Abstract
Background
Thyroid cancer-related deaths mostly result from metastasis. It was reported that the immunometabolism associated enzyme interleukin-4-induced-1 (IL4I1) was related to tumor metastasis. The present study was intended to investigate the effects of IL4I1 on thyroid cancer metastasis and its relationship with the prognosis.
Methods
Data from Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) were analyzed to find out the different mRNA expressions of IL4I1 between thyroid cancer and normal tissues. And Human Protein Atlas (HPA) was used to assess IL4I1 protein expression. To further differentiate thyroid cancer from normal tissues and estimate the impact of IL4I1 on the prognosis, the receiver operating characteristic curve (ROC) and Kaplan–Meier (KM) method was performed. The protein–protein interaction (PPI) network was established using STRING, and functional enrichment analyses were conducted by “clusterProfiler” package. Then, we assayed the correlation between IL4I1 and some related molecules. The relationship between IL4I1 and immune infiltration was performed using “Gene Set Variation Analysis (GSVA)” package in TCGA and tumor-immune system interaction database (TISIDB). Finally, we did in vitro experiments in order to further prove the bioeffects of IL4I1 on metastasis.
Results
The expression of IL4I1 mRNA and IL4I1 protein was significantly upregulated in thyroid cancer tissues. The increment of IL4I1 mRNA expression was related to high-grade malignancy, lymph node metastases and extrathyroidal extension. The ROC curve displayed the cutoff value of 0.782, with the sensitivity of 77.5% and the specificity of 77.8%. KM survival analysis showed that there was a worse PFS in patients with high IL4I1 expression than those with low IL4I1 expression (p = 0.013). Further study indicated that IL4I1 was associated with lactate, body fluid secretion, positive regulation of T cell differentiation, and cellular response to nutrients in Gene Ontology (GO) analysis. Moreover, IL4I1 was found correlated with immune infiltration. Finally, the in vitro experiments revealed the promotion of IL4I1 on cancer cell proliferation, migration and invasion.
Conclusions
The increased IL4I1 expression is markedly correlated with the immune imbalance in the tumor microenvironment (TME) and predicts poor survival in thyroid cancer. This study reveals the potential clinical biomarker of poor prognosis and the target of immune therapy in thyroid cancer.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12902-023-01407-1.
Keywords: Thyroid cancer, IL4I1, Biomarker, Poor prognosis, Immune infiltration
Introduction
In the endocrine system, thyroid cancer is the most common type of endocrine tumors, accounting for approximately 1.7% of all human malignancies [1, 2]. Based on histopathological features, thyroid cancer can be divided into papillary thyroid cancer (PTC), anaplastic thyroid cancer (ATC), follicular thyroid cancer (FTC), and medullary thyroid cancer, of which PTC is the most common type [3]. Most PTC is well-differentiated and has a good prognosis with the mortality rates of 3–10%, but the mortality rates of poorly-differentiated PTC (PDPTC) and ATC are 38–57% and ~ 100%, respectively [4]. Therefore, it is important to detect the thyroid cancer with unfavorable prognosis. Recently, active surveillance is recommended for thyroid cancer [5]. However, we still lack the diagnostic biomarkers and prognostic indices for the mechanisms underlying thyroid cancer metastasis have not been fully understood. Hence, it is pivotal to explore the molecular mechanism that drive metastasis and choose the available biomarkers for tumor surveillance and prognostic assessments in thyroid cancer.
Interleukin-4-induced-1 (IL4I1) is a secreted L-amino acid oxidase that oxidizes l-phenylalanine to corresponding α-ketoacids, H2O2, and ammonia [6, 7]. IL4I1 is reported expressed in macrophages, mature dendritic cells (DC), T cells, and B cells stimulated by interleukin (IL) -4. IL4I1 secreted by antigen presenting cells inhibits human CD4+ and CD8+ T lymphocyte activation and proliferation, in part via the production of H2O2 and some other intracellular signal pathways inside T lymphocytes [8, 9]. Besides, IL4I1 modulates B cell differentiation, especially those involved in the T-cell-dependent immune response [10–12]. Amélie et al. [13, 14] found that IL4I1 was expressed in tumor-associated macrophages (TAM) of most human malignancies, but only in rare solid tumor cells including mesotheliomas, non-small-cell carcinomas, thyroid carcinoma and ovarian carcinoma. As an immunosuppressive enzyme, IL4I1 plays an important role in tumor immune escape and predicts poor prognosis. Thus, IL4I1 could explain partially the mechanisms underlying tumor metastasis and be a potential diagnostic and prognostic biomarker of thyroid cancer.
Here, we intended to explore the correlation between IL4I1 and immune escape in thyroid cancer. In the first place, we evaluated the IL4I1 mRNA and IL4I1 protein expression, and then analyzed its relationship with clinical characteristics (criterion of AJCC 7th) using data from Cancer Genome Atlas (TCGA), Gene Expression Omnibus (GEO) and Human Protein Atlas (HPA). Moreover, given the overexpression of IL4I1 in thyroid cancer, especially in ATC, we hypothesized that IL4I1 predicted poor survival in thyroid cancer. We analyzed data from TCGA and found that IL4I1 was positively correlated to metastasis, poor prognosis and more immune infiltrates in tumor microenvironment. At last, we did some in-vitro experiments to verify IL4I1 promotion of cancer proliferation and metastasis. Here, this study reveals that the high level of IL4I1 promoted immune escape and predicted poor prognosis in thyroid cancer, which provides direction for future research into the novel target for immunotherapy, and diagnostic and unfavorable prognostic biomarker.
Methods
TCGA, and GSE50901, GSE27155 and GSE151181 datasets
TCGA database (https://genome-cancer.ucsc.edu/) [15] was used to analyze the IL4I1 gene expression and its association to the corresponding clinical information. There were 33 cancer types involved and the data was transformed from FPKM into TPM format and log2 conversion when necessary. The gene expression in thyroid cancer from TCGA comprised 502 thyroid cancer tissues and 58 peritumoral normal samples. The transcriptional expression was also analyzed in other three public databases, GSE50901 [16, 17], GSE27155 [18, 19] and GSE151181 [20]. GSE50901 was obtained from the GPL13607 Agilent-028004 SurePrint G3 Human GE 8 × 60 K Microarray, and we chose data of 3 tumor matched PTC and 3 non-tumor matched samples for analysis. GSE27155 was obtained from GPL96 [HG-U133A] Affymetrix Human Genome U133A Array, including 51 PTC, 4 ATC and 4 normal thyroid tissue samples. GSE151181 contains two parts, of which one is miRNA expression data from GPL21575, and the other is gene array from GPL23159. In the present study, only data on gene expression was processed, including 15 tumor samples from PTC before radioiodine (RAI) treatment, 11 tumor samples from PTC after RAI treatment and 6 normal thyroid tissue samples.
Human Protein Atlas (HPA)
The Human Protein Atlas (HPA) (https://proteinatlas.org/) [21, 22] includes the protein level of human genes in normal tissues, as well as in tumor tissues. The IL4I1 protein level was compared between thyroid cancer tissues and normal thyroid tissues in HPA.
Protein–Protein Interaction (PPI) network construction
The online STRING database (http://string-db.org) [23] was adopted to retrieve the genes that interact with IL4I1 gene, selecting the interaction score > 0.4 in the PPI network.
Tumor-Immune System Interaction Database (TISIDB) web
The online web TISIDB [24] (http://cis.hku.hk/TISIDB/) integrates a repository portal for tumor immune system interaction. We analyzed the IL4I1 expression and tumor-infiltrating immuocytes across human cancers using TISIDB. Besides, the correlations of IL4I1 with some tumor-infiltrating lymphocytes (TILs) were measured using the Spearman test.
Cell culture
Cell lines 8505C, TPC-1, KTC-1 and K1 [25] were given by the Department of Nuclear Medicine, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine. All cell lines were cultured at 37 °C in a humidified atmosphere with 5% CO2 using RPMI 1640 (Invitrogen) with 10% fetal bovine serum (FBS; Gibco), 100 U/ml penicillin and 100 μg/ml streptomycin.
Generation of 8505C sub-lines
Parental 8505C cells were infected with IL4I1shRNA-GFP-puro-lentivirus and GFP-puro-lentivirus in order to generate 8505C-shIL4I1 and 8505C-vector. After 72 h, 2 μg/ml puromycin was added to the transfected cells to obtain the population of anti-puro cells.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA of the cells was extracted using MolPure® Cell/Tissue Total RNA Kit (YEASEN, China). cDNA was synthesized using Hifair® III 1st Strand cDNA Synthesis SuperMix for qPCR (gDNA digester plus) (YEASEN, China). qPCR was performed using Hieff® qPCR SYBR Green Master Mix (High Rox Plus) (YEASEN, China) on ABI 7500. Primers sequences for the detection of IL4I1 were 5’-CGCCCGAAGACATCTACCAG-3’ (forward) and 5’-GATATTCCAAGAGCGTGTG CC-3’ (reverse). The primer of Gapdh was 5’-GCAGGGGGGAGCCAAAAGGG-3’ (forward) and 5’- TGCCAGCCCCAGCGTCAAAG-3’ (reverse).
Western blot
For immunoblot analyses, cells were lysed by cell lysis buffer for Western and IP containing protease inhibitor cocktail and quantified using Bradford Protein Quantification Kit (YEASEN, China). Protein was separated by SDS/PAGE, transferred to a PVDF membrane and probed with antibodies against IL4I1 (Abcepta, China) and GAPDH (Sangon Biotech, China).
Cell proliferation assay
Cell counting kit-8 (CCK8) was used to evaluate the proliferation of the 8505C with IL4I1 knocked down according to the manufacturer’s instruction (Sangon Biotech, China). In six 96-well plates, 1,000 cells per well were seeded. And 10 μl per well of CCK8 solution was added on day 1, day 2, day 3 and day 4 after seeding. Each plate was measured at 450 nm in a Multiscan MK3 plate reader (Thermo Fisher Scientific, Rochester, NY, USA).
Migration and invasion assays
8505C-shIL4I1 and its control were starved in a serum-free medium overnight. The next day, all cells were plated at equal density in the serum-free medium into 8 μm pore size 24-well transwell migration or invasion inserts (Corning). 20% FBS medium was added to the bottom well and the plates were incubated at 37 °C in a humidified atmosphere with 5% CO2 for 24 h. The upper inserts were fixed and stained with crystal violet. The non-migrated cells were removed from the inserts. The pictures were taken under the light microscope. The migrated cells were washed with 33% acetic acid and the OD value was read at 560 nm in a Multiscan MK3 plate reader (Thermo Fisher Scientific, Rochester, NY, USA).
Statistical analyses
R (V 3.6.3) was used for the statistical analysis. R package ClusterProfiler and ggplot2 were used to analyze functional enrichment and visualize expression differences. The statistical calculation was done by paired t-test and Mann–Whitney U-test. The survival curve was constructed using the KM method and the log-rank test. pROC package [26] was used to detect the cutoff of IL4I1 in the ROC curve. Cox regression was applied to evaluate the hazard ratio (HR) for the progress free interval (PFI). Two-sided t-test was used where the data met the assumptions of the test and the variance was similar between the two groups being compared. GraphPad Prism 6 was used to analyze data. All p values of less than 0.05 were considered significant.
Results
Expression profile of IL4I1 in pan-cancer and thyroid cancer
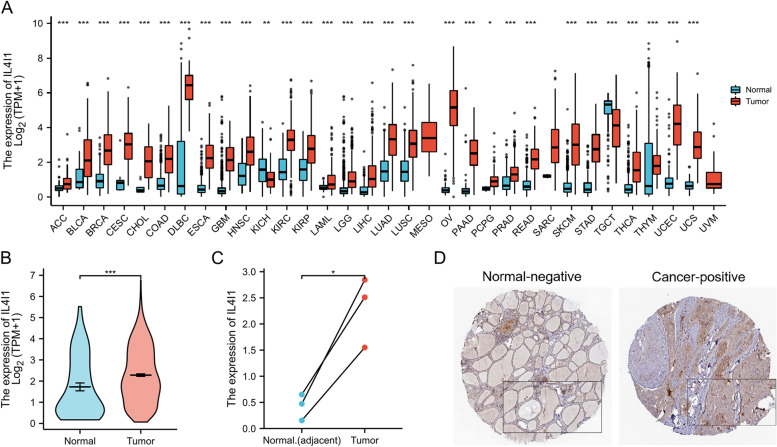
To estimate the gene expression profile of IL4I1 in various cancer types, we included 33 types of cancers containing more than three samples in both the normal and the cancer groups. As is shown in Fig. 1A, IL4I1 was expressed more in tumor groups than in the normal groups in 31 types of all 33 cancer types, including thyroid cancer (Fig. 1A and B). In the GSE50901 matched data, we obtained similar results that IL4I1 expression was higher in tumor tissues (n = 3) than that in the adjacent normal tissues (n = 3), as shown in Fig. 1C (p < 0.05). There are seven immunohistochemistry (IHC) samples stained with IL4I1 antibody from 4 patients in HPA. One patient suffered from follicular adenoma carcinoma, and the other three patients are from papillary adenocarcinoma. The IHC also showed that IL4I1 protein expression was higher in cancer tissues in thyroid cancer (Fig. 1D). All these data indicated that cancer tissues had abnormally higher expression of IL4I1 in both mRNA and protein levels in thyroid cancer.
Fig. 1.
The pattern of IL4I expression in Pan-cancers and in thyroid cancer. A The mRNA expression of IL4I1 was upregulated in 28 of 33 cancers (*** p < 0.001), in KICH (** p < 0.01) and in PCPG (* p < 0.05) compared with normal tissues. B The mRNA expression in THCA from data of TCGA. C The mRNA expression in 3 THCA and the matched-adjacent normal samples. D The protein levels of IL4I1 from data of Human Protein Atlas. ns, no significance; BLCA, bladder urothelial carcinoma; ACC, adrenocortical carcinoma; BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma; DLBC, lymphoid neoplasm diffuse large B-cell lymphoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme; HNSC, head and neck squamous cell carcinoma; KICH, kidney chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LAML, acute myeloid leukemia; LGG, brain lower grade glioma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; MESO, mesothelioma; OV, ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; PCPG, pheochromocytoma and paraganglioma; PRAD, prostate adenocarcinoma; READ, rectum adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; TGCT, testicular germ cell tumors; THCA, thyroid carcinoma; THYM, thymoma; UCEC, uterine corpus endometrial carcinoma; UCS, Uterine Carcinosarcoma; UVM, Uveal Melanoma
The relationship between high expression of IL4I1 mRNA and poor clinicopathologic characteristics of thyroid cancer
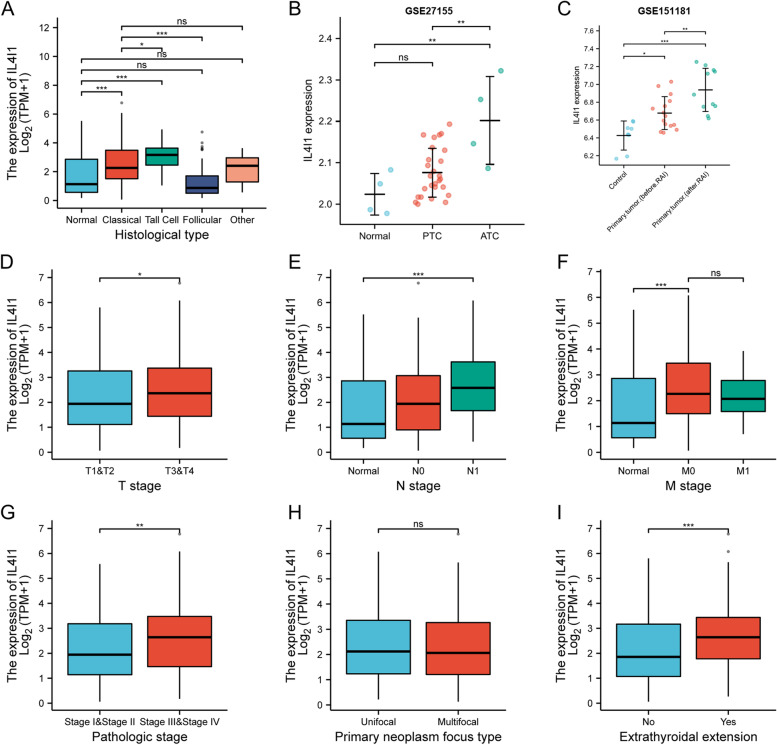
The correlation between IL4I1 expression and clinical pathological features was evaluated by Mann–Whitney U-test and logistic regression analysis. TNM stages used here are according to AJCC 7th. From Fig. 2 and Table 1, we saw that higher expression of IL4I1 is related to higher malignancy grades (Fig. 2A and B, p < 0.01), TNM stage (Fig. 2D, E and F, p < 0.05), and pathologic stage (Fig. 2G, p < 0.01) in thyroid cancer. Furthermore, patients with extrathyroidal metastasis had obvious higher IL4I1 mRNA expression (Fig. 2I, p < 0.001). It is interesting that IL4I1 was upregulated in thyroid cancer tissues (Fig. 2C, p < 0.05), and even higher after (RAI) in GSE151181 database (Fig. 2C, p < 0.01). The effects of RAI on IL4I expression were recently not clear.
Fig. 2.
The relationship between IL4I1 expression and clinical characteristics. The expression of IL4I1 was statistically correlated with histological type (A and B), RAI (C), T stage (D), N stage (E), pathologic stage (G) and extrathyroidal extension (I). And there was no significant correlation between IL4I1 expression and M stage (F) and primary neoplasm focus type (H). (ns, no significance, *p < 0.05, **p < 0.01, ***p < 0.001). PTC, papillary thyroid cancer; ATC, anaplastic thyroid cancer; RAI, radioiodine therapy
Table 1.
Clinical characteristics in thyroid cancer from TCGA data
| Characteristic | Low expression of IL4I1 | High expression of IL4I1 | p |
|---|---|---|---|
| n | 255 | 255 | |
| T stage, n (%) | 0.007* | ||
| T1 | 71 (14%) | 72 (14.2%) | |
| T2 | 100 (19.7%) | 67 (13.2%) | |
| T3 | 75 (14.8%) | 100 (19.7%) | |
| T4 | 8 (1.6%) | 15 (3%) | |
| N stage, n (%) | < 0.001* | ||
| N0 | 126 (27.4%) | 103 (22.4%) | |
| N1 | 88 (19.1%) | 143 (31.1%) | |
| M stage, n (%) | 1.000 | ||
| M0 | 129 (43.7%) | 157 (53.2%) | |
| M1 | 4 (1.4%) | 5 (1.7%) | |
| Age, meidan (IQR) | 46 (34.5, 58) | 46 (36, 58) | 0.883 |
*p < 0.05
Overexpression of IL4I1 mRNA prediction of poor progress-free interval (PFI) in the TCGA database
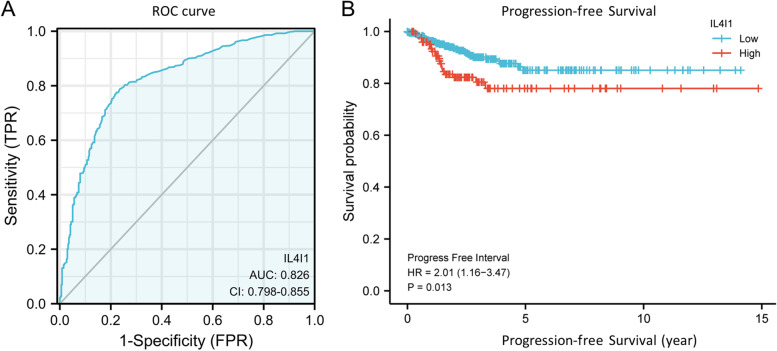
To explore the effects of IL4I1 on the prognosis of thyroid cancer, we first performed the univariate Cox analysis. The results revealed that IL4I1 was one of the risk factors in thyroid cancer based on the TCGA database (HR 1.894; 95% CI 1.083–3.310; p = 0.025; Table 2). Then, we performed ROC analysis and Fig. 3A displayed an AUC value of 0.826 (95% CI: 0.798–0.855). The cutoff value was 0.782, with a sensitivity of 77.5%, and a specificity of 77.8%. The positive predictive value was 84.1% and the negative predictive value was 69.6%. KM curves (Fig. 3B) showed that thyroid cancer patients with high-level IL4I1 had a significantly poor prognosis (HR 2.01; 95% CI 1.16, 3.47; p = 0.013). These data referred that high IL4I1 level would be a clinical biomarker indicating a poor prognosis of thyroid cancer.
Table 2.
Univariate and multivariate Cox proportional hazards analysis of IL4I1 expression
| Characteristics | Total(N) | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|---|
| Hazard ratio (95% CI) | P value | Hazard ratio (95% CI) | P value | ||
| Gender (Male vs. Female) | 510 | 1.694 (0.975–2.945) | 0.062 | 1.390 (0.767–2.519) | 0.278 |
| Age (> 45 vs. < = 45) | 510 | 1.593 (0.920–2.758) | 0.096 | 1.558 (0.856–2.835) | 0.147 |
| T stage (T3&T4 vs. T1&T2) | 508 | 2.450 (1.417–4.236) | 0.001* | 2.134 (0.925–4.925) | 0.076 |
| N stage (N1 vs. N0) | 460 | 1.658 (0.936–2.934) | 0.083 | 1.267 (0.685–2.344) | 0.450 |
| Extrathyroidal extension (Yes vs. No) | 492 | 1.874 (1.092–3.216) | 0.023* | 0.867 (0.385–1.955) | 0.731 |
| Primary neoplasm focus type (Multifocal vs. Unifocal) | 500 | 1.025 (0.595–1.764) | 0.929 | / | / |
| IL4I1 (High vs. Low) | 510 | 1.894 (1.083–3.310) | 0.025* | 1.568 (0.860–2.860) | 0.142 |
*p < 0.05
Fig. 3.
ROC and Kaplan–Meier curves for IL4I1. A ROC curve showed that IL4I1 had an AUC value of 0.826 to discriminate thyroid cancer tissues from normal controls with a cutoff of had no significant statistical difference between high IL4I1 expression and low expression (p = 0.166) indicated by Kaplan–Meier survival curves. B Kaplan–Meier survival curves showed that thyroid cancer patients with high expression of IL4I1 had shorter of PFS) (62 vs. 20 years, p = 0.013)
The interaction analysis of IL4I1 with the related molecules
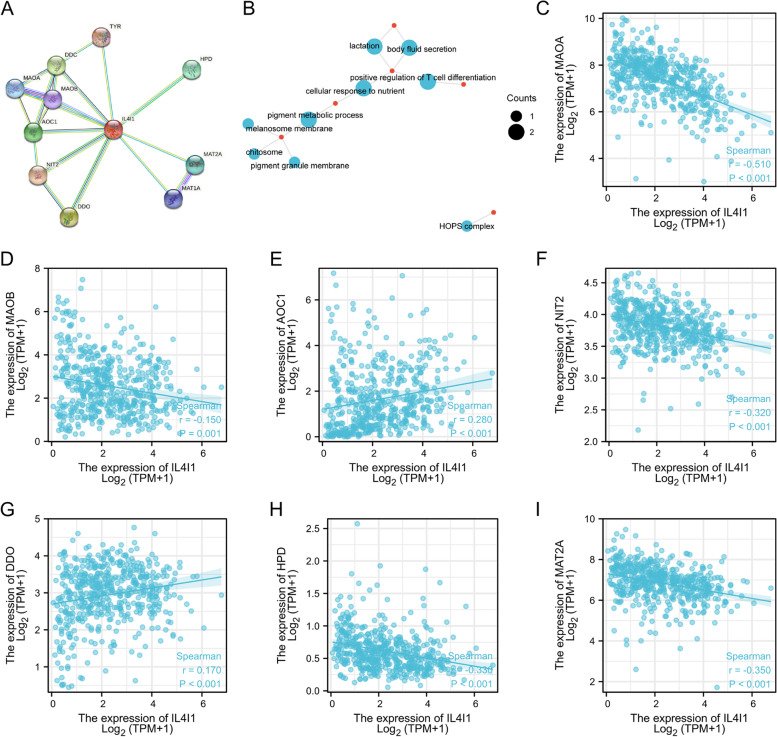
STRING database [23] was used to build protein–protein interaction (PPI) networks. Figure 4A showed the network of IL4I1 and its 10 co-expressed genes. Gene Ontology (GO) analysis revealed the biological process of IL4I1, including lactation, body fluid secretion, positive regulation of T cell differentiation, cellular response to nutrient and pigment metabolic process (Fig. 4B). The Spearman correlation analysis (Fig. 4C-I) demonstrated that there was negative relationship between IL4I1 expression and MAOA (r = -0.51), MAOB (r = -0.15), NIT2 (r = -0.32), HPD (r = -0.33), MAT2A (r = -0.35), and there was positive relationship between IL4I1 expression and AOC1 (r = 0.28,), DDO (r = 0.17) in the co-expressed genes in thyroid cancer from TCGA dataset (all p values ≤ 0.001).
Fig. 4.
PPI network and functional analysis. A A network of IL4I1 and its co-expression genes. B Functional enrichment analysis of 11 involved genes. IL4I1 was involved in lactation, positive regulation of T cell differentiation, body fluid secretion, cellular response to nutrient, pigment metabolic process. C-I The correlation analysis between IL4I1 expression and co-expressed genes in thyroid cancer. MAOA, monoamine oxidase A; MAOB, monoamine oxidase B; AOC1, amine oxidase copper containing 1; NIT2, nitrilase family member 2; DDO, D-aspartate oxidase; HPD, 4-hydroxyphenylpyruvate dioxygenase; MAT2A, methionine adenosyltransferase 2A
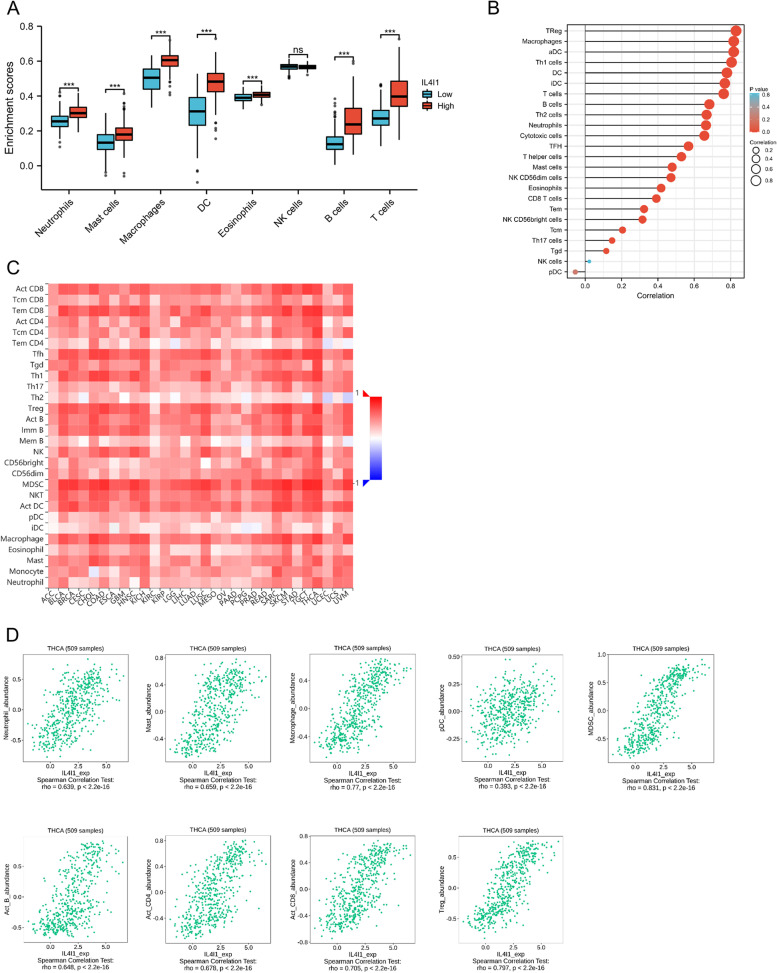
The correlation between IL4I1 level and infiltrating immunocytes
IL4I1 is an immune-associated enzyme secreted by tumor cells. Thus, we assayed the correlation of IL4I1 expression with infiltrating immunocytes around the tumor cells to investigate the changes in the tumor immune microenvironment. In TCGA database, thyroid cancer with high expression of IL4I1 mRNA had more infiltrating neutrophils, mast cells, macrophages, DC, eosinophils, B cells and T cells, but there was no difference in NK cells (Fig. 5A-B). Spearman correlation analysis revealed that IL4I1 expression was significantly related to infiltrating T cells including Treg and Th1 cells (total T cells: r = 0.763; Tregs, r = 0.831; Th1, r = 0.806; all p < 0.001), DC including active DC (aDC) and interdigitating DC (iDC) (DC, r = 0.779; aDC, r = 0.817; iDC, r = 0.768; all p < 0.001) and macrophage (r = 0.818, p < 0.001). At last, we evaluated the correlation between IL4I1 level and the abundance of 28 types of TILs in different human cancers in the TISIDB database, and the results was shown in Fig. 5C. From Fig. 5D, we saw that the high IL4I1 level was correlated with more myeloid-derived suppressor cells (MDSC, r = 0.831), Tregs cells (r = 0.797), macrophage (r = 0.77), CD8+ T cells (r = 0.705), CD4+ T cells (r = 0.678), mast cells (r = 0.659), neutrophil (r = 0.639), B cells (r = 0.648), and plasmacytoid DC (pDC, r = 0.393) (all p < 2.2x10–16). All these data illustrated that IL4I1 could play a particular part in immunocytes infiltration into the tumor microenvironment (TME) in thyroid cancer, which could provide a new perspective on immune escape in thyroid cancer.
Fig. 5.
Correlation of IL4I1 expression with immune cell infiltration level. A The high IL4I1 expression is correlated macrophage, dendritic cells (DC), T cells, B cells and neutrophils. B The correlation analysis between IL4I1 expression and the specific types of the immune cells mentioned above. C Relations between the IL4I1 expression and 28 types of tumors infiltrating lymphocytes (TILs) across human cancers. D IL4I1 expression was correlated with abundance of MDSC, Treg cells, macrophage, CD8 + T cells, CD4 + T cells, mast cells, B cells and neutrophil and DC cells
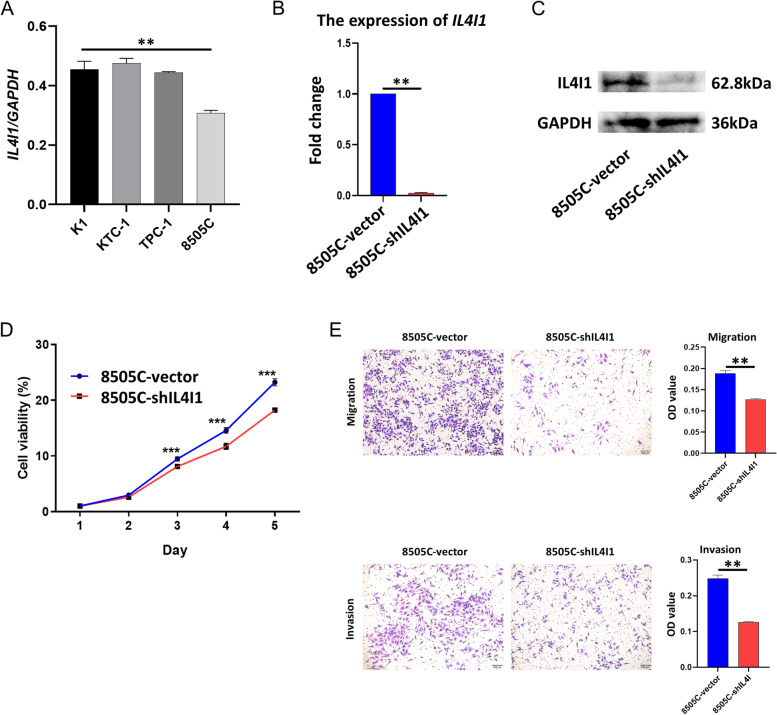
IL4I1 promoting cell proliferation, migration and invasion of thyroid cancer cells in vitro
Finally, we examined the IL4I1 expression in four thyroid cancer cell lines (K1, KTC-1, TPC-1 and 8505C). Interestingly, a much higher IL4I1 expression was found in the 8505C cells (Fig. 6A, p < 0.01). Moreover, we established stably transfected 8505C cell lines expressing shRNA targeting IL4I1 (8505C-shIL4I1) and its control, 8505C-vector (Fig. 6B, C) to study the effects of IL4I1 on cell proliferation and metastasis. The IL4I1 knockdown reduced the in vitro proliferation (Fig. 6D), and decreased the number of 8505C cells migrating across the membrane and the matrixgel invasion (Fig. 6E). Collectively, these data suggest that IL4I1 sustains the proliferation and metastasis potential of thyroid cancers cells in vitro.
Fig. 6.
The effects of IL4I1 on cell proliferation and metastasis in vitro. A 8505C cell (ATC) had the highest IL4I1 expression in the thyroid cancer cell lines K1, KTC-1, TPC-1 and 8505C tested by qPCR (** p < 0.01). B 8505C cell with IL4I1 knocked down by lentivirus containing shRNA targeting IL4I1 (8505C-shIL4I1) had lower IL4I1 expression compared with its control (8505C-vector) (** p < 0.01). C 8505C-shIL4I1 cell had lower IL4I1 protein level compared with 8505C-vector (cropping blots). D The cell proliferation assay showed the impaired proliferation without IL4I1 (*** p < 0.001). E The migration and invasion ability of 8505C-shIL4I1 cell was significantly weaken (** p < 0.01)
Discussion
The incidence of thyroid cancer has increased in the past 30 years, especially in some Asian countries, such as China. With the development of sensitive ultrasonography and computed tomography (CT) scan, we have faced more overdiagnosis for 4 ~ 11% of thyroid tumors are asymptomatic and only found in autopsy studies [27]. Though kinds of researches find the mutations in BARF gene, RAS gene and TERT gene are related to aggressiveness, these mutations may also lead to a potentially excessive treatments, such as more surgeries. Thus, it is necessary to understand the mechanism of thyroid cancer metastasis and to explore effective biomarkers to discriminate metastatic thyroid cancers with poor prognosis from benign ones. Immune escape is one of the pivotal mechanism contributing to malignancy [28]. Some immune checkpoint inhibitors, such as anti-CTLA-4 and anti-PD-L1/PD-1, have been demonstrated in helping to eliminate cancer cells [29, 30]. And some ongoing clinical trials found the therapeutic effects of immune treatment on advanced thyroid cancer [31, 32]. These studies prompted us to examine immune imbalance in aggressive thyroid cancer in depth. As an immune-associated enzyme, interleukin-4-induced-1 (IL4I1) was found overexpressed in tumor cells and to be a parameter predictive of poor prognostic in breast cancer and renal cancer [33]. However, little is known about the IL4I1 biology, including the enzymology, the substrate ranges, the role of downstream metabolites, and the association between IL4I1 and different cancers. Besides, there has been no research on the biological effects of IL4I1 on thyroid cancer. Hence, we conducted this study and found that IL4I1 expression is upregulated in thyroid cancer both in mRNA and in protein level. The upregulated IL4I1 mRNA expression is positively related to lymph node metastasis, high TNM stage and extrathyroidal extension. And high IL4I1 mRNA expression is associated with shorter PFS, which indicated that IL4I1 could be a potential biomarker for poor prognosis in thyroid cancer. What’s more, IL4I1 also predicted the specific immune infiltration in thyroid cancer. The in vitro experiments showed the high expression of IL4I1in the aggressive cell line, 8505C cells, and IL4I1 promoted the proliferation, migration and invasion of 8505C cells.
IL4I1 was first identified in B lymphoblasts which were induced by treatment with IL-4 [34]. For a long time, it was recognized as an L-amino acid oxidase specifically in leukocytes, which oxidizes aromatic amino acids. IL4I1 protein localized in lysosomal, functioned in antigen processing in B cells, and thus act in the immune response. Also, IL4I1 could be secreted into the extracellular environment regulating the immune response. The IL4I1 gene was believed to be expressed restricted to cells of the immune system previously [35]. Further studies found the expression of IL4I1 in the testis and brain both in humans and mice, and there are five isoforms of IL4I1 encoded by the human gene. Isoform 1 is expressed in lymphoid tissue and isoform 2 is expressed in the central nervous system and spermatozoa, while other isoforms are little known recently [36–38].
In this study, we found that IL4I1 was highly expressed in thyroid cancer, and IL4I1 was related to malignant tumor types, such as ATC in thyroid cancer and poor prognosis. In 2003, Copie-Bergman C, et al. [39] initially found the overexpression of IL4I1 in tumor tissues of primary mediastinal B-cell lymphoma. Then, a study on IL4I1 expression by immunochemistry in more than 30 types of human tumor tissues reported that IL4I1 expressed in almost all tumor-infiltrating macrophages, but only in specific types of cancers, such as mesothelioma and B cell lymphomas. Some researchers showed that the tumor tissues expressing IL4I1 biologically has a good prognosis with overexpression of IL4I1 [13]. On the contrary, IL4I1 was found suppressing the effective anti-tumor T-cell response in non-hematological tumors infiltrated by macrophages. As a result, the high expression of IL4I1 in tumor cells promoted the immune escape in human primary cutaneous melanoma [40]. Recent studies suggested that IL4I1 catalyzed tryptophan into indole-3-pyruvate (I3P) and promoted cancer cell motility and metastasis through. I3P-KynA/I3A metabolic pathway [41, 42]. Tiffany Horng [43] et al. found that IL4I1 promoted tumor proliferation by local anti-ferroptosis pathways via its metabolite I3P. Our study found that IL4I1 promoted the proliferation, migration and invasion of 8505C cells probably via immunometabolism of tryptophan catabolism.
The importance of IL4I1 expression in thyroid cancer metastasis and prognosis promoted us to explore the possible potential mechanism. Until recently, it is found that there were many immune cells in the microenvironment of the thyroid cancer and the type and density of the tumor-infiltrating immune cells were correlated with the prognosis in thyroid cancer patients [44]. Around the thyroid cancer cells were many immunosuppressive cells such as MDSC, Tregs (regulatory T cells), macrophages, and so on. Besides, thyroid cancer cells upregulated negative immune checkpoints, such as CTLA-4 (cytotoxic T-lymphocyte associated protein 4) and PD-L1 (programmed death-ligand 1), as well as immunosuppressive enzymes (IDO1, indoleamine 2,3-dioxygenase 1). MDSC is a type of immune cells induced by the tumor that mediates immune tolerance. Angell et al. [45] found that MDSC could be used to detect malignancy and predict the extent of disease and risk of persistence of thyroid cancer. Aggressive features of thyroid cancer and the lymph node metastasis had a high density of Treg infiltration [46, 47]. TAMs are the typical immune cells in the microenvironment of thyroid cancer and their infiltration was related to larger tumors, lymph node metastasis, capsular invasion, extrathyroidal extension and poor prognosis [48–50]. CD8+ T cells were usually considered the anti-tumor immune cells, but the enrichment of CD8+ T cells was associated with thyroid cancer recurrence, because CD8+ cells around tumor cells were found at the state of anergy [51].
What’s more, immunoregulatory enzymes, such as IDO1 and IL4I1, also created and sustained the tumor-tolerant microenvironment, which is expressed either by the tumor cells or by immune cells [7, 52]. IDO1 and IL4I1 were both oxidoreductases catalyzing the degradation of tryptophan to kynurenine. It is reported that cancers expressed high levels of IL4I1, inducing the differentiation of Tregs, promoting CD8+ T cell death, recruiting immunosuppressive TAMs and suppressing T cell proliferation and function via the Kyn-AHR axis [42]. In this study, we found that IL4I1 was positively related to infiltrating MDSC, Treg, macrophage, and CD8+ T cells, which implied that IL4I1 promoted the metastasis of thyroid cancer by inhibiting the anti-tumor immune response.
We found and verified the high expression of IL4I1 in thyroid cancer and explored the immune mechanism of its promotion in metastasis. Nevertheless, there are some limitations to this study. First, there were para-tumor tissues representing normal tissues in TCGA and there was a small sample size included in the normal group included in the analysis. Therefore, the bias was inevitable. Second, the microarray-based bioinformatic analysis is powerful in analyzing the molecular mechanisms and in verifying the effects of IL4I1 as a potential biomarker of poor prognosis in thyroid cancer, but experiments may be needed to confirm the relationship and the regulation of IL4I1 on immune cells in the tumor microenvironment. And it is interesting to perform further studies using in vitro and in vivo experiments, like flow cytometry, immunocytochemistry and conditional culture et al. on the detailed cell types and mechanisms.
Conclusions
In conclusion, IL4I1 contributed to tumor metastasis and poor prognosis of thyroid cancer integrating the analysis of TCGA and GSE datasets. As the L-amino acid oxidase, IL4I1 modulated amino acid metabolism, which is the pivotal nutrient of many tumor-infiltrating immune cells. Therefore, IL4I1 was considered to regulate the proliferation, differentiation and function of tumor cells by immune escape. Our results suggest that IL4I1 may be a potential diagnostic and prognostic marker for metastatic thyroid cancer. And further experiments are needed to prove the immunoregulating effect of IL4I1.
Supplementary Information
Acknowledgements
Not applicable.
Abbreviations
- aDC
Active dendritic cells
- AOC1
Amine oxidase copper containing 1
- ATC
Anaplastic thyroid cancer
- CCK8
Cell counting kit-8
- CT
Computed tomography
- CTLA-4
Cytotoxic T-lymphocyte associated protein 4
- DC
Dendritic cells
- DDO
D-aspartate oxidase
- FTC
Follicular thyroid cancer
- GEO
Gene Expression Omnibus
- GO
Gene Ontology
- GSVA
Gene Set Variation Analysis
- HPA
Human Protein Atlas
- HPD
4-Hydroxyphenylpyruvate dioxygenase
- HR
Hazard ratio
- iDC
Interdigitating DC
- IDO1
Indoleamine 2,3-dioxygenase 1
- IHC
Immunohistochemistry
- IL4I1
Interleukin-4-induced-1
- KM
Kaplan-Meier
- MAOA
Monoamine oxidase A
- MAOB
Monoamine oxidase B
- MAT2A
Methionine adenosyltransferase 2A
- MDSC
Myeloid-derived suppressor cells
- NIT2
Nitrilase family member 2
- PD-L1
Programmed death-ligand 1
- PDTC
Poorly differentiated thyroid cancer
- PFI
Progress free interval
- PPI
Protein-protein interaction
- PTC
Papillary thyroid cancer
- qRT-PCR
Quantitative real-time polymerase chain reaction
- RAI
Radioiodine
- RAI
Radioactive iodine therapy
- ROC
Receiver operating characteristic curve
- TAM
Tumor-associated macrophages
- TCGA
The Cancer Genome Atlas
- TILs
Tumor-infiltrating lymphocytes
- TILs
Tumor-infiltrating lymphocytes
- TISIDB
Tumor-immune system interaction database
- TME
Tumor microenvironment
- Treg
Regulatory T cells
Authors’ contributions
JH designed and supervised the study. LZ analyzed and interpreted the data. JW performed most of the experiments. JH and LZ wrote the manuscript. All authors reviewed and edited the article. LZ and JW equally contributed as first authors. LZ was chosen as the first because she analyzed and organized all data and wrote most of the article.
Funding
Ruijin Hospital Excellent Youth Training Program (Grant No. KY2021627). The research, assessment and prevention of age and age-related diseases (Grant No. XK20210060).
Availability of data and materials
The data generated and analyzed in the current study are available in GSE50901 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE50901), GSE27155 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE27155), GSE151181 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE151181), TCGA database (https://genome-cancer.ucsc.edu/), HPA (https://proteinatlas.org/), STRING database (http://string-db.org) and TISIDB (http://cis.hku.hk/TISIDB/).
Declarations
Ethics approval and consent to participate
This article has no ethic approval or consent, for there were no human participants or animals contained in this study.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Liying Zhu and Jun Wang contributed equally to this work and share first authorship.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64(1):9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Lodewijk L, Prins AM, Kist JW, Valk GD, Kranenburg O, Rinkes IH, et al. The value of miRNA in diagnosing thyroid cancer: a systematic review. Cancer Biomark. 2012;11(6):229–238. doi: 10.3233/CBM-2012-0273. [DOI] [PubMed] [Google Scholar]
- 3.Xing M. BRAF mutation in thyroid cancer. Endocr Relat Cancer. 2005;12(2):245–262. doi: 10.1677/erc.1.0978. [DOI] [PubMed] [Google Scholar]
- 4.Xu B, Ghossein R. Genomic landscape of poorly differentiated and anaplastic thyroid carcinoma. Endocr Pathol. 2016;27(3):205–212. doi: 10.1007/s12022-016-9445-4. [DOI] [PubMed] [Google Scholar]
- 5.Lowenstein LM, Basourakos SP, Williams MD, Troncoso P, Gregg JR, Thompson TC, et al. Active surveillance for prostate and thyroid cancers: evolution in clinical paradigms and lessons learned. Nat Rev Clin Oncol. 2019;16(3):168–184. doi: 10.1038/s41571-018-0116-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bod L, Lengagne R, Wrobel L, Ramspott JP, Kato M, Avril MF, et al. IL4-induced gene 1 promotes tumor growth by shaping the immune microenvironment in melanoma. Oncoimmunology. 2017;6(3):e1278331. doi: 10.1080/2162402X.2016.1278331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Molinier-Frenkel V, Prevost-Blondel A, Castellano F. The IL4I1 enzyme: a new player in the immunosuppressive tumor microenvironment. Cells. 2019;8(7):757. doi: 10.3390/cells8070757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boulland ML, Marquet J, Molinier-Frenkel V, Moller P, Guiter C, Lasoudris F, et al. Human IL4I1 is a secreted L-phenylalanine oxidase expressed by mature dendritic cells that inhibits T-lymphocyte proliferation. Blood. 2007;110(1):220–227. doi: 10.1182/blood-2006-07-036210. [DOI] [PubMed] [Google Scholar]
- 9.Aubatin A, Sako N, Decrouy X, Donnadieu E, Molinier-Frenkel V, Castellano F. IL4-induced gene 1 is secreted at the immune synapse and modulates TCR activation independently of its enzymatic activity. Eur J Immunol. 2018;48(1):106–119. doi: 10.1002/eji.201646769. [DOI] [PubMed] [Google Scholar]
- 10.Caron G, Le Gallou S, Lamy T, Tarte K, Fest T. CXCR4 expression functionally discriminates centroblasts versus centrocytes within human germinal center B cells. J Immunol. 2009;182(12):7595–7602. doi: 10.4049/jimmunol.0804272. [DOI] [PubMed] [Google Scholar]
- 11.Victora GD, Schwickert TA, Fooksman DR, Kamphorst AO, Meyer-Hermann M, Dustin ML, et al. Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell. 2010;143(4):592–605. doi: 10.1016/j.cell.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bod L, Douguet L, Auffray C, Lengagne R, Bekkat F, Rondeau E, et al. IL-4-induced gene 1: a negative immune checkpoint controlling B cell differentiation and activation. J Immunol. 2018;200(3):1027–1038. doi: 10.4049/jimmunol.1601609. [DOI] [PubMed] [Google Scholar]
- 13.Carbonnelle-Puscian A, Copie-Bergman C, Baia M, Martin-Garcia N, Allory Y, Haioun C, et al. The novel immunosuppressive enzyme IL4I1 is expressed by neoplastic cells of several B-cell lymphomas and by tumor-associated macrophages. Leukemia. 2009;23(5):952–960. doi: 10.1038/leu.2008.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lasoudris F, Cousin C, Prevost-Blondel A, Martin-Garcia N, Abd-Alsamad I, Ortonne N, et al. IL4I1: an inhibitor of the CD8(+) antitumor T-cell response in vivo. Eur J Immunol. 2011;41(6):1629–1638. doi: 10.1002/eji.201041119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomczak K, Czerwinska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn) 2015;19(1A):A68–77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barros-Filho MC, Marchi FA, Pinto CA, Rogatto SR, Kowalski LP. High diagnostic accuracy based on CLDN10, HMGA2, and LAMB3 transcripts in papillary thyroid carcinoma. J Clin Endocrinol Metab. 2015;100(6):E890–E899. doi: 10.1210/jc.2014-4053. [DOI] [PubMed] [Google Scholar]
- 17.Barros-Filho MC, de Mello JBH, Marchi FA, Pinto CAL, da Silva IC, Damasceno PKF, et al. GADD45B transcript is a prognostic marker in papillary thyroid carcinoma patients treated with total thyroidectomy and radioiodine therapy. Front Endocrinol (Lausanne) 2020;11:269. doi: 10.3389/fendo.2020.00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giordano TJ, Kuick R, Thomas DG, Misek DE, Vinco M, Sanders D, et al. Molecular classification of papillary thyroid carcinoma: distinct BRAF, RAS, and RET/PTC mutation-specific gene expression profiles discovered by DNA microarray analysis. Oncogene. 2005;24(44):6646–6656. doi: 10.1038/sj.onc.1208822. [DOI] [PubMed] [Google Scholar]
- 19.Giordano TJ, Au AY, Kuick R, Thomas DG, Rhodes DR, Wilhelm KG, Jr, et al. Delineation, functional validation, and bioinformatic evaluation of gene expression in thyroid follicular carcinomas with the PAX8-PPARG translocation. Clin Cancer Res. 2006;12(7 Pt 1):1983–1993. doi: 10.1158/1078-0432.CCR-05-2039. [DOI] [PubMed] [Google Scholar]
- 20.Colombo C, Minna E, Gargiuli C, Muzza M, Dugo M, De Cecco L, et al. The molecular and gene/miRNA expression profiles of radioiodine resistant papillary thyroid cancer. J Exp Clin Cancer Res. 2020;39(1):245. doi: 10.1186/s13046-020-01757-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 22.Uhlen M, Zhang C, Lee S, Sjostedt E, Fagerberg L, Bidkhori G, et al. A pathology atlas of the human cancer transcriptome. Science. 2017;357(6352):eaan2507. doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 23.Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49(D1):D605–D612. doi: 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ru B, Wong CN, Tong Y, Zhong JY, Zhong SSW, Wu WC, et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics. 2019;35(20):4200–4202. doi: 10.1093/bioinformatics/btz210. [DOI] [PubMed] [Google Scholar]
- 25.Meireles AM, Preto A, Rocha AS, Rebocho AP, Maximo V, Pereira-Castro I, et al. Molecular and genotypic characterization of human thyroid follicular cell carcinoma-derived cell lines. Thyroid. 2007;17(8):707–715. doi: 10.1089/thy.2007.0097. [DOI] [PubMed] [Google Scholar]
- 26.Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miranda-Filho A, Lortet-Tieulent J, Bray F, Cao B, Franceschi S, Vaccarella S, et al. Thyroid cancer incidence trends by histology in 25 countries: a population-based study. Lancet Diabetes Endocrinol. 2021;9(4):225–234. doi: 10.1016/S2213-8587(21)00027-9. [DOI] [PubMed] [Google Scholar]
- 28.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 29.Gunda V, Gigliotti B, Ndishabandi D, Ashry T, McCarthy M, Zhou Z, et al. Combinations of BRAF inhibitor and anti-PD-1/PD-L1 antibody improve survival and tumour immunity in an immunocompetent model of orthotopic murine anaplastic thyroid cancer. Br J Cancer. 2018;119(10):1223–1232. doi: 10.1038/s41416-018-0296-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gunda V, Gigliotti B, Ashry T, Ndishabandi D, McCarthy M, Zhou Z, et al. Anti-PD-1/PD-L1 therapy augments lenvatinib’s efficacy by favorably altering the immune microenvironment of murine anaplastic thyroid cancer. Int J Cancer. 2019;144(9):2266–2278. doi: 10.1002/ijc.32041. [DOI] [PubMed] [Google Scholar]
- 31.French JD. Immunotherapy for advanced thyroid cancers - rationale, current advances and future strategies. Nat Rev Endocrinol. 2020;16(11):629–641. doi: 10.1038/s41574-020-0398-9. [DOI] [PubMed] [Google Scholar]
- 32.Moretti S, Menicali E, Nucci N, Guzzetti M, Morelli S, Puxeddu E. Therapy of endocrine disease immunotherapy of advanced thyroid cancer: from bench to bedside. Eur J Endocrinol. 2020;183(2):R41–R55. doi: 10.1530/EJE-20-0283. [DOI] [PubMed] [Google Scholar]
- 33.Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14(5):518–527. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- 34.Chu CC, Paul WE. Expressed genes in interleukin-4 treated B cells identified by cDNA representational difference analysis. Mol Immunol. 1998;35(8):487–502. doi: 10.1016/S0161-5890(98)00031-5. [DOI] [PubMed] [Google Scholar]
- 35.Mason JM, Naidu MD, Barcia M, Porti D, Chavan SS, Chu CC. IL-4-induced gene-1 is a leukocyte L-amino acid oxidase with an unusual acidic pH preference and lysosomal localization. J Immunol. 2004;173(7):4561–4567. doi: 10.4049/jimmunol.173.7.4561. [DOI] [PubMed] [Google Scholar]
- 36.Chavan SS, Tian W, Hsueh K, Jawaheer D, Gregersen PK, Chu CC. Characterization of the human homolog of the IL-4 induced gene-1 (Fig1) Biochim Biophys Acta. 2002;1576(1–2):70–80. doi: 10.1016/S0167-4781(02)00295-6. [DOI] [PubMed] [Google Scholar]
- 37.Houston B, Curry B, Aitken RJ. Human spermatozoa possess an IL4I1 l-amino acid oxidase with a potential role in sperm function. Reproduction. 2015;149(6):587–596. doi: 10.1530/REP-14-0621. [DOI] [PubMed] [Google Scholar]
- 38.Wiemann S, Kolb-Kokocinski A, Poustka A. Alternative pre-mRNA processing regulates cell-type specific expression of the IL4l1 and NUP62 genes. BMC Biol. 2005;3:16. doi: 10.1186/1741-7007-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Copie-Bergman C, Boulland ML, Dehoulle C, Moller P, Farcet JP, Dyer MJ, et al. Interleukin 4-induced gene 1 is activated in primary mediastinal large B-cell lymphoma. Blood. 2003;101(7):2756–2761. doi: 10.1182/blood-2002-07-2215. [DOI] [PubMed] [Google Scholar]
- 40.Prevost-Blondel A, Richard Y. Interleukin 4-induced gene 1 as an emerging regulator of B-cell biology and its role in cutaneous melanoma. Crit Rev Immunol. 2019;39(1):39–57. doi: 10.1615/CritRevImmunol.2019030020. [DOI] [PubMed] [Google Scholar]
- 41.Wang Z, Li T, Mao C, Liu W, Tao Y. IL4I1-driven AHR signature: a new avenue for cancer therapy. Signal Transduct Target Ther. 2021;6(1):118. doi: 10.1038/s41392-021-00529-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sadik A, Somarribas Patterson LF, Ozturk S, Mohapatra SR, Panitz V, Secker PF, et al. IL4I1 is a metabolic immune checkpoint that activates the AHR and promotes tumor progression. Cell. 2020;182(5):1252–70 e34. doi: 10.1016/j.cell.2020.07.038. [DOI] [PubMed] [Google Scholar]
- 43.Zeitler L, Fiore A, Meyer C, Russier M, Zanella G, Suppmann S, et al. Anti-ferroptotic mechanism of IL4i1-mediated amino acid metabolism. Elife. 2021;10:e64806. doi: 10.7554/eLife.64806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gooden MJ, de Bock GH, Leffers N, Daemen T, Nijman HW. The prognostic influence of tumour-infiltrating lymphocytes in cancer: a systematic review with meta-analysis. Br J Cancer. 2011;105(1):93–103. doi: 10.1038/bjc.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Angell TE, Lechner MG, Smith AM, Martin SE, Groshen SG, Maceri DR, et al. Circulating myeloid-derived suppressor cells predict differentiated thyroid cancer diagnosis and extent. Thyroid. 2016;26(3):381–389. doi: 10.1089/thy.2015.0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.French JD, Weber ZJ, Fretwell DL, Said S, Klopper JP, Haugen BR. Tumor-associated lymphocytes and increased FoxP3+ regulatory T cell frequency correlate with more aggressive papillary thyroid cancer. J Clin Endocrinol Metab. 2010;95(5):2325–2333. doi: 10.1210/jc.2009-2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.French JD, Kotnis GR, Said S, Raeburn CD, McIntyre RC, Jr, Klopper JP, et al. Programmed death-1+ T cells and regulatory T cells are enriched in tumor-involved lymph nodes and associated with aggressive features in papillary thyroid cancer. J Clin Endocrinol Metab. 2012;97(6):E934–E943. doi: 10.1210/jc.2011-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qing W, Fang WY, Ye L, Shen LY, Zhang XF, Fei XC, et al. Density of tumor-associated macrophages correlates with lymph node metastasis in papillary thyroid carcinoma. Thyroid. 2012;22(9):905–910. doi: 10.1089/thy.2011.0452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fang W, Ye L, Shen L, Cai J, Huang F, Wei Q, et al. Tumor-associated macrophages promote the metastatic potential of thyroid papillary cancer by releasing CXCL8. Carcinogenesis. 2014;35(8):1780–1787. doi: 10.1093/carcin/bgu060. [DOI] [PubMed] [Google Scholar]
- 50.Ryder M, Ghossein RA, Ricarte-Filho JC, Knauf JA, Fagin JA. Increased density of tumor-associated macrophages is associated with decreased survival in advanced thyroid cancer. Endocr Relat Cancer. 2008;15(4):1069–1074. doi: 10.1677/ERC-08-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cunha LL, Marcello MA, Nonogaki S, Morari EC, Soares FA, Vassallo J, et al. CD8+ tumour-infiltrating lymphocytes and COX2 expression may predict relapse in differentiated thyroid cancer. Clin Endocrinol (Oxf) 2015;83(2):246–253. doi: 10.1111/cen.12586. [DOI] [PubMed] [Google Scholar]
- 52.Uyttenhove C, Pilotte L, Theate I, Stroobant V, Colau D, Parmentier N, et al. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nat Med. 2003;9(10):1269–1274. doi: 10.1038/nm934. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated and analyzed in the current study are available in GSE50901 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE50901), GSE27155 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE27155), GSE151181 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE151181), TCGA database (https://genome-cancer.ucsc.edu/), HPA (https://proteinatlas.org/), STRING database (http://string-db.org) and TISIDB (http://cis.hku.hk/TISIDB/).