Abstract
Interstitial lung disease (ILD) is now diagnosed by an ILD-board consisting of radiologists, pulmonologists, and pathologists. They discuss the combination of computed tomography (CT) images, pulmonary function tests, demographic information, and histology and then agree on one of the 200 ILD diagnoses. Recent approaches employ computer-aided diagnostic tools to improve detection of disease, monitoring, and accurate prognostication. Methods based on artificial intelligence (AI) may be used in computational medicine, especially in image-based specialties such as radiology. This review summarises and highlights the strengths and weaknesses of the latest and most significant published methods that could lead to a holistic system for ILD diagnosis. We explore current AI methods and the data use to predict the prognosis and progression of ILDs. It is then essential to highlight the data that holds the most information related to risk factors for progression, e.g., CT scans and pulmonary function tests. This review aims to identify potential gaps, highlight areas that require further research, and identify the methods that could be combined to yield more promising results in future studies.
Key Words: artificial intelligence, medical data analysis, interstitial lung disease, computed tomography, computer-aided diagnosis, pulmonary fibrosis, idiopathic pulmonary fibrosis
INTRODUCTION TO ARTIFICIAL INTELLIGENCE IN RADIOLOGY
In the past two decades, artificial intelligence (AI) has gained tremendous momentum within radiology. Although the underlying technology dates back to the 1940s1–3 it took more than fifty years for this to be translated into medicine.
When AI is applied to the analysis of medical images, the primary approach is based on machine learning.4 Deep learning is a form of artificial neural networks, which itself is a subset of ML. Artificial neural networks are computing systems consisting of artificial neurons which learn parametric functions.1 They are capable of running parallel computations and are used to recognise patterns in the data provided. Deep learning methods are a form of hierarchical representational learning; they go beyond machine learning by stacking multiple layers of calculations to learn nonlinear higher dimensional patterns. Regarding images as inputs, convolutional neural networks (CNNs), a type of architectures for deep learning, are translation-invariant, which means once a pattern is learned, it can recognise it anywhere in an image, regardless of its location or orientation.5 In 2012 AlexNet,6 a CNN, competed in the ImageNet competition, and since then deep learning has become dominant in image classification.
Among the different specialties in medicine, radiology was among the “early adopters” in developing, testing, and implementing various algorithms in routing diagnostic procedures. Particularly in medical images, machine learning algorithms can simplify solutions to problems by considering new patterns within pixels and voxels although these are not immediately apparent to humans.7 An advantage of radiological departments is that medical images are generated on a large scale from various modalities. Furthermore, all images are stored digitally in a unified format. DICOMs are the standard format of biomedical images throughout the world. In addition to visual data, the DICOM header stores other parameters used for the algorithmic analysis of images. In parallel to filing DICOMs data, radiology reports are simultaneously stored in a digital archive linked to the images. As digital images and reports are readily available, diagnostic imaging is ideally suited for AI approaches.
Artificial Intelligence can be used for diagnosing various medical conditions through binary or multiclass classification. While this does not represent all possible pipelines, an example of the development process of a diagnosis AI model can be seen in Figure 1. The models are trained with medical images and demographic data to learn the relationship between the features (imaging features such as radiomics, demographic and clinical features) and the class labels, which represent the different diagnoses. The choice of feature representation will greatly impact the performance of the models, so it is important to carefully consider the most informative and robust representations that capture the underlying characteristics of the disease. To ensure the accuracy of the models, they must be validated against experienced human experts in the field.
FIGURE 1.
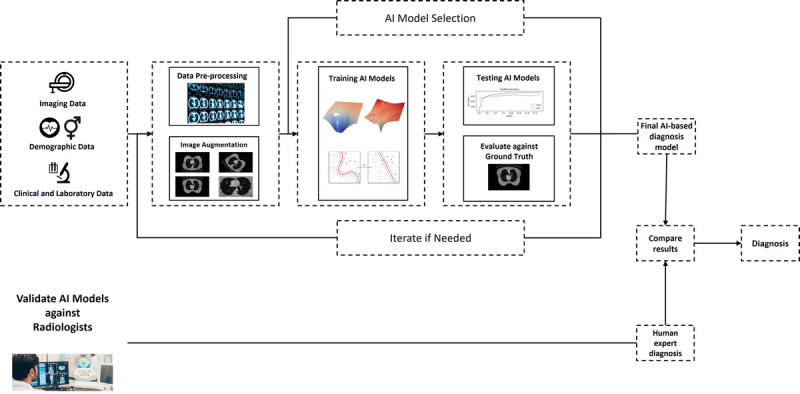
A typical workflow of a AI-based model for medical diagnosis. The first stage is to acquire data from different medical imaging modalities, patient demographic and clinical/laboratory data. This will require extensive processing to be ready for AI model development. The models developed needs to be evaluated against ground truths, and validated against human experts.
The segmentation of medical images has been used extensively in the diagnosis of a disease.8 The current state-of-the-art framework for medical image segmentation is the “nnUNet”9 framework that can achieve semantic segmentation of any biomedical image dataset, nnUNet confirmed its position as the state-of-the-art by recently winning the Abdominal Multi-Organ Segmentation (AMOS) competition in 2022, more than 4 years since its first release.10 In particular, semantic segmentation enables computers to recognise and localise objects in images and this is vital in tasks such as diagnosis and prognosis. In 2010, there were over 500 academic papers published on AI in medicine; in 2020, there were over 12000, and the exponential increase in research in this field is only expected to continue in the next ten years.11
RADIOLOGICAL REQUIREMENTS IN CHEST IMAGES
Even early development focused on chest imaging and the detection of lung nodules. The lungs are an excellent training field for automated image analysis exhibiting high tissue contrast and a binary clinical question (lung lesion, yes or no). Hence, an abundance of research started investigating AI algorithms for the purpose of automated lung lesion detection. The aim is to deploy algorithms that are capable to increase diagnostic accuracy while minimizing reading time. Especially in light of emerging screening programs, such diagnostic support algorithms are needed. Numerous research groups were able to show a significant improvement in lesion detection. Foremost deep learning models proved effective for lung lesion detection, not only on standard computed tomography (CT) scans but also in low dose exams for the purpose of lung cancer screening.12,13
In contrast to lung nodule detection conundrum, diffuse parenchymal disease poses a complex clinical problem. Lung nodule can be characterised basically in three classes, namely solid lesions, part-solid lesions and non-solid or ground-glass lesions. On the contrary, diffuse lung diseases are defined by an abundance of different patterns in various combinations and different locations. Also, diffuse lung diseases may have infectious, neoplastic, autoimmune, or idiopathic aetiologies. In addition, diffuse lung disease might affect the airways and alveoli and/or the lung interstitium. Interstitial lung diseases (ILDs are a heterogeneous group of diseases distinguished by fibrosis and inflammation of the lung parenchyma.14 In most of these diseases the pulmonary alveolar walls are infiltrated by a combination of inflammatory cells, fibrosis, and proliferation of normal cells that make up a healthy alveolar wall.15 The subset of ILDs with a fibrosing phenotype results in a decline in normal lung function, worsening symptoms and reduced quality of life.
Radiologists use a combination of CT images, clinical data, and pulmonary function tests to diagnose ILDs. AI-based methods have recently gained significant attention in the field of computational medicine. These advances can be seen in the creation of computer-aided tools for diagnosis, disease monitoring, and accurate prognostication.
This review paper aims to fill the gap in the literature by providing a comprehensive overview of the recent research using AI methods in diagnosis and prediction of the ILDs progression and prognosis, focusing on the subset of ILDs with a fibrosing phenotype due to the difficulty in their prognosis. We will summarise and evaluate the latest and most significant published methods, highlighting the strengths and weaknesses of these approaches. Specifically, we will examine the current AI methods and the data used to predict the prognosis and progression of ILDs, with a focus on data that holds the most information related to risk factors for progression, such as high-resolution CT scans and pulmonary function tests.
Through an in-depth analysis of the state of the art, we aim to identify potential gaps in research and highlight areas that require further investigation. Our review will also explore how larger datasets could help to improve procedures. By doing so, we hope to guide future researchers to create more promising outcomes in the field of AI-based methods for predicting the progression and prognosis of ILDs.
AI METHODS IN DIAGNOSIS AND PROGNOSIS OF ILDs
The central issues in the analysis of ILDs are 1) pattern detection, segmentation, and distribution; 2) diagnosis; 3) pattern quantification; 4) longitudinal evaluation and 5) disease prediction (Fig. 2). Disease phenotyping based on baseline imaging together with clinical parameters may be ultimately feasible. Yet, current models often rely solely on radiological imaging data. The most promising approach may then be to develop novel multivariate models that incorporate patient demographics, patient history, medication, lung function test, and laboratory results.
FIGURE 2.
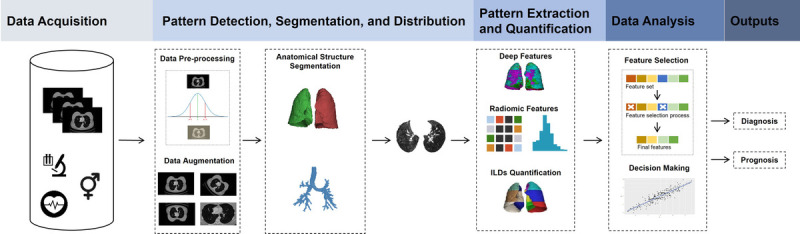
The analysis of ILDs can be separated into different stages. Firstly, segmentation of the various anatomical structures followed by detection, characterisation and quantification of pathological patterns using deep and radiomic features. The most informative features are identified and analysed with respect to clinical outcomes including diagnosis and disease progression.
Pattern Detection, Segmentation, and Distribution
It is crucial to identify the correct ILD diagnosis, as the prognosis and treatment of individual ILDs differ greatly. In general, IPF has a worse prognosis than non-IPF ILDs.14 Physicians often monitor the decline in forced vital capacity (FVC) and diffusing capacity of the lung for carbon monoxide (DLCO) as a measure of lung function. Although this is current practice, these measurements (FVC and DLCO) are unpredictable and unreliable.16 In contrast to this unreliable factor, studies have demonstrated the importance of imaging features within high-resolution CT (HRCT) scans.17 A new solution for monitoring the progression of IPF could be monitoring FVC and HRCT images at regular and fixed time intervals.
In a metanalysis, Ebner et al18 demonstrated how important is it to include demographic data in a prognostic model for pulmonary fibrosis. Patients with an unfavourable UIP/IPF diagnosis were on average 5 years older (UIP: 60 years), 21% more often male (UIP: 74%), and 19% more often smokers (UIP: 74%) than patients with non-IPF. With this simple piece of demographic information alone, the prediction of the outcome improves and this should be incorporated in a prediction model.
Radiomics involve extracting key features from medical images that radiologists cannot easily see. This is achieved by applying image segmentation and feature extraction techniques to the images, and allow the identification of key features that can provide additional insight into a patient's condition. Even with an approach that was exclusively based on radiomics approach, a recent study managed to identify two distinct clusters of systemic sclerosis patients with different outcomes.19 Current studies are attempting to extend this approach by including other clinical biomarkers and deploying automatisation by means of deep learning.
We will now discuss the data used in combination with the methodology in the selected literature, with the goal of developing frameworks that can detect patterns through feature representations and which can replicate the results of radiologists and others.
Diagnosis
The concept of computer-aided diagnosis (CAD) systems was first introduced in the 1950s. These systems can be defined as software capable of replicating the diagnostic abilities of human experts.19 Trusculescu et al21 published a comprehensive review of the application of deep learning in ILDs, with a specific focus on CAD systems. The review focuses on the use of CNNs for classifying ILDs and highlights the challenges in accurately identifying different patterns of lung tissue within HRCT images. A similar systematic review by Soffer et al8 was published more recently, and also explores the use of AI for ILD analysis and emphasizes the importance of correctly classifying fibrotic lung diseases. Both reviews stress the difficulties inherent in these tasks and believe that AI will play a crucial role in the future of ILD diagnosis. However, the accuracy of these systems is still not fully reliable. Refaee et al22 and Wenxi Yu et al23 discuss other more recent diagnostic tools utilising AI methods to classify ILDs. These recent tools also support the findings of the review papers that utilise images and deep learning to identify idiopathic pulmonary fibrosis.
We will focus on two retrospective studies that successfully developed CAD tools using AI to classify ILDs. In 2018, Walsh et al24 utilised deep learning for the classification of fibrotic lung diseases using HRCT scans. The study involved data augmentation on a database of 1,157 anonymised HRCT scans with fibrotic lung disease resulting in a final training set of 420,096 unique montages. Experienced thoracic radiologists labelled the data into three possible classes: UIP, “possible UIP”, or “inconsistent with UIP”. This supervised learning setup allowed the development of a neural network to predict one of the three classes. The algorithm was validated against 91 thoracic radiologists. The median accuracy of the radiologists was 70.7%, whilst the accuracy of the algorithm was 73.3%. The strength of this research is the sparseness of resources required to get results for individual patients once the algorithm has been trained. The main weakness in the paper is that information could be lost in the creation of the dataset; these limitations could be addressed in the future with better machines.
Furthermore, in 2019 Christe et al25 proposed a CAD tool for diagnosing pulmonary fibrosis, also using deep learning (Fig. 3). The study collected data from three providers and resulted in 307 different HRCT scans. Two experienced thoracic radiologists labelled the dataset into one of four possible categories: UIP, “possible UIP”, “indeterminate for UIP”, and “most consistent with non-IPF”. In another supervised setup, a pipeline was proposed that first segmented lung images, characterised lung tissue, and outputted a predicted class. The first two steps used CNNs to generate features for a random forest model. This study also validated results against two experienced thoracic radiologists. The CAD median F1 score was 0.80 compared to the human readers, who achieved a median of 0.79. The strength of this paper is that it was able to accurately produce results similarly to experienced radiologists; such tools could be a great asset in training new or less experienced radiologists. The annotation of the dataset was extremely costly and time consuming.
FIGURE 3.
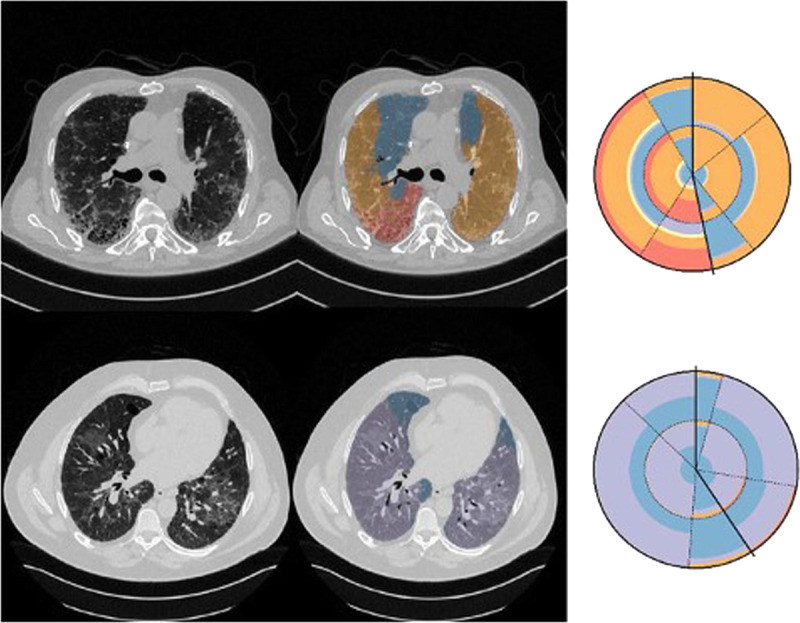
Quantification of pathological interstitial lung tissue (middle column) and visualisation using radial histograms (right column).25 Each sector denotes a region of the lung and is split into 2 parts, one for the central (inner) and one for peripheral (outer). Solid lines denote the division of left and right lungs. Top case is a typical UIP with reticulation (orange) and honeycombing (red) and bottom a non-IPF with ground glass opacity (purple).
As the basis for their ground truth labels, the latter two studies used the diagnostic criteria of the Fleischner Society for idiopathic pulmonary fibrosis.26 Three out of four studies in Table 1 have been validated against radiologists, this is vital in the development of reliable systems. As with radiologists, the studies also relied heavily on the use of imaging data. As there is no general gold standard in classifying ILDs, these tools have been created to assist radiologists with the challenges associated to ILD diagnosis. These algorithms provide significant contributions to the task of diagnosis and as technology progresses we expect results to improve in the near future. Naturally after the successful diagnosis, the next step is to explore the applicability of AI tools for prognosis and progression.
TABLE 1.
Summary of Diagnostic Approaches
| Study | Diagnosis | Metric | Results | Validated Against Human Expert |
|---|---|---|---|---|
| Refaee et al21 | Binary classification, IPF or non-IPF | Accuracy | 85.3 | Yes |
| Yu et al22 | Binary classification, IPF or non-IPF | AUC | 0.98 | No |
| Walsh et al23 | Multiclass classification, UIP, possible UIP, or inconsistent with UIP | Accuracy | 73.3 | Yes |
| Christe et al24 | Multiclass classification, UIP, possible UIP, indeterminate for UIP, and most consistent with non-IPF | F1 | 0.8 | Yes |
AUC indicates area under the receiver operating characteristic curve; IPF, Idopathic Pulmonary Fibrosis; UIP, Usual Interstitial Pneumonia.
In the past five years, there has been exponential growth in the number of FDA-approved devices for radiology,27 mainly due to the promising results AI yields. Siemens has integrated AI into their software to identify and quantify areas of interest within the lungs automatically.28 The software's major functions first offer to segment the lung tissue and then give results on lung lobe segmentation, lesion detection and measurements, and pneumonia analysis. In patients suffering with COVID-19 the software can analyse ground-glass opacities and consolidations highlighting abnormalities; this functionality should be transferrable to ILDs. This technology greatly assists radiologists in the task of diagnosis. While this is useful, it does not provide additional information on risk factors for progression in individual patients; this is still something left to radiologists; providing these extra details will reduce workloads and allow radiologists to work more efficiently. Combination of clinical data and pulmonary function tests are commonly used to identify progression in ILDs.14 In spite of the great success of CADs with ILDs, relatively little progress has been made in the prognosis and progression of ILDs.
Pattern Quantification and Follow-up
One essential element in the follow-up and prediction of fibrosing ILDs is the quantification of pathological pattern in radiological imaging. In general, a greater extent of fibrosis in chest CT correlates with overall mortality. Specific patterns, such as honeycombing, and bronchiectasis are linked to a worse prognosis.29,30 However, it has been shown that the interrater variability for the detected patterns is only moderate.31 Recent research by Humphries et al,17 showed that data-driven texture analysis can provide robust measures of disease severity. Comparison with lung function tests may indicate where the threshold lies for clinically relevant disease progression.
Longitudinal Evaluation and Disease Prognosis
Progression of clinical disease may be assessed by using AI tools that combine imaging data, clinical data, lung function tests, as well as laboratory values and those tools may perform as well as human experts. Existing AI methods for the prognosis and progression of the disease can mainly be categorised according to their output variables. With the target variable as categorising feature, AI tools can be grouped according to their desired outcome and discern the level of specificity with which previous studies have tackled the prognosis of ILDs.
Regression Tasks
One such target variable is the FVC, an extremely common variable that researchers attempt to predict. As the FVC is a continuous number, it falls into the category of regression. Two related studies,32,33 employed an end-to-end multi-modal based CNN to predict FVC decline. Experiments were run on the OSIC Pulmonary Fibrosis Progression Challenge Benchmark Dataset,34 the most popular dataset to train models for predicting patients’ severity of decline in lung function. Fibro-CoSANet extracts visual features from CT scans also by means of an attention layer, and combines such features with other clinical data.32 Then, the prediction of the FVC slope is performed by using regression. Similarly, Fibrosis-Net33 extracts image features and predicts FVC by fusing the CNN output features and demographic data. The authors also used the GSInquire method35 to make the model more explainable concerning its predictions.
Yadav et al36 designed FVC-Net, a deep learning-based architecture, to predict the progression of the disease from the patient’s CT scans and the patient’s metadata. The proposed method performs lung segmentation and returns a score for the degree of honeycombing. The model then combines the latter value with the clinical data to predict the FVC slope. The study also provides evidence that the proposed FVC-Net model can be applied and is valid in a different scenario (COVID-19 case study).
All three studies predicting FVC were performed on the same dataset, consisting of HRCT images and demographic features (including smoking status). They also utilised deep learning and measured the performance by calculating the Laplace log likelihood (LLL). FVC-Net claimed the lowest score of -6.641. A major strength of these studies is the contributions of different methods, all performed on the same dataset. However, this opens the possibility of generalising to a single dataset. FVC-net does not discuss the limitations of the research, but a weakness of the study is that FVC does not always consistently decline in patients.14
Proportional Hazards Models
There is another heterogeneous group of target variables that have been employed to characterise the evolution of ILDs and which are related to the prediction of survival/mortality. For instance, a self-supervised learning approach was developed to generate patch representations37 which are in turn employed to identify prognostic biomarkers. The training pipeline comprises three stages: 1) lung images are segmented and split into patches; 2) contrastive learning generates meaningful representations; 3) a clustering algorithm groups similar data to finally perform the survival analysis. The algorithm was tested on datasets from the Netherlands, Turkey, and the UK. One of the benefits of self-supervised learning approaches is the reduced need for annotated datasets, which can save valuable resources. Despite the limitations of the early stages of research in this area, the outcomes are still favourable and contribute to the overall advancement of the field.
Shahin et al38 propose a deep learning method that uses clinical and imaging data to predict the survival of IPF patients by means of the probability of survival. Results also show that the model trained on only imaging data outperformed the model trained using clinical and imaging data. The highlight of the survival model is the use of clinical and imaging data with an overall aim of predicant the time of death of a patient. Using 446 patients with IPF they state that clinical data, in particular FVC and DLCO, contains noisy data.
Earlier in 2022 Wu et al39 did a retrospective analysis of 232 patients diagnosed with IPF from 2011 to 2020. Based on IPF diagnosis guidelines the honeycomb lung extent was presented on CT images. A framework was established a to automatically segment area of honeycombing and calculate a fibrosis percentage based on this (Fig. 4). The results of this were validated by thoracic radiologists. Combining this with pulmonary function tests, as well as demographic, and clinical data, they created a proportional hazard model. The models developed were able to predict mortality based on this data.
FIGURE 4.
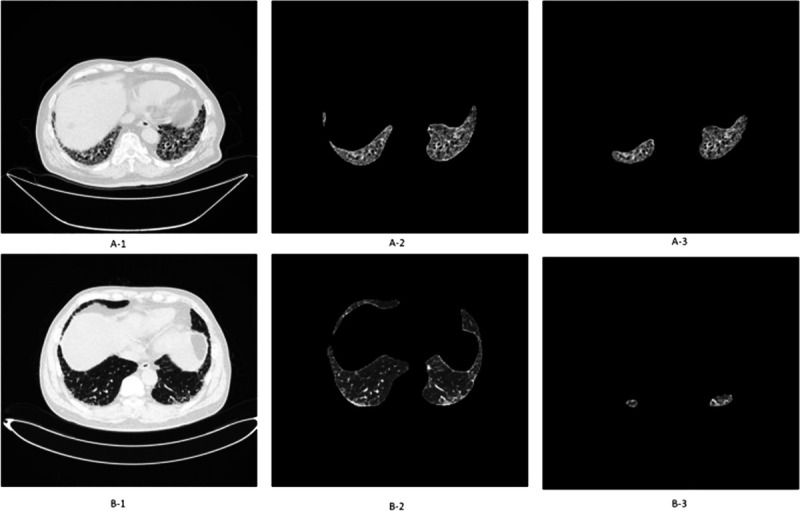
Quantifying pulmonary fibrosis I: comparing the original CT image (A-1 and B-1) with the automatically segmented honeycombing regions (A-2, A-3, B-2, B-3) to calculate the fibrosis percentage. A, Area of honeycombing is 5% to 25% of the entire lung (CT stage II). B, Area of honeycombing is <5% of the entire lung (CT stage I).39
Walsh et al.40 published the most recent study in predicting prognosis. This paper also combines deep learning methods and Cox proportional hazards to give a result. Disease progression was defined as a decline in more than 10% of FVC, death, or transplant. The data used includes: demographic, pulmonary functions, and HRCTs. Building upon the diagnosis models previously developed23 and using the same data preparation, the deep learning algorithm successfully predicted the probabilities of UIP for four individual montages of an HRCT scan. Statistical analysis was then performed by extracting these probabilities and combining them with clinical and pulmonary function data. The study aimed to predict transplant-free survival, and each patient's survival period was calculated. This paper's significant strength is building upon previous diagnostic methods. Accurate segmentation of fibrotic tissue is still a challenging task, and Walsh presented a new approach, with probabilistic outputs; In Figures 4 and 5, we note the different approaches of pattern quantification.39,40 In theory, AI research could identify patients at high risk of developing IPF, reducing the chance of developing fibrotic disease or lead to earlier specific treatments, thus improving the patient’s quality of life.
FIGURE 5.
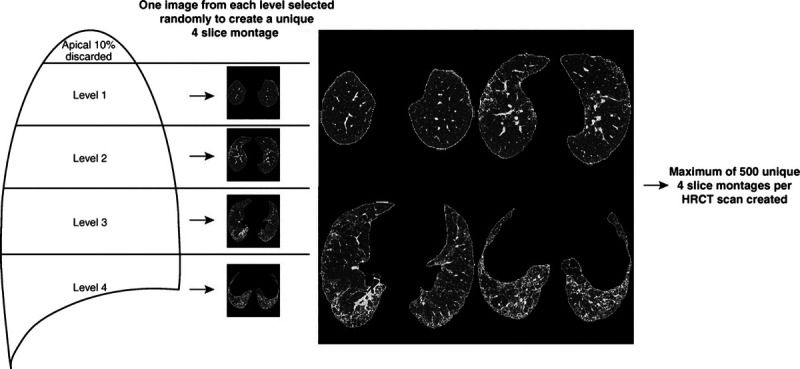
Quantifying pulmonary fibrosis II: Conversion of segmentation maps into 4 montages, then calculation of probability of UIP for each montage.40
Generative Models
A generative model is a type of ML model that learns the underlying probability distribution of a set of input data to generate new data points similar to the training data.41 Unlike supervised learning methods, which focus on predicting a specific output based on the input data, generative models can be used for tasks such as image synthesis, data augmentation, and anomaly detection.
Researchers developed an Airway Transfer Network (ATN) to synthesize realistic airways and use CT airway metrics to predict mortality.42 Although the eventual outcome was to predict mortality, the main emphasis in this paper is the use of generative models to synthesize airways. ATN performs airway synthesis by a transformation network that refines synthetic data using perceptual losses. Experiments in their research show that the proposed approach performs comparably well to other popular generative approaches, as based on CT images from 113 patients of the University Hospital Leuven, Belgium.
Quan et al43 also focus on the prediction of airway dilation, but in this case, also over time. They presented a probabilistic model to identify the regions of progressive airway dilation, given two profiles acquired at different time points. They then computed a relative change in airway volume that may be useful for quantifying IPF disease progression. They tested their approach on simulated dilation images of healthy airways and pairs of actual images of IPF-affected lungs acquired one year apart.
The generative models presented are developed without requiring large datasets or precise airway annotations. Using synthetic data, the papers were able to mimic real IPF scenarios. Quan et al43 also validated their results with labels provided by radiologists on several examples. In Pakzad A et al,42 the synthesized airways were later used for Cox regression analysis. This data alone was insufficient, so they combined this pulmonary function data (FVC and DLCO). Other models we have seen analyse the entire lung tissue; an option for future results would be to combine airway dilation with lung tissue analysis of the entire HRCT scan.
Unsupervised Learning
Unsupervised learning is a type of machine learning where the algorithm learns patterns and relationships from unlabeled data without any provided human expert task or goal.41 Pan et al44 developed another unsupervised learning model to identify predictive image markers in CT images collected in Italy and Austria. They studied 190 CT images taken from 76 patients with confirmed IPF; the dataset also included demographic and lung function tests. Principal component analysis was performed on segmentation maps to reduce image dimension, followed by clustering to group pattern signatures. To predict temporal sequences, a random forest classifier was trained on pairs of scans (longitudinal data) to predict temporal sequences. With patient follow-up scans, they formed radiological disease progression signatures; patients were clustered into 2 groups based on these, and patient survival was assessed by Kaplan-Meier analysis. The contribution of this research is beneficial in observing specific progression signatures that help predict progression. The limitations of supervised learning are that often groups of data form patterns that cannot be explained easily. In this study, the survival results should be discussed in more detail. If patients in a particular group are more likely to progress worse, it would be beneficial to prioritize them for the correct treatment plans.
DISCUSSION
The current literature suggests using AI to classify suspected ILD cases according to the latest standards instead of identifying individual diseases, primarily distinguishing between UIP, “possible UIP”, or “inconsistent with UIP”. Other approaches include classifying between fibrosing and non-fibrosing ILDs. As stated in previous reviews, AI is currently not at a high enough accuracy to rely entirely on but show great potential in assisting and training capabilities in the future of radiology.8,21 Current methods presented in diagnosis have produced comparable results to radiologists.24,25 These methods should be feasible to integrate into radiologist workstreams to help for a quicker diagnosis. More importantly, the models trained in the papers were validated against experienced specialized radiologists to ensure the safety of the software. This should be an essential step for any study to be deployed to real-life scenarios. Such steps will need to be taken to enable prognosis software to be developed.
Most of the studies that infer prognosis utilise some form of longitudinal data, i.e., data taken at different time points. This is essential in order to monitor disease progression. As a consequence, we note the heavy use of proportional hazard models to predict mortality. These attempts often had two-time points, a baseline, and a follow-up dataset. Whilst two-time points show promising results for prognosis, as in Pan et al,44 the study could benefit from looking at more time points. A visual representation of these types of studies can be seen in Figure 6. Walsh et al40 were particularly effective at applying their algorithm to HRCT scans from different institutions; this shows that the model can generalise to different populations. The last point on proportional hazard modelling is that it is prone to overfitting and does not perform well if there is missing data. In future work, it would be useful to see different AI approaches to predict progression visually over time. Ideally, in the prognostic research presented it should be feasible to combine with diagnostic models results. This was successfully shown in Walsh et al.40 If we are to develop holistic systems for diagnosis and optimal treatment, the first step must be to provide accurate diagnosis and then to provide prognosis.
FIGURE 6.
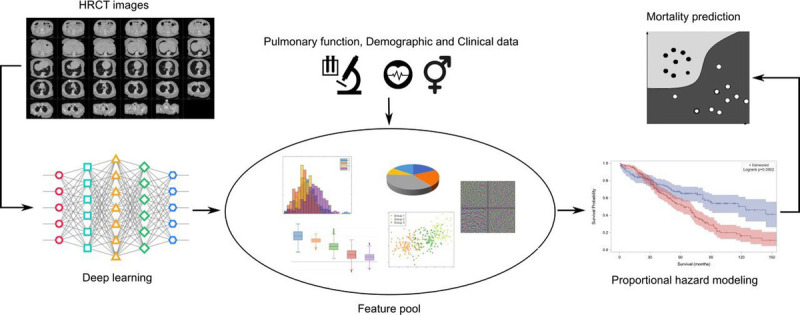
A summary of using proportional hazard modeling in prognosis of ILDs.
It is worth mentioning the self-supervised, generative models and unsupervised methods explored in the paper negate the need for radiologists to annotate large datasets, these approaches are invaluable. In general, papers which involved HRCT images, performed well. In a different study not previously mentioned, Ali S et al45 explored ensemble methods based solely on structured data; their limitations involved data processing and missing values. If the methods in the paper considered extracting corresponding image features it will have more data to learn from and replicate more closely the features a human expert looks at, this could drastically improve the results.46
Given the promising results and the even more exciting outlook, it is clear that AI will play a vital role in the future of chest radiology. To date, the landscape is dominated by the systems for detecting lung lesion detection systems, which are widely available.47–50 With the COVID-19 pandemic and given the central role of chest imaging in diagnosis and prognosis, the development of algorithms tackling diffuse parenchymal disease has exploded. Innumerable research groups have delivered remarkable results for the detection, characterisation, and prognosis of COVID-19 pneumonia.51,52 The pandemic has also spurred data availability and exchange, thus facilitating research in the field. In contrast, the investigation of ILDs by means of AI is still a minor area.
At the moment, the main directions of research in imaging of interstitial lung disease comprise pattern detection, quantification, diagnosis and prognosis. However, several limitations currently restrict the development of algorithms for these purposes. Firstly, ILDs in general are rare diseases that are most commonly referred to tertiary care centres. Secondly, there is a plethora of more than 200 different entities that might lead to pulmonary involvement characterised by an interstitial lung disease pattern. In contrast to the multitude of various ILDs in general, the CT patterns are relatively few. Hence, disease distribution and clinical history play an ever more important role. Overall, AI and ILD research suffers from a shortage of structured data. Data governance issues and restricted access to clinical information along with the scarcity of imaging exams hamper development. This was previously highlighted in 2020 by Trusculescu et al,21 but since then there have been group efforts to solve this problem.
A vital aspect in all research explored was the collaboration between engineers and healthcare providers. For successful application, data driven AI methods require large amounts of data, particularly in supervised learning, which requires the data to be annotated. Providing annotated datasets is costly and time consuming. FVC has been a particularly popular parameter for predictions, due to the availability of the well-prepared open-source dataset.34 As previously mentioned, there are suggestions that a stable FVC does not infer that the patient’s health is not declining. Prognosis inference is still challenging for physicians and AI systems alike. Utilising longitudinal data in the form of HRCT scans and pulmonary function tests is promising but requires further research. Radiomic features have been very successful in diagnostic tools22 as well as proving successful in the prognosis of systemic sclerosis ILD.19 Yet, there has been very little research in the prognosis of IPF using radiomics. Several papers performed segmentation of the HRCT scans but did not extract radiomic features based on these results.
To overcome these issues, several open access databases have been created to enable researchers to access data.34,53–57 With increasing access to CT images and clinical data, research in ILD and deployment of AI algorithms might gain momentum as in the successful introduction of AI-assisted detection systems in lung lesion detection and lung cancer screening. We hope in the future that the accessibility of the required data will no longer slow research on investigating AI in chest radiology.
In conclusion, the aim of this review was to provide an overview of current developments in the field of interstitial lung diseases and AI. Potential gaps in knowledge, areas that require further research and promising newly developed methods have been reviewed. In essence, as with all AI-driven research, the key to improving ILD classification and prognostication will be data availability and the pursuit of a multivariate approach by combining imaging and clinical information.
Footnotes
Ethan Dack, Andreas Christe, Stavroula Mougiakakou, and Lucas Ebner contributed equally to this study (shared first and shared last authorship).
Conflicts of interest and sources of funding: none declared.
Contributor Information
Ethan Dack, Email: ethan.dack@unibe.ch.
Matthias Fontanellaz, Email: matthias.fontanellaz@unibe.ch.
Lorenzo Brigato, Email: lorenzo.brigato@unibe.ch.
Johannes T. Heverhagen, Email: Johannes.Heverhagen@insel.ch.
Alan A. Peters, Email: Alan.Peters@insel.ch.
Adrian T. Huber, Email: adrian.huber@insel.ch.
Hanno Hoppe, Email: hanno.hoppe@lindenhofgruppe.ch.
Stavroula Mougiakakou, Email: stavroula.mougiakakou@unibe.ch.
Lukas Ebner, Email: lukas.ebner@insel.ch.
REFERENCES
- 1.Mcculloch W, Pitts W. A logical calculus of ideas immanent in nervous activity. Bull Math Biophys. 1943;5:127–147. [PubMed] [Google Scholar]
- 2.Turning AM. Computing machinery and intelligence. Mind. 1950;LIX:433–460. [Google Scholar]
- 3.McCarthy J Minsky ML Rochester N, et al. A proposal for the Dartmouth summer research project on artificial intelligence. August 19, 1955. https://web.archive.org/web/20080930164306/http://www-formal.stanford.edu/jmc/history/dartmouth/dartmouth.html. Accessed December 11, 2022.
- 4.Samuel AL. Some studies in machine learning using the game of checkers. IBM J Res Dev. 1959;3:210–229. [Google Scholar]
- 5.Chollet F. Deep Learning with Python. New York, NY: Manning Publications. 2017. [Google Scholar]
- 6.Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. In: Proceedings of the 25th International Conference on Neural Information Processing Systems—Volume 1 (NIPS'12) vol. 1. Red Hook, NY: Curran Associates Inc; 2012:1097–1105. [Google Scholar]
- 7.Géron A. Hands-on Machine Learning with Scikit-Learn, Keras and TensorFlow: Concepts, Tools, and Techniques to Build Intelligent Systems. 2nd ed. Gravenstein Highway North, Sebastopol, CA: O'Reilly; 2019. [Google Scholar]
- 8.Soffer S Morgenthau AS Shimon O, et al. Artificial intelligence for interstitial lung disease analysis on chest computed tomography: a systematic review. Acad Radiol. 2022;29:S226–S235. [DOI] [PubMed] [Google Scholar]
- 9.Isensee F Jaeger PF Kohl SAA, et al. nnU-net: a self-configuring method for deep learning–based biomedical image segmentation. Nat Methods. 2021;18:203–211. [DOI] [PubMed] [Google Scholar]
- 10.AMOS . About AMOS. 2022. http://www.amos.sribd.cn/about.html. Accessed December 11, 2022.
- 11.Meskó B, Görög M. A short guide for medical professionals in the era of artificial intelligence. NPJ Digit Med. 2020;3:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peters AA Huber AT Obmann VC, et al. Diagnostic validation of a deep learning nodule detection algorithm in low-dose chest CT: determination of optimized dose thresholds in a virtual screening scenario. Eur Radiol. 2022;32:4324–4332. [DOI] [PubMed] [Google Scholar]
- 13.Peters AA Decasper A Munz J, et al. Performance of an AI based CAD system in solid lung nodule detection on chest phantom radiographs compared to radiology residents and fellow radiologists. J Thorac Dis. 2021;13:2728–2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong AW, Ryerson CJ, Guler SA. Progression of fibrosing interstitial lung disease. Respir Res. 2020;21:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wuyts WA Wijsenbeek M Bondue B, et al. Idiopathic pulmonary fibrosis: best practice in monitoring and managing a relentless fibrotic disease. Respiration. 2020;99:73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guler SA Winstone TA Murphy D, et al. Does systemic sclerosis-associated interstitial lung disease burn out? Specific phenotypes of disease progression. Ann Am Thorac Soc. 2018;15:1427–1433. [DOI] [PubMed] [Google Scholar]
- 17.Humphries SM Swigris JJ Brown KK, et al. Quantitative high-resolution computed tomography fibrosis score: performance characteristics in idiopathic pulmonary fibrosis. Eur Respir J. 2018;52:1801384. [DOI] [PubMed] [Google Scholar]
- 18.Ebner L Christodoulidis S Stathopoulou T, et al. Meta-analysis of the radiological and clinical features of usual interstitial pneumonia (UIP) and nonspecific interstitial pneumonia (NSIP). PloS One. 2020;15:e0226084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schniering J Maciukiewicz M Gabrys HS, et al. Computed tomography-based radiomics decodes prognostic and molecular differences in interstitial lung disease related to systemic sclerosis. Eur Respir J. 2022;59:2004503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yanase J, Triantaphyllou E. A systematic survey of computer-aided diagnosis in medicine: past and present developments. Expert Syst Appl. 2019;138:0957–4174. [Google Scholar]
- 21.Trusculescu AA Manolescu D Tudorache E, et al. Deep learning in interstitial lung disease—how long until daily practice. Eur Radiol. 2020;30:6285–6292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Refaee T Salahuddin Z Frix AN, et al. Diagnosis of idiopathic pulmonary fibrosis in high-resolution computed tomography scans using a combination of handcrafted radiomics and deep learning. Front Med (Lausanne). 2022;9:915243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu W Zhou H Choi Y, et al. Multi-scale, domain knowledge-guided attention + random forest: a two-stage deep learning-based multi-scale guided attention models to diagnose idiopathic pulmonary fibrosis from computed tomography images. Med Phys. 2022;50:894–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walsh SLF Calandriello L Silva M, et al. Deep learning for classifying fibrotic lung disease on high-resolution computed tomography: a case-cohort study. Lancet Respir Med. 2018;6:837–845. [DOI] [PubMed] [Google Scholar]
- 25.Christe A Peters AA Drakopoulos D, et al. Computer-aided diagnosis of pulmonary fibrosis using deep learning and CT images. Invest Radiol. 2019;54:627–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lynch DA Sverzellati N Travis WD, et al. Diagnostic criteria for idiopathic pulmonary fibrosis: a Fleischner society white paper. Lancet Respir Med. 2018;6:138–153. [DOI] [PubMed] [Google Scholar]
- 27.FDA . Artificial intelligence and machine learning (AI/ML)–enabled medical devices. October 5, 2022. https://www.fda.gov/medical-devices/software-medical-device-samd/artificial-intelligence-and-machine-learning-aiml-enabled-medical-devices?utm_medium=email&utm_source=govdelivery. Accessed December 11, 2022.
- 28.Siemens Healthcare . AI-Rad Companion. 2022. https://www.siemens-healthineers.com/digital-health-solutions/digital-solutions-overview/clinical-decision-support/ai-rad-companion#:~:text=The%20AI%2DRad%20Companion%20Chest,tumor%20burden%20are%20automatically%20calculated.&text = Cinematic%20Rendering%20. Accessed December 11, 2022.
- 29.Kolb M, Vašáková M. The natural history of progressive fibrosing interstitial lung diseases. Respir Res. 2019;20:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alsumrain M De Giacomi F Nasim F, et al. Combined pulmonary fibrosis and emphysema as a clinicoradiologic entity: characterization of presenting lung fibrosis and implications for survival. Respir Med. 2019;146:106–112. [DOI] [PubMed] [Google Scholar]
- 31.Walsh SL Calandriello L Sverzellati N, et al. Interobserver agreement for the ATS/ERS/JRS/ALAT criteria for a UIP pattern on CT. Thorax. 2016;71:45–51. [DOI] [PubMed] [Google Scholar]
- 32.Al Nazi Z Rabbi Mashrur F Islam MA, et al. Fibro-CoSANet: pulmonary fibrosis prognosis prediction using a convolutional self attention network. Phys Med Biol. 2021;66:225013. [DOI] [PubMed] [Google Scholar]
- 33.Wong A Lu J Dorfman A, et al. Fibrosis-Net: a tailored deep convolutional neural network design for prediction of pulmonary fibrosis progression from chest CT images. Front Artif Intell. 2021;4:764047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.OSIC: Open Source Imaging Consortium . OSIC pulmonary fibrosis progression. May 22, 2019. https://www.osicild.org/kaggle01.html. Accessed December 11, 2022.
- 35.Lin ZQ Shafiee MJ Bochkarev S, et al. Do explanations reflect decisions? A machine-centric strategy to quantify the performance of explainability algorithms. October 29, 2019. 10.48550/arXiv.1910.07387. Accessed December 12, 2022. [DOI]
- 36.Yadav A Saxena R Kumar A, et al. FVC-NET: an automated diagnosis of pulmonary fibrosis progression prediction using honeycombing and deep learning. Comput Intell Neurosci. 2022;2022:2832400. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Zhao A, et al. Prognostic imaging biomarker discovery in survival analysis for idiopathic pulmonary fibrosis. In: Wang L Dou Q Fletcher PT, et al., eds. Medical Image Computing and Computer Assisted Intervention—MICCAI 2022. MICCAI 2022. Lecture Notes in Computer Science, Vol 13437. Cham, Switzerland: Springer; 2022. 10.1007/978-3-031-16449-1_22. [DOI] [Google Scholar]
- 38.Shahin AH Jacob J Alexander DC, et al. Survival analysis for idiopathic pulmonary fibrosis using CT images and incomplete clinical data. March 21, 2022. 10.48550/arXiv.2203.11391. Accessed December 11, 2022. [DOI]
- 39.Wu X Yin C Chen X, et al. Idiopathic pulmonary fibrosis mortality risk prediction based on artificial intelligence: the CTPF model. Front Pharmacol. 2022;13:878764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walsh SLF Mackintosh JA Calandriello L, et al. Deep learning-based outcome prediction in progressive fibrotic lung disease using high-resolution computed tomography. Am J Respir Crit Care Med. 2022;206:883–891. [DOI] [PubMed] [Google Scholar]
- 41.Bishop CM. Pattern Recognition and Machine Learning (Information Science and Statistics). Berlin, Germany: Springer-Verlag; 2006. [Google Scholar]
- 42.Pakzad A. Airway measurement by refinement of synthetic images improves mortality prediction in idiopathic pulmonary fibrosis. In: Mukhopadhyay A Oksuz I Engelhardt S, et al., eds. Deep Generative Models. DGM4MICCAI 2022. Lecture Notes in Computer Science, Vol 13609. Cham, Switzerland: Springer; 2022. 10.1007/978-3-031-18576-2_11. [DOI] [Google Scholar]
- 43.Quan K. Modelling airway geometry as stock market data using Bayesian changepoint detection. In: Suk HI Liu M Yan P, et al., eds. Machine Learning in Medical Imaging. MLMI 2019. Lecture Notes in Computer Science, Vol 11861. Cham, Switzerland: Springer; 2019. 10.1007/978-3-030-32692-0_40. [DOI] [Google Scholar]
- 44.Pan J Hofmanninger J Nenning KH, et al. Unsupervised machine learning identifies predictive progression markers of IPF. Eur Radiol. 2022;33:925–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ali S Hussain A Aich S, et al. A soft voting ensemble-based model for the early prediction of idiopathic pulmonary fibrosis (IPF) disease severity in lungs disease patients. Life (Basel). 2021;11:1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang X Zhang Y Zhang G, et al. Deep learning with radiomics for disease diagnosis and treatment: challenges and potential. Front Oncol. 2022;12:773840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ding Y Zhang J Zhuang W, et al. Improving the efficiency of identifying malignant pulmonary nodules before surgery via a combination of artificial intelligence CT image recognition and serum autoantibodies [published online ahead of print December 8, 2022]. Eur Radiol. doi: 10.1007/s00330-022-09317-x. PMID: 36480027. [DOI] [PubMed] [Google Scholar]
- 48.Ai Y Aonpong P Wang W, et al. Residual multilayer perceptrons for genotype-guided recurrence prediction of non-small cell lung cancer. Annu Int Conf IEEE Eng Med Biol Soc. 2022;2022:447–450. [DOI] [PubMed] [Google Scholar]
- 49.Lee JH Lee D Lu MT, et al. Deep learning to optimize candidate selection for lung cancer CT screening: advancing the 2021 USPSTF recommendations. Radiology. 2022;305:209–218. [DOI] [PubMed] [Google Scholar]
- 50.Jiang B Li N Shi X, et al. Deep learning reconstruction shows better lung nodule detection for ultra–low-dose chest CT. Radiology. 2022;303:202–212. [DOI] [PubMed] [Google Scholar]
- 51.Di Napoli A Tagliente E Pasquini L, et al. 3D CT-inclusive deep-learning model to predict mortality, ICU admittance, and intubation in COVID-19 patients. J Digit Imaging. 2022;1–14. doi: 10.1007/s10278-022-00734-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fontanellaz M Ebner L Huber A, et al. A deep-learning diagnostic support system for the detection of COVID-19 using chest radiographs: a multireader validation study. Invest Radiol. 2021;56:348–356. [DOI] [PubMed] [Google Scholar]
- 53.Cohen JP Morrison P Dao L, et al. COVID-19 image data collection. March 25, 2020. https://github.com/ieee8023/covid-chestxray-dataset. Accessed December 11, 2022.
- 54.Wang L Lin ZQ Wong A, et al. , DarwinAI Corp, Canada and Vision and Image Processing Research Group, University of Waterloo, Canada . Figure 1. COVID-19 Chest X-ray data initiative. May 9, 2020. https://github.com/agchung/Figure1-COVID-chestxray-dataset. Accessed December 11, 2022.
- 55.Wang L Lin ZQ Wong A, et al. , DarwinAI Corp, Canada and Vision and Image Processing Research Group, University of Waterloo, Canada . Actualmed COVID-19 chest x-ray data initiative. May 5, 2020. https://github.com/agchung/Actualmed-COVID-chestxray-dataset. Accessed December 11, 2022
- 56.COVID-19 Radiography Database. March 11, 2022. https://www.kaggle.com/tawsifurrahman/covid19-radiography-database. Accessed December 11, 2022
- 57.Radiological Society of North America . RSNA pneumonia detection challenge. December 1, 2018. https://www.kaggle.com/c/rsna-pneumonia-detection-challenge/data. December 11, 2022.


