Abstract
Purpose
To determine the proteomic profiles of exosomes derived from vitreous humour (VH) obtained from proliferative diabetic retinopathy (PDR) patients and non-diabetic controls with idiopathic macular hole/epiretinal membrane.
Methods
Vitreal exosomes were isolated using differential ultracentrifugation, followed by characterisation performed using different techniques. A label-free proteomic analysis was conducted to determine the protein profiles of the exosomes. A parallel reaction monitoring (PRM) analysis was performed to verify the identified proteins and associated functional annotations were derived by gene ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses. Receiver operating characteristic (ROC) analysis was utilised to evaluate the diagnostic value of target proteins in distinguishing PDR from controls.
Results
Exosomes were successfully isolated from VH, and were well characterised by various techniques. The results of proteomic analysis showed that a total of 758 proteins were identified and 10 proteins were screened as differentially expressed proteins, significantly changed in the PDR group containing 4 elevated proteins and 6 reduced proteins. GO analysis indicated that these differential proteins were mainly involved in many metabolic pathways, including nicotinamide adenine dinucleotide metabolism, adenosine diphosphate metabolic process and glycolytic process. The KEGG analysis enriched the top five pathways including glycolysis/gluconeogenesis, fructose and mannose metabolism, biosynthesis of amino acids, hypoxia-inducible factor 1 signalling pathway and carbon metabolism. The differential proteins, namely, lactate dehydrogenase A, ficolin 3, apolipoprotein B and apolipoprotein M, were further verified by PRM and showed a consistent trend with label-free proteomic analysis. The ROC analysis identified these proteins as promising biomarkers for PDR diagnosis.
Conclusions
Vitreal exosomes from patients with PDR contained few proteins unique to PDR; thus, exosomal proteins have great potential as disease biomarkers and therapeutic targets for PDR.
Subject terms: Predictive markers, Retinal diseases
Introduction
Proliferative diabetic retinopathy (PDR) is an advanced stage of diabetic retinopathy (DR) that can cause severe visual impairment [1]. Although modern therapies for PDR can delay vision loss, the effects are often too poor or transient. The damage to retinal blood vessels and neuronal tissue functions is irreversible despite receiving the PDR therapies [2–4]. Moreover, the pathogenesis of PDR is complicated and involves a complex interplay of biochemical, immunological and inflammatory factors, and the exact mechanisms remain unclear.
Exosomes, the most minor subtype of extracellular vesicles (EVs), have attracted considerable intense interest since their discovery, because of their broad contribution to multiple diseases [5]. They contain abundant active substances, such as lipids, proteins and microRNAs, which exert different biological functions, like promoting the repair of injured nerves, inhibiting fibrosis [6], modulating immunomodulation [7] and affecting angiogenesis. Many studies have explored the functions of exosomes under different physiologic and pathologic conditions [8]. To the best of our knowledge, although much is known about the role of exosomes in tumour development, their role in eye diseases has not been entirely determined.
In recent years, knowledge regarding the function of exosomes in ocular diseases has been increasingly broadened [9]. Exosomes have several unique features that make them ideal as targets for eye diseases. Several recent studies have identified that exosomes play an essential role in the diagnosis and treatment of age-related macular degeneration (AMD) [10], glaucoma [11], retinopathy of prematurity [12] and DR [13]. Thus, the potential use of exosomes in ocular diseases is worthy of considerable attention.
The role of exosomes in DR is gradually being recognised and little research, so far, has aimed at studying their role in the pathogenesis of DR [12–19]. The levels of IgG-laden exosomes in plasma increased and contributed to microvascular damage in diabetic patients through the activation of the classical complement pathway [13]. Peroxisome proliferator-activated receptor gamma (PPARγ), as a component of the exosomes circulating in plasma, is expressed in the aqueous humour (AH) and vitreous humour (VH) of PDR patients [18]. The concentration of PPARγ increases as the DR stage advances, indicating that PPARγ is involved in the pathogenesis of PDR. The treatment effect of exosomes on DR has also been studied. Treatment with exosomes originating from multiple sources could significantly inhibit retinal thinning and alleviate retinal ischemia in diabetic rats [14]. Therefore, exosomes could be helpful in not only providing novel insight into the pathophysiology of DR, but also have significant potential for therapeutic intervention.
Exosomes exist in bodily fluids, including AH and VH [10]. VH has a high potential to contain exosomal biomarkers for PDR. Few studies have focused on the vitreal exosomes present in uveal melanoma [20], neovascular AMD [21] and DR [22]. Liu et al. [22] demonstrated that exosomes collected from the VH of PDR patients could regulate the angiogenesis of retinal endothelial cells. However, none of these studies has discussed or explored the components of vitreal exosomes from PDR patients. PDR is the end-stage of DR and is mainly caused by retinal ischemia, with pathological neovascularization at the vitreoretinal interface. Exosomes can significantly affect angiogenesis and therefore, are a promising and effective therapeutic target for PDR. It is of great importance to obtain a more comprehensive proteomic profile of the vitreal exosomes in PDR patients, when developing some novel diagnostic or prognostic biomarkers based on exosomes. Therefore, we performed a label-free quantitative proteomics analysis to compare the protein profiles of human vitreal exosomes obtained from PDR patients and control subjects.
Subjects and methods
The study was completed at Qilu Hospital of Shandong University, China. Our research adhered to the provisions of the Declaration of Helsinki and was approved by the Ethical Review Committee of the hospital. All patients were consecutively enrolled between July 2020 and September 2021 and signed informed consent after receiving complete explanation of the nature and possible consequences of the study.
PDR subjects receiving primary pars plana vitrectomy (PPV) as treatment for vitreous haemorrhage, preretinal membranes, macular hole or retinal detachment were recruited as the PDR group. Non-diabetic controls, who were diagnosed with idiopathic macular epiretinal membrane and idiopathic macular hole, and needed PPV surgery, were recruited as the control group. DR classification was performed based on the visible ophthalmological changes and the presence of retinal neovascularization according to the results of the multicentre Early Treatment Diabetic Retinopathy Study. The detailed past medical history of each subject was collected using a standard questionnaire. We excluded people with any other ocular disease, high myopia, glaucoma, retinal diseases or history of previous ocular surgery, with the exception of mild cataract. All patients were free of other systemic disorders (e.g., malignant tumour and hyperthyroidism) and ongoing infection. In addition, patients with type 2 diabetes mellitus (T2DM) with uncontrolled systemic hypertension and subjects who had hyperlipemia and hyperproteinaemia were also excluded from this study.
Prior to initiating vitrectomy, 1 mL of non-dilute VH was harvested with careful suction before the infusion using 25-gauge vitrectomy. Non-transparent, bloody samples were not collected. Blood serum samples were obtained after an overnight fast in the early morning on the first day of admission. The samples were transferred on dry ice and stored in Eppendorf tubes at –80 °C until the assays were performed. The expression levels of triglycerides (TG), total cholesterol (TCho), high-density lipoprotein cholesterol (HDL-C), low-density lipoprotein cholesterol (LDL-C), albumin (ALB), globulin (GLB) and fasting blood glucose (FBG) in the serum were tested in the Clinical Laboratory of Qilu Hospital.
Isolation of exosomes from VH with ultracentrifugation
Differential ultracentrifugation was performed for exosome isolation from human VH samples, as described previously [23]. All centrifugation steps were performed at 4 °C. In brief, 0.5 mL of each VH sample was centrifuged at 2000×g and 10,000×g for 30 and 45 min, respectively, at 4 °C to eliminate cellular debris. The supernatant was filtered twice through 0.22 μm filters (Millipore) and then moved to new tubes. The exosomes were pelleted with ultracentrifugation at 100,000×g for 70 min in an MLA-55 fixed-angle rotor (Beckman Coulter, Brea, CA, USA). The pellet was washed in phosphate-buffered saline (PBS) to prevent protein contamination and centrifuged again at 100,000×g for 70 min. The PBS was removed, and the exosomes were re-suspended in 100 µL PBS and stored at −80 °C for further analysis. A BCA assay (Thermo Fisher Scientific, Waltham, MA, USA) was performed for the total protein of the exosomes according to the manufacturer’s protocol. Quantification of exosomes was performed using electron microscopy, nanoparticle tracking analysis (NTA) and flow cytometry. Exosome counts were used to normalise the mass spectrometer (MS) intensities for each detected m/z.
Transmission electron microscopy
Exosome morphologies were observed using an ultramicroscopic analysis. In brief, the freshly isolated exosome suspensions were fixed with 2% glutaraldehyde at 4 °C overnight and then absorbed onto a copper grid. After that, the copper grid with exosomes was fixed with 3% glutaraldehyde and then stained above the 4% uranyl acetate droplet. The grid was eventually observed with a transmission electron microscope (Thermo Fisher Scientific) at 100 kV.
Nanoparticle tracking analysis (NTA)
The size and the concentration of the vesicles were determined by NTA using NanoSight NS300 system (Malvern, UK) following the manufacturer’s protocol and without further modifications. Triplicate tests were performed and the values were averaged to derive the size distribution and concentration profiles for each sample.
Nanoscale flow cytometry
Isolated vesicles were confirmed by three typical exosomal markers, CD9, CD63 and CD81 (FITC Mouse Anti-Human antibody, Becton, Dickinson and Company, USA), using nanoscale flow cytometry (NanoFCM). The same volume of samples was used throughout the flow cytometric analysis: 30 µL of 1:4 diluted exosomes and 20 µL of FITC mouse anti-human CD9, CD63 and CD81. The MACSQuant Analyzer 10 was used for NanoFCM analysis, which was in strict accordance with the operating instructions.
Label‑free quantitative proteomic analysis of exosomes
BCA assay for the total protein of exosomes
One millilitre of RIPA lysis buffer was added to the isolated exosomes. The lysate was sonicated and then boiled for 15 min. The mixture was centrifuged at 14,000 g for 15 min at 4 °C. The supernatant was transferred to new tubes and quantified using the BCA assay (Thermo Fisher Scientific, Waltham, MA, USA).
Acetone precipitation
Equal amounts of protein (100 µg) from each sample were diluted to 1 mg/mL and alkylated with pre-chilled acetone. The proteins were shaken on ice for 30 min and centrifuged at 4 °C with a speed of 1000 g. The pellet was washed twice with 200 µL 80% chilled acetone.
Re-suspension of protein for tryptic digest
The protein pellet was mixed with 200 μL of 1% sodium deoxycholate (SDC) + 100 mM ammonium bicarbonate and sonicated for 30 min in a water bath to dissolve protein pellet. Five millilitres of tris (2-chloroethyl) phosphate was added to each sample and mixed at 55 °C for 10 min. Then, 10 mM of iodoacetamide was added after samples cooled down to room temperature (RT), which were then incubated for 15 min in darkness. Trypsin was re-suspended with re-suspension buffer to 0.5 µg/µL and incubated at RT for 5 min. Trypsin solution was added to each sample (protein: trypsin = 50: 1) and incubated at 37 °C with a thermomixer overnight. The supernatant was carefully transferred to a new tube after cleaning up of SDC.
Peptide desalting for base-RP fractionation
After desalting on the C18 trapping column (Empore™ SPE Cartridges C18, Sigma), the peptides were concentrated by vacuum centrifugation and reconstituted in 40 μL of 0.1% (v/v) formic acid for further LC–MS/MS analysis.
LC–MS/MS analysis
The LC–MS/MS analysis was performed using a Nano-HPLC (EASY-nLC1200) coupled with Q-Exactive mass spectrometry (Thermo Finnigan). Data-dependent acquisition was performed in positive ion mode. For each sample, 2 μg peptide was separated using a reversed-phase column with a 120 min gradient at a 300 nL/min flow rate. The Orbitrap analyser was set at a resolution of 70,000 at m/z 200 and a range of m/z 350–1400 for MS1; for MS2, the resolution was set to 17,500 at m/z 200. The isolation width was 2 m/z. The automatic gain control target was set to 1.0 E+6 for MS1, and 1.0 E+5 for MS2. The top 20 most intense ions were selected in the liner ion trap at a normalised collision energy of 28%; dynamic exclusion was set at 40 s.
Data analysis
Raw MS data were analysed with MaxQuant software (Version 2.0.1.0). Trypsin was chosen as a specific enzyme with up to two missed cleavages. Oxidation [M] and Acetyl [protein N-term] were set as variable modifications, Carbamidomethyl [C] was set as a fixed modification and precursor mass tolerance was set at 20 ppm. The maximum fragment mass error was 0.8 Da. The following were the criteria for protein identifications: unique protein with a false discovery rate (FDR) of less than 1% and the concurrent detection of at least two peptides.
Bioinformatics analyses
The differential proteins target genes to each term in the gene ontology (GO) database were determined by conducting a GO enrichment analysis, which included biological processes (BPs), molecular functions (MFs) and cellular components (CCs). The significant GO terms were determined using Fisher’s exact test. After the calculated p value was corrected by FDR, the threshold was corrected to p value ≤0.05. The GO term that met this criterion was regarded as a significant term that was enriched in the differentially expressed proteins. The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was used to identify their biological functions. The enriched pathway analysis was based on the KEGG pathways and hypergeometric tests were calculated to determine whether each pathway was enriched. Protein–protein interactions were performed using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database.
Parallel reaction monitoring analysis
The peptides were separated using a nano-flow ultra-performance liquid chromatography mass spectrometry system. The targeted quantification and verification of selected proteins were analysed by parallel reaction monitoring (PRM) analysis, which was performed using a Q-Exactive HF mass spectrometer (Thermo Scientific Bremen, Germany) at 120 min/sample. Transition settings were as follows: precursor charges +2, +3; ion charge +1, +2; ion type b, y; auto-selection of all matching transitions; ion match tolerance 0.005 m/z and selection of the six most intense product ions. Raw data were collected using the PRM acquisition method and normalised according to the summed intensity.
Statistical analyses
Statistical software GraphPad Prism® (GraphPad Prism® Version 5) was used for statistical analyses. Student’s t-test or Mann–Whitney test was performed, as appropriate, to compare two groups of independent samples. For categorical variables, we performed Fisher’s exact test or the χ2 test. The differentially expressed proteins between the two groups were identified with an abundance ratio (PDR/controls) ≥2 or ≤0.5, and p value of less than 0.05 was considered significant. The receiver operating characteristic (ROC) analysis and the area under the curve (AUC) were used to evaluate the diagnostic value of the index. A p value of less than 0.05 was considered significant. All data were expressed as mean ± SEM.
Results
Patient characteristics
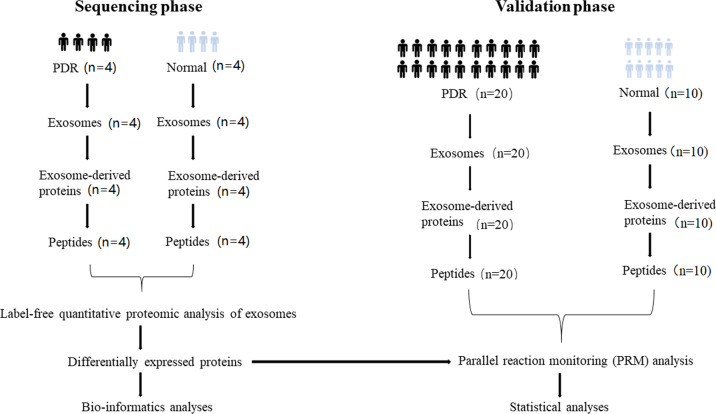
We enrolled 46 subjects in this study. As shown in Fig. 1, 16 were subjected to the sequencing phase (with 8 subjects in each group), while the rest (20 PDR patients and 10 controls) were subjected to the validation phase. Comparisons of age, gender, TG, TCho, HDL-C, LDL-C, ALB, GLB and body mass index were carried out, and no significant difference was detected. However, PDR patients had higher levels of FBG. The patient characteristics are summarised in Table 1.
Fig. 1. An overview of the experimental design.
Sequencing phase and validation phase were applied in identification and verification of exosomal proteins.
Table 1.
Clinical characteristics of the study subjects.
| Diabetic patients (diabetic group, n = 24) | Controls (control group, n = 14) | P values | |
|---|---|---|---|
| Age, years | 56.7 ± 1.5 | 61.4 ± 2.0 | 0.069 |
| Male/Female | 11/15 | 7/9 | 0.927 |
| TCho (mmol/L) | 4.2 ± 0.2 | 4.7 ± 0.2 | 0.149 |
| HDL-C (mmol/L) | 1.1 ± 0.0 | 1.2 ± 0.1 | 0.221 |
| LDL-C (mmol/L) | 2.5 ± 0.1 | 2.3 ± 0.2 | 0.286 |
| TG (mmol/L) | 1.6 ± 0.2 | 1.6 ± 0.2 | 0.856 |
| FBG (mmol/L) | 6.9 ± 0.4 | 5.3 ± 0.2 | 0.005 |
| ALB (g/L) | 39.9 ± 1.2 | 40.5 ± 0.9 | 0.721 |
| GLB (g/L) | 25.3 ± 1.0 | 24.7 ± 0.3 | 0.498 |
| Duration of DM, years | 11.2 ± 1.8 | – | – |
| BMI (kg/m2) | 22.1 ± 0.5 | 21.8 ± 0.4 | 0.595 |
| Hypertension, n (%) | 14 (53.8%) | 5 (31.3%) | 0.153 |
Data were expressed as mean ± standard errors (SE).
TCho Total cholesterol, HDL-C high-density liptein cholesterol, LDL-C low-density liptein cholesterol, TG triglyceride, FBG fasting blood glucose, ALB albumin, GLB globulin, DM diabetes mellitus, BMI body mass index.
Bold indicates significance set at p < 0.05.
Characterisation of vitreal exosomes
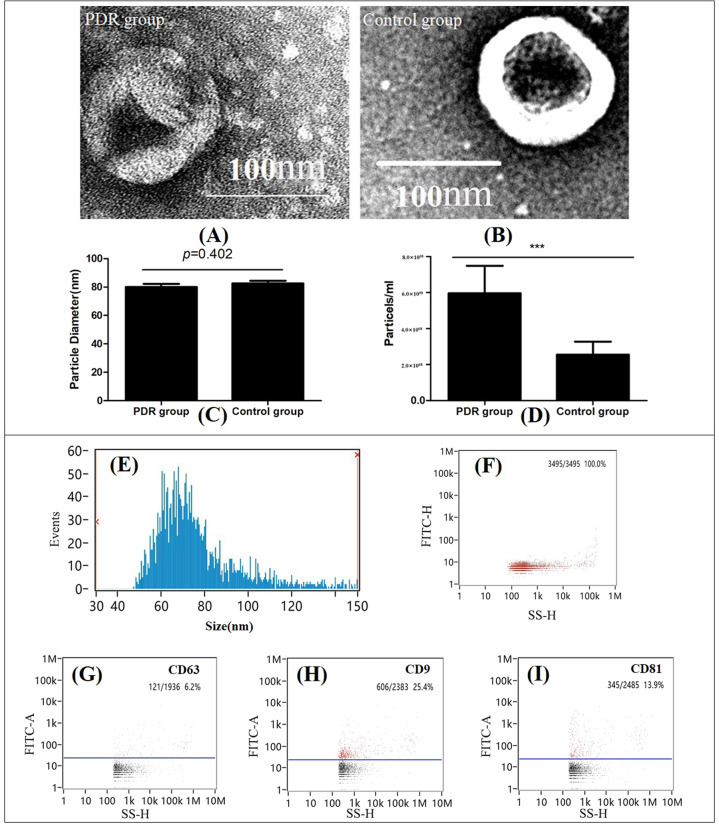
According to the electron microscopy images, the purified particles in both the PDR group and control group showed typical exosomal morphology with round and cup-like concavity (Fig. 2A, B). Three surface markers for exosomes, CD9, CD63 and CD81, were analysed using NanoFCM, and the results showed that the isolated nano-particles were positive for at least one of the canonical exosome markers. In addition, the sizes of the extracted vesicles were determined through NTA. The results indicated that the size distribution of the particles was between 30 and 120 nm and approximately 70 nm, consistent with the characteristic size range of exosomes. Moreover, the diameters of the particles were compared and no difference was detected between the two groups (Fig. 2C). These results demonstrated that the exosomes were successfully isolated from the human VH with high purity and were well characterised by various methods. We also compared the concentrations of isolated exosomes between the two groups and found a much higher concentration of exosomes in the VH of PDR patients based on statistical analysis (Fig. 2D).
Fig. 2. Characterisation of isolated vitreous exosomes across groups.
Transmission electron microscopy (TEM) of vitreous exosomes (A PDR group; B Control group). Diameter comparison of isolated vitreous exosomes between two groups (C). Concentration comparison of isolated vitreous exosomes between two groups (D). Characterisation of isolated vitreous exosomes from one male PDR patient. E Size distribution analysis. F Nanoparticle tracking analysis (NTA) of exosomes. NanoFCM analysis of the surface exosomal markers, CD9 (H), CD63 (G) and CD81 (I). ***<0.001.
Proteomics analysis and annotation of identified proteins
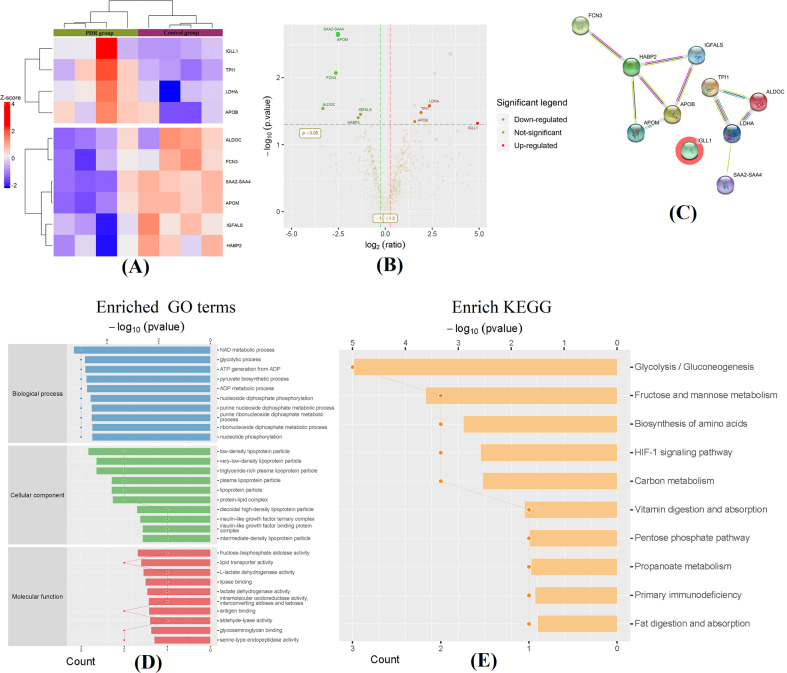
A total of 758 proteins were identified in both groups using label‑free quantitative proteomic analysis. Differential proteins with significant higher/lower amounts were filtrated with ratio A/B (PDR/controls protein abundance ratio) ≥2 or ≤0.5, unique peptide ≥2 and p value <0.05. As shown in Fig. 3, 10 proteins were screened as differentially expressed proteins between the two groups, including immunoglobulin lambda-like polypeptide 1 (IGLL1), triosephosphate isomerase 1 (TPI1), lactate dehydrogenase A (LDHA), apolipoprotein B (ApoB), apolipoprotein M (ApoM), ficolin 3 (FCN3), aldolase C (ALDOC), serum amyloid protein (SAA) 2-4, insulin-like growth factor binding protein, acid-labile subunit (IGFALS) and hyaluronan binding protein 2 (HABP2). The results of the hierarchical cluster and the volcano plot showed these differential proteins between the two groups (Fig. 3A, B). To show the potential interactions among these proteins, the web-based STRING tools were used to create comprehensive interaction networks (Fig. 3C).
Fig. 3. Hierarchical cluster and volcano plots of the differentially expressed proteins.
Fold change (FC) values >2 or <0.5 and a p value <0.05 were set as the filter criteria (A). The vertical dotted line at 0 at the X-axis delimits up- and downregulation. The red and green plots represent significant upregulated and downregulated genes with >2.0-fold change and p value <0.05 in PDR patients (B). Comprehensive interaction networks of the proteins with altered expression based on STRING tools (C). Bioinformatics analysis of the differentially expressed exosome proteins. D Gene Ontology (GO) enrichment analysis of differential proteins. Top 10 items are listed in biological process, cell component and molecular function, respectively based on their statistical significance. E KEGG pathway enrichment analysis of differential proteins. Top 10 items are listed on their statistical significance.
To investigate the functional basis of these differential exosomal proteins, GO and KEGG pathway analyses were conducted for their biological functions. The identified proteins target genes were categorised into three groups: BPs, CCs and MFs. As shown in Fig. 3D, the top 10 most statistically significant terms in each group are listed in sequence. As for the GO analysis of BP, the exosomal proteins were mainly involved in our adaptive metabolic pathways, particularly nicotinamide adenine dinucleotide metabolism, adenosine diphosphate metabolic process and glycolytic process, which indicate that vitreal exosomes may participate in the development of diabetes and its associated ocular complications. The annotated MF of the proteins revealed enrichment of vitreal exosomes related to fructose-bisphosphate aldolase activity, lipid transporter activity, glycosaminoglycan binding, etc. a GO enrichment analysis in CC was performed, and these exosomal proteins were mainly associated with various plasma lipoproteins, indicating that these vitreal exosomes were possibly originated from plasma.
To understand the affected biological pathways involved in the exosomal proteins, the proteins were further mapped with the latest KEGG pathway database. The corresponding KEGG pathways were extracted and are shown in Fig. 3E. Most proteins were involved in glycolysis/gluconeogenesis, followed by fructose and mannose metabolism, biosynthesis of amino acids and HIF-1 signalling pathway. A large majority of the identified proteins were mapped in pathways involved in DM pathogenesis.
Protein expression by PRM
All of the differentially expressed proteins based on label‑free proteomic analysis were further analysed by PRM. Another cohort of patients (20 PDR patients and 10 controls) was included in the validation phase. The expression levels of four proteins (LDHA, FCN3, ApoB and ApoM) showed a consistent trend with the quantitative label-free proteomics (Fig. 4A).
Fig. 4. Parallel reaction monitoring (PRM) analysis of the differentially expressed exosome proteins.
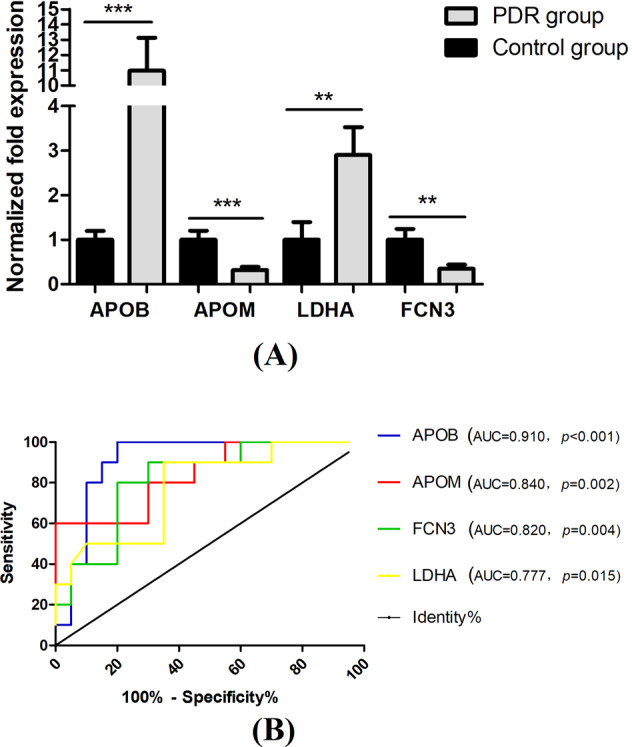
LDHA, FCN3, ApoB and ApoM showed a consistent trend with the results of proteomics analysis. **<0.01, ***<0.001 (A). Receiver operating characteristic (ROC) curve assay of these verified exosome proteins as diagnostic biomarkers (B).
ROC curves and AUC were performed to figure out whether these four proteins functioned as potential diagnostic biomarkers to distinguish PDR patients from the controls. Metabolites with AUC > 0.75 contributed to the forecast classification for the individual subjects among groups. As seen in Fig. 4B, all of the four proteins emerged as promising biomarkers for the PDR diagnosis.
Discussion
Numerous studies have now found that exosomes extensively exist in body fluids, such as blood, serum, saliva, urine, ascites, tears, AH and VH [24]. In this pilot study, we performed a comprehensive proteomic analysis of exosomes isolated from human VH. As compared to control subjects, 10 proteins that differed significantly in amount were identified in PDR patients. The results of bioinformatics analysis suggested that these differential proteins mainly affect glucose and lipid metabolism and participate in multiple signal pathways of DR. To the best of our knowledge, this is the first proteomic analysis in vitreal exosomes from PDR patients.
Vitreal exosomes analysis in PDR patients can represent a novel therapeutic drug candidate for PDR therapy [4]. In this study, we successfully isolated exosomes from the VH. Vitreal exosomes with high purity were obtained, which laid the foundation for the subsequent proteomic analysis. Exosomes carry proteins, lipids, nucleic acids and other bioactive substances that play an important role in the body’s physiological and pathological processes. Many diseases alter the proteins and RNAs of exosomes in bodily fluids [8]. In conditions of diabetes, exosomes play critical roles in the pathogenesis of diabetes and diabetic complications [25].
The proteomic analysis of vitreal exosomes from PDR patients remains unknown. In our study, a total of 758 exosomal proteins were detected in PDR patients and control subjects using label‑free proteomic analysis. Significantly changed in PDR group contained four elevated proteins and six reduced proteins. Our results showed that vitreal exosomes contain few proteins unique to PDR that participate in multiple signal pathways of DR, such as influencing metabolic signals and affecting the process of vascularisation, cell multiplication and survival.
Exosomes are the smallest subtype of Evs and can be secreted by numerous cells. After being released from donor cells, they undergo a free-floating period in which they circulate in body fluids [26]. We found that the concentration of exosomes was significantly increased in the VH of PDR patients. In biological fluid, exosome populations are highly heterogeneous and originate from various cell types. The origin of vitreal exosomes also remains unclear. Exosomes can be released from various ocular cells, such as retinal pigment epithelial cells and retinal astroglial cells [27]. Thus, it is difficult to attribute to any given cell type. We speculated that hyperglycaemia and hypoxia in the ocular cells can lead to an increased release of exosomes directly into VH. GO enrichment analysis in CC suggested that these exosomal proteins were mainly associated with various plasma lipoprotein particles. Elevated levels of circulating exosomes in diabetes have been confirmed by several studies [13]. As the exosomes can transfer substances by circulating in body fluids, the other sources of vitreal exosomes might result from serum diffusion.
Among these differential proteins, apolipoproteins form important subsets of the protein interacting network, which should be studied further. Accumulating evidence has indicated that apolipoproteins are related to the presence of DR [28]. The serum levels of apolipoproteins have been considered stronger biomarkers of DR and can thus facilitate early detection and treatment of DR [29, 30]. The enhancement of apolipoproteins is an early event in the eyes of diabetic patients [31].
Elevated levels of apoA-I and apo H in the vitreous fluid were detected in PDR patients [32]. In our study, we firstly found that exosomal ApoB levels in the VH of PDR patients were much higher than in those without diabetes. ApoB has been proposed as an ideal negative marker for plasma/serum EVs and is often co-isolated in the isolation of EVs from plasma/serum [33, 34]. Only transparent and non-bloody samples were collected in the current study. Therefore, it can be excluded that a fraction of ApoB may be associated with plasma/serum. Higher ApoB levels in VH are destructive to arterial and retinal vascular cells due to their higher lipoprotein-related toxins. In addition, the destruction of the blood-retinal barrier and neuronal apoptosis can be induced by ApoB [35]. Therefore, our findings supported the concept that exosomal ApoB is closely associated with the development of PDR and has emerged as a drug target for PDR [36].
ApoM, a relatively novel apolipoprotein, acts as a carrier for the lipid sphingosine-1-phosphate (S1P). Through delivering S1P to the S1P (1) receptor on endothelial cells, ApoM affects HDL metabolism and a wide range of biological functions, such as neuro-inflammation, angiogenesis, cell motility, cell growth, cell invasion and wound repair [37, 38]. Several studies have shown that plasma ApoM was remarkably lower in both obese mouse model of T2DM and patients with T2DM [39, 40]. Contradicting results have reported no significant differences in ApoM concentrations among T2DM and healthy individuals [41]. Our study found a reduced level of exosomal ApoM in PDR patients, which was possibly depressed by insulin and glucose. Considering the close interaction between ApoM and S1P, decreased ApoM in exosomes might exert a negative impact on DR progression. The relationship between ApoM and PDR deserves further investigation.
Apart from apolipoproteins, FCN3 and LDHA were also verified in the validation phase. FCN3 is one of the innate immunity molecules, which has been shown to activate the complement system through the lectin pathway. Abnormal serum FCN3 levels are associated with DM and diabetes complications [42, 43], rheumatic heart disease [44], susceptibility to fever and neutropenia [45], leprosy [46], systemic lupus erythematosus [47], etc. FCN3 levels were elevated in the serum and VH of PDR patients and might be used as a new therapeutic target for treatment of PDR [48]. In our study, vitreal exosomes from PDR patients showed significantly decreased FCN3 levels compared to normal subjects. The exact reason for this difference is not clear, owing to the limited data. Serum FCN3 is widely variable among healthy individuals, which may be attributed to the difference. Although the exact role of FCN3 in the progression of DR remains unclear, we speculated that different splicing isoforms of FCN3, including exosomal FCN3, can be involved in the development of DR through its related acetylation-induced complement activation.
LDHA has not been reported in the presence of DR. It is a key metabolic enzyme that preferentially catalyses the conversion of pyruvate to lactate and relates to the development of various cancers. Cancer cells usually have upregulated LDHA, which promotes a metabolic switch to aerobic glycolysis and generates lactate as a product. The downregulation of LDHA can attenuate glycolysis and suppress tumour growth in cancer models [49]. LDHA is highly expressed in many tissues and its inappropriate upregulation under diabetic conditions is well-documented [50, 51]. Interestingly, our study found that LDHA was highly expressed in vitreal exosomes of PDR patients. Although there is no prior evidence to show that LDHA affects DR, these findings suggest that LDHA plays a critical role in the pathogenesis of DR and therefore is a promising target for DR treatment.
In conclusion, we performed quantitative proteomics to profile the vitreal exosomes from PDR patients. We have demonstrated that certain exosome proteins are unique to PDR. ROC curves were constructed separately to compare the diagnostic values of these proteins to predict PDR. Our results show that vitreous exosome proteins can be detected in PDR patients, and their expression can differentiate PDR patients from healthy control subjects. Our findings are expected to expand the scope of knowledge on the function of exosomes in the pathogenesis of DR and shed light on potential exosome therapy for DR. Further studies on larger cohorts of patients are required to verify some candidate diagnostic and prognostic markers and establish exosome protein signatures indicative of PDR.
Summary
What was known before
The pathogenesis of PDR is complicated, and the role of exosomes in DR is gradually being recognised.
What this study adds
Vitreal exosomes from PDR patients contained few proteins unique to PDR; thus, exosomal proteins have great potential as diagnostic biomarkers and therapeutic targets for PDR.
Author contributions
All authors conceived of and designed the experimental protocol. JWW, ZZW and YZ collected the data. JWW and ZZW were involved in the analysis and interpretation of the data. JWW wrote the first draft of the manuscript. JQL reviewed and revised the manuscript and produced the final version. All authors read and approved the final manuscript.
Funding
This study was supported by the Jinan Clinical Medical Science and Technology Innovation Plan (202019113), Key Research and Development project of Shandong Province (2017GSF218033) and the National Natural Science Foundation of China (81700831).
Data availability
The data that support the findings of this study are not publicly available because they contain information that could compromise the privacy of patients but are available from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jiawei Wang, Zhenzhen Wang.
References
- 1.Cheung N, Mitchell P, Wong TY. Diabetic retinopathy. Lancet. 2010;376:124–36. doi: 10.1016/S0140-6736(09)62124-3. [DOI] [PubMed] [Google Scholar]
- 2.Kollias AN, Ulbig MW. Diabetic retinopathy: early diagnosis and effective treatment. Dtsch Arztebl Int. 2010;107:75–83. doi: 10.3238/arztebl.2010.0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.El Rami H, Barham R, Sun JK, Silva PS. Evidence-based treatment of diabetic retinopathy. Semin Ophthalmol. 2017;32:67–74. doi: 10.1080/08820538.2016.1228397. [DOI] [PubMed] [Google Scholar]
- 4.Nawaz IM, Rezzola S, Cancarini A, Russo A, Costagliola C, Semeraro F, et al. Human vitreous in proliferative diabetic retinopathy: characterization and translational implications. Prog Retin Eye Res. 2019;72:100756. doi: 10.1016/j.preteyeres.2019.03.002. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y, Bi J, Huang J, Tang Y, Du S, Li P. Exosome: a review of its classification, isolation techniques, storage, diagnostic and targeted therapy applications. Int J Nanomed. 2020;15:6917–34. doi: 10.2147/IJN.S264498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hervera A, De Virgiliis F, Palmisano I, Zhou L, Tantardini E, Kong G, et al. Reactive oxygen species regulate axonal regeneration through the release of exosomal NADPH oxidase 2 complexes into injured axons. Nat Cell Biol. 2018;20:307–19. doi: 10.1038/s41556-018-0039-x. [DOI] [PubMed] [Google Scholar]
- 7.Willis GR, Fernandez-Gonzalez A, Anastas J, Vitali SH, Liu X, Ericsson M, et al. Mesenchymal stromal cell exosomes ameliorate experimental bronchopulmonary dysplasia and restore lung function through macrophage immunomodulation. Am J Respir Crit Care Med. 2018;197:104–16. doi: 10.1164/rccm.201705-0925OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin C, Wu P, Li L, Xu W, Qian H. Exosomes: emerging therapy delivery tools and biomarkers for kidney diseases. Stem Cells Int. 2021;2021:7844455. doi: 10.1155/2021/7844455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klingeborn M, Dismuke WM, Bowes Rickman C, Stamer WD. Roles of exosomes in the normal and diseased eye. Prog Retin Eye Res. 2017;59:158–77. doi: 10.1016/j.preteyeres.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang GY, Bang JY, Choi AJ, Yoon J, Lee WC, Choi S, et al. Exosomal proteins in the aqueous humor as novel biomarkers in patients with neovascular age-related macular degeneration. J Proteome Res. 2014;13:581–95. doi: 10.1021/pr400751k. [DOI] [PubMed] [Google Scholar]
- 11.Takahashi E, Saruwatari J, Fujimoto T, Tanoue Y, Fukuda T, Inoue T. The effects of exosomes derived from trabecular meshwork cells on Schlemm’s canal endothelial cells. Sci Rep. 2021;11:21942. doi: 10.1038/s41598-021-01450-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moisseiev E, Anderson JD, Oltjen S, Goswami M, Zawadzki RJ, Nolta JA, et al. Protective effect of intravitreal administration of exosomes derived from mesenchymal stem cells on retinal ischemia. Curr Eye Res. 2017;42:1358–67. doi: 10.1080/02713683.2017.1319491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang C, Fisher KP, Hammer SS, Navitskaya S, Blanchard GJ, Busik JV. Plasma exosomes contribute to microvascular damage in diabetic retinopathy by activating the classical complement pathway. Diabetes. 2018;67:1639–49. doi: 10.2337/db17-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang W, Wang Y, Kong Y. Exosomes derived from mesenchymal stem cells modulate miR-126 to ameliorate hyperglycemia-induced retinal inflammation via targeting HMGB1. Invest Ophthalmol Vis Sci. 2019;60:294–303. doi: 10.1167/iovs.18-25617. [DOI] [PubMed] [Google Scholar]
- 15.Maisto R, Trotta MC, Petrillo F, Izzo S, Cuomo G, Alfano R, et al. Resolvin D1 modulates the intracellular VEGF-related miRNAs of retinal photoreceptors challenged with high glucose. Front Pharm. 2020;11:235. doi: 10.3389/fphar.2020.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang W, Dong X, Wang T, Kong Y. Exosomes derived from platelet-rich plasma mediate hyperglycemia-induced retinal endothelial injury via targeting the TLR4 signaling pathway. Exp Eye Res. 2019;189:107813. doi: 10.1016/j.exer.2019.107813. [DOI] [PubMed] [Google Scholar]
- 17.Kamalden TA, Macgregor-Das AM, Kannan SM, Dunkerly-Eyring B, Khaliddin N, Xu Z, et al. Exosomal microRNA-15a transfer from the pancreas augments diabetic complications by inducing oxidative stress. Antioxid Redox Signal. 2017;27:913–30. doi: 10.1089/ars.2016.6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katome T, Namekata K, Mitamura Y, Semba K, Egawa M, Naito T, et al. Expression of intraocular peroxisome proliferator-activated receptor gamma in patients with proliferative diabetic retinopathy. J Diabetes Complications. 2015;29:275–81. doi: 10.1016/j.jdiacomp.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 19.Tokarz A, Szuścik I, Kuśnierz-Cabala B, Kapusta M, Konkolewska M, Żurakowski A, et al. Extracellular vesicles participate in the transport of cytokines and angiogenic factors in diabetic patients with ocular complications. Folia Med Cracov. 2015;55:35–48. [PubMed] [Google Scholar]
- 20.Ragusa M, Barbagallo C, Statello L, Caltabiano R, Russo A, Puzzo L, et al. miRNA profiling in vitreous humor, vitreal exosomes and serum from uveal melanoma patients: Pathological and diagnostic implications. Cancer Biol Ther. 2015;16:1387–96. doi: 10.1080/15384047.2015.1046021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ménard C, Rezende FA, Miloudi K, Wilson A, Tétreault N, Hardy P, et al. MicroRNA signatures in vitreous humour and plasma of patients with exudative AMD. Oncotarget. 2016;7:19171–84. doi: 10.18632/oncotarget.8280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y, Yang Q, Fu H, Wang J, Yuan S, Li X, et al. Müller glia-derived exosomal miR-9-3p promotes angiogenesis by restricting sphingosine-1-phosphate receptor S1P1 in diabetic retinopathy. Mol Ther Nucleic Acids. 2022;27:491–504. doi: 10.1016/j.omtn.2021.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Théry C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;Chapter 3:Unit 3.22. [DOI] [PubMed]
- 24.Lande K, Gupta J, Ranjan R, Kiran M, Torres Solis LF, Solís Herrera A, et al. Exosomes: insights from retinoblastoma and other eye cancers. Int J Mol Sci. 2020;21:7055. [DOI] [PMC free article] [PubMed]
- 25.Chen A, Wang H, Su Y, Zhang C, Qiu Y, Zhou Y, et al. Exosomes: biomarkers and therapeutic targets of diabetic vascular complications. Front Endocrinol (Lausanne) 2021;12:720466. doi: 10.3389/fendo.2021.720466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteom. 2010;73:1907–20. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 27.Hajrasouliha AR, Jiang G, Lu Q, Lu H, Kaplan HJ, Zhang HG, et al. Exosomes from retinal astrocytes contain antiangiogenic components that inhibit laser-induced choroidal neovascularization. J Biol Chem. 2013;288:28058–67. doi: 10.1074/jbc.M113.470765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chait A, Montes VN. Apolipoproteins and diabetic retinopathy. Diabetes Care. 2011;34:529–31. doi: 10.2337/dc10-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chung JO, Park SY, Cho DH, Chung DJ, Chung MY. Associations between serum apolipoproteins, urinary albumin excretion rate, estimated glomerular filtration rate, and diabetic retinopathy in individuals with type 2 diabetes. Medicine (Baltimore) 2019;98:e15703. doi: 10.1097/MD.0000000000015703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moosaie F, Davatgari RM, Firouzabadi FD, Esteghamati S, Deravi N, Meysamie A, et al. Lipoprotein(a) and apolipoproteins as predictors for diabetic retinopathy and its severity in adults with type 2 diabetes: a case-cohort study. Can J Diabetes. 2020;44:414–21. doi: 10.1016/j.jcjd.2020.01.007. [DOI] [PubMed] [Google Scholar]
- 31.Simó R, García-Ramírez M, Higuera M, Hernández C. Apolipoprotein A1 is overexpressed in the retina of diabetic patients. Am J Ophthalmol. 2009;147:319–25.e1. doi: 10.1016/j.ajo.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 32.Simó R, Higuera M, García-Ramírez M, Canals F, García-Arumí J, Hernández C. Elevation of apolipoprotein A-I and apolipoprotein H levels in the vitreous fluid and overexpression in the retina of diabetic patients. Arch Ophthalmol. 2008;126:1076–81. doi: 10.1001/archopht.126.8.1076. [DOI] [PubMed] [Google Scholar]
- 33.Karimi N, Cvjetkovic A, Jang SC, Crescitelli R, Hosseinpour Feizi MA, Nieuwland R, et al. Detailed analysis of the plasma extracellular vesicle proteome after separation from lipoproteins. Cell Mol Life Sci. 2018;75:2873–86. doi: 10.1007/s00018-018-2773-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7:1535750. doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu M, Chen Y, Wilson K, Chirindel A, Ihnat MA, Yu Y, et al. Intraretinal leakage and oxidation of LDL in diabetic retinopathy. Invest Ophthalmol Vis Sci. 2008;49:2679–85. doi: 10.1167/iovs.07-1440. [DOI] [PubMed] [Google Scholar]
- 36.Crosby-Nwaobi R, Chatziralli I, Sergentanis T, Dew T, Forbes A, Sivaprasad S. Cross talk between lipid metabolism and inflammatory markers in patients with diabetic retinopathy. J Diabetes Res. 2015;2015:191382. doi: 10.1155/2015/191382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arkensteijn BW, Berbée JF, Rensen PC, Nielsen LB, Christoffersen C. The apolipoprotein m-sphingosine-1-phosphate axis: biological relevance in lipoprotein metabolism, lipid disorders and atherosclerosis. Int J Mol Sci. 2013;14:4419–31. doi: 10.3390/ijms14034419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blaho VA, Galvani S, Engelbrecht E, Liu C, Swendeman SL, Kono M, et al. HDL-bound sphingosine-1-phosphate restrains lymphopoiesis and neuroinflammation. Nature. 2015;523:342–6. doi: 10.1038/nature14462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Plomgaard P, Dullaart RP, de Vries R, Groen AK, Dahlbäck B, Nielsen LB. Apolipoprotein M predicts pre-beta-HDL formation: studies in type 2 diabetic and nondiabetic subjects. J Intern Med. 2009;266:258–67. doi: 10.1111/j.1365-2796.2009.02095.x. [DOI] [PubMed] [Google Scholar]
- 40.Xu N, Nilsson-Ehle P, Hurtig M, Ahrén B. Both leptin and leptin-receptor are essential for apolipoprotein M expression in vivo. Biochem Biophys Res Commun. 2004;321:916–21. doi: 10.1016/j.bbrc.2004.06.180. [DOI] [PubMed] [Google Scholar]
- 41.Cervin C, Axler O, Holmkvist J, Almgren P, Rantala E, Tuomi T, et al. An investigation of serum concentration of apoM as a potential MODY3 marker using a novel ELISA. J Intern Med. 2010;267:316–21. doi: 10.1111/j.1365-2796.2009.02145.x. [DOI] [PubMed] [Google Scholar]
- 42.Chen H, Lu J, Chen X, Yu H, Zhang L, Bao Y, et al. Low serum levels of the innate immune component ficolin-3 is associated with insulin resistance and predicts the development of type 2 diabetes. J Mol Cell Biol. 2012;4:256–7. doi: 10.1093/jmcb/mjs032. [DOI] [PubMed] [Google Scholar]
- 43.Zhang X, Hu Y, Shen J, Zeng H, Lu J, Li L, et al. Low levels of ficolin-3 are associated with diabetic peripheral neuropathy. Acta Diabetol. 2016;53:295–302. doi: 10.1007/s00592-015-0780-6. [DOI] [PubMed] [Google Scholar]
- 44.Gomaa MH, Khidr EG, Elshafei A, Hamza HS, Fattouh AM, El-Husseiny AA, et al. The clinical value of ficolin-3 gene polymorphism in rheumatic heart disease. An Egyptian adolescents study. BMC Res Notes. 2021;14:36. doi: 10.1186/s13104-021-05450-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schlapbach LJ, Aebi C, Hansen AG, Hirt A, Jensenius JC, Ammann RA. H-ficolin serum concentration and susceptibility to fever and neutropenia in paediatric cancer patients. Clin Exp Immunol. 2009;157:83–89. doi: 10.1111/j.1365-2249.2009.03957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Andrade FA, Beltrame MH, Bini VB, Gonçalves LB, Boldt AB, de Messias-Reason IJ. Association of a new FCN3 haplotype with high ficolin-3 levels in leprosy. PLoS Negl Trop Dis. 2017;11:e0005409. doi: 10.1371/journal.pntd.0005409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andersen T, Munthe-Fog L, Garred P, Jacobsen S. Serum levels of ficolin-3 (Hakata antigen) in patients with systemic lupus erythematosus. J Rheumatol. 2009;36:757–9. doi: 10.3899/jrheum.080361. [DOI] [PubMed] [Google Scholar]
- 48.Zheng B, Li T, Chen H, Xu X, Zheng Z. Correlation between ficolin-3 and vascular endothelial growth factor-to-pigment epithelium-derived factor ratio in the vitreous of eyes with proliferative diabetic retinopathy. Am J Ophthalmol. 2011;152:1039–43. doi: 10.1016/j.ajo.2011.05.022. [DOI] [PubMed] [Google Scholar]
- 49.Jin L, Chun J, Pan C, Alesi GN, Li D, Magliocca KR, et al. Phosphorylation-mediated activation of LDHA promotes cancer cell invasion and tumour metastasis. Oncogene. 2017;36:3797–806. doi: 10.1038/onc.2017.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanchez P, Khazaei M, Gatineau E, Geravandi S, Lupse B, Liu H, et al. LDHA is enriched in human islet alpha cells and upregulated in type 2 diabetes. Biochem Biophys Res Commun. 2021;568:158–66. doi: 10.1016/j.bbrc.2021.06.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li M, Wang S, Liu X, Sheng Z, Li B, Li J, et al. Cadmium exposure decreases fasting blood glucose levels and exacerbates type-2 diabetes in a mouse model. Endocrine. 2022;76:53–61. doi: 10.1007/s12020-021-02974-w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are not publicly available because they contain information that could compromise the privacy of patients but are available from the corresponding author upon reasonable request.