Abstract
Oxidative stress due to abnormal accumulation of reactive oxygen species (ROS) is an initiator of a large number of human diseases, and thus, the elimination and prevention of excessive ROS are important aspects of preventing the development of such diseases. Nuclear factor erythroid 2-related factor 2 (NRF2) is an essential transcription factor that defends against oxidative stress, and its function is negatively controlled by Kelch-like ECH-associated protein 1 (KEAP1). Therefore, activating NRF2 by inhibiting KEAP1 is viewed as a strategy for combating oxidative stress-related diseases. Here, we generated a cereblon (CRBN)-based proteolysis-targeting chimera (PROTAC), which we named SD2267, that induces the proteasomal degradation of KEAP1 and leads to NRF2 activation. As was intended, SD2267 bound to KEAP1, recruited CRBN, and induced the degradation of KEAP1. Furthermore, the KEAP1 degradation efficacy of SD2267 was diminished by MG132 (a proteasomal degradation inhibitor) but not by chloroquine (an autophagy inhibitor), which suggested that KEAP1 degradation by SD2267 was proteasomal degradation-dependent and autophagy-independent. Following KEAP1 degradation, SD2267 induced the nuclear translocation of NRF2, which led to the expression of NRF2 target genes and attenuated ROS accumulation induced by acetaminophen (APAP) in hepatocytes. Based on in vivo pharmacokinetic study, SD2267 was injected intraperitoneally at 1 or 3 mg/kg in APAP-induced liver injury mouse model. We observed that SD2267 degraded hepatic KEAP1 and attenuated APAP-induced liver damage. Summarizing, we described the synthesis of a KEAP1-targeting PROTAC (SD2267) and its efficacy and mode of action in vitro and in vivo. The results obtained suggest that SD2267 could be used to treat hepatic diseases related to oxidative stress.
Keywords: Proteolysis-targeting chimera (PROTAC), KEAP1, NRF2, Oxidative stress, Liver
Highlights
-
•
SD2267 induced CRBN-mediated proteasomal degradation of KEAP1 in hepatocytes.
-
•
SD2267 translocated NRF2 into nucleus, and increased transcriptions of its target genes such as HMOX1, NQO1, GCLC and GCLM.
-
•
SD2267 alleviated acetaminophen-induced damages in vitro and in vivo.
-
•
This is the first time that PROTACs showed anti-oxidative effect in vivo.
Abbreviation
- ALT
alanine transaminase
- APAP
acetaminophen
- AST
aspartate transaminase
- CQ
chloroquine
- CRBN
cereblon
- DART-MS
direct analysis real time – mass spectrometry.
- DCM
dichloromethane
- DIAD
diisopropyl azodicarboxylate
- DMF
N, N-dimethyl formamide
- DMSO
dimethyl sulfoxide
- EtOAc
ethyl acetate
- ESI
electrospray ionization
- FP
fluorescence polarization
- FRET
Fluorescence resonance energy transfer
- GCLC
glutamyl cysteine ligase catalytic subunit
- GCLM
glutamyl cysteine ligase modifier subunit
- GSH
glutathione
- GSSG
glutathione disulfide
- HO-1
heme oxygenase 1
- HPF
high power field
- HRMS
high resolution mass spectrometry
- ITC
isothermal titration calorimetry
- JNK
c-Jun N-terminal kinase
- KEAP1
Kelch-like ECH-associated protein 1
- MPLC
medium pressure liquid chromatography
- MSCI4
Magnetospirillum gryphiswaldense cereblon isoform 4
- NAPQI
N-acetyl-p-benzoquinone imine
- NMR
nuclear magnetic resonance
- NQO-1
NAD(P)H quinone oxidoreductase-1
- NRF2
Nuclear factor erythroid 2-related factor 2
- POI
protein of interest
- PROTAC
proteolysis-targeting chimera
- ROS
reactive oxygen species
- TEA
triethyl amine
- THF
tetrahydrofuran
- TMS
tetramethyl silane
- TUNEL
terminal deoxynucleotidyl transferase dUTP nick end labeling
1. Introduction
Reactive oxygen species (ROS) are naturally produced within the body to maintain cellular homeostasis and regulate signal transduction and gene expressions related to cell viability, migration, proliferation, and differentiation [1]. The amounts of ROS generated are controlled by redox balance between pro-oxidant and antioxidant species [2], but ROS can accumulate in cells when antioxidant defense mechanisms are defective or the cellular ROS buffering system fails, and this accumulation leads to alterations in biomolecules such as DNA, RNA, and proteins, which collectively referred to as oxidative stress [3]. In the past few decades, oxidative stress, characterized by excessive ROS, has proven to be a major contributor to many human diseases, such as drug-induced liver injury, cancers, and neurodegenerative, cardiovascular, airway, and inflammatory diseases [[4], [5], [6], [7], [8], [9], [10]]. Thus, improving oxidative stress by enhancing the cellular ROS defense system is considered a potential means of preventing the development of oxidative stress-related diseases.
Nuclear factor erythroid 2-related factor 2 (NRF2) is a key player of cellular redox homeostasis [11]. Typically, NRF2 is bound to Kelch-like ECH-associated protein 1 (KEAP1), which promotes NRF2 degradation by the ubiquitin-proteasome system [12]. In the presence of oxidative stress, NRF2 is released from KEAP1 and translocated to the nucleus, where it acts as a transcription factor. After binding to anti-oxidant response elements, NRF2 induces the expression of its target genes, which are mainly anti-oxidant genes and detoxifying enzymes that include heme oxygenase 1 (HO-1), NAD(P)H quinone oxidoreductase-1 (NQO-1), glutathione, γ-glutamyl cysteine ligase catalytic subunit (GCLC), and its modifier subunit (GCLM), among others [13]. However, several diseases occur when the degree of oxidative stress exceeds the capacity of the defense mechanism induced by the NRF2 pathway. Based on this knowledge, activating the NRF2 signaling pathway to increase anti-oxidative capacity has been suggested as an efficacious strategy to overcome several diseases mediated by oxidative stress [14,15].
Proteolysis-targeting chimera (PROTAC) technology has recently emerged as a platform technology for targeted protein degradation. PROTAC molecules possess three compartments: a ligand of the protein of interest (POI), a ligand of E3 ubiquitin ligase, and a linker that connects these two ligands [16]. These molecules bind to the POI and recruit E3 ligase, which results in the ubiquitination and proteasomal degradation of the POI. Due to this catalytic mode of action, PROTACs can be sustained in target cells even after washout, and therefore, the degradation of target proteins by PROTACs can be achieved at nanomolar concentrations. So thus, PROTACs have the unique ability to overcome the limitations of small molecule inhibitors, which can cause off-target effects when used at high concentrations in order to be effective [17].
Recently, PROTAC-based KEAP1 degraders have been synthesized [[18], [19], [20]], and their anti-oxidative effects have been examined in lung epithelial cells [19] and hepatic stellate cells [20]. However, additional studies are necessary to determine the anti-oxidative effects of KEAP1-targeting PROTACs in other cell types and their in vivo efficacies. In this study, we synthesized a KEAP1-targeting PROTAC as a novel therapeutic approach to alleviate oxidative stress in liver, and showed its in vitro and in vivo anti-oxidative effects, suggesting that SD2267 would be another type of medication to overcome human liver diseases related to oxidative stress.
2. Results and discussion
2.1. Synthesis of KEAP1-targeting PROTAC
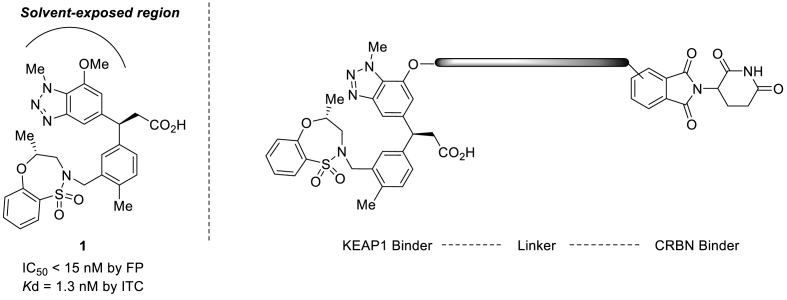
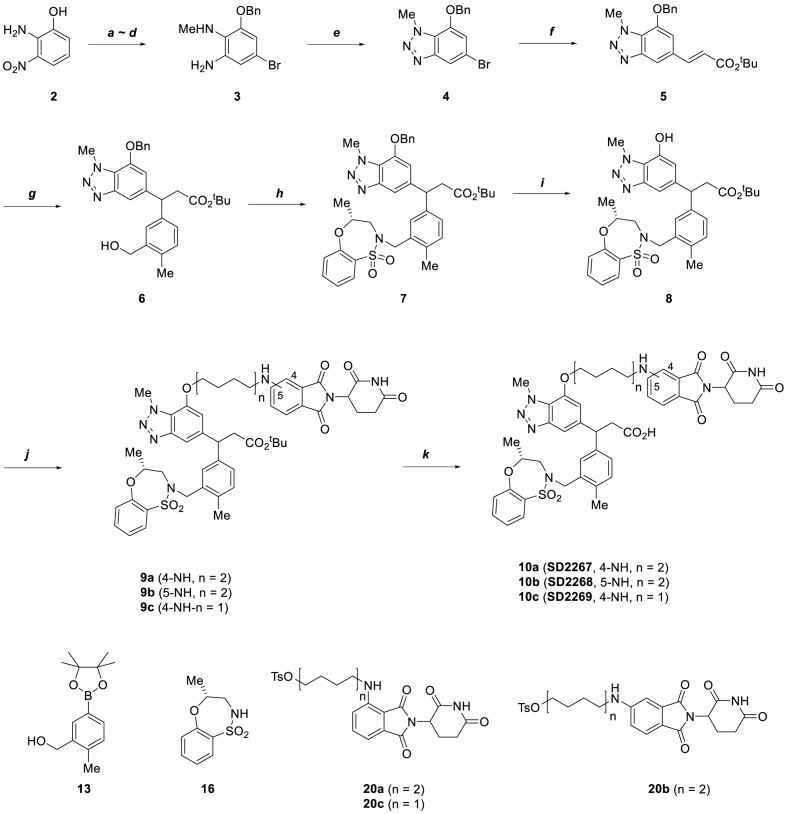
To design KEAP1 degrading PROTACs, we first selected a KEAP1 binder. Compound 1 was selected from a list of known KEAP1 inhibitors based on its binding affinity and the availability of its x-ray crystal structures. Compound 1 was reported to have the highest binding affinity by fluorescence polarization (FP) method (IC50 12.50 nM) and a Kd of 1.3 nM by isothermal titration calorimetry (ITC) method [21]. Moreover, the crystal structure (PDB ID:5FNU) of KEAP1 protein and Compound 1 complex revealed the solvent-exposed region of Compound 1 and provided an important information on the linker binding location. KEAP1-targeting PROTACs were designed by adopting cereblon (CRBN) as the E3 ligase (Fig. 1). The method used to synthesize KEAP1-targeting PROTACs is shown in Scheme 1. Compound 8 which contains a phenolic hydroxyl group as a linking position was used as the KEAP1 binder. Starting with 2-amino-3-nitrophenol 2, sequential O-benzylation of phenolic hydroxyl group with benzyl bromide and para-bromination of aniline with bromine, N-methylation of aniline with methyl iodide, and reduction of the nitro group to amine using zinc dust gave aniline 3. N-methyl-1,2-aminobenzene 3 was cyclized to benzotriazole 4 by treating it with NaNO2 and 10% H2SO4, and then the Heck reaction with tert-butyl acrylate using palladium catalyst Pd(PPh3)4 and P(o-tol)3) ligand was used to produce ester 5. The olefin was coupled with phenyl boronic acid pinacol ester 13 using a rhodium catalyst [RhCl(Cod)]2] to afford alcohol 6, which was transformed into sulfonamide 7 using the Mitsunobu reaction (DIAD, PPh3). Debenzylation of 7 using H2 and 10% Pd/C provided the key intermediate phenol compound 8. The syntheses of KEAP1-targeting PROTACs SD2267, SD2268 and SD2269 were completed by the O-alkylation of phenol hydroxyl group 8 with CRBN binder-augmented alkyl tosylates 20a, 20b and 20c, respectively, followed by tert-butyl group removal under an acidic condition.
Fig. 1.
Design of KEAP1-targeting PROTAC
Compound 1 is a reported inhibitor of KEAP1 (IC50 < 15 nM, Kd = 1.3 nM) and was used to synthesize of KEAP1-targeting PROTAC with a flexible linking vector and CRBN ligand.
Scheme 1.
Synthesis of KEAP1-targeting PROTAC.
Reagent and conditions: (a) BnBr, K2CO3, EtOH, 70 °C, 81%; (b) Br2, AcONa, AcOH, r.t, 96%; (c) MeI, NaH, DMF, 0 °C to r.t, 79%; (d) Zn dust, AcOH, 0 °C–45 °C, 59%; (e) NaNO2, 10% H2SO4 (aq.), 87%; (f) tert-Butyl acrylate, Pd(PPh3)4, P(O-Tol)3, DMF, 95 °C, 79%; (g) 13, [RhCl(Cod)]2, Et3N, Dioxane/H2O (2:1), 95 °C, 53%; (h) 16, DIAD, PPh3, anhydrous THF, r.t, 75%; (i) H2, Pd/C, MeOH, r.t, 98%; (j) 20a-c, K2CO3, DMF, 90 °C, 21–39%; (k) 4 N HCl/Dioxane, r.t, 53–88%.
To select one compound for further in vitro and in vivo evaluations, at first, KEAP1-PROTACs were subjected to a Fluorescence resonance energy transfer (FRET)-based competition assay to compare CRBN binding affinities of KEAP1 PROTACs. The Ki value of SD2267 was the lowest among that of KEAP1 PROTACs, suggesting that SD2267 showed the most powerful binding affinity to CRBN protein (Table 1).
Table 1.
FRET-based competition assay of KEAP1-targeting PROTACs. All measurement was performed in triplicate and Ki values are expressed as the mean ± standard deviation.
| Compound | Molecular weight (g/mol) | Ki value (nM) | R2 |
|---|---|---|---|
| SD2267 | 920.05 | 57.7 ± 17.5 | 0.94 |
| SD2268 | 920.05 | 8151.7 ± 5707.6 | 0.75 |
| SD2269 | 863.94 | 64.8 ± 13.2 | 0.94 |
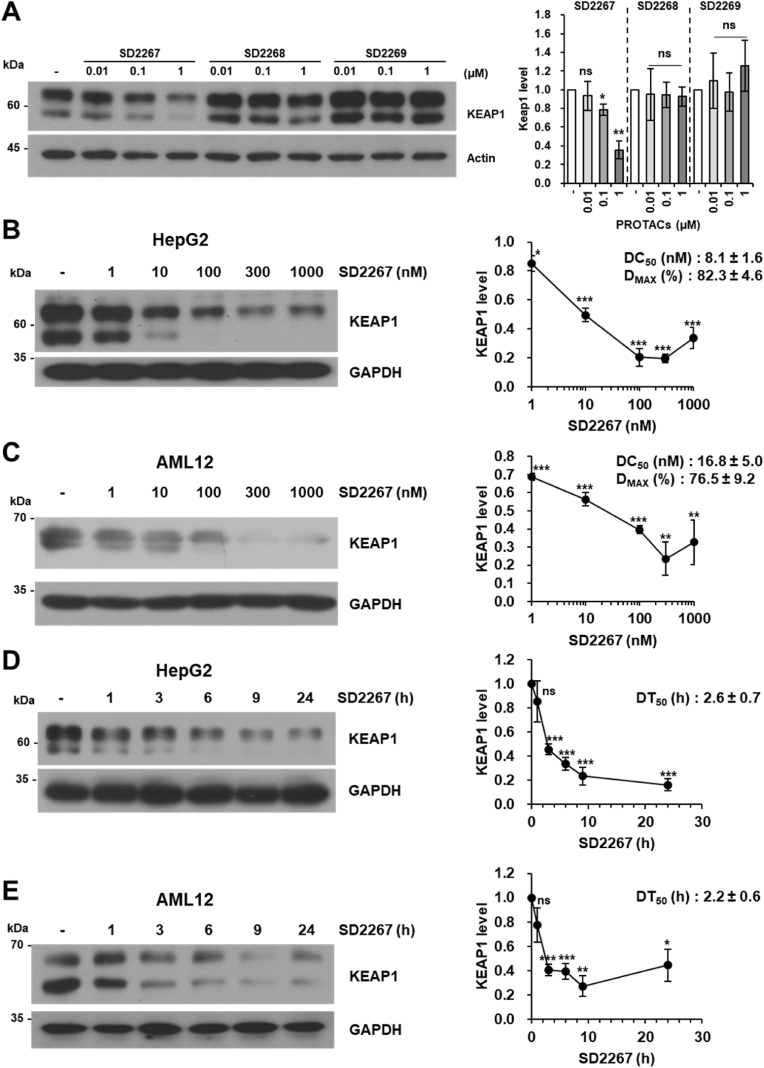
Next, to evaluate KEAP1 degradation efficacies of PROTACs, HepG2 cells were incubated with different concentrations of each PROTAC for 6 h, and then KEAP1 protein expression levels were detected by western blotting. SD2267 reduced the KEAP1 level by 64% at 1 μM, whereas SD2268 reduced it by only 7%, and SD2269 had no effect (Fig. 2A). Thus, SD2267 was chosen for further experiments. To investigate the KEAP1 degrading efficacy of SD2267 further, HepG2 and AML12 cells were treated with SD2267 at a wide range of concentrations from 1 to 1000 nM for 24 h. The KEAP1 level was decreased by SD2267 with a half-maximal degradation concentration (DC50) of 8.1 nM and a maximum degradation efficacy (Dmax) of 82.3% at 300 nM in HepG2 cells (Fig. 2B), and a DC50 of 16.8 nM and Dmax of 76.5% at 300 nM in AML12 cells (Fig. 2C). To investigate the kinetics of KEAP1 degradation by SD2267, both cell lines were incubated with SD2267 at 100 nM for different times. Half-lives (DT50) of KEAP1 in HepG2 cells and AML12 cells were 2.6 h and 2.2 h, respectively (Fig. 2D and E). These results showed that SD2267 rapidly penetrates cells and degrades KEAP1 within few hours even at nanomolar concentrations.
Fig. 2.
Selection of a KEAP1-targeting PROTAC and its efficacy in hepatocytes
(A-E) Representative image of KEAP1 protein expression in HepG2 and AML12 cells (left) and graphs of the relative mean expression of KEAP1 (right). (A) HepG2 cells were incubated with SD2267, SD2268, or SD2269 at indicated doses for 6 h. (B–E) HepG2 or AML12 cells were treated with SD2267 at different concentrations for 24 h (B and C, respectively) or at different times at 100 nM (D and E, respectively). DC50, DMAX, and DT50 value are included in the related graphs. Densitometries were performed using three independent experiments. ns; no significance, *p < 0.05, **p < 0.01, ***p < 0.001 versus untreated cells.
2.2. In vitro KEAP1 degradation machinery of SD2267 and its antioxidant properties
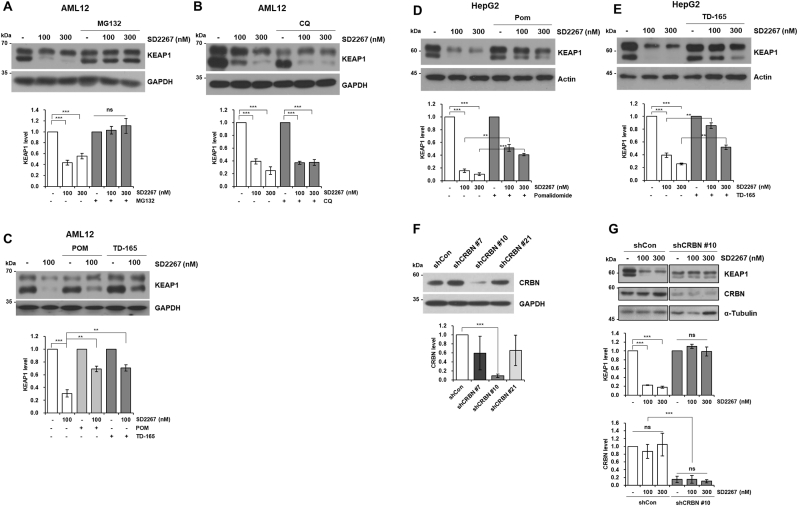
Since we designed SD2267 as a KEAP1-targeting PROTAC, we assessed in vitro machinery of SD2267 involved in KEAP1 degradation and the KEAP1 level was evaluated by western blotting. KEAP1 degradation by SD2267 was found to occur through the proteasomal degradation pathway since SD2267 could not degrade KEAP1 in the presence of MG132 (an inhibitor of proteasomal degradation) in AML12 cells (Fig. 3A). However, proteins can also be degraded by autophagy [22], and P62, acting as a cargo protein during autophagy, can bind to KEAP1 with its 349DPSTGEL355 motif and label it for autophagic degradation [23]. Therefore, to determine whether autophagy is involved in SD2267-mediated KEAP1 degradation, AML12 cells were treated with SD2267 and chloroquine (CQ), which inhibits autophagy by inhibiting autophagosome and lysosome fusion. However, autophagy inhibition did not abolish KEAP1 degradation by SD2267, suggesting that P62-mediated autophagic degradation of KEAP1 was not involved in the degradation of KEAP1 by SD2267 in AML12 cells (Fig. 3B). Next, we examined whether the proteasomal degradation of KEAP1 by SD2267 was mediated by recruiting CRBN as an E3 ligase. AML12 cells were treated with pomalidomide (a CRBN ligand) as a competitive inhibitor of CRBN or TD-165 (a CRBN-targeting von Hippel-Lindau (VHL) based PROTAC) [24]. KEAP1 degradation by SD2267 was attenuated in the presence of pomalidomide or TD-165 (Fig. 3C), and similar results were obtained from HepG2 cells (Fig. 3D and E). Also, we generated a CRBN knockdown HepG2 cell line using short-hairpin RNA targeting CRBN (shCRBN), and among three clones of shCRBN (#7, #10, and #21), shCRBN #10 was selected based on observed CRBN expression levels (Fig. 3F). While SD2267 effectively degraded KEAP1 in shCon HepG2 Cells, KEAP1 levels were not affected by SD2267 in shCRBN #10 HepG2 cells (Fig. 3G). Meanwhile, CRBN was not degraded by SD2267, although KEAP1 also acts as an E3 ligase recruiting protein (Fig. 3G). These results showed SD2267 induced KEAP1 degradation through a CRBN-mediated proteasomal degradation pathway and not a P62-mediated autophagic degradation pathway.
Fig. 3.
SD2267 degraded KEAP1 through CRBN-mediated proteasomal degradation
(A) AML12 cells were treated with SD2267 (100 or 300 nM) and MG132 (5 μM) for 4 h. (B) AML12 cells were treated with chloroquine (CQ; 20 μM) for 18 h prior to SD2267 (100 or 300 nM) treatment for 6 h. (C) AML12 cells were treated with pomalidome (POM; 1 μM) or TD-165 (1 μM) for 3 h prior to SD2267 (100 nM) treatment for 6 h. (D, E) HepG2 cells were treated with pomalidome (POM; 1 μM) or TD-165 (1 μM) for 14 h prior to SD2267 (100 or 300 nM) for 6 h. (F) Clonal selection of cereblon (CRBN)-knockdown HepG2 cells. (G) HepG2 control cells (shCon) and CRBN knockdown cells (shCRBN #10) were treated with SD2267 (100 or 300 nM) for 24 h. Protein levels were determined by western blotting, and densitometries were performed using three independent experiments. ns; no significance, **p < 0.01, ***p < 0.001.
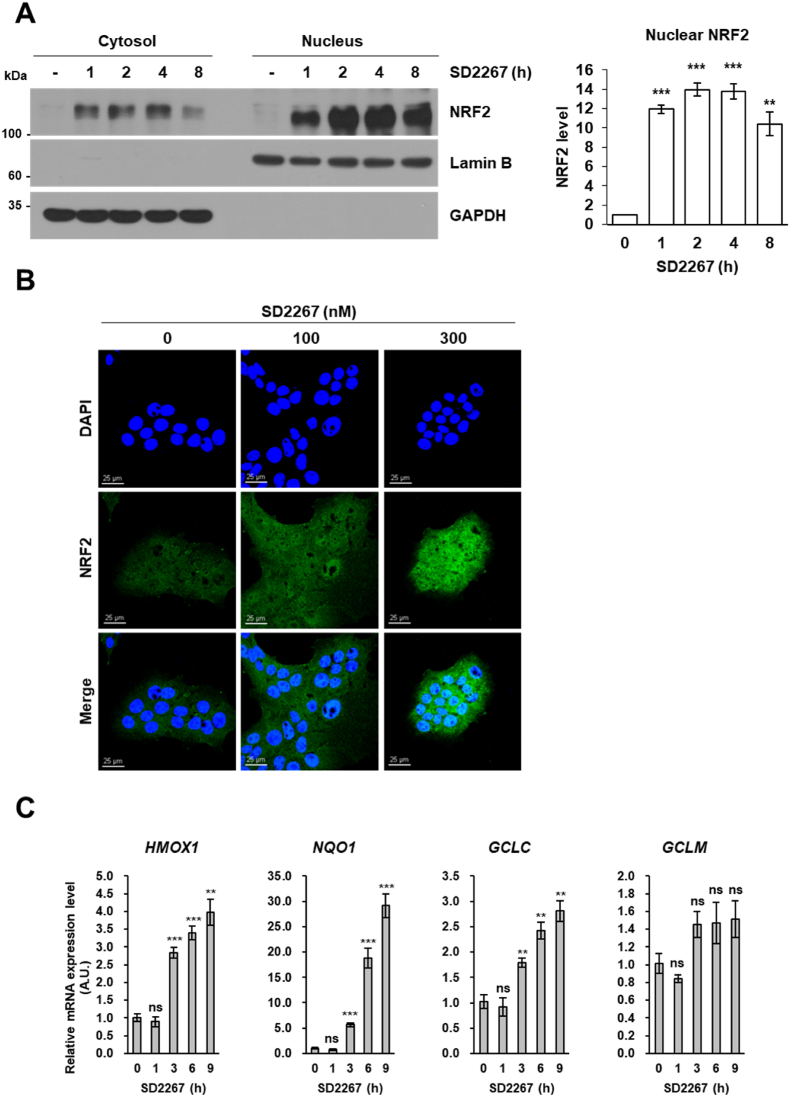
We also evaluated whether KEAP1 degradation by SD2267 activates NRF2 to induce its target genes, especially anti-oxidative genes. Since NRF2 is translocated to the nucleus after being liberated from KEAP1 [25], cells were treated with SD2267, and then cytosolic and nuclear proteins were fractionated. Nuclear NRF2 was significantly upregulated by SD2267 within 3 h in AML12 and HepG2 cells (Fig. 4A and Fig. S1A). Also, confocal microscopy showed that in immunofluorescent stained AML12 and HepG2 cells, SD2267 increased the amount and nuclear localization of NRF2 (Fig. 4B and Fig. S1B). Moreover, the mRNA expression levels of HMOX1, NQO1, GCLC and GCLM (the target genes of NRF2) were significantly increased by SD2267 in both AML12 and HepG2 cells (Fig. 4C and Fig. S1C). These results showed that SD2267 caused the dissociation of NRF2 and KEAP1 and the activations of anti-oxidative target genes by degrading KEAP1.
Fig. 4.
SD2267 promoted the nuclear translocation of NRF2 and activated NRF2 target genes in AML12 cells
(A) AML12 cells were incubated with 300 nM of SD2267 for the indicated times, and subjected to nuclear fractionation. GAPDH was used as a loading control for cytosolic proteins, and Lamin B was used as a loading control for nuclear proteins. Protein expression levels were determined by western blotting, and densitometry was performed using three independent experiments. **p < 0.01, ***p < 0.001. (B) AML12 cells were incubated with 100 or 300 nM of SD2267 for 3 h, and subjected to NRF2 immunofluorescence staining (green). DAPI was used to stain nuclei (blue). In merged NRF2 and DAPI images, nuclear NRF2 stained blue-green (Scale bar = 25 μm). (C) AML12 cells were incubated with 100 nM of SD2267 for the indicated times. Gene expression levels of HMOX1, NQO1, GCLC, and GCLM were measured by qRT-PCR. Relative expression levels are presented as mean ± SEM (n = 3 per group). **p < 0.01, ***p < 0.001, ns; not significant versus the control. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
2.3. SD2267 reduced acetaminophen induced ROS generation in mouse hepatocytes
Acetaminophen (APAP) is widely used as an anti-pyretic and analgesic agent. The recent outbreak of COVID-19 raised the demand of APAP as a recommendation for symptomatic treatment [26]. So thus, since APAP is available over the counter, accidental APAP overuse which leads liver damages should be considered. In addition, during the COVID-19 pandemic, the suspected suicidal attempts by self-poisoning were notably increased, and the most frequent single agent involved in these was APAP [27]. Therefore, medical concerns on the hepatotoxicity caused by APAP overdose are more needed than before COVID-19 pandemic.
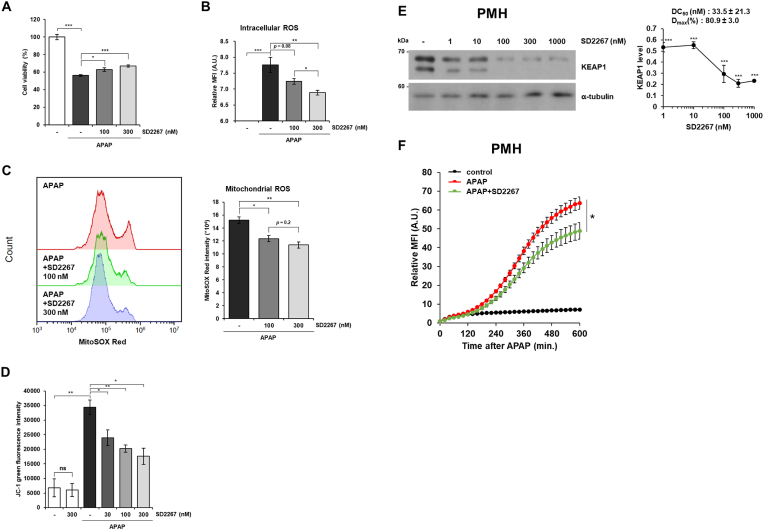
Usually, APAP is metabolized in the liver by phase II metabolisms such as glucuronidation and sulfation [28]. However, when the amount of APAP absorbed exceeds the capacity of these metabolisms, remaining APAP is metabolized by cytochrome P450 enzymes, especially CYP2E1, to N-acetyl-p-benzoquinone imine (NAPQI), which is highly reactive and generates excessive ROS leading hepatocyte death. Since it has been reported that NRF2 activation alleviates APAP-induced ROS generation and cell death [4], we examined whether SD2267 reduces ROS generation and cell death induced by APAP. The cell viability of AML12 cells was decreased by APAP treatment, and it was significantly recovered by SD2267 (Fig. 5A). Also, the relative DCF-DA fluorescence intensities were increased by APAP, and this increase was significantly attenuated by 300 nM of SD2267 (Fig. 5B), suggesting that SD2267 inhibited APAP-induced ROS generation. Since ROS generated by APAP overdose mainly comes from mitochondria and this mitochondrial ROS leads mitochondrial dysfunction such as loss of mitochondrial membrane potential [[29], [30], [31], [32]], we investigated mitochondrial ROS level and mitochondrial membrane potential in AML12 cells after APAP and/or SD2267 treatment. SD2267-treated AML12 cells showed significantly lower MitoSOX Red fluorescence intensities than AML12 cells treated with APAP only, indicating that SD2267 reduced mitochondrial ROS induced by APAP (Fig. 5C). Also, APAP significantly increased JC-1 green fluorescence intensities, and this increment was significantly reduced by SD2267, indicating that SD2267 restored the mitochondrial membrane potential which were decreased by APAP (Fig. 5D). We then investigated the anti-oxidative effect of SD2267 ex vivo using primary mouse hepatocytes. SD2267 dose-dependently degraded KEAP1 (Fig. 5E) and significantly reduced APAP-induced ROS generation (Fig. 5F). These results showed that anti-oxidative genes induced by SD2267 treatment were functionally playing its role which is reducing oxidative stress and subsequent cell death.
Fig. 5.
SD2267 reduced APAP-induced ROS generation in hepatocytes
(A) AML12 cells were treated with 15 mM of APAP in the absence or presence of SD2267 for 24 h, and the cell viability was measured by MTT assay. Data are presented as means ± SEMs (n = 5 per group). (B) AML12 cells were treated with 5 mM of APAP in the absence or presence of SD2267, and then DCF-DA fluorescence intensities were measured at 4 h after APAP treatment. (C) AML12 cells were treated with 5 mM of APAP in the absence or presence of SD2267 for 8 h, and then stained with MitoSOX Red. The intensities of MitoSOX Red were measured by flow cytometry, and presented as means ± SEMs (n = 3 per group). (D) AML12 cells were treated with 5 mM of APAP in the absence or presence of SD2267 for 8 h, and then stained with JC-1. The green fluorescence intensities of JC-1 were measured by flow cytometry, and presented as means ± SEMs (n = 3 per group). (E) Primary mouse hepatocytes (PMHs) were treated with SD2267 at various concentrations for 24 h, and KEAP1 expressions were determined by western blotting. Densitometry was performed using three independent experiments and presented as a graph with DC50 and Dmax value. (F) PMHs were treated with 100 nM of SD2267 for 6 h before APAP treatment at 20 mM, and then DCF-DA fluorescence intensities were measured every 20 min for 10 h. Data are presented as means ± SEMs (n = 4 per group). *p < 0.05, **p < 0.01, ***p < 0.001. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
2.4. In vivo pharmacokinetics profile of SD2267 in mice
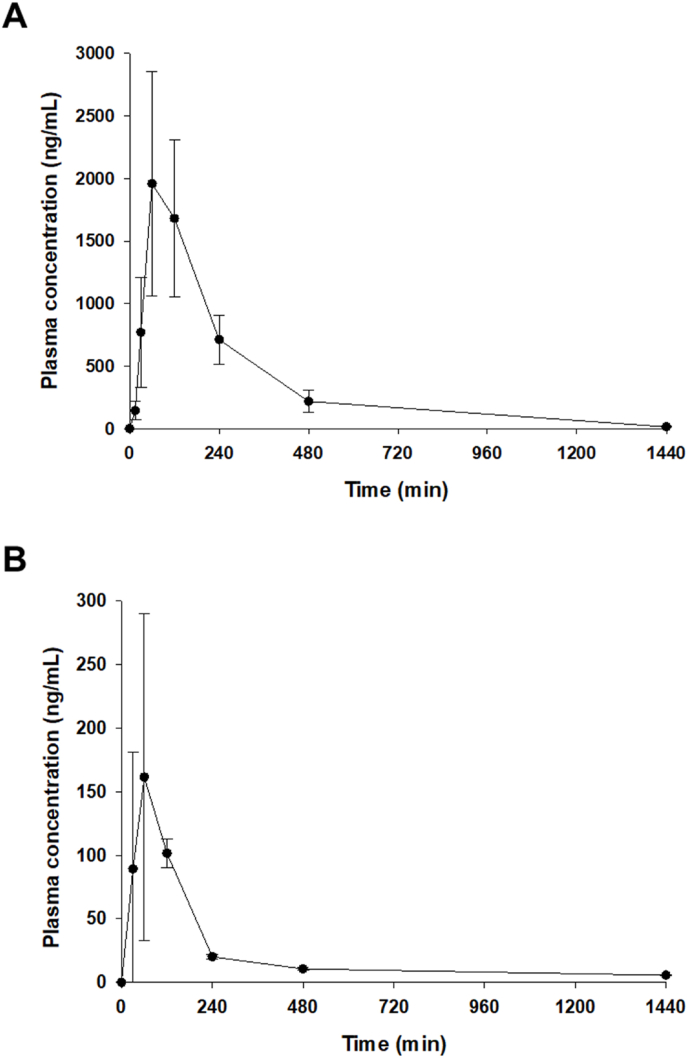
To examine the anti-oxidative effect of SD2267 in vivo, we evaluated the pharmacokinetic profile of SD2267 in ICR mice. Average plasma concentration-time profiles of SD2267 obtained after IP or PO administration at 3 mg/kg are shown in Fig. 6, and calculated pharmacokinetic parameters are presented in Table 2. After IP administration, peak plasma concentration (Cmax 1.97 μg/mL) was achieved in ∼80 min (Tmax). SD2267 then rapidly disappeared with a half-life (T1/2) of 220 min. Furthermore, the area under the plasma concentration-time curve from time 0 to infinity (AUC0∼∞) of SD2267 was 468 μg min/mL. However, after oral administration of SD2267, the Cmax of only 0.172 μg/mL was observed after ∼80 min. Furthermore, the oral AUC0∼∞ of SD2267 was 35.8 μg min/mL, indicating that IP administration resulted in much greater systemic exposure.
Fig. 6.
In vivo pharmacokinetics of SD2267 in mice
Average plasma drug concentration-time profile of SD-2267 obtained after intraperitoneal injection (A) or oral administration (B) to mice at a dose of 3 mg/kg. Data are presented as means ± SDs (n = 3 per group).
Table 2.
Pharmacokinetic parameters of SD2267 following IP or PO administrations of SD-2267 (3 mg/kg) in mice (n = 3). Data are expressed as the mean ± SD, except for Tmax, which is presented as median values with range.
| Parameter | SD2267 |
|
|---|---|---|
| IP | PO | |
| Tmax (min) | 80 [60–120] | 80 [60–120] |
| Cmax (μg/mL) | 1.97 ± 0.87 | 0.172 ± 0.116 |
| AUClast (μg·min/mL) | 463 ± 76 | 29.8 ± 8.7 |
| AUC0∼∞ (μg·min/mL) | 468 ± 76 | 35.8 ± 8.5 |
| T1/2 (min) | 220 ± 17 | 739 ± 102 |
| MRT (min) | 290 ± 76 | 727 ± 181 |
2.5. Therapeutic effect of SD2267 in acetaminophen-induced liver injury mouse model
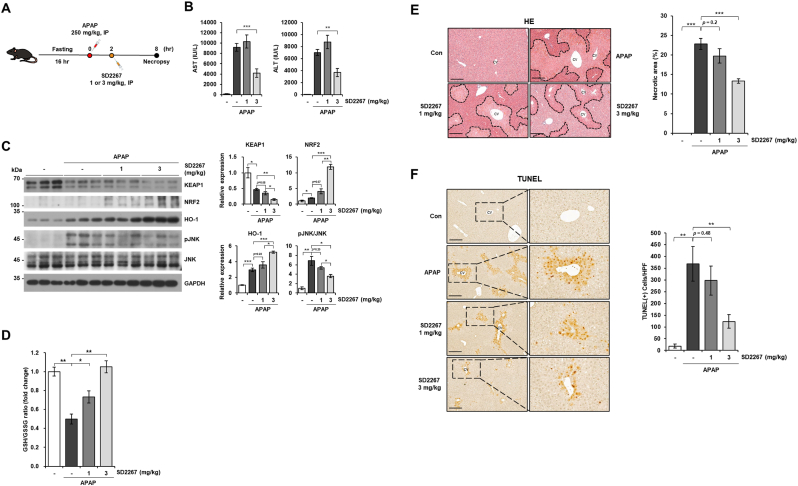
Next, we conducted APAP-induced liver injury mouse model to evaluate in vivo efficacy of SD2267 (Fig. 7A). Serum levels of aspartate transaminase (AST) and alanine transaminase (ALT) (known biomarkers of liver injury) were increased by injecting APAP (250 mg/kg, IP), but these increases were significantly decreased by ∼2.2 fold and ∼1.9 fold in mice given SD2267 at 3 mg/kg, respectively (Fig. 7B). To evaluate KEAP1 degrading effects of SD2267, we measured the protein level of KEAP1 in the liver by western blotting. In line with our in vitro results, intervention with SD2267 in the liver led to a decrease in KEAP1 protein levels compared to APAP, with statistically significant effects observed at the 3 mg/kg (p < 0.05). Although a statistically significant effect was not observed at the 1 mg/kg (p = 0.08), a trend towards a decrease was observed, suggesting that the degrading efficacy of SD2267 on KEAP1 protein level was dose-dependent (Fig. 7C). Although KEAP1 protein level was decreased even in APAP-only treated group, the subsequent increases of the protein level of NRF2 and HO-l were significantly greater at 3 mg/kg of SD2267-treated group than APAP-only treated group (Fig. 7C). Along with the activation of NRF2 pathway, decrease of glutathione (GSH) to glutathione disulfide (GSSG) ratio by APAP was significantly and dose-dependently recovered by SD2267 (Fig. 7D), suggesting that SD2267 induced NRF2 pathway and accelerated GSH replenishment.
Fig. 7.
SD2267 attenuated acetaminophen (APAP)-induced liver injury in mice
(A) Experimental schedule of the APAP-induced liver injury mouse model. After 16 h of fasting, mice were injected with APAP (250 mg/kg, IP) and 2 h later treated with SD2267 (1 or 3 mg/kg, IP) for another 6 h. (B) Serum AST and ALT levels are presented as means ± SEMs. (C) Protein expression levels of liver tissues were determined by western blotting, and densitometry results of each protein levels are presented as means ± SEMs (n = 3 per group). (D) GSH to GSSG ratio was detected from liver tissues, and presented as means ± SEMs (n = 3 per group). (E) Representative images of hematoxylin and eosin (HE) stained liver tissues (left) with measured areas of necrosis (presented as means ± SEMs) (right). Necrotic areas are marked with dashed lines in HE images. (F) Representative images of terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) stained liver tissues (left) and the number of TUNEL positive cells per high power field (HPF; x400) (presented as means ± SEMs) (right). cv; central vein. Scale bar = 100 μm *p < 0.05, **p < 0.01, ***p < 0.001.
APAP overdose results in excessive ROS and leads to mitochondrial dysfunction and activates c-Jun N-terminal kinase (JNK) pathway to amplify mitochondrial damages [[29], [30], [31], [32]]. 3 mg/kg of SD2267 significantly reduced the phosphorylation of JNK induced by APAP, indicating that the mitochondrial damage caused by APAP was ameliorated by SD2267 in vivo (Fig. 7C). Furthermore, apoptosis-inducing factor and endonuclease G are released from damaged mitochondria and induce DNA fragmentation and cell necrosis [32], which can be detected by terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining and hematoxylin and eosin (HE) staining, respectively [33], and are characteristics of APAP-induced liver injury. Therefore, we also performed HE and TUNEL staining of liver tissues from mice to evaluate whether SD2267 could alleviate APAP-induced hepatotoxicity. HE staining showed that hepatic necrosis was severely induced by 250 mg/kg of APAP which were significantly reduced by 2 h post-treatment of SD2267 at 3 mg/kg (Fig. 7E). In addition, the number of TUNEL-positive cells was significantly reduced from 369 cells per high power field (HPF; x400) in APAP-treated mice to 123 cells per HPF in APAP + 3 mg/kg of SD2267 treated mice (Fig. 7F). Taken together, SD2267 was found to effectively degrade KEAP1 and induce NRF2 pathway in vivo attenuating the hepatotoxic effects of APAP.
3. Conclusion
During the time of our research, Chen et al. reported a heterobifunctional degrader of KEAP1, evaluating in vitro activity on various cell lines (HEK293T, BEAS-2B and HCA7) demonstrating that compound 14 effectively degraded KEAP1 thereby preventing cell death induced by ROS [19]. Despite the fact that reported compound 14 (7C linker) is structurally similar to our synthesized compound SD2267 (8C linker), we considered that in order to establish a strong results and conclusions, in vivo experiments with its’ pharmacokinetic profile are much more crucial along with the supporting data of in vitro evaluation. Therefore, in this study, we developed a KEAP1-targeting PROTAC as a therapeutic strategy to alleviate liver damage induced by APAP overdose that acts by activating the NRF2 pathway.
Since the liver is damaged by excessive ROS [34], we evaluated in vitro efficacy of KEAP1-targeting PROTACs in hepatocytes. The evaluation confirmed that SD2267 induced the CRBN-mediated proteasomal degradation of KEAP1. Also, we observed that SD2267 induced the nuclear translocation of NRF2, subsequently upregulated the transcription of NRF2 target genes, and thereby ameliorated APAP-induced ROS generation. Furthermore, SD2267 exhibited therapeutic effects in an APAP-induced mouse model of liver injury.
PROTACs were mostly given through subcutaneous, intraperitoneal or intravenous injections because of their low oral bioavailability [35,36]. In our study, SD2267 showed relatively moderate Cmax and AUC with obvious efficacy in vivo via IP administration route, but SD2267 also faced to the limitation of low oral bioavailability since oral pharmacokinetic parameters were much lower than those of IP, suggesting that future studies to optimize pharmacokinetic properties including solubility, metabolic stability and permeability are still required. Although SD2267 was not fully profiled from a drug candidate perspective, its KEAP1 degrading activity and in vivo hepatoprotective effects demonstrated for the first time that KEAP1-targeting PROTACs have potential use as therapeutic agents for oxidative stress-related liver diseases.”
4. Experimental section materials and methods
4.1. General
Unless otherwise specified, all the reactions were performed in an oven-dried glassware using a magnetic stirrer under nitrogen balloon. Commercially available reagents were purchased from various companies (TCI, Sigma-Aldrich, Alfa Aesar, Junsei) that were used throughout the synthesis. Anhydrous solvents [dichloromethane (DCM), dimethyl formamide (DMF), dimethyl sulfoxide (DMSO) 1,4-dioxane] were purchased from commercial sources. Anhydrous tetrahydrofuran (THF) was prepared using sodium metal and benzophenone and anhydrous triethyl amine (TEA) using KOH pellets. The progress of reaction was checked using TLC plate Silica Gel 60 F254 by visually monitoring under UV light (254 nm and 365 nm). Products were concentrated using a Buchi rotary evaporator and purified by column chromatography using Zeochem silica gel or by Biotage Medium Pressure Liquid Chromatography (MPLC). NMR samples were prepared using CDCl3 or (CD3)2SO purchased from Cambridge Isotope Laboratories. 1H NMR spectra were obtained using a Bruker Avance III 600 MHz instrument. Chemical shifts (δ) are expressed as parts per million (ppm) versus tetramethyl silane (TMS). HRMS (ESI) data were obtained using DART-MS machine.
4.2. Chemistry
4.2.1. 6-(Benzyloxy)-4-bromo-N1-methylbenzene-1,2-diamine (3)
2-Amino-3-nitro phenol 2 (5.0 g, 32.4 mmol) was dissolved in 75 mL EtOH. Potassium carbonate (6.7 g, 48.7 mmol) was also added to the flask. Then, BnBr (4.80 mL, 40.6 mmol) was slowly added to the flask and the mixture was stirred heating at 70 °C for 4 h. The completion of the reaction was confirmed by TLC and solvent was removed by rotary evaporator. Reaction mixture was diluted with 50 mL water and extracted with 50 mL EtOAc ( × 2). EtOAc layer was collected and dried over anhydrous MgSO4 powder. EtOAc was removed under reduced pressure to get crude product. Purification was done by column chromatography EtOAc/n-Hex 1:5 to furnish benzylated yellow solid product 2a (6.40 g, 81%). 1H NMR (600 MHz, Chloroform-d) δ 7.75 (dd, J = 8.9, 1.3 Hz, 1H), 7.43–7.41 (m, 4H), 7.40–7.36 (m, 1H), 6.96 (dd, J = 7.8, 1.3 Hz, 1H), 6.59 (dd, J = 8.9, 7.7 Hz, 1H), 6.45 (s, 2H), 5.12 (s, 2H). HRMS (ESI) calculated for C13H12N2O3 (M + Na)+ 267.0745, found 267.0757.
The obtained product 2a (2.50 g, 10.2 mmol) was dissolved in 40 mL acetic acid and sodium acetate (1.34 g, 16.4 mmol) was also added to the flask. Bromine (0.60 mL, 11.3 mmol) was then added to the flask cautiously via a syringe. The final reaction mixture was stirred at r.t for 30 min and the reaction was monitored using TLC. Acetic acid was removed under reduced pressure and reaction mixture was diluted with 50 mL water followed by extraction with 50 mL DCM ( × 2). DCM layer was collected and dried over anhydrous MgSO4 powder. DCM was removed under reduced pressure to get crude product. Purification was done by column chromatography EtOAc/n-Hex 1:10 ⁓ 1:5 to furnish brominated red oily product 2b (3.17 g, 96%). 1H NMR (600 MHz, DMSO‑d6) δ 7.73 (d, J = 2.1 Hz, 1H), 7.53–7.51 (m, 2H), 7.42 (dd, J = 8.4, 6.8 Hz, 2H), 7.38–7.34 (m, 1H), 7.33 (d, J = 2.1 Hz, 1H), 7.18 (s, 2H), 5.28 (s, 2H). HRMS (ESI) calculated for C14H13BrN2O3 (M + H)+ 323.0055, found 323.0031.
The above desired brominated product 2b (3.16 g, 9.77 mmol) was dissolved in 30 mL DMF at 0 °C and sodium hydride (282 mg, 11.7 mmol) was also added to the solution. Then, the mixture was stirred for 30 min at 0 °C and then methyl iodide (0.67 mL, 10.7 mmol) was transferred to the flask containing starting material. The reaction was left to stir at r.t for 1 h and the progress of reaction was checked by TLC. DMF was removed under reduced pressure and reaction mixture was diluted with 40 mL water followed by extraction with 40 mL DCM ( × 2). DCM layer was collected and dried over anhydrous MgSO4 powder. DCM was removed under reduced pressure to get crude product. Purification was done by column chromatography EtOAc/n-Hex 1:10 to furnish N-methylated orange solid product 2c (2.62 g, 79%). 1H NMR (600 MHz, DMSO‑d6) δ 7.63 (d, J = 2.3 Hz, 1H), 7.50 (dd, J = 7.0, 2.0 Hz, 2H), 7.42 (tt, J = 8.1, 2.0 Hz, 3H), 7.32 (d, J = 2.3 Hz, 1H), 7.29 (q, J = 4.7 Hz, 1H), 5.21 (s, 2H), 2.89 (d, J = 5.3 Hz, 3H). HRMS (ESI) calculated for C14H13BrN2O3 (M + H)+ 337.0213, found 337.0187.
N-methylated product 2c (2.61 g, 7.74 mmol) was dissolved in 40 mL acetic acid. Zinc dust (3 g, 46.4 mmol) was cautiously added to the flask at 0 °C. Then, the reaction mixture was allowed to stir at 45 °C for 2 h. TLC was checked to confirm the completion of reaction. Acetic acid was removed under reduced pressure and reaction mixture was diluted with 35 mL water followed by extraction with 35 mL EtOAc ( × 2). EtOAc layer was collected and dried over anhydrous MgSO4 powder. EtOAc was removed under reduced pressure to get crude product. Purification was done by column chromatography EtOAc/n-Hex 1:10 ⁓ 1:5 ⁓1:1 to furnish reduced brown oily product 3 (1.39 g, 59%). 1H NMR (600 MHz, DMSO‑d6) δ 7.45 (d, J = 7.2 Hz, 2H), 7.39 (dd, J = 8.5, 6.8 Hz, 2H), 7.34–7.31 (m, 1H), 6.47 (d, J = 2.1 Hz, 1H), 6.45 (d, J = 2.2 Hz, 1H), 5.05 (s, 2H), 4.96 (s, 2H), 3.57 (s, 1H), 2.55 (s, 3H). HRMS (ESI) calculated for C14H15BrN2O (M + H)+ 307.0480, found 307.0484.
4.2.2. 7-(Benzyloxy)-5-bromo-1-methyl-1H-benzo[d] [1,2,3] triazole (4)
3 (1.38 g, 4.49 mmol) was dispersed in 25 mL of 10% sulfuric acid solution (aq.) and NaNO2 (436 mg, 6.28 mmol) was also added to the flask slowly. Then, after 30 min of stirring, 50 mL water was added to the flask and was left to stir for overnight. TLC plate demonstrated the accomplishment of reaction. Extraction was done with 50 mL DCM ( × 2). DCM layer was collected and dried over anhydrous MgSO4 powder. DCM was removed under reduced pressure to get crude product. Purification was done by column chromatography EtOAc/n-Hex 1:5 to furnish yellow solid product 4 (1.25 g, 87%). 1H NMR (600 MHz, DMSO‑d6) δ 7.84 (d, J = 1.4 Hz, 1H), 7.57–7.55 (m, 2H), 7.45 (t, J = 7.5 Hz, 2H), 7.40–7.37 (m, 1H), 7.24 (d, J = 1.4 Hz, 1H), 5.36 (s, 2H), 4.39 (s, 3H). HRMS (ESI) calculated for C14H12BrN3O (M + H)+ 318.0255, found 318.0242.
4.2.3. tert-Butyl (E)-3-(7-(benzyloxy)-1-methyl-1H-benzo[d] [1,2,3] triazol-5-yl) acrylate (5)
4 (1.24 g, 3.89 mmol) was dissolved in 25 mL DMF at r.t. tert-Butyl acrylate (2.81 mL, 19.6 mmol), palladium tetrakis triphenyl phosphine (223 mg, 0.190 mmol), tri(o-tolyl) phosphine (237 mg, 0.77 mmol) and DIPEA (1.69 mL, 9.72 mmol) were also added to the flask. Solvent was degassed using nitrogen balloon for 10 min and was stirred at 95 °C for 5 h. The progress of the reaction was checked by TLC and DMF was removed under reduced pressure. Reaction mixture was diluted with 40 mL water and extracted with 40 mL EtOAc ( × 2). EtOAc layer was collected and dried over anhydrous MgSO4 powder. EtOAc was removed under reduced pressure to get crude product. Purification was done by column chromatography EtOAc/n-Hex 1:6 to furnish yellow solid product 5 (1.12 g, 79%). 1H NMR (600 MHz, Chloroform-d) δ 7.72 (s, 1H), 7.67 (d, J = 15.8 Hz, 1H), 7.49–7.43 (m, 5H), 7.02 (d, J = 1.2 Hz, 1H), 6.36 (d, J = 15.9 Hz, 1H), 5.25 (s, 2H), 4.44 (s, 3H), 1.56 (s, 9H). HRMS (ESI) calculated for C21H23N3O3 (M + H)+ 366.1818, found 366.1834.
4.2.4. tert-Butyl 3-(7-(benzyloxy)-1-methyl-1H-benzo[d][1,2,3] triazol-5-yl)-3-(3(hydroxymethyl) -4-methylphenyl) propanoate (6)
5 (1.11 g, 3.29 mmol) was dissolved in a solvent mixture of 45 mL dioxane/water (2:1). Rhodium catalyst (75 mg, 0.16 mmol), and TEA (0.64 mL, 4.93 mmol) were also added to the flask and was stirred for few minutes. Then, 13 (1.14 g, 4.93 mmol) separately dissolved in 15 mL solvent mixture was transferred to the flask. The final reaction mixture was degassed using nitrogen balloon for 10 min and was stirred heating at 95 °C for 5 h. Reaction progress was monitored using TLC. Solvent was removed by rotary evaporator and reaction mixture was diluted with 35 mL water followed by extraction with 35 mL EtOAc ( × 2). EtOAc layer was collected and dried over anhydrous MgSO4 powder. EtOAc was removed under reduced pressure to get crude product. Purification was done by column chromatography EtOAc/n-Hex 1:5 to furnish yellow oily racemate product 6 (788 mg, 53%). 1H NMR (600 MHz, Chloroform-d) δ 7.50 (s, 1H), 7.40 (dt, J = 4.5, 0.7 Hz, 5H), 7.23–7.22 (m, 1H), 7.08 (s, 1H), 7.07 (d, J = 1.9 Hz, 1H), 6.69 (d, J = 1.1 Hz, 1H), 5.12 (s, 2H), 4.64 (s, 2H), 4.57 (t, J = 8.1 Hz, 1H), 4.39 (s, 3H), 3.01 (d, J = 8.5 Hz, 1H), 2.99–2.94 (m, 2H), 2.30 (s, 3H), 1.31 (s, 9H). HRMS (ESI) calculated for C29H33N3O4 (M + H)+ 488.2577, found 488.2549.
4.2.5. tert-Butyl 3-(7-(benzyloxy)-1-methyl-1H-benzo[d] [1,2,3] triazol-5-yl)-3-(4-methyl-3-(((R)-4-methyl-1,1-dioxido-3,4-dihydro-2H-benzo[b] [1,4,5] oxathiazepin-2-yl) methyl) phenyl) propanoate (7)
6 (785 mg, 1.70 mmol) and 16 (525 mg, 2.55 mmol) were dissolved in 35 mL anhydrous THF. Triphenyl phosphine (847 mg, 3.40 mmol) was also dissolved in 20 mL anhydrous THF (separate flask) and DIAD (0.66 mL, 3.40 mmol) was added to the flask, which was then left to stir at r.t till the formation of yellow-white precipitate. 10 mL of precipitate was transferred to the flask containing starting materials via a syringe and was left to stir for 3 h. The progress of reaction was checked by TLC and solvent was removed under reduced pressure. Reaction mixture was diluted with 25 mL water followed by extraction with 25 mL EtOAc ( × 2). EtOAc layer was collected and dried over anhydrous MgSO4 powder. EtOAc was removed under reduced pressure to get crude product. Purification was done by column chromatography EA/n-Hex 1:6 ⁓ 1:5 to furnish yellow solid racemate product 7 (817 mg, 75%). 1H NMR (600 MHz, DMSO‑d6) δ 7.78 (dt, J = 7.8, 1.8 Hz, 1H), 7.65 (ddt, J = 8.1, 7.4, 1.8 Hz, 1H), 7.51 (tt, J = 8.0, 1.4 Hz, 2H), 7.47 (t, J = 1.6 Hz, 1H), 7.41–7.32 (m, 5H), 7.31 (td, J = 2.9, 1.1 Hz, 1H), 7.28–7.25 (m, 1H), 7.12 (dd, J = 7.9, 2.3 Hz, 1H), 7.05 (dd, J = 21.2, 1.2 Hz, 1H), 5.27 (s, 2H), 4.48–4.36 (m, 3H), 4.34 (d, J = 5.2 Hz, 3H), 3.79 (dd, J = 14.0, 4.1 Hz, 1H), 3.62–3.55 (m, 1H), 3.08 (d, J = 8.1 Hz, 1H), 3.04 (dd, J = 15.5, 8.6 Hz, 1H), 2.73 (ddd, J = 56.2, 15.3, 1.4 Hz, 1H), 2.24 (d, J = 3.7 Hz, 3H), 1.22 (d, J = 5.3 Hz, 9H), 1.15 (d, J = 6.4 Hz, 2H), 1.03 (d, J = 6.3 Hz, 1H). HRMS (ESI) calculated for C38H42N4O6S (M + H)+ 683.2880, found 683.2870.
4.2.6. tert-Butyl 3-(7-hydroxy-1-methyl-1H-benzo[d] [1,2,3] triazol-5-yl)-3-(4-methyl-3-(((R)-4-methyl-1,1-dioxido-3,4-dihydro-2H-benzo[b] [1,4,5] oxathiazepin-2-yl) methyl) phenyl) propanoate (8)
7 (810 mg, 1.23 mmol) was dissolved in solvent mixture of 30 mL MeOH/THF (1:5) and catalytic amount of palladium (10% on Carbon) was also added to the flask. Then, vacuum was applied to the flask sealed with septum and hydrogen balloon was inserted via septum. The reaction mixture was allowed to stir for few hours and the progress of reaction was monitored by TLC. Solvent was filtered via a Celite powder washing with 20 mL EtOAc ( × 2) and obtained clear organic layer was removed under reduced pressure to furnish yellow solid racemate product 8 (685 mg, 98%). 1H NMR (600 MHz, DMSO‑d6) δ 7.77 (ddd, J = 7.8, 4.8, 1.7 Hz, 1H), 7.66–7.63 (m, 1H), 7.35 (tdd, J = 7.6, 3.9, 1.2 Hz, 1H), 7.29 (dt, J = 8.1, 1.4 Hz, 1H), 7.27 (s, 1H), 7.23 (d, J = 1.9 Hz, 1H), 7.16 (ddd, J = 8.0, 4.0, 1.9 Hz, 1H), 7.11 (dd, J = 7.9, 2.5 Hz, 1H), 6.55 (d, J = 9.6 Hz, 1H), 4.42–4.35 (m, 2H), 4.33 (d, J = 3.7 Hz, 3H), 3.80 (dd, J = 14.0, 3.0 Hz, 1H), 3.63–3.55 (m, 1H), 3.02–2.87 (m, 3H), 2.81–2.71 (m, 1H), 2.24 (s, 3H), 1.22 (d, J = 1.8 Hz, 9H), 1.17 (d, J = 6.4 Hz, 2H), 1.07 (d, J = 6.4 Hz, 1H). HRMS (ESI) calculated for C31H36N4O6S (M + H)+ 593.2435, found 593.2422.
4.2.7. General procedure for O-alkylation reaction of KEAP1 binder
20a (89 mg, 0.16 mmol), 20b (89 mg, 0.16 mmol) and 20c (80 mg, 0.16 mmol) were transferred in 20 mL DMF solution of 8 (100 mg, 0.16 mmol) and K2CO3 (33 mg, 0.24 mmol) which was stirred heating at 90 °C for 2 h. After the completion of reaction, DMF was removed under reduced pressure and reaction mixture was diluted with 25 mL water followed by extraction with 25 mL EtOAc ( × 2). EtOAc layer was collected and dried over anhydrous MgSO4 powder. Organic layer was removed under reduced pressure and purified by MPLC normal phase (EtOAc/n-Hex) to furnish green/yellow oily racemate product 9a (31 mg, 21%), 9b (57 mg, 39%) and 9c (44 mg, 30%).
4.2.8. tert-butyl 3-(7-((8-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl) amino) octyl) oxy)-1-methyl-1H-benzo[d] [1,2,3] triazol-5-yl)-3-(4-methyl-3-(((R)-4-methyl-1,1-dioxido-3,4-dihydro-2H-benzo[b][1,4,5] oxathiazepin-2-yl) methyl) phenyl) propanoate (9a)
1H NMR (600 MHz, DMSO‑d6) δ 11.10 (s, 1H), 7.79–7.75 (m, 1H), 7.65 (dddd, J = 8.1, 7.4, 3.1, 1.7 Hz, 1H), 7.56 (ddt, J = 7.7, 7.1, 0.6 Hz, 1H), 7.43 (dd, J = 2.0, 1.0 Hz, 1H), 7.37–7.32 (m, 2H), 7.31–7.25 (m, 2H), 7.11 (dd, J = 8.0, 2.8 Hz, 1H), 7.09–7.07 (m, 1H), 7.02–7.00 (m, 1H), 6.89 (dd, J = 19.9, 1.1 Hz, 1H), 6.52 (t, J = 6.0 Hz, 1H), 5.04 (dd, J = 12.9, 5.4 Hz, 1H), 4.47–4.36 (m, 3H), 4.33 (d, J = 3.8 Hz, 3H), 4.16–4.09 (m, 2H), 3.81 (dd, J = 14.1, 2.2 Hz, 1H), 3.61 (dt, J = 15.3, 10.1 Hz, 1H), 3.31–3.25 (m, 3H), 3.09–3.06 (m, 1H), 2.92–2.80 (m, 2H), 2.73 (dd, J = 15.2, 1.3 Hz, 1H), 2.63–2.54 (m, 2H), 2.23 (d, J = 3.7 Hz, 3H), 1.79 (h, J = 7.2 Hz, 2H), 1.57 (s, 3H), 1.50–1.41 (m, 3H), 1.34 (t, J = 4.8 Hz, 6H), 1.21 (d, J = 5.7 Hz, 9H), 1.08 (d, J = 6.3 Hz, 1H). HRMS (ESI) calculated for C52H61N7O10S (M + Na)+ 998.4201, found 998.4245.
4.2.9. tert-butyl 3-(7-((8-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-5-yl) amino) octyl) oxy)-1-methyl-1H-benzo[d] [1,2,3] triazol-5-yl)-3-(4-methyl-3-(((R)-4-methyl-1,1-dioxido-3,4-dihydro-2H-benzo[b] [1,4,5] oxathiazepin-2-yl) methyl) phenyl) propanoate (9b)
1H NMR (600 MHz, DMSO‑d6) δ 11.07 (s, 1H), 7.77 (ddd, J = 7.8, 4.6, 1.7 Hz, 1H), 7.65 (ddq, J = 9.4, 4.8, 1.7 Hz, 1H), 7.56 (dd, J = 8.2, 2.1 Hz, 1H), 7.44–7.43 (m, 1H), 7.37–7.33 (m, 2H), 7.29 (ddt, J = 7.6, 3.1, 1.3 Hz, 1H), 7.26 (dd, J = 7.4, 1.9 Hz, 1H), 7.12–7.09 (m, 2H), 6.94 (d, J = 2.2 Hz, 1H), 6.89 (dd, J = 19.8, 1.2 Hz, 1H), 6.84 (dt, J = 8.5, 2.7 Hz, 1H), 5.03 (dd, J = 12.8, 5.5 Hz, 1H), 4.43 (ddq, J = 22.4, 13.4, 4.1 Hz, 3H), 4.33 (d, J = 3.8 Hz, 3H), 4.12 (dq, J = 6.8, 3.8 Hz, 2H), 3.81 (dd, J = 14.0, 1.9 Hz, 1H), 3.64–3.59 (m, 1H), 3.16–3.12 (m, 3H), 3.08–3.06 (m, 1H), 3.05–3.00 (m, 1H), 2.91–2.80 (m, 2H), 2.73 (dd, J = 15.3, 1.4 Hz, 1H), 2.60–2.52 (m, 2H), 2.23 (d, J = 3.8 Hz, 3H), 1.80 (hept, J = 6.5 Hz, 2H), 1.57 (q, J = 6.9 Hz, 3H), 1.51–1.43 (m, 3H), 1.39–1.34 (m, 6H), 1.21 (d, J = 5.6 Hz, 9H), 1.09 (d, J = 6.3 Hz, 1H).). HRMS (ESI) calculated for C52H61N7O10S (M + Na)+ 998.4201, found 998.4223.
4.2.10. tert-butyl 3-(7-(4-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl) amino) butoxy)-1-methyl-1H-benzo[d] [1,2,3] triazol-5-yl)-3-(4-methyl-3-(((R)-4-methyl-1,1-dioxido-3,4-dihydro-2H-benzo[b] [1,4,5] oxathiazepin-2-yl) methyl) phenyl) propanoate (9c)
1H NMR (600 MHz, DMSO‑d6) δ 11.10 (s, 1H), 7.76 (ddd, J = 7.8, 4.2, 1.7 Hz, 1H), 7.65 (tt, J = 7.7, 1.7 Hz, 1H), 7.56 (ddd, J = 8.5, 7.2, 2.8 Hz, 1H), 7.43 (d, J = 2.7 Hz, 1H), 7.34 (tdd, J = 9.4, 4.3, 3.1 Hz, 2H), 7.30–7.25 (m, 2H), 7.13–7.09 (m, 2H), 7.03 (dd, J = 6.9, 1.3 Hz, 1H), 6.90 (dd, J = 19.1, 1.1 Hz, 1H), 6.65 (q, J = 5.8 Hz, 1H), 5.05 (dd, J = 12.8, 5.3 Hz, 1H), 4.46–4.38 (m, 3H), 4.30 (d, J = 4.2 Hz, 3H), 4.17 (t, J = 6.1 Hz, 1H), 3.81 (dd, J = 14.1, 2.5 Hz, 1H), 3.61 (dd, J = 15.3, 9.8 Hz, 1H), 3.42–3.38 (m, 2H), 3.09–3.02 (m, 2H), 2.90–2.80 (m, 2H), 2.75–2.71 (m, 1H), 2.61–2.54 (m, 2H), 2.23 (d, J = 3.7 Hz, 3H), 2.02 (d, J = 6.9 Hz, 1H), 1.90 (q, J = 7.2 Hz, 3H), 1.79 (q, J = 6.5 Hz, 2H), 1.54 (s, 1H), 1.21 (d, J = 6.1 Hz, 9H), 1.08 (d, J = 6.3 Hz, 1H).). HRMS (ESI) calculated for C48H53N7O10S (M + H)+ 920.3555, found 920.3690.
4.2.11. General procedure for tert-butyl group deprotection
9a (31 mg, 0.031 mmol), 9b (57 mg, 0.058 mmol) and 9c (44 mg, 0.047 mmol) were dissolved in 9 mL of 4 N HCl solution in dioxane at r.t and was stirred for 1 h. The progress of reaction was monitored by TLC and solvent was removed under reduced pressure washing with 10 mL MeOH ( × 2) which afforded yellow/green sticky solid racemate products 10a (16 mg, 57%), 10b (28 mg, 53%) and 10c (36 mg, 88%) respectively.
4.2.12. 3-(7-((8-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl) amino) octyl) oxy)-1-methyl-1H-benzo[d] [1,2,3] triazol-5-yl)-3-(4-methyl-3-(((R)-4-methyl-1,1-dioxido-3,4-dihydro-2H-benzo[b] [1,4,5] oxathiazepin-2-yl) methyl) phenyl) propanoic acid (10a, SD2267)
1H NMR (600 MHz, DMSO‑d6) δ 12.12 (s, 1H), 11.10 (s, 1H), 7.77 (ddd, J = 7.8, 3.4, 1.7 Hz, 1H), 7.65 (tt, J = 7.7, 1.8 Hz, 1H), 7.56 (dd, J = 8.6, 7.1 Hz, 1H), 7.42 (d, J = 7.0 Hz, 1H), 7.37–7.33 (m, 2H), 7.29 (ddd, J = 8.1, 3.0, 1.2 Hz, 1H), 7.26 (ddd, J = 10.1, 7.7, 1.9 Hz, 1H), 7.10 (d, J = 7.8 Hz, 1H), 7.08 (d, J = 8.6 Hz, 1H), 7.01 (d, J = 7.1 Hz, 1H), 6.88 (dd, J = 16.5, 1.1 Hz, 1H), 6.52 (t, J = 6.0 Hz, 1H), 5.04 (dd, J = 12.8, 5.5 Hz, 1H), 4.49 (td, J = 7.9, 4.5 Hz, 1H), 4.41 (d, J = 14.3 Hz, 2H), 4.33 (d, J = 4.0 Hz, 3H), 4.10 (ddt, J = 9.6, 5.9, 3.2 Hz, 2H), 3.80 (d, J = 14.0 Hz, 1H), 3.61 (ddd, J = 15.2, 10.2, 4.9 Hz, 1H), 3.28 (d, J = 6.7 Hz, 2H), 3.11–3.06 (m, 2H), 2.90–2.84 (m, 1H), 2.60–2.52 (m, 2H), 2.23 (d, J = 5.8 Hz, 3H), 2.04–2.00 (m, 1H), 1.79 (q, J = 7.1 Hz, 2H), 1.57 (t, J = 7.0 Hz, 2H), 1.46 (q, J = 7.0 Hz, 3H), 1.34 (t, J = 4.5 Hz, 6H), 1.23 (s, 1H), 1.18 (d, J = 6.6 Hz, 1H), 1.08 (d, J = 6.3 Hz, 1H). HRMS (ESI) calculated for C48H53N7O10S (M + Na)+ 942.3575, found 942.3534.
4.2.13. 3-(7-((8-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-5-yl) amino) octyl) oxy)-1-methyl-1H-benzo[d] [1,2,3] triazol-5-yl)-3-(4-methyl-3-(((R)-4-methyl-1,1-dioxido-3,4-dihydro-2H-benzo[b] [1,4,5] oxathiazepin-2-yl) methyl) phenyl) propanoic acid (10b, SD2268)
1H NMR (600 MHz, DMSO‑d6) δ 11.06 (s, 1H), 7.77 (ddd, J = 7.8, 4.2, 1.7 Hz, 1H), 7.65 (tt, J = 7.6, 2.0 Hz, 1H), 7.55 (d, J = 8.4 Hz, 1H), 7.44 (d, J = 2.9 Hz, 1H), 7.37–7.33 (m, 2H), 7.30 (ddd, J = 8.1, 2.4, 1.2 Hz, 1H), 7.26 (td, J = 7.7, 1.9 Hz, 1H), 7.11–7.08 (m, 2H), 6.94 (d, J = 2.1 Hz, 1H), 6.90 (dd, J = 14.4, 1.1 Hz, 1H), 6.83 (dd, J = 8.4, 2.1 Hz, 1H), 5.03 (dd, J = 12.8, 5.5 Hz, 1H), 4.52 (td, J = 8.0, 2.3 Hz, 1H), 4.43–4.39 (m, 2H), 4.33 (d, J = 3.8 Hz, 3H), 4.10 (qd, J = 5.8, 2.8 Hz, 2H), 3.80 (d, J = 14.1 Hz, 1H), 3.64–3.60 (m, 1H), 3.49 (d, J = 2.6 Hz, 3H), 3.22–3.19 (m, 1H), 3.14 (d, J = 6.3 Hz, 2H), 2.90–2.85 (m, 1H), 2.60–2.52 (m, 2H), 2.23 (d, J = 4.4 Hz, 3H), 1.79 (q, J = 6.9 Hz, 2H), 1.57 (t, J = 7.1 Hz, 2H), 1.47 (d, J = 7.5 Hz, 3H), 1.36 (s, 6H), 1.23 (s, 1H), 1.19 (d, J = 6.3 Hz, 1H), 1.09 (d, J = 6.3 Hz, 1H). HRMS (ESI) calculated for C48H53N7O10S (M + Na)+ 942.3575, found 942.3555.
4.2.14. 3-(7-(4-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl) amino) butoxy)-1-methyl-1H-benzo[d] [1,2,3] triazol-5-yl)-3-(4-methyl-3-(((R)-4-methyl-1,1-dioxido-3,4-dihydro-2H-benzo[b] [1,4,5] oxathiazepin-2-yl) methyl) phenyl) propanoic acid (10c, SD2269)
1H NMR (600 MHz, DMSO‑d6) δ 12.17 (s, 1H), 11.10 (s, 1H), 7.76 (ddd, J = 7.7, 3.1, 1.7 Hz, 1H), 7.64 (dddd, J = 8.2, 7.4, 1.7, 0.9 Hz, 1H), 7.56 (ddd, J = 9.1, 7.0, 2.5 Hz, 1H), 7.42 (d, J = 8.3 Hz, 1H), 7.36–7.33 (m, 2H), 7.29 (ddd, J = 8.1, 2.9, 1.1 Hz, 1H), 7.25 (ddd, J = 9.7, 7.7, 1.9 Hz, 1H), 7.12 (dd, J = 8.7, 5.1 Hz, 1H), 7.10 (d, J = 7.9 Hz, 1H), 7.02 (d, J = 7.0 Hz, 1H), 6.90 (dd, J = 15.8, 1.1 Hz, 1H), 6.65 (q, J = 5.7 Hz, 1H), 5.05 (dd, J = 12.9, 5.4 Hz, 1H), 4.48 (dd, J = 8.1, 5.1 Hz, 1H), 4.41 (d, J = 14.3 Hz, 2H), 4.29 (d, J = 4.2 Hz, 3H), 4.16 (t, J = 6.1 Hz, 1H), 3.80 (d, J = 14.0 Hz, 1H), 3.60 (ddd, J = 14.7, 10.2, 3.9 Hz, 1H), 3.40 (d, J = 6.5 Hz, 2H), 3.07 (t, J = 7.9 Hz, 2H), 2.91–2.80 (m, 2H), 2.73 (d, J = 15.0 Hz, 1H), 2.61–2.52 (m, 2H), 2.23 (d, J = 5.8 Hz, 3H), 2.02 (ddd, J = 12.3, 6.9, 4.5 Hz, 1H), 1.92–1.86 (m, 2H), 1.79 (q, J = 7.2 Hz, 2H), 1.23 (s, 1H), 1.18 (d, J = 6.4 Hz, 1H), 1.08 (d, J = 6.3 Hz, 1H). HRMS (ESI) calculated for C44H45N7O10S (M + Na)+ 886.2949, found 886.2979.
4.3. Competitive binding assay
4.3.1. Protein expression and purification
Mutant Magnetospirillum gryphiswaldense cereblon isoform 4 (MSCI4) for two surface tryptophan residues was prepared by molecular cloning as previously described [37]. The DNA fragment harboring residues 1 to 124 of MSCI4 and the PreScission cleavage site in front of the MSCI4 gene was synthesized by PCR and incorporated into pET-28a vector. The resultant construct contained the N-terminal hexa-histidine tag followed by the cleavage site of PreScission protease. The mutant MSCI4 (MsCI4WW/FF hereafter) was overexpressed in E. coli C41 (DE3) cells at 25 °C [38]. Protein induction was conducted by adding IPTG 0.2 mM at an OD of 0.6 overnight. Cells were harvested by centrifugation and resuspended in 20 mM Tris (pH 7.5), 100 mM NaCl, 5 mM 2-mercaptoethanol, 4 mM MgCl2, DNase I and Protease Inhibitor Cocktail (Roche). After cells were lysed by sonication, the supernatant was applied to a NiNTA agarose column. Histidine-tagged proteins were eluted with an imidazole gradient, and pooled eluents were dialyzed against 20 mM Tris (pH 7.5), 150 mM NaCl, and 5 mM 2-mercaptoethanol in the presence of PreScission protease overnight at 4 °C. Cleaved MsCI4WW/FF was concentrated using an Amicon concentrator and loaded on the S-100 size-exclusion column (Cytiva) equilibrated with 20 mM Tris (pH 7.5), 150 mM NaCl, and 5 mM 2-mercaptoethanol. Eluents were collected, concentrated, and stored at −80 °C until required.
4.3.2. Fluorescence resonance energy transfer (FRET)-based competitive binding assay
1-(N-2-MANT-imidoethyl)-uracil (MANT-uracil), which was used as a fluorescence reporter ligand of MsCI4WW/FF, was synthesized in our laboratory [37]. Fluorescence was measured using a BioTek Synergy H1 plate Reader (BioTek, VT, USA) and a black 96-well plate using excitation and emission wavelengths of 295 and 350 nm, respectively. Near the tryptophan residues, fluorescence of the bound MANT-uracil was excited at 350 nm, and FRET emissions were measured at 440 nm for all competitive binding assays. Measurements were performed in triplicate. MANT-uracil, thalidomide (positive control), and ligands stocks were prepared in DMSO and serially diluted with binding assay buffer (20 mM Tris (pH7.5), 150 mM NaCl, and 5 mM 2-mercaptoethanol) to maintain a final DMSO concentration of ≤0.02% to prevent DMSO influencing the fluorescence of MANT-uracil. The autofluorescence of MANT-uracil at appropriate concentrations was treated as a blank. Titrations of ligands for the complex of MsCI4WW/FF and MANT-uracil produced a concentration-dependent reduction in FRET signals. The MANT-uracil binding constant to MsCI4WW/FF was obtained by titrating MANT-uracil to MsCI4WW/FF, and binding resulted in higher FRET signals at 440 nm in a MANT-uracil concentration-dependent manner. The obtained Kd value was 3.26 μM, which was highly similar to the published Kd value of 3.3 μM [37]. Ki values of each ligand were calculated based on the Kd of MANT-uracil. Data processing was performed using Prizm (GraphPad). The log (competitor) response curves yielded IC50 values, which were used to obtain Ki values by non-linear fitting using a one site competition model [39].
4.4. Cell lines
HepG2 cell (human hepatocyte cell line) were purchased from the Korea Cell Line Bank (Seoul, Korea) and cultured in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% (v/v) fetal bovine serum (FBS; Welgene, Daegu, Korea), penicillin 100 U/mL, and streptomycin 100 μg/mL (1x P/S; Welgene). AML12 cells (mouse hepatocyte cell line) were purchased from the ATCC (VA, USA) and cultured in DMEM/F-12 supplemented with 10% (v/v) FBS, 1x P/S, 1x Insulin-Transferrin-Selenium-Pyruvate supplement (Welgene), and dexamethasone 40 ng/mL. Cells were sub-cultured every 3–4 days. For in vitro experiments, cells were seeded on cell-culture grade plates for 24 h, and culture media were changed to drug-containing media.
4.5. Generation of cereblon (CRBN) knockdown HepG2 cells
To establish cells lacking CRBN expression, 6 shRNA constructs in pGIPZ lentiviral vector were obtained from Horizon Discovery (Saint Louis, MO, USA). Each construct was transfected into HEK293T cells using accessory vectors to generate lentivirus. After 2 days of transfection, culture supernatants were collected and filtered to remove cell debris. Supernatants were mixed with 5 μg/mL polybrene and added to HepG2 cells. Cells expressing the vector harboring the GFP gene were isolated by FACS sorting. CRBN expressions were determined by western blotting with anti-CRBN antibodies.
4.6. Primary mouse hepatocyte isolation and culture
Primary mouse hepatocytes were isolated from 8-to 10-week-old male C57BL6/N mice by collagenase perfusion, as previously described [40] with slight modification. Briefly, mice were anesthetized with an Alfaxalone-xylazine mixture (Alfaxalone 80 mg/kg + xylazine 10 mg/kg, IP), and livers were perfused with EGTA solution followed by collagenase solution via the inferior vena cava. Isolated hepatocytes were then seeded on Corning® Primaria™ 6-well plates (Corning, NY, USA) in medium 199 (Sigma-Aldrich, St Louis, MO, USA) supplemented with 10% (v/v) FBS, P/S, 10 nM dexamethasone and 23 mM HEPES. After 4–6 h, culture media were replaced with fresh media, and cells were treated with SD2267 at concentrations from 1 to 1000 nM.
4.7. In vitro ROS measurement
Primary mouse hepatocytes were seeded on a 96-well black plate and, the following day, incubated with 100 nM of SD2267 for 6 h. Cells were then loaded with 50 μM of dichlorodihydrofluorescein diacetate (DCF-DA) (Cayman Chemical, MI, USA) in serum-free media for 30 min at 37 °C, washed with serum-free media, and treated with 20 mM of acetaminophen (APAP; Sigma-Aldrich, MO, USA). Fluorescence intensities (ex488/em525) were measured using a BioTek Synergy H1 plate Reader (BioTek) every 20 min for 10 h. AML12 cells were seeded on a 96-well black plate and, the following day, incubated with 5 mM of APAP in the absence or presence of SD2267. After 4 h, cells were stained with 50 μM of DCF-DA in serum-free media for 30 min at 37 °C, washed with serum-free media, and fluorescence intensities (ex488/em525) were measured using a BioTek Synergy H1 plate Reader. Data were analyzed using BioTek Gen5 software (BioTek).
4.8. Mitochondrial ROS and mitochondrial membrane potential measurement
AML12 cells were treated with 5 mM of APAP in the absence or presence of SD2267. After 8 h, cells were stained with 5 μM of MitoSOX Red (Invitrogen, CA, USA) or 0.25 μg/mL of JC-1 dye (MedChemExpress, NJ, USA) for measuring mitochondrial ROS or mitochondrial membrane potential, respectively. Stained cells were subjected to flow cytometry using CytoFLEX (Beckman Coulter, CA, USA). Red fluorescence intensity of MitoSOX Red stained cells was referred to mitochondrial ROS. Green fluorescence of JC-1 indicate JC-1 monomer which is increased when the mitochondrial membrane potential is decreased. Data were analyzed using software Kaluza (Beckman Coulter).
4.9. Nuclear fractionation
HepG2 and AML12 cells were seeded on 100 mm cell culture dishes and, the following day, incubated with 100 nM of SD2267, harvested using 1 mL of PBS per dish, and transferred to 1.5 mL microcentrifuge tubes, and centrifuged (4000 G, 4 °C, 5 min). Supernatants were discarded, and cell pellets were resuspended in 500 μL of cytosolic protein extraction buffer per tube, sonicated, vortexed, and centrifuged (15,000 G, 4 °C, 20 min). Supernatants (cytosolic fractions) were transferred to 1.5 mL microcentrifuge tubes and stored at −20 °C until required. Cell pellets were resuspended in 100 μL of nuclear protein extraction buffer per tube and then frozen. The next day, after vortexing and centrifugating (15,000 G, 4 °C, 20 min), supernatants (nuclear fractions) were transferred to 1.5 mL microcentrifuge tubes and stored at −20 °C required. Proteins in cytosolic and nuclear fractions were quantified by BCA assay and subjected to western blotting.
4.10. Western blotting
Cells were lysed with RIPA assay buffer, transferred to 1.5 mL microcentrifuge tubes on ice for 15 min, vortexed every 5 min, and centrifuged at 13000 rpm for 20 min at 4 °C. Supernatants were transferred to 1.5 mL microcentrifuge tubes, and BCA assayed. Protein solutions containing equal amounts of protein were mixed with 5x sample loading buffer at a volume ratio of 4:1, heated for 5 min at 95 °C, cooled, and then separated by SDS-PAGE electrophoresis. Proteins were then transferred to PVDF membranes, which were blocked with skim milk (5% in TBS-T), washed, and incubated with primary antibodies overnight at 4 °C. The following day, membranes were conjugated with respective secondary antibodies, and immunoreactive bands were detected by enhanced chemiluminescence. Immunoreactive band intensities were measured using Image J software, and target protein band intensities were normalized versus GAPDH. Primary antibodies used in this study are listed in Supplementary Table 1.
4.11. Real-time quantitative reverse transcription-polymerase chain reaction (qRT-PCR) analysis
Cells were lysed with TRIzol reagent (Invitrogen), and total RNA was extracted. Complementary DNA was synthesized from isolated RNA using a PrimeScript 1st strand cDNA Synthesis Kit (Takara, Kyoto, Japan) and used for qRT-PCR using a CFX Connect Real-Time System (Biorad Laboratories, CA, USA). The expression levels of target genes were normalized versus GAPDH, and relative expression levels were determined using the ΔΔCt method [37]. The primer sequences used in this study are listed in Supplementary Table 2.
4.12. Immunofluorescence staining
For immunofluorescence staining, HepG2 and AML12 cells were seeded on coverslips in 12-well plates, treated with SD2267 for 3 h, fixed with 4% (w/v in PBS) paraformaldehyde for 10 min at room temperature, and permeabilized with 0.1% (v/v in PBS) Triton X-100 for 10 min at room temperature. After rinsing with PBS-T (0.1% (v/v) Tween 20 in PBS), cells were blocked with 1% (w/v in PBS-T) BSA for 1 h, incubated with anti-NRF2 antibody (Proteintech, 16396-1-AP, IL, USA) (1:200 in 1% BSA) overnight at 4 °C, washed with PBS-T, and incubated with donkey anti-rabbit IgG FITC (Santa Cruz Biotechnology, SC-2090, 1:200 in 1% BSA) for 1 h at room temperature. After secondary antibody incubation, cells were washed with PBS-T and transferred to slide glass. Nuclear staining was performed using DAPI mounting medium (ImmunoBioScience Corp., AR-6501-01, WA, USA), and immunofluorescence images were obtained using a confocal laser scanning microscope (A1 plus, Nikon, Japan) and analyzed using Nikon NIS-E image analysis software.
4.13. In vivo experiments
4.13.1. Animals
The in vivo pharmacokinetics of SD2267 were evaluated in male ICR mice (7–8 weeks old, 30–35 g), which were purchased from Orient Bio (Seongnam, Republic of Korea), and the acetaminophen (APAP)-induced liver injury model was conducted using 7 weeks old C57BL/6 N male mice purchased from Koatech (Pyeongtaek, Republic of Korea). Mice were acclimatized to standardized laboratory conditions for at least one week before the experiments. During acclimatization, mice were allowed free access to food and water and maintained under a daily light/dark cycle. The animal study protocols were approved by the Institutional Animal Care and Use Committee of Gachon University (Approval # GU1-2022-IA0040 for the pharmacokinetics study and Approval # GU1-2022-IA0011 for the APAP-induced liver injury model) and conducted in accordance with ARRIVE guidelines [41].
4.13.2. Pharmacokinetic study
In vivo pharmacokinetic studies in mice were conducted, as previously described [42]. SD2267 (0.75 mg/mL) was dissolved in a mixture of Cremophor EL, polyethylene glycol 400 and double distilled water (10/40/50%, v/v), and the resultant solution was administered PO or IP to overnight-fasted mice at 3 mg/kg. Blood samples were collected at 15, 30, 60, 120, 240, 480, and 1440 min after IP administration from the retro-orbital sinus using heparinized capillaries, and at 30, 60, 120, 240, 480, and 1440 min after oral administration. Blood samples were immediately centrifuged at 14,000 rpm for 15 min at 4 °C, and supernatants (plasma) were stored at −20 °C until required.
4.13.3. LC-MS/MS analysis and pharmacokinetic parameters
Concentrations of SD2267 in plasma samples were determined by LC-MS/MS. Briefly, plasma samples (10 μL) were mixed with 10 μL methanol and 50 μL of internal standard solution (100 ng/mL of phenacetin in methanol). The resultant mixtures were vortexed for 1 min, centrifuged at 14,000 rpm for 15 min at 4 °C, and supernatants were transferred to analytical vials for LC-MS/MS analysis. A series of calibration standards were also prepared by mixing 10 μL of blank mice plasma, 10 μL of stock solutions of SD2267 (10, 50, 100, 500, 1000, 2000, and 5000 ng/mL in methanol), and 50 μL of internal standard solution.
An AB SCIEX Triple Quad™ 3500 (TQ3500) mass spectrometer (AB Sciex LLC, Framingham, MA, USA) connected to Agilent 1290 HPLC system (Agilent Technologies, Santa Clara, CA, USA) was used to quantify SD2267 concentrations in mouse plasma. The HPLC system was equipped with a Synergi™ polar reverse-phase column (pore size 80 Å, particle size 4 μm, dimensions 150 × 2 mm, Phenomenex, Torrance, CA, USA). The mobile phase used for isocratic elution consisted of acetonitrile and 0.1% aqueous formic acid (70:30, v/v) at a flow rate of 0.2 mL/min. The chromatographic column was maintained at 25 °C, and the sample injection volume used was 2 μL. The TQ3500 mass spectrometer was operated in positive electrospray ionization mode by multiple reaction monitoring (MRM). The optimized MRM conditions and MS parameters used were; m/z of precursor ion: 920.204, m/z of product ion: 679.4, declustering potential: 231 V, entrance potential: 10 V, collision energy: 55 V, collision cell exit potential: 12 V, curtain gas pressure: 25 psi, collision gas pressure: 9 psi, ion spray voltage: 5500 V, ion source temperature: 500 °C, nebulizing gas (GS1) pressure: 50 psi, and drying gas (GS2) pressure: 50 psi. The optimal MS parameters used for the internal standard (phenacetin) were; m/z of precursor ion: 180.035, m/z of product ion: 110.0, declustering potential: 76 V, entrance potential: 10 V, collision energy: 27 V, collision cell exit potential: 8 V, curtain gas pressure: 25 psi, collision gas pressure: 9 psi, ion spray voltage: 5500 V, ion source temperature: 500 °C, nebulizing gas (GS1) pressure: 50 psi, and drying gas (GS2) pressure: 50 psi. Analyst version 1.7.2 was used for instrument control, data acquisition, and analysis. The calibration curve of SD2267 in plasma in the concentration range 10–5000 ng/mL showed good linearity, as demonstrated by a high correlation coefficient (r = 0.9994 with a weighing factor of 1/x). The accuracy at all concentration standards was between 90 and 110%. Plasma drug concentrations were used to calculate pharmacokinetics parameters for PO and IP administration by non-compartmental analysis (WinNonlin® software version 8.3, Pharsight Corporation, Mountain View, CA, USA).
4.13.4. Acetaminophen (APAP)-induced liver injury model
The APAP-induced liver injury model was conducted as previously reported [43]. APAP was dissolved in PBS at 55–60 °C to a concentration of 12.5 mg/mL. SD2267 (0.25 mg/mL or 0.75 mg/mL) was dissolved in a mixture of Cremophor EL, polyethylene glycol 400, and double distilled water (10/40/50%, v/v). After fasting for 16 h, mice were administered PBS or APAP (250 mg/kg, IP), and food was allowed. PBS-injected mice were classified as (1) controls (n = 5), and APAP-injected mice were randomly divided into three groups: (2) APAP-only treatment group (n = 7), (3) APAP + 1 mg/kg of SD2267 group (n = 6), and (4) APAP + 3 mg/kg of SD2267 group (n = 7). Two hrs after PBS or APAP injection, the control group and the APAP-only treatment group were administered the SD2267 vehicle IP, whereas the two APAP + SD2267 groups were administered 1 or 3 mg/kg of SD2267 IP. Mice were sacrificed 6 h after vehicle or SD2267 injections, and biological samples (blood and liver tissues) were collected for evaluation. Serum was isolated from blood samples, and liver tissues were formalin-fixed.
4.13.5. Serum analysis and histopathological evaluation
Serum aspartate aminotransferase (AST) and alanine aminotransferase (ALT) levels were measured using HITACHI 7180 autoanalyzer (Hitachi, Ltd., Tokyo, Japan).
For histopathological evaluations, formalin-fixed liver tissues were processed, paraffin-embedded, and sectioned at 4 μm. Sections were stained with hematoxylin and eosin (HE) staining or terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining using a TUNEL assay kit (Elabscience, #E-CK-A331, Wuhan, China). Necrotic areas and the number of TUNEL-positive cells per high power field (HPF) were quantified using QuPath digital pathology software (version 0.4.1).
4.13.6. GSH and GSSG measurement
GSH and GSSG level in liver tissues were measured by recycling assay following the established protocol [44]. Briefly, liver tissues were homogenized in ice-cold 5% metaphosphoric acid and 0.6% sulfosalicylic acid mixture, and the homogenates were centrifuged at 3000 G at 4 °C for 10 min. Supernatants were used for the analysis. For total GSH (GSH+2GSSG) measurement, 20 μL of each sample were mixed with Ellman's reagent (Daejung Chemicals & Metals, Siheung, Korea), glutathione reductase (Sigma-Aldrich), and β-NADPH (TCI, Tokyo, Japan), and then the absorbance at 412 nm was read using microplate reader. For GSSG measurement, 100 μL of each sample were mixed with 2-vinylpyridine (Sigma-Aldrich) for 1 h, and neutralized with triethanolamine (Daejung Chemicals & Metals) for another 10 min. After then, the mixture (sample + 2-vinylpyridine + triethanolamine) was used for further analysis as same as GSH measurement.
4.14. Statistical analysis
The analysis was conducted using one-way ANOVA followed by the Student's t-test. Results are presented as means ± standard errors of means (SEM), except plasma drug concentration-time profile results in Fig. 6, which are presented as means ± standard deviations (SDs). Statistical significance was accepted for p values < 0.05.
Declaration of competing interest
The authors declare no competing interests.
Acknowledgements
This research was supported by grants from the National Research Foundation of Korea (NRF) funded by the Korean government Ministry of Education (2020R1A6A1A03043708), the Bio & Medical Technology Development Program (2021M3H9A1097551 and 2022M3A9B6082668) through the NRF funded by the Korean government Ministry of Science, ICT and Future Planning (MSIT).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2023.102783.
Contributor Information
Dongyun Shin, Email: dyshin@gachon.ac.kr.
Seung Hyun Oh, Email: eyeball@hanmail.net, eyeball@gachon.ac.kr.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Snezhkina A.V., Kudryavtseva A.V., Kardymon O.L., Savvateeva M.V., Melnikova N.V., Krasnov G.S., et al. ROS generation and antioxidant defense systems in normal and malignant cells. Oxid. Med. Cell. Longev. 2019;2019 doi: 10.1155/2019/6175804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pisoschi A.M., Pop A., Iordache F., Stanca L., Predoi G., Serban A.I. Oxidative stress mitigation by antioxidants - an overview on their chemistry and influences on health status. Eur. J. Med. Chem. 2021;209 doi: 10.1016/j.ejmech.2020.112891. [DOI] [PubMed] [Google Scholar]
- 3.Schieber M., Chandel N.S. ROS function in redox signaling and oxidative stress. Curr. Biol. 2014;24(10):R453–R462. doi: 10.1016/j.cub.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu D., Xu M., Jeong S., Qian Y., Wu H., Xia Q., et al. The role of Nrf2 in liver disease: novel molecular mechanisms and therapeutic approaches. Front. Pharmacol. 2018;9:1428. doi: 10.3389/fphar.2018.01428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu S., Lu H., Bai Y. Nrf2 in cancers: a double-edged sword. Cancer Med. 2019;8(5):2252–2267. doi: 10.1002/cam4.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calkins M.J., Johnson D.A., Townsend J.A., Vargas M.R., Dowell J.A., Williamson T.P., et al. The Nrf2/ARE pathway as a potential therapeutic target in neurodegenerative disease. Antioxidants Redox Signal. 2009;11(3):497–508. doi: 10.1089/ars.2008.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li J., Ichikawa T., Janicki J.S., Cui T. Targeting the Nrf2 pathway against cardiovascular disease. Expert Opin. Ther. Targets. 2009;13(7):785–794. doi: 10.1517/14728220903025762. [DOI] [PubMed] [Google Scholar]
- 8.Cho H.Y., Kleeberger S.R. Nrf2 protects against airway disorders. Toxicol. Appl. Pharmacol. 2010;244(1):43–56. doi: 10.1016/j.taap.2009.07.024. [DOI] [PubMed] [Google Scholar]
- 9.Joelsson J.P., Asbjarnarson A., Sigurdsson S., Kricker J., Valdimarsdottir B., Thorarinsdottir H., et al. Ventilator-induced lung injury results in oxidative stress response and mitochondrial swelling in a mouse model. Lab Anim Res. 2022;38(1):23. doi: 10.1186/s42826-022-00133-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saha S., Buttari B., Panieri E., Profumo E., Saso L. An overview of Nrf2 signaling pathway and its role in inflammation. Molecules. 2020;25(22) doi: 10.3390/molecules25225474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamamoto M., Kensler T.W., Motohashi H. The KEAP1-NRF2 system: a thiol-based sensor-effector apparatus for maintaining redox homeostasis. Physiol. Rev. 2018;98(3):1169–1203. doi: 10.1152/physrev.00023.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kansanen E., Kuosmanen S.M., Leinonen H., Levonen A.L. The Keap1-Nrf2 pathway: mechanisms of activation and dysregulation in cancer. Redox Biol. 2013;1(1):45–49. doi: 10.1016/j.redox.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee J.M., Li J., Johnson D.A., Stein T.D., Kraft A.D., Calkins M.J., et al. Nrf2, a multi-organ protector? Faseb. J. 2005;19(9):1061–1066. doi: 10.1096/fj.04-2591hyp. [DOI] [PubMed] [Google Scholar]
- 14.Qi J.H., Dong F.X. The relevant targets of anti-oxidative stress: a review. J. Drug Target. 2021;29(7):677–686. doi: 10.1080/1061186X.2020.1870987. [DOI] [PubMed] [Google Scholar]
- 15.Tu W., Wang H., Li S., Liu Q., Sha H. The anti-inflammatory and anti-oxidant mechanisms of the Keap1/Nrf2/ARE signaling pathway in chronic diseases. Aging Dis. 2019;10(3):637–651. doi: 10.14336/AD.2018.0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li X., Song Y. Proteolysis-targeting chimera (PROTAC) for targeted protein degradation and cancer therapy. J. Hematol. Oncol. 2020;13(1):50. doi: 10.1186/s13045-020-00885-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pettersson M., Crews C.M. PROteolysis TArgeting Chimeras (PROTACs) - past, present and future. Drug Discov. Today Technol. 2019;31:15–27. doi: 10.1016/j.ddtec.2019.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du G., Jiang J., Henning N.J., Safaee N., Koide E., Nowak R.P., et al. Exploring the target scope of KEAP1 E3 ligase-based PROTACs. Cell Chem. Biol. 2022;29(10):1470–1481 e31. doi: 10.1016/j.chembiol.2022.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen H., Nguyen N.H., Magtoto C.M., Cobbold S.A., Bidgood G.M., Meza Guzman L.G., et al. Design and characterization of a heterobifunctional degrader of KEAP1. Redox Biol. 2023;59 doi: 10.1016/j.redox.2022.102552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang F., Zhan Y., Li M., Wang L., Zheng A., Liu C., et al. Cell-permeable PROTAC degraders against KEAP1 efficiently suppress hepatic stellate cell activation through the antioxidant and anti-inflammatory pathway. ACS Pharmacol Transl Sci. 2023;6(1):76–87. doi: 10.1021/acsptsci.2c00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davies T.G., Wixted W.E., Coyle J.E., Griffiths-Jones C., Hearn K., McMenamin R., et al. Monoacidic inhibitors of the Kelch-like ECH-associated protein 1: nuclear factor erythroid 2-related factor 2 (KEAP1:NRF2) protein-protein interaction with high cell potency identified by fragment-based Discovery. J. Med. Chem. 2016;59(8):3991–4006. doi: 10.1021/acs.jmedchem.6b00228. [DOI] [PubMed] [Google Scholar]
- 22.Lilienbaum A. Relationship between the proteasomal system and autophagy. Int J Biochem Mol Biol. 2013;4(1):1–26. [PMC free article] [PubMed] [Google Scholar]
- 23.Ulasov A.V., Rosenkranz A.A., Georgiev G.P., Sobolev A.S. Nrf2/Keap1/ARE signaling: towards specific regulation. Life Sci. 2022;291 doi: 10.1016/j.lfs.2021.120111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim K., Lee D.H., Park S., Jo S.H., Ku B., Park S.G., et al. Disordered region of cereblon is required for efficient degradation by proteolysis-targeting chimera. Sci. Rep. 2019;9(1) doi: 10.1038/s41598-019-56177-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bellezza I., Giambanco I., Minelli A., Donato R. Nrf2-Keap1 signaling in oxidative and reductive stress. Biochim. Biophys. Acta Mol. Cell Res. 2018;1865(5):721–733. doi: 10.1016/j.bbamcr.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 26.Ooi E.E., Dhar A., Petruschke R., Locht C., Buchy P., Low J.G.H. Use of analgesics/antipyretics in the management of symptoms associated with COVID-19 vaccination. NPJ Vaccines. 2022;7(1):31. doi: 10.1038/s41541-022-00453-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Farah R., Rege S.V., Cole R.J., Holstege C.P. Suspected suicide attempts by self-poisoning among persons aged 10-19 Years during the COVID-19 pandemic - United States, 2020-2022. MMWR Morb. Mortal. Wkly. Rep. 2023;72(16):426–430. doi: 10.15585/mmwr.mm7216a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ben-Shachar R., Chen Y., Luo S., Hartman C., Reed M., Nijhout H.F. The biochemistry of acetaminophen hepatotoxicity and rescue: a mathematical model. Theor. Biol. Med. Model. 2012;9:55. doi: 10.1186/1742-4682-9-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramachandran A., Jaeschke H. Acetaminophen hepatotoxicity: a mitochondrial perspective. Adv. Pharmacol. 2019;85:195–219. doi: 10.1016/bs.apha.2019.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jaeschke H., Xie Y., McGill M.R. Acetaminophen-induced liver injury: from animal models to humans. J Clin Transl Hepatol. 2014;2(3):153–161. doi: 10.14218/JCTH.2014.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jaeschke H., Duan L., Nguyen N., Ramachandran A. Mitochondrial damage and biogenesis in acetaminophen-induced liver injury. Liver Res. 2019;3(3–4):150–156. doi: 10.1016/j.livres.2019.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoon E., Babar A., Choudhary M., Kutner M., Pyrsopoulos N. Acetaminophen-induced hepatotoxicity: a comprehensive update. J Clin Transl Hepatol. 2016;4(2):131–142. doi: 10.14218/JCTH.2015.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jaeschke H., Adelusi O.B., Akakpo J.Y., Nguyen N.T., Sanchez-Guerrero G., Umbaugh D.S., et al. Recommendations for the use of the acetaminophen hepatotoxicity model for mechanistic studies and how to avoid common pitfalls. Acta Pharm. Sin. B. 2021;11(12):3740–3755. doi: 10.1016/j.apsb.2021.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li S., Tan H.Y., Wang N., Zhang Z.J., Lao L., Wong C.W., et al. The role of oxidative stress and antioxidants in liver diseases. Int. J. Mol. Sci. 2015;16(11):26087–26124. doi: 10.3390/ijms161125942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watt G.F., Scott-Stevens P., Gaohua L. Targeted protein degradation in vivo with Proteolysis Targeting Chimeras: current status and future considerations. Drug Discov. Today Technol. 2019;31:69–80. doi: 10.1016/j.ddtec.2019.02.005. [DOI] [PubMed] [Google Scholar]
- 36.Pike A., Williamson B., Harlfinger S., Martin S., McGinnity D.F. Optimising proteolysis-targeting chimeras (PROTACs) for oral drug delivery: a drug metabolism and pharmacokinetics perspective. Drug Discov. Today. 2020;25(10):1793–1800. doi: 10.1016/j.drudis.2020.07.013. [DOI] [PubMed] [Google Scholar]
- 37.Boichenko I., Deiss S., Bar K., Hartmann M.D., Hernandez Alvarez B. A FRET-based assay for the identification and characterization of cereblon ligands. J. Med. Chem. 2016;59(2):770–774. doi: 10.1021/acs.jmedchem.5b01735. [DOI] [PubMed] [Google Scholar]
- 38.Hartmann M.D., Boichenko I., Coles M., Zanini F., Lupas A.N., Hernandez Alvarez B. Thalidomide mimics uridine binding to an aromatic cage in cereblon. J. Struct. Biol. 2014;188(3):225–232. doi: 10.1016/j.jsb.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 39.Cheng Y., Prusoff W.H. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 1973;22(23):3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 40.Charni-Natan M., Goldstein I. Protocol for primary mouse hepatocyte isolation. STAR Protoc. 2020;1(2) doi: 10.1016/j.xpro.2020.100086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Percie du Sert N., Ahluwalia A., Alam S., Avey M.T., Baker M., Browne W.J., et al. Reporting animal research: explanation and elaboration for the ARRIVE guidelines 2.0. PLoS Biol. 2020;18(7) doi: 10.1371/journal.pbio.3000411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee S.G., Cho K.H., Nguyen T.T., Vo D.K., Chae Y.J., Maeng H.J. Inhibitory effect of 20(S)-protopanaxadiol on cytochrome P450: potential of its pharmacokinetic interactions in vivo. Biomed. Pharmacother. 2022;153 doi: 10.1016/j.biopha.2022.113514. [DOI] [PubMed] [Google Scholar]
- 43.Kim M.W., Kang J.H., Jung H.J., Park S.Y., Phan T.H.L., Namgung H., et al. Allyl isothiocyanate protects acetaminophen-induced liver injury via NRF2 activation by decreasing spontaneous degradation in hepatocyte. Nutrients. 2020;12(11) doi: 10.3390/nu12113585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rahman I., Kode A., Biswas S.K. Assay for quantitative determination of glutathione and glutathione disulfide levels using enzymatic recycling method. Nat. Protoc. 2006;1(6):3159–3165. doi: 10.1038/nprot.2006.378. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.