered squares at branches indicate the origin of Atlantic Forest (AF)-endemic lineages. Bars at the tips represent hypervolume sizes and convex hull areas for extant species. The convex hull area is cubic root-transformed to highlight the overall linear relationship between variables among AF-endemic species. The scale at the bottom represents the time before the present (in millions of years ago). The grey band highlights the Pleistocene. Throughout, colours indicate geographical distributions as follows: blue, AF-endemic; yellow, AF and other domains; red, outside the AF domain.
Overall, hypervolume size and convex hull area had a similar distribution of values across species, with small values usually co-occurring in AF-endemic species. Nonetheless, this association did not hold mostly among non-endemic species. Species age did not affect either hypervolume size or convex hull area because shorter and longer branches had both small and large values for both variables.
The OUMV model had the lowest ΔAICc for hypervolume size and convex hull area across 100 and 74 % of the phylogenetic trees, respectively. Following our fourth prediction, the OUMV represents a selection-driven scenario, in which each geographical distribution (evolutionary regime) has its own ϴ and σ values but has a single α value (Table 1).
Considering the OUMV estimates for hypervolume size, ϴ values of AF-endemic species (ϴ = 1.8 ± 0.03, mean ± s.d.) were three times smaller than those of species spread in the AF and other domains (ϴ = 4.4 ± 0.10) and species outside the AF domain (ϴ = 4.6 ± 0.08) (Fig. 6A). The σ values of AF-endemic species (σ = 5.1 ± 3.8) were two and three times smaller than those of species spread in the AF and other domains (σ = 11.2 ± 8.5) and species outside the AF domain (σ = 15.5 ± 12), respectively (Fig. 6B). The mean hypervolume size in all geographical distributions was smaller than BM expectations (P < 0.05), but deviation from BM expectations was greater for AF-endemic species (Fig. 6C).
Fig. 6.
OUMV model estimates for hypervolume size and convex hull area considering species geographical distribution as evolutionary regimes. The ϴ (A) and σ (B) estimates for hypervolume size. The ϴ (D) and σ (E) estimates for convex hull area. Observed means (continuous lines) of hypervolume size (C) and convex hull area (F) and associated standard errors (dotted lines) for each geographical distribution and the expected means (grey distributions) under the Brownian motion (BM) model. Throughout, colours indicate geographical distributions: blue, AF-endemic; yellow, AF and other domains; red, outside the AF domain.
Considering the OUMV estimates for convex hull area, ϴ values of AF-endemic species (ϴ = 19.4 ± 7.3) were 20 and 15 times smaller than those of species spread in the AF and other domains (ϴ = 369.2 ± 358.9) and species outside the AF domain (ϴ = 283.3 ± 11.91), respectively (Fig. 6D). The σ values of AF-endemic species (σ = 98 ± 110) were 13 and eight times smaller than those of species spread in the AF and other domains (σ = 1339 ± 1574) and species outside the AF domain (σ = 795 ± 905), respectively (Fig. 6E). The mean convex hull area in all geographical distributions was smaller than BM expectations (P < 0.05), but deviation from BM expectations was greater for AF-endemic species (Fig. 6F).
Ancestral reconstruction of geographical range: AF-endemics rapidly reduced their geographical ranges after their evolution
Ancestral reconstructions of convex hull area indicated greater values at older than at more recent ages (Fig. 7). The last common ancestral of AF-endemic species had a greater convex hull area than same-age non-endemic species, but this difference rapidly inverted after the evolution of AF-endemic species. Between 8 and 6 Mya, ancestral AF-endemic species had a 52 % reduction of convex hull area, while species spread in the AF and other domains and species outside the AF domain had 23 and 0.5 % reduction of convex hull area, respectively. After this rapid inversion, ancestral AF-endemic species retained smaller convex hull areas than ancestral non-endemic species during the following ages. Other changes of convex hull area occurred during the Pleistocene. During this period, AF-endemic species and species outside the AF domain had a 1.3 and 22 % reduction of convex hull area, respectively, whereas species spread in the AF and other domains had an 8 % increase.
Fig. 7.
The convex hull area, a proxy for species geographical range size, in each geographical distribution throughout time. Ancestral values inferred by the OUMV model. Dots represent the mean estimate during a time period, and vertical lines represent the associated standard error. The scale at the bottom represents the time before the present (in millions of years). The dashed line marks the beginning of the Pleistocene. Colours indicate geographical distributions: blue, AF-endemic; yellow, AF and other domains; red, outside the AF domain.
DISCUSSION
Environmental heterogeneity imposes selective pressures favouring specialization, which, in turn, can constrain geographical range and promote the assemblage of endemic floras. The AF domain is a hotspot that harbours a highly endemic flora, and Miconia is the most diverse and remarkably endemic plant genus in this domain. Here, we have hypothesized that Miconia species have faced increased environmental heterogeneity and, consequently, have been selected towards increased specialization in the AF domain, and this increased specialization has greatly reduced species geographical range, ultimately promoting endemism. Based on this, we have made five predictions and tested them using geographical, environmental and phylogenetic data on the Miconia supersect. Discolores, a major lineage of woody angiosperms that includes AF-endemic and non-endemic species. We have found overall support for all of our predictions, as follows: (1) environmental heterogeneity was 33–60 % higher around AF-endemic species than around non-endemic species; (2) AF-endemic species were overall 60 % more specialized than non-endemic species; (3) specialization strongly predicted smaller geographical ranges among AF-endemic species, but not among non-endemic species; (4) AF-endemic species have evolved towards specialization and small geographical ranges under a selection-driven scenario; and (5) ancestral AF-endemic species underwent a 52 % reduction of geographical range immediately after their evolution, maintaining their small geographical ranges throughout historical periods. The implications of our results are discussed below.
Atlantic Forest-endemic species face great environmental heterogeneity
Environmental heterogeneity was overall 33–60 % greater around AF-endemic species than around non-endemic species (Fig. 2A). Such heterogeneity resulted from greater spatial variation in all analysed environmental gradients (Supplementary data Fig. S4). This pattern supports our first prediction that AF-endemic species should face greater environmental heterogeneity than non-endemic species. Hence, AF-endemic species are more likely to face different environments while dispersing to neighbouring sites (Brown and Pavlovic, 1992). This great environmental heterogeneity around AF-endemic species was considerably associated with elevation (Fig. 2B), indicating that montane landscapes have prompted environmental variation. The spatial complexity in montane landscapes has long been thought to increase environmental heterogeneity (Körner, 2004), and this spatial complexity probably increases with elevation. Other montane features, such as age and position, also contribute to environmental heterogeneity (Perrigo et al., 2020), but their relative roles were not our main focus.
Mountains can shape the regional climate by deflecting radiation and relocating precipitation, and mountain weathering can affect soil composition by moving sediments from highlands to lowlands (Perrigo et al., 2020). In the AF domain, mountain ranges can host more climatic zones than nearby lowland areas, ranging from wet tropical in the foothills to subtropical with short summers at mountain tops (Alvares et al., 2013). Moreover, the mountain ranges of the AF date back to different geological periods and provide different sediment types (Hiruma et al., 2010), such that soil types can vary greatly along mountain slopes (Cunha et al., 2019). These mountain-associated gradients are major drivers of plant distribution in the AF domain, because they underlie distinct vegetation types, from coastal woodlands (restingas) up to cloud dwarf forests (matinhas nebulares) (Neves et al., 2017).
Mountains can also explain a major part of the environmental heterogeneity in other Neotropical domains. For instance, the Cerrado domain encompasses a minor portion of the eastern Brazilian highlands, over which annual precipitation and soil types vary the most (Sano et al., 2019). Likewise, the Campos rupestres province displays a significant elevational variation that underlies steep gradients of drought severity and temperature oscillation (Bueno et al., 2021). Aside from mountains, environmental heterogeneity around non-endemic species can also result from the Amazon basin, which can concentrate different soil patches into relatively small areas (Quesada et al., 2020). Nonetheless, these different geological and edaphic features are not as spatially close in extra-Andean Neotropical domains as mountain ranges in the AF domain, granting the latter a higher environmental heterogeneity.
Atlantic Forest-endemic species are environmentally specialized
Specialization was 60 % greater overall among AF-endemic species than among non-endemic species (Fig. 3), reflecting a narrower niche breadth over solar radiation and soil pH (Supplementary data Fig. S6). This pattern supports our second prediction that AF-endemic species should be more specialized than non-endemic species. Increased specialization among AF-endemic species is likely to be a response to their immediate surroundings with great environmental heterogeneity. As environments become more different, they impose contrasting challenges that can necessitate trade-offs by organisms (Levins, 1962). Hence, when different environments are spatially close (i.e. great environmental heterogeneity), organisms are more likely to experience reduced fitness when dispersing to their surroundings (Brown and Pavlovic, 1992). Consequently, in an environmentally heterogeneous background, optimally exploring fewer similar environments would lead to greater fitness than exploring several different environments (Brown and Pavlovic, 1992).
The montane landscapes of the AF harbour different environments that can demand contrasting responses from plant species. Supporting this, environmental gradients predict functional beta-diversity among plant communities in the AF domain, also delimiting different plant physiognomies across montane regions (Silva et al., 2021). For instance, the carbon allocation strategy of woody Rubiaceae species changes with light conditions, varying between valley and ridge areas in the AF domain (Torres-Leite et al., 2019), revealing different plant responses across montane areas. Hence, a single generalist plant strategy would be unlikely to cope with the environmental heterogeneity imposed by the montane landscapes of the AF. Among all environmental gradients, solar radiation and soil pH vary greatly in space owing to montane features. Montane slope and aspect influence solar radiation by changing ground inclination and orientation for sunlight (Martin et al., 2019). Likewise, the montane slope influences soil pH by changing soil material flux (Cunha et al., 2019). As a likely response to this environmental variation, AF-endemic Miconia species displayed increased specialization regarding solar radiation and soil pH (Supplementary data Fig. S9). The effect of this specialization on species functional traits must be addressed elsewhere. Other environmental gradients might also favour specialization in the AF domain because they predict functional variation across plant assemblages (Silva et al., 2021), but assessing their role requires studies in other plant lineages structured by them.
Consequently, specialization can be a frequent, but still overlooked, pattern among endemic plant lineages in the AF domain. In support of this, the phylogenetic turnover of plant communities strongly follows mountain gradients (Mariano et al., 2020), indicating that closely related plant lineages are specialized to different environments. Moreover, phylogenetic endemism overall increases over the montane regions of the AF (Brown et al., 2020), reflecting a spatial accumulation of divergent plant lineages, a likely effect of specialization. Indeed, environmental gradients predict phylogenetic endemism in different taxonomic groups, including plants, in the AF domain (Paz et al., 2021), thus highlighting that endemic plant lineages are associated with specific environmental conditions. Based on this, the high plant endemism in the AF domain could hide a general pattern of plant specialization to different habitats, as recorded in other montane domains (e.g. Casazza et al., 2005; Boulangeat et al., 2012; Buira et al., 2020). Nonetheless, assessing the generality of this pattern requires further studies inferring specialization on other endemic plant systems in the AF domain.
Specialization leads to small geographical ranges, mostly for AF-endemic species
The geographical range of species was positively correlated with their niche breadth, most strongly among AF-endemic species (Fig. 4). This pattern supports our third prediction that specialization should lead to smaller geographical ranges. Specialization can decrease species geographical range by limiting suitable conditions and, consequently, reducing the inhabited areas (Brown, 1984). This process is pervasive, because specialization predicts smaller geographical ranges across different taxonomic groups, especially when related to environmental tolerances (Slatyer et al., 2013). Given that plants have low mobility during their lifespan, geographical ranges more readily reflect environmental niche breadth among plant species (e.g. Sheth and Angert, 2014). As a result, specialization greatly affects the geographical range of plants among all other range-limiting factors (Sheth et al., 2020). Such a relationship could be an artefact owing to great spatial autocorrelation, under which small-ranged species are considered specialists because their occurrences are over environmentally correlated regions (Cardillo et al., 2018). However, great spatial autocorrelation did not predict great specialization in our study system. Hence, as environmental specialization increased, Miconia species had fewer suitable areas to inhabit and, consequently, established small geographical ranges, thus prompting endemism in the AF domain.
Nonetheless, AF-endemic species might also experience other range-limiting factors that are confounded by environmental heterogeneity. The mountains of the AF impose steep environmental gradients that restrict the dispersal of bird species (Thom et al., 2021), which could, consequently, restrict dispersal of Miconia species (Messeder et al., 2021). Based on this, environmental heterogeneity would favour plant specialization and reduce plant dispersal, thus confounding different range-limiting factors. The additive effects of specialization and dispersal limitation could underlie the smaller range sizes of AF-endemic species compared with equally specialized non-endemic species (Fig. 4). However, addressing the role of dispersal limitation in plant endemism requires further investigation. Another possible range-limiting factor are historical processes, and Pleistocene climatic shifts are considered a major driver of small-ranged plant lineages in the AF domain (e.g. Bünger et al., 2016). Nonetheless, AF-endemic species underwent a minor reduction of geographical range during the Pleistocene (Fig. 7; for further discussion, see Small geographical ranges are a long-standing pattern in the AF domain. Hence, range size reduction among AF-endemic species is likely to derive from great specialization, possibly coupled with dispersal limitations.
In turn, small geographical ranges were moderately associated with specialization among non-endemic species, indicating a greater effect of different range-limiting factors in other Neotropical domains. Given that non-endemic species overall face less environmental heterogeneity than AF-endemic species, they are more likely to establish themselves successfully on surrounding sites (Brown and Pavlovic, 1992), thus forecasting geographical expansion. Hence, specialization among non-endemic species might not respond to environmental heterogeneity. Specialization is also favoured in extremely limiting but frequent conditions, which provide reduced competition with increased area availability (Futuyma and Moreno, 1988). For instance, flooded plains are typical limiting habitats in the Amazon domain, but floodplain-specialist plant species contribute little to Amazonian endemism, because they are often found in other Neotropical domains (Wittmann et al., 2013). Based on this, limiting but frequent conditions might have allowed specialization among non-endemic species without small geographical ranges. Moreover, as specialization becomes less pronounced, other range-limiting factors are likely to take place (Sheth et al., 2020). For instance, Amazonian rivers are strong dispersal barriers to small-sized birds (Hayes and Sewlal, 2004), which can, consequently, limit Miconia fruit dispersal in the Amazon basin. Likewise, short-ranging ant species have a major role as Miconia fruit dispersers in the Cerrado domain (Christianini and Oliveira, 2009). These dispersal limitations can impact geographical range size regardless of species niche breadth, which might have led some generalist non-endemic species to have small geographical ranges. Thus, non-endemic species with small geographical ranges might better reflect dispersal limitations rather than specialization.
Specialization and small geographical ranges are adaptative optima for AF-endemic species
The niche breadth and geographical range of Miconia species evolved under a selection-driven scenario (the OUMV model), with different optima (ϴ) and evolutionary rates (σ) among geographical distributions, but under a single attraction force (Beaulieu et al., 2012). The AF-endemic species have evolved towards more specialized and small-ranged optima than non-endemic species (Fig. 6A, D). Also, AF-endemic species have evolved under lower rates around their respective optima than non-endemic species (Fig. 6B, E). Observed niche breadths and geographical ranges deviated from expectations under neutral evolution for all geographical distributions (Fig. 6C, F). Accordingly, the single attraction force across geographical distributions indicates that selection intensity upon species niche breadth and geographical range are invariant to biogeographical domains. All these patterns support our fourth prediction that specialization and smaller geographical ranges among AF-endemic species should conform to a selection-driven evolutionary scenario rather than to a neutral evolutionary scenario. Nonetheless, they also indicate that selective pressure in the AF domain, as elaborated further, might also occur in other biogeographical domains, but favouring other niche breadth and geographical range values.
Specialization and small geographical ranges among AF-endemic species are likely to be an adaptative response to the great environmental heterogeneity in AF montane landscapes. With increased environmental heterogeneity, nearby areas are more likely to impose contrasting environmental demands, thus imposing selective pressures favouring repeated dispersal and specialization to a few similar environments (Brown and Pavlovic, 1992). In the AF domain, montane habitats contain locally adapted populations for different plant species (e.g. Brancalion et al., 2018; Feliciano et al., 2022; Vetö et al., 2022), thus highlighting selective pressures towards specialization in the montane landscapes of the AF. Hence, AF-endemic species have evolved towards specialization and small geographical ranges probably as an optimal adaptative response to the contrasting environments in the montane landscapes of the AF. Moreover, as environmental heterogeneity increases, selective pressures towards specialization would intensify (Levins, 1962), hence the reduced evolutionary rates for AF-endemic species can reflect a strong selective pressure impairing generalization and geographical range expansion (Beaulieu et al., 2012). Aside from selective pressures, reduced evolutionary rates can also derive from evolutionary constraints. Specialization can lead to evolutionary constraints via accumulation of mutations and antagonistic pleiotropy, and both processes impair phenotypic evolution (Poisot et al., 2011). Hence, as specialization increased, AF-endemic species might have lacked some potential to evolve towards generalization and large geographical ranges (Poisot et al., 2011). We lack the genetic data to infer evolutionary constraints properly, but our results indicate a possibility that deserves further attention. Thus, as dispersal to the AF domain or extinction outside it proceeded, Miconia species experienced a novel selective regime favouring specialization and small range sizes, thus promoting endemism.
Environmental heterogeneity has been suggested to promote plant endemism in the AF, but the underlying evolutionary processes have remained mostly assumed. Environmental heterogeneity has long been thought selectively to favour plant specialization, facilitating in situ speciation and species accumulation, ultimately promoting endemism (Asthon, 1969; Ricklefs, 1976; Gentry, 1992). For instance, Protea species have diversified in the Amazon domain under the repeated evolution of soil specialization, hence sister species are often geographically close but ecologically divergent (Fine et al., 2005). As a result of evolutionary processes, great environmental heterogeneity can predict high endemism in plant communities across different domains (e.g. Irl et al., 2015; Molina-Venegas et al., 2017; Tordoni et al., 2020). Although environmental heterogeneity has been linked to plant endemism in the AF domain (e.g. Furtado and Menini Neto, 2021), selection-driven plant specialization has been assumed rather than tested. When analysing different AF-endemic plant lineages, Leão et al. (2020) found increased diversification rates associated with small geographical ranges, a pattern assumed to reflect specialization-driven speciation. Albeit robust, this study has not inferred specialization and its mode of evolution among endemic plant lineages. As a likely example, endemic bromeliad lineages have evolved towards habitat specificity in the AF domain, and this specificity could have allowed these lineages to occupy overlapping geographical distributions (Maciel et al., 2020). To our knowledge, our study is the first to address environmental heterogeneity and consequent selective pressures favouring specialization as promoters of plant endemism in the AF. Generalizing this explanatory model, of course, requires further studies on other endemic plant systems in the AF domain.
Small geographical ranges are a long-standing pattern in the AF domain
Geographical ranges of the earliest ancestral species were greater than those of extant species regardless of geographical distribution (Fig. 7), indicating widespread ancestral species. However, the earliest ancestral AF-endemic species underwent a sharp reduction of geographical range immediately after their evolution, while ancestral non-endemic species underwent minor reductions in the same time period. After this sharp reduction, ancestral AF-endemic species maintained smaller geographical ranges than ancestral non-endemic species. Hence, this pattern supports our fifth prediction that small geographical ranges among AF-endemic species should date back to the occupation of the AF domain rather than to more recent time periods. This pattern most probably derives from the selective pressures favouring specialization and small geographical ranges in the montane landscapes of the AF, as already discussed. Given that montane ranges are ancient geographical features in the AF domain (145–23 Mya; e.g. Hiruma et al., 2010), this selection-driven reduction of geographical range would have been acting upon Miconia species since their establishment in the AF domain. Therefore, the association between specialization and small geographical ranges probably arose with the occupation of the AF domain, lingering throughout historical periods.
Nonetheless, the Pleistocene climatic shifts have probably impacted ancestral Miconia species, which underwent changes in geographical range during the Pleistocene (Fig. 7). Glacial–interglacial cycles are thought to have imposed a contraction–expansion dynamic on Neotropical forests during the Pleistocene (Prance, 1982). This historical process has been supported in the AF domain, where climatically stable forest refugia would have sheltered animal taxa during the Pleistocene (Carnaval and Moritz, 2008). Proposed refugia currently harbour greater genetic diversity for different plant species than non-refugial areas (e.g. Turchetto-Zolet et al., 2016; Leal et al., 2018; Mäder et al., 2021), thus supporting their role as climatic shelters for plants as well. One major refugium is inferred over the montane regions of the Southern AF domain (Porto et al., 2013), where AF-endemic Miconia species often occur. In the Southern AF domain, montane plant lineages are thought to have either expanded or displaced their geographical ranges during the Pleistocene (Wilson et al., 2021). These processes might also have occurred with AF-endemic Miconia species, but applied reconstruction methods are not sensible to these processes (see the Materials and Methods section). Ancestral geographical ranges are better inferred via palaeo-projections of environmental niche models followed by the empirical validation of fossil records (Svenning et al., 2011). Nonetheless, this approach assumes species with non-evolving environmental niches (Soberón and Nakamura, 2009), an assumption that may not hold at the macroevolutionary scale (Donoghue and Edwards, 2014). Despite the limitations, the applied ancestral reconstructions most probably depict group-wise trends in geographical range, whereas palaeo-projections will probably reveal species-specific changes, refining our understanding about the Pleistocene climatic shifts.
In sum, plant endemism in the AF domain most probably derives from the interaction between time-generalized ecological specialization and time-specific environmental changes, but the latter has received far more attention (e.g. Fiaschi and Pirani, 2009). On the one hand, the evolution of ecological specialization depends on spatially variable but temporally steady environmental backgrounds (Sexton et al., 2017), such that natural selection can optimize specialists to a given environment throughout generations (Poisot et al., 2011). Under this perspective, the Pleistocene climatic shifts would immediately have disfavoured ecological specialization by changing environments in the AF domain. On the other hand, geological and climatic changes are known to have increased environmental heterogeneity in the Neotropics, including within the AF domain (Hughes et al., 2013; Palma-Silva et al., 2022). Under this other perspective, the Pleistocene climatic shifts would have favoured ecological specialization by providing novel environmental niches in the AF domain. Understanding this interaction demands further studies on other endemic plant systems in the AF domain, but similar patterns have been recorded in neighbouring biogeographical units. For instance, in the montane Campos rupestres province, endemic plant lineages have diversified under different scenarios, both Pleistocene related and unrelated, consequently suggesting specialization and dispersal limitations as endemism drivers as well (Vasconcelos et al., 2020). Indeed, plant endemism is likely to result from the interaction among range-limiting factors (Sheth et al., 2020); therefore, macroevolutionary studies should address the relative roles of factors upon different plant lineages across distinct biogeographical units.
Conclusions
Environmental heterogeneity favours the selection-driven evolution of ecological specialization, constraining the geographical range of organisms and, ultimately, promoting endemism. The AF domain has a highly endemic flora and encompasses distinct environments across its montane landscapes, potentially hiding a plant specialization pattern. Here, we have found that AF-endemic plant species face greater environmental heterogeneity than closely related non-endemic species, probably owing to the montane landscapes of the AF. Furthermore, AF-endemic plant species were more specialized than non-endemic species, a likely response to their contrasting environmental surroundings. We have found a strong association between specialization and small geographical ranges among AF-endemic lineages, supporting specialization as a main range-limiting factor upon endemic lineages. Moreover, AF-endemic plant species have evolved towards specialization and small geographical ranges under a selective regime, probably imposed by the montane landscapes of the AF. We also inferred that small geographical ranges have probably evolved with the occupation of the AF domain. Based on that, we have argued that plant endemism in the AF domain derives mainly from an increased ecological specialization favoured by the great environmental heterogeneity over montane landscapes. Ecological specialization is a time-generalized process driving plant endemism in the AF domain, probably interacting with other time-specific processes.
Supplementary Material
ACKNOWLEDGEMENTS
We thank A. F. Augustin and J. Meirelles for providing molecular data and taxonomic advice. E.K.N. thanks Universidade Federal do ABC for his PhD scholarship and Brazil’s Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) for his PDSE visiting-PhD scholarship.
Contributor Information
Eduardo K Nery, Programa de Pós-Graduação em Evolução e Diversidade, Centro de Ciências Naturais e Humanas, Universidade Federal do ABC, São Bernardo do Campo – SP, Brazil.
Mayara K Caddah, Departamento de Botânica, Universidade Federal de Santa Catarina, Florianópolis – SC, Brazil.
Matheus F Santos, Centro de Ciências Naturais e Humanas, Universidade Federal do ABC, São Bernardo do Campo – SP, Brazil.
Anselmo Nogueira, Centro de Ciências Naturais e Humanas, Universidade Federal do ABC, São Bernardo do Campo – SP, Brazil.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following.
Fig. S1: principal coordinate analysis of Robison and Foulds’ distance among phylogenetic trees based on different loci.
Fig. S2: the mean-valued maximum clade credibility phylogenetic tree.
Fig. S3: total species occurrences and their inferred sampling density.
Fig. S4: Neotropical domains recognized in this study.
Fig. S5: the number of species in each category of percentage occurrence in the Atlantic Forest domain.
Fig. S6: Rao’s Q-values over the Neotropics.
Fig. S7: distribution of Rao’s Q-value per species.
Fig. S8: species Q-values for different environmental variables and their relationship with species geographical distribution.
Fig. S9: species interquartile range values over different environmental variables and their relationship with species geographical distribution.
List S1: species names researched for the taxonomic database of occurrences.
List S2: acknowledged experts on Miconia taxonomy, in alphabetical order.
Table S1: the loci used in the phylogenetic reconstruction and their respective GenBank accession numbers.
Table S2: the total number of occurrences per species and their distribution over different Neotropical domains.
Table S3: predictive performance of hypervolume models per species.
FUNDING
This work was supported by Brazil’s National Council for Scientific and Technological Development (CNPq; 423616/2016-1 to M.K.C.) and the São Paulo Research Foundation (FAPESP; grant 2019/19544-7 to A.N.).
LITERATURE CITED
- Ackerly DD, Schwilk DW, Webb CO.. 2006. Niche evolution and adaptive radiation: testing the order of trait divergence. Ecology 87: S50–S61. doi: 10.1890/0012-9658(2006)87[50:neaart]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Aiello-Lammens ME, Boria RA, Radosavljevic A, Vilela B, Anderson RP.. 2015. spThin: an R package for spatial thinning of species occurrence records for use in ecological niche models. Ecography 38: 541–545. doi: 10.1111/ecog.01132. [DOI] [Google Scholar]
- Allenspach N, Dias M.. 2012. Frugivory by birds on Miconia albicans (Melastomataceae), in a fragment of cerrado in São Carlos, southeastern Brazil. Brazilian Journal of Biology 72: 407–413. doi: 10.1590/s1519-69842012000200024. [DOI] [PubMed] [Google Scholar]
- Allouche O, Tsoar A, Kadmon R.. 2006. Assessing the accuracy of species distribution models: prevalence, kappa and the true skill statistic (TSS). Journal of Applied Ecology 43: 1223–1232. doi: 10.1111/j.1365-2664.2006.01214.x. [DOI] [Google Scholar]
- Alvares CA, Stape JL, Sentelhas PC, De Moraes Gonçalves JL, Sparovek G.. 2013. Köppen’s climate classification map for Brazil. Meteorologische Zeitschrift 22: 711–728. doi: 10.1127/0941-2948/2013/0507. [DOI] [Google Scholar]
- Anderson S. 1994. Area and endemism. The Quarterly Review of Biology 69: 451–471. doi: 10.1086/418743. [DOI] [Google Scholar]
- Anselin L. 1995. Local indicators of spatial association—LISA. Geographical Analysis 27: 93–115. [Google Scholar]
- Arellano G, Macía MJ.. 2014. Local and regional dominance of woody plants along an elevational gradient in a tropical montane forest of northwestern Bolivia. Plant Ecology 215: 39–54. [Google Scholar]
- Asthon PS. 1969. Speciation among tropical forest trees: some deductions in the light of recent evidence. Biological Journal of the Linnean Society 1: 155–196. [Google Scholar]
- Batjes NH, Ribeiro E, Van Oostrum A.. 2020. Standardized soil profile data to support global mapping and modelling (WoSIS snapshot 2019). Earth System Science Data 12: 299–320. doi: 10.5194/essd-12-299-2020. [DOI] [Google Scholar]
- Beaulieu JM, Jhwueng DC, Boettiger C, O’Meara BC.. 2012. Modeling stabilizing selection: expanding the Ornstein–Uhlenbeck model of adaptive evolution. Evolution 66: 2369–2383. doi: 10.1111/j.1558-5646.2012.01619.x. [DOI] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL.. 2005. GenBank. Nucleic Acids Research 33: D34–D38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blonder B, Lamanna C, Violle C, Enquist BJ.. 2014. The n-dimensional hypervolume. Global Ecology and Biogeography 23: 595–609. doi: 10.1111/geb.12146. [DOI] [Google Scholar]
- Blonder B, Morrow CB, Maitner B, et al. 2018. New approaches for delineating n-dimensional hypervolumes. Methods in Ecology and Evolution 9: 305–319. [Google Scholar]
- Borregaard MK, Gotelli NJ, Rahbek C.. 2012. Are range-size distributions consistent with species-level heritability? Evolution 66: 2216–2226. doi: 10.1111/j.1558-5646.2012.01581.x. [DOI] [PubMed] [Google Scholar]
- Bouckaert RR, Drummond AJ.. 2017. bModelTest: Bayesian phylogenetic site model averaging and model comparison. BMC Evolutionary Biology 17: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulangeat I, Lavergne S, Van Es J, Garraud L, Thuiller W.. 2012. Niche breadth, rarity and ecological characteristics within a regional flora spanning large environmental gradients. Journal of Biogeography 39: 204–214. [Google Scholar]
- Brancalion PHS, Oliveira GCX, Zucchi MI, et al. 2018. Phenotypic plasticity and local adaptation favor range expansion of a Neotropical palm. Ecology and Evolution 8: 7462–7475. doi: 10.1002/ece3.4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C, Burslem DFRP, Illian JB, et al. 2013. Multispecies coexistence of trees in tropical forests: spatial signals of topographic niche differentiation increase with environmental heterogeneity. Proceedings of the Royal Society B: Biological Sciences 280: 20130502. doi: 10.1098/rspb.2013.0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JH. 1984. On the relationship between abundance and distribution of species. The American Naturalist 124: 255–279. doi: 10.1086/284267. [DOI] [Google Scholar]
- Brown JL, Paz A, Reginato M, et al. 2020. Seeing the forest through many trees: multi-taxon patterns of phylogenetic diversity in the Atlantic Forest hotspot. Diversity and Distributions 26: 1160–1176. doi: 10.1111/ddi.13116. [DOI] [Google Scholar]
- Brown JS, Pavlovic NB.. 1992. Evolution in heterogeneous environments: effects of migration on habitat specialization. Evolutionary Ecology 6: 360–382. doi: 10.1007/bf02270698. [DOI] [Google Scholar]
- Bueno ML, Rezende VL, De Paula LFA, et al. 2021. Understanding how environmental heterogeneity and elevation drives the distribution of woody communities across vegetation types within the campo rupestre in South America. Journal of Mountain Science 18: 1192–1207. doi: 10.1007/s11629-020-6125-0. [DOI] [Google Scholar]
- Buira A, Cabezas F, Aedo C.. 2020. Disentangling ecological traits related to plant endemism, rarity and conservation status in the Iberian Peninsula. Biodiversity and Conservation 29: 1937–1958. doi: 10.1007/s10531-020-01957-z. [DOI] [Google Scholar]
- Bünger MDO, Mazine FF, Forest F, Bueno ML, Stehmann JR, Lucas EJ.. 2016. The evolutionary history of Eugenia sect. Phyllocalyx (Myrtaceae) corroborates historically stable areas in the southern Atlantic forests. Annals of Botany 118: 1209–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgman MA, Fox JC.. 2003. Bias in species range estimates from minimum convex polygons: implications for conservation and options for improved planning. Animal Conservation 6: 19–28. doi: 10.1017/s1367943003003044. [DOI] [Google Scholar]
- Caddah MK. 2013. Estudos taxonômicos e filogenéticos em Miconia sect. Discolor (Melastomataceae, Miconieae). Brazil: PhD Thesis, Universidade Estadual de Campinas. [Google Scholar]
- Caddah MK, Augustin AF, Goldenberg R.. 2020. Deflating Miconia (Melastomataceae) from Eastern Brazil, with 31 new synonyms and other nomenclatural issues. Phytotaxa 468: 283–295. doi: 10.11646/phytotaxa.468.3.4. [DOI] [Google Scholar]
- Caddah MK, Meirelles J, Nery EK, et al. 2022. Beneath a hairy problem: phylogeny, morphology, and biogeography circumscribe the new Miconia supersection Discolores (Melastomataceae: Miconieae). Molecular Phylogenetics and Evolution 171: 107461. doi: 10.1016/j.ympev.2022.107461. [DOI] [PubMed] [Google Scholar]
- Cardillo M, Dinnage R, McAlister W.. 2018. The relationship between environmental niche breadth and geographic range size across plant species. Journal of Biogeography 46: 97–109. doi: 10.1111/jbi.13477. [DOI] [Google Scholar]
- Carnaval AC, Moritz C.. 2008. Historical climate modelling predicts patterns of current biodiversity in the Brazilian Atlantic forest. Journal of Biogeography 35: 1187–1201. doi: 10.1111/j.1365-2699.2007.01870.x. [DOI] [Google Scholar]
- Casazza G, Barberis G, Minuto L.. 2005. Ecological characteristics and rarity of endemic plants of the Italian Maritime Alps. Biological Conservation 123: 361–371. doi: 10.1016/j.biocon.2004.12.005. [DOI] [Google Scholar]
- Christianini AV, Oliveira PS.. 2009. The relevance of ants as seed rescuers of a primarily bird-dispersed tree in the Neotropical cerrado savanna. Oecologia 160: 735–745. doi: 10.1007/s00442-009-1349-2. [DOI] [PubMed] [Google Scholar]
- Colli-Silva M, Reginato M, Cabral A, Forzza RC, Pirani JR, Vasconcelos TN da C.. 2020. Evaluating shortfalls and spatial accuracy of biodiversity documentation in the Atlantic Forest, the most diverse and threatened Brazilian phytogeographic domain. Taxon 69: 567–577. [Google Scholar]
- Cunha A de M, Fontes MPF, Lani JL.. 2019. Mineralogical and chemical attributes of soils from the Brazilian atlantic forest domain. Scientia Agricola 76: 82–92. doi: 10.1590/1678-992x-2017-0109. [DOI] [Google Scholar]
- Darroch SAF, Saupe EE.. 2018. Reconstructing geographic range-size dynamics from fossil data. Paleobiology 44: 25–39. doi: 10.1017/pab.2017.25. [DOI] [Google Scholar]
- Denslow JS, Ellison AM, Sanford RE.. 1998. Treefall gap size effects on above- and below-ground processes in a tropical wet forest. Journal of Ecology 86: 597–609. doi: 10.1046/j.1365-2745.1998.00295.x. [DOI] [Google Scholar]
- Devictor V, Clavel J, Julliard R, et al. 2010. Defining and measuring ecological specialization. Journal of Applied Ecology 47: 15–25. doi: 10.1111/j.1365-2664.2009.01744.x. [DOI] [Google Scholar]
- Díaz-Uriarte R, Garland T.. 1996. Testing hypotheses of correlated evolution using phylogenetically independent contrasts: sensitivity to deviations from Brownian motion. Systematic Biology 45: 27–47. doi: 10.1093/sysbio/45.1.27. [DOI] [Google Scholar]
- Donoghue MJ, Edwards EJ.. 2014. Biome shifts and niche evolution in plants. Annual Review of Ecology, Evolution, and Systematics 45: 547–572. doi: 10.1146/annurev-ecolsys-120213-091905. [DOI] [Google Scholar]
- Drummond AJ, Rambaut A.. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology 7: 2141–2148. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Ho SYW, Phillips MJ, Rambaut A.. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biology 4: e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar DFE, Cardoso VJM.. 2015a. Longevity of seeds and soil seed bank of the Cerrado tree Miconia chartacea (Melastomataceae). Seed Science Research 25: 386–394. [Google Scholar]
- Escobar DFE, Cardoso VJM.. 2015b. Germinación y latencia de semillas de Miconia chartacea (Melastomataceae), en respuesta a luz, temperatura y hormonas vegetales. Revista de Biologia Tropical 63: 1169–1184. [Google Scholar]
- Feliciano DC, De Godoy SM, Marques JF, et al. 2022. Landscape genetics reveal low diversity and adaptive divergence in Portulaca hatschbachii (Portulacaceae): an endangered species endemic to rocky outcrops of the Atlantic Forest. Botanical Journal of the Linnean Society 200: 116–141. [Google Scholar]
- Felsenstein J. 1988. Phylogenies and quantitative characters. Annual Review of Ecology and Systematics 19: 445–471. doi: 10.1146/annurev.es.19.110188.002305. [DOI] [Google Scholar]
- Fiaschi P, Pirani JR.. 2009. Review of plant biogeographic studies in Brazil. Journal of Systematics and Evolution 47: 477–496. doi: 10.1111/j.1759-6831.2009.00046.x. [DOI] [Google Scholar]
- Fick SE, Hijmans RJ.. 2017. Worldclim 2: new 1-km spatial resolution climate surfaces for global land areas. International Journal of Climatology 37: 4302–4315. doi: 10.1002/joc.5086. [DOI] [Google Scholar]
- Fine PVA, Daly DC, Muñoz GV, Mesones I, Cameron KM.. 2005. The contribution of edaphic heterogeneity to the evolution and diversity of Burseraceae trees in the Western Amazon. Evolution 59: 1464–1478. doi: 10.1554/04-745. [DOI] [PubMed] [Google Scholar]
- Flora do Brasil. 2020. http://floradobrasil.jbrj.gov.br/. 29 March 2021.
- Fourcade Y. 2016. Comparing species distributions modelled from occurrence data and from expert-based range maps. Implication for predicting range shifts with climate change. Ecological Informatics 36: 8–14. doi: 10.1016/j.ecoinf.2016.09.002. [DOI] [Google Scholar]
- Freitas TMS, Montag LFA, De Marco P, Hortal J.. 2020. How reliable are species identifications in biodiversity big data? Evaluating the records of a neotropical fish family in online repositories. Systematics and Biodiversity 18: 181–191. doi: 10.1080/14772000.2020.1730473. [DOI] [Google Scholar]
- Furtado SG, Menini Neto L.. 2021. What is the role of topographic heterogeneity and climate on the distribution and conservation of vascular epiphytes in the Brazilian Atlantic Forest? Biodiversity and Conservation 30: 1415–1431. doi: 10.1007/s10531-021-02150-6. [DOI] [Google Scholar]
- Futuyma DJ, Moreno G.. 1988. The evolution of ecological specialization. Annual Review of Ecology and Systematics 19: 207–233. doi: 10.1146/annurev.es.19.110188.001231. [DOI] [Google Scholar]
- Gavrutenko M, Reginato M, Kriebel R, Nicolas AN, Michelangeli FA.. 2020. Evolution of floral morphology and symmetry in the Miconieae (Melastomataceae): multiple generalization trends within a specialized family. International Journal of Plant Sciences 181: 732–747. doi: 10.1086/708906. [DOI] [Google Scholar]
- Gentry AH. 1992. Tropical forest biodiversity: distributional patterns and their conservational significance. Oikos 63: 19–28. doi: 10.2307/3545512. [DOI] [Google Scholar]
- Goldenberg R, Bacci LF.. 2020. Taxonomic notes in South American Miconia IV: the rare Miconia pennipilis Cogn. is actually a synonym of the enigmatic Miconia dura Triana. Phytotaxa 459: 296–300. doi: 10.11646/phytotaxa.459.4.6. [DOI] [Google Scholar]
- Goldenberg R, Penneys DS, Almeda F, Judd WS, Michelangeli FA.. 2008. Phylogeny of Miconia (Melastomataceae): patterns of stamen diversification in a megadiverse Neotropical genus. International Journal of Plant Sciences 169: 963–979. doi: 10.1086/589697. [DOI] [Google Scholar]
- Goldenberg R, Reginato M, Michelangeli FA.. 2018. Disentangling the infrageneric classification of megadiverse taxa from Mata Atlantica: phylogeny of Miconia section Chaenanthera (Melastomataceae: Miconieae). Taxon 67: 537–551. doi: 10.12705/673.15. [DOI] [Google Scholar]
- Grantham TA. 1995. Hierarchical approaches to macroevolution: recent work on species selection and the effect hypothesis. Annual Review of Ecology and Systematics 26: 301–321. doi: 10.1146/annurev.es.26.110195.001505. [DOI] [Google Scholar]
- Grobler BA, Cowling RM.. 2021. The composition, geography, biology and assembly of the coastal flora of the Cape Floristic Region. PeerJ 9: e11916. doi: 10.7717/peerj.11916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffer J. 1969. Speciation in Amazonian forest birds. Science 165: 131–137. doi: 10.1126/science.165.3889.131. [DOI] [PubMed] [Google Scholar]
- Hamann E, Kesselring H, Armbruster GFJ, Scheepens JF, Stöcklin J.. 2016. Evidence of local adaptation to fine- and coarse-grained environmental variability in Poa alpina in the Swiss Alps. Journal of Ecology 104: 1627–1637. doi: 10.1111/1365-2745.12628. [DOI] [Google Scholar]
- Hansen TF. 1997. Stabilizing selection and the comparative analysis of adaptation. Evolution 51: 1341–1351. doi: 10.1111/j.1558-5646.1997.tb01457.x. [DOI] [PubMed] [Google Scholar]
- Haridasan M. 1988. Performance of Miconia albicans (Sw.) Triana, an aluminum-accumulating species, in acidic and calcareous soils. Communications in Soil Science and Plant Analysis 19: 1091–1103. doi: 10.1080/00103628809367997. [DOI] [Google Scholar]
- Harmon LJ, Weir JT, Brock CD, Glor RE, Challenger W.. 2008. GEIGER: investigating evolutionary radiations. Bioinformatics 24: 129–131. doi: 10.1093/bioinformatics/btm538. [DOI] [PubMed] [Google Scholar]
- Harrison S, Noss R.. 2017. Endemism hotspots are linked to stable climatic refugia. Annals of Botany 119: 207–214. doi: 10.1093/aob/mcw248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes FE, Sewlal JAN.. 2004. The Amazon River as a dispersal barrier to passerine birds: effects of river width, habitat and taxonomy. Journal of Biogeography 31: 1809–1818. doi: 10.1111/j.1365-2699.2004.01139.x. [DOI] [Google Scholar]
- Hijmans RJ. 2021. raster: geographic data analysis and modeling. [Google Scholar]
- Hiruma ST, Riccomini C, Modenesi-Gauttieri MC, Hackspacher PC, Neto JCH, Franco-Magalhães AOB.. 2010. Denudation history of the Bocaina Plateau, Serra do Mar, southeastern Brazil: relationships to Gondwana breakup and passive margin development. Gondwana Research 18: 674–687. [Google Scholar]
- Hughes CE, Pennington RT, Antonelli A.. 2013. Neotropical plant evolution: assembling the big picture. Botanical Journal of the Linnean Society 171: 1–18. [Google Scholar]
- Hutchinson GE. 1957. Concluding remarks. Cold Spring Harbor Symposium on Quantitative Biology 22: 415–427. [Google Scholar]
- IBGE. 2012. Manual técnico da vegetação brasileira. Rio de Janeiro: Instituto brasileiro de geografia e estatística. [Google Scholar]
- Irl SDH, Harter DEV, Steinbauer MJ, et al. 2015. Climate vs. topography – spatial patterns of plant species diversity and endemism on a high-elevation island. Journal of Ecology 103: 1621–1633. doi: 10.1111/1365-2745.12463. [DOI] [Google Scholar]
- Jablonski D. 1987. Heritability at the species level: analysis of geographic ranges of Cretaceous mollusks. Science 238: 360–363. doi: 10.1126/science.238.4825.360. [DOI] [PubMed] [Google Scholar]
- Jenkins CN, Pimm SL, Joppa LN.. 2013. Global patterns of terrestrial vertebrate diversity and conservation. Proceedings of the National Academy of Sciences of the United States of America 110: E2603–E2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez I, Ricklefs RE.. 2014. Diversity anomalies and spatial climate heterogeneity. Global Ecology and Biogeography 23: 988–999. doi: 10.1111/geb.12181. [DOI] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kier G, Mutke J, Dinerstein E, et al. 2005. Global patterns of plant diversity and floristic knowledge. Journal of Biogeography 32: 1107–1116. doi: 10.1111/j.1365-2699.2005.01272.x. [DOI] [Google Scholar]
- Körner C. 2004. Mountain biodiversity, its causes and function. Ambio 33: 11–17. [PubMed] [Google Scholar]
- Leal BSS, Medeiros LR, Peres EA, et al. 2018. Insights into the evolutionary dynamics of Neotropical biomes from the phylogeography and paleodistribution modeling of Bromelia balansae. American Journal of Botany 105: 1725–1734. doi: 10.1002/ajb2.1167. [DOI] [PubMed] [Google Scholar]
- Leão TCC, Lughandha EN, Reich PB.. 2020. Evolutionary patterns in the geographic range size of Atlantic Forest plants. Ecography 43: 1510–1520. [Google Scholar]
- Legendre P. 1993. Spatial autocorrelation: trouble or new paradigm? Ecology 74: 1659–1673. doi: 10.2307/1939924. [DOI] [Google Scholar]
- Levins R. 1962. Theory of fitness in a heterogeneous environment. I. The fitness set and adaptive function. The American Naturalist 96: 361–373. doi: 10.1086/282245. [DOI] [Google Scholar]
- Lima RAF, Souza VC, Siqueira MF, ter Steege H.. 2020. Defining endemism levels for biodiversity conservation: tree species in the Atlantic Forest hotspot. Biological Conservation 252: 108825. [Google Scholar]
- MacArthur RH. 1965. Patterns of species diversity. Biological Reviews 40: 510–533. doi: 10.1111/j.1469-185x.1965.tb00815.x. [DOI] [Google Scholar]
- Maciel JR, Zizka G, Alves M.. 2020. Differential geographical and ecological dynamics allow diversification of morphologically convergent giant bromeliads in the Atlantic Forest. Journal of Biogeography 47: 2684–2697. doi: 10.1111/jbi.13961. [DOI] [Google Scholar]
- Mäder G, Zamberlan PM, Segatto ALA, Stehmann JR, Bonatto SL, Freitas LB.. 2021. When phylogeography meets niche suitability to unravel the evolutionary history of a shrub from the Brazilian Atlantic Forest. Botanical Journal of the Linnean Society 195: 77–92. [Google Scholar]
- Mariano RF, Rezende VL, Mendes CN, et al. 2020. Phylogenetic beta diversity in an upper montane Atlantic Forest along an altitudinal gradient. Plant Ecology 221: 671–682. doi: 10.1007/s11258-020-01041-0. [DOI] [Google Scholar]
- Martin TC, da Rocha HR, Joly CA, Freitas HC, Wanderley RL, da Silva JM.. 2019. Fine-scale climate variability in a complex terrain basin using a high-resolution weather station network in southeastern Brazil. International Journal of Climatology 39: 218–234. [Google Scholar]
- Maruyama PK, Borges MR, Silva PA, Burns KC, Melo C.. 2013. Avian frugivory in Miconia (Melastomataceae): contrasting fruiting times promote habitat complementarity between savanna and palm swamp. Journal of Tropical Ecology 29: 99–109. doi: 10.1017/s0266467413000138. [DOI] [Google Scholar]
- Matzke NJ. 2013. Probabilistic historical biogeography: new models for founder-event speciation, imperfect detection, and fossils allow improved accuracy and model-testing. Frontiers of Biogeography 5: 242–248. [Google Scholar]
- Meirelles J. 2015. Filogenia de Miconia seção Miconia subseção Seriatiflorae e revisão taxonômica do clado Albicans (Melastomataceae, Miconieae). Brazil: PhD Thesis, Universidade Estadual de Campinas. [Google Scholar]
- Messeder JVS, Silveira FAO, Cornelissen TG, Fuzessy LF, Guerra TJ.. 2021. Frugivory and seed dispersal in a hyperdiverse plant clade and its role as a keystone resource for the Neotropical fauna. Annals of Botany 127: 577–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer J-Y, Florence J.. 1996. Tahiti’s native flora endangered by the invasion of Miconia calvescens DC. (Melastomataceae). Journal of Biogeography 23: 775–781. doi: 10.1111/j.1365-2699.1996.tb00038.x. [DOI] [Google Scholar]
- Michelangeli FA, Goldenberg R, Almeda F, et al. 2019. Nomenclatural novelties in Miconia (Melastomataceae: Miconieae). Brittonia 71: 82–121. [Google Scholar]
- Mod HK, Scherrer D, Luoto M, Guisan A.. 2016. What we use is not what we know: environmental predictors in plant distribution models. Journal of Vegetation Science 27: 1308–1322. doi: 10.1111/jvs.12444. [DOI] [Google Scholar]
- Molina-Venegas R, Aparicio A, Lavergne S, Arroyo J.. 2017. Climatic and topographical correlates of plant palaeo- and neoendemism in a Mediterranean biodiversity hotspot. Annals of Botany 119: 229–238. doi: 10.1093/aob/mcw093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro JAF, Prado CHBA.. 2006. Apparent carboxylation efficiency and relative stomatal and mesophyll limitations of photosynthesis in an evergreen cerrado species during water stress. Photosynthetica 44: 39–45. doi: 10.1007/s11099-005-0156-1. [DOI] [Google Scholar]
- Myers N, Mittermeier RA, da Fonseca GAB, Kents J.. 2000. Biodiversity hotspots for conservation priorities. Biodiversity and Conservation 16: 853–858. [DOI] [PubMed] [Google Scholar]
- Neves DM, Dexter KG, Pennington RT, et al. 2017. Dissecting a biodiversity hotspot: the importance of environmentally marginal habitats in the Atlantic Forest Domain of South America. Diversity and Distributions 23: 898–909. doi: 10.1111/ddi.12581. [DOI] [Google Scholar]
- Oliveira AKM, Mota CMG, Agnes DC.. 2014. Efeito de diferentes temperaturas na germinação de sementes e no crescimento inicial de plântulas de Miconia albicans (Melastomataceae). Revista Brasileira de Plantas Medicinais 16: 755–759. doi: 10.1590/1983-084x/11_147. [DOI] [Google Scholar]
- Olson DM, Dinerstein E, Wikramanayake ED, et al. 2001. Terrestrial ecoregions of the world: a new map of life on Earth. BioScience 51: 933–938. doi: 10.1641/0006-3568(2001)051[0933:teotwa]2.0.co;2. [DOI] [Google Scholar]
- Ostroski P, Saiter FZ, Amorim AM, Fiaschi P.. 2020. Angiosperm endemism in a Brazilian Atlantic Forest biodiversity hot-point. Revista Brasileira de Botanica 43: 397–404. [Google Scholar]
- Palma-Silva C, Turchetto-Zolet AC, Fay MF, Vasconcelos T.. 2022. Drivers of exceptional Neotropical biodiversity: an updated view. Botanical Journal of the Linnean Society 199: 1–7. doi: 10.1093/botlinnean/boac005. [DOI] [Google Scholar]
- Paz A, Brown JL, Cordeiro CLO, et al. 2021. Environmental correlates of taxonomic and phylogenetic diversity in the Atlantic Forest. Journal of Biogeography 48: 1377–1391. [Google Scholar]
- Pebesma E. 2018. Simple features for R: standardized support for spatial vector data. R Journal 10: 439–446. [Google Scholar]
- Perrigo A, Hoorn C, Antonelli A.. 2020. Why mountains matter for biodiversity. Journal of Biogeography 47: 315–325. [Google Scholar]
- Phillips SJ, Dudík M, Elith J, et al. 2009. Sample selection bias and presence-only distribution models: implications for background and pseudo-absence data. Ecological Applications 19: 181–197. doi: 10.1890/07-2153.1. [DOI] [PubMed] [Google Scholar]
- Pinheiro J, Bates D, DebRoy S, Sarkar D, R Core Team. 2021. nlme: linear and non-linear mixed effects models. https://CRAN.R-project.org/package=nlme. Accessed 01 January 2021. [Google Scholar]
- Poisot T, Bever JD, Nemri A, Thrall PH, Hochberg ME.. 2011. A conceptual framework for the evolution of ecological specialisation. Ecology Letters 14: 841–851. doi: 10.1111/j.1461-0248.2011.01645.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponder WF, Carter GA, Flemons P, Chapman RR.. 2001. Evaluation of museum collection data for use in biodiversity assessment. Conservation Biology 15: 648–657. doi: 10.1046/j.1523-1739.2001.015003648.x. [DOI] [Google Scholar]
- Porto TJ, Carnaval AC, da Rocha PLB.. 2013. Evaluating forest refugial models using species distribution models, model filling and inclusion: a case study with 14 Brazilian species. Diversity and Distributions 19: 330–340. [Google Scholar]
- Potts MD, Davies SJ, Bossert WH, Tan S, Supardi MNN.. 2004. Habitat heterogeneity and niche structure of trees in two tropical rain forests. Oecologia 139: 446–453. doi: 10.1007/s00442-004-1525-3. [DOI] [PubMed] [Google Scholar]
- Prance GT. 1982. A review of the phytogeographic evidences for Pleistocene climate changes in the Neotropics. Annals of the Missouri Botanical Garden 69: 594–624. doi: 10.2307/2399085. [DOI] [Google Scholar]
- Quesada CA, Paz C, Mendoza EO, Phillips OL, Saiz G, Lloyd J.. 2020. Variations in soil chemical and physical properties explain basin-wide Amazon forest soil carbon concentrations. Soil 6: 53–88. [Google Scholar]
- R Development Core Team. 2021. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing. https://www.R-project.org/. Accessed 01 January 2021. [Google Scholar]
- Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA.. 2018. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Systematic Biology 67: 901–904. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ree RH, Smith SA.. 2008. Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Systematic Biology 57: 4–14. doi: 10.1080/10635150701883881. [DOI] [PubMed] [Google Scholar]
- Reginato M, Michelangeli FA.. 2016. Untangling the phylogeny of Leandra s.str. (Melastomataceae, Miconieae). Molecular Phylogenetics and Evolution 96: 17–32. doi: 10.1016/j.ympev.2015.11.015. [DOI] [PubMed] [Google Scholar]
- Renner SS. 1989. A survey of reproductive biology in Neotropical Melastomataceae and Memecylaceae. Annals of the Missouri Botanical Garden 76: 496–518. doi: 10.2307/2399497. [DOI] [Google Scholar]
- Ricklefs RE. 1976. Environmental heterogeneity and plant species diversity: a hypothesis. The American Naturalist 111: 376–381. [Google Scholar]
- Robinson DF, Foulds LR.. 1981. Comparison of phylogenetic trees. Mathematical Biosciences 53: 131–147. doi: 10.1016/0025-5564(81)90043-2. [DOI] [Google Scholar]
- Rocchini D, Marcantonio M, Ricotta C.. 2017. Measuring Rao’s Q diversity index from remote sensing: an open source solution. Ecological Indicators 72: 234–238. doi: 10.1016/j.ecolind.2016.07.039. [DOI] [Google Scholar]
- Rocchini D, Thouverai E, Marcantonio M, et al. 2021. rasterdiv—an information theory tailored R package for measuring ecosystem heterogeneity from space: to the origin and back. Methods in Ecology and Evolution 12: 1093–1102. doi: 10.1111/2041-210X.13583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royer-Carenzi M, Didier G.. 2016. A comparison of ancestral state reconstruction methods for quantitative characters. Journal of Theoretical Biology 404: 126–142. doi: 10.1016/j.jtbi.2016.05.029. [DOI] [PubMed] [Google Scholar]
- Sano EE, Rodrigues AA, Martins ES, et al. 2019. Cerrado ecoregions: a spatial framework to assess and prioritize Brazilian savanna environmental diversity for conservation. Journal of Environmental Management 232: 818–828. doi: 10.1016/j.jenvman.2018.11.108. [DOI] [PubMed] [Google Scholar]
- Santos AMO, Jacobi CM, Silveira FAO.. 2017. Frugivory and seed dispersal effectiveness in two Miconia (Melastomataceae) species from ferruginous campo rupestre. Seed Science Research 27: 65–73. doi: 10.1017/s0960258517000071. [DOI] [Google Scholar]
- Schliep K, Potts AJ, Morrison DA, Grimm GW.. 2017. Intertwining phylogenetic trees and networks. Methods in Ecology and Evolution 8: 1212–1220. doi: 10.1111/2041-210x.12760. [DOI] [Google Scholar]
- Sexton JP, Montiel J, Shay JE, Stephens MR, Slatyer RA.. 2017. Evolution of ecological niche breadth. Annual Review of Ecology, Evolution, and Systematics 48: 183–206. doi: 10.1146/annurev-ecolsys-110316-023003. [DOI] [Google Scholar]
- Sheth SN, Angert AL.. 2014. The evolution of environmental tolerance and range size: a comparison of geographically restricted and widespread Mimulus. Evolution 68: 2917–2931. doi: 10.1111/evo.12494. [DOI] [PubMed] [Google Scholar]
- Sheth SN, Morueta-Holme N, Angert AL.. 2020. Determinants of geographic range size in plants. New Phytologist 226: 650–665. [DOI] [PubMed] [Google Scholar]
- Silva HV, Alves-Silva E, Santos JC.. 2016. On the relationship between fluctuating asymmetry, sunlight exposure, leaf damage and flower set in Miconia fallax (Melastomataceae). Tropical Ecology 57: 419–427. [Google Scholar]
- Silva JLA, Souza AF, Vitória AP.. 2021. Historical and current environmental selection on functional traits of trees in the Atlantic Forest biodiversity hotspot. Journal of Vegetation Science 32: 1–12. [Google Scholar]
- Silveira FAO, Fernandes GW, Lemos-Filho JP.. 2013. Seed and seedling ecophysiology of Neotropical Melastomataceae: implications for conservation and restoration of savannas and rainforests. Annals of the Missouri Botanical Garden 99: 82–99. doi: 10.3417/2011054. [DOI] [Google Scholar]
- Slatyer RA, Hirst M, Sexton JP.. 2013. Niche breadth predicts geographical range size: a general ecological pattern. Ecology Letters 16: 1104–1114. doi: 10.1111/ele.12140. [DOI] [PubMed] [Google Scholar]
- Soberón J, Nakamura M.. 2009. Niches and distributional areas: concepts, methods, and assumptions. Proceedings of the National Academy of Sciences of the United States of America 106: 19644–19650. doi: 10.1073/pnas.0901637106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staude IR, Navarro LM, Pereira HM.. 2020. Range size predicts the risk of local extinction from habitat loss. Global Ecology and Biogeography 29: 16–25. [Google Scholar]
- Svenning JC, Fløjgaard C, Marske KA, Nógues-Bravo D, Normand S.. 2011. Applications of species distribution modeling to paleobiology. Quaternary Science Reviews 30: 2930–2947. doi: 10.1016/j.quascirev.2011.06.012. [DOI] [Google Scholar]
- The Brazil Flora Group. 2021. Brazilian Flora 2020: leveraging the power of a collaborative scientific network. Taxon 71: 178–198. [Google Scholar]
- Thom G, Gehara M, Smith BT, Miyaki CY, do Amaral FR.. 2021. Microevolutionary dynamics show tropical valleys are deeper for montane birds of the Atlantic Forest. Nature Communications 12: 6269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tordoni E, Casolo V, Bacaro G, Martini F, Rossi A, Boscutti F.. 2020. Climate and landscape heterogeneity drive spatial pattern of endemic plant diversity within local hotspots in South-Eastern Alps. Perspectives in Plant Ecology, Evolution and Systematics 43: 125512. doi: 10.1016/j.ppees.2020.125512. [DOI] [Google Scholar]
- Torres-Leite F, Cavatte PC, Garbin ML, et al. 2019. Surviving in the shadows: light responses of co-occurring Rubiaceae species within a tropical forest understory. Flora: Morphology, Distribution, Functional Ecology of Plants 261: 151487. [Google Scholar]
- Turchetto-Zolet AC, Salgueiro F, Turchetto C, et al. 2016. Phylogeography and ecological niche modelling in Eugenia uniflora (Myrtaceae) suggest distinct vegetational responses to climate change between the southern and the northern Atlantic Forest. Botanical Journal of the Linnean Society 182: 670–688. doi: 10.1111/boj.12473. [DOI] [Google Scholar]
- Vasconcelos TNC, Alcantara S, Andrino CO, et al. 2020. Fast diversification through a mosaic of evolutionary histories characterizes the endemic flora of ancient Neotropical mountains. Proceedings of the Royal Society B: Biological Sciences 287: 20192933. doi: 10.1098/rspb.2019.2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables WN, Ripley BD.. 2002. Modern applied statistics with S. New York: Springer. [Google Scholar]
- Vetö NM, Postolache D, Guzman Escudero FL, et al. 2022. Population structure and signals of local adaptation in Eugenia uniflora (Myrtaceae), a widely distributed species in the Atlantic Forest. Botanical Journal of the Linnean Society 201: 100–113. doi: 10.1093/botlinnean/boac012. [DOI] [Google Scholar]
- Wijesinghe DK, Hutchings MJ.. 1999. The effects of environmental heterogeneity on the performance of Glechoma hederacea: the interactions between patch contrast and patch scale. Journal of Ecology 87: 860–872. doi: 10.1046/j.1365-2745.1999.00395.x. [DOI] [Google Scholar]
- Wilson OJ, Mayle FE, Walters RJ, Lingner DV, Vibrans AC.. 2021. Floristic change in Brazil’s southern Atlantic Forest biodiversity hotspot: from the Last Glacial Maximum to the late 21st Century. Quaternary Science Reviews 264: 107005. doi: 10.1016/j.quascirev.2021.107005. [DOI] [Google Scholar]
- Wittmann F, Householder E, Piedade MTF, et al. 2013. Habitat specificity, endemism and the neotropical distribution of Amazonian white-water floodplain trees. Ecography 36: 690–707. [Google Scholar]
- Young KR, Ulloa CU, Luteyn JL, Knapp S.. 2002. Plant evolution and endemism in Andean South America: an introduction. Botanical Review 68: 4–21. doi: 10.1663/0006-8101(2002)068[0004:peaeia]2.0.co;2. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.