Abstract
After spinal cord injury (SCI), use chronic urinary catheters for bladder management is common, making these patients especially vulnerable to catheter-associated complications. Chronic catheterization is associated with bacterial colonization and frequent catheter-associated urinary tract infections (CAUTI). One determinant of infection success and treatment resistance is production of catheter-associated biofilms, composed of microorganisms and host- and microbial-derived components. To better understand the biofilm microenvironment, we performed proteomics analysis of catheter-associated biofilms and paired urine samples from four people with SCI with chronic indwelling urinary catheters. We developed a novel method for the removal of adhered cellular components on catheters that contained both human and microbial homologous proteins. Proteins from seven microbial species were identified including: Escherichia coli, Klebsiella species (spp), Enterococcus spp, Proteus mirabilis, Pseudomonas spp, Staphylococcus spp, and Candida spp. Peptides identified from catheter biofilms were assigned to 4,820 unique proteins, with 61% of proteins assigned to the biofilm-associated microorganisms, while the remainder were human-derived. Contrastingly, in urine, only 51% were assigned to biofilm-associated microorganisms and 4,554 proteins were identified as a human-derived. Of the proteins assigned to microorganisms in the biofilm and paired urine, Enterococcus, Candida spp, and P. mirabilis had greater associations with the biofilm phase, whereas E. coli and Klebsiella had greater associations with the urine phase, thus demonstrating a significant difference between the urine and adhered microbial communities. The microbial proteins that differed significantly between the biofilm and paired urine samples mapped to pathways associated with amino acid synthesis, likely related to adaptation to high urea concentrations in the urine, and growth and protein synthesis in bacteria in the biofilm. Human proteins demonstrated enrichment for immune response in the catheter-associated biofilm. Proteomic analysis of catheter-associated biofilms and paired urine samples has the potential to provide detailed information on host and bacterial responses to chronic indwelling urinary catheters and could be useful for understanding complications of chronic indwelling catheters including CAUTIs, urinary stones, and catheter blockages.
Keywords: Catheter, biofilm, urine, proteomics, microbial proteins, spinal cord injury
Introduction
Catheter-associated urinary tract infections (CAUTIs) account for up to 40% of all nosocomial infections and are therefore the most common cause of hospital-acquired infections [1]. While duration of catheterization is the most important determinant of bacteriuria [2], the presence of a catheter also perturbs host defenses, provides direct access of pathogens to the bladder, and like all foreign bodies in the urinary tract, incurs biofilm formation [3]. These catheter-associated biofilms are complex organic microenvironments consisting of microorganisms growing colonies within an extracellular mucopolysaccharide matrix comprised of host and microbial proteins and urinary solutes, including magnesium and calcium ions. The mechanisms driving the development of these biofilms result from a combination of factors such as the catheter’s physical and chemical properties as well as its insertion into the body. For example, catheter surfaces have been shown to have disordered and pitted topographies as well as surface charges that facilitate bacterial adhesion [4,5]. These characteristics, combined with the constant supply of nutrients from urine in an otherwise nutrient-poor environment, enhance key regulatory pathways involving interbacterial interactions, nutrient scavenging, and appendage attachment to surfaces. Further aiding this process is the placement of the urinary catheter [6,7] (Flores-Mirales, A. et al. 2015, Nat Rev Microbiol; Pelling, H. et al. 2019, Appl Microbiol Lett). Following implantation, the bladder is often irritated, resulting in inflammation and the release of host fibrinogen. The fibrinogen adheres to the catheter and subsequently provides a niche food source for uropathogens (e.g. Enterococcus faecalis) as well as offers pili-mediated binding sites [6] (Flores-Mirales, A. et al. 2015, Nat Rev Microbiol). Escherichia coli is the most common infectious microorganism detected in catheter-associated biofilms [2]; followed by other frequently isolated genera such as, Enterobacteriaceae, Enterococci spp, coagulase-negative Staphylococcus, Pseudomonas aeruginosa, and non-fermenter Candida spp [8]. Within catheter-associated biofilms, crystalline biofilms are of particular importance due to the common clinical complications that they are associated with during long-term urethral catheterization. Crystalline biofilms are produced by microorganisms equipped with ureases, which are capable of hydrolyzing urea into carbon dioxide and ammonia, thus increasing the pH of the urine. Ultimately, this urine alkalization generates calcium crystals and magnesium precipitates that block the catheter lumen and outflow as well as protects the microbial community against host immune responses. For example, urea-splitting bacteria such as Proteus mirabilis have unique importance in individuals with chronic catheterization because of their association with urinary stone formation, a difficult to manage catheter complication [9].
Between 20% and 50% of people with spinal cord injury (SCI) use a chronic indwelling urinary catheter for bladder management [10-12], making these patients especially vulnerable to CAUTIs and other complications such as stone formation, and catheter obstruction due to proteinaceous and mineral encrustation. Approximately half of patients with chronic indwelling catheters experience catheter blockage at some time, while some patients experience recurrent obstruction [13,14]. Additionally, recent attention has been brought to the fact that current standard methods for detecting UTIs rely on urine samples, which poses a significant challenge because the dominant microbial species existing in a planktonic state often differ from those found in a biofilm [15,16]. Relying solely on urine analyses presents a bias towards planktonically growing cells compared to adhered. This is not ideal because catheter-associated UTIs are often preceded by biofilm formation which includes predominantly attached cells rather than planktonic [17]. Therefore, it is advantageous to understand the most prevailing genera and species that make up each of these communities to ultimately assist in the diagnosis and reduction of CAUTI infections.
Given the frequency of catheter-related complications in people with SCI, we sought to gain a deeper understanding of the catheter biofilm- and urine-associated features that might underlie these pathologies. We chose to analyze paired urine and catheter biofilm specimens using a novel catheter-associated extraction method in conjunction with shotgun proteomics to evaluate host and bacterial features in urine as well as biofilm of chronically catheterized patients.
Materials and methods
Clinical sample collection
Participants in this study had SCI and accompanying neurogenic bladder requiring management with chronic indwelling urinary Foley (14 Fr) catheters. Subjects were recruited from October 2018 until April 2019 from the SCI/D inpatient or outpatient service. They were initially identified by their attending physician and if aggregable in principle to participate were approached by the research personal for formal enrollment in the study. All participants answered a short survey about catheter-related complications and participant demographics were obtained via chart review. All patients signed an informed consent and HIPAA consent for use of their data and catheter samples under an approved Veterans Affair Office of Research and Development and Stanford University IRB research protocol (protocol number 44986). Inclusion criteria for participants included those diagnosed with SCI, used a chronic indwelling catheter with a history of blockages, and were 18 years of age or older. Individuals were excluded if diagnosed with multiple sclerosis (MS) or amyotrophic lateral sclerosis (ALS), bladder cancer, received antibiotics treatment for a UTI, were intermittently catheterized for bladder management, or did not use an indwelling catheter for bladder management. Catheters were routinely exchanged using sterile technique every four weeks unless there was a catheter blockage event, in which case a catheter exchange was performed at that time. Catheters were collected at the time 4-week exchange event along with accompanying urine samples that were collected after the new catheter was placed. Approximately the distal three inches of the catheter were cut off and placed directly into a sterile container as soon as the catheter was removed from the patient. Catheter and urine specimens were stored at -20°C until analysis.
Catheter and urine sample preparation
We performed shotgun proteomics on urinary catheter biofilms and paired urine samples from four people with SCI using a label-free proteomics approach. Proteins were removed by modifying two previously reported extraction protocols [18,19] and optimizing them for catheter associated biofilms. Specifically, 0.5 inch catheter segments, void of urine, were incubated in 4 mL of 2% sodium dodecyl sulfate solution (SDS, Thermo Fisher Scientific) (% weight/volume) containing 1X protease inhibitors (Sigma-Aldrich) and sonicated with a Branson probe sonicator (Fisher Scientific) for 30 minutes at a frequency of 40 kHz. The extracted protein material was then sonicated to shear DNA and RNA for 3 cycles of 15 seconds with the amplitude set to 40%. The extracted proteins were quantified by using a Bicinchoninic acid (BCA) protein assay (Thermo Scientific). For each sample, a 25 µg aliquot of protein was prepared for shotgun proteomic analysis by reducing disulfide bonds on cysteines with 5 µL of 200 mM Tris (2-carboxyethyl) phosphine (TCEP) (Sigma-Aldrich) in a 100 µL sample volume and incubated at 65°C for 1.5 hours. Then, free sulfhydryl groups were capped with 7.5 µL 200 mM iodoacetamide (Acros Organics) and incubated for 45 minutes at room temperature in the dark. Proteins were precipitated with 1 mL of cold acetone (Fisher Scientific) and stored at -20°C overnight. The precipitated proteins were pelleted by centrifuging samples at 14,000 g for 10 minutes at 4°C, acetone was removed, and pelleted proteins were left to dry down for 5 minutes at room temperature. Proteins were digested with 1 µg of sequencing grade modified trypsin enzyme (Thermo Fisher Scientific) in 50 µL of 50 mM ammonium bicarbonate digestion buffer and incubated at 37°C overnight. The resulting tryptic peptides were dried down using a speed vacuum and reconstituted in 50 µL of 0.1% formic acid (Fisher Scientific) in HPLC MS grade water (Fisher Scientific) for LC/MS analysis.
For paired urine analyses, samples were aspirated from patient catheters after removal. 15 mL of urine was concentrated 10-fold to 1.5 mL by using a 4 mL Amicon centrifugal filter (Sigma Aldrich). A 100 µL aliquot of the concentrated urine was used to performed shotgun proteomics following a mini S-trap protocol provided by the manufacturer (Protifi). Briefly, 100 µL of 10% SDS solution was added to each sample and sonicated with a probe sonicator using the same conditions described above. Extracted proteins were reduced with 10 µL of 200 mM TCEP solution, incubated for 1.5 hours at room temperature and alkylated with 15 µL of 200 mM iodoacetamide in the dark at room temperature for 45 minutes. The lysate was then acidified with 20 µL of 12% phosphoric acid, diluted 7-fold using S-trap binding buffer consisting of 90% methanol and 100 mM Triethylammonium bicarbonate (TEAB), and loaded onto the S-trap column. Following a series of washes, proteins were digested with 2 µg of trypsin in 50 mM ammonium bicarbonate buffer and incubated overnight at 37°C. Peptides were eluted with 80 µL of 0.2% formic acid followed by a second 80 µL aliquot of 0.2% formic acid in 50% acetonitrile. Samples were dried down using a speed vacuum and then dissolved in 50 µL of 0.1% formic acid in water for LC/MS analysis.
For proteomic analysis of non-catheter-derived urine samples, three pools of normal human urine were purchased from Innovative Research. 5 mL of each pool was centrifuged at 3000 g for 5 min and then concentrated using an Amicon filter. Ammonium bicarbonate was then added to each aliquot and subsequent reduction, alkylation, and trypsin digestion was performed as described above.
LC/MS analysis
A Dionex Ultimate Rapid Separation Liquid Chromatography system (Thermo Fisher Scientific) was used to load 3 µL of the reconstituted tryptic peptides onto a C18 trap column (Thermo Fisher Scientific) with the flow rate set at 5 µL/min for 10 minutes. Tryptic peptides were separated by reversed-phase chromatography on a 25 cm C18 analytical (New Objective) packed with Magic C18 AQ resin (Michrom Bioresources). Peptides were eluted by changing the mixture of mobile phase A (0.1% formic acid in water) and mobile phase B (0.1% formic acid in acetonitrile). The gradient program entailed holding mobile phase B at 2% for the first 10 minutes, followed by a gradual ramp to 35% over the next 100 minutes, and then an increase to 85% over 7 minutes with a 5 minute hold. The analytical column was re-equilibrated for 10 minutes prior to each sample injection. The flow rate throughout the gradient was set to 0.5 µL/min, and each sample was analyzed in triplicate. Eluted peptides were analyzed using a LTQ Orbitrap Elite mass spectrometer (Thermo Fisher Scientific). The top 10 most abundant ions per MS1 scan were selected for higher energy collision induced dissociation using a 35 eV voltage in a data-dependent mode. MS1 resolution was set to 60,000, FT AGC target was set at 1e6, and the m/z scan range was set from m/z=400-1800. MS2 AGC target at 3e4 and dynamic exclusion was enabled for 30 seconds.
Peptide identification, quantification, data processing and statistical analysis
For each LC-MS run, the resulting raw data file was searched using Byonic 2.11.0 (Protein Metrics, San Carlos, CA) against the curated Swiss-Prot database of the six most common bacteria attributed to catheter-acquired urinary infections [20] (E. coli: 2018; 23,110 entries, Klebsiella spp.: 2018; 1,808 entries, Enterococcus spp.: 2018; 596 entries, P. mirabilis: 2018; 608 entries, Pseudomonas spp.: 2018; 11,854 entries, and Staphylococcus spp.: 2018; 13,420 entries) as well as the most common causes of fungal infections [21] (Candida spp.: 2018; 1,493 entries and the human reference proteome 2017; 20,496). Database search parameters included trypsin digestion with a maximum of two missed cleavages, the MS1 precursor mass tolerance set to 10 ppm, and the fragment mass tolerance of 0.5 Da. Fixed cysteine carbamidomethylation (+57.021 Da) and variable methionine oxidation (+15.994 Da) and asparagine deamination (+0.984 Da), were also specified. Peptide identifications were filtered to exclude those with a > 1% false discovery rate (FDR) and those identified by less than two spectra. Peptides identified in only one of the two databases were also removed to be conservative. All proteins of each species with non-homologous peptides were analyzed using an in-house R script using the three technical replicates from each experimental condition. Quantitative information was extracted from MS1 spectra of all identified peptides using an in-house R script based on the MSnbase package [22] as the AUC of the extracted ion current (XIC) of all remaining peptides after the alignment of the chromatographic runs. The protein abundances were calculated in two ways. To perform a global comparison between species: protein abundances were calculated in terms of iBAQ, as the log of the sum of the intensities of every identified peptide divided by the number of identifiable peptides, with two missed cleavages and within a m/z range from 400 to 1800 Th. To compare protein levels between samples, the quantitative information was expressed as Z-scores at protein level as described previously [23], in which the log2 ratios, calculated comparing the AUC of the peptides in each sample against the average signal across all the samples, are considered using their corresponding statistical weight, calculated at the spectrum level, according to the WSPP model [23]. Peptide quantities are then rescaled and standardized to a normal distribution N(0,1). The validity of the null hypothesis at each of the levels (spectrum, peptide and protein) was checked by plotting the cumulative distributions. The final statistical comparison was performed using a Student’s-t test.
Gene enrichment analysis and functional treemap
Statistically significant human proteins were used to test for enrichment of biological pathways by performing an overrepresentation analysis using the Reactome pathway annotation database (65 Released 2019-12-22) [24]. Pathways were considered as a significant when they included four or more proteins with a FDR lower than 1% based on Fisher’s Exact test. To characterize the biological functions related to microorganism proteins detected in the biofilm, we annotated the entire list of proteins using TIGRfam [25], and quantified the total contribution of the proteins to main class using the average of iBAQ values. Data available upon request within the limits of approval and oversight from the Department of Veterans Affairs.
Results
Participant data
Four adult men with cervical or upper thoracic level SCI (injury duration 354 days-8 years) who used a chronic indwelling urinary catheter were recruited for the study. Three of the participants used a transurethral Foley catheter and one participant used a suprapubic catheter. Two participants reported a history of frequent (more than once a month) or constant (weekly or more) catheter blockage events while the other two participants reported rare catheter blockage (a few times a year). All four participants reported rarely requiring antibiotic therapy for UTIs. None of the participants had a symptomatic UTI at the time of urine and catheter sample collection.
Metaproteomics analysis of catheter biofilms and urine
Tryptic peptides of catheter-associated biofilms and paired aspirated urine were analyzed using liquid chromatography - tandem mass spectrometry (LC-MS/MS) with three technical replicates per sample. The raw data were searched using Byonic (2.11.0 version) against the curated Swiss-Prot database of the most common bacteria associated with catheter-acquired urinary infections [20]. After a conservative removal of homolog peptides, the spectra acquired from the catheter biofilm and urine samples were assigned to a total of 4,820 and 4,554 proteins, respectively (Figure 1A). Protein levels were quantified based on iBAQ values, demonstrating that many proteins are found at high abundance, and that intermediate and low abundance proteins can also be readily detected with confidence. Additionally, proteins could be distinguished by their source or origin as well as host or microbial (Figure 1A). In the biofilm, there were a higher number of microorganism proteins (2,939) compared to human proteins (1,881). Contrastingly in the aspirated urine, there were a higher number of human proteins (2,114) compared to microorganism proteins (2,440) (Figure 1B, top).
Figure 1.

Summary of Identified Proteins. A. Cumulative protein masses annotated by source of origin expressed in terms of log10 of iBAQ. B. Protein distribution by human (yellow) vs microorganism (purple) populations in catheter biofilms (left) and urine (right). The bottom bar graphs show percentage of proteins assigned to microorganism in catheter and urine. C. Bar graphs of protein mass for individual matched catheterized patient urine and biofilm (catheter) samples, and three urine samples collected from non-catheterized patients, broken down by source of origin.
Quantified metaproteomic data allowed assessment of the relative proportions of the microorganisms in catheter biofilms and demonstrated that they differed from those found in the urine (Figure 1B, bottom). The predominant microorganismal proteins found in the catheter biofilms were E. coli and Klebsiella spp, with 28% and 21% of all protein signal (human plus microorganisms) detected, respectively. In the urine, E. coli and Klebsiella spp only accounted for 17% and 13% or proteins, respectively. Conversely, the most common microbial proteins in urine were Pseudomonas spp and Candida spp with 30% and 13% of the total protein signal, respectively, compared to only 23%, and 4% in the catheter biofilms. Furthermore, of the percentage of proteins assigned to microorganisms in the biofilm and paired urine, we identified protein masses associated individual microbial species for each patient (Figure 1B, bottom), showing a low variability between the four patients analyzing meaning a high consistency of the microorganism’s populations detected. Finally, we tested three pools of urine samples collected from individuals without catheterization, showing that human protein content accounts to up 97% of the identified peptides and more than 90% of the total MS signal detected (Figure 1C).
Differences between urine and catheter biofilm microorganism proteins
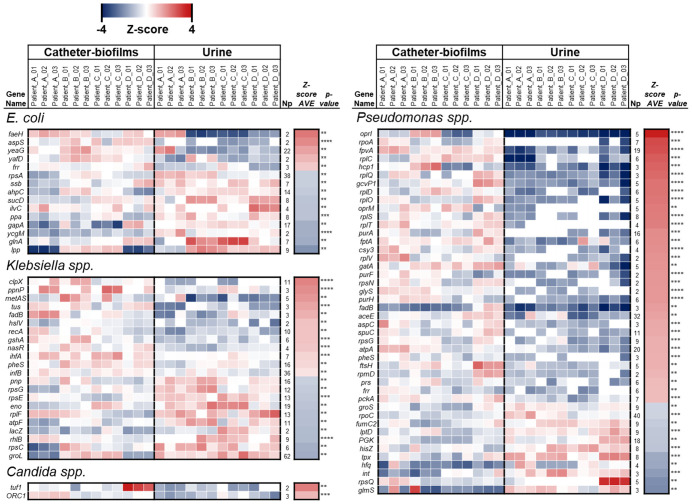
To understand the differences observed between protein profiles in the urine and catheter-associated biofilms, we analyzed the relative quantification of the non-homologous peptides from the four microorganism species whose proportions changed most significantly: E. coli, Klebsiella spp, Pseudomonas spp, and Candida spp. Across the four individual patient samples, we were able to detect 15 proteins belonging to E. coli, 22 proteins from Klebsiella spp, 2 proteins from Candida spp, and 44 proteins from Pseudomonas spp that could be quantified in at least 80% of the technical and biological replicates, were identified with more than one peptide, and were significantly different between urine and biofilm samples (P < 0.01 based of Student’s-t test) (Figure 2).
Figure 2.
Heat map of microorganism proteins that differ significantly between urine and catheter biofilms (P < 0.01, Np > 1). The intensity of the color represents the concentration change (log2) from -4 (blue) to 4 (red), determined by label-free quantification.
In the proteins derived from Candida spp, we detected decreased levels of Tuf1 and ORC1 in the paired urine samples compared to the corresponding catheter biofilms. Tuf1 is a translation elongation factor whose levels decrease in stationary phase cells [27]. Similarly, ORC1, a component of the origin recognition complex (ORC) that binds origins of replication and is critical to initiation of the cell cycle, was also significantly decreased in urine samples. ORC1 has a role in both chromosomal replication and mating type transcription silencing by similarity of yeast (S. cerevisiae) Orc1p.
Based on these interesting findings in Candida spp, we performed an ortholog annotation of all proteins in Figure 2 from E. Coli, Klebsiella spp, and Pseudomonas spp into TIGRfam [25] and quantified the total contribution of every protein to Main Class using the average of iBAQ values (Figure 3). Based on this analysis, we observed significant differences in several pathways in the bacterial species. In E. coli, the protein signatures suggested a large increase in amino acid biosynthesis in microorganisms in the urine compared to the same bacteria in catheter biofilms (Figure 3). Notably, we observed that the glutamine synthetase enzyme, glnA, levels were significantly increased in the urine compared to catheter biofilms. A treemap of E. coli proteins found in catheter biofilms compared to urine shows alterations in numerous pathways. The biofilm proteins show enrichment for energy metabolism, protein synthesis, regulatory functions, transcription, and purine/pyrimidine nucleoside biosynthesis (Figure 3).
Figure 3.

Treemap of orthogonal protein classification according to TIGRFAM of the three populations of microorganisms that differ most between paired catheter (left) and urine (right) specimens based on protein mass.
For Klebsiella spp, the treemap (Figure 3) comparing catheter biofilm and urine differed significantly from that of E. coli in that fewer bacterial pathways differed significantly between the urine and catheter-associated biofilm. The biological functions for Klebsiella spp enriched in the urine included energy metabolism, and protein synthesis and are consistent with a sustained growth of Klebsiella spp. in urine. We also observed increased urinary levels Klebsiella spp groL (Figure 2), which prevents misfolding and promotes the refolding and proper assembly of unfolded polypeptides generated under stress conditions. In catheter biofilms, there was enrichment for proteins related with protein synthesis and DNA metabolism. Interestingly, catheter biofilms showed high levels of fadB (Figure 2), fatty acid oxidation complex subunit alpha. This enzyme is involved in the aerobic and anaerobic degradation of long-chain fatty acids via the beta-oxidation cycle and suggests that Klebsiella spp could use fatty acids as an energy source in the biofilm, and it was also reported in other infective microenviroments such as lung surfactant biofilms [28].
For Pseudomonas spp, we observed enrichment of proteins related with protein synthesis in catheter biofilms compared to urine (Figure 3), including the ribosomal proteins rplO, rplS, rplV, rpsG, and rpmD (Figure 2). Like Klebsiella spp, there were fewer pathways that differed between urine and the catheter biofilm, and these were mostly observed in the catheter biofilm and showed increased protein biosynthetic pathway activation. As in E. coli, there were increases in amino acid biosynthesis pathways in the urine compared to the biofilm. For example, in the urine, high levels of hisZ were observed, an enzyme required for the first step of histidine biosynthesis. In addition, urinary proteins showed high levels of Pseudomonas spp glmS, which catalyzes the first step in hexosamine metabolism, a key process in N-linked protein glycosylation that initiates bacterial cell envelope biosynthesis [29].
Human proteins and pathways in urine and catheter biofilms
Combining the human derived proteins from the catheter biofilms (1,881 unique proteins) and urine samples (2,114 proteins), we were able to quantify 2,801 unique human proteins. After removing homologs, we identified 84 proteins that differed significantly between catheter biofilms and urine using similar criteria to those used for bacterial proteins; namely, present in at least 80% of the MS analyses, identified with two or more peptides, significant (P < 0.01) based on Student’s T test (Figure 4A, 4B). Interestingly, the majority of the human proteins with different levels between sample types were identified in the catheter biofilms, rather than in the urine. An overrepresentation analysis using the Reactome pathway annotation database (65 Released 2019-12-22) [24], identified nine functional categories of proteins (FDR < 0.01), each with five or more proteins (Figure 4C). From highest to lowest enrichment, these pathways include: RUNX regulated genes involved in differentiation of hematopoietic stem cells (HSCs), antigen processing-cross presentation, apoptosis, platelet degranulation, response to elevated platelet cytosolic Ca2+, neutrophil degranulation, M phase, innate immune system, and cellular responses to stress. These proteins characterize several types of host response to the catheter, as a foreign body in the urinary tract, as well as host response to the accompanying bacterial colonization/infection.
Figure 4.
Proteins with significant differences in levels between catheter-associated biofilms and paired urine samples. (A) Heatmap of human proteins (P < 0.01, Np > 1) with higher levels in catheter-associated biofilms compared to urine, and (B) higher levels in urine compared to catheter-associated biofilms. (C) Overrepresentation analysis demonstrating functional pathways according to Panther Pathways. Significant Panther Pathways (FDR < 0.05), according to Fisher’s Exact test and corrected using FDR with four or more proteins.
Discussion
To better understand the bacterial and host factors associated with chronic indwelling catheters, we applied a label-free shotgun proteomics to paired catheter-associated biofilms and urine samples in four people with SCI. We were able to provide a comprehensive catalogue of both human and microbial protein signatures with 4,820 unique proteins found in the catheter biofilm and 4,554 proteins found in the urine samples. Specifically, in the catheter biofilm, 61% of the proteins (2,940) were assigned to biofilm-associated microorganisms, whereas in the urine, only 51% of the proteins (2,440) were annotated as a biofilm-associated, thereby indicating a greater number of human proteins in the urine compared to biofilm (Figure 1B). These results align with a study that utilized metaproteomics to elucidate host and pathogen protein expression during CAUTIs from suprapubic catheters [30]. They similarly found a greater number of unique proteins associated with the biofilm (1,064) compared to urine (896) and that the biofilm had a larger number of microbial-associated proteins (612) compared to urine (387). While these results are in agreement with our work, the studies differ in that only 2 cm of the distal portion of suprapubic catheter tips were analyzed in patients with asymptomatic UTIs, in contrast to this study, which analyzed 3-inches of the catheter tip in patients with suprapubic as well as urethral catheters with no UTI symptoms. Additionally, our results not only aligned with catheter associated biofilms, but also oral biofilms. For example, a study that evaluated host-biofilm interactions in an oral infection model using proteomic profiling, found a greater concentration of microbial-associated proteins in oral biofilms compared to human proteins. Likewise, they found that more human-derived proteins resided in gingival tissue alone than when present with a biofilm [31]. Establishing the distribution of human and microbial proteins within biofilms and urine enhances our ability to know where these cellular components localize as well as aids the identification of microbial hotspots.
Bioinformatics analysis allowed species-specific mapping to identify seven different microorganisms with high confidence. By quantifying protein levels, we were able to determine the relative contribution of microorganisms and the hosts to the proteins in the urine and biofilm, as well as perform pathway analysis to understand the state of the organisms and the nature of the host response. In addition, comparison of the biofilm and paired urine protein outputs identified pathways differentially active in the urine and biofilm and could provide insights into selective pressures in each of these microenvironments. Our approach demonstrated the feasibility of in-depth proteomics analysis, revealing the diversity of proteins in the biofilm and allowing classification of functional pathways in the host and microorganisms underlying biofilm composition.
Comparison of biofilm and urinary protein reveals differences in pathways that might suggest mechanisms activated by bacteria to facilitate survival in the different environments. For example, both E. coli and Pseudomonas spp. showed an enrichment for amino acid biosynthesis when present in the urine compared to catheter biofilm. This is of particular interest because although nitrogen is present at high concentrations in urine in form of urea, E. coli and most Pseudomonas spp, lack the urea-splitting enzyme, urease, thus restricting their ability to access urine as a nitrogen source. Furthermore, the urine showed high levels of glmS, which a key enzyme in the synthesis of the bacterial cell envelope [29], an observation consistent with the inability of the bacterial species to use urea as a source of nitrogen [29]. In conditions of low bioavailable nitrogen and amino acids, such as in urine, microorganisms activate amino acid synthesis pathways as a survival strategy. As seen in our results, amino acid biosynthesis was significantly enriched in the urine samples along with the activation of nitrogen-regulated genes such as glnA in E. coli [32] and hisZ in Pseudomonas spp. This result was also observed in a study that tracked global gene expression profiling of E. coli during biofilm growth in human urine [33]. Similar to our work, genes associated with amino acid biosynthesis, cell membrane biogenesis, and carbohydrate metabolism were down regulated in E. coli strains during biofilm growth. In biofilms, these enzymes are expressed at much lower levels, and the change likely represents a more favorable microenvironment where the cells can shift their metabolic pathways from amino acid synthesis to protein production in order to support synthesis of biofilm matrix proteins.
Comprehensive classification of the proteins in catheter biofilms has been used to a limited extent. Comprehensive analyses of biofilm and urine proteomes using LC-MS/MS approaches and metaproteomics have been carried out on small numbers of patient samples [30,34,35]. In a recent study by Yu et al., profiles of nine patient samples revealed similar numbers of host and bacterial proteins as observed in our study, as well as the presence of multiple microbial species in biofilms. In addition, analysis of the proteomic data revealed several important factors in bacterial success, including modulators of transitional metal ions, persistence of multiple bacterial species, mixed acid fermentation in resident bacteria, adaptations to a hypoxic milieu and epithelial adherence pathways [36]. With improvements in technology as well as development of comprehensive datasets for microorganismal and human proteins, as well as new analytic approaches that facilitate blending of these datasets and parsing data, there are opportunities for in-depth classification of catheter-associated biofilms. In addition, comparison of paired biofilm and urine samples could generate insights and testable hypothesis for understanding the selective pressures and adaptive responses of both the microorganisms and host to growth in these environments. This could provide insights into the role biofilms in bacterial persistence, infections, and clinical complications such as CAUTI, catheter encrustation, and catheter plugging.
Our study has shortcomings that should be noted. The small sample size of the study limits the conclusions that can be drawn regarding bacterial adaptation and host responses. However, the study does demonstrate that large amounts of meaningful data can be generated that allows for robust pathway analysis. Our study was limited to available databases for seven available microbial species, and we therefore could not characterize the full microbial diversity. As microbial protein databases expand and improve, more comprehensive analyses will be possible. Since this was a proof-of-concept study, we also did not perform validation of microbial type with other approaches, such as 16S RNA sequencing, nor did we attempt to validate individual protein expression by another means. In future studies, critical pathways and microbe identity will need to be confirmed before firm conclusions can be made about functional aspects of the microenvironment, bacterial adaptions, or clinical utility of proteins, such as for predictive biomarkers.
Despite these shortcomings, we established a method for comprehensive analysis of catheter-associated biofilms using shotgun proteomics. This method can be applied to study catheter-associated complications in patients with catheter blockages due to mucoid or crystalline biofilm as well as provides a basis for investigating catheter-associated urinary tract infections. Future directions include deploying this method in discrete cohorts of patients with these catheter-based complications.
Conclusion
In summary, proteomic analysis of paired urine and biofilms in people with SCI with chronic indwelling urinary catheters revealed the complexity of bacterial and host proteins that comprise catheter biofilms and demonstrate the ability of LC-MS/MS to characterize these factors, including bacterial speciation, and quantification. In addition, analysis of protein pathways that differ between the urine and biofilm proteome can provide insights into the bacterial adaptions to these particular environments. Complex proteomic analysis could be applied to diverse patients with discrete catheter-associated complications including UTI, encrustation, urinary tract stones, and obstruction to understand the factors underlying these important clinical events and provide biomarkers of risk for their occurrence. Improved understanding could facilitate development of new approaches for preventing and treating these complications.
Acknowledgements
Supported by T32DK007357 to ALP and in part by U54 DK130065 to JDB. Unfunded research support from the Department of Veterans Affairs.
Disclosure of conflict of interest
None.
References
- 1.Magill SS, Edwards JR, Bamberg W, Beldavs ZG, Dumyati G, Kainer MA, Lynfield R, Maloney M, McAllister-Hollod L, Nadle J, Ray SM, Thompson DL, Wilson LE, Fridkin SK Emerging Infections Program Healthcare-Associated Infections and Antimicrobial Use Prevalence Survey Team. Multistate point-prevalence survey of health care-associated infections. N Engl J Med. 2014;370:1198–1208. doi: 10.1056/NEJMoa1306801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hooton TM, Bradley SF, Cardenas DD, Colgan R, Geerlings SE, Rice JC, Saint S, Schaeffer AJ, Tambayh PA, Tenke P, Nicolle LE Infectious Diseases Society of America. Diagnosis, prevention, and treatment of catheter-associated urinary tract infection in adults: 2009 International Clinical Practice Guidelines from the Infectious Diseases Society of America. Clin Infect Dis. 2010;50:625–663. doi: 10.1086/650482. [DOI] [PubMed] [Google Scholar]
- 3.Stickler DJ. Bacterial biofilms in patients with indwelling urinary catheters. Nat Clin Pract Urol. 2008;5:598–608. doi: 10.1038/ncpuro1231. [DOI] [PubMed] [Google Scholar]
- 4.Wilks SA, Koerfer VV, Prieto JA, Fader M, Keevil CW. Biofilm development on urinary catheters promotes the appearance of viable but nonculturable bacteria. mBio. 2021;12:e03584-20. doi: 10.1128/mBio.03584-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng S, Bawazir M, Dhall A, Kim HE, He L, Heo J, Hwang G. Implication of surface properties, bacterial motility, and hydrodynamic conditions on bacterial surface sensing and their initial adhesion. Front Bioeng Biotechnol. 2021;9:643722. doi: 10.3389/fbioe.2021.643722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol. 2015;13:269–284. doi: 10.1038/nrmicro3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pelling H, Nzakizwanayo J, Milo S, Denham EL, MacFarlane WM, Bock LJ, Sutton JM, Jones BV. Bacterial biofilm formation on indwelling urethral catheters. Lett Appl Microbiol. 2019;68:277–293. doi: 10.1111/lam.13144. [DOI] [PubMed] [Google Scholar]
- 8.Nicolle LE. Urinary catheter-associated infections. Infect Dis Clin North Am. 2012;26:13–27. doi: 10.1016/j.idc.2011.09.009. [DOI] [PubMed] [Google Scholar]
- 9.Matsukawa M, Kunishima Y, Takahashi S, Takeyama K, Tsukamoto T. Bacterial colonization on intraluminal surface of urethral catheter. Urology. 2005;65:440–444. doi: 10.1016/j.urology.2004.10.065. [DOI] [PubMed] [Google Scholar]
- 10.Weld KJ, Dmochowski RR. Effect of bladder management on urological complications in spinal cord injured patients. J Urol. 2000;163:768–772. [PubMed] [Google Scholar]
- 11.Singh R, Rohilla RK, Sangwan K, Siwach R, Magu NK, Sangwan SS. Bladder management methods and urological complications in spinal cord injury patients. Indian J Orthop. 2011;45:141–147. doi: 10.4103/0019-5413.77134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sekar P, Wallace DD, Waites KB, DeVivo MJ, Lloyd LK, Stover SL, Dubovsky EV. Comparison of long-term renal function after spinal cord injury using different urinary management methods. Arch Phys Med Rehabil. 1997;78:992–997. doi: 10.1016/s0003-9993(97)90063-0. [DOI] [PubMed] [Google Scholar]
- 13.Kohler-Ockmore J, Feneley RC. Long-term catheterization of the bladder: prevalence and morbidity. Br J Urol. 1996;77:347–351. doi: 10.1046/j.1464-410x.1996.09074.x. [DOI] [PubMed] [Google Scholar]
- 14.Getliffe KA. The characteristics and management of patients with recurrent blockage of long-term urinary catheters. J Adv Nurs. 1994;20:140–149. doi: 10.1046/j.1365-2648.1994.20010140.x. [DOI] [PubMed] [Google Scholar]
- 15.Polasko AL, Ramos P, Kaner RB, Mahendra S. A multipronged approach for systematic in vitro quantification of catheter-associated biofilms. J Hazard Mater Lett. 2021;2:100032. [Google Scholar]
- 16.Hola V, Ruzicka F, Horka M. Microbial diversity in biofilm infections of the urinary tract with the use of sonication techniques. FEMS Immunol Med Microbiol. 2010;59:525–528. doi: 10.1111/j.1574-695X.2010.00703.x. [DOI] [PubMed] [Google Scholar]
- 17.Percival SL, Suleman L, Vuotto C, Donelli G. Healthcare-associated infections, medical devices and biofilms: risk, tolerance and control. J Med Microbiol. 2015;64:323–334. doi: 10.1099/jmm.0.000032. [DOI] [PubMed] [Google Scholar]
- 18.Nguyen BVG, Nagakubo T, Toyofuku M, Nomura N, Utada AS. Synergy between sophorolipid biosurfactant and SDS increases the efficiency of P. aeruginosa biofilm disruption. Langmuir. 2020;36:6411–6420. doi: 10.1021/acs.langmuir.0c00643. [DOI] [PubMed] [Google Scholar]
- 19.Mandakhalikar KD, Rahmat JN, Chiong E, Neoh KG, Shen L, Tambyah PA. Extraction and quantification of biofilm bacteria: method optimized for urinary catheters. Sci Rep. 2018;8:8069. doi: 10.1038/s41598-018-26342-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nicolle LE. Catheter associated urinary tract infections. Antimicrob Resist Infect Control. 2014;3:23. doi: 10.1186/2047-2994-3-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bagshaw SM, Laupland KB. Epidemiology of intensive care unit-acquired urinary tract infections. Curr Opin Infect Dis. 2006;19:67–71. doi: 10.1097/01.qco.0000200292.37909.e0. [DOI] [PubMed] [Google Scholar]
- 22.Gatto L, Lilley KS. MSnbase-an R/Bioconductor package for isobaric tagged mass spectrometry data visualization, processing and quantitation. Bioinformatics. 2012;28:288–289. doi: 10.1093/bioinformatics/btr645. [DOI] [PubMed] [Google Scholar]
- 23.Navarro P, Trevisan-Herraz M, Bonzon-Kulichenko E, Nunez E, Martinez-Acedo P, Perez-Hernandez D, Jorge I, Mesa R, Calvo E, Carrascal M, Hernaez ML, Garcia F, Barcena JA, Ashman K, Abian J, Gil C, Redondo JM, Vazquez J. General statistical framework for quantitative proteomics by stable isotope labeling. J Proteome Res. 2014;13:1234–1247. doi: 10.1021/pr4006958. [DOI] [PubMed] [Google Scholar]
- 24.Fabregat A, Sidiropoulos K, Viteri G, Forner O, Marin-Garcia P, Arnau V, D’Eustachio P, Stein L, Hermjakob H. Reactome pathway analysis: a high-performance in-memory approach. BMC Bioinformatics. 2017;18:142. doi: 10.1186/s12859-017-1559-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haft DH, Loftus BJ, Richardson DL, Yang F, Eisen JA, Paulsen IT, White O. TIGRFAMs: a protein family resource for the functional identification of proteins. Nucleic Acids Res. 2001;29:41–43. doi: 10.1093/nar/29.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perez-Riverol Y, Bai J, Bandla C, Garcia-Seisdedos D, Hewapathirana S, Kamatchinathan S, Kundu DJ, Prakash A, Frericks-Zipper A, Eisenacher M, Walzer M, Wang S, Brazma A, Vizcaino JA. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022;50:D543–D552. doi: 10.1093/nar/gkab1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kusch H, Engelmann S, Bode R, Albrecht D, Morschhauser J, Hecker M. A proteomic view of Candida albicans yeast cell metabolism in exponential and stationary growth phases. Int J Med Microbiol. 2008;298:291–318. doi: 10.1016/j.ijmm.2007.03.020. [DOI] [PubMed] [Google Scholar]
- 28.Willsey GG, Ventrone S, Schutz KC, Wallace AM, Ribis JW, Suratt BT, Wargo MJ. Pulmonary surfactant promotes virulence gene expression and biofilm formation in klebsiella pneumoniae. Infect Immun. 2018;86:e00135-18. doi: 10.1128/IAI.00135-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khan MA, Gorke B. A multifunctional small RNA binding protein for sensing and signaling cell envelope precursor availability in bacteria. Microb Cell. 2020;7:139–142. doi: 10.15698/mic2020.05.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lassek C, Burghartz M, Chaves-Moreno D, Otto A, Hentschker C, Fuchs S, Bernhardt J, Jauregui R, Neubauer R, Becher D, Pieper DH, Jahn M, Jahn D, Riedel K. A metaproteomics approach to elucidate host and pathogen protein expression during catheter-associated urinary tract infections (CAUTIs) Mol Cell Proteomics. 2015;14:989–1008. doi: 10.1074/mcp.M114.043463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bao K, Belibasakis GN, Selevsek N, Grossmann J, Bostanci N. Proteomic profiling of host-biofilm interactions in an oral infection model resembling the periodontal pocket. Sci Rep. 2015;5:15999. doi: 10.1038/srep15999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Snyder JA, Haugen BJ, Buckles EL, Lockatell CV, Johnson DE, Donnenberg MS, Welch RA, Mobley HL. Transcriptome of uropathogenic escherichia coli during urinary tract infection. Infect Immun. 2004;72:6373–6381. doi: 10.1128/IAI.72.11.6373-6381.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hancock V, Klemm P. Global gene expression profiling of asymptomatic bacteriuria escherichia coli during biofilm growth in human urine. Infect Immun. 2007;75:966–976. doi: 10.1128/IAI.01748-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu Y, Zielinski MD, Rolfe MA, Kuntz MM, Nelson H, Nelson KE, Pieper R. Similar neutrophil-driven inflammatory and antibacterial responses in elderly patients with symptomatic and asymptomatic bacteriuria. Infect Immun. 2015;83:4142–4153. doi: 10.1128/IAI.00745-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu Y, Tsitrin T, Singh H, Doerfert SN, Sizova MV, Epstein SS, Pieper R. Actinobaculum massiliense proteome profiled in polymicrobial urethral catheter biofilms. Proteomes. 2018;6:52. doi: 10.3390/proteomes6040052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu Y, Singh H, Tsitrin T, Bekele S, Lin YH, Sikorski P, Moncera KJ, Torralba MG, Morrow L, Wolcott R, Nelson KE, Pieper R. Urethral catheter biofilms reveal plasticity in bacterial composition and metabolism and withstand host immune defenses in hypoxic environment. Front Med (Lausanne) 2021;8:667462. doi: 10.3389/fmed.2021.667462. [DOI] [PMC free article] [PubMed] [Google Scholar]