Abstract
For the digestive system, there exists one common malignant tumor, known as gastric cancer. It is the third most prevalent type of tumor among different tumors worldwide. It has been reported that long noncoding RNAs (lncRNAs), participate in various biological processes of gastric cancer. However, there are still many lncRNAs with unknown functions, and we discovered a novel lncRNA designated as FBXO18-AS. Whether lncRNA FBXO18-AS participates in gastric cancer progression is still unknown. Bioinformatic analysis, immunohistochemistry, Western blotting, and qPCR were carried out to explore FBXO18-AS and TGF-β1 expression. In addition, EdU, MTS, migration and transwell assays were performed to investigate the invasion, proliferation and migration of gastric cancer in vitro. We first discovered that FBXO18-AS expression was upregulated in gastric cancer and linked to poorer outcomes among patients with gastric cancer. Then, we confirmed that FBXO18-AS promoted the proliferation, invasion, migration, and an EMT-like process in gastric cancer in vivo and in vitro. Mechanistically, FBXO18-AS was found to be involved in the progression of gastric cancer by modulating TGF-β1/Smad signaling. Therefore, it might offer a possible biomarker for gastric cancer diagnosis and an effective strategy for clinical treatment.
Key words: LncRNA, FBXO18-AS, gastric cancer, TGF-β1
Introduction
Gastric cancer is regarded as one of the most pervasive and malignant neoplasms in the digestive system and an important disease because of its advanced stage at diagnosis.1,2 At present, the primary methods to treat gastric cancer are still radical surgical resection and chemoradiotherapy.3With the improvement in surgical techniques and progress in traditional radiotherapy and chemotherapy, the 5-year survival rate of patients with previous gastric cancer can approach >95%.2 However, most patients are diagnosed with advanced cancer at the time of gastric cancer diagnosis and therefore have missed the best surgical window and encounter large challenges for treatment.4 Therefore, moleculartargeted therapy may become an effective treatment for advanced gastric cancer. The specific mechanisms that facilitate gastric cancer progression have been confirmed recently. For example, different noncoding RNAs (ncRNAs), such as long noncoding RNAs (lncRNAs) and microRNAs (miRNAs), participate in the development of gastric cancer.5 LncRNAs are an array of RNAs with a length of over 200 nucleotides that participate in various genetic phenomena, including transcriptional, posttranscriptional and epigenetic processes.6-8 Moreover, lncRNAs have been considered modulators in the induction and progression of gastric cancer and have been found to be involved in tumor promotion, radioresistance, chemoresistance, and sensitivity to target treatment.9,10. For example, LINC01857 facilitates gastric cancer cell proliferation, invasion and migration by modulating miR-4731-5p/HOXC6.11 LncRNA CRYM-AS1 inhibits gastric cancer progression by epigenetically regulating CRYM.12 However, there are still vast numbers of lncRNAs with unknown functions, which need to be further investigated.
In this study, we report the discovery of a new lncRNA, FBXO18-AS (ENSG00000232807/NONHSAT156604.1), which has no previous research report, from the TCGA gastric dataset. The expression of lncRNA FBXO18-AS in gastric cancers was upregulated compared with that in normal gastric tissues. Furthermore, we also found that FBXO18-AS promoted the proliferation, invasion, migration and epithelial-mesenchymal transition (EMT) of gastric cancer. TGF-β/Smad signaling is one of the most pivotal signaling pathways involved in regulating the EMT process in gliomas.13,14 Based on GSEA (gene set enrichment analysis), our research also discovered that TGF-β signaling was increased with the expression of FBXO18-AS, suggesting that FBXO18-AS may adjust the gastric cancer EMT process through TGF-β/Smad signaling. This may also offer a potential target for gastric cancer treatment.
Materials and Methods
Patient samples and ethical approval
We used 60 clinical specimens from gastric cancer patients. Simultaneously, neighboring normal cells were gathered from 2014 to 2016 at Wenzhou Hospital of Integrated Traditional Chinese and Western Medicine. This research acquired the support of the Ethics Committee of Wenzhou Hospital of Integrated Traditional Chinese and Western Medicine (No. 2022-K067), and all patients provided written informed consent.
Cell culture
The gastric cancer cell lines SGC7901, MKN-45, HGC-27 and AGS and the normal gastric epithelial cell line GSE-1 were obtained from the Chinese Academy of Sciences Cell Bank (Shanghai, China) and cultured in RPMI-1640 medium (HyClone) supplemented with 10% fetal bovine serum (FBS; Gibco, Carlsbad, CA, USA) and 1% penicillin/streptomycin (Gibco) at 37°C with 5% CO2.
Lentiviral vector construction and transfection
We carried out efficacy assessment, along with lentivirus transfection, as previously described.15 Lentivirus-based vectors for FBXO18-AS overexpression and RNAi-mediated FBXO18-AS knockdown, along with their controls, were generated by Gene- Chem (Shanghai, China). The sequences of the siRNAs were as follows: FBXO18-AS-KD1: forward 5’- AUUUUCCUUCCAGUUACUGAG -3’, reverse 5’- CAGUAACUGGAAGGAAAAUGA -3’ and forward 5’- UUCAGUUUCAUUUUCCUUCCA -3’, reverse 5’- GAAGGAAAAUGAAACUGAAAA -3’. qPCR was used to determine the transfection efficacy.
RNA extraction and qPCR
RNA extraction and qPCR assays were implemented as previously described.15 Initially, based on the manufacturer’s instructions, the Mini-BEST Universal RNA Extraction kit (TaKaRa, Kyoto, Japan) was used to extract total RNA from the gastric cancer tissues and gastric cancer cells. For FBXO18-AS and TGF-β1, the RNA was reverse transcribed into cDNA via the Prime Script RT Master Mix reagent kit (TaKaRa). Then, qPCR assays were performed using SYBR Green Master Mix (TaKaRa) with a PCR LightCycler480 (Roche Diagnostics, Basel, Switzerland). β-Actin was used as the internal control. Primers were as follows: FBXO18-AS: F’: 5’- TGGTGTCCTCTTCCTCAG-3’, R’: 5’- TGCTCTTCCTCGTCTTCT-3’; TGF-β1: F’: 5’- GGCCAGATCCTGTCCAAGC- 3’, R’: 5’- GTGGGTTTCCACCATTAGCAC- 3’; β-actin: F’: 5’- CATGTACGTTGCTATCCAGGC-3’, R’: 5’- CTCCTTAATGTCACGCACGAT-3’.
Western blotting
We carried out Western blotting as previously described.15 First, a cellular protein extraction kit (KeyGen Biotechnology, Nanjing, China) was used to isolate total proteins from the gastric cancer tissues and gastric cancer cells. After SDS-polyacrylamide gel electrophoresis (SDS-PAGE), the total protein of every sample was transferred to polyvinylidene difluoride (PVDF) membranes. Then, at room temperature, the PVDF membranes were blocked with 2% bovine serum albumin (KeyGen Biotechnology) for 2 h, and then all membranes were incubated at 4 °C overnight with the primary antibodies against the following proteins: E-cadherin (1:1,000; Abcam, Cambridge, UK), vimentin (1:1,000; Abcam), TGF-β1 (1:2,000; Abcam), p-SMAD2 (1:1,000; Cell Signaling Technology, Danvers, MA, USA), SMAD2 (1:1,000; Cell Signaling Technology), Snail (1:1,000; Cell Signaling Technology), Slug (1:1,000; Cell Signaling Technology), and β-actin (1:2000; Abcam). Finally, after 2 h of secondary antibody incubation, the bands were visualized using a chemiluminescence ECL kit (Beyotime Biotechnology, Beijing, China) and quantified by ImageJ software (National Institutes of Health, Bethesda, MD, USA). β-Actin was used as the internal control.
Cell viability assay
Cellular viability assays were carried out as previously described.15 First, the gastric cancer cell lines MKN-45, SGC7901, AGS, and HGC-27 treated with different conditions were placed into 96-well plates at a density of 1 × 103 cells/well and cultured for 0, 24, 48, 72, 96, and 120 h. Next, cellular viability was explored by the CellTiter 96® Aqueous Non-Radioactive Cell Proliferation Assay Kit (Promega, Madison, WI, USA) based on the manufacturer’s instructions.
EdU assay
EdU assays were carried out as previously described15. Briefly, based on the manufacturer’s instructions, the proliferation of SGC7901, MKN-45, AGS, and HGC-27 gastric cancer cells under different treatment conditions was determined by an EdU assay kit (Beyotime Biotechnology). Initially, the gastric cancer cells were placed into 24-well plates at a 1×105 cells/well density for 24 h. Then, 10 μM EDU reagent was added to the medium and incubated for 2 h at 37 °C with 5% CO2. After being fixed with 4% paraformaldehyde (Solarbio) and permeabilized with 0.5% Triton X-100 (Solarbio), the gastric cancer cells treated with different conditions were counterstained. Finally, the proportion of EdUpositive cells was assessed with a laser scanning confocal microscope (Olympus).
Cell migration and transwell assays
For the cellular migration assay, gastric cancer cells under different treatment conditions were resuspended in serum-free medium (HyClone) at a density of 2 × 105 cells/mL. Next, 600 μl RPMI- 1640 medium with 10% FBS was added to the lower chamber. A 100 μl cellular suspension was seeded into the upper chamber (Costar, Corning, NY, USA). After incubation at 37 °C for 24 h, the gastric cancer cells were fixed with 4% paraformaldehyde (Solarbio) for 10 min at room temperature and stained with 1% crystal violet solution (Solarbio) for 20 min. Finally, the cell amounts were counted by determining the mean of 5 random domains through an inverted microscope (Olympus). For transwell assays, the 8 μm pore dimension polycarbonate membrane was covered with 100 μL of 50 ng/μL Matrigel solution (BD, Franklin Lakes, NJ, USA). The subsequent steps were identical to those of the cellular migration assay.
ELISA
We carried out ELISAs as previously described.16 The TGF-β1 concentrations in the media supernatants of SGC7901, MKN-45, AGS, and HGC-27 gastric cancer cells under different treatment conditions were determined by the TGF-β1 Quantikine ELISA Kit (R&D Systems, Minneapolis, MN, USA). In the control groups, the ELISA readings were standardized to the concentration of the protein.
Xenograft experiments
In line with the procedures of the Animal Care Committee of Wenzhou Hospital of Integrated Traditional Chinese and Western Medicine (No. xmsq2022-0667), xenograft tests were carried out as previously described.15 In short, 6-week-old female BALB/c nude mice (Beijing Vital River Laboratory Animal Technology, Beijing, China) were divided into the FBXO18-AS-EV group and the FBXO18-AS-OE group. Every group had 5 mice, which were bred in the Laboratory Animal Center of Wenzhou Hospital of Integrated Traditional Chinese and Western Medicine in special pathogen-free situations. Then, SGC7901 gastric cancer cells under different treatment conditions were injected into the back flanks of the nude mice (5 × 105 cells per mouse). The mice were sacrificed by cervical spine dislocation at day 35 after implantation, and the tumors were weighed and photographed. The neoplasm dimension and tumor weight were determined. A Vernier caliper was used to detect the tumor size, and its volume was computed by means of the following formula: V = (D × d)/2 mm, in which D is the longest diameter and d is the shortest diameter.
Figure 1.
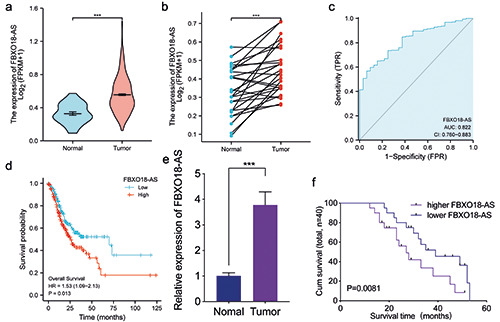
LncRNA FBXO18-AS is expressed at higher levels in gastric cancer and linked to poor outcomes in patients. a,b) TCGA database showed that gastric cancer FBXO18-AS expression was upregulated in comparison to the normal control (p<0.001, Student's t-test). c) ROC dissection of FBXO18-AS in gastric cancers according to the TCGA database. d) Kaplan-Meier analysis indicated the predictive significance of low FBXO18-AS expression vs high FBXO18-AS expression in gastric cancer patients in the TCGA database (p=0.013). e) qPCR illustrated that FBXO18-AS expression was increased in gastric cancer cells compared to normal control cells (p<0.001, Student's t-test). f ) Kaplan-Meier analysis showed the predictive significance of higher FBXO18-AS expression versus lower FBXO18-AS expression in 40 gastric cancer patients as determined by qPCR (p=0.0081; log-rank test). The overall data are presented as the mean ± SD (three independent tests); ***p<0.001; **p<0.01; *p<0.05.
Immunohistochemistry staining
We implemented immunohistochemistry (IHC) staining as described previously using an immunohistochemical labeling kit (MaxVision Biotechnology, Fuzhou, China).15 In brief, gastric cancer cells were placed in paraffin and cut into slices with a thickness of 4 μm. Then, the sections were deparaffinized in xylene and rehydrated in gradient ethanol, followed by antigen retrieval using 0.01 mol/L citrate buffer (pH 6.0) with a microwave at 95°C for 15 min. Then, 3% hydrogen peroxide was used to block endogenous peroxidase activity for 10 min. The nonspecific binding was blocked with normal goat serum for 15 min. The sections were labeled at 4°C overnight in a humidified chamber with primary antibodies against the following proteins: E-cadherin (1:100; Abcam), Ki-67 (1:100; Abcam), and vimentin (1:100; Abcam). After washing with PBS three times, the sections were incubated with biotinylated goat anti-rabbit IgG at room temperature for 15 min, followed by 3,3′-diaminobenzidine (DAB) staining for 20 s to 1 min. Finally, the slides were counterstained with hematoxylin for 3 min, dehydrated and mounted with coverslips. The slides were imaged using a light microscope (Olympus, Tokyo, Japan). The expression levels, along with the staining intensity, were assessed based on the German immunohistochemical score (GIS).17 The percentage of positive cells was classified as 0 (negative), 1 (< 10% positive cells), 2 (11-50% positive cells), 3 (51-80% positive cells), or 4 (>80% positive cells). Staining intensity was classified as 0 (no staining), 1 (weak), 2 (moderate), or 3 (strong). The final immunoreactive GIS was defined as the product of both grading results (percentage of positive cells × staining intensity). Additionally, we conducted negative controls by excluding the primary antibody and positive controls using the available human cell line A-431. Supplementary Figure 1 displays the results for the negative and positive control of IHC staining for Ki-67, E-cadherin, and vimentin.
Bioinformatics analysis
The data on FBXO18-AS expression in gastric cancer patients originated in TCGA (The Cancer Genome Atlas) (http://cancergenome. nih.gov). The enrichment of signaling pathways between the low and high FBXO18-AS expression groups was explored by GSEA (http://www.broadinstitute.org/gsea/index.jsp).
Statistical analysis
The results were reported as the mean ±SD, and the tests were performed a minimum of 3 times. The statistical analysis was performed by SPSS 25.0 software (SPSS, Chicago, IL, USA). A oneway analysis of variation evaluated the statistical significance in three or more groups. However, the comparisons of two independent groups were assessed by a two-tailed Student’s t-test and chisquare testing. The survival rates of each group were analyzed via Kaplan-Meier analysis and log-rank testing; p<0.05 were considered significant.
Figure 2.
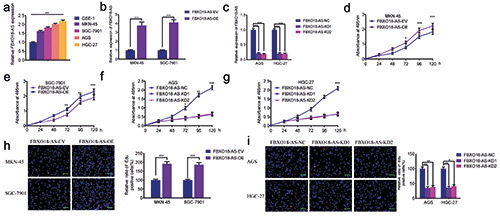
FBXO18-AS promoted gastric cancer cell proliferation. a) FBXO18-AS expression in different gastric cancer cell tissues was determined by qPCR (p<0.001; one-way ANOVA). b,c) qPCR validated FBXO18-AS expression in MKN-45 and SGC7901 cells after FBXO18-AS overexpression and in AGS and HGC-27 cells after FBXO18-AS knockdown (FBXO18-AS overexpression: p<0.001, Student’s t-test; FBXO18-AS knockdown: p<0.001; one-way ANOVA). d-g) MTS assays revealed that overexpression of FBXO18-AS impacted the viability of MKN-45 (d) and SGC7901 (e) cells, and FBXO18-AS knockdown affected the viability of AGS (f ) and HGC- 27 cells (g) (MKN-45: p<0.05; SGC7901: p<0.01; AGS: p<0.05; HGC-27: p<0.01; one-way ANOVA). h,i) EdU assays indicated that FBXO18-AS overexpression impacted the proliferation of MKN-45 and SGC7901 cells (h), and FBXO18-AS knockdown affected the proliferation of AGS and HGC-27 cells (i). Scale bars: 100 μm (MKN-45: p<0.001; SGC7901: p<0.001; Student’s t-test; AGS: p<0.001; HGC-27: p<0.001; one-way ANOVA). The overall data are presented as the mean ± SD (three independent tests);. ***p<0.001; **p<0.01; *p<0.05.
Results
FBXO18-AS was expressed at higher levels in gastric cancer and correlated with shorter patient survival
In line with TCGA databases, FBXO18-AS expression in gastric cancer was found to be higher than that in normal gastric tissues and adjacent normal gastric tissues (Figure 1 a,b). Analysis of ROC curves found that FBXO18-AS may be a potential biomarker of gastric cancer (AUC = 82.2%) (Figure 1c). We also found that the survival of gastric cancer patients with high FBXO18-AS expression was shorter than that of gastric cancer patients with low FBXO18- AS expression in TCGA databases (Figure 1d). In addition, we explored FBXO18-AS expression in 60 clinical gastric cancer tissues and discovered that the expression of FBXO18-AS in gastric cancer tissues was higher than that in normal gastric tissues via qPCR (Figure 1e). Among these, only 40 cases had complete prognostic and survival information. Kaplan-Meier survival analysis also revealed that the survival time of gastric cancer patients with higher FBXO18-AS expression was shorter than that of patients with lower FBXO18-AS expression (Figure 1f). Taken together, these results show that FBXO18-AS was expressed at higher levels in gastric cancer and was linked to shorter survival of patients.
FBXO18-AS promoted the proliferation of gastric cancer cells
We further confirmed by qPCR that FBXO18-AS expression in different gastric cancer cells was greater than that in the common gastric cell line GSE-1 (Figure 2a). Then, we selected the gastric cancer cell lines SGC7901 and MKN-45, which had lower FBXO18-AS expression, to perform FBXO18-AS overexpression experiments, while the gastric cancer cell lines HGC-27 and AGS with higher FBXO18-AS expression were selected to perform FBXO18-AS silencing experiments. qPCR was used to validate the efficiency of FBXO18-AS knockdown and overexpression (Figure 2 b,c). In addition, MTS assays illustrated that MKN-45 and SGC7901 cell viability was increased after FBXO18-AS overexpression but decreased in AGS and HGC-27 cells after FBXO18-AS knockdown (Figure 2 d-g). EdU assays also revealed that the corresponding rate of EDU-positive cells in the SGC7901 and MKN-45 cells was increased after FBXO18-AS overexpression but decreased in AGS and HGC-27 cells after FBXO18-AS knockdown (Figure 2 h,i). Taken together, these results show that FBXO18-AS could boost gastric cancer proliferation.
FBXO18-AS promoted the invasion, migration and EMT-like process of gastric cancer cells
We further confirmed whether FBXO18-AS could regulate gastric cancer cell invasion, migration, and an EMT-like process. GSEA showed that the FBXO18-AS higher expression group was enriched in epithelial-mesenchymal transition (Figure 3a). We also analyzed the correlation between FBXO18-AS and EMT markers in the TCGA dataset. The results showed that there was a positive correlation between FBXO18-AS and vimentin and N-cadherin, while there was a negative correlation between FBXO18-AS and E-cadherin (Figure 3b). Both migration and transwell assays indicated that the migration and invasion of MKN-45 and SGC7901 cells were increased after FBXO18-AS overexpression but decreased in AGS and HGC-27 cells after FBXO18-AS knockdown (Figure 3 c-h). The expression of the EMT-like process marker was detected by Western blotting, and the results showed that the expression of E-cadherin decreased and that of vimentin increased in FBXO18-AS-overexpressing MKN-45 and SGC7901 cells, while the opposite results were obtained in FBXO18-ASsilenced AGS and HGC-27 cells (Figure 3 i,j). Therefore, we showed that FBXO18-AS also promoted gastric cancer cell invasion, migration, and an EMT-like process.
Figure 3.
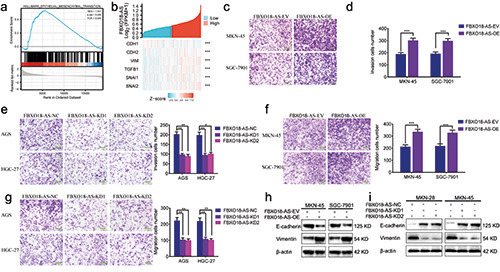
FBXO18-AS promoted gastric cancer cell migration, invasion and an EMT-like process. a) GSEA elucidated the enrichment of epithelial-mesenchymal transition in the FBXO18-AS higher expression group. b) The correlation between FBXO18-AS and markers of EMT and TGF-β1/Smad signaling in the TCGA dataset. c-e) Transwell assays showed the invasion of MKN-45 and SGC7901 cells after FBXO18-AS overexpression (c,e) and that of AGS and HGC-27 cells after FBXO18-AS knockdown (d); scale bars: 50 μm. f-h) Migration assays showed the migration of AGS and HGC-27 cells after FBXO18-AS knockdown (f ) and that of MKN-45 and SGC7901 cells after FBXO18-AS overexpression (g,h); scale bars: 50 μm. i,j) The expression of vimentin and E-cadherin in FBXO18- AS-overexpressing MKN-45 and SGC7901 cells and FBXO18-AS-silenced AGS and HGC-27 cells was determined by Western blotting. All data are presented as the mean ± SD (three independent tests); ***p<0.001; **p<0.01; *p<0.05.
FBXO18-AS promoted the expression of TGF-β1 and activated TGF-β1/Smad signaling
We also found that FBXO18-AS might regulate TGF-β1 expression and be associated with TGF-β signaling according to the GSEA (Figure 4a). The correlation between FBXO18-AS and genes downstream of TGF-β1/Smad signaling was analyzed in the TCGA dataset. The correlation analysis illustrated that there was an active relationship between FBXO18-AS and TGF-β1, Snail and Slug in the TCGA dataset (Figure 3b). Then, we confirmed by qPCR that TGF-β1 expression in FBXO18-AS-overexpressing SGC7901 and MKN-45 cells increased in comparison to the control group (Figure 4b). Nevertheless, the opposite results were observed in FBXO18-AS-silenced AGS and HGC-27 cells (Figure 4c). In addition, ELISAs also confirmed that TGF-β1 expression in FBXO18-AS-overexpressing MKN-45 and SGC7901 cells was increased, while it was decreased in FBXO18-AS-silenced AGS and HGC-27 cells (Figure 4 d,e). Additionally, Western blotting was carried out to explore the expression of proteins downstream of TGF-β1/Smad signaling, which showed that Slug, Snail and p- Smad2 were altered in FBXO18-AS-overexpressing MKN-45 and SGC7901 cells. However, the opposite results were obtained in FBXO18-AS-silenced AGS and HGC-27 cells (Figure 4 f,g). Moreover, to determine whether FBXO18-AS could bind to the TGF-β1 protein, we performed catRAPID analysis and found that FBXO18-AS may bind to the TGF-β1 protein at two sites (26-77 bp and 1001-1075 bp) (Figure 4h). Taken together, these results show that FBXO18-AS promoted TGF-β1 expression and activated TGF-β1/Smad signaling.
Figure 4.
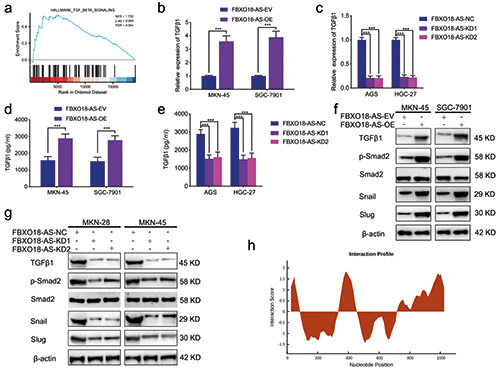
FBXO18-AS promoted TGF-β1 expression and activated TGF-β1/Smad signaling. a) FBXO18-AS expression was linked to TGF-β1/Smad signaling via GSEA based on TCGA databases. b,c) qPCR showed that TGF-β1 expression was changed in FBXO18-ASoverexpressing MKN-45 and SGC7901 cells (b) and FBXO18-AS-silenced AGS and HGC-27 cells (c) (MKN-45 and SGC7901 cells: p<0.001; Student’s t-test; AGS and HGC-27 cells: p<0.001; one-way ANOVA). d,e) ELISA tests showed that TGF-β1 expression was changed in FBXO18-AS-overexpressing MKN-45 and SGC7901 cells and FBXO18-AS-silenced AGS and HGC-27 cells (MKN-45 and SGC7901 cells: p<0.001; Student’s t-test; AGS and HGC-27 cells: p<0.001; one-way ANOVA). f,g) Western blotting showed that TGF- β1 expression and the proteins downstream of the TGF-β1/Smad signaling pathway were changed in FBXO18-AS-overexpressing MKN- 45 and SGC7901 cells and FBXO18-AS-silenced AGS and HGC-27 cells. h) CatRAPID analysis showed that FBXO18-AS may bind to the TGF-β1 protein at two sites (26-77 bp and 1001-1075 bp). The overall data are presented as the mean ± SD (three independent tests); ***p<0.001; **p<0.01; *p<0.05.
Figure 5.
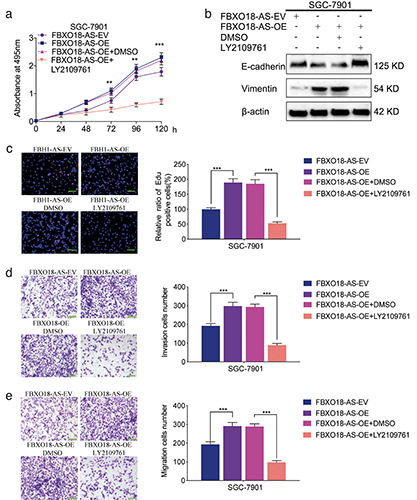
FBXO18-AS promoted cancer cell proliferation and invasion via TGF-β1/Smad signaling. a) MTS assays indicated that FBXO18-AS overexpression impacted the survival of SGC7901 cells, while this change was reversed after LY2109761 treatment (oneway ANOVA; p<0.01). b) Western blotting illustrated that FBXO18-AS overexpression affected vimentin and E-cadherin expression in SGC7901 cells; however, this change was reversed after LY2109761 treatment. c) EdU assays revealed that FBXO18-AS overexpression impacted SGC7901 cell proliferation, while this change was reversed after LY2109761 treatment; scale bars: 100 μm (p<0.001; Student’s t-test). d,e) Transwell (d) and migration assays (e) showed that FBXO18-AS overexpression affected SGC7901 cell invasion and migration, whereas this variation was reversed after LY2109761 treatment; scale bars: 50 μm (p<0.001; Student’s t-assay). The overall data are presented as the mean ± SD (three independent tests); ***p<0.001; **p<0.01; *p<0.05.
FBXO18-AS promoted the proliferation and invasion of gastric cancer via TGF-β1/Smad signaling
As previously described, we showed that FBXO18-AS promoted the expression of TGF-β1 and activated TGF-β1/Smad signaling. We further studied whether FBXO18-AS can facilitate gastric cancer proliferation and invasion via TGF-β1/Smad signaling. We performed MTS assays and found that the viability of SGC7901 cells was increased after FBXO18-AS overexpression but decreased after inhibition of TGF-β1/Smad signaling by LY2109761 treatment (Figure 5a). EdU assays also showed that the EdU-positive cell rate among SGC7901 cells was increased after FBXO18-AS overexpression and decreased following treatment with the TGF-β1/Smad signaling inhibitor LY2109761 (Figure 5c). Then, migration and Transwell assays showed that SGC7901 cell migration and invasion increased after FBXO18-AS overexpression but decreased after treatment with the TGF- β1/Smad signaling inhibitor LY2109761 (Figure 5 d,e). In addition, the expression of an EMT-like process marker was detected by Western blotting, and the results illustrated that the expression of E-cadherin decreased and that of vimentin increased among FBXO18-AS-overexpressing SGC7901 cells, while these changes were reversed following treatment with the TGF-β1/Smad signaling inhibitor LY2109761 (Figure 5b). Therefore, these results show that FBXO18-AS promoted gastric cancer proliferation and invasion through TGF-β1/Smad signaling.
Figure 6.
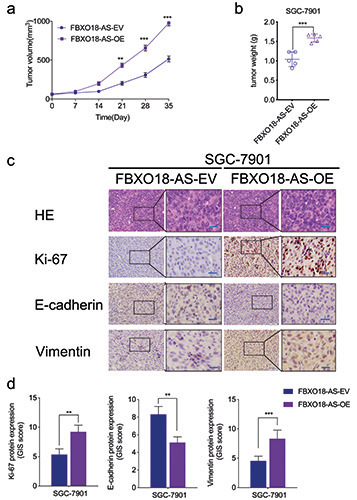
FBXO18-AS promoted the growth of gastric cancer in vivo. a,b) The measured tumor volumes (a), along with tumor weights (b), were increased after FBXO18-AS overexpression (one-way ANOVA; tumor volumes: p<0.01; neoplasm weights: p<0.001; Student’s t-test). c) Typical immunohistochemical staining elucidated the variations in vimentin, E-cadherin and Ki-67 in the negative control and FBXO18-AS overexpression groups in the patterns of the orthotopic xenografts; scale bars: 50 μm. d) Quantitative analysis of IHC staining according to the German immunohistochemical scoring system. The overall data are expressed as the mean ± SD (three independent tests); ***p<0.001; **p<0.01; *p<0.05.
FBXO18-AS promoted gastric cancer growth in vivo
Xenografts were used to verify the impacts of FBXO18-AS on the tumorigenesis and progression of gastric cancer in vivo. In comparison to the control group, tumor volumes and weights were increased in the FBXO18-AS overexpression group (Figure 6 a,b). Immunohistochemistry staining illustrated that Ki-67 and vimentin expression was increased in the FBXO18-AS overexpression group in comparison to the control group. Nevertheless, E-cadherin expression was reduced in the FBXO18-AS overexpression group (Figure 6 c,d). Together, these results show that FBXO18-AS promoted the tumorigenesis and progression of gastric cancer in vivo.
Discussion
Gastric cancer is regarded as a common and malignant tumor and is the third highest cause of death among all cancers worldwide. 18 Clinical treatment options for gastric cancer are available, such as radical surgical resection, radiotherapy, and chemotherapy, but the mortality of patients with gastric cancer remains high.19 The specific mechanisms of gastric cancer occurrence and progression are not completely understood and are still significantly valuable to study. It has been confirmed that the incidence of gastric cancer is related to changes in corresponding RNA expression in gastric cells.20 Therefore, the specific molecular mechanism should be evaluated to study the possible diagnostic markers and clinical therapeutic targets of gastric cancer.
LncRNAs are regarded as one kind of ncRNA whose length exceeds 200 nt and does not possess any potential for protein coding. They also play an essential role in altering various cell processes in diverse cancers.5,21,22 It has been reported that many lncRNAs are abnormally expressed in gastric cancer and participate in chemoresistance by regulating disparate target genes.23-25 For example, lncRNA MALAT1 was confirmed to regulate PTX resistance in gastric cancer cells by targeting miR-23b-3p, along with ATG12.26 LncRNA ZFAS1 was revealed to improve PTX resistance in the gastric cancer cell line SGC7901 by varying EMT marker expression, cell cycle-related proteins, and the Wnt/β- catenin signaling pathway.27
Based on existing research, we initially discovered that lncRNA FBXO18-AS expression was increased in gastric cancer and linked to poorer outcomes in patients with gastric cancer. Furthermore, we confirmed that FBXO18-AS promoted the proliferation of gastric cancer cells by MTS assays and EdU assays. Interestingly, we also confirmed that FBXO18-AS promoted the migration, invasion, and an EMT-like process of gastric cancer cells in vitro by migration assays, Western blotting, and transwell assays to explore EMT marker expression. Therefore, it is suggested that altering the expression of FBXO18-AS in gastric cancer could impact the development of gastric cancer in patients.
To study the specific molecular mechanism by which FBXO18-AS regulates the progression of gastric cancer, we performed GSEA and further confirmed that FBXO18-AS could regulate TGF-β1 expression and activate TGF-β1/Smad signaling. Previous studies have confirmed that TGF-β/Smad signaling is one of the most pivotal signaling pathways in altering the EMT process of tumors.13,14 As an example, circ_0006089 facilitates the growth of gastric cancer, glycolysis, metastasis, and angiogenesis by altering miR-361-3p/TGFB1.28 LncRNA SND1-IT1 boosts TGF-β1- induced EMT through the miR-124/COL4A1 axis in gastric cancer. 2 In our study, TGF-β1/Smad signaling was also confirmed to participate in gastric cancer cell proliferation and invasion, which were regulated using FBXO18-AS. After treatment with the TGF- β1/Smad signaling inhibitor LY2109761, the promoting effects of FBXO18-AS overexpression on gastric cancer were reversed. In addition, we obtained the same conclusion in vivo. Therefore, our study highlights FBXO18-AS as a clinical diagnosis marker and possible medicinal target in gastric cancer.
Our study found a novel lncRNA, FBXO18-AS, that was overexpressed in gastric cancer and linked to poor outcomes. FBXO18- AS could increase the proliferation, migration, invasion and an EMT-like process in gastric cancer in vitro and in vivo. Mechanistically, FBXO18-AS promoted TGF-β1 expression and activated TGF-β1/Smad signaling. Therefore, we discovered a novel mechanism of lncRNAs in the emergence and progression of gastric cancer. This brings FBXO18-AS to the forefront in the diagnosis and prognosis of gastric cancer, and it may become a molecular target for the treatment of gastric cancer.
Funding Statement
Funding: this work was supported by the Public Welfare Technology Research Plan / Social Development Project of Zhejiang, China (No. LGF20H070003) and Wenzhou basic science research project (No. Y20220889).
References
- 1.Smyth EC, Nilsson M, Grabsch HI, Van Grieken NCT, Lordick F. Gastric cancer. Lancet 2020;396:635-48. [DOI] [PubMed] [Google Scholar]
- 2.Song Z, Wu Y, Yang J, Yang D, Fang X. Progress in the treatment of advanced gastric cancer. Tumour Biol 2017. 39:1010428317714626. [DOI] [PubMed] [Google Scholar]
- 3.Feng W, Ding Y, Zong W, Ju S. Non-coding RNAs in regulating gastric cancer metastasis. Clin Chim Acta 2019;496:125-33. [DOI] [PubMed] [Google Scholar]
- 4.Peng Z, Wang CX, Fang EH, Wang G B, Tong Q. Role of epithelial-mesenchymal transition in gastric cancer initiation and progression. World J Gastroenterol 2014;20:5403-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei L, Sun J, Zhang N, Zheng Y, Wang X, Lv L, et al. Noncoding RNAs in gastric cancer: implications for drug resistance. Mol Cancer 2020;19:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang XQ, Leung G K. Long non-coding RNAs in glioma: functional roles and clinical perspectives. Neurochem Int 2014;77:78-85. [DOI] [PubMed] [Google Scholar]
- 7.Chen M, Wu X, Ma W, Zhou Q, Wang X, Zhang R, et al. Decreased expression of lncRNA VPS9D1-AS1 in gastric cancer and its clinical significance. Cancer Biomark 2017;21:23-8. [DOI] [PubMed] [Google Scholar]
- 8.Cech T R, Steitz J A. The noncoding RNA revolution-trashing old rules to forge new ones. Cell 2014;157:77-94. [DOI] [PubMed] [Google Scholar]
- 9.Zhang F, Li Y, Xu W, He L, Tan Y, Xu H. Long non-coding RNA ZFAS1 regulates the malignant progression of gastric cancer via the microRNA-200b-3p/Wnt1 axis. Biosci Biotechnol Biochem 2019;83:1289-99. [DOI] [PubMed] [Google Scholar]
- 10.Ghafouri-Fard S, Taheri M. Long non-coding RNA signature in gastric cancer. Exp Mol Pathol 2020;113:104365. [DOI] [PubMed] [Google Scholar]
- 11.Chen S, Jundi D, Wang W, Ren C. LINC01857 promotes the proliferation, migration, and invasion of gastric cancer cells via regulating miR-4731-5p/HOXC6. Can J Physiol Pharmacol 2022;100:689-701. [DOI] [PubMed] [Google Scholar]
- 12.Zhang P, Chen C, Zhang J, Yu X. LncRNA CRYM-AS1 inhibits gastric cancer progression via epigenetically regulating CRYM. Ann Clin Lab Sci 2022;52:249-59. [PubMed] [Google Scholar]
- 13.Jiang Y, Zhou J, Hou D, Luo P, Gao H, Ma Y, et al. Prosaposin is a biomarker of mesenchymal glioblastoma and regulates mesenchymal transition through the TGF-β1/Smad signaling pathway. J Pathol 2019;249:26-38. [DOI] [PubMed] [Google Scholar]
- 14.Guo Z, Li G, Bian E, Ma CC, Wan J, Zhao B. TGF-β-mediated repression of MST1 by DNMT1 promotes glioma malignancy. Biomed Pharmacother 2017;94:774-80. [DOI] [PubMed] [Google Scholar]
- 15.Jiang Y, Zhou J, Zhao J, Hou D, Zhang H, Li L, et al. MiR-18adownregulated RORA inhibits the proliferation and tumorigenesis of glioma using the TNF-alpha-mediated NF-kappaB signaling pathway. EBioMedicine 2020;52:102651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang Y, Han S, Cheng W, Wang Z, Wu A. NFAT1-regulated IL6 signalling contributes to aggressive phenotypes of glioma. Cell Commun Signal 2017;15:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang Y, Zhou J, Luo P, Gao H, Ma Y, Chen YS, et al. Prosaposin promotes the proliferation and tumorigenesis of glioma through toll-like receptor 4 (TLR4)-mediated NFkappaB signaling pathway. EBioMedicine 2018;37:78-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torre L A, Bray F, Siegel R L, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. [DOI] [PubMed] [Google Scholar]
- 19.Chen H, Guan R, Lei Y, Chen J, Ge Q, Zhang X, et al. Lymphangiogenesis in gastric cancer regulated through Akt/mTOR-VEGF-C/VEGF-D axis. BMC Cancer 2015;15:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. ceRNA in cancer: possible functions and clinical implications. J Med Genet 2015;52:710-8. [DOI] [PubMed] [Google Scholar]
- 21.Li Z, Lü M, Zhou Y, Xu L, Jiang Y, Liu Y, et al. Role of long non-coding RNAs in the chemoresistance of gastric cancer: a systematic review. Onco Targets Ther 2021;14:503-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye Y, Yang S, Han Y, Sun J, Xv L, Wu L, et al. HOXD-AS1 confers cisplatin resistance in gastric cancer through epigenetically silencing PDCD4 via recruiting EZH2. Open Biol 2019;9:190068. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Guo Y, Yue P, Wang Y, Chen G, Li Y. PCAT-1 contributes to cisplatin resistance in gastric cancer through miR-128/ZEB1 axis. Biomed Pharmacother 2019;118:109255. [DOI] [PubMed] [Google Scholar]
- 24.Wu Q, Ma J, Wei J, Meng W, Wang Y, Shi M. FOXD1-AS1 regulates FOXD1 translation and promotes gastric cancer progression and chemoresistance by activating the PI3K/AKT/mTOR pathway. Mol Oncol 2021;15:299-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tian X, Zhu X, Yan T, Yu C, Shen C, Hong J, et al. Differentially expressed lncRNAs in gastric cancer patients: a potential biomarker for gastric cancer prognosis. J Cancer 2017;8:2575-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yiren H, Yingcong Y, Sunwu Y, Keqin L, Xiaochun T, Senrui C, et al. Long noncoding RNA MALAT1 regulates autophagy associated chemoresistance via miR-23b-3p sequestration in gastric cancer. Mol Cancer 2017;16:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu W, He L, Li Y, Tan Y, Zhang F, Xu H. Silencing of lncRNA ZFAS1 inhibits malignancies by blocking Wnt/β-catenin signaling in gastric cancer cells. Biosci Biotechnol Biochem 2018;82:456-65. [DOI] [PubMed] [Google Scholar]
- 28.Zhou Y, Zhang Q, Liao B, Qiu X, Hu S, Xu Q. circ_0006089 promotes gastric cancer growth, metastasis, glycolysis, and angiogenesis by regulating miR-361-3p/TGFB1. Cancer Sci 2022;113:2044-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu YZ, Hu ZL, Liao TY, Li Y, Pan YL. LncRNA SND1-IT1 facilitates TGF-β1-induced epithelial-to-mesenchymal transition via miR-124/COL4A1 axis in gastric cancer. Cell Death Discov 2022;8:73. [DOI] [PMC free article] [PubMed] [Google Scholar]


