Abstract
Background:
The World Health Organization (WHO) classification of central nervous system (CNS) tumors necessitates testing of isocitrate dehydrogenase (IDH) 1/2 gene mutation in patients with adult-type diffuse glioma (ADG) for better disease management. In clinical practice, the testing of IDH1 is primarily achieved using immunohistochemistry (IHC) specific to IDH1-R132, which carries a sensitivity of 80% and specificity of 100%. However, in some cases, non-specific background staining or regional heterogeneity in the protein expression of IDH1 may necessitate confirmatory genetic analysis. Robust and reliable assays are needed for IDH1/2 mutation testing. The aim of the current study was to detect IDH1 mutation in cfDNA and tissue of adult-type diffuse glioma with allele-specific qPCR.
Materials and Methods:
In the current study, IDH1-R132H mutation was analyzed in tumor tissue with paired cell-free DNA (cfDNA) in patients with ADG (n = 45) using IHC and competitive allele-specific Taqman PCR (CAST-PCR). Genomic DNA was extracted from formalin-fixed paraffin-embedded (FFPE) tissue and matched serum for cfDNA using commercially available kits. CAST-PCR with IHC for the detection of IDH1-R132H mutation was also compared.
Results:
The IDH1-R132H mutation was detected in 46.67% (21/45) cases and 57.78% (26/45) cases using IHC and allele-specific CAST-PCR. In cfDNA of matched IDH1-mutant FFPE tissue DNA, IDH1-R132H mutation was detected in 11.54% (3/26) using CAST-PCR. The concordance rate for IDH1-R132Hmutation between IHC and CAST-PCR was 80.77% (21/26).
Conclusion:
The CAST-PCR assay is more precise and sensitive for IDH1-R132Hdetection than traditional IHC, and IDH1-R132H mutation detection using cfDNA may add to the current methods of glioma genomic characterization.
Key Words: Adult, type Diffuse Glioma, IDH1/2, Cell-free DNA, CAST-PCR, Mutation diagnostics
Introduction
Adult-type diffuse gliomas (ADG) are characterized predominantly by astrocytic or oligodendroglial morphology (Rodriguez et al., 2016). Molecular parameters were included in the classification of central nervous system (CNS) tumors in the update 2016 CNS World Health Organization (WHO), which has broken the century-old rule of diagnosis dependent only on microscopy (Louis et al., 2016), and the last update in 2021 has further increased the diagnostic use of genetic alterations (Brat et al., 2020; Louis et al., 2021). Molecular markers in the CNS tumor have both diagnostic and prognostic importance. Various clinical decisions are being made based on the results of these biomarkers, whether for diagnosis, treatment, or prediction monitoring (Fan et al., 2020; P. Yang et al., 2016). The inclusion of isocitrate dehydrogenase (IDH) 1/2 as the “molecular signature” of gliomas and mutations in either IDH1 or IDH2 as the primary prognostic factor and molecular diagnostic criterion for ADG in the 2016 WHO classification marked a significant departure from the previous morphology-alone classification (Louis et al., 2016; Parsons et al., 2008). IDH1/2 mutation is a feature of ‘oligodendroglioma, IDH-mutant, and 1p19q-codeleted’ and ‘astrocytoma IDH-mutant’ while IDH-wildtype is a key diagnostic marker for ‘Glioblastoma, IDH-wildtype’ and ‘Diffuse pediatric-type high-grade glioma, H3-wildtype, and IDH-wildtype’ as recommended by the 2021 WHO classification of CNS (Brat et al., 2020; Louis et al., 2021).
Determining IDH1 mutation is crucial for diagnosis and selecting an appropriate treatment strategy. Typically, the first step in treating a glioma is to perform the safest radical resection to provide enough tumor tissue for a reliable diagnosis. Regardless of tumor grade, any glioma expressing IDH-wildtype should be regarded as glioblastoma, IDH-wildtype (Louis et al., 2021) and treated with aggressive chemoradiotherapy according to the Stupp protocol (Stupp et al., 2005, 2014). The treatment of gliomas expressing mutated variations of IDH should be guided by the presentation of clinical and molecular features. For radically resected low-grade tumors exhibiting both the 1p/19q co-deletion and an IDH mutation, one might even consider omitting oncotherapy altogether and recommend watchful follow-up (Weller et al., 2017; Weller et al., 2021).
In clinical practice, the testing of IDH1/2 mutation is primarily based on immunohistochemistry (IHC) specific for IDH1/2 protein expression, which is limited in terms of sensitivity, and cross-reactivity. In some cases, non-specific background staining or regional heterogeneity in IDH1-R132H protein expression may necessitate confirmatory genetic analysis. Assays such as competitive allele-specific TaqMan Polymerase chain reactions (CAST-PCR) are characterized by their high sensitivity and specificity to detect minimal amounts of mutated DNA in a sample containing large amounts of normal wildtype DNA (Barbano et al., 2015; Bolton et al., 2015). It can robustly detect mutant alleles at values as low as 0.1% in a wildtype background and has >99% concordance with other technologies, including technology based on digital PCR and Sanger sequencing (Yang et al., 2018).
In recent years, various approaches have been developed, including “liquid biopsies” (nucleic acid extracted from biological fluids such as plasma, urine, and cerebrospinal fluid (CSF)) for detecting the IDH mutation (Satomi et al., 2022); D2HG detection in body fluids; and advanced MRI imaging with specific D2HG detection by magnetic resonance spectroscopy (MRS) (Fujita et al., 2022; Mithraprabhu et al., 2021; Tuna et al., 2022). However, none of them is currently used in clinical practice. A non-invasive, rapid, sensitive, and cost-effective method for IDH mutation analysis is needed to enhance diagnosis and predict survival. Analysis of cell-free nucleic acids has entered clinical practice in tumors like lung and colon cancer (Kolenčík et al., 2020; Kwapisz, 2017). The current study evaluated the IDH1-R132H mutation in cfDNA and respective tissue of adult-type diffuse glioma using a CAST-PCR assay. Further, the sensitivity and effectiveness of the CAST-PCR assay were compared with IHC for IDH1-R132H.
Materials and Methods
Study samples
Our study included histologically confirmed ADG patients (n=45) according to the WHO 2021 CNS classification. When the study commenced, cases were diagnosed based on the WHO 2016 classification of CNS tumors (Louis et al., 2016). They have been reclassified as per WHO 2021 CNS classification, and the staining of IDH1-IHC was used for IDH1 status in all cases (Louis et al., 2020; Louis et al., 2021). All IDH-mutated astrocytomas have been categorized as astrocytoma IDH-mutant. IDH-wildtype cases were further classified on the histological features, including proliferation, necrosis, and mitosis. The Institutional ethics committee cleared the study (IEC No.26/18), and all study participants gave informed consent and have therefore been performed under the ethical standards of the Declaration of Helsinki (World Medical Association, 2013)
Sample collection
Formalin-fixed paraffin-embedded (FFPE) tumor tissue blocks were obtained from the departmental tumor archive after histopathological diagnosis, and peripheral blood (3.0ml) was collected in silica gel vials (BD Vacutainer, UK) from post-op patients who underwent biopsy or with remnant tumor. Samples were collected within 22-65 days (Q1-Q3) after the procedure. The serum was separated by centrifugation at 4,000 rpm for 10 minutes and stored at −80°C until further processing. All the samples were processed within 02 hours of collection.
Immunohistochemistry (IHC) of Isocitrate Dehydrogenase 1 (IDH1)
Immunohistochemical analysis of IDH1 was performed using Anti-IDH1 (R132H) (Dianova, USA Clone: H09-unconj) in a dilution of 1:50. All Immunohistochemical assays were performed on a VENTANA BenchMark XT automated staining instrument according to the manufacturer’s instructions with onboard deparaffinization, retrieval, and staining (Ventana Medical Systems, Inc., Tucson, USA). An expert pathologist (NH) independently analyzed all immunohistochemically stained sections.
Staining interpretation of IDH1
The staining interpretation of IDH was as follows: Mutant: intense cell cytoplasm and nucleus of tumor cells, wildtype: weak diffuse staining of cytoplasm and staining of macrophages. Normal/residual glial and vascular endothelial cells were internal negative controls.
DNA extraction from formalin-fixed paraffin-embedded (FFPE) tissue and serum
FFPE tumor tissue was used to extract genomic DNA. The tumor area was defined on a Hematoxylin and Eosin (HE) stained slide and a corresponding section marked for DNA isolation. DNA was extracted using the QIAamp DNA FFPE Tissue Kit (Qiagen, Germany, Cat No. #56404). Serum cell-free DNA (cfDNA) was extracted using the ChargeSwitch® gDNA 1 mL Serum Kit (Invitrogen, USA) as per the manufacturer’s instructions. The quality and quantity of the DNA were measured using Nanodrop (DeNoVix, USA, Model #DS-11).
IDH1 mutation detection by CAST-PCR
IDH1-R132H mutation was determined in 45 FFPE tissue DNA with paired cfDNA using CAST-PCR assay (TaqMan® Mutation Detection Assay). Each case was amplified using a reference assay (#Hs00001019_rf) and a mutation assay (#Hs00000981_mu) in a 20µl reaction, using a 10µl of genotyping master mix (#4371353), 1µl of primer and probe for reference and mutation assay, 1-9 µl of template DNA (up to 50ng) and volume were brought to 20μl by nuclease-free water. Real-time amplification was performed using the AriaMx Real-Time PCR system (Agilent Technologies, USA) as per the manufacturer’s instructions. In each batch, NTC was run to check for cross-contamination (Figure 1i). ΔCT values were calculated by CT (mutant allele assay)– CT (gene reference assay). A case was defined as mutated if the ΔCT values were ≤9.96 (TaqMan Mutation Detection Assays, #4467012, Applied Biosystems, USA).
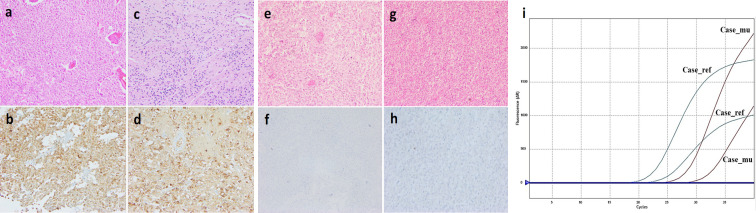
Figure 1.
Immunohistochemical Analysis by Anti-IDH Antibody against Glioma tissue a,b) Oligodendroglioma, IDH-mutant, 1p19q co-deleted, 2021 WHO CNS Grade 3; c,d) Oligodendroglioma, IDH-mutant, 1p19q co-deleted, 2021 WHO CNS Grade 2; e,f) Glioblastoma, IDH-wildtype, 2021 WHO CNS Grade 4; g,h) Glioblastoma, IDH-wildtype 2021 WHO CNS Grade 4; i) Amplification plots of Competitive Allele-Specific TaqMan qPCR
Statistical analysis
The IBM SPSS v22 Statistical Package for Social Sciences (SPSS, IBM, USA) was used for the statistical analysis. Cohen’s kappa statistics tested the agreement between IHC and CAST-PCR in FFPE and cfDNA. Reverse Kaplan-Meier was used to compute the median follow-up time. Kaplan-Meier and log-rank analyses were performed to assess survival concerning each parameter; differences were considered significant when p ≤ 0.05.
Results
Patient’s characteristics
Histologically confirmed oligodendroglioma (n=10), diffuse astrocytoma and (n=11) glioblastoma (n=24) were enrolled. As shown in Table 1, most patients (24/45) were ≤40 years old, with a male predominance (36/45). Most of the patients underwent GTE (36/45), the site of the tumor was frontal (19/45), temporal (05/45), parietal (02/45), and multiple (19/45), and all patients underwent a planned regimen. The median follow-up duration was 34 months (95%CI: 32-36). Among 45 patients, 28 (62.22%) died during the follow-up. The average OS was 22 months [95% CI: 18–26].
Table 1.
The Distribution of ADG Cases According to Clinical Findings
| Characteristics | N (%) | |
|---|---|---|
| Age | ≤40 Years | 24 (53.33) |
| >40 Years | 21 (46.67) | |
| Median (Q1-Q3) | 40.00 (32.25-54.00) | |
| Gender | Males | 36 (80.00) |
| Females | 09 (20.00) | |
| Karnofsky Performance Score | ≤80 | 31 (68.89) |
| >80 | 14 (31.11) | |
| Frontal | 19 (42.22) | |
| Temporal | 05 (11.11) | |
| Tumor Site | Parietal | 02 (4.44) |
| Multiple | 19 (42.22) | |
| Extent of Excision | Gross Total Excision | 36 (80.00) |
| Partial Excision/Biopsy | 09 (20.00) | |
| WHO CNS 2021 | Grade 2 | 13 (28.89) |
| Grade | Grade 3 | 07 (15.55) |
| Grade 4 | 25 (55.55) | |
| WHO CNS 2021 | Astrocytoma IDH-mutant | 11 (24.44) |
| Classification | Oligodendroglioma IDH-mutant 1p19q Deleted | 10 (22.22) |
| Glioblastoma IDH-wildtype | 24 (53.33) | |
| Adjuvant Radiotherapy | Radiotherapy | 15 (33.33) |
| Radiotherapy+Chemotherapy | 30 (66.67) | |
| Survival | Exitus | 28 (62.22) |
| Alive | 17 (37.78) |
IDH1 mutation in formalin-fixed paraffin-embedded (FFPE) tissue
Out of 45 cases, IHC successfully detected IDH1-R132H mutation in 46.67% (21/45) of the cases. An IDH1-R132H mutation was determined in FFPE tissue DNA and cfDNA. The CAST-PCR assay successfully detected the IDH1-R132H mutation in 26/45 (57.78%) of FFPE DNA. Five cases showed mutations in the CAST-PCR test, but IHC showed they were all wildtype (Figure 1e-h). The rate of concordance for IDH1-R132H mutation between IHC and CAST-PCR was 80.77% (21/26) (p= 0.000; k= 0.780) (Table 2 and Supplementary Table 1).
Table 2.
Agreement Test of IDH1-R132H between Immunohistochemistry and CAST-PCR Assay
| CAST-PCR Assay for IDH1 | Total | p-Value | Kappa | |||
|---|---|---|---|---|---|---|
| Mutant | Wildtype | |||||
| Mutant | 21 | 0 | 21 | |||
| IHC for IDH1 | Wildtype | 5 | 19 | 24 | 0.000 | 0.780 |
| Total | 26 | 19 | 45 | |||
IDH1 mutation in cell-free DNA (cfDNA)
In the CAST-PCR assay, 03/26 (11.54%) cfDNA samples were found as IDH1-R132H mutated (Figure 1a-d). Total cfDNA level of IDH1-wildtype (n =19) and mutant cases (n = 26) were 470.30±687.30ng/ml, and 175.00±206.90ng/ml respectively (p=0.04). Out of 26 mutated cases, IDH1-mutated cfDNA (n = 03) had mean cfDNA values of 189.70 (±235.80) ng/ml while wild type cfDNA (n = 23) have mean cfDNA value of 173.10 (±208.70) ng/ml (p = 0.89). There was slight agreement between the cfDNA and FFPE DNA for IDH1 mutation detection (p =0.125; k = 0.099) (Tables 3, 4 and Supplementary Table 1).
Table 3.
Agreement Test of CAST-PCR Assay between FFPE DNA and cfDNA
| CAST-PCR Assay for IDH1 in FFPE DNA | Total | p-Value | Kappa | |||
|---|---|---|---|---|---|---|
| Mutant | Wildtype | |||||
| CAST-PCR Assay for IDH1 in cfDNA | Mutant | 3 | 0 | 3 | ||
| Wildtype | 23 | 19 | 42 | 0.125 | 0.099 | |
| Total | 26 | 19 | 45 | |||
Table 4.
Distribution of IDH Mutation in cfDNA and FFPE Tumor Tissue of ADG
| Adult-type diffuse glioma | WHO Grade | IHC-IDH (MT/WT) |
CAST-PCR-IDH (MT/WT) |
Mean cfDNA (ng/mL) |
IDH in cfDNA (MT/WT) |
|---|---|---|---|---|---|
| Astrocytoma IDH-mutant | Grade 2 (n=6) | 6/0 | 6/0 | 280.80±361.80 | 0/6 |
| Grade 3 (n=4) | 4/0 | 4/0 | 79.57±50.55 | 0/4 | |
| Grade 4 (n=1) | 1/0 | 1/0 | 66.70±0.00 | 0/1 | |
| Oligodendroglioma IDH-mutant 1p19q co-deleted |
Grade 2 (n=7) | 7/0 | 7/0 | 151.50±143.90 | 1/6 |
| Grade 3 (n=3) | 3/0 | 3/0 | 293.20±203.20 | 1/2 | |
| Glioblastoma IDH-wildtype | Grade 4 (n=24) | 0/24 | 5/19 | 394.80±626.90 | 1/23 |
Survival
The median follow-up duration was 34 months (95%CI 32-36). Among 45 patients, 28 (62.22%) died during the follow-up. The average OS was 22 months [95% CI 18–26]. Patients in the lower age group (40 years) had better survival than patients in the higher age group (> 40years) in the Kaplan-Meier survival analysis (log-rank p = 0.010). Increased survival trends were observed with an increase in KPS score (log-rank p= 0.001), while survival decreased with an increase in WHO grade (log-rank p=0.001). IDH1-mutant patients had better survival than IDH1-wildtype patients (log-rank p=0.000). Gender (log-rank p= 0.332) and extent of excision (log-rank p=0.429) did not show a relationship to the patient’s overall survival, as depicted in Table 5 & Figure 2.Patients with IDH-mutation in cfDNA showed OS survival of 25 months (95% CI; 11-39) and IDH-wildtype cfDNA showed OS of 22 months (95% CI; 18-26) (log-rank p=0.35).
Table 5.
Predictors of Time to Overall Survival in Patients with Adult-Type Diffuse Glioma
| Variables | N (%) | Overall Survival | ||
|---|---|---|---|---|
| Median (Months) | p-Value (Log-rank) | |||
| Age | ≤44 Years | 24 (53.33) | 31 | 0.01 |
| >44 Years | 21 (46.67) | 12 | ||
| Gender | Male | 36 (80.00) | 22 | 0.332 |
| Female | 09 (20.00 | 15 | ||
| Karnofsky Performance Score |
70 | 15 (33.33) | 10 | 0.001 |
| 80 | 16 (35.56) | 17 | ||
| 90 | 14 (31.11) | - | ||
| Excision | GTE | 36 (80.00) | 22 | 0.429 |
| PE/Biopsy | 09 (20.00) | 14 | ||
| WHO CNS 2021 Type | Astrocytoma IDH-mutant | 11 (24.44) | - | 0.002 |
| Oligodendroglioma IDH-mutant 1p19q Deleted | 10 (22.22) | 24 | ||
| Glioblastoma IDH-wildtype | 24 (53.33) | 12 | ||
| WHO CNS 2021 Grade | 2 | 13 (28.89) | - | 0.001 |
| 3 | 07 (15.55) | 31 | ||
| 4 | 25 (55.55) | 13 | ||
| IHC-IDH | Mutant | 21 (46.67) | - | 0.000 |
| Wildtype | 24 (53.33) | 12 | ||
| CAST-PCR-IDH | Mutant | 26 (57.78) | 31 | 0.003 |
| Wildtype | 19 (42.22) | 12 | ||
GTE, Gross Total Excision; PE, Partial Excision; RANO, Response Assessment in Neuro-Oncology; IDH1, Isocitrate Dehydrogenase- 1
Figure 2.
Kaplan-Meier Curves. Overall survival rate (OS) for patients with adult-type diffuse glioma. a) OS according to age. b) OS according to gender. c) OS according to excision extent. d) OS according to KPS. e) OS according to WHO 2021 grade. f) OS according to IHC-IDH1 expression. g) OS according to WHO 2021 adult-type diffuse glioma. h) OS according to CAST-PCR-IDH1 expression. The log-rank test calculated p-values
Discussion
It has been found that IDH1 gene mutations are common in diffuse gliomas (Parsons et al., 2008). Based on the mutation profiles of glioma subtypes and primary and recurrent tumors, IDH1 mutations are considered one of the most significant genetic modifications in gliomagenesis (Johnson et al., 2014; Yan et al., 2009) that can aid in the diagnosis and prognosis. In addition, patients with known IDH1 mutations may benefit from new IDH1-targeted chemotherapeutic regimens under development (Golub et al., 2019; Karpel-Massler et al., 2019; Mellinghoff et al., 2020).
A study by Matthias Preusser et al., (2011) showed the need for confirmatory genetic analysis in cases with non-specific background staining and/or regional heterogeneity of IDH1-R132H expression using the DIA H09 antibody (Preusser et al., 2011). Although Sanger sequencing is the “gold standard” for detecting mutations because of its low rate of false positives and high specificity, it has several drawbacks, including low sensitivity, long assay times, the necessity for high-quality tissue samples, and manual interpretation (Gao et al., 2016). Furthermore, next-generation sequencing (NGS), which is used to detect various mutations, has the drawback of taking too much time and being expensive to discover a single genetic variant. An alternative technique for mutation detection is a real-time CAST-PCR assay (Roma et al., 2013). Competitive allele-specific TaqMan PCR allows the selective amplification of minor alleles and blocks the amplification of non-mutant alleles.
In a significant subset of patients, immunohistochemical reactivity is not detected; it is likely but not certain that the tumor is IDH-wildtype. The study by Andrews and Prayson (2020) recommends PCR testing for all patients whose tumor is negative by IHC. However, this is not always performed, both because it is expensive and in some patient groups (e.g., elderly patients with tumors demonstrating necrosis), it is almost always negative (i.e., these are almost invariably glioblastoma). So, keeping this in mind, we decided to test the CAST PCR in IDH-negative tumors of all age groups.
In the current study, real-time PCR, combined with an MGB-blocking oligonucleotide to suppress the normal allele, was used to detect IDH1-R132H gene mutations in gliomas. The CAST-PCR identified 26/45 cases as IDH1 mutated, and IHC detected 21/45 cases with an IDH1 mutation. While both IHC and CAST-PCR assays confirmed 21 cases as IDH1 mutants. A total of 05 cases were reported as mutated in CAST-PCR while were wild type in IHC. The reason leading to this inconsistency may be due to the sensitivity and specificity of antibodies (Preusser et al., 2011).
Our results showed that the IDH1 mutation detection rate in gliomas was significantly different (p=0.000) between IHC and CAST-PCR (κ=0.780). A study by Agarwal et al., (2013) compared the performance of IHC and DNA sequencing for IDH1 mutation and found a concordance rate of 88% (44/50) (Agarwal et al., 2013). Similarly, in our study, the concordance rate between CAST-PCR and IHC in detecting IDH1 mutation was 80.77%. Moreover, the CAST-PCR assay was more sensitive than IHC in identifying the IDH1 mutation. Further, cfDNA and FFPE DNA show concordant results only in 11.54% of cases; this low concordance may be due to a lack of tumor-derived DNA or a low copy number of mutated DNA. All cases with cfDNA-positive for IDH1 on CAST-PCR were <55 years, including two cases of oligodendroglioma, IDH-mutant, 1p19q co-deleted, CNS WHO grade 2 and 3, respectively, and one case of Glioblastoma, IDH-wildtype CNS WHO grade 4 by IHC analysis.
We have included only the IDH1-R132H mutation in both IHC and CAST-PCR, and other mutations of IDH1 & IDH2 were not analyzed. However, these mutation types become secondary due to low frequency because their testing costs may burden the patients. Sanger sequencing is considered the gold standard for IDH1 & IDH2 mutation detection; however, we could not validate our results using the sequencing method.
Finally, the CAST-PCR technique for detecting glioma IDH1 gene mutations has high sensitivity, good reproducibility, ease of use, and accurate results. It offers a more precise method for detecting mutations in the IDH1 gene in formalin-fixed paraffin-embedded tissue samples but lacks sensitivity using an alternative source, cfDNA, in the case of tissue scarcity. Despite the small sample size, evidence suggests that CAST-PCR assays outperform IHC. More sensitive techniques may be required to detect IDH mutations in cfDNA.
Author Contribution Statement
Conceptualization, Funding acquisition, Project administration, Resources, Software, Supervision, Validation, Writing - original draft, Writing - review & editing: Nuzhat Husain; Data curation, Investigation, Methodology, Writing - original draft, Writing - review & editing: Adil Husain; Methodology, Writing - review & editing: Sridhar Mishra; Resources, Software, Supervision, Writing - review & editing: Mohammed Haris Siddiqui.
Acknowledgements
Funding statement
The authors would like to acknowledge the University Grant Commission, New Delhi, Government of India, for the JRF-NET Fellowship to Adil Husain (Fellowship No. NOV-2017-343185) and Dr. Ram Manohar Lohia Institute of Medical Sciences, Lucknow, for an Intramural Research Grant to Prof. Nuzhat Husain (26/18).
Ethics Approval
Institutional Ethics Committee of Dr. Ram Manohar Lohia Institute of Medical Sciences (RMLIMS), Lucknow (IEC-26/18) and Integral University, Lucknow (Manuscript Communication Number-IU/R&D/2022-MCN0001618).
Conflict of Interest
None.
References
- Agarwal S, Sharma MC, Jha P, et al. Comparative study of IDH1 mutations in gliomas by immunohistochemistry and DNA sequencing. Neurooncology. 2013;15:718–26. doi: 10.1093/neuonc/not015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews C, Prayson RA. IDH mutations in older patients with diffuse astrocytic gliomas. Ann Diagn Pathol. 2020;49:151653. doi: 10.1016/j.anndiagpath.2020.151653. [DOI] [PubMed] [Google Scholar]
- Barbano R, Pasculli B, Coco M, et al. Competitive allele-specific TaqMan PCR (Cast-PCR) is a sensitive, specific and fast method for BRAF V600 mutation detection in Melanoma patients. Sci Rep. 2015;5:1–11. doi: 10.1038/srep18592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton L, Reiman A, Lucas K, Timms J, Cree IA. KRAS Mutation Analysis by PCR: A Comparison of Two Methods. PLoS One. 2015;10:e0115672. doi: 10.1371/journal.pone.0115672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brat DJ, Aldape K, Colman H, et al. cIMPACT-NOW Update 5: Recommended Grading Criteria and Terminologies for IDH-mutant Astrocytomas. Acta Neuropathol. 2020;139:603–8. doi: 10.1007/s00401-020-02127-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Z, Liu Y, Li S, et al. Association of tumor growth rates with molecular biomarker status: a longitudinal study of high-grade glioma. Aging. 2020;12:7908–26. doi: 10.18632/aging.103110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Nunez-Rubiano L, Dono A, et al. IDH1 p R132H ctDNA and D-2-hydroxyglutarate as CSF biomarkers in patients with IDH-mutant gliomas. J Neurooncol. 2022;159:261–70. doi: 10.1007/s11060-022-04060-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Wu H, Wang L, et al. Validation of targeted next-generation sequencing for RAS mutation detection in FFPE colorectal cancer tissues: comparison with Sanger sequencing and ARMS-Scorpion real-time PCR. BMJ Open. 2016;6:e009532. doi: 10.1136/bmjopen-2015-009532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub D, Iyengar N, Dogra S, et al. Mutant Isocitrate Dehydrogenase Inhibitors as Targeted Cancer Therapeutics. Front Oncol. 2019;9:417. doi: 10.3389/fonc.2019.00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BE, Mazor T, Hong C, et al. Mutational Analysis Reveals the Origin and Therapy-Driven Evolution of Recurrent Glioma. Science. 2014;343:189–93. doi: 10.1126/science.1239947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpel-Massler G, Nguyen TTT, Shang E, Siegelin MD. Novel IDH1-Targeted Glioma Therapies. CNS Drugs. 2019;33:1155–66. doi: 10.1007/s40263-019-00684-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolenčík D, Shishido SN, Pitule P, et al. Liquid Biopsy in Colorectal Carcinoma: Clinical Applications and Challenges. Cancers. 2020;12:1376. doi: 10.3390/cancers12061376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwapisz D. The first liquid biopsy test approved Is it a new era of mutation testing for non-small cell lung cancer? Ann Transl Med. 2017;5:46. doi: 10.21037/atm.2017.01.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131:803–20. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- Louis DN, Wesseling P, Aldape K, et al. cIMPACT-NOW update 6: new entity and diagnostic principle recommendations of the cIMPACT-Utrecht meeting on future CNS tumor classification and grading. Brain Pathol. 2020;30:844–56. doi: 10.1111/bpa.12832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis DN, Perry A, Wesseling P, et al. The 2021 WHO Classification of Tumors of the Central Nervous System: a summary. Neurooncology. 2021;23:1231–51. doi: 10.1093/neuonc/noab106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellinghoff IK, Ellingson BM, Touat M, et al. Ivosidenib in Isocitrate Dehydrogenase 1–Mutated Advanced Glioma. J Clin Oncol. 2020;38:3398–06. doi: 10.1200/JCO.19.03327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mithraprabhu S, Chen M, Savvidou I, Reale A, Spencer A. Liquid biopsy: an evolving paradigm for the biological characterisation of plasma cell disorders. Leukemia. 2021;35:2771–83. doi: 10.1038/s41375-021-01339-6. [DOI] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, et al. An Integrated Genomic Analysis of Human Glioblastoma Multiforme. Science. 2008;321:1807–12. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preusser M, Wöhrer A, Stary S, et al. Value and Limitations of Immunohistochemistry and Gene Sequencing for Detection of the IDH1-R132H Mutation in Diffuse Glioma Biopsy Specimens. J Neuropath Exp Neur. 2011;70:715–23. doi: 10.1097/NEN.0b013e31822713f0. [DOI] [PubMed] [Google Scholar]
- Rodriguez FJ, Vizcaino MA, Lin MT. Recent Advances on the Molecular Pathology of Glial Neoplasms in Children and Adults. J Mol Diagn. 2016;18:620–34. doi: 10.1016/j.jmoldx.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roma C, Esposito C, Rachiglio AM, et al. Detection of EGFR Mutations by TaqMan Mutation Detection Assays Powered by Competitive Allele-Specific TaqMan PCR Technology. BioMed Res Int. 2013;2013:385087. doi: 10.1155/2013/385087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satomi K, Yoshida A, Matsushita Y, et al. Clinical application of a highly sensitive digital PCR assay to detect a small fraction of IDH1 R132H-mutant alleles in diffuse gliomas. Brain Tumor Pathol. 2022;39:210–7. doi: 10.1007/s10014-022-00442-5. [DOI] [PubMed] [Google Scholar]
- Stupp R, Weller M, Belanger K, et al. Radiotherapy plus Concomitant and Adjuvant Temozolomide for Glioblastoma. N Engl J Med. 2005;352:987. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- Stupp R, Brada M, van den Bent MJ, Tonn JC, Pentheroudakis G. High-grade glioma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2014;25:93–101. doi: 10.1093/annonc/mdq187. [DOI] [PubMed] [Google Scholar]
- Tuna G, Dal-Bekar NE, Akay A, et al. Minimally Invasive Detection of IDH1 Mutation With Cell-Free Circulating Tumor DNA and D-2-Hydroxyglutarate, D/L-2-Hydroxyglutarate Ratio in Gliomas. J Neuropath Exp Neur. 2022;81:502–10. doi: 10.1093/jnen/nlac036. [DOI] [PubMed] [Google Scholar]
- Weller M, van den Bent M, Tonn JC, et al. European Association for Neuro-Oncology (EANO) guideline on the diagnosis and treatment of adult astrocytic and oligodendroglial gliomas. Lancet Oncol. 2017;18:315–29. doi: 10.1016/S1470-2045(17)30194-8. [DOI] [PubMed] [Google Scholar]
- Weller M, van den Bent M, Preusser M, et al. EANO guidelines on the diagnosis and treatment of diffuse gliomas of adulthood. Nat Rev Clin Oncol. 2021;18:170–86. doi: 10.1038/s41571-020-00447-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Medical Association. World Medical Association Declaration of Helsinki: Ethical Principles for Medical Research Involving Human Subjects. JAMA. 2013;310:2191–4. doi: 10.1001/jama.2013.281053. [DOI] [PubMed] [Google Scholar]
- Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 Mutations in Gliomas. N Engl J Med. 2009;360:765–73. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Cai J, Yan W, et al. Classification based on mutations of TERT promoter and IDH characterizes subtypes in grade II/III gliomas. Neurooncology. 2016;18:1099–08. doi: 10.1093/neuonc/now021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Meng Y, Zhang H, et al. Detection of EGFR and BRAF mutations by competitive allele-specific TaqMan polymerase chain reaction in lung adenocarcinoma. Oncol Lett. 2018;15:3295–04. doi: 10.3892/ol.2017.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]