In a recent paper, Braun et al.1 compared the ability of five different methods to detect well-known protein-protein interactions (PPIs). The remarkable outcome of this study was that each of the tested methods detected a different subset of interactions with no single method detecting more than 36% of the tested gold-standard interactions (Fig. 1). Use of all five methods was required to find 62 of 92 interactions (67.4%), whereas individual methods detected only 28.8 out of 92 interactions on average (31.3%). Here we report a similar result when using multiple variants of the yeast two-hybrid (Y2H) system instead of multiple different methods.
Figure 1 |.
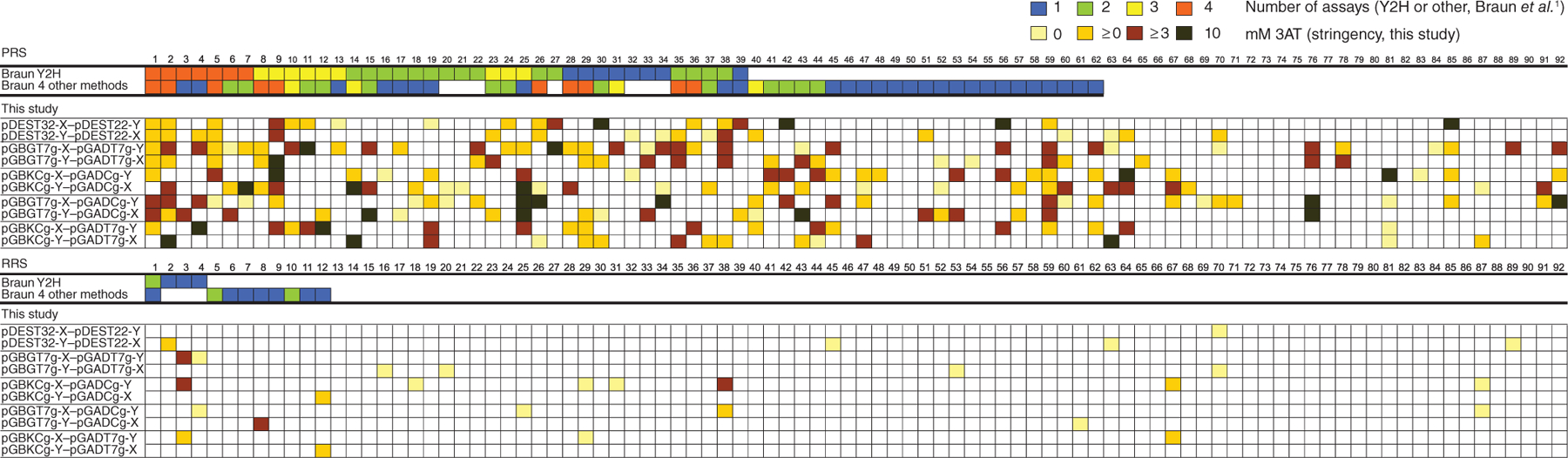
Comparison of protein interaction methods used by Braun et al.1 and exhaustive Y2H testing. PRS, positive reference set; RRS, random reference set. X and Y indicate bait and prey proteins, respectively. For each of 92 interactions, the box color indicates the number of assays (out of four Y2H assays (Braun Y2H) or four other assays (Braun 4 other methods)) that detect the interaction for the Braun et al.1 study and the stringency of the assay that detected the interaction for our study. White color indicates that the interaction was not detected.
We have shown that different two-hybrid systems detect markedly different subsets of interactions in the same interactome2,3. Therefore we determined whether the five different methods used by Braun et al.1 could be entirely replaced by variants of the Y2H system. This would have the benefit that only one method needs to be established, and the results of the variants are directly comparable. The set of protein pairs of Braun et al.1 provides a perfect benchmark for such a comparison.
We cloned the human positive reference set (PRS) and the random reference set (RRS) from Braun et al.1 (92 protein pairs each) into the following yeast two-hybrid bait-prey vectors: pGBGT7g-pGADCg, pGBGT7g-pGADT7g, pDEST32-pDEST22, pGBKCg-pGADT7g and pGBKCg-pGADCg2,3 (Supplementary Data). In addition to each vector pair, we tested each protein both as activation (prey) and DNA-binding domain fusion (bait), including C-terminal fusions in pGBKCg and pGADCg. This way, we tested each protein pair in ten different configurations (Fig. 1 and Supplementary Table 1).
According to our results (Fig. 1), and similar to the multiple methods tested by Braun et al.1, each Y2H variant detected a subset of interactions. The number of detected PPIs from the PRS ranged from 15 (16.3%) with pGBKC-pGADT7g to 37 (40.2%) with pGBG-pGADT7g, which is almost the same range as that found in Braun et al.1 (who found 19 PPIs with a modified version of the nucleic acid programmable protein array (wNAPPA) and 39 with Y2H). On average, our Y2H screens detected 23.3 PRS interactions, compared to 28.8 interactions detected by all methods of Braun et al.1. Overall, we detected 73 out of 92 (79.3%) PRS interactions. None of these 92 interactions were detected by all ten Y2H variants, and 14 were detected by only one (Fig. 1).
The utility of any method depends on its signal-to-noise ratio. In the best-case scenario Braun et al.1 had suppressed all false positives in their CEN plasmid–based Y2H assays, but they had still detected 23 true positive interactions. Similarly, under our most stringent conditions (using 10 mM 3-amino-1,2,4-triazole (3AT)), we detected 57 PRS interactions with no false positives. At our lowest stringency (0 mM 3AT) we detected 73 true positives but 20 false positives overall; this was again similar to the result in Braun et al.1, where the authors found 62 true positives and 12 false positives when they combined all assays.
Here we demonstrated that the Y2H system alone can yield interaction data of similar quality as five different methods as long as a variety of Y2H vectors is used. Because of the dramatic variability of PPI detection methods, future interactome projects must incorporate multiple protocols, either different methods as shown in Braun et al.1 or variations of the same method as we showed here. Our results show that a single method can be as informative and reliable as five different methods when it is used exhaustively and with proper controls and adequate stringency.
Supplementary Material
ACKNOWLEDGEMENTS
P. Braun (Dana-Farber Cancer Institute, Boston) kindly provided the gold-standard clones used in this project. S. Shtivelband helped in the initial phase of this project. S. Fields, T. Hazbun and M. Koegl made helpful comments on the manuscript. This project was supported by the Landesstiftung Baden-Württemberg (Germany), the US National Institutes of Health (RO1GM79710) and the European Union (HEALTH-F3-2009-223101).
Footnotes
Note: Supplementary information is available on the Nature Methods website.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Braun P et al. Nat. Methods 6, 91–97 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rajagopala SV, Hughes KT & Uetz P Proteomics 9, 5296–5302 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stellberger T et al. Proteome Sci 8, 8 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


