Abstract
Even though Helicobacter pylori (H. pylori) is a serious pathogen, its origin is unknown. Poultry (Chicken, Turkey, Quebec, Goose, and Ostrich) are consumed as a regular protein source by a large number of people across the world; therefore, sanitary ways of delivering poultry for food are important for global health. As a result, we looked at the distribution of the pathogenicity cagA, vacA, babA2, oipA, and iceA in H. pylori isolates in poultry meat, as well as their antimicrobial resistance. Wilkins Chalgren anaerobic bacterial medium was used to cultivate 320 raw poultry specimens. Disk diffusion and Multiplex-PCR were used to investigate antimicrobial resistance and genotyping patterns, separately. H. pylori was found in 20 of 320 (6.25%) raw poultry samples. The highest incidence of H. pylori was found in chicken raw meat (15%), whereas the fewest was found in Goose and Quebec (0.00%). Resistance to ampicillin (85%), tetracycline (85%), and amoxicillin (75%) were greatest in H. pylori isolates. The percentage of H. pylori isolates with a MAR value of more than 0.2 was 17/20 (85%). The most prevalent genotypes discovered were VacA s1a (75%), m1a (75%), s2 (70%) and m2 (65%), and cagA (60%). The most typically discovered genotype patterns were s1am1a (45%), s2m1a (45%), and s2m2 (30%). BabA2, OipA + , and OipA− genotypes were found in 40%, 30%, and 30% of the population. In summary, the poultry flesh was polluted by H. pylori, with the babA2, vacA, and cagA genotypes being more prevalent. The simultaneous occurrence of vacA, cagA, iceA, oipA, and babA2 genotypes in antibiotic-resistant H. pylori bacteria implies a serious public health concern about raw poultry eating. In the future, researchers should look into H. pylori's resistance to multiple antibacterial drugs in Iran.
Subject terms: Microbiology, Applied microbiology, Bacteria
Introduction
Poultry (Chicken, Turkey, Quebec, Goose, and Ostrich) is an essential source of proteins for people1. Chickens are killed, skinned, and torn to pieces by hand in regulated slaughtering operations. The corpse is drained, the visceral contents are separated, and the liver, heart, and intestines are gathered during the evisceration process2. The discharge of digestive contents might contaminate these tissues. The corpses are cleaned with water after excoriation, which might be a major cause of bacterial infection3. Poultry is consumed by millions of people around the world every day as a source of animal protein, hence sanitary techniques of delivering hens for food are critical to public healthcare4.
Helicobacter genera are Gram-negative helical coccoid flagellar bacteria that range in length from 2 to 4 μm and breadth from 0.5 to 1.0 μm5. Helicobacter may be quite harmful, and it has been found in the biliary tract and the stomach of a variety of mammals6. These microorganisms are classed as stomach or enterohepatic based on their preferred colonization location7. These two different types of germs are referred to as zoonotic bacteria8. The gastrointestinal Helicobacter colonizes the stomachs in particular; the enterohepatic Helicobacter family colonizes the proximal region of the digestive tract and the biliary duct especially9. Helicobacter pullorum (H. pullorum), a species of the enterohepatic Helicobacter family, was initially derived from the cecum of seemingly healthy domesticated birds10. In addition, poultry was infectedwith Helicobacter hepaticus (H.hepaticus), Helicobacter canis (H. canis), Helicobacter bilis (H. bilis), and Helicobacter cinaedi (H. cinaedi) too11.
Helicobacter pylori (H. pylori) is an opportunistic pathogen linked to stomach cancer and intestinal perforation in humans12. Information on the frequency and distribution of H. pylori is critical for controlling the disease's distribution and identifying high-risk patients, especially in areas where gastritis and stomach cancer are uncommon13. While H. pylori isolates have been identified from a variety of meals, the relevance of animal-derived products in the development of H. pylori infection is unknown14. H. pylori pathogenesis is linked to virulence genes. H. pylori is evolutionarily changeable, according to studies15,16, and particular virulence genes are only found in certain groups. H. pylori has been derived from a variety of clinical specimens and identified using a multiplex polymerase chain reaction (Multiplex-PCR). Several virulence genes in H. pylori isolates have been reported, including Vacuolating cytotoxin A (VacA)17, cytotoxin-associated A (cagA)17, restriction endonuclease A (IceA)18, Outer inflammatory protein A (OipA)19, and blood-group antigen-binding adhesin (BabA2)19. These pathogenicity genes may have a role in the progression of H. pylori. The cagA gene was detected in around half of all H. pylori strains and is involved in intestinal mucosal inflammation, IL-8 generation, and stomach cancer etiology20. Furthermore, researchers discovered that the vacA gene is present overall in H. pylori strains and is involved in the development of stomach cancer and ulceration by destroying the mucous membrane. Different signaling domains and mid-regions make up the vacA genome, which is polymorphic. The s-region is divided into two types: s1 and s2, and the m-region is divided into two types: m1 and m2. The s1 variety is divided into s1a, s1b, and s1c subgroups, while the m1 variety is divided into m1a and m1b subgroups21. The operational state of oipA is controlled by a repair process dependent on CT dinucleotide repetitions, which affect the reading frame and hence determine whether the gene is functional or not22. In H. pylori collected from PUD and gastritis individuals, the iceA gene was discovered. The iceA gene has at least two variants, iceA1, and iceA2. Some research has found that iceA (iceA1/iceA2) is substantially related to digestive tract diseases, whereas others have found the opposite23. In H. pylori, the babA2 gene encodes a membrane protein that helps the bacteria attach to the stomach mucosa24. As a result, multiplex-PCR molecular genotyping of H. pylori is regarded an intense approach to detecting virulence. Among the most efficient ways for studying relationships between H. pylori strains from diverse samples is genotyping using the virulence genes (cagA, vacA, babA2, oipA, and iceA)25.
Medication is another important technique to minimize the transmission of bacteria in the community, given H. pylori's surprising resistance to multiple antimicrobial drugs26. H. pylori’s resistance to antibacterial treatments varies by geography and tends to be rising over time in many areas27–30. Moreover, employing multiple antibiotic resistance (MAR) scores to identify pathogen sources is thought to be a cost-effective and efficient strategy. Krumperman (1983) looked into this index and found that a value of 0.2 implies a greater frequency of illness in areas where antibacterial drugs are commonly utilized31.
There have been no published enough investigations on the antibiotic resistance of H. pylori obtained from edible and non-edible raw poultry (Chicken, Turkey, Quebec, Goose, and Ostrich) in Iran. H. pylori's importance and prevalence in Iran are yet unknown. Human H. pylori infection can be prevented and controlled by eating animal-derived products, particularly fowl (chicken, turkey, Quebec, goose, and ostrich). As a result, the recent study looked at the propagation of the cagA, vacA, babA2, oipA, and iceA pathogenicity genotypes in H. pylori isolates derived from the meat of broiler chickens, turkeys, Quebec goose, and ostriches in vitro, as well as their resistance to multiple antibiotics.
Methods
Sample origin
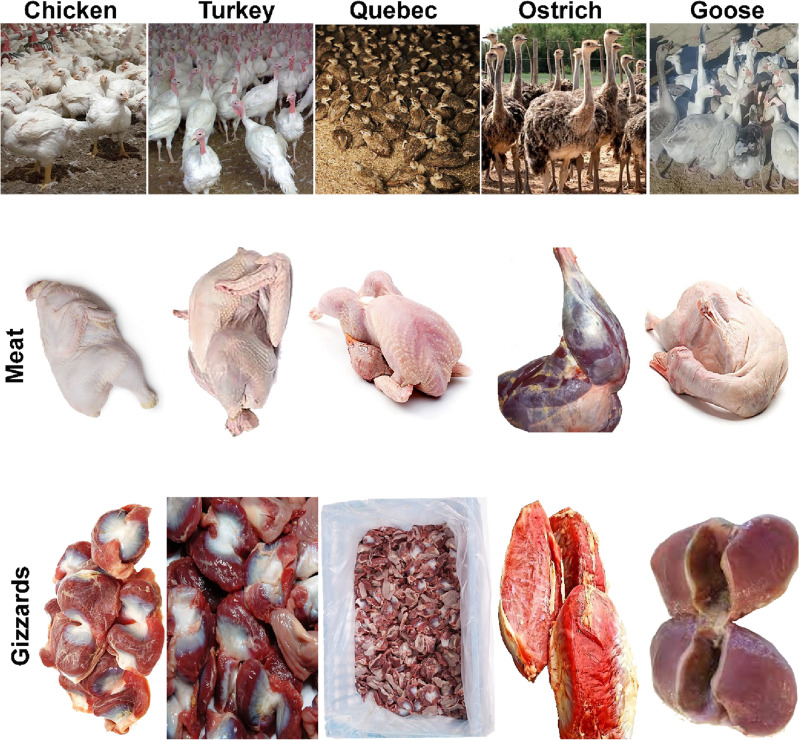
From April to July 2020, 320 Poultry samples were randomly gathered from farms, retail shops, supermarkets, abattoirs…. etc. in the Shahrekord region, Iran, containing specimens of Chicken (n = 60), Turkey (n = 55), Quebec (n = 65), Goose (n = 65), and Ostrich (n = 75). All sample was stored in a specific sterile Ziploc bag that was water-resistant. For isolation and molecular characterization by Multiplex-PCR, samples were gathered from meat, livers, and gizzards, including the jejunum, cecum, and colon (Fig. 1). Till further analysis, all samples were kept at − 80 °C. A statement to confirm that all methods are reported in accordance with ARRIVE guidelines (https://arriveguidelines.org) (PLoS Bio 8(6), e1000412,2010). All studies were conducted per the National Institutes of Health's Guide for the Clinical and Laboratory Standards Institute Animals (NIH Publications No. 8023). The university's Ethics Committee approved them for Animal Care (Iran). The study was approved by the Ethics Committee of the Islamic Azad University of Shahrekord Branch in Iran (IR.IAU.SHK.REC).
Figure 1.
Meat and gizzard samples were collected from chicken, turkey, Quebec, goose, and ostrich from Shahrekord farms.
Helicobacter genus determination
Colony morphology, gram staining, and Biochemical analysis of H. pylori
H. pylori Specific Peptone (HPSP) agar media revealed normal Helicobacter colonies as distinct, round colonies with a width of 0.5–2 mm after 5–7 days of incubation. The gram-negative, S- or C-shaped bacteria were seen by transferring the colonies on slants and staining them with gram. It was discovered that rod and coccoid forms exist. Biochemical tests were performed on the purified cultures to confirm their identity 3. As a control strain, the H. pylori ATCC 700392 strain was used. The urease assay was used to quickly detect H. pylori. On a urea agar medium, a single colony of the investigated microorganism was streaked across the whole surface. For 18–24 h, the samples were incubated at 37 °C in the surrounding atmosphere. H. pylori, which generates cytochrome oxidase enzyme, was also biochemically identified using the oxidase assay. Utilizing oxidase testing kits, all strains' oxidase activities (blue/purple hue) were assessed (Sigma-Aldrich, USA). Furthermore, the collected isolates' catalase test was performed using the drop method. In a nutshell, a purified colony was treated with hydrogen peroxide (H2O2) before being put on a slide. The creation of oxygen bubbles was thought to be a good thing.
Genotypical identification of H. pylori by 16S rRNA-based PCR confirmation
The Helicobacter genus was identified using 16S rRNA (Table 1). Lactofeed Biotech Group approved oligonucleotide sequences (Iran). Wilkins Chalgren anaerobe broth enhanced colonies were sub-cultured. Using a DNA extraction kit, genomic DNA was isolated from bacteria (Cinna-colon, Iran). The process was carried out according to the manufacturer's instructions. The extracted DNA's quality (A260/A280) and quantity were then tested (NanoDrop, Thermo Scientific, Waltham, MA, USA). On a 2% agarose gel dyed with ethidium bromide (0.5 g/mL), the DNA's veracity was evaluated (Thermo Fisher Scientific, St. Leon-Rot, Germany). A PCR thermal cycler (Eppendorf Co., Hamburg, Germany) was used to execute the polymerase chain reaction (PCR) according to the Piri-Gharaghie et al.32 protocol.
Table 1.
Oligonucleotide sequences, product length, and cycling conditions of H. pylori virulence genotypes.
| Target gene | Oligonucleotide sequence (5′—> 3′) | Size (bp) | Tm (°C) | References |
|---|---|---|---|---|
| 16S rRNA | F: CTATGACGGGTATCCGGC | 1026 | 53 | This study |
| R: ATTCCACCTACCTCTCCCA | ||||
| VacA | ||||
| s1a | F: CTCTCGCTTTAGTAGGAGC | 213 | 64 | This study |
| R: CTGCTTGAATGCGCCAAAC | ||||
| s1b | F: AGCGCCATACCGCAAGAG | 187 | 64 | This study |
| R: CTGCTTGAATGCGCCAAAC | ||||
| s1c | F: CTCTCGCTTTAGTGGGGYT | 213 | 64 | This study |
| R: CTGCTTGAATGCGCCAAAC | ||||
| s2 | F: GCTAACACGCCAAATGATCC | 199 | 64 | This study |
| R: CTGCTTGAATGCGCCAAAC | ||||
| m1a | F: GGTCAAAATGCGGTCATGG | 290 | 64 | 52 |
| R: CCATTGGTACCTGTAGAAAC | ||||
| m1b | F: GGCCCCAATGCAGTCATGGA | 291 | 64 | This study |
| R: GCTGTTAGTGCCTAAAGAAGCAT | ||||
| m2 | F: GGAGCCCCAGGAAACATTG | 352 | 64 | This study |
| R: CATAACTAGCGCCTTGCA | ||||
| CagA | ||||
| CagA | F: GATAACAGCCAAGCTTTTGAGG | 300 | 56 | 52 |
| R: CTGCAAAAGATTGTTTGGCAGA | ||||
| IceA | ||||
| IceA1 | F: GTGTTTTTAACCAAAGTATC | 247 | 56 | This study |
| R: CTATAGCCASTYTCTTTGCA | ||||
| IceA2 | F: GTTGGGTATATCACAATTTAT | 229/334 | 56 | This study |
| R: TTRCCCTATTTTCTAGTAGGT | ||||
| OipA | F: GTTTTTGATGCATGGGATTT | 401 | 56 | This study |
| R: GTGCATCTCTTATGGCTTT | ||||
| BabA2 | F: CCAAACGAAACAAAAAGCGT | 105–124 | 57 | This study |
| R: GCTTGTGTAAAAGCCGTCGT | ||||
H. pylori antibiotic sensitivity pattern
There seem to be no generally recognized standardized methods for checking H. pylori antibiotic susceptibilities, and therefore procedures shown in this research were focused on Ranjbar et al.5 and Performance Standards for Antibiotic Sensitivity Testing- Clinical and Laboratory Standards Institute—30th ed CLSI supplement M100. To inoculate Muller Hinton agar plates, bacterial solutions were diluted to 0.5 Mcfarland (equal to 1–2 × 108 CFU/ml). The current study employed antibiotic discs with varied doses to investigate the in vitro susceptibility of H. pylori isolates to antimicrobial drugs routinely used to treat H. pylori. Antimicrobial discs (amoxicillin (10 μg), ampicillin (10 μg), metronidazole (5 μg), streptomycin (10 μg), cefsulodin (30 μg), erythromycin (5 μg), levofloxacin (5 μg), trimethoprim (25 μg), furazolidone (1 μg), clarithromycin (2 μg), rifampin (30 μg), tetracycline (30 μg), and spiramycin (100 μg) (Mast, UK) were used, and the plates were incubated at 35 °C for 16–18 h under anaerobic condition. The standard technique was used to assess and analyze the inhibition zone induced by each antibiotic. H. pylori ATCC 26695 and ATCC 43,504 were used as quality management isolates. The following formula was used to calculate the MAR index of each strain:
Genotyping analysis
Multiplex-PCR was used to determine the prevalence of the cagA, vacA, babA2, oipA, and iceA alleles19–22. The primers and PCR conditions used to genotype the cagA, vacA, babA2, oipA, and iceA alleles are listed in Table 1. Initially, all specimens were subjected to pre-tests to determine the best reaction time, temperature, and volume. In all PCR operations, a programmed DNA thermo-cycler was employed. Positively and negatively controlled were PCR-grade water and H. pylori standard strains (ATCC 43504), respectively. The total volume of 25 µl consisted of 5 µl of deoxy-nucleoside triphosphate mix, 2.5 µl of 10X PCR buffer, 0.25 µl of the primer, and 1 µl of the DNA template, was performed. Ethidium bromide (Sigma, USA) has been used to dye ten microliters of PCR product electrophoresed in a 2 percent agarose gel in 1X TBE buffer at 80 V for 30 min. The UVI doc gel documentation devices (Grade GB004, Jencons PLC, London, UK) were used for image processing.
Analytical statistics
The IBM Statistical Package for the Social Sciences (SPSS) software, version 20.0 for Windows, was used to conduct the statistical study. The information was given in the form of a mean, standard deviation, or percentage. For categorical variables, the Chi-squared test was utilized. At < 0.05, the P value was significant.
Ethical approval
The study was conducted according to the National Academy of Sciences guide for the care and use of laboratory animals and in compliance with best practices of veterinary care. A statement to confirm that all methods are reported in accordance with ARRIVE guidelines (https://arriveguidelines.org).
Results
Helicobacter spp. prevalence in poultry based on morphological and biochemical analysis
In 320 cases of poultry flesh, the presence of H. pylori was evaluated. Table 2 shows the prevalence of Helicobacter spp. in poultry flesh. The urease, oxidase, and catalase assays were used to rapidly diagnose Helicobacter spp. The 20 positive Helicobacter spp. were detected by urease, oxidase, and catalase assays after 4 h of incubation, respectively, by a purple hue, a blue/purple color, and the generation of oxygen bubbles. Helicobacter spp. were found in 20 of 320 (6.25%) poultry meat specimens. According to findings, 9 (15.00%) Chicken specimens, 7 (12.72%) Turkey specimens, 0 (0%) Quebec specimens, 0 (0%) Goose specimens, and 4 (5.33%) Ostrich specimens were all infected with Helicobacter spp.
Table 2.
Prevalence of H. pylori in different types of raw poultry meat samples.
| Raw meat samples | No samples collected | N (%) of H. pylori-positive samples | H. pylori 16SrRNA PCR confirmation (%) |
|---|---|---|---|
| Chicken | 60 | 9 (15.00) | 9 (15.00) |
| Turkey | 55 | 7 (12.72) | 7 (12.72) |
| Quebec | 65 | 0 | 0 |
| Goose | 65 | 0 | 0 |
| Ostrich | 75 | 4 (5.33) | 4 (5.33) |
| Total | 320 | 20 (6.25) | 20 (6.25) |
H. pylori was identified via PCR amplification of the 16SrRNA
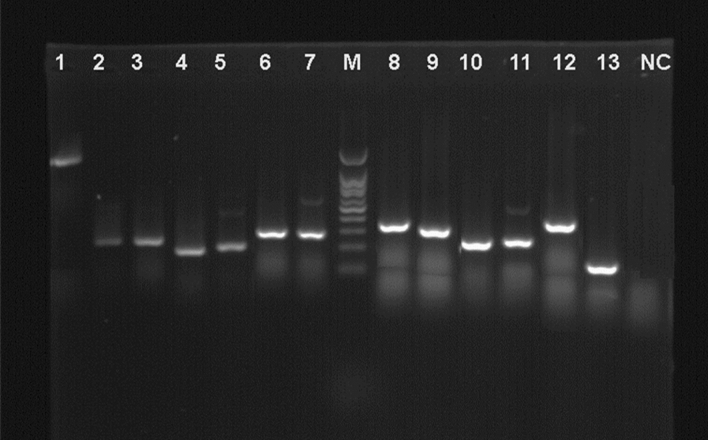
The 16SrRNA gene PCR amplification was used to confirm all of the strains. The electrophoretically displayed PCR results from 20 Helicobacter spp. identified from 320 poultry flesh specimens. H. pylori was recognized as Helicobacter spp. with a 1026-bp PCR product of 16S rRNA in 20/20 (100%). According to PCR results, all 20 (100%) isolates belonged to H. pylori (Fig. 2). The largest incidence of H. pylori bacteria was found in chicken (15.00%) and turkey (12.72%) meat specimens, while the fewest were found in Quebec and goose (0.00%). Between the specimens and the frequency of H. pylori isolates, a significant statistical variation (P < 0.05) was found.
Figure 2.
Gel electrophoresis for the DNA products of each gene. 1: 16SrRNA, 2: VacA s1a, 3: VacA s1c, 4: VacA s1b, 5: VacA s2, 6: VacA m1a, 7: VacA m1b, 8: VacA m2, 9: CagA, 10: IceA1, 11: IceA2, 12: OipA, 13: BabA2, NC: negative control for PCR, M: 100 bp DNA ladder.
H. pylori sensitivity to antibiotics and the MAR index
Antimicrobial resistance profiles of H. pylori isolates recovered from various kinds of specimens collected are depicted in Table 3. Antimicrobial resistance was found to be most common in H. pylori isolates ampicillin (85%), tetracycline (85%), and amoxicillin (75%). H. pylori isolates also had the lowest rate of resistance to furazolidone (5%), spiramycin (30%), cefsulodin (30%), and levofloxacin (30%). Furthermore, resistance to metronidazole (50%) and streptomycin (50%) was common, as was 40% resistance to erythromycin, rifampin, and 35% resistance to trimethoprim and clarithromycin. Results showed that 17/20 (85%) of the H. pylori isolates obtained from poultry samples were resistant to at least three antibiotics. In fact, these isolates showed the Multi-Drag Resistant (MDR) phenotype. There was a statistical difference between the specimens and antimicrobial resistance incidence (P < 0.05).
Table 3.
Antibiotic resistance pattern of H. pylori strains isolated from different types of raw poultry meat samples.
| Type of raw meat samples (N of H. pylori strains) | N (%) isolates resistant to each antibiotic | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AM10a | Met5 | ER5 | CLR2 | AMX 10 | Tet30 | Lev5 | S10 | RIF30 | Cef30 | TRP25 | FZL1 | Spi100 | |
| Chicken (9) | 8 (88.88) | 6 (66.66) | 4 (44.44) | 4 (44.44) | 7 (77.77) | 7 (77.77) | 3 (33.33) | 2 (22.22) | 3 (33.33) | 2 (22.22) | 3 (33.33) | 2 (22.22) | 3 (33.33) |
| Turkey (7) | 5 (71.42) | 2 (28.57) | 3 (42.85) | 2 (28.57) | 5 (71.42) | 7 (100) | 2 (28.57) | 7 (100) | 4 (57.14) | 4 (57.14) | 3 (42.85) | 3 (42.85) | 3 (42.85) |
| Ostrich (4) | 4 (100) | 2 (50) | 1 (25) | 1 (25) | 3 (75) | 3 (75) | 1 (25) | 1 (25) | 1 (25) | – | 1 (25) | – | – |
| Total (20) | 17 (85) | 10 (50) | 8 (40) | 7 (35) | 15 (75) | 17 (85) | 6 (30) | 10 (50) | 8 (40) | 6 (30) | 7 (35) | 5 (25) | 6 (30) |
AM10: ampicillin (10 μg), Met5: metronidazole (5 μg), ER5: erythromycin (5 μg), CLR2: clarithromycin (2 μg), AMX10: amoxicillin (10 μg), Tet30: tetracycline (30 μg), Lev5: levofloxacin (5 μg), S10: streptomycin (10 μg), RIF30: rifampin (30 μg), Cef30: cefsulodin (30 μg), TRP25: trimethoprim (25 μg), FZL1: furazolidone (1 μg) and Spi100: spiramycin (100 μg).
The MAR index of 20 H. pylori isolates in poultry flesh is shown in Table 4. All H. pylori isolates had an average MAR index of 0.622. 17 of the 20 H. pylori isolates tested positive for antibiotic resistance (MDR phenotype), with MAR indexes varying from 0.230 to 1. Strains No. 1 and 2 were highly resistant to all antibacterial agents (MAR index of 1.0), whereas strains Nos. 3–5 were resistant to 12 of the 13 tested antibiotics (MAR index of 0.923). The MAR index for strains 6–7 was 0.846. The MAR scores for Nos. 8–17 ranged from 0.23 to 0.769. Nos. 19 and 20 had the lowest MAR score (0.076). The percentage of H. pylori isolates with a MAR value of more than 0.2 was 17/20 (85%); the frequency with a MAR value of less than 0.2 was 3/20 (15%). As a result, H. pylori is extremely resistant to numerous antibacterial drugs that have been evaluated and have large MAR index values.
Table 4.
Antimicrobial resistance profile of H. pylori strains (n = 20).
| No. | Antimicrobial resistance profile | MAR index |
|---|---|---|
| 1 | AM10, Met5, ER5, CLR2, AMX10, Tet30, Lev5, S10, RIF30, Cef30, TRP25, FZL1, Spi100 | 1 |
| 2 | AM10, Met5, ER5, CLR2, AMX10, Tet30, Lev5, S10, RIF30, Cef30, TRP25, FZL1, Spi100 | 1 |
| 3 | AM10, Met5, ER5, CLR2, AMX10, Tet30, Lev5, S10, RIF30, Cef30, TRP25, FZL1 | 0.923 |
| 4 | AM10, Met5, ER5, CLR2, AMX10, Tet30, Lev5, S10, RIF30, Cef30, TRP25, FZL1 | 0.923 |
| 5 | AM10, Met5, ER5, CLR2, AMX10, Tet30, Lev5, S10, RIF30, Cef30, TRP25, FZL1 | 0.923 |
| 6 | AM10, Met5, ER5, CLR2, AMX10, Tet30, Lev5, S10, RIF30, Cef30, TRP25 | 0.846 |
| 7 | AM10, Met5, ER5, CLR2, AMX10, Tet30, Lev5, S10, RIF30, Cef30, TRP25 | 0.846 |
| 8 | AM10, Met5, ER5, CLR2, AMX10, Tet30, Lev5, S10, RIF30, Cef30 | 0.769 |
| 9 | AM10, Met5, ER5, CLR2, AMX10, Tet30, Lev5, S10, RIF30, Cef30 | 0.769 |
| 10 | AM10, Met5, ER5, CLR2, AMX10, Tet30, Lev5, S10, RIF30, Cef30 | 0.769 |
| 11 | AM10, Met5, ER5, CLR2, AMX10, Tet30, Lev5, S10, RIF30 | 0.692 |
| 12 | AM10, Met5, ER5, CLR2, AMX10, Tet30, Lev5, S10 | 0.615 |
| 13 | AM10, Met5, ER5, CLR2, AMX10, Tet30, Lev5, S10 | 0.615 |
| 14 | AM10, Met5, ER5, CLR2, AMX10, Tet30, Lev5 | 0.538 |
| 15 | AM10, Met5, ER5, CLR2, AMX10 | 0.384 |
| 16 | AM10, Met5, ER5, CLR2 | 0.307 |
| 17 | AM10, Met5, ER5 | 0.230 |
| 18 | AM10, Met5 | 0.153 |
| 19 | AM10 | 0.076 |
| 20 | AM10 | 0.076 |
| Average | 0.622 |
Genotype distribution among H. pylori isolates obtained from various kinds of poultry samples
The genotype distribution of H. pylori isolates recovered from various kinds of poultry specimens is shown in Table 5. The most commonly found genotypes among the H. pylori bacteria isolated from various sorts of poultry specimens were VacA s1a (75%), m1a (75%), s2 (70%), and m2 (65%), and cagA (60%). The H. pylori isolates identified from several sorts of poultry samples with the lowest frequency were VacA s1c (5%) and IceA2 (15%). VacAs1b, VacAm1b, and OipA genes were also found in 25% of Helicobacter pylori strains from various poultry specimens. IceA1 and BabA2 genes were distributed in 40% of the population. Between the kinds of specimens and the incidence of genotypes, there was a statistical difference (P < 0.05).
Table 5.
Distribution of genotypes amongst the H. pylori strains isolated from different types of raw poultry meat samples.
| Type of raw meat samples (N of H. pylori strains) | N (%) isolates harbor each genotype | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VacA | CagA | IceA | OipA | BabA2 | ||||||||
| s1a | s1b | s1c | s2 | m1a | m1b | m2 | IceA1 | IceA2 | ||||
| Chicken (9) | 7 (77.77) | 3 (33.33) | 1 (11.11) | 6 (66.66) | 7 (77.77) | 3 (33.33) | 6 (66.66) | 6 (66.66) | 4 (44.44) | 2 (22.22) | 3 (33.33) | 4 (44.44) |
| Turkey (7) | 6 (85.71) | 2 (28.57) | – | 6 (85.71) | 6 (85.71) | 1 (14.28) | 5 (71.42) | 5 (71.42) | 3 (42.85) | 1 (14.28) | 2 (28.57) | 3 (42.85) |
| Ostrich (4) | 2 (50) | – | – | 2 (50) | 2 (50) | 1 (25) | 2 (50) | 1 (25) | 1 (25) | – | – | 1 (25) |
| Total (20) | 15 (75) | 5 (25) | 1 (5) | 14 (70) | 15 (75) | 5 (25) | 13 (65) | 12 (60) | 8 (40) | 3 (15) | 5 (25) | 8 (40) |
H. pylori strains' genotyping patterns
The genotyping frequency of H. pylori isolates recovered from varying sorts of poultry specimens is shown in Table 6. The most commonly found genotyping patterns of the vacA alleles of H. pylori bacteria originating from various kinds of poultry fresh meat specimens were s1am1a (45%), s2m1a (45%), and s2 m2 (30%). BabA2, OipA + , and OipA− genotypes were distributed 40 percent, 30 percent, and 30 percent, respectively. we discovered that iceA1/iceA2 genotyping was present in 10% of H. pylori isolates. Among the diverse genotyping profiles of H. pylori isolates, S1cm1b (0%), S1 cm2 (5%), S1cm1a (5%), and CagA + (5%) exhibited the lowest frequency. The distribution of other genotypes including s1am1b (15%), s1 am2 (25%), s1bm1a (15%), s1bm1b (10%), s1bm2 (10%), s2m1b (15%), CagA− (25%), BabA2 + (25%) and IceA1/IceA2 (10%) were moderate.
Table 6.
Genotyping pattern of H. pylori strains isolated from different types of raw poultry meat samples.
| Type of raw meat samples (N of H. pylori strains) | Genotyping pattern (%) | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| s1am1a | s1am1b | s1 am2 | s1bm1a | s1bm1b | s1bm2 | s1cm1a | s1cm1b | s1 cm2 | s2m1a | s2m1b | s2 m2 | CagA + | CagA− | IceA1/IceA2 | OipA + | OipA− | BabA2 + | BabA2- | |
| Chicken (9) | 5 (55.55) | 2 (22.22) | 4 (44.44) | 2 (22.22) | 1 (11.11) | 2 (22.22) | 1 (11.11) | – | 1 (11.11) | 4 (44.44) | 2 (22.22) | 3 (33.33) | 6 (66.66) | 3 (33.33) | 1 (11.11) | 3 (33.33) | 6 (66.66) | 4 (44.44) | 5 (55.55) |
| Turkey (7) | 3 (42.85) | 1 (14.28) | 1 (14.28) | 1 (14.28) | 1 (14.28) | - | – | – | – | 3 (42.85) | 1 (14.28) | 2 (28.57) | 3 (42.85) | 2 (28.57) | 1 (14.28) | 2 (28.571) | – | 1 (14.28) | 2 (28.57) |
| Ostrich (4) | 1 (25) | – | – | – | – | – | – | – | – | 2 (50) | – | 1 (25) | 1 (25) | – | – | 1 (25) | – | – | 1 (25) |
| Total (20) | 9 (45) | 3 (15) | 5 (25) | 3 (15) | 2 (10) | 2 (10) | 1 (5) | – | 1 (5) | 9 (45) | 3 (15) | 6 (30) | 10 (5) | 5 (25) | 2 (10) | 6 (30) | 6 (30) | 5 (25) | 8 (40) |
Discussion
Too far, there is little indication that poultry is a major reservoir for the H. pylori bacteria prevalent in people. As a result, the H. pylori bacteria separated from commercial broiler flesh in this investigation are likely to have been acquired during shooting and/or processing. Since men are the bacterium's primary natural host, butcher employees were most probably the principal cause of H. pylori infection in our specimens collected. In the present study, 20 (6.25 percent) H. pylori strains were discovered in 320 commercial poultry samples, indicating that this bacterium poses a risk to humans. Even though the root cause of such a result is unknown, cross-contamination of poultry meat is considered to be a significant source of H. pylori infection in the poultry meat industry. The three primary operations that may increase the incidence of H. pylori infection include cutting, keeping, and shipping poultry meat. Ranjbar et al. discovered that H. pylori can survive in water in a separate study5,32. As a result, an additional cause for the occurrence of H. pylori in the poultry sample obtained is the use of polluted water in the meat industry. Furthermore, contaminated slaughtering personnel and equipment, like blades, may contribute to a higher prevalence of this pathogen33. Generally, our findings are similar to those of Meng et al. (2008), who used Multiplex-PCR to analyze 11 fresh chicken specimens (total chicken including skin) and discovered that 4 (36%) were H. pylori-positive, although our ratios (6.25%) were significantly smaller. H. pylori is also a foodborne organism that may be transferred to humans, according to these investigators34. El Dairouty et al. (2016) reported that 5% of ground beef, raw bird, and sandwich meat specimens tested positive for H. pylori35.
Genomic approaches have subsequently been employed by several studies to discover the various genotypes of H. pylori, which also are strongly connected to its distribution. Multiplex-PCR is a commonly used test for genotyping and identifying homologous genes in H. pylori isolates obtained from clinical specimens35,36. The 16S rRNA gene was employed as a reference gene in this investigation. The frequency of this gene was 100 percent, indicating that the 16S rRNA gene is a good candidate for identifying different H. pylori isolate. Similar findings were achieved by Piri-Gharaghie and El Dairouty et al.32,35. The 16S rRNA genetic code is a unique gene for recognizing bacterial species recovered from specimens, according to these scientists, when contrasted to the other reference genes. H. pylori's proliferation and cell wall formation is dependent on the 16S rRNA gene. As a result, this gene has indeed been widely used to diagnose H. pylori infections36. Our research looks at the incidence of the virulence genes IceA, babA2, OipA, vacA, and cagA. In H. pylori isolates collected from edible and non-edible tissues from the poultry meat industry, the IceA (27.5 percent), babA2 (40 percent), OipA (25 percent), vacA (48.57 percent), and cagA (60 percent) genes were all found. As a result, certain virulence genes, notably cagA, were found in larger numbers in commercial poultry flesh, which is regarded as ready-to-eat human food. The major impediments of H. pylori in the human digestive system are thought to be increased by these genotypes. Bibi et al. earlier hypothesized a relationship between the existence of the H. pylori babA2/cagA + /vacAs1 genotype and the prevalence of gastroenteritis, stomach carcinoma, and ulcerative colitis25. In H. pylori isolates recovered from clinical specimens of human and animal populations, a high incidence of vacA, cagA, iceA1, oipA, and babA2 genotypes has also been described25–38. Moreover, H. pylori isolates recovered from varying sorts of dietary specimens have shown a significant frequency of these genes39,40. Previous studies have linked the H. pylori genotypes vacA, cagA, iceA, oipA, and babA2 to interleukin-8 and cytotoxin exudation, attachment to gastric epithelial cells, increase in the frequency of inflammatory impact, vacuolization, apoptosis process in gastric epithelial cells, stomach ulcers ulceration, increased intense neutrophilic incursion38–42. Consumption of fresh poultry meat infected with virulent isolates of H. pylori would increase duodenum ulcers, gastric epithelium shrinkage, and stomach carcinoma because the H. pylori strain in this experiment carried the vacA, cagA, iceA, oipA, and babA2 genes. Furthermore, certain H. pylori strains tested positive for multiple genotypes at the same time, indicating that they are more harmful43.
Another noteworthy result in the ongoing investigation is the high prevalence of antibiotic resistance among H. pylori isolates. H. pylori bacteria showed significant resistance to antimicrobials ampicillin (85%), tetracycline (85%), and amoxicillin (75%) in this study. Similar findings were found by Mousavi et al. (2014). These researchers discovered that H. pylori bacteria in meat were resistant to ampicillin (84.4%), tetracycline (76.6%), erythromycin (70.5%), and metronidazole (70.5%)44. In addition, previous researchers found that H. pylori in food products showed high levels of resistance to amoxicillin, metronidazole, ampicillin, and oxytetracycline. Furthermore, epidemiologic studies in different countries found that H. pylori isolate in healthcare specimens had a high level of resistance to antimicrobials like metronidazole, ampicillin, tetracycline, and amoxicillin which is consistent with our results39–45. According to with MAR index, 85 percent of the H. pylori isolates tested positive for 3 or more antibiotic medicines employed in the study, indicating a large chance of infection in poultry. Antibiotic resistance may have become more common as a result of the nonselective use of such antibacterial medicines, according to our findings. Many researchers have looked at the incidence of H. pylori resistance to multiple antibiotics, but some of them have run into problems, notably with the number of isolates studied 46,47. Antimicrobial resistance testing revealed that H. pylori were transmitted from infectious poultry samples to meat. The lesser resistance of H. pylori isolates to metronidazole (50%) and streptomycin (50%) was common, as was 40% resistance to erythromycin (40%), rifampin (40%), trimethoprim (35%), and clarithromycin (35%) was also discovered in our investigation, which might be attributed to the antibiotic medications being prescribed less often. There has been some speculation about a link between virulence genes and antibiotic resistance. According to research done in Ireland in 2009, the lack of cagA could be a potential risk for acquiring metronidazole sensitivity48. Other research has linked clarithromycin susceptibility changes to the less pathogenic vacA genotypes49. Some other studies identified a link between cagE and vacA S1 and clarithromycin and metronidazole susceptibility50, whereas others reported no link between cagA or vacA and susceptibility51–53. As a result, it's crucial to determine whether there's a link between the existence of pathogenic indicators and antimicrobial resistance within H. pylori isolates.
Conclusions
H. pylori infection in humans could be spread through raw poultry flesh. This analysis revealed that poultry is another reservoir of pathogenic H. pylori isolates. As a result, slaughterhouses and butchering sanitary measures are critical in reducing the risk of H. pylori infection from poultry meat spreading to humans. Furthermore, the H. pylori isolates showed great resistance to ampicillin (85%), tetracycline (85%), and amoxicillin (75%), as well as having high MAR index values. on the other hand, H. pylori demonstrated lower resistance to metronidazole (50%) and streptomycin (50%) as well as erythromycin (40%), rifampin (40%), trimethoprim (35%), and clarithromycin (35%); hence, we propose utilizing these antibiotic drugs in Iran to combat H. pylori. Also, Our research looks at the incidence of the virulence genes IceA, babA2, OipA, vacA, and cagA. In H. pylori isolates collected from edible and non-edible tissues from the poultry meat industry, the IceA (27.5 percent), babA2 (40 percent), OipA (25 percent), vacA (48.57 percent), and cagA (60 percent) genes were all found. As a result, certain virulence genes, notably cagA, were found in larger numbers in commercial poultry flesh, which is regarded as ready-to-eat human food. The major impediments of H. pylori in the human digestive system are thought to be increased by these genotypes.
Acknowledgements
The authors would like to thank the staff members of the Biotechnology Research Center of the Islamic Azad University of East-Tehran Branch in Iran.
Abbreviations
- Multiplex-PCR
Multiplex polymerase chain reaction
- VacA
Vacuolating cytotoxin A
- cagA
Cytotoxin-associated A
- IceA
Restriction endonuclease A
- OipA
Outer inflammatory protein A
- BabA2
Blood-group antigen-binding adhesin
Author contributions
T.P.G and G.G.: wrote the main draft of the manuscript, M.A.S, and S.T.S.Y: prepared tables, S.KH.D.: sample collection. The authors read and approved the final manuscript.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
This article has been retracted. Please see the retraction notice for more detail: https://doi.org/10.1038/s41598-024-60110-w
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
4/23/2024
This article has been retracted. Please see the Retraction Notice for more detail: 10.1038/s41598-024-60110-w
References
- 1.Oz F, Celik T. Proximate composition, color and nutritional profile of raw and cooked goose meat with different methods. J. Food Process. Preserv. 2015;39(6):2442–2454. doi: 10.1111/jfpp.12494. [DOI] [Google Scholar]
- 2.Piri Gharaghie T, Doosti A, Mirzaei SA. Prevalence and antibiotic resistance pattern of Acinetobacter spp. infections in Shahrekord medical centers. Dev. Biol. 2021;13(4):35–46. [Google Scholar]
- 3.Ricci C, Holton J, Vaira D. Diagnosis of Helicobacter pylori: Invasive and non-invasive tests. Best Pract. Res. Clin. Gastroenterol. 2007;21(2):299–313. doi: 10.1016/j.bpg.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 4.Wong JT, de Bruyn J, Bagnol B, Grieve H, Li M, Pym R, Alders RG. Small-scale poultry and food security in resource-poor settings: A review. Glob. Food Sec. 2017;15:43–52. doi: 10.1016/j.gfs.2017.04.003. [DOI] [Google Scholar]
- 5.Ranjbar R, Farsani FY, Dehkordi FS. Phenotypic analysis of antibiotic resistance and genotypic study of the vacA, cagA, iceA, oipA and babA genotypes of the Helicobacter pylori strains isolated from raw meat. Antimicrob. Resist. Infect. Control. 2018;7(1):1–4. doi: 10.1186/s13756-018-0409-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Waskito LA, Salama NR, Yamaoka Y. Pathogenesis of Helicobacter pylori infection. Helicobacter. 2018;23:e12516. doi: 10.1111/hel.12516. [DOI] [PubMed] [Google Scholar]
- 7.Camilo V, Sugiyama T, Touati E. Pathogenesis of Helicobacter pylori infection. Helicobacter. 2017;22:e12405. doi: 10.1111/hel.12405. [DOI] [PubMed] [Google Scholar]
- 8.Piri Gharaghie T, Hajimohammadi S. Comparison of anti-candida effects of aqueous, ethanolic extracts and essential oil of E. angustifolia with fluconazole on the growth of clinical strains of Candida. New Cell. Mol. Biol. J. 2021;11(43):25–38. [Google Scholar]
- 9.Zarinnezhad A, Shahhoseini MH, Piri GT. Evaluating the relative frequency of fungal infections in the serum of patients with multiple sclerosis and healthy subjects using PCR. BJM. 2021;10(37):37–50. [Google Scholar]
- 10.Javed S, Gul F, Javed K, Bokhari H. Helicobacter pullorum: An emerging zoonotic pathogen. Front. Microbiol. 2017;8:604. doi: 10.3389/fmicb.2017.00604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamada M, Elbehiry A, Marzouk E, Moussa IM, Hessain AM, Alhaji JH, Heme HA, Zahran R, Abdeen E. Helicobacter pylori in a poultry slaughterhouse: Prevalence, genotyping and antibiotic resistance pattern. Saudi J. Biol. Sci. 2018;25(6):1072–1078. doi: 10.1016/j.sjbs.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chehelgerdi M, Doosti A. Effect of the cagW-based gene vaccine on the immunologic properties of BALB/c mouse: An efficient candidate for Helicobacter pylori DNA vaccine. J. Nanobiotechnol. 2020;18(1):1–6. doi: 10.1186/s12951-020-00618-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sepulveda AR. Helicobacter, inflammation, and gastric cancer. Curr. Pathobiol. Rep. 2013;1(1):9–18. doi: 10.1007/s40139-013-0009-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Öztekin M, Yılmaz B, Ağagündüz D, Capasso R. Overview of Helicobacter pylori infection: Clinical features, treatment, and nutritional aspects. Diseases. 2021;9(4):66. doi: 10.3390/diseases9040066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kishk RM, Soliman NM, Anani MM, Nemr N, Salem A, Attia F, Allithy AN, Fouad M. Genotyping of Helicobacter pylori virulence genes caga and vaca: Regional and national study. Int. J. Microbiol. 2021 doi: 10.1155/2021/5540560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Syam AF, Waskito LA, Rezkitha YA, Simamora RM, Yusuf F, Danchi KE, Bakry AF, Mulya E, Siregar GA, Sugihartono T, Maulahela H. Helicobacter pylori in the Indonesian Malay’s descendants might be imported from other ethnicities. Gut Pathog. 2021;13(1):1. doi: 10.1186/s13099-021-00432-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mahmoudi Vashian Z, Doosti A. Cloning and gene expression of ureG gene as a DNA vaccine candidate against Helicobacter pylori. J. GUMS. 2017;26(102):20–29. [Google Scholar]
- 18.Kisiala M, Kowalska M, Pastor M, Korza HJ, Czapinska H, Bochtler M. Restriction endonucleases that cleave RNA/DNA heteroduplexes bind dsDNA in A-like conformation. Nucleic Acids Res. 2020;48(12):6954–6969. doi: 10.1093/nar/gkaa403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rizzato C, Kato I, Plummer M, Muñoz N, Stein A, Jan van Doorn L, Franceschi S, Canzian F. Risk of advanced gastric precancerous lesions in Helicobacter pylori infected subjects is influenced by ABO blood group and cagA status. IJC. 2013;133(2):315–322. doi: 10.1002/ijc.28019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keikha M, Karbalaei M. EPIYA motifs of Helicobacter pylori cagA genotypes and gastrointestinal diseases in the Iranian population: A systematic review and meta-analysis. NMNI. 2021;41:100865. doi: 10.1016/j.nmni.2021.100865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghorbani F, Gheisari E, Dehkordi FS. Genotyping of vacA alleles of Helicobacter pylori strains recovered from some Iranian food items. Trop. J. Pharm. Res. 2016;15(8):1631–1636. doi: 10.4314/tjpr.v15i8.5. [DOI] [Google Scholar]
- 22.Farzi N, Yadegar A, Aghdaei HA, Yamaoka Y, Zali MR. Genetic diversity and functional analysis of oipA gene in association with other virulence factors among Helicobacter pylori isolates from Iranian patients with different gastric diseases. Infect. Genet. Evol. 2018;60:26–34. doi: 10.1016/j.meegid.2018.02.017. [DOI] [PubMed] [Google Scholar]
- 23.Abu-Taleb AM, Abdelattef RS, Abdel-Hady AA, Omran FH, El-Korashi LA, El-hady AA, El-Gebaly AM. Prevalence of Helicobacter pylori cagA and iceA genes and their association with gastrointestinal diseases. Int. J. Microbiol. 2018 doi: 10.1155/2018/4809093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doohan D, Rezkitha YA, Waskito LA, Yamaoka Y, Miftahussurur M. Helicobacter pylori BabA–SabA key roles in the adherence phase: The synergic mechanism for successful colonization and disease development. Toxins. 2021;13(7):485. doi: 10.3390/toxins13070485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bibi F, Alvi SA, Sawan SA, Yasir M, Sawan A, Jiman-Fatani AA, Azhar EI. Detection and genotyping of Helicobacter pylori among gastric ulcer and cancer patients from Saudi Arabia. PJMHS. 2017;33(2):320. doi: 10.12669/pjms.332.12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cardos IA, Zaha DC, Sindhu RK, Cavalu S. Revisiting therapeutic strategies for H. pylori treatment in the context of antibiotic resistance: Focus on alternative and complementary therapies. Molecules. 2021;26(19):6078. doi: 10.3390/molecules26196078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mégraud F. H pylori antibiotic resistance: Prevalence, importance, and advances in testing. Gut. 2004;53(9):1374–1384. doi: 10.1136/gut.2003.022111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ansari H, Doosti A, Kargar M, Bijanzadeh M, Jafarinya M. Antimicrobial resistant determination and prokaryotic expression of smpA gene of Acinetobacter baumannii isolated from admitted patients. Jundishapur J.Microbiol. 10(11), (2017).
- 29.Khademi F, Poursina F, Hosseini E, Akbari M, Safaei HG. Helicobacter pylori in Iran: A systematic review on the antibiotic resistance. IJBMS. 2015;18(1):2. [PMC free article] [PubMed] [Google Scholar]
- 30.Alexander SM, Retnakumar RJ, Chouhan D, Devi TN, Dharmaseelan S, Devadas K, Thapa N, Tamang JP, Lamtha SC, Chattopadhyay S. Helicobacter pylori in human stomach: The inconsistencies in clinical outcomes and the probable causes. Front. Microbiol. 2021;12:2277. doi: 10.3389/fmicb.2021.713955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krumperman PH. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 1983;46(1):165–170. doi: 10.1128/aem.46.1.165-170.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tohid GP, Shandiz SAS. The inhibitory effects of silver nanoparticles on bap gene expression in antibiotic-resistant Acientobacter bumanni isolates using real-time PCR. JIUMS. 2018;26(4):175–85. doi: 10.29252/sjimu.26.4.175. [DOI] [Google Scholar]
- 33.Gilani A, Razavilar V, Rokni N, Rahimi E. VacA and cagA genotypes of Helicobacter pylori isolated from raw meat in Isfahan province, Iran. Vet. Res. Forum. 2017;8:1–75. [PMC free article] [PubMed] [Google Scholar]
- 34.Meng X, Zhang H, Law J, Tsang R, Tsang T. Detection of Helicobacter pylori from food sources by a novel multiplex PCR assay. J. Food Saf. 2008;28(4):609–619. doi: 10.1111/j.1745-4565.2008.00135.x. [DOI] [Google Scholar]
- 35.El Dairouty RK, Murad HA, El Shenawy MA, Hosny IM, Okda AY, El Shamy SM. Helicobacter pylori and its interrelations with other foodborne pathogenic bacteria in Egyptian meat and some meat products. Curr. Sci. Int. 2016;5(2):139–146. [Google Scholar]
- 36.Talimkhani A, Mashak Z. Prevalence and genotyping of Helicobacter pylori isolated from meat, meat and vegetable in Iran. Jundishapur J.Microbiol. 10(11), (2017).
- 37.Momtaz H, Dabiri H, Souod N, Gholami M. Study of Helicobacter pylori genotype status in cows, sheep, goats and human beings. BMC Gastroenterol. 2014;14(1):1–7. doi: 10.1186/1471-230X-14-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dabiri H, Jafari F, Baghaei K, Shokrzadeh L, Abdi S, Pourhoseingholi MA, Mohammadzadeh A. Prevalence of Helicobacter pylori vacA, cagA, cagE, oipA, iceA, babA2 and babB genotypes in Iranian dyspeptic patients. Microb. Pathog. 2017;105:226–230. doi: 10.1016/j.micpath.2017.02.018. [DOI] [PubMed] [Google Scholar]
- 39.Khaji L, Banisharif G, Alavi I. Genotyping of the Helicobacter pylori isolates of raw meat and traditional dairy products. Microbiol. Res. 2017;8(2):43–46. doi: 10.4081/mr.2017.7288. [DOI] [Google Scholar]
- 40.Hemmatinezhad B, Momtaz H, Rahimi E. VacA, cagA, iceA and oipA genotypes status and antimicrobial resistance properties of Helicobacter pylori isolated from various types of ready to eat foods. Ann. Clin. Microbiol. 2016;15(1):1–9. doi: 10.1186/s12941-015-0115-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Torkan S, Shahreza MH. VacA, CagA, IceA and OipA genotype status of Helicobacter pylori isolated from biopsy samples from Iranian dogs. Trop. J. Pharm. Res. 2016;15(2):377–384. doi: 10.4314/tjpr.v15i2.22. [DOI] [Google Scholar]
- 42.Podzorski RP, Podzorski DS, Wuerth A, Tolia V. Analysis of the vacA, cagA, cagE, iceA, and babA2 genes in Helicobacter pylori from sixty-one pediatric patients from the Midwestern United States. Diagn. Microbiol. Infect. 2003;46(2):83–88. doi: 10.1016/S0732-8893(03)00034-8. [DOI] [PubMed] [Google Scholar]
- 43.Alexander SM, Retnakumar RJ, Chouhan D, Devi TN, Dharmaseelan S, Devadas K, Thapa N, Tamang JP, Lamtha SC, Chattopadhyay S. Helicobacter pylori in human stomach: The inconsistencies in clinical outcomes and the probable causes. Front. Microbiol. 2021;2277:713955. doi: 10.3389/fmicb.2021.713955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Souod N, Kargar M, Doosti A, Ranjbar R, Sarshar M. Genetic analysis of cagA and vacA genes in Helicobacter pylori isolates and their relationship with gastroduodenal diseases in the west of Iran. Iran. Red Crescent Med. J. 2013;15(5):371. doi: 10.5812/ircmj.3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mousavi S, Dehkordi FS. Virulence factors and antibiotic resistance of Helicobacter pylori isolated from raw meat and unpasteurized dairy products in Iran. JVATiTD. 2015;20:1–7. doi: 10.1186/1678-9199-20-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yahaghi E, Khamesipour F, Mashayekhi F, Safarpoor Dehkordi F, Sakhaei MH, Masoudimanesh M, Khameneie MK. Helicobacter pylori in vegetables and salads: Genotyping and antimicrobial resistance properties. Biomed Res. Int. 2014;2014:757941. doi: 10.1155/2014/757941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li J, Deng J, Wang Z, Li H, Wan C. Antibiotic resistance of Helicobacter pylori strains isolated from pediatric patients in Southwest China. Front. Microbiol. 2021;11:621791. doi: 10.3389/fmicb.2020.621791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sabbagh P, Mohammadnia-Afrouzi M, Javanian M, Babazadeh A, Koppolu V, Vasigala VR, Nouri HR, Ebrahimpour S. Diagnostic methods for Helicobacter pylori infection: Ideals, options, and limitations. EJCMID. 2019;38(1):55–66. doi: 10.1007/s10096-018-3414-4. [DOI] [PubMed] [Google Scholar]
- 49.Taneike I, Nami A, O’connor A, Fitzgerald N, Murphy P, Qasim A, O’connor H, O’morain C. Analysis of drug resistance and virulence-factor genotype of Irish Helicobacter pylori strains: Is there any relationship between resistance to metronidazole and cagA status. AP&T. 2009;30(7):784–90. doi: 10.1111/j.1365-2036.2009.04095.x. [DOI] [PubMed] [Google Scholar]
- 50.Boyanova L, Markovska R, Yordanov D, Gergova G, Mitov I. Clarithromycin resistance mutations in Helicobacter pylori in association with virulence factors and antibiotic susceptibility of the strains. Microb. Drug Resist. 2016;22(3):227–232. doi: 10.1089/mdr.2015.0199. [DOI] [PubMed] [Google Scholar]
- 51.Ghotaslou R, Leylabadlo HE, Asl YM. Prevalence of antibiotic resistance in Helicobacter pylori: A recent literature review. World J. Methodol. 2015;5(3):164. doi: 10.5662/wjm.v5.i3.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Doorn LJ, Glupczynski Y, Kusters JG, Mégraud F, Midolo P, Maggi-Solcà N, Queiroz DM, Nouhan N, Stet E, Quint WG. Accurate prediction of macrolide resistance in Helicobacter pylori by a PCR line probe assay for detection of mutations in the 23S rRNA gene: multicenter validation study. Antimicrob. Agents Chemother. 2001;45(5):1500–1504. doi: 10.1128/AAC.45.5.1500-1504.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hou P, Tu ZX, Xu GM, Gong YF, Ji XH, Li ZS. Helicobacter pylori vacA genotypes and cagA status and their relationship to associated diseases. World J. Gastroenterol. 2000;6(4):605. doi: 10.3748/wjg.v6.i4.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.