Abstract
Dietary green tea epigallocatechin-3-gallate (EGCG) could attenuate experimental autoimmune encephalomyelitis via the modification of the balance of CD4+ T helper (Th) cells. Moreover, EGCG administration in vitro has a direct impact on the regulatory cytokines and differentiation of CD4+ T cells. Here, we aim to determine whether EGCG directly affects the cell division and progression in naive CD4+ T cells. We first investigate the effect of EGCG on naïve CD4+ T cell division and progression in vitro. An integrated analysis of network pharmacology and molecular docking was utilized to further identify the targets of EGCG for T cell-mediated autoimmune diseases and multiple sclerosis (MS). EGCG treatment prevented naïve CD4+ T cells from progressing through the cell cycle when stimulated with anti-CD3/CD28 antibodies. This was achieved by increasing the proportion of cells arrested in the G0/G1 phase by 8.6% and reducing DNA synthesis activity by 51% in the S phase. Furthermore, EGCG treatment inhibited the expression of cyclins (cyclin D1, cyclin D3, cyclin A, and cyclin B1) and CDKs (CDK2 and CDK6) during naïve CD4+ T cell activation in response to anti-CD3/CD28 stimulation. However, EGCG inhibited the decrease of P27Kip1 (CDKN1B) during naïve CD4+ T cell activation, whereas it inhibited the increase of P21Cip1 (CDKN1A) expression 48 h after mitogenic stimulation. The molecular docking analysis confirmed that these proteins (CD4, CCND1, and CDKN1A) are the primary targets for EGCG, T cell-mediated autoimmune diseases, and MS. Finally, target enrichment analysis indicated that EGCG may affect the cell cycle, T cell receptor signaling pathway, Th cell differentiation, and NF-κB signaling pathway. These findings reveal a crucial role of EGCG in the division and progression of CD4+ T cells, and underscore other potential targets of EGCG in T cell-mediated autoimmune diseases such as MS.
Keywords: EGCG, Network pharmacology, Molecular docking, Cell cycle, T cell-mediated autoimmune diseases, multiple sclerosis
Graphical abstract

Highlights
-
•
EGCG directly inhibited activated naïve T cell division and progression via targeting cell cycle-related proteins.
-
•
There were 64 core targets, including CD4 and cell cycle-related proteins among EGCG, T cell-mediated autoimmune diseases and multiple sclerosis.
-
•
CD4, CCND1, and CDKN1A are the primary targets for EGCG in the treatment of T cell-mediated autoimmune diseases, and MS.
1. Introduction
T cells are critical components of the immune responses to specific infections. Autoimmune diseases caused by abnormal T-cell responses include multiple sclerosis (MS), rheumatoid arthritis (RA), and ulcerative colitis (Ponticelli, 2011). T cell activation can be induced following stimulation of the T cell receptor (TCR) and costimulatory molecules (Shi et al., 2009); additionally, cytokines such as IL-2 have been shown to promote lymphocyte proliferation (Smith, 1980). Accordingly, studies have shown that blocking T-cell activation and proliferation can control T-cell-mediated immunopathogenesis (Schlöder et al., 2022; Xu et al., 2022). Several immunosuppressive drugs are available in clinics, including rapamycin (Brown et al., 1994), FK506 (Liu et al., 1991), and cyclosporine (Liu et al., 1991), for preventing graft rejection and autoimmune diseases by suppressing T cell activation and proliferation. However, their application is limited due to their toxicity and proclivity to induce tolerance (Marcen, 2009). Thus, it is crucial to develop novel suppressive drugs with satisfactory safety and efficacy.
As the most biologically active component of green tea, epigallocatechin-3-gallate (EGCG) has potential health benefits (El-Mowafy et al., 2010), including antioxidation, anti-tumor, and anti-inflammation. In addition to the aforementioned characteristics, EGCG has been shown to exhibit immunomodulating activity (Wu et al., 2009; Kuo et al., 2014; Schwager et al., 2021). We and others reported that EGCG administration could attenuate the severity of experimental autoimmune encephalomyelitis (EAE), an animal model for human MS, by influencing T cell-mediated immune responses (Aktas et al., 2004; Wang et al., 2012b; Janssen et al., 2015). Our serial studies (Pae et al., 2010; Wang et al., 2012a, Wang et al., 2013; Wu et al., 2009) have shown that EGCG directly inhibits T cell function, particularly CD4+ T cells, including CD4+ T cell proliferation, differentiation, and cytokine secretion. The mechanistic investigation reveals that EGCG inhibits IL-2R expression, which impairs IL-2/IL-2R downstream signaling, leading to reduced T cell division and cell cycle. These findings indicate that EGCG inhibits T cell-mediated responses by altering cytokine IL-2 signaling, thereby impairing T cell proliferation and expansion. To date, there has been no evidence reported on the exact mechanisms underlying EGCG's impact on the T cell cycle.
Network pharmacology is a promising approach for predicting the critical targets and biomarkers of a given drug among its multiple targets and biomarkers (Wang et al., 2021; Liu et al., 2022). This study used network pharmacology and experimental evidence to predict EGCG's multiple targets in T-cell-mediated autoimmune diseases and MS. Our findings will shed light on the potential mechanisms underlying EGCG's alternative action on the suppression of cell cycle-related functions in naïve CD4+ T cells.
2. Materials and methods
2.1. Animals
Female C57BL/6 mice, aged 6–8 weeks, were obtained from Charles River (Wilmington, MA). Mice were kept in a 12-h light: dark cycle with a constant temperature and humidity. Ad libitum water and diet were provided. The Animal Care and Use Committee of the Jean Mayer USDA Human Nutrition Research Center on Aging at Tufts University approved all conditions and handling, which were conducted following the NIH Guidelines for the Care and Use of Laboratory Animals.
2.2. Naïve CD4+ T cell purification
After mice were sacrificed by CO2, the spleens were aseptically removed, and the single-cell suspension was prepared. The CD4+CD62L + T Cell Isolation Kit II (Miltenyi Biotec) was used to isolate naïve CD4+ T cells according to the manufacturer's protocol. The purity of naïve CD4+ T cells was higher than 95%. Purified naïve CD4+ T cells were cultured in 24-well culture plates in RPMI 1640 medium supplemented with 5% FBS, 25 mmol/L HEPES, 2 mmol/L glutamine, 100 kU/L penicillin, and 100 mg/L streptomycin (all from Gibco Invitrogen).
2.3. Cell viability
EGCG was added to naïve CD4+ T cells at concentrations of 0 and 10 μmol/L for 72 h. Subsequently, 10 μL of CCK-8 solution was added to each well and incubated at 37 °C for 1 h. Using a microplate reader (SpectraMax i3x, Molecular Devices, United States), the absorbance at 450 nm was measured. The formula (absorbance of treated cells/absorbance of control cells) × 100 was utilized to determine cell viability.
2.4. T cell proliferation
Purified naïve CD4+ T cells were stained with 0.5 μM carboxyfluorescein succinimidyl ester (CFSE) and then pre-incubated with or without EGCG (10 μmol/L) (Sigma-Aldrich) for 2 h. The cells were subsequently stimulated with immobilized anti-CD3 Ab (5 mg/L) and soluble anti-CD28 Ab (1 mg/L) (Both from BD Biosciences) (anti-CD3/CD28) in 24-well culture plates at 37 °C for 72 h. This concentration of EGCG did not affect the viability of naïve CD4+ T cells, and was selected based on our previous studies (Wu et al., 2009; Pae et al., 2010; Wang et al., 2012a, 2013). CFSE can enter cells and interact with amine groups, leading to the production of fluorescence. This fluorescence remains stable within cells, and it is evenly distributed to both daughter cell populations during cell division. Flow cytometry was used to determine cell division in an Accuri C6 flow cytometer. Acquired data were analyzed using FlowJo 7.6 software.
2.5. Ki-67 and p-RB expression for cell proliferation
Naïve CD4+ T cells were pre-incubated with or without EGCG for 2 h and then stimulated for 48 h with anti-CD3/CD28 Abs, before being blocked with anti-moused CD16/CD32 Ab and then stained with FITC-conjugated anti-CD4 Ab (both from BD Bioscience). They were then fixed and permeabilized using Foxp3 Fixed/Perm buffer (BD Bioscience) and then stained with PE-conjugated anti-Ki-67 (eBioscience) and Alexa Fluor® 647 conjugated anti-RB (pS807/pS811) (Cell Signaling Technology, Inc.) was performed. Data were acquired using an Accuri C6 flow cytometer and FlowJo 7.6 software was used for analysis.
2.6. Cell cycle analysis
After naïve CD4+ T cells pre-incubated with or without EGCG for 2 h and then stimulated with anti-CD3/CD28 Abs in 24-well culture plates for 48 h, cell cycle analysis was carried out by flow cytometry using the APC BrdU Flow kit (BD Biosciences). Briefly, cells were treated with 10 μmol/L BrdU 45 min before being harvested. After cells were stained with fluorescent conjugated anti-CD4 Ab, they were fixed, permeabilized, and then labeled with the APC conjugated anti-BrdU Ab following the manufacturer's instructions (BD Biosciences). The cell cycle properties were then analyzed using BrdU incorporation and 7-AAD staining. Data were analyzed using FlowJo 7.6 software after flow cytometry was performed on an Accuri C6 flow cytometer.
2.7. Western blot
For the Western blot analysis, cells were incubated with or without EGCG for 2 h and then stimulated with anti-CD3/CD28 Abs for the indicated time. The cells were then lysed in RIPA cell lysis buffer, which contained a protease inhibitor cocktail (Roche Applied Science) and a phosphatase inhibitor cocktail (Sigma-Aldrich). Protein concentration was determined using the Bradford method (Bio-Rad protein assay dye), and equal amounts of protein were loaded onto SDS polyacrylamide gels for electrophoretic separation. After that, the protein was transferred to a nitrocellulose membrane. Subsequently, the membrane was blocked with 5% non-fat milk before being incubated with specific primary antibodies targeting the following proteins: P21Cip1 and P27Kip1 (1:1000, both from BD Biosciences), CDK2, Cyclin A, Cyclin B1 (1:1000, all from Santa Cruz), CDK6, Cyclin D1, Cyclin D3 (1:1000, all from Cell Signaling Technologies) or β-actin (1:10000, Sigma). Finally, the membrane was exposed to ECL (Pierce, Rockford, IL, USA) after being incubated with HRP-conjugated secondary antibodies.
2.8. Prediction of candidate targets of EGCG, T-cell-mediated autoimmune diseases, and MS
The EGCG molecular structure was obtained from PubChem Database and its target was predicted utilizing the Swiss Target Prediction database (http://www.swisstargetprediction.ch/index.php), Comparative Toxicogenomics Database (CTD) (http://ctdbase.org/) and Traditional Chinese Medicine Systems Pharmacology (TCMSP) database (https://tcmspw.com/tcmsp.php). Furthermore, gene-disease molecular targets for T-cell-mediated autoimmune diseases and MS were collected using CTD, GeneCards (https://www.genecards.org/), and DisgeNet (https://www.disgenet.org/).
2.9. Protein-protein interaction (PPI) analysis
String Database (https://string-db.org/) (Szklarczyk et al., 2019) was used to establish the PPI analysis for the intersection targets of EGCG, T cell-mediated autoimmune diseases, and MS. The results were presented using Cytoscape 3.7.2 (Shannon et al., 2003).
2.10. Gene ontology (GO) and kyoto encyclopedia of genes and genomes (KEGG) pathway enrichment analysis
The predicted targets of Homo sapiens were imported into the DAVID database (Database for Annotation, Visualization, and Integrated Discovery, https://david.ncifcrf.Gov) to perform GO and KEGG pathway enrichment analysis. The top 10 significantly enriched biological process (BP), cellular component (CC), and molecular function (MF) terms, as well as the top 20 significantly enriched KEGG pathways, were used by the R project to generate a bar graph and bubble plot, respectively, based on a P-value of less than 0.05.
2.11. Molecular docking
The molecular docking was conducted as previously described (Liu et al., 2022). Briefly, the RCSB Protein Data Bank (PDB) and PubChem databases were used to obtain the structure of core targets and the 2D molecular form of EGCG. The 2D molecular structure of EGCG was then converted into a 3D model using the ChemBio3D Ultra 14.0 software. The molecular docking was conducted using PyMoL and the AutoDockTools-1.5.6 software.
2.12. Statistical analysis
Results were presented as mean ± SEM of repeated experiments, as indicated in the figure legends, with the cells from different mice used for each repeat. Prism 6.0 statistical software was used to perform a one-way analysis of variance (NAOVA) test for three and more groups, followed by Tukey's HSD post hoc test. The significance level was set at P < 0.05.
3. Results
3.1. EGCG inhibits cell division and proliferative response in mitogen-activated naïve CD4+ T cells
Our previous report found that EGCG supplementation modulates the proliferation, cytokine production, and differentiation of CD4+ T cells that contributes to attenuating CD4+ T cell-mediated autoimmune diseases (Pae et al., 2010). However, it is unknown whether EGCG may directly inhibit naïve CD4+ T cell proliferation. We initially assessed the impact of EGCG at 10 μM on the viability of naïve CD4+ T cells through the use of a CCK8 kit. However, no significant differences were observed in cell viability between 0 and 10 μM EGCG (100.7 ± 0.69 vs 99.71 ± 0.72). We subsequently examined the effect of EGCG on naïve CD4+ T cell division. As shown in Fig. 1A–B, EGCG inhibited naïve CD4+ T cell division after stimulation with anti-CD3/CD28 Abs. Furthermore, we found that EGCG inhibited the expression of Ki-67 (Fig. 1C–E), an antigen associated with nuclear cell proliferation that is expressed during all active stages of the cell cycle (Schluter et al., 1993).
Fig. 1.
EGCG inhibits cell division (A and B) and a nuclear cell proliferation-associated antigen, Ki-67 expression (C to E) in anti-CD3/CD28-stimulated naïve CD4+ T cells. The histograms show a representative experiment (A and C), and the bar figures are mean ± SEM of three independent experiments (B, D and E). Means without a common letter differ at P < 0.05. Unsti: unstimulated naïve CD4+ T cells; -EGCG: anti-CD3/CD28-stmulated naïve CD4+ T cells without EGCG; +EGCG: anti-CD3/CD28-stmulated naïve CD4+ T cells with EGCG; MFI: mean fluorescence intensity.
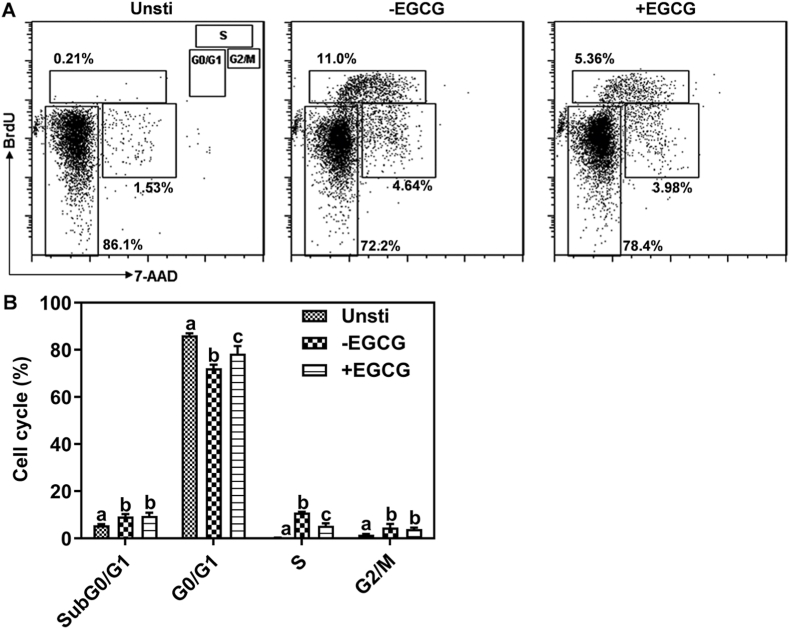
3.2. EGCG induced cell cycle arrest in mitogen-activated naïve CD4+ T cells
Next, we investigated how EGCG affects naïve CD4+ T cell proliferation following anti-CD3/CD28 stimulation. The cell cycle process of naïve CD4+ T cells was impeded by EGCG as evidenced by increased cell arrest in G0/G1 phase by 8.6%, decreased DNA synthesis activity in S phase by 51%, and no effect on the progression into G2/M phage (Fig. 2).
Fig. 2.
EGCG inhibits cell progression in anti-CD3/CD28-stimulated naïve CD4+ T cells. The dot plots show a representative experiment (A), and the bar figures are mean ± SEM of three independent experiments (B). Means without a common letter differ at P < 0.05. Unsti: unstimulated naïve CD4+ T cells; -EGCG: anti-CD3/CD28-stmulated naïve CD4+ T cells without EGCG; +EGCG: anti-CD3/CD28-stmulated naïve CD4+ T cells with EGCG.
3.3. EGCG inhibits cell cycle-regulatory protein expression in mitogen-activated naïve CD4+ T cells
The retinoblastoma protein (RB) is responsible for a critical G0/G1 checkpoint that prevents S-phase entry and cell growth. When cells are ready to divide, RB is phosphorylated, rendering it inactive, and allowing the cell cycle to progress. Therefore, we wanted to determine whether EGCG treatment altered RB phosphorylation (p-RB) in anti-CD3/CD28-stimulated CD4+ T cells. Anti-CD3/CD28 stimulation demonstrably increased RB phosphorylation in naïve CD4+ T cells (Fig. 3A–C). However, such effect was reversed by EGCG administration.
Fig. 3.
EGCG inhibits cell cycle-related protein expression in anti-CD3/CD28-stimulated naïve CD4+ T cells. (A–C) Flow cytometry detected the retinoblastoma protein phosphorylation in anti-CD3/CD28-stimulated naïve CD4+ T cells. The histograms show a representative experiment (A), and the bar figures are mean ± SEM of three independent experiments (B and C). (D) Western blot detected the cell cyclin-related protein expression in anti-CD3/CD28-stimulated naïve CD4+ T cells. Data are representatives of three independent experiments. Means without a common letter differ at P < 0.05. Unsti: unstimulated naïve CD4+ T cells; -EGCG: anti-CD3/CD28-stmulated naïve CD4+ T cells without EGCG; +EGCG: anti-CD3/CD28-stmulated naïve CD4+ T cells with EGCG; MFI: mean fluorescence intensity.
Furthermore, the transverse of G0/G1 and entry into the S phase is controlled by the sequential activation of complexes containing D cyclins, CDKs, cyclin A, and cyclin B. Here, we first used Western blot to determine the protein expression of cyclins and cyclin-dependent kinases (CDK). We found that anti-CD3/CD28 stimulation upregulated the expression of cyclins (cyclin D1, cyclin D3, cyclin A, and cyclin B1) and CDKs (CDK2 and CDK6) in naïve CD4+ T cells. However, EGCG prevented this effect (Fig. 3D).
Mitogenic stimulation downregulates the expression of P27kip1, a CDK inhibitor that accumulates in quiescent cells. As shown in Fig. 3D, P27kip1 expression was high in naïve CD4+ T cells that were downregulated by anti-CD3/CD28 stimulation. However, EGCG prevented the decrease of P27kip1 during naïve CD4+ T cell activation. According to network pharmacology prediction, P21Cip1 (CDKN1A), another inhibitor of CDKs, is a core target of EGCG, T-cell-mediated autoimmune diseases, and MS, we also evaluated the effect of EGCG on P21Cip1 expression in anti-CD3/CD28-activated naïve CD4+ T cells. We found that it has marginal expression in naïve CD4+ T cells and activated naïve CD4+ T cells 24 h following anti-CD3/CD28 stimulation. However, P21Cip1 expression increased 48 h after mitogenic stimulation, whereas EGCG treatment decreased its expression (Fig. 3D).
3.4. EGCG's target prediction and protein-protein regulation network construction
Fig. 4A showed the two-dimensional structure of EGCG obtained from PubChem Database, and 124 targets of EGCG were identified utilizing the Swiss Target Prediction database, CTD and TCMSP database. We further constructed a protein regulatory network with 125 link nodes representing the EGCG's potential targets (Fig. 4B).
Fig. 4.
The 2D structure of EGCG (A) and its target proteins (B).
3.5. Construction of the protein-protein regulation network of the predictive targets of EGCG, T cell-mediated autoimmune diseases, and MS
We identified 3011 targets for T cell-mediated autoimmune disorders and 1005 targets for MS using Genecards and the CTD Database (Fig. 5A). The intersection of the 797 targets was defined as the candidate targets of T cell-mediated autoimmune disorders and MS. Venn diagram analysis showed the intersection of 64 targets in EGCG, T cell-mediated autoimmune diseases, and MS (Fig. 5B), and these targets were used to construct a protein-protein regulatory network with 64 link nodes (Fig. 5C). We further used a string database to draw a PPI network of 64 targets (Fig. 5D) and analyzed PPI with Cytoscape 3.7.2 software to determine the most critical indicators related to T cells, cell cycle and inflammation (Fig. 5E), including CD4, CCND1, CDNK1A, IL-6, STAT1, STAT3, and TNF. The indicators are more critical because of the brighter color and the larger area (Fig. 5E).
Fig. 5.
Construction of the protein-protein regulation network of the predictive targets of EGCG, T cell-mediated autoimmune diseases, and MS. (A) Venn diagram shows the intersection targets between MS and T cell-mediated autoimmune diseases. (B) Venn diagram shows the targets that EGCG and T cell-mediated autoimmune diseases intersect with MS. (C) PPI network of EGCG, T cell-mediated autoimmune diseases and MS. (D) PPI network of 64 targets of EGCG, T cell-mediated autoimmune diseases and MS. (E) PPI network diagram of potential target degree value. The node size is proportional to the degree. PPI, protein-protein interaction.
3.6. GO and KEGG pathway enrichment analysis of the predictive targets of EGCG, T cell-mediated autoimmune diseases, and MS
We used DAVID to enrich GO terms (BP, CC, and MF) with P < 0.05. The top 10 terms for BP, CC, and MF were presented as bar plots (Fig. 6A). These terms include positive regulation of smooth muscle cell proliferation, positive regulation of gene expression, positive regulation of cell migration, aging, and other terms. Furthermore, the top twenty KEGG pathways presented in Fig. 6B revealed that the main enriched pathways of EGCG in modulating T cell-mediated autoimmune diseases and MS includes cell cycle, MAPK signaling pathway, chemokine signaling pathway, Th cell (Th1, Th2, and Th17) differentiation, NF-κB signaling pathway, and other pathways. A HIF-1 signaling pathway is associated with the cell cycle-related protein P21Cip1 (CDKN1A).
Fig. 6.
GO (A) and KEGG pathway (B) enrichment analysis of EGCG, T cell-mediated autoimmune diseases, and MS. The larger green bubbles indicate more significant P-values for the enriched target genes. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.7. Molecular docking analysis of the predictive targets of EGCG, T cell-mediated autoimmune diseases, and MS
Molecular docking is commonly used to design new drugs by investigating the interactions of receptor biomacromolecules and their ligands. Furthermore, molecular docking can predict binding affinity and assess ligand confirmation. The activity of compound-target protein binding can be predicted by binding energy. For a compound, the lower the binding energy it has, the more tightly it binds to its target protein, and the more stable its conformation is. Binding energy less than −5 kcal/mol indicates good binding ability, while binding energy less than −7 kcal/mol suggests high activity (Pinzi and Rastelli, 2019). To learn more about the interaction between EGGC and its various targets (CD4, TNF, CDKN1A, STAT1, STAT3, CCND1 and IL-6), the molecular docking verification was performed as described above in the “Materials and Methods” section. As shown in Fig. 7, all the targets and EGCG had binding energies that were less than −5 kcal/mol, suggesting that EGCG has a strong binding effect on its various target proteins and has a high degree of matching. The strongest binding ability of EGCG was demonstrated with CD4, TNF, CDKN1A, STAT1 and STAT3, all of which have binding energies of less than −7 kcal/mol. This finding suggests the interaction between EGCG and T cell-mediated autoimmune diseases such as MS via these target proteins.
Fig. 7.
Binding modes (A) and docking score (B) of 7 related protein targets and EGCG.
4. Discussion
Autoimmunity is defined as an overactive immune system that attacks the host's self-tissues. Autoimmune disorders can manifest as a wide range of autoimmune diseases in different organs, tissues, and systems. Although the etiology of autoimmune diseases is still unclear, T cells, especially CD4+ T cells, are crucial in their pathogenesis. MS is one of these autoimmunity disorders that is characterized by inflammatory cell infiltration in the CNS, multifocal white matter demyelination, axonal damage, and oligodendrocyte destruction. EAE is a commonly used experimental animal model that exhibits MS pathological characteristics. A series of studies by us (Wu et al., 2009; Pae et al., 2010; Wang et al., 2012a, 2012b, 2013) have demonstrated that EGCG administration could improve the clinical symptoms of EAE, which is associated with suppression in CD4+ T cell proliferation, proinflammatory Th1/Th17 cell differentiation, TNF-α production, TNF signaling pathway, Th1/Th2 cell differentiation, and NFκB signaling pathway, as well as by a reversal of IL-6-induced inhibition of Treg cell differentiation via inhibiting IL-6/JAK/STAT3 signaling pathway, which could be predicted by network pharmacology and molecular docking in this study.
The generation of a pool of daughter cells from a small number of naïve T cells is a necessary step in mounting an adaptive immune response against pathogens. However, abnormal T-cell division can cause various autoimmune diseases (Ponticelli, 2011). Thus, strict regulation of cell proliferation is required to maintain the immune system's delicate balance. EGCG blocks T cell proliferation and modulates the balance among CD4+ T cell subsets, including inhibition of pro-inflammatory Th1 and Th17 cells and promotion of Treg cells, resulting in alleviated clinical symptoms of EAE (Wang et al., 2012a, 2013). However, the change in the balance of Th cell subsets cannot explain T cells' inability to proliferate in response to EGCG treatment. We thus decided to investigate the mechanisms by which EGCG blocks T-cell proliferation.
Network pharmacology predicted that CD4 is the intersection among EGCG, T-cell-mediated autoimmune diseases and MS. Thus, we focused on examining EGCG's impact on CD4+ T cell responses. We found that impairment of naive T-cell division was accompanied by an inability to upregulate the expression of Ki-67, a nuclear cell proliferation-associated marker (Schluter et al., 1993).
During cell division, activated T cells go through a cell cycle process. To better understand the action of EGCG on naïve CD4+ T cell division, we characterized the T cell cycle and found that following stimulation with anti-CD3/CD28 Abs, a significantly higher proportion of EGCG-treated T cells was in the G0/G1 phase than untreated T cells. The cell cycle is comprised of four distinct phases: M, G0/G1, S and G2 phase (Golias et al., 2004). Following stimulation with anti-CD3/CD28 Abs, IL-2 can promote T cell cycle progression via JAK3/STAT5 pathways (Shi et al., 2009). It has been shown that EGCG inhibits IL-2 binding to its receptor and limits STAT5 phosphorylation in primarily activated T cells (Wang et al., 2012a). These findings suggest that EGCG's potent inhibitory effect on STAT5 activation may be related to its inhibitory effect on T cell cycle and proliferation.
Retinoblastoma protein (Rb) regulates the cell cycle by repressing the transcriptional activator E2F, which controls the expression of cell cycle-regulatory proteins, such as cyclin E, cyclin A, Cdc25A, and E2F itself (Wu et al., 1995; Rotgers et al., 2014). The repression is lifted by the phosphorylation of Rb to progress through the G0/G1 phase. Thus, lowering p-Rb helps maintain cells in the G0/G1 phase. EGCG has been shown to inhibit the Rb phosphorylation in estrogen-induced MCF cancer cells (Huang et al., 2008). Our findings are consistent with this study in that EGCG inhibits Rb's phosphorylation in naïve CD4+ T cells following stimulation with TCR/CD28.
Cell cycle regulatory proteins such as cyclin A, cyclin B, and cyclin D can be upregulated after Rb's repression is removed by phosphorylation. Due to a lack of enzymatic activity, cyclins act by forming cyclin/CDK complexes with their respective CDKs (Wells and Morawski, 2014). Thus, cyclins and CDKs are critical regulators of cell growth. Following stimulation with mitogens and growth factors, cells produce cyclin D, cyclin A, cyclin E, and CDKs like CDK2 and CDK6 (Rowell et al., 2006; Rodriguez et al., 2007). Consistent with these findings, we found that naïve T cells increased expression of cyclin A, cyclin D1, cyclin D3, and cyclin B1 in response to anti-CD3/CD28 Abs. Evidence has shown that CDKs promote T cell proliferation and induce T cell activation and differentiation. CDK2 and is a pivotal regulator supporting T cells from G1→S transit and effector T cell differentiation (Ekholm and Reed, 2000; Veiga-Fernandes and Rocha, 2004; Chunder et al., 2012; Kreslavsky et al., 2012; Zhang et al., 2013). Here, we showed that TCR/CD28 engagement increased the expression of CDK2 and CDK6.
In contrast, co-administration of EGCG with TCR/CD28 reduced the expression of cell cycle-regulatory proteins. EGCG has been shown to reduce cyclin D3 levels in estradiol-treated MCF7 cells while not affecting cyclin A and CDK2 protein levels (Huang et al., 2008). This difference could be attributed to the difference in cell type and treatment. However, it remains to be determined whether EGCG impacts CDK activity in the presence of TCR/CD28 engagement. Future research using CDK knockout mice would also aid in better characterizing the target(s) of EGCG in T cell-mediated immune responses.
In addition to CDKs, cyclin-dependent kinase inhibitors (CDKIs) help maintain cell cycle progression (Vidal and Koff, 2000). Cell cycle progression is facilitated by cyclins’ binding to CDKs to form active holoenzymes (Vidal and Koff, 2000). CDKIs such as P21Cip1 and P27Kip1 inhibit these holoenzymes, particularly, P27kip1, which is expressed constitutively in the resting naïve T cells and inhibits cyclin D/CDK6 and cyclin E/CDK2 activity (Rowell and Wells, 2006). Evidence suggests that P27kip1 controls T cell proliferation and mediates immune tolerance in gene knockout and transgenic mice (Zhang et al., 2000; Shen and Kaplan, 2002; Rowell et al., 2006). Levels of P27Kip1 are reduced in response to stimulation of both Ag receptors and cytokine signaling (Kwon et al., 1997). Indeed, using anti-CD3/CD28 co-stimulation, naïve CD4+ T cells constitutively express P27kip1, which is down-regulated after 24 and 48 h of culture. However, the administration of EGCG prevents the decrease of the P27kip1 protein. Thus, these findings suggest that treatment of EGCG treatment increases protein levels of P27kip1 resulting in inhibited T cell proliferation.
Within hours, the cellular levels of P27Kip1 are primarily modulated via post-translation (Pagano et al., 1995). IL-2 or TCR stimulation causes T cell proliferation and growth (Appleman et al., 2000). We have previously shown that EGCG can block IL-2/IL-2R signaling (Wang et al., 2012a). Thus, these findings suggest that reducing cytokine-mediated signaling may be an approach to preventing EGCG's ability to reduce P27kip1. EGCG can also prevent P27kip1 degradation (Huang et al., 2008). Furthermore, CDKN1A (P21Cip1), a key target of EGCG, T-cell-mediated autoimmune disorders and MS, negatively regulates long-term stimulated T cell proliferation, and it is crucial for preserving tolerance toward nuclear antigens (Balomenos et al., 2000). However, in this study, EGCG reversed the upregulation of P21Cip1 in naïve CD4+ T cells after stimulation, suggesting that P21Cip1 may not be involved in EGCG's effect on T cell function. Therefore, further investigation is necessary to understand the underlying mechanisms.
This study focuses on the critical role of EGCG in the progression of activated naive CD4+ T cells in mice and measures the expression of cell-cycle-related proteins. These proteins are core targets of EGCG, which may be beneficial in treating T-cell-mediated autoimmune diseases and MS. There are a few limitations of this study. First, we did not determine whether EGCG binds directly to these targets to regulate T cell division and progression. Second, we did not determine whether EGCG modulates the expression of these targets in human T cell division and MS. Finally, we only measured CDKIs P21Cip1 and P27Kip1, however, there are other CDKIs, such as P16ink4 and P18ink4c (Rowell et al., 2006), that affect T cell proliferation and differentiation, and whether these CDKIs are impacted by EGCG should be investigated in the future studies.
In summary, network pharmacology prediction supported the effect of ECGC's targets on T cell-mediated autoimmune diseases and MS. Importantly, we also verified the role of EGCG in the control of naïve CD4+ T cell proliferation in vitro via affecting cell cycle and cell cycle-regulatory protein expression. This effect on the cell cycle is attributed to the effect of EGCG on T cell proliferation. Further investigation on T cell cycle-regulatory protein levels in TCR/CD28 engagement suggests a primary role for cyclins, CDKs, and CDIs in T cells, which may help in identifying other relevant targets involved in the antiproliferative effect of EGCG.
CRediT authorship contribution statement
Xinli Niu: Data curation, Formal analysis, Investigation, Writing – original draft. Zejin Liu: Data curation, Formal analysis, Investigation, Writing – original draft. Junpeng Wang: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Supervision, Writing - original draft, Writing - review & editing. Dayong Wu: Conceptualization, Investigation, Supervision, Writing - original draft, Writing - review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (U2004104).
Handling Editor: Professor A.G. Marangoni
Contributor Information
Junpeng Wang, Email: jpwangchina@henu.edu.cn.
Dayong Wu, Email: dayong.wu@tufts.edu.
Data availability
Data will be made available on request.
References
- Aktas O., Prozorovski T., Smorodchenko A., Savaskan N.E., Lauster R., Kloetzel P.M., Infante-Duarte C., Brocke S., Zipp F. Green tea epigallocatechin-3-gallate mediates T cellular NF-kappa B inhibition and exerts neuroprotection in autoimmune encephalomyelitis. J. Immunol. 2004;173(9):5794–5800. doi: 10.4049/jimmunol.173.9.5794. 173/9/5794. [DOI] [PubMed] [Google Scholar]
- Appleman L.J., Berezovskaya A., Grass I., Boussiotis V.A. CD28 costimulation mediates T cell expansion via IL-2-independent and IL-2-dependent regulation of cell cycle progression. J. Immunol. 2000;164(1):144–151. doi: 10.4049/jimmunol.164.1.144. ji_v164n1p.144. [DOI] [PubMed] [Google Scholar]
- Balomenos D., Martín-Caballero J., García M.I., Prieto I., Flores J.M., Serrano M., Martínez-A C. The cell cycle inhibitor p21 controls T-cell proliferation and sex-linked lupus development. Nat. Med. 2000;6(2):171–176. doi: 10.1038/72272. [DOI] [PubMed] [Google Scholar]
- Brown E.J., Albers M.W., Shin T.B., Ichikawa K., Keith C.T., Lane W.S., Schreiber S.L. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature. 1994;369(6483):756–758. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- Chunder N., Wang L., Chen C., Hancock W.W., Wells A.D. Cyclin-dependent kinase 2 controls peripheral immune tolerance. J. Immunol. 2012;189(12):5659–5666. doi: 10.4049/jimmunol.1202313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekholm S.V., Reed S.I. Regulation of G(1) cyclin-dependent kinases in the mammalian cell cycle. Curr. Opin. Cell Biol. 2000;12(6):676–684. doi: 10.1016/s0955-0674(00)00151-4. S0955-0674(00)00151-4. [DOI] [PubMed] [Google Scholar]
- El-Mowafy A.M., Al-Gayyar M.M., Salem H.A., El-Mesery M.E., Darweish M.M. Novel chemotherapeutic and renal protective effects for the green tea (EGCG): role of oxidative stress and inflammatory-cytokine signaling. Phytomedicine. 2010;17(14):1067–1075. doi: 10.1016/j.phymed.2010.08.004. [DOI] [PubMed] [Google Scholar]
- Golias C.H., Charalabopoulos A., Charalabopoulos K. Cell proliferation and cell cycle control: a mini review. Int. J. Clin. Pract. 2004;58(12):1134–1141. doi: 10.1111/j.1742-1241.2004.00284.x. [DOI] [PubMed] [Google Scholar]
- Huang H.C., Way T.D., Lin C.L., Lin J.K. EGCG stabilizes p27kip1 in E2-stimulated MCF-7 cells through down-regulation of the Skp2 protein. Endocrinology. 2008;149(12):5972–5983. doi: 10.1210/en.2008-0408. [DOI] [PubMed] [Google Scholar]
- Janssen A., Fiebiger S., Bros H., Hertwig L., Romero-Suarez S., Hamann I., Chanvillard C., Bellmann-Strobl J., Paul F., Millward J.M., Infante-Duarte C. Treatment of chronic experimental autoimmune encephalomyelitis with epigallocatechin-3-gallate and glatiramer acetate alters expression of heme-oxygenase-1. PLoS One. 2015;10(6) doi: 10.1371/journal.pone.0130251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreslavsky T., Gleimer M., Miyazaki M., Choi Y., Gagnon E., Murre C., Sicinski P., von Boehmer H. beta-Selection-induced proliferation is required for alphabeta T cell differentiation. Immunity. 2012;37(5):840–853. doi: 10.1016/j.immuni.2012.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo C.L., Chen T.S., Liou S.Y., Hsieh C.C. Immunomodulatory effects of EGCG fraction of green tea extract in innate and adaptive immunity via T regulatory cells in murine model. Immunopharmacol. Immunotoxicol. 2014;36(5):364–370. doi: 10.3109/08923973.2014.953637. [DOI] [PubMed] [Google Scholar]
- Kwon T.K., Buchholz M.A., Ponsalle P., Chrest F.J., Nordin A.A. The regulation of p27Kip1 expression following the polyclonal activation of murine G0 T cells. J. Immunol. 1997;158(12):5642–5648. [PubMed] [Google Scholar]
- Liu J., Farmer J.D., Jr., Lane W.S., Friedman J., Weissman I., Schreiber S.L. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell. 1991;66(4):807–815. doi: 10.1016/0092-8674(91)90124-h. 0092-8674(91)90124-H. [DOI] [PubMed] [Google Scholar]
- Liu Z., Niu X., Wang J. Naringenin as a natural immunomodulator against T cell-mediated autoimmune diseases: literature review and network-based pharmacology study. Crit. Rev. Food Sci. Nutr. 2022;1–18 doi: 10.1080/10408398.2022.2092054. [DOI] [PubMed] [Google Scholar]
- Marcen R. Immunosuppressive drugs in kidney transplantation: impact on patient survival, and incidence of cardiovascular disease, malignancy and infection. Drugs. 2009;69(16):2227–2243. doi: 10.2165/11319260-000000000-000004. [DOI] [PubMed] [Google Scholar]
- Pae M., Ren Z., Meydani M., Shang F., Meydani S.N., Wu D. Epigallocatechin-3-gallate directly suppresses T cell proliferation through impaired IL-2 utilization and cell cycle progression. J. Nutr. 2010;140(8):1509–1515. doi: 10.3945/jn.110.124743. [DOI] [PubMed] [Google Scholar]
- Pagano M., Tam S.W., Theodoras A.M., Beer-Romero P., Del Sal G., Chau V., Yew P.R., Draetta G.F., Rolfe M. Role of the ubiquitin-proteasome pathway in regulating abundance of the cyclin-dependent kinase inhibitor p27. Science. 1995;269(5224):682–685. doi: 10.1126/science.7624798. [DOI] [PubMed] [Google Scholar]
- Pinzi L., Rastelli G. Molecular docking: shifting paradigms in drug discovery. Int. J. Mol. Sci. 2019;20(18):4331. doi: 10.3390/ijms20184331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponticelli C. Present and future of immunosuppressive therapy in kidney transplantation. Transplant. Proc. 2011;43(6):2439–2440. doi: 10.1016/j.transproceed.2011.06.025. [DOI] [PubMed] [Google Scholar]
- Rodriguez P.C., Quiceno D.G., Ochoa A.C. L-arginine availability regulates T-lymphocyte cell-cycle progression. Blood. 2007;109(4):1568–1573. doi: 10.1182/blood-2006-06-031856. blood-2006-06-031856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotgers E., Rivero-Muller A., Nurmio M., Parvinen M., Guillou F., Huhtaniemi I., Kotaja N., Bourguiba-Hachemi S., Toppari J. Retinoblastoma protein (RB) interacts with E2F3 to control terminal differentiation of Sertoli cells. Cell Death Dis. 2014;5 doi: 10.1038/cddis.2014.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowell E.A., Wang L., Hancock W.W., Wells A.D. The cyclin-dependent kinase inhibitor p27kip1 is required for transplantation tolerance induced by costimulatory blockade. J. Immunol. 2006;177(8):5169–5176. doi: 10.4049/jimmunol.177.8.5169. 177/8/5169. [DOI] [PubMed] [Google Scholar]
- Rowell E.A., Wells A.D. The role of cyclin-dependent kinases in T-cell development, proliferation, and function. Crit. Rev. Immunol. 2006;26(3):189–212. doi: 10.1615/critrevimmunol.v26.i3.10. 3355ca53351ee89f,0d07ca0139b1446a. [DOI] [PubMed] [Google Scholar]
- Schlöder J., Shahneh F., Schneider F.J., Wieschendorf B. Boosting regulatory T cell function for the treatment of autoimmune diseases - that's only half the battle. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.973813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter C., Duchrow M., Wohlenberg C., Becker M.H., Key G., Flad H.D., Gerdes J. The cell proliferation-associated antigen of antibody Ki-67: a very large, ubiquitous nuclear protein with numerous repeated elements, representing a new kind of cell cycle-maintaining proteins. J. Cell Biol. 1993;123(3):513–522. doi: 10.1083/jcb.123.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwager J., Seifert N., Bompard A., Raederstorff D., Bendik I. Resveratrol, EGCG and vitamins modulate activated T lymphocytes. Molecules. 2021;26(18):5600. doi: 10.3390/molecules26185600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen R., Kaplan M.H. The homeostasis but not the differentiation of T cells is regulated by p27(Kip1) J. Immunol. 2002;169(2):714–721. doi: 10.4049/jimmunol.169.2.714. [DOI] [PubMed] [Google Scholar]
- Shi M., Lin T.H., Appell K.C., Berg L.J. Cell cycle progression following naive T cell activation is independent of Jak3/common gamma-chain cytokine signals. J. Immunol. 2009;183(7):4493–4501. doi: 10.4049/jimmunol.0804339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K.A. T-cell growth factor. Immunol. Rev. 1980;51:337–357. doi: 10.1111/j.1600-065x.1980.tb00327.x. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P., Jensen L.J., Mering C.V. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veiga-Fernandes H., Rocha B. High expression of active CDK6 in the cytoplasm of CD8 memory cells favors rapid division. Nat. Immunol. 2004;5(1):31–37. doi: 10.1038/ni1015. [DOI] [PubMed] [Google Scholar]
- Vidal A., Koff A. Cell-cycle inhibitors: three families united by a common cause. Gene. 2000;247(1–2):1–15. doi: 10.1016/s0378-1119(00)00092-5. S0378-1119(00)00092-5[pii] [DOI] [PubMed] [Google Scholar]
- Wang J., Pae M., Meydani S.N., Wu D. Epigallocatechin-3-gallate inhibits expression of receptors for T cell regulatory cytokines and their downstream signaling in mouse CD4+ T cells. J. Nutr. 2012;142(3):566–571. doi: 10.3945/jn.111.154419. [DOI] [PubMed] [Google Scholar]
- Wang J., Pae M., Meydani S.N., Wu D. Green tea epigallocatechin-3-gallate modulates differentiation of naive CD4(+) T cells into specific lineage effector cells. J. Mol. Med. 2013;91(4):485–495. doi: 10.1007/s00109-012-0964-2. [DOI] [PubMed] [Google Scholar]
- Wang J., Ren Z., Xu Y., Xiao S., Meydani S.N., Wu D. Epigallocatechin-3-gallate ameliorates experimental autoimmune encephalomyelitis by altering balance among CD4+ T-cell subsets. Am. J. Pathol. 2012;180(1):221–234. doi: 10.1016/j.ajpath.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.Y., Chu F.H., Gu N.N., Wang Y., Feng D., Zhao X., Meng X.D., Zhang W.T., Li C.F., Chen Y., Wei S.S., Ma Z.Q., Lin R.C., Zhao C.J., Zou D.X. Integrated strategy of LC-MS and network pharmacology for predicting active constituents and pharmacological mechanisms of Ranunculus japonicus Thunb. for treating rheumatoid arthritis. J. Ethnopharmacol. 2021;271 doi: 10.1016/j.jep.2021.113818. [DOI] [PubMed] [Google Scholar]
- Wells A.D., Morawski P.A. New roles for cyclin-dependent kinases in T cell biology: linking cell division and differentiation. Nat. Rev. Immunol. 2014;14(4):261–270. doi: 10.1038/nri3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.L., Zukerberg L.R., Ngwu C., Harlow E., Lees J.A. In vivo association of E2F and DP family proteins. Mol. Cell Biol. 1995;15(5):2536–2546. doi: 10.1128/mcb.15.5.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., Guo Z., Ren Z., Guo W., Meydani S.N. Green tea EGCG suppresses T cell proliferation through impairment of IL-2/IL-2 receptor signaling. Free Radic. Biol. Med. 2009;47(5):636–643. doi: 10.1016/j.freeradbiomed.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Xu L., He D., Zhang C., Bai Y., Zhang C. The regulate function of polysaccharides and oligosaccharides that with sulfate group on immune-related disease. J. Funct.Foods. 2022;88 doi: 10.1016/j.jff.2021.104870. [DOI] [Google Scholar]
- Zhang S., Lawless V.A., Kaplan M.H. Cytokine-stimulated T lymphocyte proliferation is regulated by p27Kip1. J. Immunol. 2000;165(11):6270–6277. doi: 10.4049/jimmunol.165.11.6270. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Liu Q., Leskov K.S., Wu X., Duan J., Zhang G.L., Hall M., Rosenbaum J.T. Roscovitine suppresses CD4+ T cells and T cell-mediated experimental uveitis. PLoS One. 2013;8(11) doi: 10.1371/journal.pone.0081154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.