Summary
Background
Male infertility is a worldwide population health concern, but its aetiology remains largely understood. Although CFAP70 variants have already been reported in two oligo-astheno-teratozoospermia (OAT) individuals by sequencing, animal evidence to support CFAP70 as a credible OAT-pathogenic gene is lacking.
Method
Cfap70-KO mice were generated to explore the physiological role of CFAP70. CFAP70 variants were detected in infertile men with OAT by whole exome sequencing and Sanger sequencing confirmation. Cfap70-truncated mice were further generated to explore the pathogenicity of the nonsense variant of CFAP70 identified in the proband.
Findings
Here, we demonstrate that Cfap70-KO mice are sterile mainly due to OAT and further identify a Chinese infertile man carrying a homozygous nonsense variant (c.2962C > T/p.R988X) of CFAP70. Cfap70-truncated mice lacking 5–8 tetratricopeptide repeats (TPRs) mimic the patient's symptoms. CFAP70 is required for the biogenesis of spermatid flagella partially by regulating the expression of OAT-associated proteins (e.g., QRICH2), assisting the cytoplasmic preassembly of the calmodulin- and radial spoke-associated complex (CSC), and controlling the manchette localization of axoneme-related proteins. Moreover, we suggest that CFAP70-associated male infertility could be overcome by intracytoplasmic sperm injection (ICSI) treatment.
Interpretation
Overall, we demonstrate that CFAP70 is necessary to assemble spermatid flagella and that CFAP70 gene could be used as a diagnostic target for male infertility with OAT in the clinic.
Funding
This study was supported by the National Key Research and Development Project (2019YFA0802101 to S.C), Open Fund of Key Laboratory of Cell Proliferation and Regulation Biology, Ministry of Education (to S.C), Central Government to Guide Local Scientific and Technological Development (ZY21195023 to B.W), and Basic Research Projects of Central Scientific Research Institutes (to B.W).
Keywords: Male infertility, Oligo-astheno-teratozoospermia, CFAP70, Knockout mice, Sperm flagellar assembly
Research in context.
Evidence before this study
FAP70, a homolog of the mammalian CFAP70, resides at the axonemal structure in Chlamydomonas. Whole exome sequencing identifies two CFAP70 variants in two unrelated OAT individuals. However, the animal evidence (e.g., Cfap70-KO mice to reveal the physiological role of CFAP70, and Cfap70-mutant mice to mimic the situation in patients) to support CFAP70 as an OAT-pathogenic gene are lacking. The potential mechanisms underlying the regulation of sperm flagella by CFAP70 are also unclear.
Added value of this study
Here, we show that Cfap70-KO male mice are sterile due to OAT. Furthermore, we identify a homozygous nonsense variant of CFAP70 in an infertile man with OAT and generate a Cfap70-truncated mice to mimic the patient's symptom. We also explore the potential mechanisms underlying the regulation of spermatid flagellar biogenesis by CFAP70.
Implications of all the available evidence
CFAP70 mutations could be used as a diagnostic target for infertile male patients primarily with OAT.
Introduction
Infertility affects 10–15% of couples worldwide, and male factors account for approximately half of these cases.1 However, genetic causes of male infertility remain largely elusive and unexplained. Azoospermia/oligozoospermia (no sperm or decreased sperm concentration), asthenozoospermia (lower sperm motility), and teratozoospermia (higher ratio of morphologically abnormal sperm) are common types of male infertility.2 Azoospermia/oligozoospermia is likely due to spermatogenetic defects (e.g., meiotic arrest or spermiogenic arrest), whereas asthenozoospermia is generally caused by the deficiency of sperm flagellar function. Deformation of spermiogenesis, the last step of spermatogenesis, generates different subtypes of teratozoospermia, including globozoospermia (round-headed sperm), multiple morphological abnormalities of the sperm flagella (MMAF, abnormal flagellar phenotypes), and acephalic spermatozoa syndrome (ASS, disrupted sperm head–tail junction).3, 4, 5 Our understanding of the genetic causes of male infertility has been improved by the development of high-throughput sequencing in cohorts of sterile men; however, the vast majority of infertile men remain undiagnosed.6
Both motile cilia and sperm flagella are highly conserved, microtubule-based subcellular organelles, and they share similar ‘9 + 2’ axonemal structures. PCD (MIM: 244400) is a multisystem disorder (e.g., chronic airway diseases, situs inversus, hydrocephalus, and/or sterility) caused by dysfunction of motile cilia/flagella. MMAF (MIM: 617576) is a subtype of astheno-teratozoospermia proposed in 2014 and is characterized by short, coiled, absent, and/or irregular flagella without any other symptoms of PCD.7 As a result of severe flagellar abnormalities, MMAF impairs sperm motility and can lead to total sperm immotility. Electron microscope analyses have revealed that MMAF results from ultrastructural disorders in the axoneme and accessory structures of flagella, including the central pair (CP) of microtubules, outer and inner dynein arms (ODA and IDA), nexin–dynein regulatory complex (DRC), radial spokes (RS), fibrous sheath (FS), and mitochondrial sheath (MS). In both male infertile patients and corresponding mouse models, members of the cilia and flagella associated protein (CFAP) family, such as CFAP43, CFAP44,8,9 CFAP47,10 CFAP58,11 CFAP61,12 CFAP65,13 CFAP69,14,15 CFAP91,16 CFAP206,17,18 and CFAP251,19,20 have been demonstrated to be MMAF-associated genes.
Cryo-electron tomography reveals that Chlamydomonas FAP70 localizes at the base of the ODA and is a component of the central apparatus projection C2a.21,22 Knockout of FAP70 in Chlamydomonas causes a reduction in cilia motility without affecting cilia length.21 Intriguingly, Beurois et al. identified two individuals carrying homozygous CFAP70 variants (c.1723-1G > T, c.178 T > A) from two unrelated MMAF-affected infertile men.23 However, whether CFAP70 is truly an MMAF-associated gene remains unclear because Cfap70-KO and Cfap70-mutant animal models have not yet been constructed to reveal the physiological roles of CFAP70 in mammalian male reproduction and the pathogenicity of point variants in CFAP70, respectively.
In this study, we report that Cfap70-KO mice exhibit male infertility with OAT and that CFAP70 is required for the assembly of flagella in mouse early spermatids. A homozygous nonsense variant (c.2962C > T/p.R988X) in CFAP70 was identified in a Chinese infertile man with OAT. This nonsense variant generates a truncated CFAP70 protein lacking the C-terminal 5–8 tetratricopeptide repeats (TPRs). We further generated Cfap70-truncated mice lacking 5–8 TPRs that recapitulate the OAT phenomena of the patient.
Methods
Ethics approval and consent to participate
One OAT patient (proband, II-1) and his fertile brother (II-2) from a Chinese family were enrolled in this study. The human study was approved by the Ethics Committee of the National Research Institute for Family Planning (#2021010), and informed consent was obtained. The OAT patient (II-1) was 33 years old with normal physical development. The couple had regular sexual intercourse with endovaginal ejaculation and no contraception, and his wife had no obvious fertility-related abnormalities. However, his wife has been unable to conceive for 5 years. The results of the patient's physical examination revealed normal development of male external genitalia, normal bilateral testicular size, no abnormality in the bilateral spermatic veins, normal hormone levels and no PCD-related symptoms. The proband exhibited a normal karyotype (46, XY), and no microdeletions were found on the Y chromosome. Five millilitres of peripheral blood was collected from the proband (II-1) and his brother (II-2).
Semen characteristics analysis
Fresh semen samples were collected from the proband (II-1) and his brother (II-2) via masturbation after 2–7 days of sexual abstinence and evaluated after liquefaction for 30 min at 37 °C. Semen analyses were conducted in accordance with World Health Organization (WHO) guidelines.24 At least 200 spermatozoa were examined to evaluate the percentages of morphologically normal spermatozoa. Sperm motility was assessed by a computer-assisted sperm analysis (CASA) system (Cyto-S, VideoTesT, Switzerland). The morphological abnormalities of sperm flagella were classified into five categories: (1) absent, (2) short, (3) coiled flagella, (4) angulation, and (5) irregular calibre. One spermatozoon was classified in only one morphological category according to its major flagellar abnormality.
WES and bioinformatic analysis
Genomic DNA was extracted from peripheral blood samples of the proband (II-1) using a QIAamp DNA Blood Mini Kit (QIAGEN, Germany). The exomes of the subject were captured by Agilent SureSelect Human All Exon V6 Enrichment kits (Agilent, CA, USA) and then sequenced on a NovaSeq platform (Illumina, CA, USA) according to the manufacturer's instructions. All reads were mapped to the human reference genome (UCSC Genome Browser hg19) using Burrows‒Wheeler Alignment.25 Single nucleotide variants and indels were detected using the Genome Analysis Toolkit,26 and then annotated by ANNOVAR.27 Variants that met the following criteria were retained for further analysis: i) missense, nonsense, frameshift, nonframeshift or splicing site variants; ii) missense variants predicted to be deleterious by at least two of the following online software programs: SIFT, PolyPhen-2 and MutationTaster28, 29, 30; iii) variants that were absent or rare (variants with a minor allele frequency <0.1%) from East Asia and the total population in the gnomAD dataset. All candidate variants derived from WES are listed in Table S1. The nonsense variant in CFAP70 was verified by Sanger sequencing using the following primers: 5′-AGTGAGCGGAGATTGAGC-3′ and 5′-CAGCCTGCCTGAGTTTGA-3’.
Mouse models
Animal experiments were approved by the Animal Care and Use Committee of the College of Life Sciences, Beijing Normal University (2022-SW-017). The Cfap70 gene (NCBI: NM_001163638.1; Ensembl: ENSMUSG00000039543) is located on mouse chromosome 14. A total of 29 exons were identified, with the ATG start codon in exon 2 and the TGA stop codon in exon 29. Cfap70-KO and Cfap70-truncated mice were generated by CRISPR/Cas9 technology. Exons 3–8 of the Cfap70-203 transcript were targeted: gRNAs (5’→3′) TATACATGTTAACGGGACAG (PAM: TGG) and GCCCATGGGCATGCACTAAG (PAM: GGG). To generate Cfap70-truncated mice, exon 25 was selected as the target site. Exon 25 starts approximately 85.79% into the coding region, and deletion of a small fragment within exon 25 is expected to generate a truncated CFAP70 lacking TPRs 5–8: gRNA sequence (5’→3’) AGCATTCTTTGGCCTCAGCA (PAM: TGG). The CRISPR/Cas9-mediated gene deletion procedure has been described previously.31 The gRNAs were synthesized by Sangon Biotech (Shanghai, China). The two complementary DNA oligos of each gRNA target were annealed and ligated to the pUC57-sgRNA plasmid (Addgene, MA, USA) for cloning. The recombinant plasmid was transformed into DH5α competent cells, and the positive clones were screened based on kanamycin resistance and sequencing. The recombinant plasmid was linearized and purified by phenol chloroform extraction. Transcripts of the gRNAs in vitro were performed using the MEGAshortscript Kit and purified using the MEGAclear Kit (Ambion, TX, USA). Mouse zygotes were coinjected with an RNA mixture of Cas9 mRNA (50 ng/μL; TriLink BioTechnologies, CA, USA) and sgRNA (30 ng/μL). The injected zygotes were transferred into pseudopregnant recipients to obtain the F0 generation. DNA was extracted from tail tissues from 7-day-old offspring, and PCR amplification was performed with genotyping primers (Table S2) using the Mouse Tissue Direct PCR Kit (Tiangen Biotech, Beijing, China). A stable F1 generation (heterozygous mice) was obtained by mating positive F0 generation mice with wild-type C57BL/6JG-pt mice.
Fertility test
To confirm the fertility of the mice, natural mating tests were conducted.32 Briefly, sexually mature Cfap70-KO male mice and their littermate wild-type male mice were individually caged with sexually mature wild-type females for 2 months. The vaginal plugs of the mice were examined every morning. Then, the female mice with vaginal plugs were separately housed, and the litter sizes were recorded.
Histological analysis of mouse tissues
Testes were dissected from adult Cfap70-KO mice and their littermate wild-type mice and fixed in Bouin's solution (Bedebio, Beijing, China) for 4 h at 4 °C. The heart, liver, spleen, lung, kidney, retina, intestine, trachea, epididymis, and oviduct were fixed in 4% PFA overnight at 4 °C. Fixed tissues were embedded in paraffin, sectioned (5 μm thick), dewaxed, and rehydrated. The sections of testes were stained with Periodic Acid Schiff's solution (Solarbio, Beijing, China), and the sections of other tissues were stained with H&E solution before imaging using a Leica DM-500 optical microscope (Leica Microsystems, German).
Mouse sperm count and sperm motility
Semen samples of adult Cfap70-KO and their littermate wild-type mice were collected from the cauda epididymis, diluted in 1 mL sperm rinse (Vitrolife, Sweden), and examined after incubation for 15 min at 37 °C. Sperm counts were determined using a Fertility Counting Chamber (Makler, Israel) under a light microscope, and sperm mobility was assessed via the application of the CASA system (Hamilton Thorne-TOX IVOS, USA).
Papanicolaou staining of sperm
Sperm suspensions from humans (II-1 and II-2) and mice (Cfap70-KO and WT) were mounted on a glass slide, air-dried, and fixed with 4% PFA for 20 min at room temperature. The slides were stained with Papanicolaou solution (Solarbio) and observed using a Leica DM-500 optical microscope (Leica Microsystems).
Intracytoplasmic sperm injection (ICSI)
Eight-week-old C57BL/6 J female mice were superovulated by administration of 10 IU PMSG combined with 10 IU hCG (48 h later). Oocytes were obtained from the ampulla of the uterine tube at 14 h after hCG injection. Sperm heads of adult Cfap70-KO mice and their littermate wild-type mice were separated from sperm tails and injected into mouse oocytes using a Piezo-driven pipette (PrimeTech, Osaka, Japan). The injected oocytes were cultured in KSOM medium at 37 °C under 5% CO2. Two-cell embryos and blastocysts were counted 20 h and 96 h later, respectively. All reagents were purchased from Nanjing Aibei Biotechnology (Naijing, China). Blastocysts were transferred into pseudopregnant recipients to obtain offspring. DNA was extracted from tail tissues from 7-day-old offspring, and PCR amplification was carried out with genotyping primers (Table S2) using the Mouse Tissue Direct PCR Kit.
Expression plasmids and transient transfection
Mouse Cfap70 cDNA was chemically synthesized (Sangon) and inserted into Flag- or HA-tagged pCMV vectors (Beyotime, Shanghai, China). The QRICH2-Myc plasmid was a gift from Dr. Ying Shen (West China Second University Hospital, Sichuan University, China)33; the CFAP61-Myc and CFAP91-Myc plasmids were gifts from Dr. Ming-Xi Liu (State Key Laboratory of Reproductive Medicine, Nanjing Medical University, China).34,35 Full-length cDNAs encoding DNAI1, DNAI2, ODAD3, DNALI1, SPAG6, RSPH1, RSPH3, RSPH9, DRC2, and DRC4 were amplified by PCR using mouse testis cDNA as the template and cloned into Myc-tagged pCMV vectors (Beyotime). The primers for plasmid construction are listed in Table S3. The construction of expression plasmids in this study was confirmed by sequencing (Sango Biotech). HEK293T cells (ATCC, VA, USA) were cultured at 37 °C in a 5% CO2 incubator with Dulbecco's modified Eagle's medium (DMEM) with 10% foetal bovine serum (FBS) and 1% penicillin–streptomycin (HyClone, UT, USA). Transient transfection of HEK293T cells was performed using Lipofectamine 3000 transfection reagent (Thermo Fisher Scientific, CA, USA) following the manufacturer's protocol. Cells were then harvested 48 h after transfection. The cycloheximide (CHX) used in protein degradation analysis was purchased from MedChemExpress (MCE, Shanghai, China).
Enrichment of haploid spermatids by flow sorting
The testes from Cfap70-KO mice and their littermate wild-type mice were decapsulated, and seminiferous tubules were mechanically dispersed. The seminiferous tubules were incubated with 1 mg/mL collagenase IV in PBS for approximately 15 min on a shaker and washed twice with PBS to roughly remove Leydig cells. The sediment was further digested with 1 mg/mL collagenase IV, 1 mg/mL hyaluronidase, 1 mg/mL trypsin, and 0.5 mg/mL DNase I for 15 min on a shaker. This digestion step produced a testicular cell suspension. All enzymes were purchased from Solarbio (Beijing, China). Hoechst 33,342 at a final concentration of 2 μg/mL was added to the DMEM of testicular cells, followed by incubate at 34 °C for 1 h in the dark. Samples were placed in a tube rotator every 10 min. The cell suspension was filtered with a prewetted 40 μm strainer, and the filtered solution was kept on ice and in the dark until FACS using a FACS Arial II flow cytometer (BD Company, CA, USA). Cell debris was excluded based on the FSC vs. SSC plot pattern. The DNA content gate was set by plotting a histogram of cell counts based on Hoechst blue fluorescence level. Three peaks with increasing concentrations of Hoechst blue fluorescence should appear, representing haploid (1C), diploid (2C), and tetraploid (4C) cells. Three gates were set, one for each peak. Using a 70 μm nozzle, the cells were sorted at a rate of 1000–2000 cells/second. The selected subpopulation gates were collected into the collection tubes with 1 mL DMEM. The collected cells were centrifuged at 600 g at 4 °C for 10 min, and the cell pellet was resuspended in 1 mL DMEM. Approximately 40 μL of washed cells were pipetted onto a clean glass slide and fixed with 4% PFA for 20 min. The purity of isolated haploid spermatids was confirmed by staining with the acrosome dye PNA-FITC (Sigma–Aldrich, MO, USA). Cells were counterstained with DAPI. Photos were taken by a Zeiss LSM780 confocal microscope (Carl Zeiss, Germany).
iTRAQ quantification proteomics
Proteins were extracted from the testes of adult Cfap70-KO mice and their littermate wild-type mice (n = 3 each group) using RIPA lysis buffer (Applygen, Beijing, China) containing 1 mM PMSF and protease inhibitors on ice. The supernatants were collected following centrifugation at 12,000 g for 20 min. Protein concentrations were calculated by Bradford quantification and SDS‒PAGE. According to a trypsin enzyme (μg): substrate protein (μg) ratio of 1:20, enzyme solution was added to 100 μg protein samples, vortexed, centrifuged at low speed for 1 min, and incubated at 37 °C for 4 h. The peptide liquid obtained after salt removal was freeze-dried. The peptide sample was dissolved in 0.5 M TEAB and added to the corresponding iTRAQ labelling reagent, followed by storage at room temperature for 2 h. The Shimadzu LC-20AB liquid phase system was used, and the separation column was a 5 μm 4.6 × 250 mm Gemini C18 column for liquid phase separation of the sample. The dried peptide samples were reconstituted with mobile phase A (2% ACN, 0.1% FA) and centrifuged at 20,000 g for 10 min, and the supernatant was taken for injection. Separation was performed by UltiMate 3000 UHPLC (Thermo Fisher). The sample was first enriched in a trap column and desalted, and then entered a self-packed C18 column. The peptides separated by liquid phase chromatography were ionized by a nanoESI source and then passed to a tandem mass spectrometer Q-Exactive HF X (Thermo Fisher) for DDA (Data Dependent Acquisition) mode detection. Raw data were converted to mgf files for bioinformatics analysis, and protein identification from tandem mass spectra was performed by database searching (UniProt). The protein quantification process includes the following steps: protein identification, tag impurity correction, data normalization, missing value imputation, protein ratio calculation, statistical analysis, and results presentation. Proteins with a 1.5-fold change and p value (using Student's t test) less than 0.05 were defined as differentially expressed proteins. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the iProX partner repository36 with the dataset identifier PXD033133.
Quantitative RT‒PCR
Total RNA was extracted from the spermatids of Cfap70-KO mice and their littermate wild-type mice using an RNA Easy Fast Tissue/Cell Kit (Tiangen Biotech). Approximately 0.3 mg total RNA was converted into cDNA with a FastKing One-Step RT‒PCR Kit (Tiangen Biotech) according to the manufacturer's instructions. The cDNAs were individually diluted 10-fold to be used as templates for the subsequent real-time fluorescence quantitative PCR with RealUniversal Colour PreMix (SYBR Green) (Tiangen Biotech). Mouse Gapdh was used as an internal control. Qrich2 and Ttll5 mRNA expression was quantified according to the 2−ΔΔCt method. The primers for qRT‒PCR are listed in Table S4.
Co-immunoprecipitation (co-IP)
As previously described,31 48 h after transfection, HEK293T cells were lysed with Pierce™ IP Lysis Buffer (Thermo Fisher) with protease inhibitor cocktail (Beyotime) for 30 min at 4 °C and then centrifuged at 12,000 g for 10 min. To prepare input samples, 30 μL protein lysates (∼25 μg) were collected and boiled for 5 min in 1.2 × SDS loading buffer. The lysates were precleared with 10 μL Pierce™ Protein A/G-conjugated Agarose (Thermo Fisher) for 1 h at 4 °C. Precleared lysates were incubated overnight with 2 μg anti-Flag antibody (Abmart, Shanghai, China) at 4 °C. The lysates were then incubated with 20 μL Pierce™ Protein A/G-conjugated Agarose for 2 h at 4 °C. The agarose beads were washed four times with Pierce™ IP Lysis Buffer and boiled for 5 min in 1.2 × SDS loading buffer. Input and IP samples were analysed by Western blotting using anti-Flag, anti-Myc, or anti-HA antibodies. For endogenous co-IP, testes from adult Cfap70-KO mice and their littermate wild-type mice were lysed with Pierce™ IP Lysis Buffer. Precleared lysates were separated into two groups: one group was treated with 2 μg anti-DNAI2 antibody (Proteintech, Wuhan, China) or anti-CFAP61 antibody (a gift from Dr. Ming-Xi Liu), and another group (negative control) was treated with 2 μg rabbit IgG (Beyotime). Detailed information on the antibodies used in the IP experiments is provided in Table S5, and RRID tags are provided.
Western blotting
As previously described,31 proteins from HEK293T cells or tissues form Cfap70-KO mice and their littermate wild-type mice were extracted using RIPA lysis buffer (Applygen) containing 1 mM PMSF and protease inhibitors on ice. The supernatants were collected following centrifugation at 12,000 g for 20 min. Proteins were electrophoresed in 10% SDS–PAGE gels and transferred to nitrocellulose membranes (GE Healthcare, WI, US). The blots were blocked in 5% milk and incubated with primary antibodies overnight at 4 °C, followed by incubation with anti-rabbit or mouse IgG H&L (HRP) (Abmart) at a 1/10,000 dilution for 1 h. The signals were evaluated using Super ECL Plus Western Blotting Substrate (Applygen) and a chemiluminescence imaging system (Tanon, Shanghai, China). The antibodies used in WB are listed in Table S5, and RRID tags are provided.
Immunofluorescence
After permeabilization with 1% Triton X-100 for 30 min, the slides of testicular spermatids from adult Cfap70-KO mice and their littermate wild-type mice were blocked with 5% goat serum for 45 min. Samples were stained with primary antibodies overnight at 4 °C. After washing with PBS three times, slides were incubated with Alexa Fluor-labelled goat anti-rabbit IgG (H + L) (Beyotime) at room temperature for 1 h. PNA-FITC (Sigma–Aldrich, MO, USA) was used to stain the acrosome, and cells were counterstained with DAPI. Photos were taken by a Zeiss LSM780 confocal microscope (Carl Zeiss, Germany). The antibodies used in the IF are listed in Table S5, and RRID tags are provided.
Scanning electron microscopy (SEM)
As previously described,32 spermatozoa from the cauda epididymis and a piece of the lateral ventricle from adult Cfap70-KO mice and their littermate wild-type mice were fixed in 2.5% phosphate-buffered glutaraldehyde (GA) (Zhongjingkeyi Technology, Beijing, China) at room temperature for 30 min and then deposited on coverslips. The coverslips were dehydrated via an ascending gradient of 50%, 70%, 95%, and 100% ethanol and air-dried. Specimens were then attached to specimen holders and coated with gold particles using an ion sputter coater before being viewed with a JSM-IT300 scanning electron microscope (JEOL, Tokyo, Japan).
Transmission electron microscopy (TEM)
Precipitation of sperm as well as tissues of testis and brain (∼1 mm3) from adult Cfap70-KO mice and their littermate wild-type mice were fixed with 2.5% (vol/vol) glutaraldehyde in 0.1 M phosphate buffer (PB) (pH 7.4), washed two times in PB and two times in ddH2O. Then, tissues were immersed in 1% (wt/vol) OsO4 and 1.5% (wt/vol) potassium ferricyanide aqueous solution at 4 °C for 2 h. After washing, the samples were dehydrated through graded alcohol (30%, 50%, 70%, 80%, 90%, 100%, 100%, 10 min each) into pure acetone (10 min twice). Samples were infiltrated in a graded mixture (3:1, 1:1, 1:3) of acetone and SPI-PON812 resin (21 mL SPO-PON812, 13 mL DDSA and 11 mL NMA), and the pure resin was changed. The specimens were embedded in pure resin with 1.5% BDMA and polymerized for 12 h at 45 °C and 48 h at 60 °C. The ultrathin sections (70 nm thick) were sectioned with a microtome (Leica EM UC6), double-stained with uranyl acetate and lead citrate, and examined by a transmission electron microscope (FEI Tecnai Spirit120kV). All reagents were purchased from Zhongjingkeyi Technology (Beijing, China).
Statistical analysis
Sample sizes were determined based on the means and variation of preliminary data to achieve at least 80% power and allow for a 5% type I error. Calculation was performed using an online sample size calculator (https://clincalc.com/stats/samplesize.aspx). In this study, data were presented as the mean ± standard deviation (SD). A Q–Q plot ("Q" stands for quantile) test was utilized to evaluate the normality of parameters. Statistical significance was performed using GraphPad Prism version 5.01 (Graph Pad Software, San Diego, CA, USA). Bartlett's test was used to analyse the homogeneity of variance. Student's t test (two-tail, unpaired) was used for the statistical analyses. In the case of violations, Welch's t test was applied.
Role of funders
Funders had no role in the study design, data collection, data analyses, interpretation, or writing of the report.
Results
Generation of Cfap70-KO mice
The CFAP70 protein is highly conserved among different species (https://www.ncbi.nlm.nih.gov/homologene/17048). According to the transcriptome database, both human CFAP70 mRNA and mouse Cfap70 mRNA are predominantly expressed in testis tissues (Figure S1a and b). We also observed that mouse CFAP70 protein was predominantly expressed in the testis and brain by Western blot analysis (Figure S1c. According to the single-cell RNA-seq database, both human CFAP70 mRNA and mouse Cfap70 mRNA are predominantly restricted to early/late spermatids within the testes (Figure S1d and e).
To characterize the physiological roles of CFAP70, its knockout mice were created using the CRISPR‒Cas9 system. The Cfap70 gene is located at chromosome 14 and has 6 transcripts. According to the structure of the Cfap70 gene, exon 3 to exon 8 of the Cfap70-203 (ENSMUST00000056073.13) transcript was selected as the knockout region (Fig. 1a). This region contains an 824-bp coding sequence, and deletion of this region is expected to completely disrupt protein function. Cfap70-heterozygous mice were interbred to obtain Cfap70-KO mice and were confirmed by genotyping PCR. A complete absence of the CFAP70 protein in testis tissues was confirmed by Western blotting (Fig. 1a). Some Cfap70-KO pups died before 4 weeks due to hydrocephalus, characterized by enlarged dome-shaped skulls and enlargement of the ventricles of the brain (Figure S2a). Haematoxylin-eosin (H&E) staining and scanning electron microscopy (SEM) images of the ependymal epithelium revealed sparse ependymal cilia in Cfap70-KO mice (Figure S2b and c). Loss of CFAP70 expression in ependymal cilia was revealed by immunofluorescence staining of brain sections (Figure S2d). Transmission electron microscopy (TEM) analysis revealed disruption of the axoneme structures of ependymal cilia in Cfap70-KO mice (Figure S2e). Moreover, histological analysis of Cfap70-KO mice revealed the generally normal morphology of the heart, liver, spleen, lung, kidney, retina, intestine, trachea, and oviduct (Figure S3).
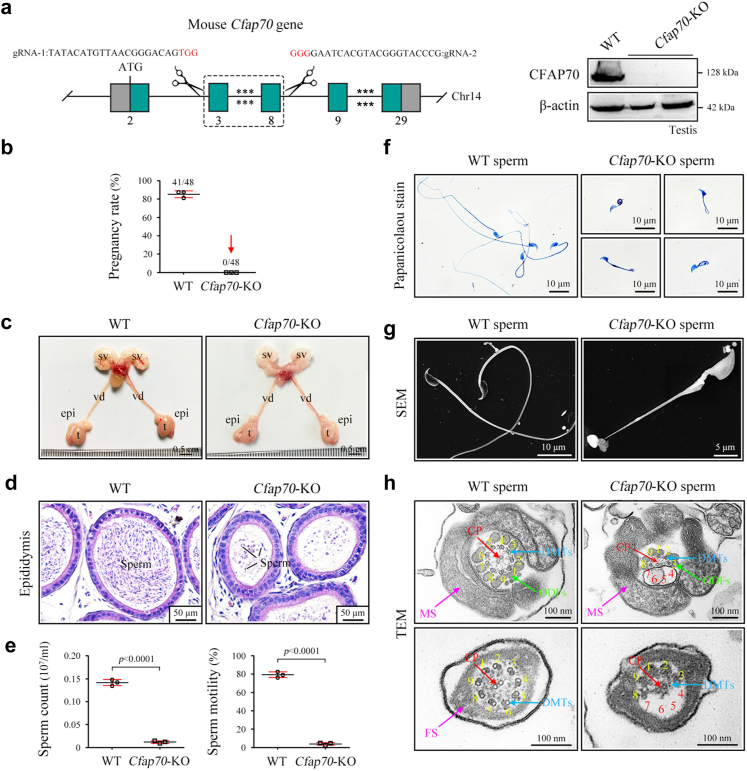
Fig. 1.
Cfap70-KO male mice were infertile with the OAT phenotype. (a) Schematic illustration of the targeting strategy for generating Cfap70-KO mice. Immunoblotting of CFAP70 was performed in testis protein lysates of adult wild-type (WT) and Cfap70-KO mice and their littermate wild-type (WT) mice (n = 3 each group). β-actin served as a loading control. Representative images were shown. (b) Pregnancy rate of Cfap70-KO male mice and their littermate WT male mice. No females mated with Cfap70-KO male mice were pregnant. Data were presented as the mean ± SD (n = 3 each group). (c) Representative morphology of the male reproductive system from Cfap70-KO mice and their littermate WT mice. t, testis; epi, epididymis; vd, vas deferens; sv, seminal vesicle. (d) Representative histological morphology of the cauda epididymis from Cfap70-KO mice and their littermate WT mice by H&E staining. Scale bars, 50 μm. (e) Sperm count ( × 107/mL) and total sperm motility (%) in Cfap70-KO mice and their littermate WT mice. Data were presented as the mean ± SD (n = 3 each group). Statistical significance was determined by two-tailed, unpaired Student's t test; for sperm count, p < 0.0001; for total sperm motility, p < 0.0001. (f) Representative sperm morphology of Cfap70-KO mice and their littermate WT mice as revealed by Papanicolaou staining. Scale bars, 10 μm. (g) Representative SEM images of spermatozoa from Cfap70-KO mice and their littermate WT mice. Scale bar, 5 or 10 μm. (h) Representative TEM images of spermatozoa from Cfap70-KO mice and their littermate WT mice. CP, central pair; DMTs, peripheral doublet microtubules; ODFs, outer dense fibres; MS, mitochondrial sheath. Scale bars, 100 nm.
Cfap70-KO mice exhibit OAT and male infertility
Cfap70-KO male mice were completely infertile because no pups were born from Cfap70-KO males during the 2-month fertility test (Fig. 1b). In contrast, Cfap70-KO female mice are fertile. There was no difference in the reproductive system between Cfap70-KO and their littermate wild-type male mice (Fig. 1c). Histological examination revealed only a few spermatozoa in the cauda epididymis of Cfap70-KO mice (Fig. 1d). We further histologically examined the process of spermatogenesis in the seminiferous tubules using Periodic Acid-Schiff staining. The differentiation from spermatogonia to spermatids in the Cfap70-KO testis was generally normal except for fewer and short-tailed spermatids in stage V-VIII seminiferous epithelia (Figure S4. Semen characteristics and sperm morphology were then investigated with spermatozoa collected from the cauda epididymis. Both sperm count and total motility were much lower in Cfap70-KO mice than in their littermate wild-type mice (Fig. 1e). Morphologic analysis of spermatozoa using Papanicolaou staining (Fig. 1f) and SEM (Fig. 1g) revealed that Cfap70-KO spermatozoa were malformed with short and coiled flagella. Thus, Cfap70-KO male mice perfectly manifest the OAT or MMAF phenotypes in humans. TEM analysis revealed that the very few remaining spermatozoa in the Cfap70-KO cauda epididymis exhibited axoneme malformations. In the flagella of wild-type spermatozoa, the axoneme is primarily composed of a “9 + 2” structure, including nine pairs of peripheral doublet microtubules (DMTs) and the central pair (CP) of microtubules. Nonetheless, more cross sections of flagella exhibiting DMTs 4–7 missing with a concomitant loss of the associated outer dense fibres (ODFs) were present in Cfap70-KO mice (Fig. 1h and Figure S5). Taken together, Cfap70-KO mice produce very few spermatozoa in the cauda epididymis, and the remaining immotile spermatozoa exhibit short and coiled flagella with defective axoneme structures.
CFAP70 is required for the biogenesis of spermatid flagella within testes
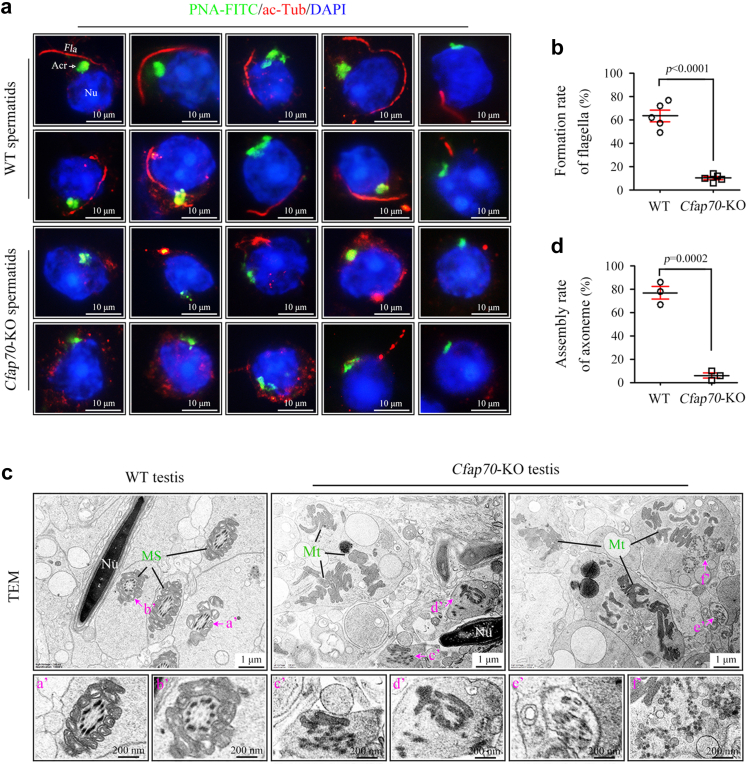
To understand the physiological role of CFAP70 in flagella formation, we isolated spermatids from Cfap70-KO and their littermate wild-type testes using flow cytometry (Figure S6) and analysed flagella formation during the early stage of spermiogenesis by immunostaining of acetylated tubulin. We found that microtubule-based axoneme structure formation was either blocked or seriously defective in Cfap70-KO round spermatids (Fig. 2a). The formation rate of flagellar axonemes was significantly lower in Cfap70-KO spermatids than in wild-type spermatids (Fig. 2b). TEM analysis was further applied to identify the axoneme ultrastructure in spermatids within wild-type and Cfap70-KO testes. Typical “9 + 2” axonemes surrounded by well-arranged accessory structures (e.g., mitochondrial sheath) were identified everywhere in the TEM sections of wild-type testes. In contrast, axoneme structures were rarely observed in Cfap70-KO spermatids, and the mitochondria exhibited a dispersed cytoplasmic distribution (Fig. 2c, upper panel). The rare axoneme structures identified in Cfap70-KO spermatids exhibited severe deformation of the “9 + 2” axoneme (Fig. 2c, lower panel). The ratio of axoneme assembly in Cfap70-KO spermatids was significantly lower than that in their littermate wild-type spermatids (Fig. 2d). Accordingly, we conclude that CFAP70 is required for the biogenesis of the flagellar axoneme in early spermatids rather than the stability of the assembled axoneme.
Fig. 2.
A failure of flagella biogenesis in Cfap70-KO spermatids. (a) Spermatids were isolated from testicular cells using flow cytometry after Hoechst 33,342 staining. Representative immunofluorescence staining of acetylated-tubulin (ac-Tub) (red) and PNA-FITC (green) in round spermatids from Cfap70-KO mice and their littermate WT mice. Nu, nucleus; Acr, acrosome; Fla, flagella. Scale bar, 50 μm. (b) As revealed by ac-Tub staining, the percentage of round spermatids with normal flagella formation was analysed in Cfap70-KO mice and their littermate WT mice. Data were presented as the mean ± SD (n = 5 each group). At least 50 round spermatids were counted for each mouse. Statistical significance was determined by two-tailed, unpaired Student's t test; p < 0.0001. (c) Representative TEM images of testis sections in Cfap70-KO mice and their littermate WT mice. In WT spermatids, a well-organized “9 + 2” axoneme could be easily observed and the midpiece of the flagella was surrounded by the mitochondrial sheath (MS). However, assembled axoneme structures were rarely observed in Cfap70-KO spermatids, and mitochondria (Mt) were scattered. If observed, the axoneme was also severely disorganized. The lower panel (a’-f’) was the enlarged images. Nu, nuclei. Scale bar, 1 μm. (d) As revealed by TEM, the percentage of spermatids with assembled axonemes was analysed in the testes of Cfap70-KO mice and their littermate WT mice. Data were presented as the mean ± SD (n = 3 each group). At least 30 spermatids were counted for each mouse. Statistical significance was determined by two-tailed, unpaired Student's t test; p = 0.0002.
CFAP70 regulates the stability of the MMAF-associated protein QRICH2
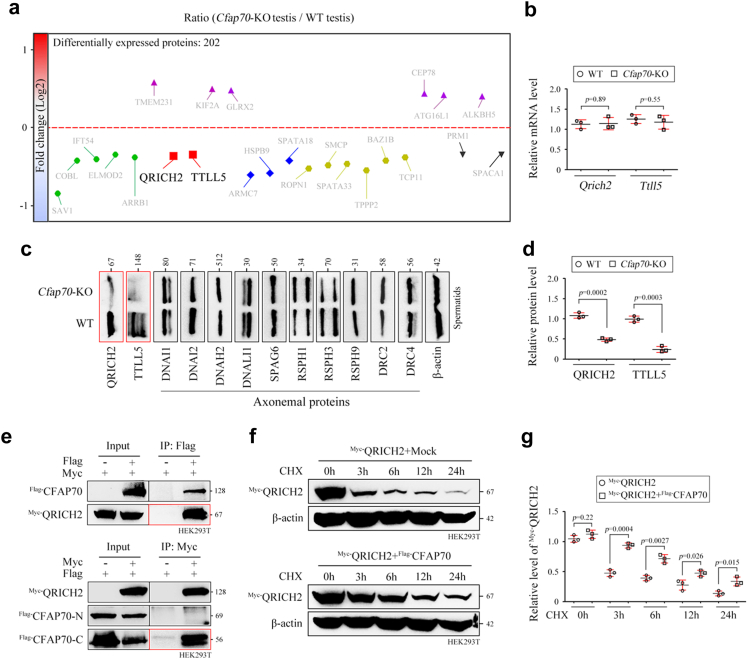
To determine the molecular basis of the failure of flagellar assembly in Cfap70-KO spermatids, quantitative iTRAQ (isobaric tags for relative and absolute quantification) and mass spectrometry were applied to identify the proteome of wild-type testes and Cfap70-KO testes (n = 3 each group) (Fig. 3a). A moderate number of differentially expressed (DE) proteins were obtained when a cut-point of 1.5-fold change and a p value (student's t test) less than 0.05 was selected. Compared with wild-type testes, CFAP70 deletion caused a decrease of 96 and an increase of 106 proteins. Among DE proteins, glutamine rich 2 (QRICH2) and tubulin tyrosine ligase-like family member 5 (TTLL5) attracted our attention because they exhibit sperm-specific expression and because knockout of these genes in mice produces male infertility with the MMAF phenotype.34,37 The cross-section of short sperm flagella from Qrich2-KO mice reveals disorganized and incomplete DMTs, CP, and ODFs.34 Severe disruption of microtubule polymers and missing doublet 4 from the “9 + 2” axoneme are observed in Ttll5-KO sperm flagella.37 The mRNA levels of Qrich2 and Ttll5 in testis tissues were similar between wild-type and Cfap70-KO mice, indicating that CFAP70 does not transcriptionally regulate the expression of Qrich2 and Ttll5 (Fig. 3b). Surprisingly, no axoneme structural proteins were identified in the list of DE proteins. Given that testes possess many cell types other than spermatids, isolated spermatids may be more suitable than whole testes for DE protein screening. We further isolated haploid spermatids (1 N) from testes of Cfap70-KO mice and their littermate wild-type mice using flow cytometry. The protein levels of QRICH2, TTLL5, and ten known axoneme structural proteins, including DNAI1, DNAI2, DNAH2, DNALI1, SPAG6, RSPH1, RSPH3, RSPH9, DRC2, and DRC4 (Fig. 3c), were examined by Western blotting. The expression of ten axoneme structural proteins was not significantly altered after the loss of CFAP70, indicating that CFAP70 may not directly affect the expression of axoneme structural proteins in spermatids. As expected, the protein levels of QRICH2 and TTLL5 were significantly lower in Cfap70-KO spermatids than in wild-type spermatids (Fig. 3c and d).
Fig. 3.
CFAP70 regulates the expression of QRICH2 and TTLL5 but not axonemal proteins. (a) Quantitative proteomics of testis protein lysates from Cfap70-KO mice and WT mice (n = 3 each group). (b) Relative mRNA levels of Qrich2 and Ttll5 in testes of Cfap70-KO mice and their littermate WT mice, as revealed by qRT‒PCR. Data were presented as the mean ± SD (n = 3 each group). Statistical significance was determined by two-tailed, unpaired Student's t test; for Qrich2, p = 0.89; for Ttll5, p = 0.55. (c) Representative immunoblots of QRICH2, TTLL5, and ten selected axoneme proteins (DNAI1, DNAI2, DNAH2, DNALI1, SPAG6, RSPH1, RSPH3, RSPH9, DRC2, and DRC4) in the protein lysates of spermatids from Cfap70-KO mice and their littermate WT mice. β-actin served as a loading control. (d) Relative protein levels of QRICH2 and TTLL5 in spermatids of Cfap70-KO mice and their littermate WT mice. Data were presented as the mean ± SD (n = 3 each group). Statistical significance was determined by two-tailed, unpaired Student's t test; for QRICH2 level, p = 0.0002; for TTLL5 level, p = 0.0003. (e) Coimmunoprecipitation (co-IP) assay indicated that Flag-tagged CFAP70 interacted with Myc-tagged QRICH2 in HEK293T cells. A co-IP assay further revealed that Myc-tagged QRICH2 interacted with the Flag-tagged C-terminus of CFAP70 but not N-terminus of CFAP70. The co-IP experiments were repeated for three times and representative blots were shown. (f) The effect of Flag-tagged CFAP70 coexpression on the stability of Myc-tagged QRICH2 was examined using protein stability assays in HEK293T cells. Protein samples were harvested at the indicated times after treatment with 100 μg/mL cycloheximide (CHX) and representative blots were shown. (g) Relative protein level of QRICH2 in the QRICH2 group and the QRICH2+CFAP70 group at 0 h, 3 h, 6 h, 12 h, and 24 h after CHX treatment. Data were presented as the mean ± SD (n = 3 each group) and band intensities were normalized to β-actin. Statistical significance was determined by two-tailed, unpaired Student's t test; for 0 h, p = 0.22; for 3 h, p = 0.0004; for 6 h, p = 0.0027; for 12 h, p = 0.026; for 24 h, p = 0.015.
CFAP70 harbours eight tetratricopeptide repeats (TPRs), and TPR proteins typically function as chaperones or cofactors to mediate protein–protein interactions.38 We demonstrated that Flag-tagged CFAP70 could interact with Myc-tagged QRICH2 via its C-terminal region (containing TPRs) in HEK293T cell lysates by coimmunoprecipitation (co-IP) (Fig. 3e). To determine whether CFAP70 regulated the expression of its interacting partner QRICH2 through a mechanism of protein stability, HEK293T cells overexpressing QRICH2 with or without CFAP70 were treated with cycloheximide (100 μg/mL) for up to 24 h to block protein synthesis. Only ∼45% of QRICH2 remained after 3 h of treatment; however, this value was increased to ∼90% when CFAP70 was coexpressed with QRICH2 (Fig. 3f). The degradation rate of QRICH2 was slowed down in the coexpression group compared with QRICH2 alone (Fig. 3g). These results indicated that CFAP70 promotes the assembly of spermatid flagella partially by stabilizing some of its interacting partners, such as the MMAF-associated protein QRICH2, at the protein level.
CFAP70 regulates the cytoplasmic preassembly of CFAP61 and CFAP91
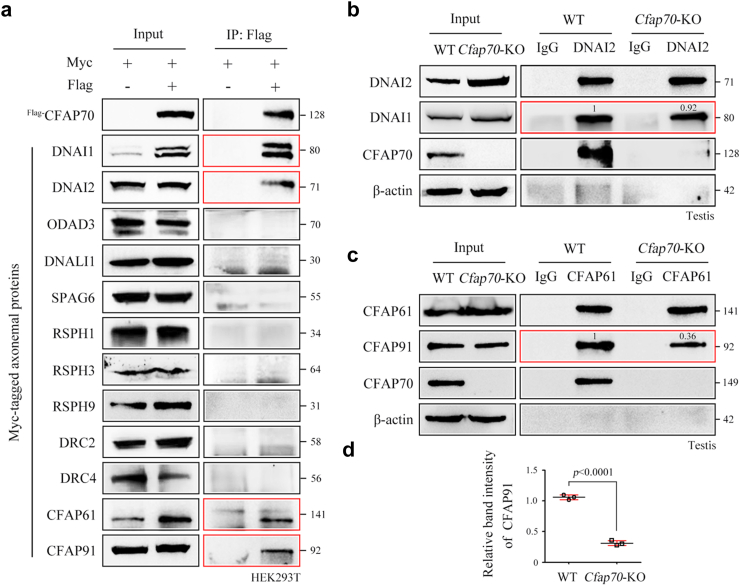
To further explore the mechanisms underlying the regulation of spermatid flagellar assembly by CFAP70, we screened CFAP70-associated axoneme proteins. IP-mass spectrometry using the CFAP70 antibody in testis protein lysates is a direct method to explore CFAP70-interacting partners, but unfortunately, all available antibodies for CFAP70 were not suitable for endogenous IP. We chose an alternative approach to construct fifteen plasmids for tag-labelled axoneme proteins and performed candidate screening of CFAP70-interacting partners. We found that Flag-tagged CFAP70 coimmunoprecipitated with Myc-tagged DNAI1, DNAI2, CFAP61, and CFAP91 but not ODAD3 (an ODA-docking complex protein), DNALI1 (an IDA protein), SPAG6 (a CP protein), RSPH1/3/9 (RS proteins), or DRC2/4 (DRC proteins) in HEK293T cell extracts (Fig. 4a). DNAI1 and DNAI2 encode the intermediate chains of ODA of the ciliary apparatus, and their variants are known genetic causes of PCD.33,39 The interaction between CFAP70 and DNAI1/2 is consistent with a cryo-electron tomography study revealing that Chlamydomonas FAP70 localizes at the base of the ODA.21 CFAP61, CFAP91, and CFAP251 are conserved components of the calmodulin- and radial spoke-associated complex (CSC) of cilia, which is located at the base of the RS and interacts with the DRC and IDA.40,41 Variants of CFAP61,35,42 CFAP91,16 and CFAP25119,20,43 have been identified in MMAF patients. We hypothesized that CFAP70 promotes spermatid flagellar assembly partially by regulating the cytoplasmic preassembly of ODA and/or CSC. To examine these possibilities, we immunoprecipitated endogenous DNAI2 or CFAP61 from testis extracts using their specific antibodies. DNAI2 coprecipitated DNAI1 at similar levels from both wild-type and Cfap70-KO testis extracts (Fig. 4b), indicating that loss of CFAP70 does not primarily impact ODA subunit heterodimerization. In contrast, we observed significantly reduced immunoprecipitation of CFAP91 by CFAP61 in Cfap70-KO testis extracts (∼0.6-fold reduction, normalized to corresponding levels in wild-type testis extracts) (Fig. 4c and d). Although the loss of CFAP70 does not influence the protein levels of CFAP61 and CFAP91, CFAP70 regulates their cytoplasmic preassembly.
Fig. 4.
CFAP70 regulates the cytoplasmic preassembly of the CSC components CFAP61 and CFAP91. (a) Myc-tagged plasmids of twelve axoneme proteins were constructed by PCR using testis cDNA template and confirmed by sequencing. Flag-tagged CFAP70 was immunoprecipitated with Myc-tagged DNAI1, DNAI2, CFAP61, and CFAP91 but not Myc-tagged ODAD3, DNALI1, SPAG6, RSPH1, RSPH3, RSPH9, DRC2, and DRC4 in HEK293T cells using co-IP assays. The co-IP experiments were repeated for three times and representative blots were shown. (b) Endogenous immunoprecipitation of DNAI2 from testis extracts reveals no defects in DNAI2 association with its heterodimeric partner DNAI1 in Cfap70-KO mice. (c) Endogenous immunoprecipitation of CFAP61 from testis extracts revealed partial disruption in its association with CFAP91 in Cfap70-KO mice compared to WT mice, as revealed by the intensities of the coprecipated bands of CFAP91. The grey values of the protein bands in b and c were measured by ImageJ software (https://imagej.en.softonic.com/). (d) The relative amount of CFAP91 immunoprecipitated by CFAP61 in the testis protein lysates from Cfap70-KO mice and their littermate WT mice. Data were presented as the mean ± SD (n = 3 each group). Statistical significance was determined by two-tailed, unpaired Student's t test; p < 0.0001.
Intramanchette transport (IMT), a cargo transport system, plays critical roles in the formation of spermatid flagella.44 This transport machinery consists of motor kinesin-II and IFT-A and -B multisubunit particles. IFT88 is localized along the manchette of spermatids, and Ift88-deficient sperm display taillessness.45 Accordingly, we further examined the manchette localization of IFT88 and its potential cargo in wild-type and Cfap70-KO spermatids. In most of wild-type elongating spermatids, IFT88, DNAI1, and CFAP61 were observed to perfectly colocalize with the acetylated tubulin-labelled manchette (Figure S7a–c). In contrast, IFT88, DNAI1, and CFAP61 were partially mislocalized in the cytoplasm of Cfap70-KO elongating spermatids (Figure S7a–c). The percentage of Cfap70-KO spermatids exhibited matched localization of IFT88, DNAI1, and CFAP61 with acetylated tubulin was lower than in wild-type spermatids (Figure S7d–f). Same immunofluorescence double staining was performed in other MMAF-associated mouse models,13,46 to show that this defect is specific for Cfap70-KO mice.
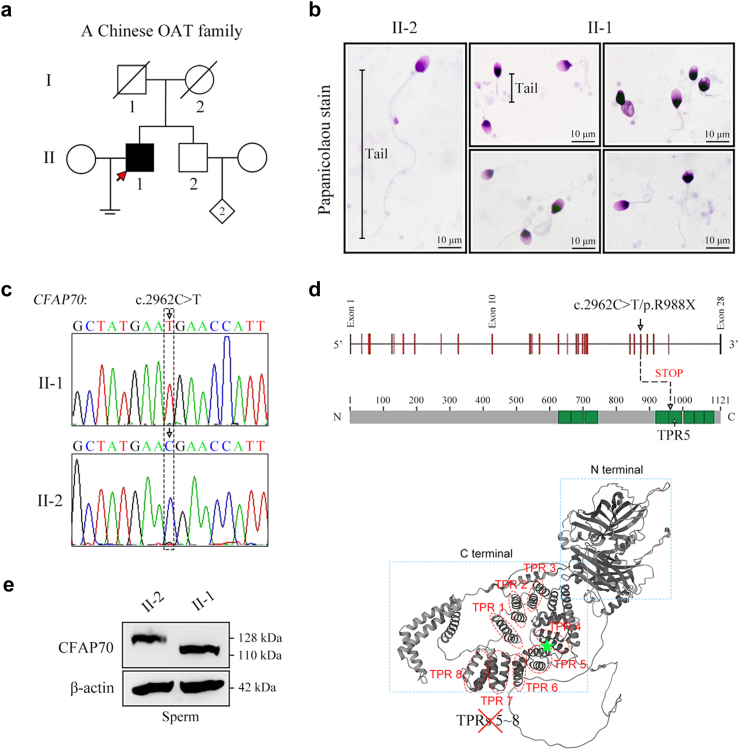
A homozygous nonsense variant in CFAP70 is identified in a Chinese infertile man with OAT
A 33-year-old man (II-1) who was diagnosed with primary infertility for five years and his fertile brother (II-2) were recruited for our study (Fig. 5a). The somatic cell karyotype (46, XY), bilateral testicular size, secondary sex characteristics, and hormone levels were normal in the proband (II-1). No microdeletions on the Y chromosome were observed in the proband (II-1). A semen analysis of the proband (II-1) revealed normal semen volume (1.8 mL), extremely low sperm concentration (2.8 × 106 spermatozoa/mL), no progressive mobile sperm, and a low percent of morphologically normal sperm (2%) (Table 1). Papanicolaou staining of his spermatozoa revealed severe sperm flagellar abnormalities, which were characterized by coiled, short, absent, irregular and bent flagella (Fig. 5b). To investigate the genetic cause of OAT, we performed WES on the proband (II-1) to reveal variants in 26 candidate genes (Table S1). A homozygous nonsense variant c.2962C > T/p.R988X in CFAP70 attracted our attention because CFAP70 is the only OAT-associated gene based on the literature.23 The variant is absent in the East Asian population and extremely rare (MAF = 0.000032) in the global population of the gnomAD database. Sanger sequencing of the CFAP70 gene revealed that the proband (II-1) carried the homozygous nonsense variant, and his fertile brother (II-2) harboured the wild-type allele (Fig. 5c). Sanger sequencing of this variant could not be performed in the parents of the proband because they had passed away. CFAP70 harbours eight tetratricopeptide repeat (TPR) motifs, in which 34 amino acid repeats are arranged in two antiparallel α-helices.38 The nonsense variant c.2962C > T/p.R988X in CFAP70 occurs at the fifth TPR of the CFAP70 protein and is expected to lead to the loss of TPRs 5–8 (Fig. 5d). Indeed, a truncated CFAP70 protein (∼110 kDa) was observed in the sperm protein extracts of the proband (II-1), whereas his fertile brother (II-2) exhibited a full-length CFAP70 (128 kDa) (Fig. 5e).
Fig. 5.
OAT phenotype of a CFAP70-mutated patient from a Chinese family. (a) Identification of a homozygous nonsense variant, c.2962C > T/p.R988X, of the CFAP70 gene in an OAT patient (II-1). His brother (II-2) is fertile. The pedigree of this family is presented, and the proband (II-1) is indicated by the arrow. (b) Representative sperm morphology in the proband (II-1) and his brother (II-2) by Papanicolaou staining under light microscopy. The spermatozoa of the proband (II-1) exhibited multiple flagellar malformations, including short, coiled, and absent flagella. Scale bars, 10 μm. (c) The DNA samples of the proband (II-1) and his brother (II-2) were subjected to Sanger sequencing. The chromatogram of CFAP70 in the proband (homozygous c.2962C > T) and his brother (wild-type allele). The arrow denotes the variant site. (d) The location of the variant (c.2962C > T/p.R988X) in the intron‒exon structure of the CFAP70 gene and the protein domain map of CFAP70. Eight tetratricopeptide repeats (TPRs) are annotated with green squares. The variant was located in the fifth TPR domain of CFAP70 and is expected to generate a truncated CFAP70 protein lacking 5–8 TPRs. The structure of CFAP70 was predicted by AlphaFold (https://alphafold.ebi.ac.uk/), and eight TPRs were shown. (e) The proteins of sperm samples were extracted from the proband (II-1) and his brother (II-2). Western blot analysis indicated a full-length CFAP70 protein (128 kDa) in II-2 and a truncated CFAP70 protein (∼110 kDa) in II-1. β-actin served as a loading control. Western blotting experiments were repeated for three times and representative blots were shown.
Table 1.
Semen characteristics of the proband.
| Semen parameters | Proband (II-1) | Reference limitsa |
|---|---|---|
| Sperm volume (mL) | 1.8 | >1.5 |
| Sperm concentration (106/mL) | 2.8 | >15 |
| Total motility (%) | 2.5 | >40 |
| Progressive motility (%) | 0 | >32 |
| Normal spermatozoa (%) | 2 | >4 |
| Coiled flagella (%) | 39.41 | – |
| Short flagella (%) | 24.63 | – |
| Absent flagella (%) | 24.14 | – |
| Flagella of irregular calibre (%) | 0.99 | – |
| Bent flagella (%) | 0.49 | – |
Reference limits according to the WHO standards.
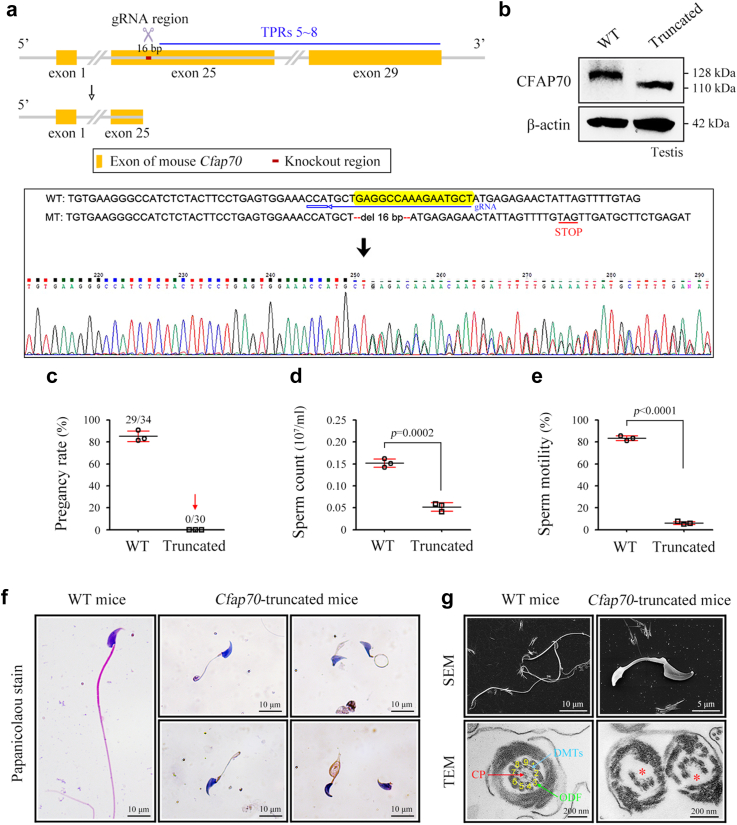
Cfap70-truncated mice lacking TPRs 5–8 exhibit the OAT phenotype
The amino acid 1008 in mouse CFAP70 is the homologous position of the 988th amino acid in human CFAP70 (Figure S8). To investigate whether the c.2962C > T/p.R988X variant in CFAP70 represented the primary cause of OAT, we deleted a 16-bp within exon 25 of mouse Cfap70 to generate a stop codon after amino acid 1007, expecting to generate a truncated CFAP70 protein also lacking TPRs 5–8 (Fig. 6a). We did not construct Cfap70-mutant mice carrying the variant equivalent to that in the patient because the 1008th amino acid in mice is extremely hard to edit (only one gRNA exists, and it is ∼23-bp from the edit site). As expected, our knockout strategy generated a truncated protein (∼110 kDa) of CFAP70, as indicated by Western blotting using testis lysates (Fig. 6b). Thus, Cfap70-truncated mice achieve the same effect as Cfap70-mutant mice.
Fig. 6.
CFAP70-truncated mice exhibiting the OAT phenotype. (a) Schematic illustration of the targeting strategy for generating Cfap70-truncated mice. A 16-bp sequence within exon 25 was deleted using the CRISPR‒Cas9 technology. Sanger sequencing of founder mice and gRNA sequence are presented. (b) Immunoblotting of CFAP70 was performed in protein samples of mouse testis tissues. A full-length CFAP70 (∼128 kDa) and a truncated CFAP70 protein (∼110 kDa) were identified in testis samples of WT mice and Cfap70-truncated mice, respectively. β-actin served as a loading control. Western blotting experiments were repeated for three times and representative blots were shown. (c) Pregnancy rate of Cfap70-truncated male mice and their littermate WT male mice for 2 months. No females mated with Cfap70-truncated male mice were pregnant. Data are presented as the mean ± SD (n = 3 each group). (d) Sperm counts ( × 107/mL) of Cfap70-truncated mice and their littermate WT mice. Data are presented as the mean ± SD (n = 3 each group). Statistical significance was determined by two-tailed, unpaired Student's t test; p = 0.0002. (e) Total sperm motility (%) of Cfap70-truncated mice and their littermate WT mice. Data are presented as the mean ± SD (n = 3 each group). Statistical significance was determined by two-tailed, unpaired Student's t test; p < 0.0001. (f) Sperm morphology in WT mice and Cfap70-truncated mice was observed by Papanicolaou staining. Experiments were repeated for three times and representative images were shown. Scale bars, 10 μm. (g) Representative SEM and TEM images of sperm from Cfap70-truncated mice and their littermate WT mice. CP, central pair; DMTs, peripheral doublet microtubules; ODFs, outer dense fibres. Scale bars in SEM, 10 μm; scale bars in TEM, 200 nm.
Cfap70-truncated male mice were infertile because no pups were born from Cfap70-truncated males during the 2-month fertility test (Fig. 6c). Both the sperm count and sperm motility were significantly lower in Cfap70-truncated mice than in their littermate wild-type mice (Fig. 6d and e). Papanicolaou staining and SEM of spermatozoa indicated that Cfap70-truncated mice produced sperm with short, coiled, and absent flagella (Fig. 6f). TEM analysis further revealed an absence of CP and a partial loss of DMTs in sperm flagella of Cfap70-truncated mice (Fig. 6g). Taken together, animal evidence indicates that this nonsense variant in CFAP70 constituted the primary cause of OAT in the patient.
ICSI outcomes of CFAP70-associated male infertility are evaluated in mice and humans
Intracytoplasmic sperm injection (ICSI) has been reported to be efficient for most OAT-associated patients.10,47,48 To examine whether CFAP70-associated male infertility could be overcome via ICSI, we conducted experiments using spermatozoa from Cfap70-KO mice. In general, the infertility of Cfap70-KO male mice was recovered by ICSI because healthy offspring could be obtained using spermatozoa from Cfap70-KO mice. However, the rates of two-cell embryos and blastocysts were reduced upon ICSI treatment using spermatozoa from Cfap70-KO mice compared with those using wild-type spermatozoa (Table 2). ICSI using the spermatozoa from the CFAP70-mutated patient (II-1) was applied for two cycles and successfully resulted in a live birth (Table 3). Our data suggest that ICSI could serve as a generally promising treatment for infertile men harbouring CFAP70 pathogenic variants.
Table 2.
ICSI experiments using sperm from Cfap70-KO mice.
| WT sperm | Cfap70-KO sperm | |||||
|---|---|---|---|---|---|---|
| No. oocytes for injection | 59 | 85 | 67 | 79 | 74 | 80 |
| No. 2-cell embryos | 48 | 67 | 53 | 47 | 27 | 44 |
| Rate of 2-cell embryosa | 81% | 79% | 79% | 59% | 36% | 55% |
| No. blastocysts | 31 | 42 | 32 | 16 | 9 | 12 |
| Rate of blastocystsb | 65% | 63% | 60% | 34% | 33% | 27% |
| Litter size after transfer |  |
 |
 |
6 | 2 | 6 |
| Genotype of litters |  |
 |
 |
+/+: 3 +/−:3 −/−:0 |
+/+: 2 +/−:0 −/−:0 |
+/+: 2 +/−:4 −/−:0 |
Data were presented as the mean ± SD (n = 3 each group). Two-tailed, unpaired Student's t test.
p = 0.014.
p = 0.0002.
Table 3.
Clinical outcomes of ICSI treatment using sperm from the proband.
| Female age (year) | 34 |
| Length of primary infertility history (year) | 7 |
| BMI (kg/m2) | 21.88 |
| Basal hormones | |
| FSH (IU/L) | 8.60 |
| LH (IU/L) | 6.50 |
| E2 (pg/mL) | 57.79 |
| PRL (ng/mL) | 16.84 |
| P (ng/mL) | 0.28 |
| T (ng/mL) | 0.17 |
| Cycle 1 | |
| Protocol | Long-acting GnRH agonist |
| E2 level on the trigger day (pg/mL) | 4154 |
| No. of follicles ≥14 mm on the trigger day | 8 |
| No. of follicles ≥18 mm on the trigger day | 4 |
| No. of oocytes retrieved | 16 |
| ICSI | |
| No. of MII oocytes | 7 |
| Oocytes injected | 7 |
| Fertilization rate | 5/7 |
| Cleavage rate | 5/5 |
| 6-cell formation rate | 3/5 |
| 8-cell formation rate | 1/5 |
| Blastocyst formation rate | 0/0 |
| Cycle 2 | |
| Protocol | GnRH antagonist |
| E2 level on the trigger day (pg/mL) | 2482 |
| No. of follicles ≥14 mm on the trigger day | 6 |
| No. of follicles ≥18 mm on the trigger day | 3 |
| No. of oocytes retrieved | 15 |
| ICSI | |
| No. of MII oocytes | 10 |
| Oocytes injected | 10 |
| Fertilization rate | 9/10 |
| Cleavage rate | 9/9 |
| 6-cell formation rate | 9/9 |
| 8-cell formation rate | 8/9 |
| Blastocyst formation rate | 7/7 |
| No. of blastocyst transferred | 1 |
| Implantation rate | 1/1 |
| Clinical pregnancy rate | 1/1 |
| Offspring | A healthy boy |
In the first ICSI cycle, only one 8-cell embryo was obtained and transplantation was not performed. In the second cycle, seven blastocysts were obtained and a healthy boy was born after transplantation. The detailed information was described in this table.
Discussion
Sperm motility is the primary determinant of successful fertilization. Accordingly, studies on sperm flagellar development are of great importance to understand the reasons and mechanisms underlying astheno-teratozoospermia which is characterized by decreased sperm motility and structural abnormalities of the sperm flagella. MMAF (MIM: 617576), a subtype of astheno-teratozoospermia, is characterizing by short, coiled, absent, and/or irregular flagella.7 A previous high-throughput sequencing study identified two unrelated MMAF individuals with very low sperm counts carrying two homozygous variants (c.178 T > A/p.F60I and c.1723-1G > T) in CFAP70, implying that CFAP70 may be a novel MMAF-associated gene.23 However, the physiological role of CFAP70 in mammalian sperm flagellar development and the pathogenicity of the two identified CFAP70 variants are unknown. The conclusion that CFAP70 variants are pathogenic factors that cause male infertility cannot be drawn at the current stage primarily due to the lack of Cfap70-knockout/mutant animal models and more patients identified with CFAP70 pathogenic variants.
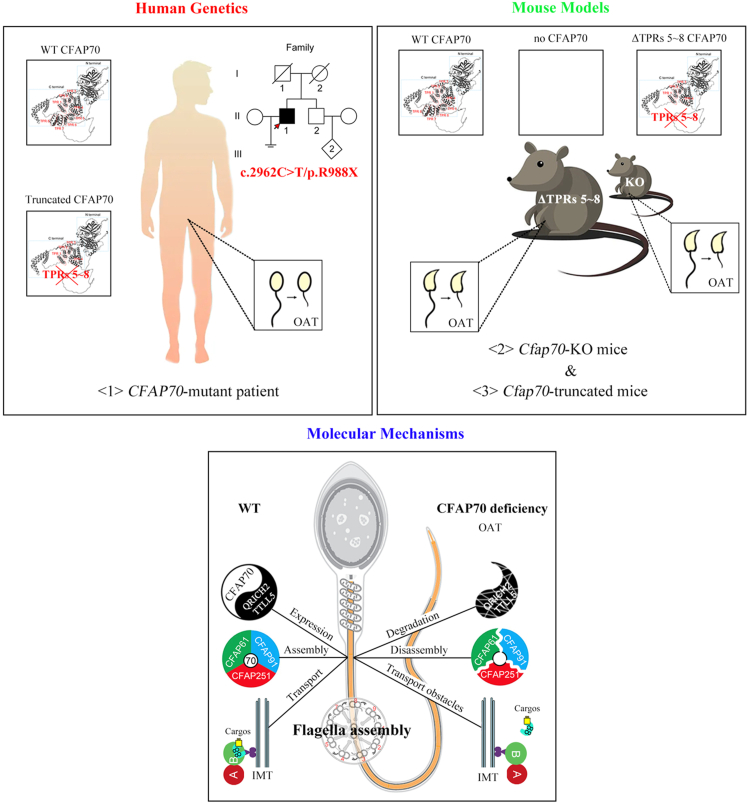
Here, we provide animal evidence to confirm that CFAP70 is indeed an OAT-associated gene, as Cfap70-KO mice exhibited male sterility with OAT. We further describe a homozygous nonsense CFAP70 variant (c.C2962 > T/p.R988X), which generates a truncated CFAP70 protein lacking C-terminal TPRs 5–8 in a Chinese OAT patient. Importantly, Cfap70-truncated mice (also lacking TPRs 5–8) mimicked the OAT phenotype of the patient, confirming the pathogenicity of this nonsense variant on CFAP70 function. The main findings in our current study were summarized in Fig. 7.
Fig. 7.
Schematic diagram of male infertility induced by CFAP70 deficiency. A homozygous nonsense mutation (c.2962C > T/p.R988X) in the CFAP70 gene was identified in a Chinese infertile man with OAT. This nonsense mutation produces a truncated CFAP70 lacking 5–8 tetratricopeptide repeats (TPRs). Two Cfap70 mouse models were generated by using CRISPR/Cas9 technology. Cfap70-KO mice (complete loss of CFAP70) and Cfap70-truncated mice (lacking 5–8 TPR at the C-terminus of CFAP70) perfectly mimic the OAT phenotype of the patient harboring CFAP70 mutation. CFAP70 is required for the assembly of sperm flagella through i) promoting the stability of the MMAF-associated proteins (e.g., QRICH2 and TTLL5), ii) assisting the cytoplasmic preassembly of CFAP61 and CFAP91, and iii) regulating the manchette localization of axoneme-associated proteins (e.g., IFT88, DNAI1, and CFAP61).
We suggest that the C-terminal TPRs 5–8 are critical for CFAP70 function because Cfap70-truncated mice lacking TPRs 5–8 exhibited male infertility with OAT (quite similar to Cfap70-KO mice). Beurois J et al. identified two variants (c.178 T > A/p.F60I and c.1723-1G > T) in CFAP70.23 These two variants localize at the N-terminal region of CFAP70 (before TPR domains). Unfortunately, the pathogenicity of the missense variant is uncertain because missense variants may or may not affect protein function. We could not exclude the possibility that variants in the N-terminal or middle region of CFAP70 may also be important for the full function of CFAP70.
High-throughput sequencing technologies, such as WES, in cohorts of infertile individuals have been successfully utilized to identify causative genes associated with human male infertility.4 We would like to call your attention that knockout mice of candidate genes are necessary to explore their physiological role in spermatogenesis, and mutant mice carrying candidate variants equivalent to those in patients are also essential to determine the pathogenicity of identified variants.6 Without animal evidence, the candidate gene variant(s) identified by WES cannot be directly linked with male infertility. In our study, Cfap70-KO mice were generated to reveal the physiological role of CFAP70 in spermatid flagellar biogenesis. Cfap70-truncated mice are further implied to confirm the pathogenicity of the nonsense variant in CFAP70 identified from the OAT patient. From a clinical standpoint, only those genes whose variants are identified by independent laboratories with a relatively high incidence in infertile patients could be applied for genetic diagnosis. To date, c.178 T > A/p.F60I and c.1723-1G > T23 as well as c. C2962 > T/p.R988X (this study) in CFAP70 have been identified in OAT patients from different ethnic groups. Further exploration of pathogenic variants in the CFAP70 gene in a large cohort of infertile men primarily with OAT will be helpful to evaluate the diagnostic value of CFAP70 variants as a genetic cause of OAT.
Neonatal hydrocephalus is a common disorder affecting the human nervous system with an estimated incidence of 1–3 per 1,000 live births, creating a health care burden of $2 billion annually.49 In addition to OAT, Cfap70-KO mice suffer from neonatal hydrocephalus, which leads to a high ratio of premature lethality. These phenomena were not surprising because motile cilia (e.g., ependymal cilia and trachea cilia) and sperm flagella shared similar ‘9 + 2’ axoneme structures and protein compositions. CFAP70 is predominantly expressed in the testis and brain, and very low levels of CFAP70 have been identified in the trachea. The combined phenotypes of male infertility and cilia disorders have also been shown in knockout mouse models of other CFAP family genes. Cfap43-KO mice exhibited both MMAF and early-onset hydrocephalus, and CFAP206 is required for sperm motility, mucociliary clearance of the airways and brain development.17,18,50 Patients carrying CFAP70 variants in Charles Coutton's study23 and our current study have no symptoms other than male sterility, indicating no obvious role or functional redundancy of CFAP70 in the development of ependymal cilia in humans. Another explanation is that mice are more sensitive to CFAP70 deficiency or that hydrocephalus development is easier in mice than in humans. We could not exclude the possibility that CFAP70 deficiency may also lead to hydrocephalus in some patients. Further clinical studies are needed to evaluate the pathological outcomes of CFAP70 variants.
One of the major knowledge gaps in the field of MMAF is how MMAF-associated proteins participate in sperm flagellar organization. DNAH1, DNAH2, DNAH8, DNAH10, and DNAH17 are heavy chains of ODAs or IDAs.7,51, 52, 53, 54 WDR63 is an intermediate chain of IDAs.48 CFAP61, CFAP91, and CFAP251 are conserved components of the CSC of cilia.16,20,35 TTC21A (IFT139A), TTC29, WDR19 (IFT144), and SPEF2 are IFT-associated proteins.55, 56, 57, 58 Beyond this information, the regulatory mechanisms of axoneme formation in sperm flagella by a majority of MMAF-associated proteins are largely unknown. Here, our study demonstrates that CFAP70 is critical for flagellar biogenesis/assembly in early spermatids and reveals some interesting mechanisms. First, quantitative proteomics revealed significantly reduced protein levels of QRICH2 and TTLL5 in Cfap70-KO mice. Deficiency of QRICH2 or TTLL5 leads to MMAF and male infertility in both humans and mice.34,37,59 CFAP70 interacts with QRICH2 via its C-terminal TPR motifs and stabilizes the expression of QRICH2. These findings favour the notion that TPR proteins typically function as chaperones or cofactors to mediate protein–protein interactions and protein stability.38 Second, co-IP screening indicated that DNAI1, DNAI2, CFAP61, and CFAP251 are CFAP70-binding partners. DNAI1 and DNAI2 are orthologues of Chlamydomonas ODA intermediate chains (IC1 and 2), and they are preassembled in the cytoplasm before they are transported into motile cilia/flagella.60, 61, 62 CFAP61, 91, and 251 are mammalian orthologues of Chlamydomonas CSC subunits, and they build the basal substructures of RS3.41,63 CFAP70 deletion does not affect the interaction of DNAI1 and DNAI2 but reduces the preassembly of CFAP61 with CFAP91. Third, CFAP70 influences the manchette distributions of IMT component IFT88 and its cargo proteins DNAI1 and CFAP61 in elongating spermatids. The Ift88 mouse mutant also exhibits a failure of sperm flagella formation due to disrupted IMT.45 Both CFAP70 and IFT88 are TPR proteins, and TPR motifs of different TPR proteins may interact directly.64
Cryo-electron tomography reveals that Chlamydomonas FAP70 localizes at the base of the ODA and the central apparatus projection C2a.21,22 We suggest that the physiological roles of CFAP70/FAP70 between mammalian spermatozoa and Chlamydomonas flagella may be different because knockout of FAP70 in Chlamydomonas causes a reduction in cilia motility (but has no effect on flagella length), whereas CFAP70 deficiency in mice and humans leads to a failure of flagellar assembly.21
Several recent studies, including our current work on CFAP70, highlight the critical role of TPR proteins in sperm flagellar formation. Biallelic variants in TTC21A or TTC29 induce male in/subfertility with OAT in both humans and mice.55,65 DNAH2, DNAH10 and DNAH17 are heavy chains of ODAs or IDAs, and evidence from mouse models and clinical relevance indicates their involvement in MMAF.53,66,67 Among IFT proteins, IFT88, IFT139, IFT140, IFT144 (WDR19), and IFT172 harbour TPR motifs, and abnormalities in sperm flagellar formation are observed in Ift88, Ift140, and Ift172 mutant/knockout mice, as well as WDR19-mutated patients.45,57,68,69
Our study was primarily conducted using Cfap70-KO/truncated mice and their littermate wild-type mice. In most experiments, three animals were used in each group. Although those animal experiments are not very dependent on statistical analysis, we could not thoroughly exclude some of our results and their interpretation will be confounded due to unknown factors.
In conclusion, our study provide solid animal evidence to suggest that CFAP70 is necessary to assemble spermatid flagella and CFAP70 gene could be used as a diagnostic target for male infertility with OAT in clinical practice. It will be exciting to see more pathogenic mutations in CFAP70 to be reported in patients suffering from male infertility.
Contributors
S.C. supervised the study and wrote the manuscript. H.J. designed experiments and performed molecular and biochemical experiments. J.W., C.W., and B.W. recruited patients and performed WES. S.C. performed animal experiments. H.J., and X.G. contributed to the cell culture, plasmid construction, and animal breeding. S.C., and B.W. verified the underlying data. All authors read and approved the final version of the manuscript.
Data sharing statement
Proteomics data have been deposited to the ProteomeXchange Consortium via the iProX partner repository with the dataset identifier PXD033133. Data supporting this study are available on request; please contact our corresponding author.
Declaration of interests
The authors declare no competing interests.
Acknowledgements
This study was supported by the National Key Research and Development Project (2019YFA0802101 to S.C), Central Government to Guide Local Scientific and Technological Development (ZY21195023 to B.W), Open Fund of Key Laboratory of Cell Proliferation and Regulation Biology, Ministry of Education (to S.C), and Basic Research Projects of Central Scientific Research Institutes (to B.W).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104675.
Contributor Information
Bin-Bin Wang, Email: wbbahu@163.com.
Su-Ren Chen, Email: chensr@bnu.edu.cn.
Appendix A. Supplementary data
References
- 1.Tuttelmann F., Ruckert C., Ropke A. Disorders of spermatogenesis: perspectives for novel genetic diagnostics after 20 years of unchanged routine. Med Genet. 2018;30:12–20. doi: 10.1007/s11825-018-0181-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ray P.F., Toure A., Metzler-Guillemain C., et al. Genetic abnormalities leading to qualitative defects of sperm morphology or function. Clin Genet. 2017;91:217–232. doi: 10.1111/cge.12905. [DOI] [PubMed] [Google Scholar]
- 3.Dam A.H., Feenstra I., Westphal J.R., et al. Globozoospermia revisited. Hum Reprod Update. 2007;13:63–75. doi: 10.1093/humupd/dml047. [DOI] [PubMed] [Google Scholar]
- 4.Toure A., Martinez G., Kherraf Z.E., et al. The genetic architecture of morphological abnormalities of the sperm tail. Hum Genet. 2021;140:21–42. doi: 10.1007/s00439-020-02113-x. [DOI] [PubMed] [Google Scholar]
- 5.Wu B., Gao H., Liu C., Li W. The coupling apparatus of the sperm head and tail. Biol Reprod. 2020;102:988–998. doi: 10.1093/biolre/ioaa016. [DOI] [PubMed] [Google Scholar]
- 6.Jiao S., Yang Y., Chen S. Molecular genetics of infertility: loss-of-function mutations in humans and corresponding knockout/mutated mice. Hum Reprod Update. 2021;27:154–189. doi: 10.1093/humupd/dmaa034. [DOI] [PubMed] [Google Scholar]
- 7.Ben Khelifa M., Coutton C., Zouari R., et al. Mutations in DNAH1, which encodes an inner arm heavy chain dynein, lead to male infertility from multiple morphological abnormalities of the sperm flagella. Am J Hum Genet. 2014;94:95–104. doi: 10.1016/j.ajhg.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang S., Wang X., Li W., et al. Biallelic Mutations in CFAP43 and CFAP44 cause male infertility with multiple morphological abnormalities of the sperm flagella. Am J Hum Genet. 2017;100:854–864. doi: 10.1016/j.ajhg.2017.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coutton C., Vargas A.S., Amiri-Yekta A., et al. Mutations in CFAP43 and CFAP44 cause male infertility and flagellum defects in Trypanosoma and human. Nat Commun. 2018;9:686. doi: 10.1038/s41467-017-02792-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu C., Tu C., Wang L., et al. Deleterious variants in X-linked CFAP47 induce asthenoteratozoospermia and primary male infertility. Am J Hum Genet. 2021;108:309–323. doi: 10.1016/j.ajhg.2021.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He X., Liu C., Yang X., et al. Bi-allelic loss-of-function variants in CFAP58 cause flagellar axoneme and mitochondrial sheath defects and asthenoteratozoospermia in humans and mice. Am J Hum Genet. 2020;107:514–526. doi: 10.1016/j.ajhg.2020.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang Y., Liu C., Li M., et al. Absence of murine CFAP61 causes male infertility due to multiple morphological abnormalities of the flagella. Sci Bull. 2020;65:854–864. doi: 10.1016/j.scib.2020.01.023. [DOI] [PubMed] [Google Scholar]
- 13.Li W., Wu H., Li F., et al. Biallelic mutations in CFAP65 cause male infertility with multiple morphological abnormalities of the sperm flagella in humans and mice. J Med Genet. 2020;57:89–95. doi: 10.1136/jmedgenet-2019-106344. [DOI] [PubMed] [Google Scholar]
- 14.Dong F.N., Amiri-Yekta A., Martinez G., et al. Absence of CFAP69 causes male infertility due to multiple morphological abnormalities of the flagella in human and mouse. Am J Hum Genet. 2018;102:636–648. doi: 10.1016/j.ajhg.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He X., Li W., Wu H., et al. Novel homozygous CFAP69 mutations in humans and mice cause severe asthenoteratospermia with multiple morphological abnormalities of the sperm flagella. J Med Genet. 2019;56:96–103. doi: 10.1136/jmedgenet-2018-105486. [DOI] [PubMed] [Google Scholar]
- 16.Martinez G., Beurois J., Dacheux D., et al. Biallelic variants in MAATS1 encoding CFAP91, a calmodulin-associated and spoke-associated complex protein, cause severe astheno-teratozoospermia and male infertility. J Med Genet. 2020;57:708–716. doi: 10.1136/jmedgenet-2019-106775. [DOI] [PubMed] [Google Scholar]
- 17.Shen Q., Martinez G., Liu H., et al. Bi-allelic truncating variants in CFAP206 cause male infertility in human and mouse. Hum Genet. 2021;140:1367–1377. doi: 10.1007/s00439-021-02313-z. [DOI] [PubMed] [Google Scholar]
- 18.Beckers A., Adis C., Schuster-Gossler K., et al. The FOXJ1 target Cfap206 is required for sperm motility, mucociliary clearance of the airways and brain development. Development. 2020;147:dev188052. doi: 10.1242/dev.188052. [DOI] [PubMed] [Google Scholar]
- 19.Kherraf Z.E., Amiri-Yekta A., Dacheux D., et al. A homozygous ancestral SVA-insertion-mediated deletion in WDR66 induces multiple morphological abnormalities of the sperm flagellum and male infertility. Am J Hum Genet. 2018;103:400–412. doi: 10.1016/j.ajhg.2018.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Auguste Y., Delague V., Desvignes J.P., et al. Loss of calmodulin- and radial-spoke-associated complex protein CFAP251 leads to immotile spermatozoa lacking mitochondria and infertility in men. Am J Hum Genet. 2018;103:413–420. doi: 10.1016/j.ajhg.2018.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shamoto N., Narita K., Kubo T., Oda T., Takeda S. CFAP70 is a novel axoneme-binding protein that localizes at the base of the outer dynein arm and regulates ciliary motility. Cells. 2018;7:124. doi: 10.3390/cells7090124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hou Y., Zhao L., Kubo T., et al. Chlamydomonas FAP70 is a component of the previously uncharacterized ciliary central apparatus projection C2a. J Cell Sci. 2021;134:jcs258540. doi: 10.1242/jcs.258540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beurois J., Martinez G., Cazin C., et al. CFAP70 mutations lead to male infertility due to severe astheno-teratozoospermia. A case report. Hum Reprod. 2019;34:2071–2079. doi: 10.1093/humrep/dez166. [DOI] [PubMed] [Google Scholar]
- 24.Cooper T.G., Noonan E., von Eckardstein S., et al. World Health Organization reference values for human semen characteristics. Hum Reprod Update. 2010;16:231–245. doi: 10.1093/humupd/dmp048. [DOI] [PubMed] [Google Scholar]
- 25.Li H., Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McKenna A., Hanna M., Banks E., et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protocols. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 29.Adzhubei I.A., Schmidt S., Peshkin L., et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwarz J.M., Cooper D.N., Schuelke M., Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 31.Zhang X., Wei L., Zhang X., Jin H., Chen S. Loss of perinuclear theca ACTRT1 causes acrosome detachment and severe male subfertility in mice. Development. 2022;149:dev200489. doi: 10.1242/dev.200489. [DOI] [PubMed] [Google Scholar]
- 32.Zhang X., Wei L., Jin H., Zhang X., Chen S. The perinuclear theca protein Calicin helps shape the sperm head and maintain the nuclear structure in mice. Cell Rep. 2022;40 doi: 10.1016/j.celrep.2022.111049. [DOI] [PubMed] [Google Scholar]
- 33.Guichard C., Harricane M.C., Lafitte J.J., et al. Axonemal dynein intermediate-chain gene (DNAI1) mutations result in situs inversus and primary ciliary dyskinesia (Kartagener syndrome) Am J Hum Genet. 2001;68:1030–1035. doi: 10.1086/319511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen Y., Zhang F., Li F., et al. Loss-of-function mutations in QRICH2 cause male infertility with multiple morphological abnormalities of the sperm flagella. Nat Commun. 2019;10:433. doi: 10.1038/s41467-018-08182-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu S., Zhang J., Kherraf Z.E., et al. CFAP61 is required for sperm flagellum formation and male fertility in human and mouse. Development. 2021;148:dev199805. doi: 10.1242/dev.199805. [DOI] [PubMed] [Google Scholar]
- 36.Ma J., Chen T., Wu S., et al. iProX: an integrated proteome resource. Nucleic Acids Res. 2019;47:D1211–D1217. doi: 10.1093/nar/gky869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee G.S., He Y., Dougherty E.J., et al. Disruption of Ttll5/stamp gene (tubulin tyrosine ligase-like protein 5/SRC-1 and TIF2-associated modulatory protein gene) in male mice causes sperm malformation and infertility. J Biol Chem. 2013;288:15167–15180. doi: 10.1074/jbc.M113.453936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blatch G.L., Lassle M. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays. 1999;21:932–939. doi: 10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 39.Loges N.T., Olbrich H., Fenske L., et al. DNAI2 mutations cause primary ciliary dyskinesia with defects in the outer dynein arm. Am J Hum Genet. 2008;83:547–558. doi: 10.1016/j.ajhg.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dymek E.E., Smith E.F. A conserved CaM- and radial spoke associated complex mediates regulation of flagellar dynein activity. J Cell Biol. 2007;179:515–526. doi: 10.1083/jcb.200703107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heuser T., Dymek E.E., Lin J., Smith E.F., Nicastro D. The CSC connects three major axonemal complexes involved in dynein regulation. Mol Biol Cell. 2012;23:3143–3155. doi: 10.1091/mbc.E12-05-0357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu T., Meng L., Tan C., et al. Biallelic CFAP61 variants cause male infertility in humans and mice with severe oligoasthenoteratozoospermia. J Med Genet. 2022 doi: 10.1136/jmedgenet-2021-108249. jmedgenet-20210108249. [DOI] [PubMed] [Google Scholar]
- 43.Li W., He X., Yang S., et al. Biallelic mutations of CFAP251 cause sperm flagellar defects and human male infertility. J Hum Genet. 2019;64:49–54. doi: 10.1038/s10038-018-0520-1. [DOI] [PubMed] [Google Scholar]
- 44.Pleuger C., Lehti M.S., Dunleavy J.E., Fietz D., O'Bryan M.K. Haploid male germ cells-the Grand Central Station of protein transport. Hum Reprod Update. 2020;26:474–500. doi: 10.1093/humupd/dmaa004. [DOI] [PubMed] [Google Scholar]
- 45.Kierszenbaum A.L., Rivkin E., Tres L.L., et al. GMAP210 and IFT88 are present in the spermatid golgi apparatus and participate in the development of the acrosome-acroplaxome complex, head-tail coupling apparatus and tail. Dev Dyn. 2011;240:723–736. doi: 10.1002/dvdy.22563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang X., Zheng R., Liang C., et al. Loss-of-function mutations in CEP78 cause male infertility in humans and mice. Sci Adv. 2022;8 doi: 10.1126/sciadv.abn0968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wambergue C., Zouari R., Fourati Ben Mustapha S., et al. Patients with multiple morphological abnormalities of the sperm flagella due to DNAH1 mutations have a good prognosis following intracytoplasmic sperm injection. Hum Reprod. 2016;31:1164–1172. doi: 10.1093/humrep/dew083. [DOI] [PubMed] [Google Scholar]
- 48.Lu S., Gu Y., Wu Y., et al. Bi-allelic variants in human WDR63 cause male infertility via abnormal inner dynein arms assembly. Cell Discov. 2021;7:110. doi: 10.1038/s41421-021-00327-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wallmeier J., Nielsen K.G., Kuehni C.E., et al. Motile ciliopathies. Nat Rev Dis Primers. 2020;6:77. doi: 10.1038/s41572-020-0209-6. [DOI] [PubMed] [Google Scholar]
- 50.Rachev E., Schuster-Gossler K., Fuhl F., et al. CFAP43 modulates ciliary beating in mouse and Xenopus. Dev Biol. 2020;459:109–125. doi: 10.1016/j.ydbio.2019.12.010. [DOI] [PubMed] [Google Scholar]
- 51.Li Y., Sha Y., Wang X., et al. DNAH2 is a novel candidate gene associated with multiple morphological abnormalities of the sperm flagella. Clin Genet. 2019;95:590–600. doi: 10.1111/cge.13525. [DOI] [PubMed] [Google Scholar]
- 52.Liu C., Miyata H., Gao Y., et al. Bi-allelic DNAH8 variants lead to multiple morphological abnormalities of the sperm flagella and primary male infertility. Am J Hum Genet. 2020;107:330–341. doi: 10.1016/j.ajhg.2020.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tu C., Cong J., Zhang Q., et al. Bi-allelic mutations of DNAH10 cause primary male infertility with asthenoteratozoospermia in humans and mice. Am J Hum Genet. 2021;108:1466–1477. doi: 10.1016/j.ajhg.2021.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Whitfield M., Thomas L., Bequignon E., et al. Mutations in DNAH17, encoding a sperm-specific axonemal outer dynein arm heavy chain, cause isolated male infertility due to asthenozoospermia. Am J Hum Genet. 2019;105:198–212. doi: 10.1016/j.ajhg.2019.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu W., He X., Yang S., et al. Bi-allelic mutations in TTC21A induce asthenoteratospermia in humans and mice. Am J Hum Genet. 2019;104:738–748. doi: 10.1016/j.ajhg.2019.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lores P., Dacheux D., Kherraf Z.E., et al. Mutations in TTC29, encoding an evolutionarily conserved axonemal protein, result in asthenozoospermia and male infertility. Am J Hum Genet. 2019;105:1148–1167. doi: 10.1016/j.ajhg.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ni X., Wang J., Lv M., et al. A novel homozygous mutation in WDR19 induces disorganization of microtubules in sperm flagella and nonsyndromic asthenoteratospermia. J Assist Reprod Genet. 2020;37:1431–1439. doi: 10.1007/s10815-020-01770-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu C., Lv M., He X., et al. Homozygous mutations in SPEF2 induce multiple morphological abnormalities of the sperm flagella and male infertility. J Med Genet. 2020;57:31–37. doi: 10.1136/jmedgenet-2019-106011. [DOI] [PubMed] [Google Scholar]
- 59.Kherraf Z.E., Cazin C., Coutton C., et al. Whole exome sequencing of men with multiple morphological abnormalities of the sperm flagella reveals novel homozygous QRICH2 mutations. Clin Genet. 2019;96:394–401. doi: 10.1111/cge.13604. [DOI] [PubMed] [Google Scholar]
- 60.Cho K.J., Noh S.H., Han S.M., et al. ZMYND10 stabilizes intermediate chain proteins in the cytoplasmic pre-assembly of dynein arms. PLoS Genet. 2018;14 doi: 10.1371/journal.pgen.1007316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mali G.R., Yeyati P.L., Mizuno S., et al. ZMYND10 functions in a chaperone relay during axonemal dynein assembly. eLife. 2018;7 doi: 10.7554/eLife.34389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Omran H., Kobayashi D., Olbrich H., et al. Ktu/PF13 is required for cytoplasmic pre-assembly of axonemal dyneins. Nature. 2008;456:611–616. doi: 10.1038/nature07471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Urbanska P., Song K., Joachimiak E., et al. The CSC proteins FAP61 and FAP251 build the basal substructures of radial spoke 3 in cilia. Mol Biol Cell. 2015;26:1463–1475. doi: 10.1091/mbc.E14-11-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kierszenbaum A.L. Intramanchette transport (IMT): managing the making of the spermatid head, centrosome, and tail. Mol Reprod Dev. 2002;63:1–4. doi: 10.1002/mrd.10179. [DOI] [PubMed] [Google Scholar]
- 65.Liu C., He X., Liu W., et al. Bi-allelic mutations in TTC29 cause male subfertility with asthenoteratospermia in humans and mice. Am J Hum Genet. 2019;105:1168–1181. doi: 10.1016/j.ajhg.2019.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hwang J.Y., Nawaz S., Choi J., et al. Genetic defects in DNAH2 underlie male infertility with multiple morphological abnormalities of the sperm flagella in humans and mice. Front Cell Dev Biol. 2021;9 doi: 10.3389/fcell.2021.662903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang B., Ma H., Khan T., et al. A DNAH17 missense variant causes flagella destabilization and asthenozoospermia. J Exp Med. 2020;217 doi: 10.1084/jem.20182365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang S., Liu Y., Huang Q., et al. Murine germ cell-specific disruption of Ift172 causes defects in spermiogenesis and male fertility. Reproduction. 2020;159:409–421. doi: 10.1530/REP-17-0789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang Y., Liu H., Li W., et al. Intraflagellar transporter protein 140 (IFT140), a component of IFT-A complex, is essential for male fertility and spermiogenesis in mice. Cytoskeleton. 2018;75:70–84. doi: 10.1002/cm.21427. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.