Graphical abstract
Keywords: Colorectal cancer, Gut virome, Diagnostic virome signature, Risk assessment model, Machine learning, Meta-analysis
Highlights
-
•
The gut virome research in colorectal cancer and adenoma includes >1,200 samples.
-
•
Siphoviridae and Microviridae viruses were notably different between controls and CRC patients.
-
•
The viral markers of colorectal cancer and adenoma were identified.
-
•
For CRC patients, our model had better predictive ability than other bacteria-based models.
-
•
The virome analysis achieved an optimal AUC of 0.772 to distinct adenoma patients and controls.
Abstract
Introduction
Viruses have been reported as inducers of tumorigenesis. Little studies have explored the impact of the gut virome on the progression of colorectal cancer. However, there is still a problem with the repeatability of viral signatures across multiple cohorts.
Objectives
The present study aimed to reveal the repeatable gut vial signatures of colorectal cancer and adenoma patients and decipher the potential of viral markers in disease risk assessment for diagnosis.
Methods
1,282 available fecal metagenomes from 9 published studies for colorectal cancer and adenoma were collected. A gut viral catalog was constructed via a reference-independent approach. Viral signatures were identified by cross-cohort meta-analysis and used to build predictive models based on machine learning algorithms. New fecal samples were collected to validate the generalization of predictive models.
Results
The gut viral composition of colorectal cancer patients was drastically altered compared with healthy, as evidenced by changes in some Siphoviridae and Myoviridae viruses and enrichment of Microviridae, whereas the virome variation in adenoma patients was relatively low. Cross-cohort meta-analysis identified 405 differential viruses for colorectal cancer, including several phages of Porphyromonas, Fusobacterium, and Hungatella that were enriched in patients and some control-enriched Ruminococcaceae phages. In 9 discovery cohorts, the optimal risk assessment model obtained an average cross-cohort area under the curve of 0.830 for discriminating colorectal cancer patients from controls. This model also showed consistently high accuracy in 2 independent validation cohorts (optimal area under the curve, 0.906). Gut virome analysis of adenoma patients identified 88 differential viruses and achieved an optimal area under the curve of 0.772 for discriminating patients from controls.
Conclusion
Our findings demonstrate the gut virome characteristics in colorectal cancer and adenoma and highlight gut virus-bacterial synergy in the progression of colorectal cancer. The gut viral signatures may be new targets for colorectal cancer treatment. In addition, high repeatability and predictive power of the prediction models suggest the potential of gut viral biomarkers in non-invasive diagnostic tests of colorectal cancer and adenoma.
Introduction
Colorectal cancer: Global statistics and socio-economic burden
Colorectal cancer (CRC) is one of the most common cancers with an increasing global incidence and mortality rate, especially in many low- and middle-income countries [1]. In 2018, a worldwide total of 1.85 million people were diagnosed with CRC [2]. Based on population aging and population growth projections, the number of new cases of colorectal cancer is expected to reach 3.2 million by 2040 [3]. To date, the exact etiology of colorectal cancer remains unclear, but genetic and environmental factors are thought to be important. Most colorectal cancer cases are sporadic and <25 % are inherited [4]. Although the prognosis of colorectal cancer treatment is generally favorable, the increasing number of cases and the rising incidence in the younger generation still bring a heavy financial burden and a huge public health challenge [5].
Bacteriome and virome as the disease risk factors
With the rise of next-generation sequencing technology, a large number of studies have confirmed that the dysbiosis of gut microbiome is related to the development of colorectal cancer and suggest that the gut microbiome can be used as biomarkers to build disease prediction models to predict colorectal cancer [6], [7], [8], [9]. Recent studies showed that Fusobacterium nucleatum was enriched in the human colon cancer tissues and stool samples of CRC patients compared to the healthy controls [10]. F. nucleatum recruited tumor-infiltrating myeloid cells in the mouse model, which accelerated carcinogenesis [10]. Pathogenic mechanisms of F. nucleatum in CRC have been widely reported, such as promoting CRC cell glucose metabolism and inducing formate secretion [11], [12]. These results provide insight into potential pathogenic mechanisms of gut bacterial communities in CRC. Interestingly, given that colonoscopy for CRC screening is an invasive diagnostic tool, many studies have evaluated the potential of bacterial CRC biomarkers as non-invasive diagnostic tests [6], [7], [13]. Through the meta-analysis of cross-cohort studies, Thomas et al. [6] and Wirbel et al. [7] identified accurate and reproducible CRC biomarker species in human fecal metagenomes, such as F. nucleatum, Parvimonas micra, Gemella morbillorum, Peptostreptococcus stomatis, Solobacterium moorei, and Porphyromonas asaccharolytica. Based on these biomarkers, the prediction models demonstrated good discrimination in distinguishing CRC patients from healthy controls. Additionally, a multi-cohort study for colorectal adenoma was also performed based on 16S rRNA gene sequencing [14], and revealed the applicability of adenoma-specific bacteria for diagnosing colorectal adenoma. These findings suggest a high diagnostic potential of gut bacterial communities in CRC and colorectal adenoma.
Viruses are crucial members of the human gut microbial ecosystem and are often underemphasized in previous studies. Recently, several human systemic diseases, such as rheumatoid arthritis, inflammatory bowel disease, and irritable bowel syndrome [15], [16], [17], [18], [19], have been associated with both gut bacteria and viruses. Although some studies have found a link between specific viruses and CRC [20], [21], [22], [23], only a few of them have been implicated in the CRC gut virome thus far. Hannigan et al. reported that alpha and beta diversities of gut virome were not significantly different between CRC patients and healthy controls [24]. The relative abundance of multiple gut viruses had a significant difference between CRC patients and healthy controls [24], [25], [26]. Furthermore, in three studies [24], [25], [26], prediction models based on CRC-associated viruses demonstrated an acceptable potential for classifiability of controls vs patients (0.802). Among three studies, the viral markers in Nakatsu et al.’s study failed to correctly classify CRC patients vs health controls among an additional three independent cohorts [25]. The reproducibility and predictive accuracy of microbial markers cannot be validated across multiple studies. This is because many biological confounders (e.g., host clinical parameters) can lead to false positives and the heterogeneity of data generation and processing can decrease the stability and reliability of results. To reduce the effect of biological and technical factors, previous studies performed meta-analyses on the gut bacteriome and identified CRC-associated changes that were consistent across populations [6], [7]. However, large-scale and cross-cohort studies focused on the CRC gut virome are still lacking. Thus, it is crucial to perform meta-analyses across studies to avoid biased associations between the gut virome and CRC.
The workflow and objective of the study
In this study, we downloaded 1,282 fecal metagenomes from 9 published cohorts, generated a nonredundant viral catalog, and profiled the gut viromes of each to characterize the gut viral signatures in CRC. By referring to the methods of the previous studies [6], [7], we performed meta-analyses to identify accurate CRC-associated gut viral signatures across 9 cohorts of CRC patients and healthy controls. To figure out the diagnostic potential of these viral biomarkers for CRC, we performed intra-dataset and cross-dataset prediction and validation based on these 9 cohorts. We further validated the efficiency of viral biomarkers in our newly recruited cohort and another recently published independent cohort.
Material and methods
Data collection
The publicly available datasets of 9 CRC studies, covering fecal metagenomes from 554 CRC patients, 182 adenomas patients, and 546 healthy controls, were downloaded from the NCBI SRA database. The following accession IDs: PRJEB6070 for Zeller_2014, PRJEB10878 for Yu_2015, PRJEB7774 for Feng_2015, PRJEB12449 for Vogtmann_2016, PRJNA389927 for Hannigan_2018, PRJNA447983 for Thomas_a_2019 and Thomas_b_2019, PRJEB27928 for Wirbel_2019, and PRJDB4176 for Yachida_2019 were used to download datasets from NCBI SRA database. The metadata of samples was obtained from the basic research studies or extracted from the NCBI BioSample database. Fecal metagenomes of the recently published validation cohort, Yang_2021, were downloaded from the NCBI SRA database with accession ID PRJNA763023.
Recruitment of an independent cohort
To verify the accuracy of our virus marker model in other data, a newly recruited independent cohort was adopted as the validation cohort. Individuals were recruited at Dalian University Affiliated Xinhua Hospital and Dalian Medical University between 2020 and 2021. A total of 27 CRC patients and 28 healthy controls were included.
Ethics statement
Written informed consent was obtained from all participants in this study. Ethical approval for this study was obtained from the Ethics Committees in the Dalian University Affiliated Xinhua Hospital [Approval no. 2022–04-01]. All procedures followed were in accordance with the Helsinki Declaration of 1975, as revised in 2008.
Metagenomic sequencing
DNA was extracted from fecal samples using a TIANamp Stool DNA Kit (TIANGEN, China). The quality of DNA was assessed with Qubit 2.0. The extracted DNA samples were stored at −80℃ until use. A sequencing library was generated using the NEB Next® Ultra™ DNA Library Prep Kit (NEB, USA) following the manufacturer’s recommendations, and index codes were added to each sample. Library quality was confirmed with an Agilent 2100. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using the Illumina PE Cluster Kit (Illumina, USA) according to the manufacturer’s instructions. After cluster generation, the DNA libraries were sequenced on the Illumina NovaSeq platform and 150 bp paired-end reads were generated.
Preprocessing and assembly
Raw reads were qualified via fastp v0.20.1 [27] with the options ‘-u 30 -q 20 -l 90 -y --trim_poly_g’, and human reads were further removed by matching quality-filtered reads against the human genome GRCh38 with bowtie2 v2.4.1 [28]. The remaining clean reads of each sample were assembled into contigs using Megahit v1.2.9 with the options ‘--k-list 21, 41, 61, 81, 101, 121, 141′[29].
Analyses of viral sequences
All assembled contigs (≥5 kb) were used to identify viral sequences in each sample. The detection, decontamination, and clustering of viral sequences were performed as described in our previous study[30]. Taxonomic and functional annotation of viral sequences were also implemented based on the criteria described in our previous study[30]. Virus-host prediction was performed based on the Unified Human Gastrointestinal Genome (UHGG) database[31] using two methods that included CRISPR-spacer matches and prophage blasts (see the methods of our previous study for the detailed flow[30]).
Taxonomic profiling and diversity
Clean reads in each sample were mapped to the non-redundant viral catalog of 37,030 vOTUs using bowtie2 with the options ‘--end-to-end --fast -–no-head -–no-unal -–no-sq. The abundance profile of vOTUs in each sample was generated by aggregating the number of reads mapped to each vOTU. Random subsampling of each sample was implemented to achieve a parity number of reads that equals the minimum number of mapped reads among all samples. After subsampling, the relative abundance of vOTUs was its abundance divided by the number of total mapped reads in each sample. The relative abundance profile at the family level was generated by aggregating the relative abundance of vOTUs assigned to the same family. Alpha diversity indexes were assessed based on the relative abundance profiles at the vOTU level. The Shannon index was calculated using the function diversity with option ‘index = shannon’ in the R platform. The number of observed vOTUs was the count of unique vOTUs in each sample. In addition, the taxonomic profiling of the bacteriome was performed based on 4,644 prokaryotic genomes from UHGG [31] by the aforementioned methods. The alpha diversity indexes of the bacteriome were calculated based on the relative abundance profiles at the species level.
Identification of the biomarkers
Based on vOTUs profile, the statistically different vOTUs among the groups were selected from each of the nine public fecal shotgun CRC datasets by a Wilcoxon rank-sum test. Among these, we retained 516 vOTUs that existed in more than four datasets with consistent trends between patients and controls and had a P value of < 0.05 for Wilcoxon rank-sum test. To overcome the limitations of single research, these vOTUs were further filtered by meta-analysis. First, the relative abundances of 516 vOTUs were transformed using the arcsine-square root-transformation method. Then we compared the transformed abundances of each vOTU between healthy controls and patients based on Hedges' g effect sizes using the escalc function with the parameter ‘measure=SMD’ in R platform. Heterogeneity across studies was defined based on Cochran’s Q test and I2 statistics using the rma function in R platform. P values were adjusted by Benjamini–Hochberg procedure. We observed 407 vOTUs with adjusted P values of < 0.01.
To avoid confounding due to intra-individual variation (i.e., gender, age, and BMI), the associations between 407 vOTUs and CRC were tested further by linear regression analysis. In short, the vOTUs profiles were log10 transformed after adding a pseudocount of 1 × 10-10, and then used to fit linear models with adjustment for gender, age, and BMI via the lmFit function in R platform. After getting the coefficient and corresponding standard error, the meta-analysis was performed to calculate the pooled coefficient and their 95 % confidence interval. Finally, 405 vOTUs were kept as final CRC biomarkers with the pooled coefficients with 95 % CI that did not contain zero after adjusting for confounders.
Correlation network analysis
We used three methods to evaluate whether there is a relationship between CRC-associated vOTUs and prokaryotes. 1) The aforementioned host assignment. 2) Based on the SparCC algorithm[32], co-abundance relationships were established on the read count profiles of vOTUs and prokaryotes using fastspar v0.0.10 [33] with the option ‘--iterations 20′, while fastspar_pvalues v0.0.10 was used to calculate p-value by 1,000 bootstrap datasets derived from fastspar_bootstrap v0.0.10. The co-abundance relationships with the threshold of the correlation coefficient > 0.35 and q-value < 0.01 (p-value adjusted by the function p.adjust with the option “method = BH”) were retained. 3) To identify co-occurrence relationships, the presence and absence of each vOTU and prokaryote were treated as binary traits in each sample, and then compiled into a contingency table that displayed the numbers of microbial populations. The pairwise co-occurrence relationship was assessed based on the contingency table using Fisher's exact test by the function fisher.test in R platform, while p-value was adjusted by the function p.adjust with the option “method = BH” in the R platform. If the odds ratio of co-occurrence pair was>100 and the q-value was<0.0001, the two microbial populations were considered to be a co-occurrence. Finally, these relationships between CRC-associated vOTUs and prokaryotes were visualized using Cytoscape v3.8.2[34].
Prediction modeling
The Random forest model (randomForest package in the R platform, ntree = 2,000) and the L1-regularized (LASSO) logistic regression model (SIAMCAT[35] in the R platform, the same parameter setting derived from Wirbel’s study [7]) were used to build the classifier based on the vOTUs abundance profile. In LASSO model, the relative abundance of vOTUs were log10-transformed after adding a pseudocount of 1 × 10-10, and then standardized into z-scores. In random forest model, the profile was used without any pretreatment process.
The performances of prediction models were quantified by calculating AUC scores using the pROC package in R platform. In short, the data were divided into training and testing sets for five times repeated, fivefold stratified cross-validation. For each split, a machine learning model was trained on the training set, which was then used to predict the test set. In cross-cohort prediction, a model built from one dataset is used to respectively predict the other eight datasets. In the LOCO setting, one cohort serves as the test set and the remaining eight datasets serve as the training set by using five times-repeated fivefold stratified cross-validations.
Data and code availability
The raw metagenomic sequencing data from the independent cohorts has been submitted to the China Nucleotide Sequence Archive with the accession code CNP0002641. The authors declare that all other data supporting the findings of the study are available in the paper and supplementary materials, or from the corresponding authors upon request.
Results
Collection of datasets
In this meta-analysis study, we included 9 published datasets that used whole-metagenome shotgun sequencing to characterize the fecal microbial communities of patients with colorectal cancer or adenoma (Table 1; Table S1). Participants from all studies were diagnosed by colonoscopy or alternative methods, and the controls were confirmed after the absence of disease. Healthy subjects with a history of colorectal surgery in Yachida et al.’s study were excluded.
Table 1.
Summary of sample characteristics of data sets included in this study.
| Dataset | Country | Number of samples |
Data per sample (Gbp) | Total data amount (Gbp) | NCBI accession ID | ||
|---|---|---|---|---|---|---|---|
| CRC | Adenoma | Control | |||||
| Zeller_2014[36] | France | 33 | 33 | 35 | 1.5 ± 1.2 | 152.8 | PRJEB6070 |
| Yu_2015[37] | China | 74 | – | 54 | 4.6 ± 1.0 | 581.9 | PRJEB10878 |
| Feng_2015[38] | Austria | 46 | 47 | 63 | 4.2 ± 0.7 | 654.4 | PRJEB7774 |
| Vogtmann_2016[39] | USA | 23 | – | 16 | 1.1 ± 0.3 | 43.2 | PRJEB12449 |
| Hannigan_2018[24] | USA, Canada | 16 | 8 | 16 | 0.8 ± 0.3 | 30.0 | PRJNA389927 |
| Thomas_a_2019[6] | Italy | 29 | 27 | 24 | 4.6 ± 2.7 | 365.5 | PRJNA447983 |
| Thomas_b_2019[6] | Italy | 32 | – | 28 | 3.9 ± 1.7 | 235.0 | PRJNA447983 |
| Wirbel_2019[7] | Germany | 43 | – | 60 | 2.5 ± 1.3 | 259.5 | PRJEB27928 |
| Yachida_2019[8] | Japan | 258 | 67 | 250 | 6.3 ± 1.6 | 3,535.3 | PRJDB4176 |
| Overall | 554 | 182 | 546 | 4.6 ± 2.3 | 5,857.5 | ||
To ensure consistency, the metagenomic data from these studies were processed using a consistent protocol, and fecal metagenomes were excluded if the proportion of human DNA sequences exceeded 10 % or the number of high-quality reads was<4 million. A total of 1,282 samples, including 554 colorectal cancer (CRC) patients, 182 adenoma patients, and 546 healthy controls, containing nearly 5.9 Tbp of data, were retained for further analysis (Table 1).
Diversity and overall structure of gut viral community in relation to CRC and adenoma.
To characterize the gut viral communities, we performed de novo assembly of the fecal metagenomes (generating a total of 5.5 million contigs at a minimum length of 5,000 bp; Table S1) and identified 208,048 viruses from the assembled contigs based on the homolog-based and machine learning-based methods (see Methods). A nonredundant viral catalog of 37,030 viral operational taxonomic units (vOTUs; average length of 30,111 bp; N50 length of 44,627 ranging from 5,000 bp to 410,947 bp; Fig. S1A) was then generated under the species-level nucleotide similarity threshold of 95 %[40], [41]. The quality levels of these vOTU sequences were estimated by CheckV[42], which resulted in 6.7 % complete, 21.1 % high-quality, 19.3 % medium-quality, and 52.8 % low-quality viral genomes, and 0.2 % quality-undetermined sequences (Fig. S1B). Taxonomically, 45.9 % of these vOTUs were assigned to a known viral family, the majority of them consisted of Siphoviridae, Myoviridae, Podoviridae, Quimbyviridae, and crAss-like viruses (Fig. S1C). The currently available collections of human gut virome, including the Gut Virome Database[40] (covering 13.97 % vOTUs in this study), Gut Phage Database[41] (covering 35.72 % vOTUs in this study), and Metagenomic Gut Virus catalog[43] (covering 25.93 % vOTUs), identified 43.86 percent of 37,030 vOTUs (Fig. S1D).
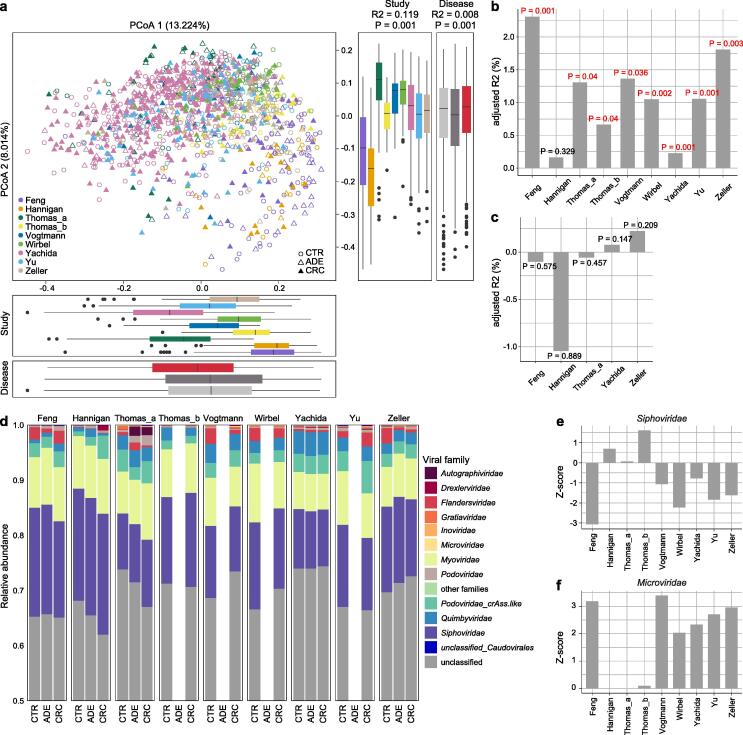
Fig. 1.
Viral community variation among the nine study populations. (A) Principal coordinates analysis (PCoA) based on the Bray-Curtis distance at the vOTU level. CTR, healthy controls; ADE, adenoma patients; CRC, colorectal carcinoma patients. Effect size (R2) and statistical significance were obtained by PERMANOVA (adonis). (B to C), Effect size (adjusted R2) of CRC status (B) and adenoma status (C) versus healthy controls on gut viral composition in each dataset. (D) The family-level composition of gut virome in each dataset. (E to F), Comparison of relative abundance of Siphoviridae (E) and Myoviridae (F) between healthy controls and CRC patients in each dataset. Absolute z-score above 2 was considered as statistically significant. Z-score above 0 was considered CRC-enriched, while Z-score below 0 was considered control-enriched.
Two parameters, the observed number of vOTUs and the Shannon index, were used to estimate the within-sample (alpha) diversity of the gut viral community of 9 analyzed datasets. The observed number of vOTUs in the virome of CRC patients was significantly higher in two datasets (Wilcoxon rank-sum test, p < 0.001 in Feng_2015, p = 0.006 in Thomas_b_2019), but showed no consistent trend in other studies; while the Shannon index showed no significant difference between two groups in all datasets. (Fig. S2A). Likewise, both diversity parameters were approximately equal in the viromes of adenoma patients and healthy controls (Fig. S2B). Furthermore, we found that the viral diversity parameters of samples were highly consistent with their bacterial diversities (Fig. S2C-D), suggesting an extensive connection between the virome and the bacterial microbiome.
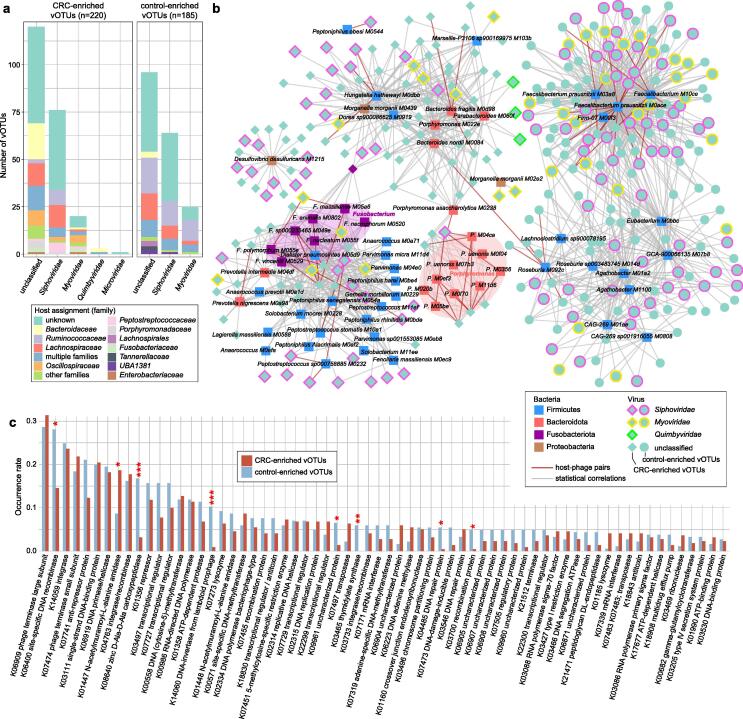
Fig. 2.
The CRC-associated viral signatures. (A) The family-level taxonomy and host assignment of CRC-associated vOTUs. The vOTUs are grouped at the family level, and their hosts are shown at the family level. The numbers of vOTUs that had more than one predicted host are colored in blue (multiple families). (B) The interaction network between CRC-associated viruses and bacteria. The network was constructed based on host-phage pairs and statistical co-abundance (SparCC r > 0.35 and q < 0.01) or co-occurrence (Fisher’s exact test, odd ratio > 100 and q < 0.01) correlations between viruses and bacteria. (C) The comparison of the occurrence rate of KOs detected in no<10 CRC-associated vOTUs between CRC-enriched and control-enriched vOTUs. Statistical test was performed using Fisher’s exact test, and adjusted using the FDR method. * q < 0.05, ** q < 0.01, *** q < 0.001.
Principal coordinates analysis (PCoA) showed a substantial difference in viral composition among the nine study populations (permutational multivariate analysis of variance [PERMANOVA] R2 = 11.9 %, p < 0.001; Fig. 1A). Despite that, the disease status of subjects still had a significant impact on the overall viral composition (PERMANOVA R2 = 0.8 %, p < 0.001). We then quantified the effect size of disease status on gut virome within each study and discovered that, with the exception of Hannigan et al.'s study, CRC status was significantly associated with viral composition in almost all cohorts (Fig. 1B). However, the adenoma status had no significant effect on viral composition in all cohorts (Fig. 1C). Furthermore, because individual heterogeneity has been shown to closely correlate with viral taxon variants, we assessed the effect sizes of host characteristics such as age, gender, and body mass index (BMI) on gut virome based on all data. BMI showed a considerable impact on the overall viral composition (PERMANOVA R2 = 1.1 %, p < 0.001), whereas the effect sizes of age and gender were relatively lower (R2 = 0.4 % and 0.3 %, respectively; Table S2). Moreover, adjusting the host’s age, gender, and BMI didn’t visibly change the size of the effect of disease status on the gut virome, suggesting that there was little interaction between them.
Finally, we did a compositional analysis of CRC and adenoma patients and healthy controls at the viral family level and excluded the unclassified vOTUs at the family level, which accounted for nearly 67 % of total sequences. For all datasets, the gut virome was dominated by Siphoviridae, Myoviridae, Podoviridae, and Quimbyviridae (Fig. 1D). Compared with the healthy controls, Siphoviridae was significantly depleted in CRC patients in study Feng_2015 (q = 0.015) and approached significant level in study Wirbel_2019 (q = 0.087), and with meta-analysis coefficient estimate (μ) = -0.14 (q = 0.26; Fig. 1E; Fig. S3A). Microviridae, a family of small ssDNA viruses, were significantly enriched in the CRC patients compared with healthy controls in 5 of 9 studies, with a meta-analysis μ = 0.34 (q = 0.02; Fig. 1F; Fig. S3A). Autographiviridae and Gratiaviridae, were also considerably enriched in the CRC patients (meta-analysis q < 0.05; Fig. S3A). In addition, the differential families between adenoma patients and healthy controls were few, except for two clades, Podoviridae and unclassified_Caudovirales, which showed nearly significant differences between them (meta-analysis q < 0.10; Fig. S3B).
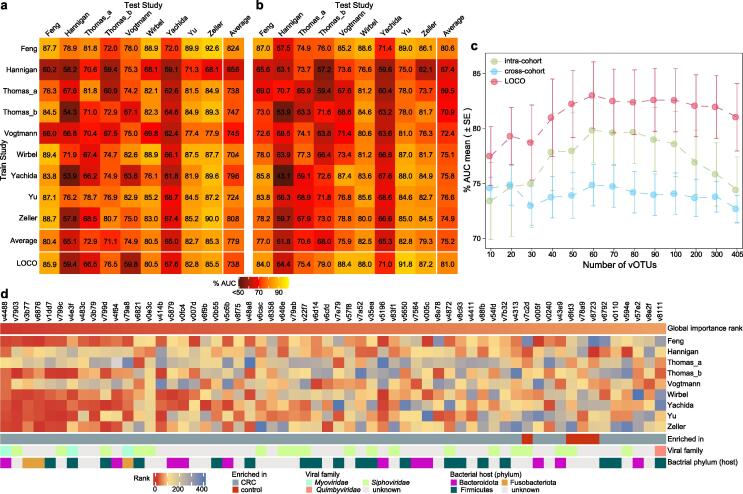
Fig. 3.
The prediction model of CRC status based on relative abundances of 405 CRC-associated vOTUs. (A to B) Performance assessment as AUC scores of intra-dataset and cross-dataset predictions using least absolute shrinkage and selection operator (A) and random forest models (B) in predicting CRC status. The model of intra-dataset prediction (diagonal) was validated using five repeats of fivefold cross-validations. The model of cross-dataset prediction (off-diagonal) was built on the dataset corresponding to each row and validated on the dataset corresponding to each column. The LOCO row refers to leave-one-cohort-out (LOCO) analysis in which models were built on eight datasets combined and validated on the remaining one corresponding to each column. (C) Average AUC values for different numbers of CRC-associated vOTUs using random forest models. (D) Heatmap showing the 60 most important ranking vOTUs in the random forest model. The global importance rank refers to the mean rank of each vOTU for all studies. The feature importance was calculated by the “mean decrease accuracy” method.
Identification of CRC-associated viral signatures
Given the large effect of study heterogeneity on viral shifts, we performed a more accurate meta-analysis approach to identify CRC-associated viral biomarkers across nine datasets. For each study, a Wilcoxon rank-sum test was performed on the vOTUs relative abundance profile between patients and controls. In most studies, a substantial enrichment of a set of vOTUs with very small P values was observed as compared to the expected distribution under the null hypothesis (Fig. S4A-B), indicating that some of these vOTUs are actual CRC-associated viral signatures. Based on this, we selected 516 vOTUs that had a significant abundance difference (p < 0.05 in the Wilcoxon rank-sum test) and a consistent trend between patients and controls in at least 4 of 9 studies. Then, we pooled evidence of differential abundance across datasets by random effects meta-analysis and further identified 407 vOTUs at a false discovery rate (FDR) < 0.01. Finally, 405 CRC-associated vOTUs were identified after adjusting for confounding variables including age, BMI, and gender (Fig. S4C; Table S3).
Fig. 4.
Validation of 405 CRC-associated vOTUs in independent cohorts. (A and C) Volcano plots showing fold change and statistical significance in CRC-associated vOTUs abundance between CRC patients and healthy controls recruited in this study (A) and Yang’s study (C). (B and D) Performance assessment as AUC scores for CRC-associated vOTUs using random forest models in the cohort from this study (B) and Yang’s study (D), respectively. The model was built by combining nine published datasets and was validated by independent cohorts. Blue curve, the model based on all CRC-associated vOTUs abundances. Red curve, the model based on 60 most important ranking vOTUs abundances.
Over 85 %, 77 %, and 74.8 % of the 405 CRC-associated vOTUs were independently significant in Yu_2015, Wirbel_2019, and Yachida_2019 datasets, respectively (Fig. S4D), indicating that these three studies are the primary contributors to CRC viral signatures. Inversely, only<20 % of the CRC-associated vOTUs were independently significant within datasets Hannigan_2018 and Thomas_b_2019.
In the CRC patients, 220 of the 405 biomarkers were more abundant, while 185 of them were enriched in healthy controls. The CRC-enriched vOTUs included 76 members of Siphoviridae, 20 Myoviridae, 3 Quimbyviridae, and 1 Microviridae, and 120 unclassified viruses, while the control-descending vOTUs were composed of 64 Siphoviridae, 25 Myoviridae, and 96 unclassified viruses (Fig. 2A; Table S3). We performed a host assignment of the vOTUs based on their homology or CRISPR spacers to the 4,644 prokaryotic genomes from the Unified Human Gastrointestinal Genome (UHGG) collections[31]. The analysis assigned 54.3 % of the CRC-associated vOTUs to one or more prokaryotic hosts. The control-enriched vOTUs had a large proportion (23.2 %) of Ruminococcaceae phages, whereas only 4.5 % of the CRC-enriched vOTUs were those (Fisher’s exact test q < 0.001; Fig. 2A; Table S3). Inversely, the CRC-enriched vOTUs contained significantly higher proportions of Bacteroidaceae, Oscillospiraceae, and Peptostreptococcaceae phages than the control-enriched viruses (Fisher’s exact test q < 0.05). At the genus level, 37 control-enriched vOTUs were Faecalibacterium phages and 6 were Roseburia phages (Table S3). These two taxa are well-known SCFA-producers and have shown beneficial effects on multiple common disorders[44], [45], though they are not usually deficient in the gut microbiome of CRC patients. Several other viruses that were predicted to infect species of Porphyromonas (5 vOTUs), Fusobacterium (4 vOTUs), and Hungatella (3 vOTUs) were enriched in the CRC virome, in agreement with previous studies showing that these taxa are overgrown in the CRC bacteriome[6], [7].
To further investigate the interactions between CRC-associated viruses and bacteria, we identified 83 CRC-associated bacterial species from the datasets using the same approach as with virome (meta-analysis q < 0.01; Table S4) and performed correlation analysis with 405 vOTU biomarkers. We revealed a large virus-bacterium interaction network (Fig. 2B), consisting of a total of 1,331 interactions that included 62 host-phage pairs and 1,269 statistical co-abundance or co-occurrence correlations. Diverse groups of bacteria, including the CRC-enriched Fusobacterium spp., Porphyromonas spp., Peptoniphilus spp., Hungatella hathewayi, Bacteroides fragilis, and Desulfovibrio desulfuricans and the control-enriched Faecalibacterium prausnitzii, Roseburia spp., and Agathobacter spp., drove the major correlations in the network (Table S4), suggesting that they may play keystone roles in the CRC gut ecosystem. The network spanned 310 vOTUs, which are potential bacterium-dependent viruses that may impact host CRC status in cooperating with the corresponding bacteria. In addition, the remaining 85 out of 95 vOTUs might independently act on disease, as they were enriched in CRC patients (Table S3). To characterize the functional potential of the CRC-associated viruses, we annotated the proteins of 405 vOTUs using the KEGG (Kyoto Encyclopedia of Genes and Genomes) database[46]. The KEGG orthology (KO) approach was used to analyze 19.6 % (4,398/22,410) of the viral proteins that covered a total of 1,225 KOs for analysis. Statistically,14 KOs had significantly differed in frequency between the CRC-enriched and control-enriched vOTUs (Fisher’s exact test, q < 0.05; Table S5). Several enzymes, including K08640 (zinc d-Ala-d-Ala carboxypeptidase), K14060 (DNA-invertase from lambdoid prophage), K03465 (thymidylate synthase), K06400 (site-specific DNA recombinase), K04485 (DNA repair protein), and K03700 (recombination protein), were more frequently encoded in the control group-enriched viruses, whereas K01447 (N-acetylmuramoyl-l-alanine amidase) was more abundant in the CRC-enriched viruses (Fig. 2C).
Constructing the CRC predictive model using viral signatures
To detect the diagnostic ability of gut viral signatures in CRC, we performed intra-dataset and cross-dataset prediction and validation on the overall set of 554 CRC and 546 control samples based on their relative abundances of 405 vOTUs. Two machine learning algorithms-least absolute shrinkage and selection operator (LASSO) and random forest (RF) were used for modeling and testing (see Methods). In intra-dataset, we observed performances ranging in the area under the receiver operating characteristic curve (AUC) score from 0.582 to 0.900 (average 0.779) for the LASSO algorithm and from 0.631 to 0.870 (average 0.752) for the RF algorithm (Fig. 3A-B). The Hannigan_2018 study obtained the lowest AUCs in both algorithms, which could potentially be explained by its small sample size. In cross-dataset prediction and validation, we obtained pairwise AUCs ranging from 0.539 to 0.926 (average 0.753) for the LASSO algorithm and from 0.539 to 0.890 (average 0.737) for the RF algorithm.
To overcome the sample size limitations on single studies, we performed a leave-one-cohort-out (LOCO) analysis, in which models were built on eight datasets combined and validated on the left-out dataset, for each dataset in turn. The LOCO AUCs ranged from 0.594 to 0.859 (average 0.779) and from 0.644 to 0.918 (average 0.810) for nine studies using the LASSO and RF models, respectively (Fig. 3A-B). In addition, adding the host properties (i.e., age, BMI, and gender) didn’t improve the predicting performance for both algorithms (Fig. S5). Taken together, our intra-dataset, cross-dataset, and combined-dataset analyses suggest that the gut viral signatures are efficient for distinguishing CRC patients from healthy controls.
Finally, to generate a minimal set of vOTU signatures, we calculated the global importance ranks of 405 vOTUs from the RF models of all studies. Using the RF algorithm, CRC was accurately identified with an average cross-dataset AUC 0.798 and LOCO AUC 0.830 when using a subset of 60 top importance vOTUs (Fig. 3C; Fig. S6). Notably, 56 of the top importance vOTUs were CRC-enriched biomarkers, which included 4 Peptostreptococcus, 3 Fusobacterium, and 3 Porphyromonas phages (Fig. 3D; Table S3).
Validation of CRC viral markers in independent cohorts
To validate the efficiency of CRC viral signatures in the independent cohort, we recruited 27 CRC patients having matched age, BMI, and gender and healthy controls and performed whole-metagenome shotgun sequencing of their fecal samples (Table S6). In this new cohort, 207 out of the 405 CRC-associated vOTUs, including 46 CRC-enriched and 161 control-enriched vOTUs, were significantly different in relative abundance between patients and controls, with a consistent trend with the meta-analysis of nine studies (p < 0.05 in Wilcoxon rank-sum test; Fig. 4A; Table S3). Compared with the control-enriched vOTUs, the lower rate of CRC-enriched vOTUs might be contributed by their lower occurrence rate in fecal samples of the new cohort as well as in samples of the above nine studies (Fig. S7). Using the RF algorithm, we trained two models based on the abundances of 405 vOTUs and 60 top importance vOTUs in the original nine cohorts. These models obtained the AUC of 0.906 and 0.877, respectively, in distinguishing between CRC and controls in the independent new cohort (Fig. 4B). We also validated the efficiency of CRC viral signatures in the recently published cohort of 100 onset CRC patients and 100 healthy controls (Yang_2021)[13]. The models obtained an acceptable diagnostic efficacy in distinguishing onset CRC patients and controls (Fig. 4D). These findings suggest high repeatability and predictive power of CRC viral markers in independent cohorts.
Gut viral signatures in adenoma
Lastly, we profiled the gut viral profiles of adenoma patients (n = 182) from five studies and compared them with those of corresponding healthy controls (n = 388; Table 1) to explore adenoma viral signatures. A meta-analysis based on the aforementioned approach identified 88 significant adenoma-associated vOTUs (meta-analysis q < 0.05; Table S7). In adenoma patients, 47 of these vOTUs were more abundant, including 20 Siphoviridae, 3 Myoviridae, a Quimbyviridae, and a crAss-like viruses; while 41 vOTUs were enriched in controls, including 20 Siphoviridae, 3 Myoviridae, and 1 Flandersviridae viruses. Host assignment showed that the adenoma-enriched vOTUs included 4 Enterobacteriaceae, 3 Bacteroidaceae, and 3 Oscillospiraceae phages, whereas 3 of control-enriched vOTUs were Bifidobacteriaceae phages (Table S7). Additionally, adenoma and CRC shared 4 differential viral markers, including 2 unclassified adenoma/CRC-enriched and 2 control-enriched vOTUs that belonged to Siphoviridae.
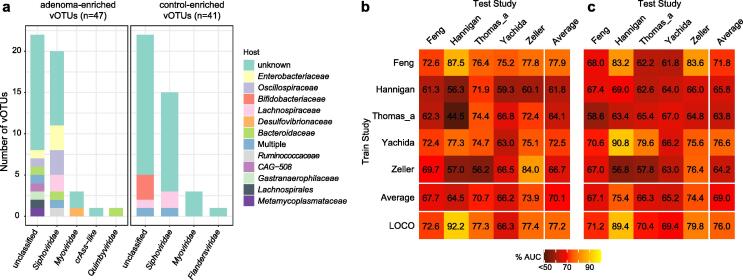
We performed machine learning for adenoma prediction and observed the intra-dataset, cross-dataset, and LOCO average AUCs of 0.700, 0.686, and 0.772, respectively. In five adenoma studies using the LASSO algorithm (Fig. 5a), average AUCs of 0.690, 0.698, and 0.760 using the RF algorithm (Fig. 5b), suggesting the validity of adenoma-specific viral markers.
Fig. 5.
The adenoma-associated viral signatures. (A) The family-level taxonomy and host assignment of adenoma-associated vOTUs. The vOTUs were grouped at the family level, and their hosts are shown at the family level. The number of vOTUs that has more than one predicted hosts are colored in blue (multiple families). (B to C) Performance assessment as AUC scores of intra-dataset and cross-dataset predictions using LASSO (B) and random forest (C) model in predicting adenoma status. The model of intra-cohort prediction (diagonal) was validated using five repeats of 5-fold cross-validations. The model of cross-cohort prediction (off-diagonal) was built on the dataset corresponding to each row and validated on the dataset corresponding to each column. The LOCO row refers to leave-one-cohort-out (LOCO) analysis in which models were built on eight datasets combined and validated on the remaining one corresponding to each column.
Discussion
Virus genomic database
Although a series of studies indicated intestinal dysbacteriosis in CRC patients, most of them focused on the characterization of the gut bacteriome [6], [7], [8], [13], [14], and only a few studies explored a limited number of known viruses [24], [25]. In this study, we introduced a more comprehensive database of viral genomes that contained 37,030 non-redundant viral genomes derived from the de novo assembly of 1,282 fecal metagenomes from 9 published cohorts. Over half of the genomes in this database were undetected by the current available viral databases, which supported that our database was necessary to gain insight into the alteration of gut viral communities in CRC patients. On the basis of this database, we found that the gut virome diversities of CRC patients were comparable to those of healthy controls in most of the cohorts collected in this study, consistent with the findings of previous studies [24], [26]. The viral diversity parameters of samples were highly consistent with their bacterial diversities, which might be explained by the fact that most of the viruses in this study resided within bacterial cells.
Identification of CRC-associated markers and their potential mechanisms
We identified 405 reproducible vOTUs associated with CRC using meta-analysis based on 9 independent cohorts. The majority of these vOTUs are classified as viral dark matter because of the absence of referral genomes. We found that most of these vOTUs (310/405) were closely connected with bacterial biomarkers for CRC, and they aggregated into a multi-centric virus-bacteria interaction network (Fig. 2B). The bacterial biomarkers were central members within the network, which suggested that these viruses might indirectly influence CRC development by altering bacterial biomarkers. Among this network, the central members of two hubs were mainly the butyrate-producing bacteria from the genera Faecalibacterium, Roseburia, and Agathobacter. Some studies implied that butyrate producers, Faecalibacterium prausnitzii in particular, were potential probiotics with anti-tumorigenic properties and might contribute to preventing CRC development [13], [47]. Our results showed that a group of viruses, reduced in CRC patients, were mainly connected with butyrate-producing bacteria, which suggested that these viruses may contribute to tumorigenesis by modulating the butyrate-producing bacteria in the human gut. Another hub in this network involved a variety of common oral bacteria from the genera Anaerococcus, Fusobacterium, Peptostreptococcus, Parvimonas, and Solobacterium. Many studies have reported that these bacteria are significantly enriched in fecal samples from CRC patients [6], [8]. In particular, Fusobacterium nucleatum contains a highly conserved virulence factor FadA that binds to the extracellular domain of epithelial cadherin to promote cancer cell proliferation through the Wnt signaling pathway [48]. Translocation of oral microbes to the gut caused an increased abundance of them in cancer patients. In this study, we observed that one N-acetylmuramoyl-l-alanine amidase (K01447) was more abundant in the CRC-enriched viruses than in the CON-enriched viruses. This enzyme can digest the peptidoglycan of the bacterial cell wall and disrupt the biofilms [49], [50]. CRC-enriched viruses, coexisting with bacterial biomarkers commonly present in the oral cavity, might affect CRC development by disrupting bacterial biofilms to promote dispersal of these bacteria. Overall, our results partly explained and put a spotlight on how CRC-associated viruses can influence cancer progression. However, our findings are data-driven, and further studies will be necessary to uncover the linkage between viruses, bacteria, and carcinogenesis by in vitro and in vivo experimental validation.
Accuracy and repeatability of predictive models based on viral signatures
Furthermore, gut bacteria-based CRC prediction models have demonstrated high diagnostic potential; however, the models are not available in all cohorts [6], [7], implying that the identification of new potential predictors was required. Although gut viruses have not been regarded as effective CRC predictors for a long time [24], [25], a recent study reported the potential of gut viruses for the classifiability of controls versus patients with CRC [6]. In this study, we performed viral biomarker-based random forest models with LOCO validation and were able to distinguish CRC patients from healthy controls with an average performance of 0.81 AUC which was comparable to the performance of models based on bacterial biomarkers (Fig. 3B). Importantly, we also validated the efficiency of CRC viral signatures in two independent cohorts. For the newly-recruited cohort in this study, CRC viral signatures displayed excellent potential for classifiability of controls versus patients (Fig. 4B). For other independent cohorts, the models only obtained acceptable diagnostic efficacy in distinguishing onset CRC patients and controls (Fig. 4D). Alteration of certain CRC viral signatures is induced by therapeutic approaches. Among CRC viral signatures, a phage (v4488) infecting Bacteroides xylanisolvens was ranked as the top contributor to CRC prediction models (Fig. 3D). But Bacteroides xylanisolvens has not been reported as the CRC biomarker so far, which suggests that certain gut phages play an irreplaceable role in CRC prediction. In addition, several top-ranking important biomarkers infected the CRC-associated species of the genera, including Peptostreptococcus, Fusobacterium, and Porphyromonas (Fig. 3D), suggesting that these viruses might be the CRC predictors with similar performances as their hosts. Taken together, our findings showed that viral biomarkers would be the new efficient predictors for CRC diagnosis.
Viral signatures for adenomas are also predictive
Moreover, colorectal adenomas are regarded as precursor lesions of CRC. The detection of colorectal adenomas could reduce the risk of CRC and improve survival rates. Therefore, we also identified a series of adenomas-associated viruses and assessed the performance of prediction models for adenoma status. Both LASSO and RF algorithms with LOCO approaches demonstrated that adenomas patients and healthy controls could be distinguished with acceptable accuracy (average AUC > 0.75). Similar results were observed in the bacteria-based prediction models for adenoma status [14]. These findings indicate the dysbiosis of intestinal viral communities in patients with colorectal adenomas, highlighting the potential role of viral biomarkers in the diagnosis of colorectal adenomas.
Conclusions
We comprehensively observed shifts in intestinal viral composition in CRC patients and also described interactions between viral biomarkers and bacteria. Although the precise mechanisms of how these viruses cause tumorigenesis are still unclear, our works provide several potential explanations. Furthermore, the present findings indicate that gut viromes could be used to improve microbiome-based CRC diagnostics. Our analysis strongly suggests that more research is needed to determine the precise role of the omitted gut virome in CRC development.
Phages are recognized as cancer diagnostic and therapeutic tools [51], [52] and have a strong potential in the treatment of CRC. Several gut bacteria, especially the members of Fusobacterium, were associated with CRC tumorigenesis. A series of gut viruses in this study were closely connected with CRC-associated bacteria and may be potential targets to eliminate harmful bacteria via phage therapy [53]. In addition, a study has tried to use fecal virome transplantation (FVT) as a therapeutic strategy for type 2 diabetes and obesity [54]. Identification of CRC-associated viral markers serves as a guide for future studies involving FVT for the treatment of CRC. On the other hand, prediction models exhibited high repeatability and predictive power of gut viral markers in colorectal cancer and colorectal adenoma. Compared with colonoscopy, fecal metagenome-based model predictions are more patient-friendly and easier to accept, which improves the diagnosis of colorectal cancer in the absence of clinical symptoms.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
This work was financially supported by National Natural Science Foundation of China (No. 82225048 and No. 81902037), Distinguished professor of Liaoning Province (XLYC2002008), and Dalian Science and Technology Leading Talents Project (2019RD15).
Data and materials availability
All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Compliance with Ethics Requirements
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (Dalian University Affiliated Xinhua Hospital [NO 2022-04-01]) and with the Helsinki Declaration of 1975, as revised in 2008. Informed consent was obtained from all patients for being included in the study.
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2022.09.012.
Contributor Information
Qiulong Yan, Email: qiulongy1988@163.com.
Xiaochi Ma, Email: maxc1978@163.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Arnold M., Sierra M.S., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66(4):683–691. doi: 10.1136/gutjnl-2015-310912. [DOI] [PubMed] [Google Scholar]
- 2.Sharma R. An examination of colorectal cancer burden by socioeconomic status: evidence from GLOBOCAN 2018. EPMA Journal. 2020;11(1):95–117. doi: 10.1007/s13167-019-00185-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xi Y., Xu P. Global colorectal cancer burden in 2020 and projections to 2040. Transl Oncol. 2021;14(10) doi: 10.1016/j.tranon.2021.101174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carethers J.M., Jung B.H. Genetics and Genetic Biomarkers in Sporadic Colorectal Cancer. Gastroenterology. 2015;149(5):1177–1190 e1173. doi: 10.1053/j.gastro.2015.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keum N., Giovannucci E. Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat Rev Gastroenterol Hepatol. 2019;16(12):713–732. doi: 10.1038/s41575-019-0189-8. [DOI] [PubMed] [Google Scholar]
- 6.Thomas A.M., Manghi P., Asnicar F., Pasolli E., Armanini F., Zolfo M., et al. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat Med. 2019;25(4):667–678. doi: 10.1038/s41591-019-0405-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wirbel J., Pyl P.T., Kartal E., Zych K., Kashani A., Milanese A., et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat Med. 2019;25(4):679–689. doi: 10.1038/s41591-019-0406-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yachida S., Mizutani S., Shiroma H., Shiba S., Nakajima T., Sakamoto T., et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat Med. 2019;25(6):968–976. doi: 10.1038/s41591-019-0458-7. [DOI] [PubMed] [Google Scholar]
- 9.Liu N.-N., Jiao N.a., Tan J.-C., Wang Z., Wu D., Wang A.-J., et al. Multi-kingdom microbiota analyses identify bacterial–fungal interactions and biomarkers of colorectal cancer across cohorts. Nat Microbiol. 2022;7(2):238–250. doi: 10.1038/s41564-021-01030-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kostic A., Chun E., Robertson L., Glickman J., Gallini C., Michaud M., et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14(2):207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hong J., Guo F., Lu S.-Y., Shen C., Ma D., Zhang X., et al. F. nucleatum targets lncRNA ENO1-IT1 to promote glycolysis and oncogenesis in colorectal cancer. Gut. 2021;70(11):2123–2137. doi: 10.1136/gutjnl-2020-322780. [DOI] [PubMed] [Google Scholar]
- 12.Ternes D., Tsenkova M., Pozdeev V.I., Meyers M., Koncina E., Atatri S., et al. The gut microbial metabolite formate exacerbates colorectal cancer progression. Nature metabolism. 2022;4(4):458–475. doi: 10.1038/s42255-022-00558-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Y., Du L., Shi D., Kong C., Liu J., Liu G., et al. Dysbiosis of human gut microbiome in young-onset colorectal cancer. Nat Commun. 2021;12(1):6757. doi: 10.1038/s41467-021-27112-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu Y., Jiao N.a., Zhu R., Zhang Y., Wu D., Wang A.-J., et al. Identification of microbial markers across populations in early detection of colorectal cancer. Nat Commun. 2021;12(1) doi: 10.1038/s41467-021-23265-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo R, Li S, Zhang Y, Zhang Y, Wang G, Ma Y, Yan Q: Dysbiotic oral and gut viromes in untreated and treated rheumatoid arthritis patients. bioRxiv 2021. [DOI] [PMC free article] [PubMed]
- 16.Clooney A.G., Sutton T.D.S., Shkoporov A.N., Holohan R.K., Daly K.M., O’Regan O., et al. Whole-Virome Analysis Sheds Light on Viral Dark Matter in Inflammatory Bowel Disease. Cell Host Microbe. 2019;26(6):764–778.e5. doi: 10.1016/j.chom.2019.10.009. [DOI] [PubMed] [Google Scholar]
- 17.Zhang X., Zhang D., Jia H., Feng Q., Wang D., Liang D.i., et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med. 2015;21(8):895–905. doi: 10.1038/nm.3914. [DOI] [PubMed] [Google Scholar]
- 18.Franzosa E.A., Sirota-Madi A., Avila-Pacheco J., Fornelos N., Haiser H.J., Reinker S., et al. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol. 2019;4(2):293–305. doi: 10.1038/s41564-018-0306-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li M.o., Wang C., Guo Q., Xu C., Xie Z., Tan J., et al. More Positive or More Negative? Metagenomic Analysis Reveals Roles of Virome in Human Disease-Related Gut Microbiome. Front Cell Infect Microbiol. 2022;12 doi: 10.3389/fcimb.2022.846063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Damin D.C., Ziegelmann P.K., Damin A.P. Human papillomavirus infection and colorectal cancer risk: a meta-analysis. Colorectal Dis. 2013;15(8):e420–e428. doi: 10.1111/codi.12257. [DOI] [PubMed] [Google Scholar]
- 21.Bedri S., Sultan A.A., Alkhalaf M., Al Moustafa A.E., Vranic S. Epstein-Barr virus (EBV) status in colorectal cancer: a mini review. Hum Vaccin Immunother. 2019;15(3):603–610. doi: 10.1080/21645515.2018.1543525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su F.-H., Le T.N., Muo C.-H., Te S.A., Sung F.-C., Yeh C.-C. Chronic Hepatitis B Virus Infection Associated with Increased Colorectal Cancer Risk in Taiwanese Population. Viruses. 2020;12(1):97. doi: 10.3390/v12010097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen H.-P., Jiang J.-K., Lai P.-Y., Chen C.-Y., Chou T.-Y., Chen Y.-C., et al. Tumoral presence of human cytomegalovirus is associated with shorter disease-free survival in elderly patients with colorectal cancer and higher levels of intratumoral interleukin-17. Clin Microbiol Infect. 2014;20(7):664–671. doi: 10.1111/1469-0691.12412. [DOI] [PubMed] [Google Scholar]
- 24.Hannigan G.D., Duhaime M.B., Ruffin M.T., Koumpouras C.C., Schloss P.D., Miller J.F. Diagnostic Potential and Interactive Dynamics of the Colorectal Cancer Virome. Diagnostic Potential and Interactive Dynamics of the Colorectal Cancer Virome. 2018;9(6) doi: 10.1128/mBio.02248-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakatsu G., Zhou H., Wu W.K.K., Wong S.H., Coker O.O., Dai Z., et al. Alterations in Enteric Virome Are Associated With Colorectal Cancer and Survival Outcomes. Gastroenterology. 2018;155(2):529–541.e5. doi: 10.1053/j.gastro.2018.04.018. [DOI] [PubMed] [Google Scholar]
- 26.Shen S., Huo D., Ma C., Jiang S., Zhang J., Xu Z.Z., et al. Expanding the Colorectal Cancer Biomarkers Based on the Human Gut Phageome. Microbiol Spectr. 2021;9(3) doi: 10.1128/Spectrum.00090-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen S., Zhou Y., Chen Y., Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li D., Liu C.M., Luo R., Sadakane K., Lam T.W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31(10):1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 30.Li S, Guo R, Zhang Y, Li P, Chen F, Wang X, Li J, Jie Z, Lv Q, Jin H: A Catalogue of 48,425 Nonredundant Viruses From Oral Metagenomes Expands the Horizon of the Human Oral Virome. under review, 48. [DOI] [PMC free article] [PubMed]
- 31.Almeida A., Nayfach S., Boland M., Strozzi F., Beracochea M., Shi Z.J., et al. A unified catalog of 204,938 reference genomes from the human gut microbiome. Nat Biotechnol. 2021;39(1):105–114. doi: 10.1038/s41587-020-0603-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Friedman J., Alm E.J. Inferring correlation networks from genomic survey data. PLoS Comput Biol. 2012;8(9):e1002687. doi: 10.1371/journal.pcbi.1002687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watts S.C., Ritchie S.C., Inouye M., Holt K.E. FastSpar: rapid and scalable correlation estimation for compositional data. Bioinformatics. 2019;35(6):1064–1066. doi: 10.1093/bioinformatics/bty734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wirbel J., Zych K., Essex M., Karcher N., Kartal E., Salazar G., et al. Microbiome meta-analysis and cross-disease comparison enabled by the SIAMCAT machine learning toolbox. Genome Biol. 2021;22(1):1–27. doi: 10.1186/s13059-021-02306-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zeller G., Tap J., Voigt A.Y., Sunagawa S., Kultima J.R., Costea P.I., et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol Syst Biol. 2014;10:766. doi: 10.15252/msb.20145645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu J., Feng Q., Wong S.H., Zhang D., Liang Q.Y., Qin Y., et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut. 2017;66(1):70–78. doi: 10.1136/gutjnl-2015-309800. [DOI] [PubMed] [Google Scholar]
- 38.Feng Q., Liang S., Jia H., Stadlmayr A., Tang L., Lan Z., et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat Commun. 2015;6:6528. doi: 10.1038/ncomms7528. [DOI] [PubMed] [Google Scholar]
- 39.Vogtmann E., Hua X., Zeller G., Sunagawa S., Voigt A.Y., Hercog R., et al. Colorectal Cancer and the Human Gut Microbiome: Reproducibility with Whole-Genome Shotgun Sequencing. PLoS ONE. 2016;11(5):e0155362. doi: 10.1371/journal.pone.0155362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gregory A.C., Zablocki O., Zayed A.A., Howell A., Bolduc B., Sullivan M.B. The Gut Virome Database Reveals Age-Dependent Patterns of Virome Diversity in the Human Gut. Cell Host Microbe. 2020;28(5):724–740 e728. doi: 10.1016/j.chom.2020.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Camarillo-Guerrero L.F., Almeida A., Rangel-Pineros G., Finn R.D., Lawley T.D. Massive expansion of human gut bacteriophage diversity. Cell. 2021;184(4):1098–1109 e1099. doi: 10.1016/j.cell.2021.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nayfach S., Camargo A.P., Schulz F., Eloe-Fadrosh E., Roux S., Kyrpides N.C. CheckV assesses the quality and completeness of metagenome-assembled viral genomes. Nat Biotechnol. 2020 doi: 10.1038/s41587-020-00774-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nayfach S., Paez-Espino D., Call L., Low S.J., Sberro H., Ivanova N.N., et al. Metagenomic compendium of 189,680 DNA viruses from the human gut microbiome. Nat Microbiol. 2021;6(7):960–970. doi: 10.1038/s41564-021-00928-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Machiels K., Joossens M., Sabino J., De Preter V., Arijs I., Eeckhaut V., et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut. 2014;63(8):1275–1283. doi: 10.1136/gutjnl-2013-304833. [DOI] [PubMed] [Google Scholar]
- 45.Zhou L., Zhang M., Wang Y., Dorfman R.G., Liu H., Yu T., et al. Faecalibacterium prausnitzii Produces Butyrate to Maintain Th17/Treg Balance and to Ameliorate Colorectal Colitis by Inhibiting Histone Deacetylase 1. Inflamm Bowel Dis. 2018;24(9):1926–1940. doi: 10.1093/ibd/izy182. [DOI] [PubMed] [Google Scholar]
- 46.Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(D1):D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen J., Vitetta L. Inflammation-modulating effect of butyrate in the prevention of colon cancer by dietary fiber. Clinical colorectal cancer. 2018;17(3):e541–e544. doi: 10.1016/j.clcc.2018.05.001. [DOI] [PubMed] [Google Scholar]
- 48.Gur C., Ibrahim Y., Isaacson B., Yamin R., Abed J., Gamliel M., et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42(2):344–355. doi: 10.1016/j.immuni.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Domenech M., García E., Moscoso M. In vitro destruction of Streptococcus pneumoniae biofilms with bacterial and phage peptidoglycan hydrolases. Antimicrob Agents Chemother. 2011;55(9):4144–4148. doi: 10.1128/AAC.00492-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vermassen A., Leroy S., Talon R., Provot C., Popowska M., Desvaux M. Cell wall hydrolases in bacteria: insight on the diversity of cell wall amidases, glycosidases and peptidases toward peptidoglycan. Front Microbiol. 2019;10:331. doi: 10.3389/fmicb.2019.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barbu E.M., Cady K.C., Hubby B. Phage Therapy in the Era of Synthetic Biology. Cold Spring Harb Perspect Biol. 2016;8(10) doi: 10.1101/cshperspect.a023879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abbaszadeh F., Leylabadlo H.E., Alinezhad F., Feizi H., Mobed A., Baghbanijavid S., et al. Bacteriophages: cancer diagnosis, treatment, and future prospects. Journal of Pharmaceutical Investigation. 2021;51(1):23–34. [Google Scholar]
- 53.Dolgin E. Fighting cancer with microbes. Nature. 2020;577(7792):S16–S. doi: 10.1038/d41586-020-00199-x. [DOI] [PubMed] [Google Scholar]
- 54.Rasmussen T.S., Mentzel C.M.J., Kot W., Castro-Mejía J.L., Zuffa S., Swann J.R., et al. Faecal virome transplantation decreases symptoms of type 2 diabetes and obesity in a murine model. Gut. 2020;69(12):2122–2130. doi: 10.1136/gutjnl-2019-320005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw metagenomic sequencing data from the independent cohorts has been submitted to the China Nucleotide Sequence Archive with the accession code CNP0002641. The authors declare that all other data supporting the findings of the study are available in the paper and supplementary materials, or from the corresponding authors upon request.