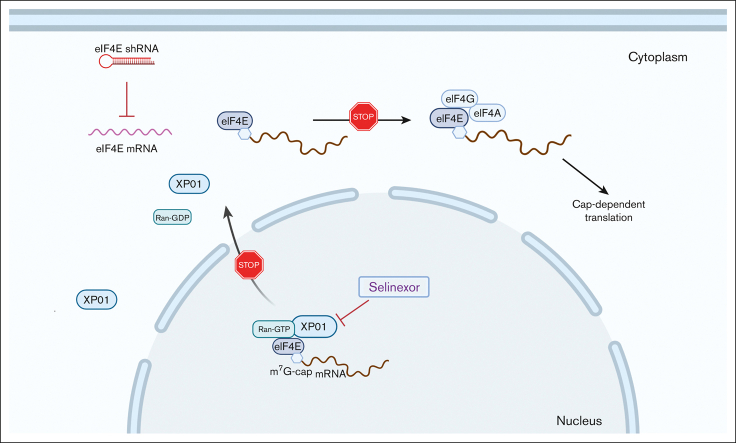
Key Points
-
•
Knockdown of eIF4E sensitizes resistant MM cell lines to selinexor.
-
•
These data provide a rationale for further evaluating the combination of selinexor and elF4E inhibitors in MM.
Visual Abstract
Abstract
Selinexor (KPT-330) is a small molecule inhibitor of XPO1, which mediates the transport of tumor suppressor proteins, oncogene messenger RNAs, and other proteins involved in governing cell growthfrom the cell nucleus to the cytoplasm. It is overexpressed in many cancer types. Because eukaryotic translation initiator factor 4E (eIF4E) plays a critical role in protein translation in cancer cells in multiple myeloma (MM), we evaluated the effectiveness of combined inhibition of protein translation and nuclear export in MM. Selinexor, an inhibitor of nuclear protein export, dose-dependently decreased eIF4E, IKZF1, and c-MYC protein levels. Using a doxycycline-inducible–pLKO-Tet-On vector, knockdown of eIF4E significantly enhanced the antiproliferative effects of selinexor, sensitized resistant MM cells to selinexor, and increased apoptosis in MM cells. Immunofluorescent analysis of MM cells showed that the combined treatment increased the localization of residual eIF4E to the nucleus compared with selinexor-only treatment. The overexpression of eIF4E at least partially rescued the effects of selinexor in MM cells by reducing G1 cell cycle arrest and increasing the selinexor-IC50 10-fold. Moreover, the combination of selinexor with pharmacologic inhibitors of protein translation showed synergistic anti-MM effects. These results suggest a synergistic anti-MM effect of selinexor combined with eIF4E inhibitors in vitro. Our work provides a better understanding of the potential mechanism of resistance to selinexor and a rationale for combining selinexor with eIF4E inhibitors for the treatment of MM.
Introduction
Multiple myeloma (MM) is the second most commonly diagnosed blood cancer, after non-Hodgkin lymphoma.1,2 Despite the Food and Drug Administration (FDA) approval of multiple new therapies with different mechanisms of action for the treatment of MM over the past 16 years, the disease remains an incurable plasma cell malignancy, and novel treatments, especially for relapsed/refractory MM, are urgently needed. Selinexor (KPT-330) is a small molecule inhibitor of XPO1, which received FDA approval for use in previously treated MM and accelerated the approval for treating diffuse large B-cell lymphoma.3,4 In addition, clinical and preclinical administration of selinexor alone and as drug combinations continue to be assessed in many other hematologic and solid cancers. Selinexor binds covalently to XPO1 cysteine-528, which is located in the cargo-binding pocket, abrogating XPO1-mediated nuclear export of cargo proteins, such as tumor suppressor proteins (TSPs) TP53, p21, BRCA1/2, pRB, FOXO, IκB, and others.5 Being a selective inhibitor of nuclear export (SINE) compound, selinexor locks TSPs in the nucleus. Additionally, eukaryotic translation initiation factor 4E (eIF4E) is an XPO1 cargo that shuttles oncogene messenger RNA (mRNA) species, including oncogenes such as MYC, BCL6, BCL2, and MCL1, from the nucleus to the cytoplasm.6 Selinexor treatment prevents the shuttling of eIF4E and blocks the expression of these oncogenes by sequestering their mRNAs in the nucleus. This leads to the reactivation of cell cycle checkpoints, attenuation of oncoprotein-driven growth, induction of cell cycle arrest, and apoptosis of cancer cells.3,7
Recently, eIF4E was found to be a critical mediator of tumorigenesis and cancer progression, resulting in an increased interest in therapeutic agents that directly or indirectly target eIF4E activation in cancer.8, 9, 10, 11, 12 It has been proposed that the translational efficiency of mRNA with highly complex 5′-untranslated regions is dependent on eIF4E levels.13 An increase in eIF4E levels or activity does not lead to higher rates of global translation but rather results in increased translation of mRNAs with highly complex 5′-untranslated regions.14 Several genes involved in tumorigenesis, including MYC, CEBPB, CCND1, and vascular endothelial growth factor (VEGF), are regulated at the translational level by eIF4E,15, 16, 17 suggesting that eIF4E might be an attractive target for antimyeloma treatment.
Previously, we demonstrated that eIF4E levels are significantly elevated in primary myeloma cells compared with healthy plasma cells, and the transcription factor C/EBPβ (CCAAT enhancer binding protein beta) is regulated at the translational level by eIF4E.18 Consequently, multiple C/EBPβ downstream transcription factors, such as IRF4, XBP-1, and Blimp-1, are affected. Hence, eIF4E knockdown (KD) by an inducible eIF4E–short hairpin RNA (eIF4E-shRNA) in human MM xenograft mouse models abrogates MM tumor growth.19 We reasoned that the inhibition of eIF4E combined with SINE compounds (XPO1 inhibitors) such as selinexor would therefore enhance MM cell death by further decreasing the export and translation of eIF4E-target mRNAs, representing a promising therapeutic approach. Here, we investigate the direct effects of selinexor on protein translation in MM cells alone and in combination with eIF4E KD.
Methods
Cell culture and selection
MM cell lines (MMCLs) MM.1S, H929, and U266 were purchased from the American Type Culture Collection, and LP1 and JJN3 were purchased from the German Collection of Microorganisms and Cell Cultures and cultured as previously described.18 Selinexor (Karyopharm Therapeutics) was dissolved in dimethyl sulfoxide at a final concentration of 1 mM. Rocaglamide is an eIF4A inhibitor (selleckchem) and was dissolved in dimethyl sulfoxide at a final concentration of 1 mM. Primary human MM cells were isolated from patient bone marrow samples using ficoll (GE Healthcare) followed by magnetic separation using CD138+ antibody-specific microbeads per the the manufacturer’s instructions (Miltenyi Biotech). Human bone marrow stromal cells were cultured as previously reported.19 All primary samples were procured after informed consent was obtained. All studies were approved by the institutional review board of Columbia University Medical Center, New York.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and WB analysis
Protein was extracted from the cells using RIPA buffer (radioimmunoprecipitation assay buffer) containing Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific) and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and western blotting (WB) as previously described.20 Nuclear and cytosolic fractions were obtained from U266 and H929 cells using NE-PER nuclear and cytoplasmic extraction reagents (Thermo Fisher Scientific) per the manufacturer’s protocol. The following antibodies were used for immunoblotting: anti-IKZF1 (Abcam), anti-β-actin (Sigma Aldrich), anti-XPO1, anti-c-MYC, anti-eIF4E, anti-PARP, anti-Bax, and anti-LaminB1 (Cell Signaling Technology).
Quantitative real-time polymerase chain reaction analysis
For the determination of mRNA levels of eIF4E, total RNA was isolated from the cells using Trizol (Invitrogen), following the manufacturer's instructions. Total RNA was converted into complementary DNA using Superscript III RT (Invitrogen). Quantitative real-time polymerase chain reaction was performed as previously described.18 The following primer sets were used: for eIF4E, 5′-ACAAGTCAGTCTGAAACCATCGAAC-3′ and 5′-CTTCATCCTCTTCGGCCACTCCTCC -3′; and for β-actin, 5′-GGACTTCGAGCAAGAGATGG-3′ and 5′-AGCACTGTGTTGGCGTACAG-3′.
Immunolabeling for fluorescence microscopy
Immunofluorescent staining was performed as previously described.21 Treated cells were attached to poly-L-lysine–coated coverslips via centrifugation at 1000 rpm for 10 minutes and were subsequently fixed using 4% paraformaldehyde for 10 minutes at room temperature. For whole-cell staining, cells were washed 3 times with phosphate-buffered saline (PBS), permeabilized with 0.1% Triton X-100, and blocked with 3% bovine serum albumin in PBST (0.1% Triton X-100 in PBS) for 1 hour. Cells were stained with anti-eIF4E antibodies (Sigma Aldrich) and counterstained with 4′,6-diamidino-2-phenylindole (Cell Signaling Technology) for nuclear visualization. Images were captured using a Leica DMI 6000B microscope.
Immunohistochemistry analysis of eIF4E and IKZF1
Formalin-fixed, paraffin-embedded tissue samples of healthy and MM human tissue were obtained from US Biomax Inc. Antigen retrieval was achieved using a citrate-based retrieval solution at pH 6.1 (Dako). Slides were stained using the EnVision kit (Dako) and coverslipped with a Permount mounting medium (Thermo Fisher Scientific). The following 2 immunohistochemistry antibodies were used: anti-eIF4E (polyclonal; Cell Signaling Technology) and anti-IKZF1 (polyclonal; Abcam). Images were captured using a Leica DMI 6000B microscope.
Transduction of pCDH-GFP and pCDH-GFP-eIF4E
The expression vectors for full-length wild-type eIF4E were generated by inserting the coding region into the pCDH-GFP vector. Briefly, full-length human eIF4E complementary DNA was excised from pHA-eIF4E (Addgene)22 via HindIII/XhoI double digestion followed by filling in of 5′ overhangs using DNA polymerase I large (Klenow) fragment to produce blunt ends. Similarly, pCDH-CMV-MCS-EF1-COPGFP (System Biosciences) was digested by EcoRI after filling in of 5′ overhangs and then ligated with eIF4E fragment using T4 ligase (NEB). Lentivirus was packaged and concentrated using PEG-it (System Biosciences) from 293TN cell supernatant after cotransfection of pCDH-CMV-eIF4E-EF1-COPGFP with pPACKH1-packaging plasmids (System Biosciences). Myeloma cells (2 × 106) were infected with an empty vector (EV) lentiviral control pCDH-GFP(EV) or pCDH-GFP-eIF4E lentivirus. The efficiency of virus infection was determined via green fluorescent protein coexpression. The infection rate of H929 cells was >70% at 72 hours. Infected cells were selected using Influx cell sorter (BD Bioscience) and analyzed via WB or cell proliferation assay.
eIF4E shRNA KD assay
Lentiviral shRNA was used to knock down eIF4E expression in MM cells. In brief, the lentiviral pLKO.1-tet-on-sh-eIF4E19 target sequence was produced as follows: 5′-ACTCTGTAATAGTTCAGTA-3′. U266 cells were incubated with lentivirus particles and polybrene (8 μg/mL) for 16 hours and then washed with media. Infected cells were maintained in 10% tetracycline-free fetal bovine serum (Bio-Techne). Cells were selected after 3 days in puromycin (5 μg/mL). The efficiency of virus infection was determined by checking puromycin resistance, with an infection rate >60% in U266 cells. Selected cells were treated with or without doxycycline (dox; 200 ng/mL) for 3 days to induce the KD of eIF4E.
Cell proliferation assay
MM cells were incubated in 96-well plates with the drug treatment for the indicated time. 20 μL of CellTiter 96 AQueous One Solution Cell Proliferation Assay (MTS, Promega) was added for 3 hours. The absorbance of the samples at 490 nm against the background control was performed using the Synergy HT Multidetection Microplate Reader (Biotek Instruments Inc.).
Human MM xenograft mouse model
Female severe combined immunodeficient (SCID) Beige (CB17.Cg-PrkdcscidLystbg-J/Crl) mice were purchased from Charles River Laboratories (Wilmington, MA) at ages between 6 and 8 weeks and with a weight between 20 and 25 g. Tet-on shCNTL or tet-on sheIF4E U266 myeloma cells (2 × 106) in 100 μL PBS with an equal volume of Matrigel basement membrane matrix (BD Biosciences) were injected subcutaneously. After 12 days, mice bearing tumors with an average size >100 mm3 were randomly separated into 4 groups and treated with dox (via drinking water) and selinexor (10 mg/kg; via oral gavage) every 3 days for the duration of the study with (1) vehicle control, (2) dox alone (to induce KD of eIF4E), (3) selinexor alone, or (4) a combination of dox and selinexor. Tumor sizes were measured using a caliper, and the volume was calculated using the following formula 0.5 × width2 × length, representing the 3-dimensional volume of an ellipse. Tumor sizes were measured every 3 days.19 Mice were weighed every 3 days and observed daily for diarrhea or any changes in behavior and condition. All the animal procedures were approved by the institutional animal care and the use committee of Columbia University.
Primary myeloma cell viability assay
Bone marrow–derived stromal cells (2 × 103 per well) were seeded on a 96-well plate overnight. After the incubation, primary MM cells (2 × 105 per well) were added and treated with rocaglamide and/or selinexor for 2 days. 20 μL of CellTiter 96 AQueous One Solution Cell Proliferation Assay (MTS, Promega) was added, and cells were incubated for 3 hours. The absorbance of the samples at 490 nm against the background control was performed using the Synergy HT Multidetection Microplate Reader (Biotek Instruments Inc).
Statistical analyses
Each experiment was repeated at least 3 times, and all quantitative data were presented as mean ± standard deviation. Statistical differences were determined using t test. The results were considered statistically significant if P < .05.
Results
Selinexor abrogates eIF4E, IKZF1, and c-MYC protein expression and induces growth inhibition in MM
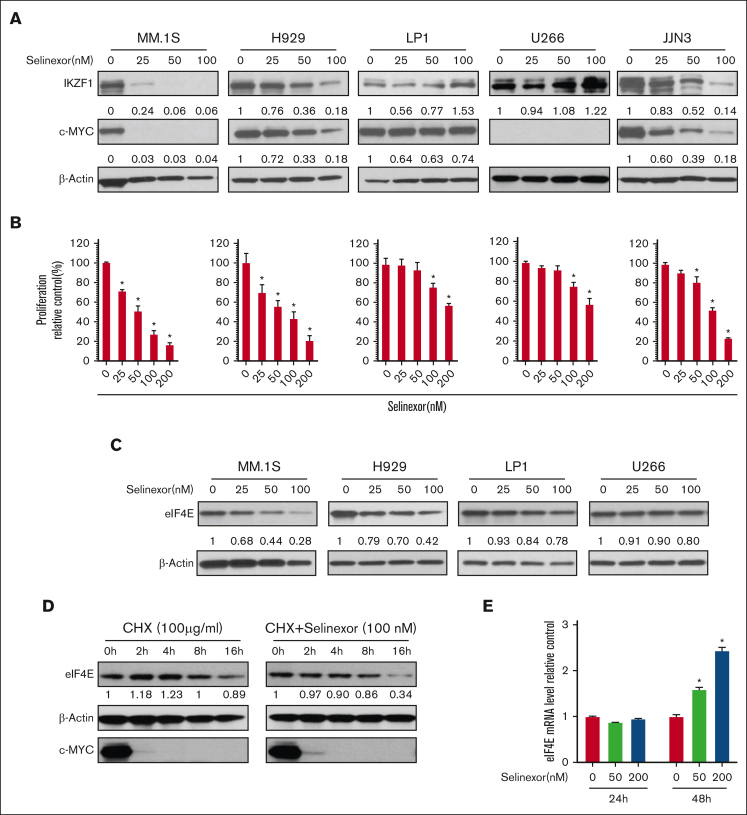
To investigate the anti-MM effects of selinexor in vitro, we tested the dose-dependent effects of selinexor on a panel of MMCLs including H929, MM.1S, JJN3, LP1, and U266. Transcriptional factors IKZF1 and c-MYC are essential for MM growth and survival.23,24 Selinexor decreased IKZF1 and c-MYC protein levels in MM.1S, H929, and JJN3 (Figure 1A) and suppressed cell proliferation in a dose-dependent manner (P < .01; Figure 1B). Compared with H929, JJN3, and MM.1S, U266 and LP1 cells were less sensitive to selinexor in terms of IKZF1 downregulation and inhibition of cell growth. In H929 and MM.1S, selinexor treatment also decreased protein levels of eIF4E, which is a known XPO1 cargo (Figure 1C). Next, we investigated how selinexor reduced the eIF4E protein level. We exposed H929 cells to the protein synthesis inhibitor cycloheximide for different durations to investigate eIF4E and c-MYC stability. We observed that in comparison with elFE, the degradation of c-MYC protein was more rapid. The combined treatment of cycloheximide with selinexor dramatically reduced eIF4E protein stability (Figure 1D). We further analyzed whether selinexor affects the levels of eIF4E mRNA. We found that selinexor did not affect eIF4E mRNA at 24 hours but increased eIF4E mRNA at 48 hours, although the protein level had decreased at this time point (Figure 1E). These findings indicate that selinexor-induced eIF4E downregulation via protein degradation.
Figure 1.
Selinexor abrogates IKZF1 and c-MYC protein expression and induces growth inhibition in MM. (A) MMCLs were treated with selinexor at indicated doses for 48 hours. IKZF1 and c-MYC levels were detected using WB using β-actin as a loading control. The densities of the bands were measured using NIH ImageJ and normalized against β-actin. The immunoblot is representative of 3 independent experiments. (B) Selinexor inhibited MMCL proliferation in a dose-dependent manner. MMCLs were treated with different doses of selinexor for 4 days and the proliferation was detected using AQueous One Solution Cell Proliferation Assay (MTS). ∗P < .05 (C) MMCLs were treated with selinexor at indicated doses for 48 hours. eIF4E levels were detected using WB using β-actin as a loading control. The densities of the bands were measured using NIH ImageJ and normalized against β-actin. Immunoblot representative of 3 independent experiments is shown. (D) H929 cells were treated with cycloheximide (CHX; 100 μg/mL) or CHX and selinexor (100 nM) for the indicated times. Immunoblot analysis of eIF4E, c-MYC, or β-actin was performed. (E) H929 cells were treated with selinexor (50 and 200 nM) for the indicated times. Treated cells were used for mRNA extraction and reverse transcription. eIF4E mRNA levels were compared using real-time PCR. ∗P < .05. PCR, polymerase chain reaction.
eIF4E KD sensitizes resistant MMCLs to selinexor
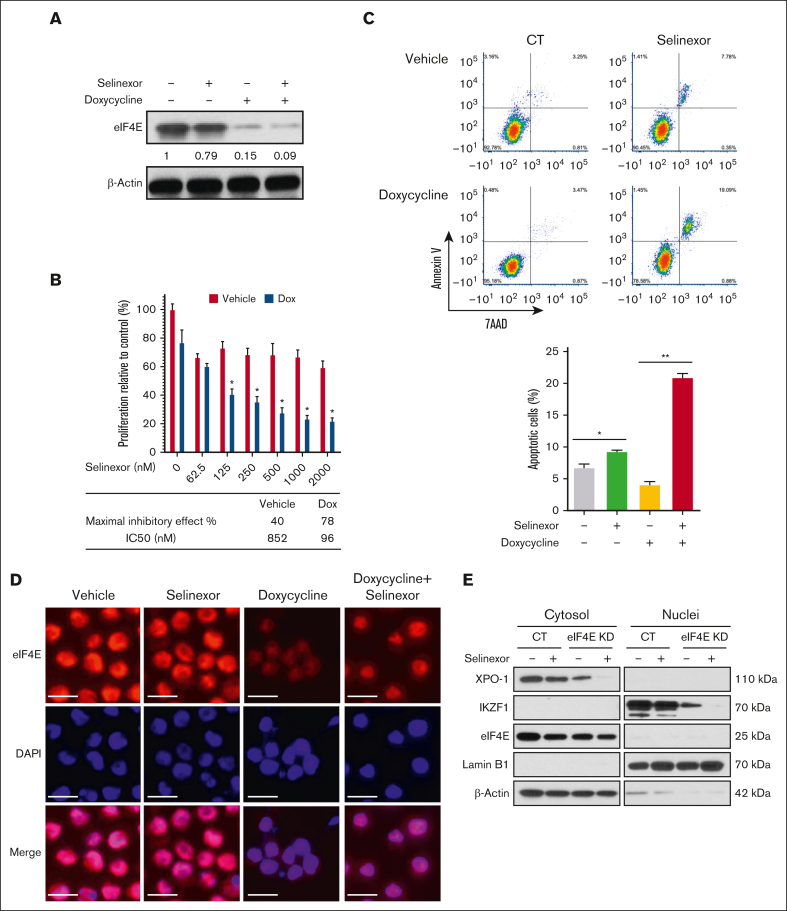
Compared with MM.1S and H929, U266 and LP1 MMCLs were resistant to selinexor in terms of IKZF1 and eIF4E downregulation and inhibition of cell growth. A previous study also showed similar results, with a higher IC50 value of selinexor in U266 than that in MM.1S cells.25 Therefore, to examine the effects of selinexor on eIF4E-dependent oncogene translation and MM cell growth, we transduced U266 cells with a dox-inducible KD of eIF4E (Figure 2A). After 72 hours of dox treatment, eIF4E KD became detectable but did not affect cell proliferation and apoptosis. In contrast, when combined with selinexor, the KD of eIF4E for 72 hours significantly inhibited cell growth (selinexor alone vs with eIF4E KD: 40.4% vs 77.9%), decreased the IC50 of selinexor (selinexor alone vs with eIF4E KD: 852 vs 96 nM) (Figure 2B), and increased selinexor-induced apoptosis (selinexor alone vs with eIF4E KD: 7.8% vs 19.1%) (Figure 2C). As visualized using immunofluorescence, the treatment of eIF4E KD cells caused more residual eIF4E to be localized to the nucleus compared with the treatment of selinexor cells (Figure 2D). To confirm the effects of selinexor on the nuclear export of critical proteins, we treated eIF4E KD cells with selinexor and then performed cellular fractionation to separate the cytoplasm and nuclear compartments (Figure 2E). Selinexor eliminated the cytoplasmic expression of XPO1 and reduced the nuclear expression of IKZF1. The eIF4E protein level was undetectable in the nucleus compared with that in the cytoplasm.
Figure 2.
eIF4E KD and selinexor synergistically inhibit MM cell proliferation. Dox-inducible eIF4E KD U266 cells were treated with selinexor (200 nM, unless otherwise indicated) and/or dox (500 ng/mL) for 72 hours. (A) The protein expression of eIF4E level was detected using WB with β-actin as a loading control. The densities of the bands were measured using NIH ImageJ and normalized for β-actin. The immunoblot is representative of 3 independent experiments. (B) Cell proliferation was assessed and the combination of doxycycline and selinexor was compared with selinexor alone. ∗P < .05. Quantification of the maximal inhibitory effects of selinexor is shown underneath the bar graph. (C) Example dot plots (top) and quantification of replicates (bottom) for cell apoptosis were detected via flow cytometry. (D) Immunofluorescence microscopy for eIF4E (red) and with 4′,6-diamidino-2-phenylindole for nuclear counterstaining (blue). Scale bar, 20 μM. (E) Cytoplasmic and nuclear fractions were extracted and analyzed using WB using antibodies against XPO1, eIF4E, IKZF1, and c-MYC. LaminB1 and β-actin were blotted as the markers for nuclear and cytoplasmic fractions, respectively.
Overexpression of eIF4E induces resistance to selinexor in MM cells
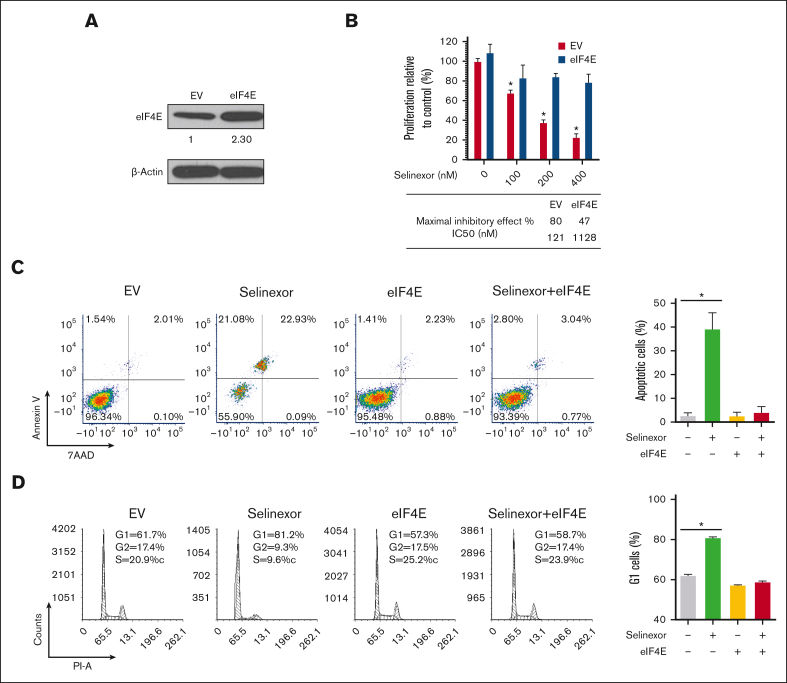
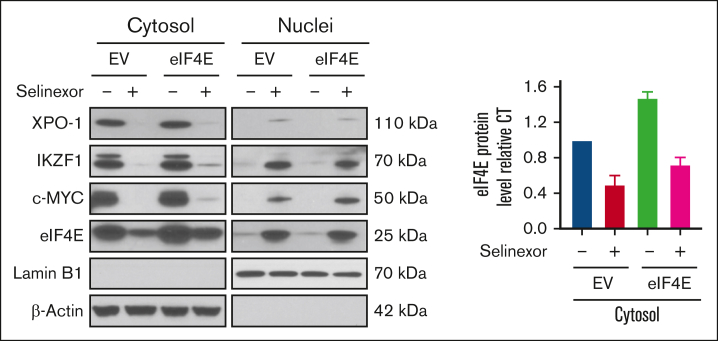
To investigate the causal role of eIF4E inhibition in the antimyeloma effect of selinexor, we facilitated the overexpression of eIF4E in MM cells. H929 is sensitive to selinexor, and its eIF4E level is downregulated by selinexor. A previous study showed that H929 cells transfected with eIF4E were significantly more resistant to PI3Kð inhibitors and proteasome inhibitors compared with control cells.26 Therefore, we selected H929 cells for eIF4E overexpression experiments (Figure 3A). As predicted, the overexpression of eIF4E reversed the inhibitory effects of selinexor, resulting in a relatively higher IC50 value. Notably, the IC50 value of selinexor increased by 10-fold when eIF4E was overexpressed compared with the control cells (Figure 3B). Selinexor-induced apoptosis and cell cycle arrest were also completely inhibited by eIF4E’s overexpression (Figure 3C-D).
Figure 3.
Overexpression of eIF4E induces resistance to selinexor. (A) eIF4E was overexpressed in H929 MM cells and confirmed using WB. The densities of the bands were measured using NIH ImageJ and normalized for β-actin. The immunoblot is representative of 4 independent experiments. (B) EV- or eIF4E-overexpressing (eIF4E) H929 cells were treated with different doses of selinexor, and cell proliferation was analyzed. ∗P < .05. Quantification of the maximal inhibitory effect of selinexor on H929 proliferation is shown. (C) EV- or eIF4E-transduced H929 cells were treated with selinexor and evaluated via cell apoptosis assay. (D) EV- or eIF4E-transduced H929 cells were treated with selinexor followed by assessment with a cell cycle assay. Right panel values represent the mean ± SEM for 2 independent experiments. SEM, standard error of the mean.
To confirm the effects of selinexor on the nuclear export of critical proteins, we treated either EV- or eIF4E-overexpressing H929 cells with selinexor and then performed cellular fractionation to separate the cytoplasm and nuclear compartments (Figure 4). Selinexor eliminated and reduced the cytoplasmic expressions of XPO1 and eIF4E, respectively, while increasing the nuclear accumulation of eIF4E (consistent with Figure 2D) in both EV and eIF4E-overexpressing cells (Figure 4).
Figure 4.
The effects of selinexor on the nuclear export of critical proteins. EV- or eIF4E-overexpressing H929 cells were treated without or with selinexor. Cytoplasmic and nuclear fractions were extracted followed by WB using antibodies against XPO1, eIF4E, IKZF1, and c-MYC. LaminB1 and β-actin were blotted as the markers for nuclear and cytoplasmic fractions, respectively. (Right) Band density analysis of eIF4E in cytoplasmic fractions. eIF4E protein levels were quantified using NIH ImageJ. Values represent the mean ± SEM for 2 independent experiments.
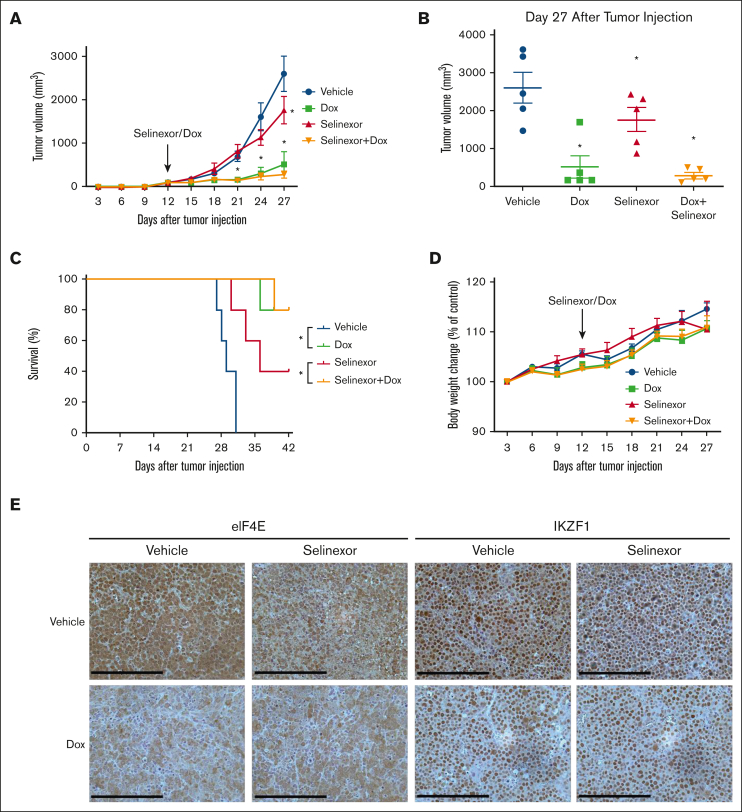
Combined targeting of eIF4E and selinexor blocks tumor progression in vivo
To investigate the combined effects of eIF4E KD and selinexor on MM tumor growth in vivo, we generated subcutaneous MM xenografts in nonobese diabetic SCID mice using inducible eIF4E shRNA U266 cells. Dox-treated animals (to activate eIF4E KD) were presented with significantly reduced (P < .001) tumor volume compared with controls (Figure 5A-B). Selinexor-treated (moderate dose) animals with eIF4E KD had smaller tumors than selinexor-treated animals without the KD (Figure 5A-B). In addition, we found that survival was significantly prolonged in the eIF4E KD groups compared with the controls or the group that received selinexor without dox (Figure 5C). When combining eIF4E silencing with selinexor in vivo, we did not observe an enhanced or synergistic antitumor effect because eIF4E KD alone is so potent that additional effects of selinexor could not be investigated. Selinexor and/or dox treatment did not result in toxicity and did not affect the body weight (Figure 5D). Performing immunohistochemistry on the tumor sections confirmed the decrease of eIF4E expression in dox- and/or selinexor-treated mice bearing U266-tet-on-sh-eIF4E inducible tumors compared with vehicle-treated tumors (Figure 5E, left panel). In accordance with our WB findings, we observed a decreased expression of IKZF1 in eIF4E KD tumors (Figure 5E, right panel).
Figure 5.
Combined targeting of eIF4E and selinexor blocks tumor progression in vivo. Tet-on-sh-eIF4E U266 cells were subcutaneously injected into SCID/bg mice. (A) Subcutaneous tumor growth was measured using calipers and calculated with the following volume formula: 0.5 × long diameter × square of the short diameter. Each bar represents the mean ± SEM (n = 5). ∗P < .05. (B) Tumor volume was measured after 27 days. (C) Kaplan-Meier survival analysis of mice during 6 weeks of follow-up (n = 5 per group). Using a Log-Rank test, a survival benefit was observed for selinexor vs vehicle (P < .05) and selinexor + dox vs vehicle (P < .05). (D) Body weights were monitored every 3 days. No significant differences were observed between different groups. (E) Tumors harvested at the end of the study were fixed in 10% formalin and subsequently processed for immunohistochemical staining for eIF4E and IKZF1. The staining was observed using a Leica DMI 6000B microscope. Scale bar, 200 μM.
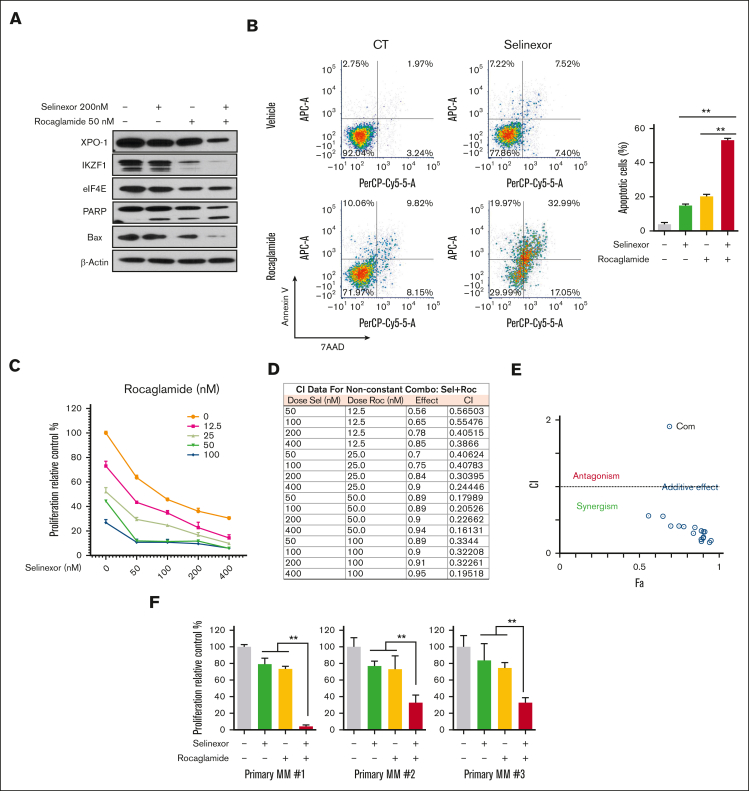
Pharmacological blockage of protein translation sensitizes resistant MMCLs to selinexor
To investigate the pharmacological dual targeting of protein translation and nuclear protein export, we evaluated the combined effects of selinexor and rocaglamide (an eIF4A inhibitor that regulates the protein translation complex) in U266 cells. We found that the combination enhanced the downregulation of IKZF1 compared with selinexor treatment alone in WB assays. PARP cleavage and Bax expression as markers of apoptosis were also analyzed. Dual-drug treatment increased PARP cleavage and decreased Bax expression compared with the single-agent treatment (Figure 6A). Similar to eIF4E silencing, rocaglamide combined with selinexor increased selinexor-induced apoptosis (Figure 6B) and dose-dependent inhibition of proliferation (Figure 6C). The combined effect of selinexor and rocaglamide on cell proliferation was analyzed using CompuSyn software analysis of the combination index (CI) of the combined action.27 The quantitative definition of drug combinations is CI = 1 for additive effect, CI < 1 for synergism, and CI > 1 for antagonism. The combination of selinexor (50, 100, 200, and 400 nM) and rocaglamide (12.5, 25, 50, 100 nM) showed a synergistic effect at all dosages (CI < 1; Figure 6D). CI was also used in the action point diagram (Figure 6E) to quantitatively describe the synergism and antagonism of combined drugs at a given dose-effect level. We further evaluated the combined effects of selinexor and rocaglamide in primary myeloma cells, showing that their combination significantly enhanced the anti-MM effect of selinexor alone in primary myeloma cells (Figure 6F). These data suggest that the combination of selinexor with the pharmacologic inhibitors of eIF4A has synergistic anti-MM effects.
Figure 6.
Pharmacological blockage of protein translation sensitizes resistant MM cells to selinexor. (A) U266 cells were treated with selinexor (200 nM, unless otherwise indicated) and/or rocaglamide (50 nM) for 24 hours. The protein expression level of XPO-1, IKZF1, PARP, eIF4E, and Bax were detected via WB, using β-actin as a loading control. The immunoblot is representative of 3 independent experiments. (B) U266 cells were treated with the combination of rocaglamide and selinexor at indicated doses for 4 days. Cell apoptosis was detected using flow cytometry, example dot plots (left) and quantification of replicates (right) for ∗∗P < .01. (C) Cell proliferation was assessed and the combination of rocaglamide and selinexor at indicated doses for 4 days. (D) The CI value was analyzed using CompuSyn software. (E) Dot plot of the combined action of selinexor and rocaglamide. (F) Primary MM cells were cocultured with bone marrow stromal cells and treated with rocaglamide (25 nM) and/or selinexor (50 nM) for 2 days. Cell viability was analyzed using MTS assay. ∗P < .05.
Discussion
Despite the availability of therapeutic modalities with varying mechanisms of action, most patients with MM develop relapsed or refractory disease. The underlying mechanisms of disease progression/relapse include clonal outgrowth of cells with complex genetic and epigenetic alterations, including overexpression of oncoproteins and/or inactivation of TSPs. Thus, novel approaches to better understand and circumvent mechanisms of resistance toward existing therapies are needed to improve outcomes and quality of life among patients with MM.
The nuclear export system is a promising target for anticancer therapy. Of the 8 mammalian proteins that mediate macromolecular transport from the nucleus to the cytoplasm, XPO1 is the exclusive exporter for TSPs, the glucocorticoid receptor, and eIF4E-bound oncoprotein mRNAs.28, 29, 30, 31 XPO1 is overexpressed in many cancers, including MM, and correlates with poor survival.32, 33, 34, 35, 36, 37, 38, 39 Abnormal or disrupted nuclear-cytoplasmic trafficking is oncogenic and a mechanism for evading cell cycle checkpoint controls and chemotherapeutic resistance.40 Thus, targeting XPO1 with SINE compounds, such as selinexor, forces the nuclear retention and functional activation of TSPs and growth regulators, thereby inducing cancer cell apoptosis.
Selinexor was first approved by the US FDA in July 2019 in combination with dexamethasone for patients with heavily pretreated MM41 and is a SINE.42, 43, 44 In preclinical studies, the selinexor-dexamethasone combination blocked the nuclear export of phosphorylated glucocorticoid receptor and repressed the mammalian target of rapamycin signaling.45,46 The clinical benefit of selinexor has been investigated in numerous clinical trials both as a single agent and in combination with conventional MM therapies, such as dexamethasone, bortezomib, carfilzomib, pomalidomide, lenalidomide, and others.
Multiple cancers, including MM, display elevated levels of translation initiation factors,47,48 and eIF4E’s overexpression induces malignant transformation and drug resistance.49 Furthermore, several eIF4E targets are upregulated in MM, including MMP9, Bcl-2, Bcl-XL, survivin, cyclin D1, c-MYC, and VEGF.50,51 A study by Attar-Schneider et al demonstrated that VEGF drives eIF4E activation via an Akt-dependent mechanism and that eIF4E KD results in a significant antimyeloma effect.52 The role of eIF4E in MM tumorigenesis was confirmed in a murine xenograft model, showing that the KD of eIF4E inhibits tumor growth, whereas its overexpression accelerates tumor progression.19 The use of inhibitors that target eIF4E has exhibited promising effects in MM. Translation inhibitor 4EGI-1 impaired the assembly of the eIF4F complex and dose-dependently increased the expression of ER stress–related proteins.53 For example, Zismanov et al52 assessed the eIF4E inhibitory effects of ribavirin, an antiviral drug that acts as a competitive inhibitor to eIF4E by mimicking the 5′ cap present on human transcripts.11,54, 55, 56
It has been established that XPO1 forms a complex with eIF4E to transport 7-methyl-guanine–capped mRNAs, including multiple oncoprotein mRNAs (such as MYC, CCND1, and MDM2) to the cytoplasm.30 Our previous studies show that a high, or at least a critical threshold level, of eIF4E is required for MM cell growth to maintain sufficient protein translation of cap-dependent transcription factors. Selinexor decreased IKZF1, c-MYC, and eIF4E protein levels and suppressed cell proliferation in a dose-dependent manner in multiple MMCLs. Compared with H929, JJN3, and MM.1S, U266 and LP1 cells were less sensitive to selinexor in terms of IKZF1 downregulation and cell growth inhibition. The detailed mechanism of resistance to selinexor remains unclear. However, published data have correlated the sensitivity to XPO1 depletion with KRAS (Kirsten rat sarcoma virus) mutation status.57 Consistently, both LP1 and U266 are RAS (rat sarcoma virus) wild-type MMs. The silencing of eIF4E in MM cells reduced the translation of mRNAs with highly complex 5′-untranslated regions, such as IKZF1 and MYC, and augmented eIF4E silencing-induced MM growth inhibition. eIF4E is localized in both the nucleus and cytoplasm, where it facilitates the nuclear export of the mRNA and the cytoplasmic translation of a specific subset of mRNAs.58 It is known that eIF4E itself is a cargo of XPO1.6 In this study, we showed that selinexor blocked the export of eIF4E into the cytoplasm and reduced the levels of c-MYC and IKZF1. In addition, selinexor treatment combined with eIF4E KD resulted in reduced total eIF4E protein levels, greater residual eIF4E localization to the nucleus, and decreased levels of proteins derived from capped mRNAs associated with eIF4E, compared with selinexor treatment alone. Furthermore, we observed the synergistic effect of combining rocaglamide with selinexor in vitro. The combination treatment increased PARP cleavage and decreased the Bax and IKZF1 expression. Previous results have shown that the KD of IKZF1 leads to c-MYC and IRF4 downregulation and induces apoptosis in MM cells.59 However, the functional significance of these perturbations of transcriptional factors in cell death remains to be determined. Similar to eIF4E silencing, rocaglamide combined with selinexor resulted in significant dose-dependent inhibition of proliferation and an increased selinexor-induced apoptosis. These data suggest that the combination of selinexor with pharmacologic inhibitors of eIF4A has synergistic anti-MM effects in vitro. In addition, the introduction of ectopic eIF4E increased eIF4E in the cytoplasm. Upon selinexor treatment, cytoplasmic eIF4E remained at a relatively high level, comparable with the cytoplasmic eIF4E level in nontreated control cells. Taken together, overexpression eIF4E induced resistance to selinexor, suggesting a significant clinical interest to investigate the possible role of eIF4E as a biomarker for the response to selinexor.
In summary, our findings demonstrate that the combination of eIF4E KD with selinexor results in enhanced anticancer effects in MM. Selinexor dose-dependently downregulated oncoproteins such as IKZF1 and c-MYC and augmented eIF4E silencing-induced MM growth inhibition. These data, along with the clinically validated treatment responses of patients with refractory MM treated with selinexor in combination with proteasome inhibitors, immunomodulatory agents, or monoclonal antibodies to CD38 and SLAMF7, suggest that targeting eIF4E may be yet another approach to improving outcomes in this population.
Conflict-of-interest disclosure: C.J.W. is an employee of Karyopharm Therapeutics. M.Y.M. reports research funding from and is a consultant for Ossium Health. Y.L. is a former employee of Karyopharm Therapeutics. S. Li was the chief scientific adviser and a shareholder of Caelum Biosciences until 5 October 2021; is an adviser for Janssen, Regeneron, Adaptive, Pfizer, Bristol Myers Squibb, Sanofi, and GlaxoSmithKline; and receives research grant funding from Sanofi and Zentalis. The remaining authors declare no competing financial interests.
Acknowledgments
The graphical abstract was created with BioRender.com.
S. Li, J.F., and S. Lentzsch were supported by National Institutes of Health grant R01CA175313. M.Y.M. was supported by National Institutes of Health grant R01HL93716. The research reported in this publication was performed in part in the CCTI Flow Cytometry Core, Columbia University funded by the Office of the Director of the National Institutes of Health under awards S10RR027050 and S10OD020056. Karyopharm Therapeutics, Inc. supported the research project. JetPub Scientific Communications LLC assisted with the manuscript preparation in accordance with the Good Publication Practice (GPP3) guidelines.
Authorship
Contribution: S. Li and J.F. conducted experiments; J.Y. contributed to the WB assays; C.J.W., Y.L., D.B., R.C., N.M., and M.Y.M. critically revised the manuscript; and S. Li., J.F., and S. Lentzsch designed the research studies, analyzed data, and wrote the manuscript.
Footnotes
Data are available on request from the corresponding author, Suzanne Lentzsch (sl3440@cumc.columbia.edu).
References
- 1.Anderson KC. Progress and paradigms in multiple myeloma. Clin Cancer Res. 2016;22(22):5419–5427. doi: 10.1158/1078-0432.CCR-16-0625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumar SK, Rajkumar V, Kyle RA, et al. Multiple myeloma. Nat Rev Dis Primers. 2017;3:17046. doi: 10.1038/nrdp.2017.46. [DOI] [PubMed] [Google Scholar]
- 3.Bianchi G, Richardson PG, Anderson KC. Promising therapies in multiple myeloma. Blood. 2015;126(3):300–310. doi: 10.1182/blood-2015-03-575365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kalakonda N, Maerevoet M, Cavallo F, et al. Selinexor in patients with relapsed or refractory diffuse large B-cell lymphoma (SADAL): a single-arm, multinational, multicentre, open-label, phase 2 trial. Lancet Haematol. 2020;7(7):e511–e522. doi: 10.1016/S2352-3026(20)30120-4. [DOI] [PubMed] [Google Scholar]
- 5.Xu D, Farmer A, Chook YM. Recognition of nuclear targeting signals by karyopherin-β proteins. Curr Opin Struct Biol. 2010;20(6):782–790. doi: 10.1016/j.sbi.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Culjkovic B, Topisirovic I, Skrabanek L, Ruiz-Gutierrez M, Borden KLB. eIF4E is a central node of an RNA regulon that governs cellular proliferation. J Cell Biol. 2006;175(3):415–426. doi: 10.1083/jcb.200607020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tai YT, Landesman Y, Acharya C, et al. CRM1 inhibition induces tumor cell cytotoxicity and impairs osteoclastogenesis in multiple myeloma: molecular mechanisms and therapeutic implications. Leukemia. 2014;28(1):155–165. doi: 10.1038/leu.2013.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hagner PR, Schneider A, Gartenhaus RB. Targeting the translational machinery as a novel treatment strategy for hematologic malignancies. Blood. 2010;115(11):2127–2135. doi: 10.1182/blood-2009-09-220020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tamburini J, Green AS, Chapuis N, et al. Targeting translation in acute myeloid leukemia: a new paradigm for therapy? Cell Cycle. 2009;8(23):3893–3899. doi: 10.4161/cc.8.23.10091. [DOI] [PubMed] [Google Scholar]
- 10.Robert F, Pelletier J. Translation initiation: a critical signalling node in cancer. Expert Opin Ther Targets. 2009;13(11):1279–1293. doi: 10.1517/14728220903241625. [DOI] [PubMed] [Google Scholar]
- 11.Assouline S, Culjkovic B, Cocolakis E, et al. Molecular targeting of the oncogene eIF4E in acute myeloid leukemia (AML): a proof-of-principle clinical trial with ribavirin. Blood. 2009;114(2):257–260. doi: 10.1182/blood-2009-02-205153. [DOI] [PubMed] [Google Scholar]
- 12.Hsieh AC, Ruggero D. Targeting eukaryotic translation initiation factor 4E (eIF4E) in cancer. Clin Cancer Res. 2010;16(20):4914–4920. doi: 10.1158/1078-0432.CCR-10-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sonenberg N. Translation factors as effectors of cell growth and tumorigenesis. Curr Opin Cell Biol. 1993;5(6):955–960. doi: 10.1016/0955-0674(93)90076-3. [DOI] [PubMed] [Google Scholar]
- 14.Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433(7025):477–480. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- 15.Graff JR, Konicek BW, Vincent TM, et al. Therapeutic suppression of translation initiation factor eIF4E expression reduces tumor growth without toxicity. J Clin Invest. 2007;117(9):2638–2648. doi: 10.1172/JCI32044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mezquita P, Parghi SS, Brandvold KA, Ruddell A. Myc regulates VEGF production in B cells by stimulating initiation of VEGF mRNA translation. Oncogene. 2005;24(5):889–901. doi: 10.1038/sj.onc.1208251. [DOI] [PubMed] [Google Scholar]
- 17.Kevil’ CG, De Benedetti A, Payne DK, Coe LL, Laroux’ FS, Alexander JS. Translational regulation of vascular permeability factor by eukaryotic initiation factor 4E: implications for tumor angiogenesis. Int J Cancer. 1996;65(6):785–790. doi: 10.1002/(SICI)1097-0215(19960315)65:6<785::AID-IJC14>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 18.Li S, Pal R, Monaghan SA, et al. IMiD immunomodulatory compounds block C/EBPβ translation through eIF4E down-regulation resulting in inhibition of MM. Blood. 2011;117(19):5157–5165. doi: 10.1182/blood-2010-10-314278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li S, Fu J, Lu C, et al. Elevated translation initiation factor eIF4E is an attractive therapeutic target in multiple myeloma. Mol Cancer Ther. 2016;15(4):711–719. doi: 10.1158/1535-7163.MCT-15-0798. [DOI] [PubMed] [Google Scholar]
- 20.Feng R, Oton A, Mapara MY, Anderson G, Belani C, Lentzsch S. The histone deacetylase inhibitor, PXD101, potentiates bortezomib-induced anti-multiple myeloma effect by induction of oxidative stress and DNA damage. Br J Haematol. 2007;139(3):385–397. doi: 10.1111/j.1365-2141.2007.06772.x. [DOI] [PubMed] [Google Scholar]
- 21.Li S, Fu J, Wang H, et al. IMiD compounds affect CD34+ cell fate and maturation via CRBN-induced IKZF1 degradation. Blood Adv. 2018;2(5):492–504. doi: 10.1182/bloodadvances.2017010348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okumura F, Zou W, Zhang DE. ISG15 modification of the eIF4E cognate 4EHP enhances cap structure-binding activity of 4EHP. Genes Dev. 2007;21(3):255–260. doi: 10.1101/gad.1521607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jovanović KK, Roche-Lestienne C, Ghobrial IM, Facon T, Quesnel B, Manier S. Targeting MYC in multiple myeloma. Leukemia. 2018;32(6):1295–1306. doi: 10.1038/s41375-018-0036-x. [DOI] [PubMed] [Google Scholar]
- 24.Krönke J, Udeshi ND, Narla A, et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science. 2014;343(6168):301–305. doi: 10.1126/science.1244851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu F, Chen XQ, Li XP, et al. Drug resistance biomarker ABCC4 of selinexor and immune feature in multiple myeloma. Int Immunopharmacol. 2022;108:108722. doi: 10.1016/j.intimp.2022.108722. [DOI] [PubMed] [Google Scholar]
- 26.Deng C, Lipstein MR, Scotto L, et al. Silencing c-Myc translation as a therapeutic strategy through targeting PI3Kδ and CK1ε in hematological malignancies. Blood. 2017;129(1):88–99. doi: 10.1182/blood-2016-08-731240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chou TC. Drug combination studies and their synergy quantification using the chou-talalay method. Cancer Res. 2010;70(2):440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 28.Vogl DT, Dingli D, Cornell RF, et al. Selective inhibition of nuclear export with oral Selinexor for treatment of relapsed or refractory multiple myeloma. J Clin Oncol. 2018;36(9):859–866. doi: 10.1200/JCO.2017.75.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gravina GL, Senapedis W, McCauley D, Baloglu E, Shacham S, Festuccia C. Nucleo-cytoplasmic transport as a therapeutic target of cancer. J Hematol Oncol. 2014;7(1):85. doi: 10.1186/s13045-014-0085-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Culjkovic-Kraljacic B, Baguet A, Volpon L, Amri A, Borden KLB. The oncogene eIF4E reprograms the nuclear pore complex to promote mRNA export and oncogenic transformation. Cell Rep. 2012;2(2):207–215. doi: 10.1016/j.celrep.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richter J, Madduri D, Richard S, Chari A. Selinexor in relapsed/refractory multiple myeloma. Ther Adv Hematol. 2020;11(Mm) doi: 10.1177/2040620720930629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parikh K, Cang S, Sekhri A, Liu D. Selective inhibitors of nuclear export (SINE)-a novel class of anti-cancer agents. J Hematol Oncol. 2014;7(1):78. doi: 10.1186/s13045-014-0078-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Senapedis WT, Baloglu E, Landesman Y. Clinical translation of nuclear export inhibitors in cancer. Semin Cancer Biol. 2014;27:74–86. doi: 10.1016/j.semcancer.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 34.Noske A, Weichert W, Niesporek S, et al. Expression of the nuclear export protein chromosomal region maintenance/exportin 1/Xpo1 is a prognostic factor in human ovarian cancer. Cancer. 2008;112(8):1733–1743. doi: 10.1002/cncr.23354. [DOI] [PubMed] [Google Scholar]
- 35.Shen A, Wang Y, Zhao Y, Zou L, Sun L, Cheng C. Expression of CRM1 in human gliomas and its significance in p27 expression and clinical prognosis. Neurosurgery. 2009;65(1):153–159. doi: 10.1227/01.NEU.0000348550.47441.4B. discussion 159-60. [DOI] [PubMed] [Google Scholar]
- 36.Yao Y, Dong Y, Lin F, et al. The expression of CRM1 is associated with prognosis in human osteosarcoma. Oncol Rep. 2009;21(1):229–235. [PubMed] [Google Scholar]
- 37.Lapalombella R, Sun Q, Williams K, et al. Selective inhibitors of nuclear export show that CRM1/XPO1 is a target in chronic lymphocytic leukemia. Blood. 2012;120(23):4621–4634. doi: 10.1182/blood-2012-05-429506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kojima K, Kornblau SM, Ruvolo V, et al. Prognostic impact and targeting of CRM1 in acute myeloid leukemia. Blood. 2013;121(20):4166–4174. doi: 10.1182/blood-2012-08-447581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmidt J, Braggio E, Kortuem KM, et al. Genome-wide studies in multiple myeloma identify XPO1/CRM1 as a critical target validated using the selective nuclear export inhibitor KPT-276. Leukemia. 2013;27(12):2357–2365. doi: 10.1038/leu.2013.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hill R, Cautain B, De Pedro N, Link W. Targeting nucleocytoplasmic transport in cancer therapy. Oncotarget. 2014;5(1):11–28. doi: 10.18632/oncotarget.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chari A, Vogl DT, Gavriatopoulou M, et al. Oral selinexor-dexamethasone for triple-class refractory multiple myeloma. N Engl J Med. 2019;381(8):727–738. doi: 10.1056/NEJMoa1903455. [DOI] [PubMed] [Google Scholar]
- 42.Neggers JE, Vercruysse T, Jacquemyn M, et al. Identifying drug-target selectivity of small-molecule CRM1/XPO1 inhibitors by CRISPR/Cas9 genome editing. Chem Biol. 2015;22(1):107–116. doi: 10.1016/j.chembiol.2014.11.015. [DOI] [PubMed] [Google Scholar]
- 43.Sun Q, Carrasco YP, Hu Y, et al. Nuclear export inhibition through covalent conjugation and hydrolysis of leptomycin B by CRM1. Proc Natl Acad Sci U S A. 2013;110(4):1303–1308. doi: 10.1073/pnas.1217203110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jardin F, Pujals A, Pelletier L, et al. Recurrent mutations of the exportin 1 gene (XPO1) and their impact on selective inhibitor of nuclear export compounds sensitivity in primary mediastinal B-cell lymphoma. Am J Hematol. 2016;91(9):923–930. doi: 10.1002/ajh.24451. [DOI] [PubMed] [Google Scholar]
- 45.Argueta C, Kashyap T, Klebanov B, et al. Selinexor synergizes with dexamethasone to repress mTORC1 signaling and induce multiple myeloma cell death. Oncotarget. 2018;9(39):25529–25544. doi: 10.18632/oncotarget.25368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muqbil I, Aboukameel A, Elloul S, et al. Anti-tumor activity of selective inhibitor of nuclear export (SINE) compounds, is enhanced in non-Hodgkin lymphoma through combination with mTOR inhibitor and dexamethasone. Cancer Lett. 2016;383(2):309–317. doi: 10.1016/j.canlet.2016.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Agnelli L, Fabris S, Bicciato S, et al. Upregulation of translational machinery and distinct genetic subgroups characterise hyperdiploidy in multiple myeloma. Br J Haematol. 2007;136(4):565–573. doi: 10.1111/j.1365-2141.2006.06467.x. [DOI] [PubMed] [Google Scholar]
- 48.Barnhart BC, Simon MC. Taking aim at translation for tumor therapy. J Clin Invest. 2007;117(9):2385–2388. doi: 10.1172/JCI33107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu K Da, Zhou L, Burtrum D, Ludwig DL, Moore MAS. Antibody targeting of the insulin-like growth factor I receptor enhances the anti-tumor response of multiple myeloma to chemotherapy through inhibition of tumor proliferation and angiogenesis. Cancer Immunol Immunother. 2007;56(3):343–357. doi: 10.1007/s00262-006-0196-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pratt G. Molecular aspects of multiple myeloma. Mol Pathol. 2002;55(5):273–283. doi: 10.1136/mp.55.5.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramírez-Valle F, Braunstein S, Zavadil J, Formenti SC, Schneider RJ. eIF4GI links nutrient sensing by mTOR to cell proliferation and inhibition of autophagy. J Cell Biol. 2008;181(2):293–307. doi: 10.1083/jcb.200710215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Attar-Schneider O, Drucker L, Zismanov V, Tartakover-Matalon S, Rashid G, Lishner M. Bevacizumab attenuates major signaling cascades and eIF4E translation initiation factor in multiple myeloma cells. Lab Invest. 2012;92(2):178–190. doi: 10.1038/labinvest.2011.162. [DOI] [PubMed] [Google Scholar]
- 53.Wu M, Zhang C, Li XJ, Liu Q, Wanggou S. Anti-cancer effect of cap-translation inhibitor 4EGI-1 in human glioma U87 cells: involvement of mitochondrial dysfunction and ER stress. Cell Physiol Biochem. 2016;40(5):1013–1028. doi: 10.1159/000453158. [DOI] [PubMed] [Google Scholar]
- 54.Matassa DS, Amoroso MR, Agliarulo I, et al. Translational control in the stress adaptive response of cancer cells: a novel role for the heat shock protein TRAP1. Cell Death Dis. 2013;4(10):e851. doi: 10.1038/cddis.2013.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Issur M, Bougie I, Despins S, Bisaillon M. Enzymatic synthesis of RNAs capped with nucleotide analogues reveals the molecular basis for substrate selectivity of RNA capping enzyme: impacts on RNA metabolism. PLoS One. 2013;8(9):e75310. doi: 10.1371/journal.pone.0075310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tan K, Culjkovic B, Amri A, Borden KLB. Ribavirin targets eIF4E dependent Akt survival signaling. Biochem Biophys Res Commun. 2008;375(3):341–345. doi: 10.1016/j.bbrc.2008.07.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim J, McMillan E, Kim HS, et al. XPO1-dependent nuclear export is a druggable vulnerability in KRAS-mutant lung cancer. Nature. 2016;538(7623):114–117. doi: 10.1038/nature19771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Carroll M, Borden KLB. The oncogene eIF4E: using biochemical insights to target cancer. J Interferon Cytokine Res. 2013;33(5):227–238. doi: 10.1089/jir.2012.0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bjorklund CC, Lu L, Kang J, et al. Rate of CRL4CRBN substrate Ikaros and Aiolos degradation underlies differential activity of lenalidomide and pomalidomide in multiple myeloma cells by regulation of c-Myc and IRF4. Blood Cancer J. 2015;5(10) doi: 10.1038/bcj.2015.66. e354-e354. [DOI] [PMC free article] [PubMed] [Google Scholar]