Abstract
Introduction: The neonate exposed to opioids in utero faces a constellation of withdrawal symptoms postpartum commonly called neonatal opioid withdrawal syndrome (NOWS). The incidence of NOWS has increased in recent years due to the opioid epidemic. MicroRNAs (miRNAs) are small non-coding RNA molecules that play a crucial role in gene regulation. Epigenetic variations in microRNAs (miRNAs) and their impact on addiction-related processes is a rapidly evolving area of research.
Methods: The Illumina Infinium Methylation EPIC BeadChip was used to analyze DNA methylation levels of miRNA-encoding genes in 96 human placental tissues to identify miRNA gene methylation profiles as-sociated with NOWS: 32 from mothers whose prenatally opioid-exposed infants required pharmacologic management for NOWS, 32 from mothers whose prenatally opioid-exposed infants did not require treat-ment for NOWS, and 32 unexposed controls.
Results: The study identified 46 significantly differentially methylated (FDR p-value ≤ 0.05) CpGs associated with 47 unique miRNAs, with a receiver operating characteristic (ROC) area under the curve (AUC) ≥0.75 including 28 hypomethylated and 18 hypermethylated CpGs as potentially associated with NOWS. These dysregulated microRNA methylation patterns may be a contributing factor to NOWS pathogenesis.
Conclusion: This is the first study to analyze miRNA methylation profiles in NOWS infants and illustrates the unique role miRNAs might have in diagnosing and treating the disease. Furthermore, these data may provide a step toward feasible precision medicine for NOWS babies as well.
Keywords: MicroRNAs, opioid use disorder (OUD), neonatal opioid withdrawal syndrome (NOWS), methylation, biomarkers
Introduction
Opioid use disorder (OUD) is a global health crisis that has led to a sharp increase in drug overdose deaths. Regular use of opioids during pregnancy can lead to Neonatal Opioid Withdrawal Syndrome (NOWS). This illness is associated with increased morbidity and mortality in infancy and is a significant risk factor with negative impacts on the neurodevelopment of infants. Every 15 min a baby is born to a mother with an OUD (Honein et al., 2019), and 8.7 million children in the US have a parent with OUD (Lipari and Van Horn, 2013). Many in-utero opioid-exposed neonates are born prematurely below 32 weeks of gestational age (Cleary et al., 2011) with a wide range of neurobiological symptoms. Acutely, neonates experiencing NOWS have autonomic nervous system dysfunction, insomnia, feeding difficulty, inconsolable crying, irritability, and seizures; they may require opioid medication and/or extended hospitalization. Longer-term sequalae of NOWS can include learning and cognitive disabilities (Baldacchino et al., 2015). The most common pharmacotherapies of choice for pregnant women with OUD include methadone or buprenorphine (Meyer et al., 2015). Previous genome-wide DNA methylation and transcriptome studies reported differentially methylated and expressed genes and multiple dysregulated biological pathways closely associated with mothers of infants with NOWS (Radhakrishna et al., 2021a; Radhakrishna et al., 2021b). Furthermore, gene-specific methylation variation profiles in mothers with OUD were reported in comparison to normal controls (Chorbov et al., 2011; Wachman et al., 2013; Wachman et al., 2018) and genetic variants were associated with NOWS risk and severity (Oei et al., 2012; Wachman et al., 2013; Wachman et al., 2015; Mactier et al., 2017; Wachman et al., 2017; Metpally et al., 2019).
Non-coding RNAs (ncRNAs) are emerging as key regulators of cellular processes such as genome integrity, cell growth, proliferation, differentiation, development, chromatin organization, gene expression, and signal transduction (Puvvula, 2019; Li et al., 2020). There are many classes of ncRNAs, with a few functionally important types including microRNA (miRNA), long non-coding RNA (lncRNA), circular RNA (circRNA), piwi-interfering RNA (piRNA), and small nucleolar RNA (snoRNAs). Dysregulation of these ncRNAs is involved in many cancers and also drug abuse (Zhang K. et al., 2016). MicroRNAs (miRNAs) are indeed one of the most prominent and extensively studied classes of non-coding RNAs (ncRNAs). They are small, single-stranded non-coding RNAs, and are highly conserved post-transcriptional negative regulators of gene expression by binding to the 3’ untranslated region of target mRNA (Bartel, 2004; Zhang et al., 2017). MicroRNAs have gained significant attention in the field of molecular biology and genetics due to their involvement in various biological processes, including gene regulation and disease pathogenesis (Contreras and Rao, 2012; Banerjee and Sen, 2015; Iacomino and Siani, 2017; Sliwinska et al., 2017; Zhou et al., 2018). Many diseases and conditions are influenced by variations in miRNA gene expression. Variations in miRNAs expression can result from deletions and duplications of larger sequences or chromosomes, mutations involving miRNA loci, or epigenetic methylation, among other factors (Ali Syeda et al., 2020).
Epigenetic mechanisms such as DNA methylation play an important role in regulating miRNA expression. Most of the miRNA genes were found in CpG-rich regions (Bianchi et al., 2017) and therefore DNA methylation may play an important role in altered miRNA expression. miRNAs can directly target epigenetic factors, such as DNA methyltransferases or histone deacetylases, thus regulating chromatin structure. These miRNAs are useful not only as diagnostic biomarkers for drug exposure but may also be neuroprotective in the context of drug use (Saugstad, 2010).
Since the 1993 discovery of miRNAs in Caenorhabditis elegans (Lee et al., 1993), there are now approximately 2500 defined human miRNAs, though many are without experimental validation (Chiang et al., 2010; Kozomara and Griffiths-Jones, 2014). The miRNAs do not require precise complementarity for target recognition (Christopher et al., 2016), a single miRNA can regulate the expression of multiple genes as its targets, while one gene may be targeted by many miRNAs (Adlakha and Saini, 2014). miRNA-based regulation is implicated in many disease etiologies and has been studied for treatment. Hence, miRNAs serve as an ideal unifying molecular marker to better understand the pathophysiological processes that may regulate gene expression. However, miRNAs association in placental tissues of mothers of in-utero opioid-exposed infants born with NOWS has not yet been fully addressed.
The placenta is a temporary fetal organ, which allows the exchange of nutrients and gases between the mother and the developing fetus. The placenta governs the development of fetal organs including the brain (Burton and Fowden, 2015). During pregnancy, miRNAs of placental origin are released continually in the maternal circulatory system, indicating that these miRNAs might serve as biomarkers for placental function during pregnancy and in cellular communication (Mouillet et al., 2015). Prior reports indicate that most addictive drugs easily cross the placenta and can affect fetal brain development (Ross et al., 2015).
As such, placenta-derived miRNAs may be assessed as surrogate markers for brain health at birth, following in-utero opioid exposure, and may predict the severity of NOWS before the emergence of physiological signs of withdrawal. However, the potential roles of placental miRNAs in OUD-pregnancy and NOWS outcomes are unknown. This study was undertaken to identify methylation differences in miRNA-encoding genes in the placentas of mothers with OUD who gave birth to infants with NOWS, which may help to identify potential biomarkers of NOWS.
Materials and methods
This study was approved by the Institutional Ethics Committee of William Beaumont Health System, Royal Oak, MI, USA (2019–086) and informed consent was waived as the research utilized paraffin blocks with archived records. The study was carried out under the principles of the Declaration of Helsinki. The study participants included only European Americans in the United States and details on the study cohort have been previously published (Radhakrishna et al., 2021a; Radhakrishna et al., 2021b). Inclusion criteria were the diagnosis of infants with NOWS born to opioid-misusing mothers. Mothers were evaluated for OUD using the Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition, or DSM-5, assessment criteria (Hasin et al., 2013) by physicians/psychiatrists. Newborns with NOWS were studied by neonatologists based on ICD-10 clinical criteria (ICD-10 P96.1). All infants born to mothers with OUD were monitored in the inpatient unit for 4–5 days to observe for signs of NOWS. The infant was scored using the Finnegan Neonatal Abstinence Scoring Tool (FNAST), which was used to determine the use of pharmacological management with morphine. This scoring was done by the postpartum nurses and/or NICU nurses.
To identify DNA methylation patterns of miRNA encoding genes and their effect on expression, we analyzed 96 Formalin-Fixed Paraffin-Embedded (FFPE) archived placental tissue specimens for which DNA methylation and RNA sequencing (RNA-seq) data was recently published by our NOWS consortium (Radhakrishna et al., 2021a; Radhakrishna et al., 2021b). Placental specimens were obtained 2 cm away from the umbilical cord insertion site on the mother’s side of the placenta and were divided into 3 groups. Group 1, consisted of 32 infants prenatally exposed to opioids who received pharmacologic treatment for NOWS symptoms (+Opioids/+NOWS), and 32 infants, prenatally exposed to opioids that did not require pharmacologic therapy for NOWS (+Opioids/-NOWS) and 32 unexposed controls. Among 64 opioid-exposed mothers, 61 (95%) had a history of tobacco smoking. Among 32 mothers (unexposed controls) consisted of individuals who did not have any opioid use and did not have infants diagnosed with NOW (-Opioids/-NOWS, control), 11 (34%) were smokers.
DNA extraction, bisulfite conversion, and restoration
DNA was extracted from the FFPE placental blocks using a QIAamp DNA FFPE tissue kit (Qiagen, catalog no. 56404) according to the manufacturer’s protocol. Infinium HD FFPE Restore kit (Illumina, San Diego, CA) was used for DNA restoration (Siegel et al., 2014) followed by bisulfite conversion of 500 ng of DNA using the EZ DNA Methylation kit (Zymo Research, Irvine, CA) according to the manufacturer’s instructions. The methodology has been detailed earlier (Radhakrishna et al., 2016; Radhakrishna et al., 2021b).
Illumina Infinium Methylation EPIC BeadChip
Genome-wide DNA methylation profiling was performed using the Infinium Methylation EPIC BeadChip array (Illumina Inc., San Diego, CA) with bisulfite-treated genomic DNA according to the manufacturer’s protocol (Vishweswaraiah et al., 2019; Radhakrishna et al., 2021b). The Methylation EPIC BeadChip microarray provides quantitative measurement of 853,307 CpG sites, including the methylation of 9,961 CpG site regulators of miRNA-encoding genes present in the array (Moran et al., 2016). The sample placement in each chip was randomized to avoid confounding and to achieve successful microarray experiments (Verdugo et al., 2009). During the preprocessing of the methylation data, CpG-sites annotated to X and Y chromosomes and/or containing SNPs near or within the probe sequence (within 10 bp of the CpG site), probes that lacked beta values, and probes with minor allele frequency exceeding 0.05 were excluded from the analysis (Chen et al., 2013; Liu et al., 2013; Wilhelm-Benartzi et al., 2013; Daca-Roszak et al., 2015), followed by the analysis of the beta values for the remaining CpG sites. SNPs within 10 bp of the microRNA binding site may affect microRNA and mRNA interactions and reduce binding affinity (Li et al., 2016).
Network analysis of miRNAs
To establish associations between miRNAs and the biological processes they regulate in OUD/NOWs, we performed a gene ontology (GO) analysis. Pathway analysis was carried out using Ingenuity Pathway Analysis (IPA®) using differentially expressed miRNAs at FDR p-value <0.05. MiRNAs were removed from the analysis if they were duplicated or unrecognized by IPA. The most statistically significant miRNAs identified in the NOWS were used in canonical pathways.
Results
The present cohort comprised 96 infants including 64 diagnosed with NOWS born to mothers with OUD and 32 unexposed controls (Table 1). We initially generated genome-wide methylation and gene expression measurements in the cohort mentioned above and published them (Radhakrishna et al., 2021a; Radhakrishna et al., 2021b). In the follow-up study, we aimed to identify miRNA genes in the proximity of CpG sites, in which modifications of the epigenetic profile are associated with NOWS. We analyzed the methylation of 9,961 CpG site regulators of miRNA-encoding genes present in the Illumina epic array.
TABLE 1.
Demographic characteristics of the study subjects: opioid-exposed NOWS newborns that need treatment (+Opioids/+NOWS), mothers with prenatally opioid-exposed infants that did not require treatment for NOWS (+Opioids/-NOWS), and unexposed mothers with normal controls (-Opioids/-NOWS).
| Number of subjects | Maternal age in years- Mean (SD) | Gestational age at delivery in weeks– Mean (SD) | Alcohol history | Tobacco history | Birth weight (g) –Mean (SD) | NICU admission | NOWS development | ||
|---|---|---|---|---|---|---|---|---|---|
| (+Opioids/+NOWS) versus (+Opioids/-NOWS) | Cases | 32 | 31 (4.7) | 37.94 (3.16) | 4 (12%) | 30 (93%) | 2800 (780.7) | 32 (100%) | 32 (100%) |
| Controls | 32 | 28 (4.9) | 37.49 (2.96) | 3 (9%) | 31 (96%) | 2752 (671.2) | 11 (34%) | 0 | |
| q-value (FDR) | NA | 0.9947 | 0.9947 | 0.9947 | 0.9947 | 0.9947 | <0.00001 | <0.00001 | |
| +Opioids/+NOWS plus +Opioids/-NOWS versus -Opioids/-NOWS | Cases | 64 | 29.5 (5.1) | 37.72 (3.07) | 7 (11%) | 61 (95%) | 2776 (729.7) | 43 (67%) | 32 (50%) |
| Controls | 32 | 32.5 (4.1) | 38.09 (3.37) | 3 (9%) | 11 (34%) | 3117 (760.6) | 3 (9%) | 0 | |
| q-value (FDR) | NA | 0.8514 | 0.0053 | 0.0237 | 0.0643 | 0.0168 | 0.0052 | 0.0948 | |
| +Opioids/+NOWS versus -Opioids/-NOWS | Cases | 32 | 31 (4.7) | 37.94 (3.16) | 4 (12%) | 30 (93%) | 2800 (780.7) | 32 (100%) | 32 (100%) |
| Controls | 32 | 32.5 (4.02) | 38.09 (3.37) | 3 (9%) | 11 (34%) | 3117 (760.6) | 3 (9%) | 0 | |
| q-value (FDR) | NA | 0.5747 | 0.821 | 0.2522 | 0.024 | 0.1243 | <0.00001 | <0.00001 |
We identified 47 significantly differentially methylated (FDR p-value ≤0.05) CpGs associated with 46 unique miRNAs, with a receiver operating characteristic (ROC) area under the curve (AUC) ≥0.75 which were used in the follow-up study. Detailed hypomethylated and hypermethylated miRNAs were identified in the placentas of infants with NOWS (Tables 2; Table 3; Table 4). Table 2 describes the miRNA dataset from +Opioids/+NOWS, which was compared against + Opioids/-NOWS. miRNAs such as miR-301, miR-573, and miR-548 among others were found significantly differentially methylated. Table 3 describe data distinguishing the combined two OUD groups from unexposed controls (+Opioids/+NOWS plus + Opioids/-NOWS versus -Opioids/-NOWS, control). The differentially methylated miRNAs identified include miR-181, miR-34, miR-129, and miR-548. Finally, Table 4 defines + Opioids/+NOWS versus unexposed controls (-opioids/-NOWS), yielded the following differentially methylated miRNAs: miR-34, miR-10, miR-548, miR-518, miR-1909, and miR-1178.
TABLE 2.
The analysis of +Opioids/+NOWS versus + Opioids/-NOWS. Significantly differentially methylated microRNAs based on FDR adjusted-p values < 0.05 provided with (AUC) ≥0.75 for NOWS detection. Details of corresponding CpG loci, chromosomes, and methylation status.
| TargetID | miR | CHR | p-Val | FDR p-Val | % Methylation | CI | Methylation | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Control | Difference | AUC | lower | upper | ||||||
| cg11210410 | miR-1268A | 9 | 3.12539E-38 | 2.70346E-32 | 74.62 | 64.79 | 9.83 | 0.82 | 0.72 | 0.93 | Hyper |
| cg14929554 | miR-5095 | 16 | 2.56839E-14 | 2.22166E-08 | 60.20 | 70.37 | −10.17 | 0.76 | 0.65 | 0.88 | Hypo |
| cg16769912 | miR-558 | 2 | 1.85302E-13 | 1.60286E-07 | 60.52 | 70.36 | −9.83 | 0.78 | 0.67 | 0.90 | Hypo |
| cg13790797 | miR-548G | 3 | 4.61626E-11 | 3.99307E-05 | 49.01 | 58.67 | −9.66 | 0.76 | 0.64 | 0.88 | Hypo |
| cg04312413 | miR-548F1 | 1 | 6.42158E-11 | 5.55466E-05 | 71.41 | 78.91 | −7.50 | 0.77 | 0.66 | 0.89 | Hypo |
| cg02656609 | miR-301A | 17 | 1.18711E-10 | 0.000102685 | 76.05 | 82.73 | −6.68 | 0.81 | 0.70 | 0.92 | Hypo |
| cg10046367 | miR-548W | 17 | 1.30176E-10 | 0.000112602 | 69.25 | 76.95 | −7.70 | 0.83 | 0.73 | 0.93 | Hypo |
| cg26481500 | miR-1278 | 1 | 1.48897E-10 | 0.000128796 | 77.09 | 83.57 | −6.48 | 0.78 | 0.67 | 0.89 | Hypo |
| cg23632539 | miR-5096 | 1 | 6.57986E-10 | 0.000569158 | 78.51 | 71.87 | 6.64 | 0.74 | 0.61 | 0.86 | Hyper |
| cg15350946 | miR-1273H | 4 | 6.59135E-10 | 0.000570152 | 45.23 | 54.40 | −9.18 | 0.73 | 0.61 | 0.85 | Hypo |
| cg06769231 | miR-2117 | 17 | 6.72269E-10 | 0.000581513 | 64.17 | 72.20 | −8.03 | 0.76 | 0.64 | 0.87 | Hypo |
| cg22395021 | miR-548H3 | 6 | 7.95169E-10 | 0.000687821 | 61.28 | 69.57 | −8.29 | 0.70 | 0.57 | 0.83 | Hypo |
| cg00071872 | miR-548AE2 | 16 | 9.20749E-10 | 0.000796448 | 14.56 | 9.27 | 5.29 | 0.72 | 0.60 | 0.85 | Hyper |
| cg18363417 | miR-573 | 4 | 2.94005E-09 | 0.00254314 | 73.90 | 80.45 | −6.54 | 0.77 | 0.65 | 0.88 | Hypo |
| cg21254731 | miR-939 | 8 | 3.15349E-09 | 0.002727773 | 81.93 | 87.17 | −5.24 | 0.86 | 0.77 | 0.95 | Hypo |
| cg02790122 | miR-1296 | 10 | 4.68281E-09 | 0.00405063 | 77.11 | 83.10 | −5.99 | 0.71 | 0.58 | 0.84 | Hypo |
| cg26042267 | miR-3663HG | 10 | 5.19928E-09 | 0.004497379 | 35.81 | 28.35 | 7.47 | 0.72 | 0.59 | 0.84 | Hyper |
| cg25249290 | miR-181A1HG | 1 | 8.31433E-09 | 0.007191897 | 61.56 | 69.35 | −7.79 | 0.73 | 0.61 | 0.86 | Hypo |
| cg19350059 | miR-548F5 | 13 | 8.80098E-09 | 0.007612844 | 18.63 | 13.02 | 5.61 | 0.74 | 0.62 | 0.87 | Hyper |
| cg00224335 | miR-4327 | 21 | 1.13961E-08 | 0.009857667 | 52.54 | 44.68 | 7.86 | 0.75 | 0.63 | 0.87 | Hyper |
| cg17777998 | miR-1183 | 7 | 1.58092E-08 | 0.013674961 | 38.38 | 46.76 | −8.38 | 0.71 | 0.59 | 0.84 | Hypo |
| cg13641189 | miR-658 | 22 | 2.75754E-08 | 0.02385271 | 11.27 | 6.90 | 4.37 | 0.79 | 0.68 | 0.91 | Hyper |
| cg03425468 | miR-92B | 1 | 3.25876E-08 | 0.028188258 | 11.44 | 7.06 | 4.38 | 0.75 | 0.63 | 0.87 | Hyper |
| cg24289256 | miR-548F3 | 1 | 4.69893E-08 | 0.040645767 | 72.95 | 79.16 | −6.21 | 0.70 | 0.58 | 0.83 | Hypo |
| cg03135982 | miR-3650 | 5 | 4.94006E-08 | 0.042731525 | 76.26 | 82.01 | −5.75 | 0.74 | 0.62 | 0.87 | Hypo |
TABLE 3.
Analysis of (+Opioids/+NOWS), + (+Opioids/-NOWS), versus (-Opioids/-NOWS, control). Differentially methylated microRNAs and the corresponding methylated CpG sites with Target ID, Gene ID, chromosome location, p-value, FDR p-value, and % methylation change are given.
| Target ID | miR | CHR | p-Val | FDR p-Val | % Methylation | AUC | CI | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Control | Difference | lower | upper | Methylation | ||||||
| cg11479035 | miR-1276 | 15 | 3.00299E-37 | 2.59759E-31 | 86.38 | 79.64 | 6.74 | 0.83 | 0.73 | 0.93 | Hyper |
| cg13767940 | miR-34B | 11 | 2.67438E-17 | 2.31334E-11 | 9.13 | 16.99 | −7.86 | 0.72 | 0.59 | 0.85 | Hypo |
| cg23632539 | miR-5096 | 1 | 2.20382E-13 | 1.9063E-07 | 80.63 | 73.11 | 7.51 | 0.82 | 0.72 | 0.92 | Hyper |
| cg19782652 | miR-125A | 19 | 1.19367E-09 | 0.001032525 | 78.09 | 84.18 | −6.09 | 0.92 | 0.85 | 0.99 | Hypo |
| cg19069367 | miR-6850 | 8 | 1.61143E-09 | 0.001393891 | 9.86 | 5.38 | 4.48 | 0.82 | 0.72 | 0.92 | Hyper |
| cg21913981 | miR-1178 | 12 | 2.14827E-09 | 0.001858257 | 73.86 | 66.75 | 7.10 | 0.79 | 0.68 | 0.90 | Hyper |
| cg19756622 | miR-518A1; miR-518E | 19 | 7.21604E-09 | 0.006241877 | 81.17 | 75.24 | 5.93 | 0.84 | 0.74 | 0.94 | Hyper |
| cg24774002 | miR-1909 | 19 | 3.6236E-08 | 0.031344152 | 89.10 | 92.79 | −3.69 | 0.83 | 0.73 | 0.93 | Hypo |
| cg13477253 | miR-548G | 3 | 5.62137E-08 | 0.048624868 | 15.48 | 10.50 | 4.98 | 0.77 | 0.66 | 0.89 | Hyper |
TABLE 4.
Analysis of (+Opioids/+NOWS), versus (-Opioids/-NOWS, control). Details of CpG targets significantly differentially methylated microRNAs in NOWS. Differentially methylated CpG sites with Target ID, Gene ID, chromosome location, p-value, FDR p-value, and % methylation change details are given.
| CpG target | miR | CHR | p-value | FDR p-Val | % Methylation | AUC | CI | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Control | diffreence | lower | upper | Methylation | ||||||
| cg20276377 | miR-548G | 3 | 1.98166E-23 | 1.71414E-17 | 15.95839 | 25.9444 | −9.98601 | 0.65 | 0.53 | 0.77 | Hypo |
| cg01514668 | miR-129-2 | 11 | 8.5878E-13 | 7.42844E-07 | 12.95384 | 19.57678 | −6.62294 | 0.58 | 0.46 | 0.70 | Hypo |
| cg18576861 | miR-3621 | 9 | 4.85176E-11 | 4.19677E-05 | 23.36382 | 30.90311 | −7.53929 | 0.74 | 0.63 | 0.86 | Hypo |
| cg20351875 | miR-548C | 12 | 1.0169E-10 | 8.79621E-05 | 16.31689 | 22.83412 | −6.51723 | 0.67 | 0.55 | 0.79 | Hypo |
| cg25147193 | miR-181C | 19 | 1.44145E-10 | 0.000124685 | 8.817696 | 13.97549 | −5.157794 | 0.64 | 0.52 | 0.76 | Hypo |
| cg11479035 | miR-1276 | 15 | 2.1566E-10 | 0.000186546 | 84.82172 | 79.64312 | 5.1786 | 0.77 | 0.66 | 0.87 | Hyper |
| cg06871184 | miR-1238 | 19 | 4.3656E-10 | 0.000377624 | 64.27575 | 57.02732 | 7.24843 | 0.72 | 0.60 | 0.83 | Hyper |
| cg13781167 | miR-8081 | 9 | 1.58717E-09 | 0.001372905 | 64.99973 | 71.75161 | −6.75188 | 0.70 | 0.58 | 0.81 | Hypo |
| cg19756622 | miR-518A1 | 19 | 3.09143E-09 | 0.002674085 | 80.68514 | 75.23586 | 5.44928 | 0.82 | 0.72 | 0.92 | Hyper |
| cg17540499 | miR-592 | 7 | 3.8092E-09 | 0.00329496 | 31.95645 | 39.38867 | −7.43222 | 0.59 | 0.47 | 0.71 | Hypo |
| cg13767940 | miR-34B | 11 | 1.06906E-08 | 0.009247326 | 11.87235 | 16.98866 | −5.11631 | 0.64 | 0.52 | 0.76 | Hypo |
| cg26438516 | miR-1256 | 10 | 2.79234E-08 | 0.024153783 | 66.7636 | 60.35334 | 6.41026 | 0.71 | 0.60 | 0.83 | Hyper |
Pathway analysis of significant miRNAs
Gene ontology analysis found major dysregulated pathways affected by methylation changes on miRNAs associated with NOWS and OUD-related outcomes. A comparison was made between three groups i. Distinguishing NOWS from prenatal opioid-exposure without NOWS (+Opioids/+NOWS, versus + Opioids/-NOWS), ii. Distinguishing prenatal opioid use versus unexposed controls (OUD detection) (+Opioids/+NOWS and +Opioids/-NOWS versus -Opioids/-) and iii. distinguishing NOWS versus unexposed controls (+Opioids/+NOWS versus + Opioids/-NOWS). Significant functional biological processes regulated by these miRNAs were identified with the plausible molecular mechanism of differentially expressed miRNAs. Network interaction between miRNAs and their target genes was provided in Figure 1, Figure 2, and Figure 3.
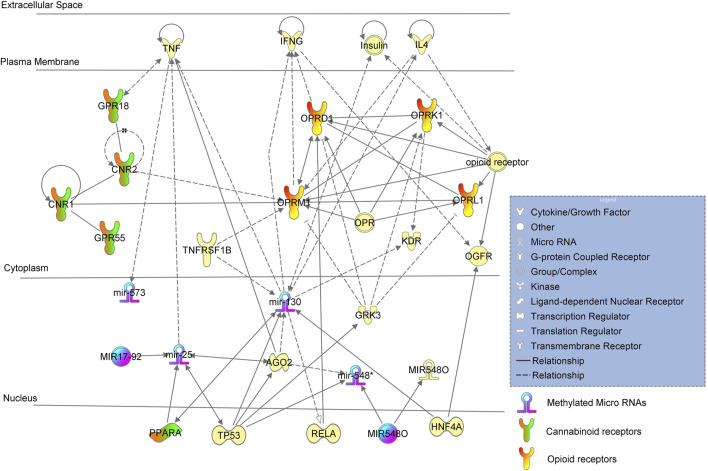
FIGURE 1.
Ingenuity pathway analysis (IPA) of significant methylation regulators of miRNA-encoding genes and network analysis with a p-value <0.05 are depicted using + Opioids/+NOWS versus + Opioids/-NOWS.
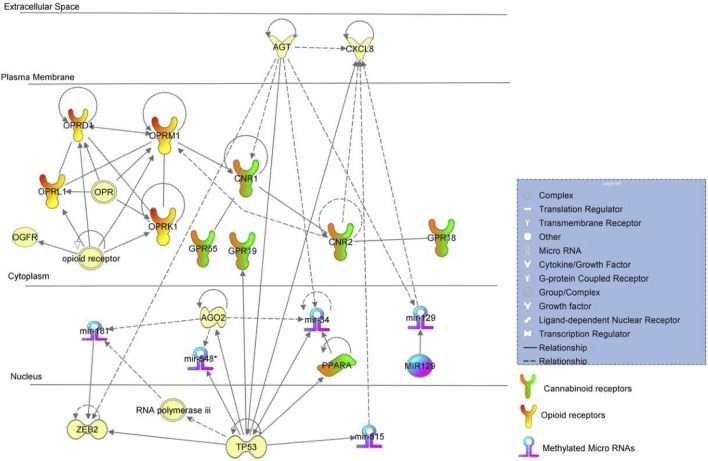
FIGURE 2.
Pathway analysis of +Opioids/+NOWS and +Opioids/-NOWS versus -Opioids/-.
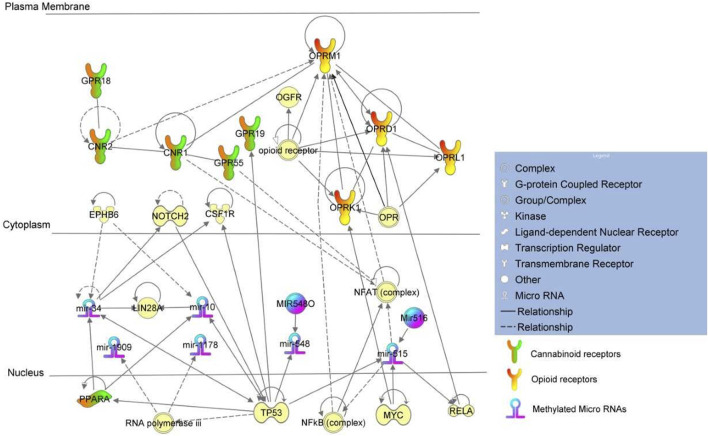
FIGURE 3.
Pathway analysis of +Opioids/+NOWS versus -Opioids/-NOWS.
Target identification of differentially methylated miRNAs
MicroRNA Target Prediction and Functional Study Database (miRDB) server (http://miRdb.org) were used to predict putative targets of aberrantly methylated miRNAs. With a set target score of ≥60 (Wang, 2016), we searched the miRDB database for predictions of targets of some of these significant miRNAs. From the miRDB, we retrieved the target genes for 47 miRNAs. From this, we discovered multiple predicted targets for each miRNA (Adlakha and Saini, 2014). These predicted 47 miRNA targets were compared with the differentially methylated genes identified in our genome-wide methylation data of the same sample cohort (Radhakrishna et al., 2021b). The results showed that multiple predicted targets (genes) of these miRNAs were also found to be differentially methylated in our patients. Supplementary Tables S1 to S15 provide examples of miRNA targets discussed in the manuscript that are important and available in the miRDB database.
Discussion
Description of opioid use with NOWS vs. opioid use without NOWS
(opioid-exposed infants who required pharmacologic management for NOWS versus opioid-exposed infants that did not require treatment for NOWS) (+Opioids/+NOWS versus + Opioids/-NOWS) (Table 2). The miR-130 family includes four members (miR-130a, miR-130b, miR-301a, and miR-301b) with the same source sequences and which perform similar biological functions (Wang et al., 2020). The miR-130 mature miRNA is known to increase the expression of PPARA mRNA (Papi et al., 2013). PPARA is a nuclear hormone receptor that responds to certain types of cannabinoids (O'Sullivan, 2016).
MiR-130 is regulated by TP53, HNF4A, AGO2, TNFRFS1B, and insulin, and was found to regulate the expressions of KDR, IL4, IFNG, TNF, and RELA. This regulation is likely dampened to a certain extent due to methylation, resulting in higher protein expressions and activations of the miRNA. MiR-130 regulates IL4, which in turn regulates the mu-opioid receptor (MOR), whose principal ligands include opioid peptides and analgesics. The MOR also has an important role in dependence on other drugs of abuse, such as nicotine, cocaine, and alcohol via its modulation of the dopamine system.
Interestingly, miR-130 putatively targets several other genes that are preferentially involved in brain disorders. For instance, miR-130, in combination with other miRNAs, jointly regulates ATXN1, which causes spinocerebellar ataxia type 1 (SCA1). SCA1 causes seizures, slurred speech, slowness of movement, and cognitive impairments. Similar neurological signs such as slurred speech or slowed movements are seen in OUD subjects, and conditions such as tremors and seizures are common in both OUD and infants with NOWS. Our genome-wide methylation analysis of NOWS revealed hypermethylation of the ATXN1 gene (Radhakrishna et al., 2021b).
Inhibition of miR-130 leads to decreased translocation of phosphorylated RELA protein to nuclei (Hazra et al., 2019). RELA protein (Chen et al., 2006) plays a significant role in the downstream activation of the OPRD1 (also known as DOR, δ-opioid receptor, or delta-opioid receptor) is a paralog of OPRM1 associated with opioid dependence gene that encodes the delta-opioid receptor (DOR) protein in humans. In the central nervous system, DOR and mu-opioid receptors (MOR) can interact and modulate each other’s and are involved in the regulation of pain (Beaudry et al., 2015; Olesen et al., 2018). DOR is responsible for reducing the intensity of pain signals, while MOR is responsible for reducing the frequency of pain signals. Heterodimerization of MOR and DOR can indeed impact the recruitment of beta-arrestin 2 (ARRB2) protein, which subsequently influences downstream intracellular signaling (Rozenfeld and Devi, 2007).
Description of opioid exposure vs. unexposed controls
(opioid-exposed infants who required pharmacologic management for NOWS + opioid-exposed infants that did not require treatment for NOWS versus unexposed controls. (+Opioids/+NOWS plus + Opioids/-NOWS versus -Opioids/-NOWS). Table 3 describes the miRNA dataset from mothers with a history of opioid usage. This cohort was compared against normal unexposed controls and showed significant methylation differences in miR-34, miR-129, miR-181, and miR-548 among others.
The binding of the AGO2 protein was found to occur with miR-34 mature miRNAs (Wu et al., 2011). The presence of AGT proteins was found to increase expressions of mouse miR-34 and miR-129 mature miRNAs indicating positive regulation by AGT (Jin et al., 2012; Huo et al., 2019). AGT protein increases paracrine activation of CNR1 protein (Turu et al., 2009), followed by clustering of CNR1 with CNR2. Activated CNR2 protein increases the expression of OPRM1 (Borner et al., 2006) possibly resulting in an OPRM1-OPRD1-OPRL1-OPRK1 feedback loop.
The binding of the TP53 response element from the miR-34 promoter and TP53 protein was found to occur followed by targeting of TP53 mRNA by miR-34 mature miRNA. TP53 protein further increases the transcription of the miR-34 gene and miR-34 mature miRNA was found to, in turn, upregulate the expression of acetylated (K382) p53 protein followed by increasing activity of p53 protein (Feng et al., 2011). Protein modification of AGO2 leads to an increased expression of miR-548 and miR-181 (Jin et al., 2012; Garibaldi et al., 2016). Homozygous experimental p53 gene deletion was found to decrease the expression of miR-548 mature miRNA (Shin et al., 2009). Experimental interference of human p53 mRNA by shRNA led to an increased expression of miR-519 mature miRNA (Garibaldi et al., 2016). Binding of 3′UTR from ZEB2 mRNA and miR-181A mature miRNA occurs, indicating probable regulation of ZEB2 mRNA. Negative regulation of TP53 in this pathway leads to activations of several genes in the pathway including PPARA which is a nuclear hormone receptor capable of taking certain cannabinoid ligands.
Women who used illicit or unprescribed opioids during pregnancy have a higher risk of fetal growth restriction and preterm delivery (Maghsoudlou et al., 2017). Recent studies have shown differential miR-548 expression profile was associated with spontaneous preterm births (Gray et al., 2017). Placental DNA methylation of miR-548 and WWTR1 genes influence insulin sensitivity during pregnancy, and preterm birth and is linked to insulin resistance (Hofman et al., 2004; Mathai et al., 2012). Interestingly, nine of the 47 dysregulated miRNAs belonged to the miR-548 family, including miR548G, miR548F1, miR548W, miR548H3, miR548AE2, miR548F5, miR548F3, miR548C, and miR548C, all of which were hypermethylated. Moreover, WWTR1 is also hypermethylated in the present study.
NOWS vs. unexposed controls
(opioid-exposed infants who required pharmacologic management for NOWS versus unexposed controls (+Opioids/+NOWS versus -opioids/-NOWS, control) (Table 4). MiR-515 is the largest miRNA gene cluster in humans (Zhang M. et al., 2016), the family members of miR-515 include miR-516 and miR-518 (Zhang M. et al., 2016). MiRNA-515 and miR-34 are found to be connected directly to opioid and cannabinoid receptors respectively. MiR-515 was found to regulate opioid genes OPRM1, and OPRD1, while miR-34 was found to regulate cannabinoid genes GPR19 and PPARA via Notch and TP53 proteins.
Mature miR-515 is known to decrease the activity of the human nuclear factor kappa B (NFkB) complex (Keklikoglou et al., 2012), however, in the absence of active miR-515 due to methylation as seen in the current cohort, the Nfkb complex is actively expressed leading to continued expression of OPRM1 mRNA. Thus, methylation of miR-515 results in the transcription of human OPRM1 mRNA which otherwise would be under the strict regulation of the NFkB complex (Kraus et al., 2003; Memet, 2006).
RELA mRNA is targeted by miR-515 mature miRNA leading to its decreased expression (Keklikoglou et al., 2012). Interference of Rela mRNA by siRNA decreases the expression of Oprd1 mRNA. RELA protein increases activation of the reporter gene with a promoter fragment (-262–1) from the OPRD1 gene (Chen et al., 2006). Methylation of miR-515 leads to the absence of regulation leading to an increased expression of RELA. Increased RELA subsequently increases its downstream partner OPRD1. The OPRM1 and OPRD1 genes were also found to be distinctively methylated in the previously described datasets. OPRM1 was found hypomethylated in mothers who used opioids during pregnancy and delivered NOWS newborns which required immediate medical care when compared to mothers who had used opioids during pregnancy and delivered newborns without NOWS. In contrast, OPRD1 was found hypermethylated in mothers with a history of opioid usage who delivered babies without NOWS when compared against unexposed controls. Both OPRD1 and OPRM1 were found to be distinctly methylated in the current dataset of mothers who used opioids and delivered NOWS babies when compared against unexposed controls. However, PPARA activation status differed across these three datasets.
Role of PPARA in conjunction with opioid and cannabinoid receptors
The absence of active miR-130 in the +Opioid/+NOWS group may have led to decreased expression of PPARA mRNA, therefore, leading to reduced production of oxidation enzymes to effectively scavenge for free radicals generated by oxidative stress from opioid use. This may have resulted in mothers delivering newborns with NOWS that required medical intervention. Interestingly, when the Opioid+/NOWS- group was compared against unexposed controls, PPARA was found to be under the positive regulation of TP53 leading to its activation. This protective role of PPARA was absent in the +Opioids/+NOWS cohort of mothers when compared against unexposed controls. This is possibly due to the active status of miR-134 and/or to a different set of regulatory partners Notch and TP53 whose role in this context is not known and can only be speculated. MiR-34 was found to regulate cannabinoid genes including PPARA via Notch and TP53 proteins. Though the anti-oxidation activity of PPARA was probably negated by the extensive and consistent activations of opioid genes OPRM1, and OPRD1, unlike seen in any of the other two cohorts and thus resulting in mothers delivering NOWS newborns that required medical care. Moreover, miR-34 has a functional role in the regulation of stress-induced anxiety, depression with suicidal ideation (Haramati et al., 2011). Anxiety and depression are common and normal phenomena, in mothers of infants experiencing NOWS (Corr et al., 2020).
SIRT1 which is hypomethylated in the current cohort study (FDR) p ≤ 0.05) (Radhakrishna et al., 2021b), is a nicotinamide adenosine dinucleotide (NAD)-dependent deacetylase that removes acetyl groups from various proteins. SIRT1 normally functions to limit the expression of miR-134, which targets the critical activity-dependent transcription factor (CREB) essential for learning and memory (Lv et al., 2013; O'Neal-Moffitt et al., 2014; Liu et al., 2017). SIRT1 exerts neuroprotection against ischemic injury and various neurodegenerative disorders (Xu et al., 2018). Dysregulation of brain SIRT1 activity can have devastating brain consequences including neurological dysfunction (Xu et al., 2018).
Experimental interference of human p53 mRNA by shRNA led to an increased expression of miR-519 mature miRNA (Garibaldi et al., 2016). Homozygous experimental p53 gene deletion was found to decrease the expression of miR-548 mature miRNA (Shin et al., 2009). MiR-10 which belongs to the miR-125A family member (Tehler et al., 2011) was found to decrease the activation of p53 (Joo et al., 2013). Studies on mouse mmu-miR-10 mature miRNA were found to decrease the expression of mouse p53 protein (Sen et al., 2014). Targeting of LIN28 mRNA by miR-10 and miR-34 mature miRNAs was found to occur leading to decreased translation of LIN28 (Zhong et al., 2010; Jain et al., 2012). p53 (TP53) protein was found to decrease the binding of BRF1 protein and RNA polymerase iii complexes (Crighton et al., 2003). Experimental inhibition of active RNA polymerase iii complexes was found to result in increased expression of miR-1909 while decreasing the expression of miR-1178 mature miRNA (Koo et al., 2015) demonstrating the role of transcriptional regulation by RNA polymerase iii.
Limitations
The study has some limitations, namely, a lack of racial or ethnic diversity, given that it focused on individuals of European origin. A second limitation is that we have not replicated our findings in an independent study cohort, and therefore, we will continue to carry out in vitro and in vivo experiments to validate the conclusions in the future.
Summary and future directions
To our knowledge, this is the first report about miRNA methylation levels of NOWS on a previously well-documented large cohort of placental tissue specimens with genome-wide methylation profiles and gene expression profiles. NOWS is associated with an altered miRNA methylation pattern in the placenta, suggesting that miRNA deregulation is involved in the pathogenesis of NOWS. The current experimental data suggest that epigenetic variations in the placental tissue can serve as surrogate markers for brain health at birth, and thus infant micro-RNA “signatures” can predict the severity of NOWS even before withdrawal symptoms begin. The differential methylation of several miRNAs has been found in association with NOWS. The findings have biological plausibility, and they include several known biological pathways and genes with possible mechanisms associated with NOWS development. The identified dysregulated pathways and genes may therefore provide important opportunities for the development of novel future miRNA-based therapeutic approaches for NOWS. Several preclinical and clinical trials have been reported for miRNA-based therapeutics and few reports indicate that miRNA expression levels can be modified by modulating the miRNA processing pathway. However, validation with large NOWS cohorts and using robust techniques for DNA methylation analysis of miRNA genes are warranted before the application in clinical practice.
Funding Statement
The work was supported by both anonymous donors and Beaumont Health System, Royal Oak, MI.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
Conceptualization UR and RB-S; investigation, UR, RB-S, SN, WB, and RM, resources, UR, RB-S, and SN; funding acquisition, UR, SN, and RB-S; data curation, UR, SN, LU, AF, SM, RC, WB, RM, AV, SV, and RB-S; writing—original draft preparation, UR and AV; and writing—Review and editing, UR, SN, LU, AF, SM, RC, WB, RM, AV, SV, and RB-S; All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1215472/full#supplementary-material
Abbreviations
Hypo, Hypomethylation; Hyper; Hypermethylation.
References
- Adlakha Y. K., Saini N. (2014). Brain microRNAs and insights into biological functions and therapeutic potential of brain enriched miRNA-128. Mol. Cancer 13, 33. 10.1186/1476-4598-13-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali Syeda Z., Langden S. S. S., Munkhzul C., Lee M., Song S. J. (2020). Regulatory mechanism of MicroRNA expression in cancer. Int. J. Mol. Sci. 21 (5), 1723. 10.3390/ijms21051723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldacchino A., Arbuckle K., Petrie D. J., McCowan C. (2015). Erratum: Neurobehavioral consequences of chronic intrauterine opioid exposure in infants and preschool children: A systematic review and meta-analysis. BMC Psychiatry 15, 134. 10.1186/s12888-015-0438-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee J., Sen C. K. (2015). microRNA and wound healing. Adv. Exp. Med. Biol. 888, 291–305. 10.1007/978-3-319-22671-2_15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. P. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 116 (2), 281–297. 10.1016/s0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- Beaudry H., Gendron L., Moron J. A. (2015). Implication of delta opioid receptor subtype 2 but not delta opioid receptor subtype 1 in the development of morphine analgesic tolerance in a rat model of chronic inflammatory pain. Eur. J. Neurosci. 41 (7), 901–907. 10.1111/ejn.12829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi M., Renzini A., Adamo S., Moresi V. (2017). Coordinated actions of MicroRNAs with other epigenetic factors regulate skeletal muscle development and adaptation. Int. J. Mol. Sci. 18 (4), 840. 10.3390/ijms18040840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner C., Hollt V., Kraus J. (2006). Cannabinoid receptor type 2 agonists induce transcription of the mu-opioid receptor gene in Jurkat T cells. Mol. Pharmacol. 69 (4), 1486–1491. 10.1124/mol.105.018325 [DOI] [PubMed] [Google Scholar]
- Burton G. J., Fowden A. L. (2015). The placenta: A multifaceted, transient organ. Philos. Trans. R. Soc. Lond B Biol. Sci. 370 (1663), 20140066. 10.1098/rstb.2014.0066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. A., Lemire M., Choufani S., Butcher D. T., Grafodatskaya D., Zanke B. W., et al. (2013). Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics 8 (2), 203–209. 10.4161/epi.23470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. L., Law P. Y., Loh H. H. (2006). Sustained activation of phosphatidylinositol 3-kinase/Akt/nuclear factor kappaB signaling mediates G protein-coupled delta-opioid receptor gene expression. J. Biol. Chem. 281 (6), 3067–3074. 10.1074/jbc.M506721200 [DOI] [PubMed] [Google Scholar]
- Chiang H. R., Schoenfeld L. W., Ruby J. G., Auyeung V. C., Spies N., Baek D., et al. (2010). Mammalian microRNAs: Experimental evaluation of novel and previously annotated genes. Genes. Dev. 24 (10), 992–1009. 10.1101/gad.1884710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chorbov V. M., Todorov A. A., Lynskey M. T., Cicero T. J. (2011). Elevated levels of DNA methylation at the OPRM1 promoter in blood and sperm from male opioid addicts. J. Opioid Manag. 7 (4), 258–264. 10.5055/jom.2011.0067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopher A. F., Kaur R. P., Kaur G., Kaur A., Gupta V., Bansal P. (2016). MicroRNA therapeutics: Discovering novel targets and developing specific therapy. Perspect. Clin. Res. 7 (2), 68–74. 10.4103/2229-3485.179431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary B. J., Donnelly J. M., Strawbridge J. D., Gallagher P. J., Fahey T., White M. J., et al. (2011). Methadone and perinatal outcomes: A retrospective cohort study. Am. J. Obstet. Gynecol. 204 (2), e1–e9. 10.1016/j.ajog.2010.10.004 [DOI] [PubMed] [Google Scholar]
- Contreras J., Rao D. S. (2012). MicroRNAs in inflammation and immune responses. Leukemia 26 (3), 404–413. 10.1038/leu.2011.356 [DOI] [PubMed] [Google Scholar]
- Corr T. E., Schaefer E. W., Hollenbeak C. S., Leslie D. L. (2020). One-year postpartum mental health outcomes of mothers of infants with neonatal abstinence syndrome. Matern. Child. Health J. 24 (3), 283–290. 10.1007/s10995-019-02839-9 [DOI] [PubMed] [Google Scholar]
- Crighton D., Woiwode A., Zhang C., Mandavia N., Morton J. P., Warnock L. J., et al. (2003). p53 represses RNA polymerase III transcription by targeting TBP and inhibiting promoter occupancy by TFIIIB. EMBO J. 22 (11), 2810–2820. 10.1093/emboj/cdg265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daca-Roszak P., Pfeifer A., Zebracka-Gala J., Rusinek D., Szybinska A., Jarzab B., et al. (2015). Impact of SNPs on methylation readouts by Illumina Infinium HumanMethylation450 BeadChip array: Implications for comparative population studies. BMC Genomics 16 (1), 1003. 10.1186/s12864-015-2202-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z., Zhang C., Wu R., Hu W. (2011). Tumor suppressor p53 meets microRNAs. J. Mol. Cell. Biol. 3 (1), 44–50. 10.1093/jmcb/mjq040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garibaldi F., Falcone E., Trisciuoglio D., Colombo T., Lisek K., Walerych D., et al. (2016). Mutant p53 inhibits miRNA biogenesis by interfering with the microprocessor complex. Oncogene 35 (29), 3760–3770. 10.1038/onc.2016.51 [DOI] [PubMed] [Google Scholar]
- Gray C., McCowan L. M., Patel R., Taylor R. S., Vickers M. H. (2017). Maternal plasma miRNAs as biomarkers during mid-pregnancy to predict later spontaneous preterm birth: A pilot study. Sci. Rep. 7 (1), 815. 10.1038/s41598-017-00713-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramati S., Navon I., Issler O., Ezra-Nevo G., Gil S., Zwang R., et al. (2011). MicroRNA as repressors of stress-induced anxiety: The case of amygdalar miR-34. J. Neurosci. 31 (40), 14191–14203. 10.1523/JNEUROSCI.1673-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasin D. S., O'Brien C. P., Auriacombe M., Borges G., Bucholz K., Budney A., et al. (2013). DSM-5 criteria for substance use disorders: Recommendations and rationale. Am. J. Psychiatry 170 (8), 834–851. 10.1176/appi.ajp.2013.12060782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazra B., Chakraborty S., Bhaskar M., Mukherjee S., Mahadevan A., Basu A. (2019). miR-301a regulates inflammatory response to Japanese encephalitis virus infection via suppression of NKRF activity. J. Immunol. 203 (8), 2222–2238. 10.4049/jimmunol.1900003 [DOI] [PubMed] [Google Scholar]
- Hofman P. L., Regan F., Jackson W. E., Jefferies C., Knight D. B., Robinson E. M., et al. (2004). Premature birth and later insulin resistance. N. Engl. J. Med. 351 (21), 2179–2186. 10.1056/NEJMoa042275 [DOI] [PubMed] [Google Scholar]
- Honein M. A., Boyle C., Redfield R. R. (2019). Public health surveillance of prenatal opioid exposure in mothers and infants. Pediatrics 143 (3), e20183801. 10.1542/peds.2018-3801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo K. G., Richer C., Berillo O., Mahjoub N., Fraulob-Aquino J. C., Barhoumi T., et al. (2019). miR-431-5p knockdown protects against angiotensin II-induced hypertension and vascular injury. Hypertension 73 (5), 1007–1017. 10.1161/HYPERTENSIONAHA.119.12619 [DOI] [PubMed] [Google Scholar]
- Iacomino G., Siani A. (2017). Role of microRNAs in obesity and obesity-related diseases. Genes. Nutr. 12, 23. 10.1186/s12263-017-0577-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A. K., Allton K., Iacovino M., Mahen E., Milczarek R. J., Zwaka T. P., et al. (2012). p53 regulates cell cycle and microRNAs to promote differentiation of human embryonic stem cells. PLoS Biol. 10 (2), e1001268. 10.1371/journal.pbio.1001268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W., Reddy M. A., Chen Z., Putta S., Lanting L., Kato M., et al. (2012). Small RNA sequencing reveals microRNAs that modulate angiotensin II effects in vascular smooth muscle cells. J. Biol. Chem. 287 (19), 15672–15683. 10.1074/jbc.M111.322669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo M. S., Lee C. G., Koo J. H., Kim S. G. (2013). miR-125b transcriptionally increased by Nrf2 inhibits AhR repressor, which protects kidney from cisplatin-induced injury. Cell. Death Dis. 4, e899. 10.1038/cddis.2013.427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keklikoglou I., Koerner C., Schmidt C., Zhang J. D., Heckmann D., Shavinskaya A., et al. (2012). MicroRNA-520/373 family functions as a tumor suppressor in estrogen receptor negative breast cancer by targeting NF-κB and TGF-β signaling pathways. Oncogene 31 (37), 4150–4163. 10.1038/onc.2011.571 [DOI] [PubMed] [Google Scholar]
- Koo C. X., Kobiyama K., Shen Y. J., LeBert N., Ahmad S., Khatoo M., et al. (2015). RNA polymerase III regulates cytosolic RNA:DNA hybrids and intracellular microRNA expression. J. Biol. Chem. 290 (12), 7463–7473. 10.1074/jbc.M115.636365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A., Griffiths-Jones S. (2014). miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42, D68–D73. 10.1093/nar/gkt1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus J., Borner C., Giannini E., Hollt V. (2003). The role of nuclear factor kappaB in tumor necrosis factor-regulated transcription of the human mu-opioid receptor gene. Mol. Pharmacol. 64 (4), 876–884. 10.1124/mol.64.4.876 [DOI] [PubMed] [Google Scholar]
- Lee R. C., Feinbaum R. L., Ambros V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 75 (5), 843–854. 10.1016/0092-8674(93)90529-y [DOI] [PubMed] [Google Scholar]
- Li D., Zhang H., Ma L., Han Y., Xu M., Wang Z., et al. (2016). Associations between microRNA binding site SNPs in FGFs and FGFRs and the risk of non-syndromic orofacial cleft. Sci. Rep. 6, 31054. 10.1038/srep31054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Shan G., Teng Z. Q., Wingo T. S. (2020). Editorial: Non-Coding RNAs and human diseases. Front. Genet. 11, 523. 10.3389/fgene.2020.00523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipari R. N., Van Horn S. L. (2013). “Children living with parents who have a substance use disorder,” in The CBHSQ report (Rockville (MD)), 1–7. [PubMed] [Google Scholar]
- Liu Y., Aryee M. J., Padyukov L., Fallin M. D., Hesselberg E., Runarsson A., et al. (2013). Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat. Biotechnol. 31 (2), 142–147. 10.1038/nbt.2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Ni C., Li Z., Yang N., Zhou Y., Rong X., et al. (2017). Prophylactic melatonin attenuates isoflurane-induced cognitive impairment in aged rats through hippocampal melatonin receptor 2 - cAMP response element binding signalling. Basic Clin. Pharmacol. Toxicol. 120 (3), 219–226. 10.1111/bcpt.12652 [DOI] [PubMed] [Google Scholar]
- Lv J., Xin Y., Zhou W., Qiu Z. (2013). The epigenetic switches for neural development and psychiatric disorders. J. Genet. Genomics 40 (7), 339–346. 10.1016/j.jgg.2013.04.007 [DOI] [PubMed] [Google Scholar]
- Mactier H., McLaughlin P., Gillis C., Osselton M. D. (2017). Variations in infant CYP2B6 genotype associated with the need for pharmacological treatment for neonatal abstinence syndrome in infants of methadone-maintained opioid-dependent mothers. Am. J. Perinatol. 34 (9), 918–921. 10.1055/s-0037-1600917 [DOI] [PubMed] [Google Scholar]
- Maghsoudlou S., Cnattingius S., Montgomery S., Aarabi M., Semnani S., Wikstrom A. K., et al. (2017). Opium use during pregnancy and risk of preterm delivery: A population-based cohort study. PLoS One 12 (4), e0176588. 10.1371/journal.pone.0176588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathai S., Cutfield W. S., Derraik J. G., Dalziel S. R., Harding J. E., Robinson E., et al. (2012). Insulin sensitivity and beta-cell function in adults born preterm and their children. Diabetes 61 (10), 2479–2483. 10.2337/db11-1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memet S. (2006). NF-kappaB functions in the nervous system: From development to disease. Biochem. Pharmacol. 72 (9), 1180–1195. 10.1016/j.bcp.2006.09.003 [DOI] [PubMed] [Google Scholar]
- Metpally R. P., Krishnamurthy S., Moran K. M., Weller A. E., Crist R. C., Reiner B. C., et al. (2019). The imperative of clinical and molecular research on neonatal opioid withdrawal syndrome. Mol. Psychiatry 24 (11), 1568–1571. 10.1038/s41380-019-0522-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M. C., Johnston A. M., Crocker A. M., Heil S. H. (2015). Methadone and buprenorphine for opioid dependence during pregnancy: A retrospective cohort study. J. Addict. Med. 9 (2), 81–86. 10.1097/ADM.0000000000000092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran S., Arribas C., Esteller M. (2016). Validation of a DNA methylation microarray for 850,000 CpG sites of the human genome enriched in enhancer sequences. Epigenomics 8 (3), 389–399. 10.2217/epi.15.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouillet J. F., Ouyang Y., Coyne C. B., Sadovsky Y. (2015). MicroRNAs in placental health and disease. Am. J. Obstet. Gynecol. 213 (4), S163–S172. 10.1016/j.ajog.2015.05.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesen A. E., Nielsen L. M., Feddersen S., Erlenwein J., Petzke F., Przemeck M., et al. (2018). Association between genetic polymorphisms and pain sensitivity in patients with hip osteoarthritis. Pain Pract. 18 (5), 587–596. 10.1111/papr.12648 [DOI] [PubMed] [Google Scholar]
- O'Neal-Moffitt G., Pilli J., Kumar S. S., Olcese J. (2014). Genetic deletion of MT₁/MT₂ melatonin receptors enhances murine cognitive and motor performance. Neuroscience 277, 506–521. 10.1016/j.neuroscience.2014.07.018 [DOI] [PubMed] [Google Scholar]
- O'Sullivan S. E. (2016). An update on PPAR activation by cannabinoids. Br. J. Pharmacol. 173 (12), 1899–1910. 10.1111/bph.13497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oei J. L., Xu H. X., Abdel-Latif M. E., Vunnam K., Al-Amry A., Clews S., et al. (2012). Dopamine D2 receptor gene polymorphisms in newborn infants of drug-using women. Arch. Dis. Child. Fetal Neonatal Ed. 97 (3), F193–F198. 10.1136/archdischild-2011-300235 [DOI] [PubMed] [Google Scholar]
- Papi A., Storci G., Guarnieri T., De Carolis S., Bertoni S., Avenia N., et al. (2013). Peroxisome proliferator activated receptor-α/hypoxia inducible factor-1α interplay sustains carbonic anhydrase IX and apoliprotein E expression in breast cancer stem cells. PLoS One 8 (1), e54968. 10.1371/journal.pone.0054968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puvvula P. K. (2019). LncRNAs regulatory networks in cellular senescence. Int. J. Mol. Sci. 20 (11), 2615. 10.3390/ijms20112615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishna U., Albayrak S., Alpay-Savasan Z., Zeb A., Turkoglu O., Sobolewski P., et al. (2016). Genome-wide DNA methylation analysis and epigenetic variations associated with congenital aortic valve stenosis (AVS). PLoS One 11 (5), e0154010. 10.1371/journal.pone.0154010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishna U., Nath S. K., Vishweswaraiah S., Uppala L. V., Forray A., Muvvala S. B., et al. (2021a). Maternal opioid use disorder: Placental transcriptome analysis for neonatal opioid withdrawal syndrome. Genomics 113, 3610–3617. 10.1016/j.ygeno.2021.08.001 [DOI] [PubMed] [Google Scholar]
- Radhakrishna U., Vishweswaraiah S., Uppala L. V., Szymanska M., Macknis J., Kumar S., et al. (2021b). Placental DNA methylation profiles in opioid-exposed pregnancies and associations with the neonatal opioid withdrawal syndrome. Genomics 113 (3), 1127–1135. 10.1016/j.ygeno.2021.03.006 [DOI] [PubMed] [Google Scholar]
- Ross E. J., Graham D. L., Money K. M., Stanwood G. D. (2015). Developmental consequences of fetal exposure to drugs: What we know and what we still must learn. Neuropsychopharmacology 40 (1), 61–87. 10.1038/npp.2014.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenfeld R., Devi L. A. (2007). Receptor heterodimerization leads to a switch in signaling: beta-arrestin2-mediated ERK activation by mu-delta opioid receptor heterodimers. FASEB J. 21 (10), 2455–2465. 10.1096/fj.06-7793com [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saugstad J. A. (2010). MicroRNAs as effectors of brain function with roles in ischemia and injury, neuroprotection, and neurodegeneration. J. Cereb. Blood Flow. Metab. 30 (9), 1564–1576. 10.1038/jcbfm.2010.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen A., Prizant H., Light A., Biswas A., Hayes E., Lee H. J., et al. (2014). Androgens regulate ovarian follicular development by increasing follicle stimulating hormone receptor and microRNA-125b expression. Proc. Natl. Acad. Sci. U. S. A. 111 (8), 3008–3013. 10.1073/pnas.1318978111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S., Cha H. J., Lee E. M., Jung J. H., Lee S. J., Park I. C., et al. (2009). MicroRNAs are significantly influenced by p53 and radiation in HCT116 human colon carcinoma cells. Int. J. Oncol. 34 (6), 1645–1652. 10.3892/ijo_00000295 [DOI] [PubMed] [Google Scholar]
- Siegel E. M., Berglund A. E., Riggs B. M., Eschrich S. A., Putney R. M., Ajidahun A. O., et al. (2014). Expanding epigenomics to archived FFPE tissues: An evaluation of DNA repair methodologies. Cancer Epidemiol. Biomarkers Prev. 23 (12), 2622–2631. 10.1158/1055-9965.EPI-14-0464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sliwinska A., Kasinska M. A., Drzewoski J. (2017). MicroRNAs and metabolic disorders - where are we heading? Arch. Med. Sci. 13 (4), 885–896. 10.5114/aoms.2017.65229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tehler D., Hoyland-Kroghsbo N. M., Lund A. H. (2011). The miR-10 microRNA precursor family. RNA Biol. 8 (5), 728–734. 10.4161/rna.8.5.16324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turu G., Varnai P., Gyombolai P., Szidonya L., Offertaler L., Bagdy G., et al. (2009). Paracrine transactivation of the CB1 cannabinoid receptor by AT1 angiotensin and other Gq/11 protein-coupled receptors. J. Biol. Chem. 284 (25), 16914–16921. 10.1074/jbc.M109.003681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdugo R. A., Deschepper C. F., Munoz G., Pomp D., Churchill G. A. (2009). Importance of randomization in microarray experimental designs with Illumina platforms. Nucleic Acids Res. 37 (17), 5610–5618. 10.1093/nar/gkp573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishweswaraiah S., Swierkowska J., Ratnamala U., Mishra N. K., Guda C., Chettiar S. S., et al. (2019). Epigenetically dysregulated genes and pathways implicated in the pathogenesis of non-syndromic high myopia. Sci. Rep. 9 (1), 4145. 10.1038/s41598-019-40299-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachman E. M., Hayes M. J., Brown M. S., Paul J., Harvey-Wilkes K., Terrin N., et al. (2013). Association of OPRM1 and COMT single-nucleotide polymorphisms with hospital length of stay and treatment of neonatal abstinence syndrome. JAMA 309 (17), 1821–1827. 10.1001/jama.2013.3411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachman E. M., Hayes M. J., Sherva R., Brown M. S., Davis J. M., Farrer L. A., et al. (2015). Variations in opioid receptor genes in neonatal abstinence syndrome. Drug Alcohol Depend. 155, 253–259. 10.1016/j.drugalcdep.2015.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachman E. M., Hayes M. J., Sherva R., Brown M. S., Shrestha H., Logan B. A., et al. (2017). Association of maternal and infant variants in PNOC and COMT genes with neonatal abstinence syndrome severity. Am. J. Addict. 26 (1), 42–49. 10.1111/ajad.12483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachman E. M., Hayes M. J., Shrestha H., Nikita F. N. U., Nolin A., Hoyo L., et al. (2018). Epigenetic variation in OPRM1 gene in opioid-exposed mother-infant dyads. Genes. Brain Behav. 17 (7), e12476. 10.1111/gbb.12476 [DOI] [PubMed] [Google Scholar]
- Wang J., Zhao L., Peng X., Liu K., Zhang C., Chen X., et al. (2020). Evaluation of miR-130 family members as circulating biomarkers for the diagnosis of bladder cancer. J. Clin. Lab. Anal. 34 (12), e23517. 10.1002/jcla.23517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. (2016). Improving microRNA target prediction by modeling with unambiguously identified microRNA-target pairs from CLIP-ligation studies. Bioinformatics 32 (9), 1316–1322. 10.1093/bioinformatics/btw002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm-Benartzi C. S., Koestler D. C., Karagas M. R., Flanagan J. M., Christensen B. C., Kelsey K. T., et al. (2013). Review of processing and analysis methods for DNA methylation array data. Br. J. Cancer 109 (6), 1394–1402. 10.1038/bjc.2013.496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., So J., Davis-Dusenbery B. N., Qi H. H., Bloch D. B., Shi Y., et al. (2011). Hypoxia potentiates microRNA-mediated gene silencing through posttranslational modification of Argonaute2. Mol. Cell. Biol. 31 (23), 4760–4774. 10.1128/MCB.05776-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Jackson C. W., Khoury N., Escobar I., Perez-Pinzon M. A. (2018). Brain SIRT1 mediates metabolic homeostasis and neuroprotection. Front. Endocrinol. (Lausanne) 9, 702. 10.3389/fendo.2018.00702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Jing X., Wang G. (2016). MicroRNAs as regulators of drug abuse and immunity. Cent. Eur. J. Immunol. 41 (4), 426–434. 10.5114/ceji.2016.65142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Muralimanoharan S., Wortman A. C., Mendelson C. R. (2016). Primate-specific miR-515 family members inhibit key genes in human trophoblast differentiation and are upregulated in preeclampsia. Proc. Natl. Acad. Sci. U. S. A. 113 (45), E7069–E7076. 10.1073/pnas.1607849113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Tang C., Yu T., Zhang R., Zheng H., Yan W. (2017). MicroRNAs control mRNA fate by compartmentalization based on 3' UTR length in male germ cells. Genome Biol. 18 (1), 105. 10.1186/s13059-017-1243-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X., Li N., Liang S., Huang Q., Coukos G., Zhang L. (2010). Identification of microRNAs regulating reprogramming factor LIN28 in embryonic stem cells and cancer cells. J. Biol. Chem. 285 (53), 41961–41971. 10.1074/jbc.M110.169607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S. S., Jin J. P., Wang J. Q., Zhang Z. G., Freedman J. H., Zheng Y., et al. (2018). miRNAS in cardiovascular diseases: potential biomarkers, therapeutic targets and challenges. Acta Pharmacol. Sin. 39 (7), 1073–1084. 10.1038/aps.2018.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.