Significance
Topoisomerases are essential enzymes that break and reseal DNA to relieve torsional stress arising from cellular processes such as transcription and DNA replication. Chemotherapeutic compounds that target topoisomerases can corrupt DNA cleavage activity to promote cell death but can also lead to the evolution of topoisomerase-mediated DNA damage. Here, we use genetic, biochemical, and computational tools to characterize mutant forms of human topoisomerase IIβ (hTOP2β) that are hypersensitive to the anticancer agent etoposide. Our findings demonstrate that some hTOP2β mutations identified in cancer cell samples can exert deleterious activities in cellular contexts, underscoring the ability of topoisomerase dysfunction to disrupt genetic integrity.
Keywords: topoisomerase, chemotherapeutic, cancer, DNA damage, protein dynamics
Abstract
Type II topoisomerases transiently cleave duplex DNA as part of a strand passage mechanism that helps control chromosomal organization and superstructure. Aberrant DNA cleavage can result in genomic instability, and how topoisomerase activity is controlled to prevent unwanted breaks is poorly understood. Using a genetic screen, we identified mutations in the beta isoform of human topoisomerase II (hTOP2β) that render the enzyme hypersensitive to the chemotherapeutic agent etoposide. Several of these variants were unexpectedly found to display hypercleavage behavior in vitro and to be capable of inducing cell lethality in a DNA repair–deficient background; surprisingly, a subset of these mutations were also observed in TOP2B sequences from cancer genome databases. Using molecular dynamics simulations and computational network analyses, we found that many of the mutations obtained from the screen map to interfacial points between structurally coupled elements, and that dynamical modeling could be used to identify other damage-inducing TOP2B alleles present in cancer genome databases. This work establishes that there is an innate link between DNA cleavage predisposition and sensitivity to topoisomerase II poisons, and that certain sequence variants of human type II topoisomerases found in cancer cells can act as DNA-damaging agents. Our findings underscore the potential for hTOP2β to function as a clastogen capable of generating DNA damage that may promote or support cellular transformation.
Type II DNA topoisomerases are essential cellular enzymes that physically move one double-stranded DNA segment through another (1, 2). DNA strand passage, which requires the transient formation of covalent enzyme–DNA breaks, allows type II topoisomerases to manipulate the topological state of chromosomes and is necessary for supporting essential processes such as transcription, DNA replication, and chromosomal segregation (3, 4). In addition to their cellular utility, type IIA topoisomerases are targets of a number of clinically validated antibacterial and anticancer agents (5, 6). The most commonly used class of topoisomerase antagonists are compounds known as “poisons,” which stabilize cleaved-DNA intermediates and convert the enzyme into a genotoxic agent (7, 8).
In humans, it had been suggested that the anticancer activity of etoposide and other related antitopoisomerase agents might arise from the preferential targeting of a particular type II topoisomerase isoform, human topoisomerase IIα (hTOP2α) (9). However, biochemical and cellular studies have shown that these drugs can also act on a second topoisomerase II paralog, human topoisomerase IIβ (hTOP2β) (10–13), and that this activity can drive chromosomal rearrangements and the onset of diseases such as therapy-related leukemias (14, 15). Exposure to topoisomerase II poisons promotes cell death and can not only result in alterations to DNA repair pathways (16) but to the emergence of resistant topoisomerase mutants as well (17–19). Studies of the biochemical properties of topoisomerase variants have been useful for understanding innate catalytic function but have additionally shown how selective pressures can generate enzymes with aberrant activities and cleavage propensities (20, 21). Although much is now known about how and where antitopoisomerase poisons bind to their target enzymes (22), studies have shown that it is possible to alter the sensitivity of type II topoisomerases to chemical therapeutics by acquiring mutations far outside the site of drug binding (23, 24). Why both drug-resistance and drug-sensitizing alterations can arise at diverse loci and how they perturb therapeutic efficacy are often unclear from biochemical analyses alone but has important implications for our understanding of drug mechanism and consequences. Mapping and characterizing the nature of these changes is important for comprehending and predicting the consequences of topoisomerase variants that can emerge following chemotherapeutic treatment.
Here, we have sought to define how type II topoisomerases can become sensitized to anticancer agents. Using hTOP2β/etoposide interactions as a model system, we isolated and characterized several topoisomerase mutants that confer drug hypersensitivity both to cells expressing these enzymes and to protein activity in vitro. Interestingly, several of the isolated mutants were found biochemically and in vivo to exhibit elevated levels of DNA cleavage in the absence of any drug. Moreover, we discovered that a subset of the mutations recovered from our screen are also present in cancer genome database sequences of the human TOP2B gene. To better understand the structural factors that modulate etoposide sensitivity and elevated levels of DNA cleavage, we used computational simulations of an hTOP2β•ATP•DNA ternary model to construct a dynamic interaction network of the enzyme. This analysis revealed physical connections between discrete regions of the protein that were indicative of an allosteric communication pathway capable of modulating enzyme function. In silico modeling revealed that a cohort of our spatially distributed, damage-prone hTOP2β mutations were part of this pathway and also enabled the curation of hTOP2β variants present in large sequence datasets (such as cancer genome databases) to identify mutants with a similar dynamical signature and examine potential damage-prone phenotypes. Our studies demonstrate that inherently damage-prone hTOP2β variants are present in certain human cancers, an outcome that highlights the potential of the enzyme to act as a clastogen capable of maintaining or driving cellular transformation.
Results
Screening for Etoposide-Sensitizing Mutations in hTOP2β.
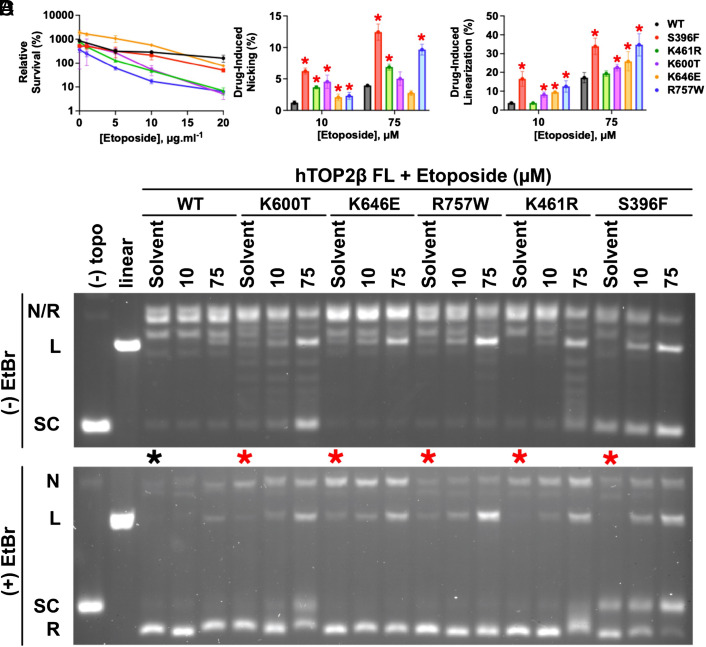
To identify topoisomerase mutants sensitized to poisoning by etoposide, we first constructed a single-copy vector that constitutively expresses full-length hTOP2β from the yeast TPI1 promoter in budding yeast. This plasmid is similar to previously constructed hTOP2β plasmids (25, 26), and complements the temperature-sensitive top2-4 allele of Saccharomyces cerevisiae (SI Appendix, Fig. S1A), allowing for the selection of active hTOP2-containing transformants at a nonpermissive temperature. Mutants of hTOP2β were generated by error-prone PCR, introduced into the hTOP2β expression vector by in vivo recombination, and then screened in yeast by replica plating on media containing 10 µg mL−1 etoposide (Methods). After this preliminary screen, potential hypersensitive mutants were examined using a semi-quantitative assay for growth on plates containing different concentrations of drug to verify etoposide hypersensitivity (SI Appendix, Fig. S1B). Positives were further tested to quantitatively determine levels of sensitivity by exposure to varying concentrations of etoposide in liquid media for 24 h. Mutants that showed cell killing after exposure to concentrations ≤20 µg mL−1 of drug were identified by Sanger sequencing of amplicons from the mutagenized region of hTOP2β; this panel conferred moderate-to-strong sensitivity to etoposide as compared to cells expressing the wild-type enzyme (SI Appendix, Table S1). After mapping the mutations onto a homology-modeled structure of full-length hTOP2β, five were selected for further study because of their distributed locations throughout the enzyme (S396F, K461R, K600T, K646E, and R757W, SI Appendix, Fig. S2); these alterations were next individually placed into fresh TPI1-driven hTOP2β expression plasmids using site-directed mutagenesis and retested for etoposide sensitivity (S396F was chosen for study on its own despite it being originally present as a double mutant (along with E475D), since the second change was a conservative substitution). As determined by colony counts and relative levels of survival, moderate (5- to 10-fold; K646E, S396F)-to-strong (>20-fold; K461R, K600T, R757W) etoposide hypersensitivity was observed for the reconstructed mutants (Fig. 1A).
Fig. 1.
Characterization of etoposide-hypersensitive hTOP2β mutants. (A) The top2-4 host strain YMM10t2-4 was transformed with hTOP2β plasmids containing the indicated alleles. Transformants were grown to mid-log phase in SC-Ura medium at 34 °C, after which the indicated concentration of etoposide was added. Incubation was continued for an additional 24 h before plating cells for survival. The survival plotted is relative to that at the time of etoposide addition. Error bars are ±SEM. (B and C) Summary of DNA linearization and nicking activity by EtopHS hTOP2β mutants induced by etoposide, as quantified by densitometry of agarose-gel-resolved cleavage products. Error bars represent the SD from three experimental replicates. Mutants showing elevated nicking or linearization relative to wild-type hTOP2β (as determined by a one-tailed t test with a P-value threshold of 0.05) are denoted with a red asterisk. (D) Representative agarose gels of supercoil relaxation and cleavage reactions conducted in the presence of etoposide. The activities of five candidate mutants from the EtopHS screen are shown in the presence of two different drug concentrations (10 and 75 μM) as compared to no drug (“Solvent”). Gels were electrophoresed in the absence (Upper) or presence (Lower) of ethidium bromide (“EtBr"). Control reactions [“(−) topo” (no enzyme) and “linear”] are included at the left of each gel. The innate cleavage propensities of the constructs can be seen in lanes denoted with asterisks (black—WT (wild type), red—mutant). The positions of different DNA species as they migrate into the gels are noted: “N/R”—nicked/relaxed; “L”—linear; “SC”—supercoiled.
Having identified human hTOP2β mutations that confer etoposide hypersensitivity (EtopHS) in vivo, we next set out to characterize the effects of these alterations on drug efficacy in vitro. The five retested hTOP2β enzymes were expressed in and purified from yeast (Methods). Purified wild-type and mutant hTOP2β enzymes were compared for their sensitivity to two concentrations of etoposide (10 or 75 μM), using native agarose-gel electrophoresis conducted in the presence or absence of ethidium bromide to aid in the identification of relaxed, nicked, and linear products (see Fig. 1 B and C for quantification of representative gel-based data shown in Fig. 1D). The mutant proteins exhibited a wide range of effects in the presence of etoposide. For three variants, supercoil relaxation was moderately inhibited by the drug (hTOP2βS396F, hTOP2βK600T, and hTOP2βK461R), whereas for the other two, it was not (hTOP2βK646E and hTOP2βR757W—compare the distribution of topoisomers present in the “75-”µM drug lanes in Fig. 1 D, Upper). hTOP2βS396F and hTOP2βR757W showed the strongest increase in DNA nicking and cleavage responses to etoposide relative to their drug-free reaction response (Fig. 1 B–D, Lower). The other three mutants were also sensitive to etoposide, albeit to a lesser effect, displaying elevated levels of either nicking or cleavage depending on the drug concentration used in the assay. Importantly, all mutants showed elevations in either nicking or double-strand cleavage at both etoposide concentrations tested. Thus, the selected mutant proteins not only exhibited elevated sensitivity to etoposide in vivo but also in vitro.
Etoposide-Sensitizing Mutations in hTOP2β Can Substantially Enhance Innate DNA Cleavage Propensity.
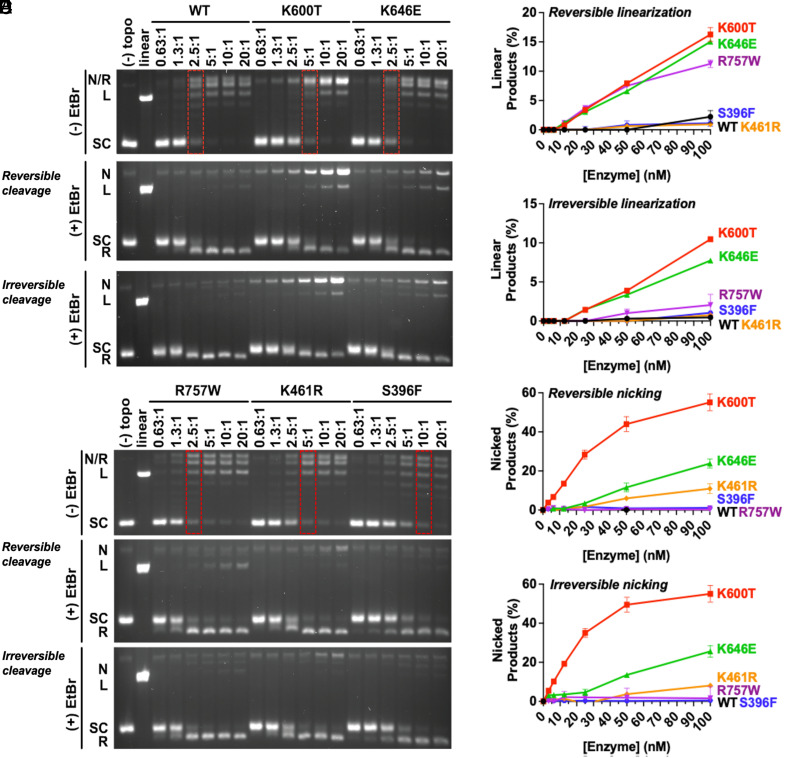
While examining the in vitro cleavage data, we noted that four of our hTOP2β mutants (S396F being the exception) appeared to nick and/or cleave the DNA at low or moderate levels even when etoposide was absent (compare “solvent-only” lanes with asterisks, Fig. 1 D, Lower). We therefore set out to determine the extent to which our mutants possessed innate differences in their drug-free biochemical activities compared to the native enzyme. We first assessed supercoil-relaxation activity, conducted in the absence of etoposide, using native gel electrophoresis. Four of the five hTOP2β mutants tested here were highly active for supercoil relaxation (see red dotted boxes, Fig. 2 A and B, Upper), with hTOP2βK646E and hTOP2βR757W displaying near wild-type levels of activity, and hTOP2βK600T and hTOP2βK461R showing only an ~twofold reduction in catalytic function. By comparison, the activity of hTOP2βS396F was reduced nearly eightfold compared to the wild-type protein.
Fig. 2.
EtopHS hTOP2β variants display elevated nicking and linearization activities in vitro in the absence of drug. (A and B) Representative agarose gels of supercoil relaxation and cleavage reactions conducted in the absence of etoposide for purified mutant proteins identified from the EtopHS screen. Enzyme titrations are denoted as the molar ratio of hTOP2β dimer to plasmid DNA. Red dotted boxes indicate enzyme concentrations necessary to achieve levels of supercoil relaxation comparable to wild type (WT). The Middle panels of (A) and (B) were quenched to reveal reversible cleavage, whereas the bottom panels were quenched to reveal irreversible cleavage. (C–F) Nicking and linearization of DNA during the relaxation of negatively supercoiled plasmid by EtopHS hTOP2β variants are compared to wild-type hTop2β. Reactions were stopped by (C and E) 1% SDS alone to trap reversible cleavage complexes, or by (D and F) 1% SDS and 50 mM EDTA to isolate irreversible cleavage complexes. Plots show a summary of DNA linearization and nicking activity by EtopHS hTOP2β mutants, as quantified by densitometry of agarose-gel-resolved cleavage reactions. Reversible and irreversible cleavage complex formation is enhanced by several etoposide-hypersensitive hTOP2β mutations. Error bars represent the SD between three experimental replicates.
We next repeated the supercoil relaxation assays but stopped the reactions either with 1% SDS alone (to trap both reversible and irreversible cleavage complexes), or with 25 mM EDTA followed by 1% SDS thirty seconds later (to isolate only irreversible cleavage complexes) and analyzed the products on agarose gels containing ethidium bromide. The distinction between reversible and irreversible cleavage is important to identify when a covalent hTOP2β–DNA intermediate has entered into a hyperstable (i.e., persistent) state: Covalent complexes generated by wild-type topoisomerase II in the presence of etoposide can be reversed by high salt, elevated temperature, or chelating agents (27, 28), whereas irreversible covalent complexes can be formed by certain etoposide-hypersensitive (EtopHS) mutant proteins in the presence of the drug (29). Here, in assays conducted in the absence of etoposide, three of the five EtopHS mutants—hTOP2βK600T, hTOP2βK646E, and hTOP2βR757W—were found to form high levels of reversible linearized species as compared to the wild-type enzyme (Fig. 2 A and B, Middle; Fig. 2C); hTOP2βK600T and hTOP2βK646E also showed a propensity for irreversibly linearizing DNA (Fig. 2 A and B, Lower; Fig. 2D). Several mutants additionally showed innately elevated levels of reversible and/or irreversible nicking activity relative to the wild-type enzyme (hTOP2βK600T, hTOP2βK646E, and to lesser extent hTOP2βK461R) (Fig. 2 A, B, E, and F). In contrast to the other four mutants, hTOP2βS396F displayed no appreciable innate (i.e., drug free) nicking/linearization activities in the absence of drug (Fig. 2 B and F).
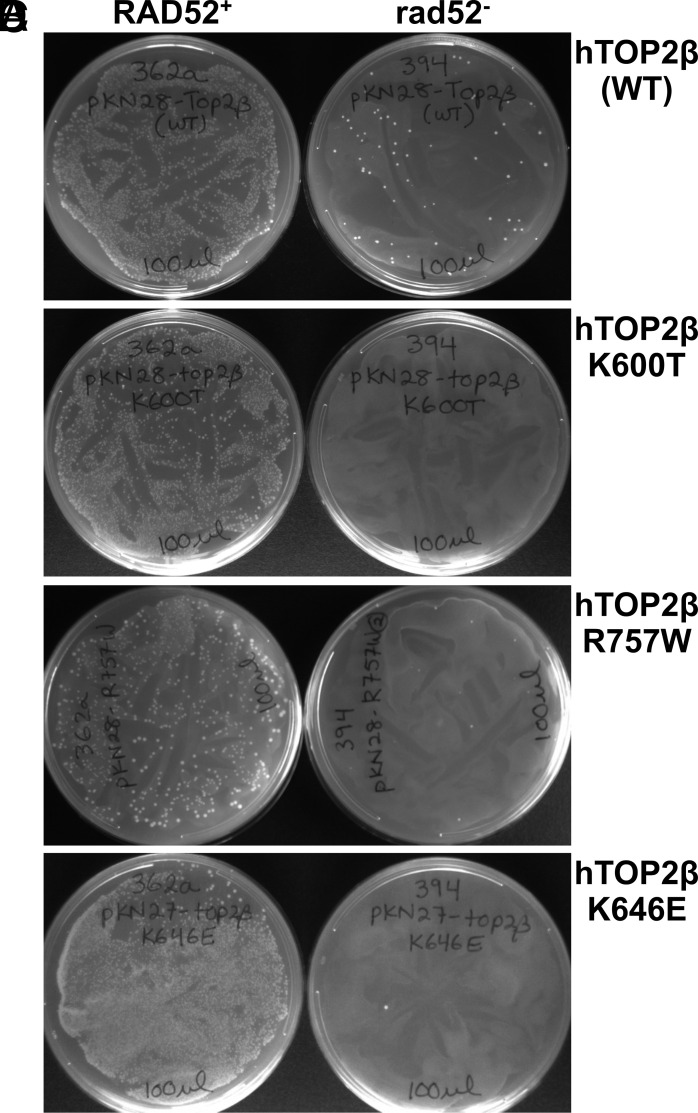
Each of the five mutant enzymes tested was able to complement the top2-4 allele (SI Appendix, Table S1), confirming that the proteins are active in vivo. However, this outcome did not reveal whether the intrinsic cleavage activities observed in vitro might also manifest in cells. To assess the DNA-damage properties of the mutants in vivo, the five different hTOP2β constructs were transformed into a top2-4 rad52–deficient yeast strain, which is unable to repair DNA breaks (30). Remarkably, the resultant analysis revealed that expression of all the five variants led to a marked loss of transformation potential in rad52− cells (Fig. 3 and SI Appendix, Fig. S3). Collectively, this outcome establishes a strong link between etoposide sensitivity and DNA-damaging activity in vivo.
Fig. 3.
Expression of hTOP2βK600T, hTOP2βK646E, and hTOP2βR757W is not viable in rad52-deficient yeast strains. (A-B) The hTOP2β expression vector pKN28 was generated to facilitate introduction of point mutations into constructs that express hTOP2β from the TPI1 promoter. pKN28 with wild-type hTOP2β and pKN28 expressing either hTOP2βK600T or hTOP2βR757W were transformed into JN362a (RAD52+) or JN394 (rad52−) strains. While transformation efficiencies were similar in RAD52+ strains, a greatly reduced number of colonies in the rad52− strain were seen with pKN28 hTOP2β wild type, and no colonies were seen with pKN28 hTOP2βK600T or hTOP2βR757W. (C, D) A similar analysis was carried out with pKN27 encoding hTOP2βK646E and hTOP2βR757W, demonstrating that these alleles also could not be introduced into rad52− strains. All transformation plates were incubated at 30 °C for 3 d. The plates were then photographed by conventional photography.
Etoposide-Sensitizing Mutations Localize to Allosterically Important Residues.
Many of the cleavage-prone mutations identified for hTOP2β from the etoposide hypersensitivity screen map outside of the catalytic sites (ATPase and DNA cleavage) of the enzyme (SI Appendix, Fig. S2). To better understand why these amino acid changes might impact drug sensitivity, we turned to molecular dynamics (MD) simulations and dynamic network analysis [“MD-DNA,” (31)]. MD-DNA uses MD trajectories to identify, with high precision and confidence, intramolecular contacts that persist over the course of a simulation, and then couples this information with network theory to assess the potential allosteric significance of these contacts within the global context of the macromolecular system.
To carry out MD-DNA, a homology model of full-length hTOP2β in complex with adenosine triphosphate (ATP) and double-stranded DNA was first generated from a structure of budding yeast Top2 and used as a template for this approach (32) (Methods). Long-timescale simulations (500 ns) were then performed on the complex to maximize conformational sampling (Movie S1). Like any other macromolecular system, the topoisomerase II dimer can be conceptualized as a network of interconnected points, or “nodes,” which are defined by a combined set of both alpha-carbon atoms and hydrogen-bond donors/acceptors for each amino acid side chain. To identify contacts between nodes in the network, coupled motions were first captured in a crosscorrelation matrix of atomic fluctuations over the duration of the simulation. Contacts were deemed as being meaningful if they occurred between pairs of nodes whose alpha-carbons or H-bond donors/acceptors stayed within a maximal cutoff distance of 5 Å during >80% of the trajectory (31).
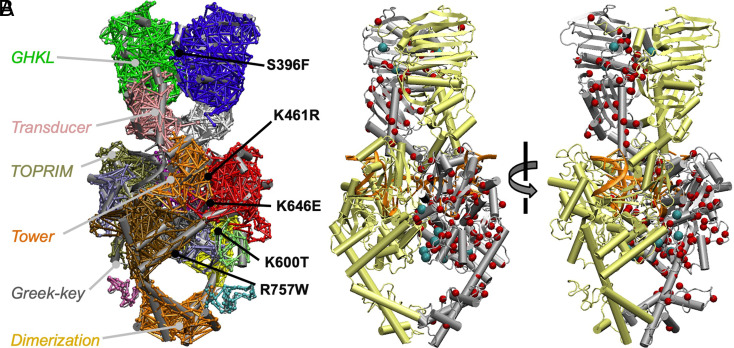
A protein’s self-interaction network can be reduced to a set of substructures (termed “communities”) within the network (31). Communities represent clusters of nodes that are densely interconnected with one another but sparsely connected to nodes in other communities, and analysis of the communication between these substructures provides insights not readily available from static structural images. The community structure for the hTOP2β•ATP•DNA complex was determined using the Girvan–Newman algorithm (33), which split the network into 18 communities of greater than 20 residues (Fig. 4A and Movie S2). Most communities obtained from this analysis turned out to correspond to known structural folds present in type II topoisomerases. For example, in the ATPase domain, the community organization overlaps reasonably well with the structurally and phylogenetically defined boundaries of the GHKL (Gyrase, Hsp90, Histidine Kinase, MutL) ATP–binding domain and its adjacent conformational “transducer” domain (an RNaseP fold). However, within the DNA-binding and -cleavage core, there was some divergence between community assignment and structural domains; for example, the Mg2+ ion–binding topoisomerase-primase (TOPRIM) fold could be subdivided into two distinct communities. Additionally, a small Greek-key element, which projects from the TOPRIM fold, is indicated as a distinct community on one subunit but is merged with that fold on the other. Lastly, the two globular domains that form the C-terminal dimerization interface appeared to behave as a single dynamic community.
Fig. 4.
Identifying prospective hypercleavage mutants of hTop2β computationally. (A) Network communities obtained for an hTOP2β•AMPPMP•DNA ternary complex homology model. The positions of the EtopHS mutations studied here are indicated. Nodal (interresidue) interactions are shown as bars connecting different parts of a cartoon rendering of the complex and are colored by community assignment: GHKLa (green), GHKLb (blue), transducera (pink), transducerb (white), TOPRIMa1 (red), TOPRIMa2 (ice blue), TOPRIM+GreekKeya (tan), TOPRIMb (red), GreekKeya (lime), WHDa (black), WHDb (gray), towera (purple), towerb (dark orange), shouldera (yellow), shoulderb (ochre), C-terminal loopa (cyan), C-terminal loopb (mauve), C-terminal dimerization interface (light orange). (B) Cartoon diagram of an hTOP2β•AMPPNP•DNA ternary complex homology model with cancer dataset mutation sites indicated as red spheres. Sites chosen for further study are indicated as cyan spheres.
Although most nodal interactions occur within a community, some nodes reside on interfaces between communities (31). Residues participating in such interfacial contacts often serve as critical contact points for the flow of allosteric information between different structural regions. Surprisingly, when we mapped the five mutations identified from the etoposide screen onto the network model, all but one (K600T) clearly mapped to a critical node on a community boundary (Fig. 4A and SI Appendix, Fig. S4); interestingly, close inspection of K600T shows that this residue maps to a position where it might aid in modulating the conformation of the Greek-key domain, which has a well-documented role in the cleavage activity of type IIA topoisomerases (34). The overall correlation between sites of etoposide hypersensitivity and intercommunal node position suggests that many of the mutations obtained from the screen may act by perturbing the communication between protein regions and/or key ligands (e.g., ATP or DNA) that help regulate enzyme dynamics. Consistent with this reasoning, four of the amino acid alterations obtained from the hypersensitivity screen appear to introduce additional steric bulk (S396F, R757W) or disrupt salt bridge contacts (K600T, K646E, R757W) at intersubunit/interdomain surfaces, while the fifth (K461R) occurs at a contact point between the enzyme and a bound G-segment DNA (SI Appendix, Fig. S2).
Mutations to Critical Node Residues Are Overrepresented in the Population of hTOP2β Mutations Present in Cancer Genomes.
Because several critical node alterations were found not only to sensitize hTOP2β to etoposide poisoning, but also to elevate innate DNA cleavage propensity, we sought to evaluate whether analogous intercommunity mutations might be present in human cell populations. Cancer genome databases are a particularly rich source of amino acid variant information. Sequence data for hTOP2β genes present in the CBioPortal (35) and COSMIC (36) databases were collated and cleaned by removing SNPs, mutations from hypermutated samples, and mutations residing in the unstructured N- and C-terminal regions of the enzyme (Methods); Fig. 4B shows this resultant panel of 129 mutations mapped onto the homology model of hTOP2β used for the network analysis. Interestingly, examination of the data (as compared to Monte Carlo simulations of random mutation networks) revealed that human cancer genome repositories collectively contain an enrichment of mutations that either directly correspond to or are proximal to (i.e., ≤5 Å) a critical node (P < 0.05, SI Appendix, Fig. S5 and Methods). Remarkably, we found that two mutations independently pulled from our etoposide-hypersensitive screen, K600T and S396F, are also present in the genome databases (albeit with K600T identified in a hypermutated cancer).
Given the apparent enrichment of cancer cell mutations at sites next to or overlapping with critical node positions, we next sought to use the dynamic network model to filter through the cancer cell TOP2B gene sequences and search for mutations that might confer hypercleavage phenotypes similar to those found from our drug screen. Our set of candidate mutations was first curated for alterations at amino acid positions that either correspond to or directly interact with residues identified as critical nodes. This group was then further filtered for substitutions that appeared sterically or electrostatically disruptive. The remaining set of alterations (10 in total) were found to be scattered throughout the hTOP2β catalytic core (Fig. 4B); these mutant proteins were subsequently cloned, expressed, purified, and tested for elevated levels of intrinsic DNA cleavage.
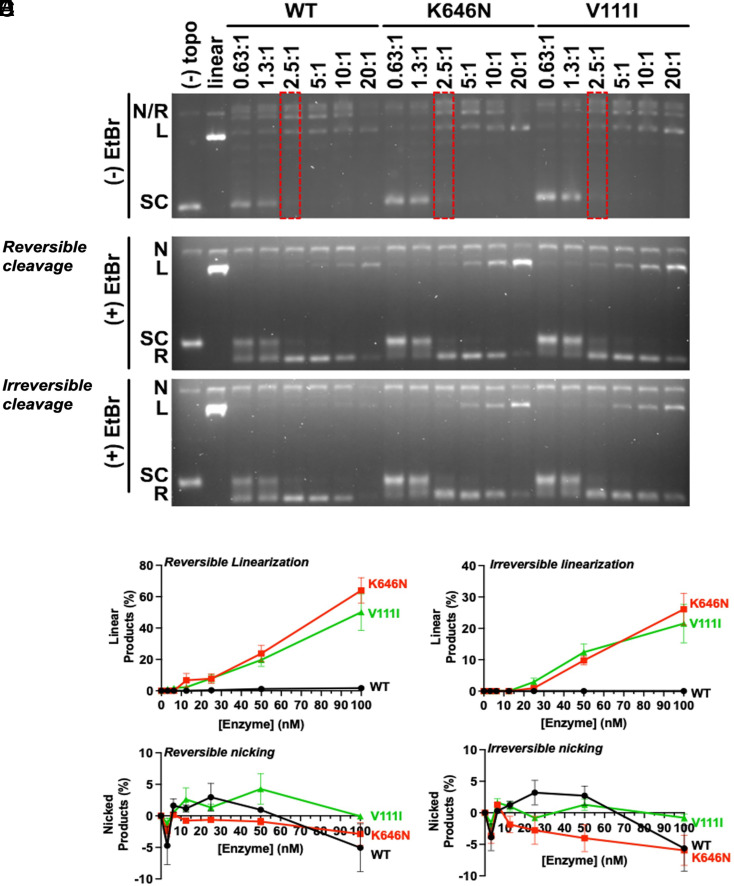
Six of the ten mutants (R67C, D173G, K600N, F755L, E774K, and Y862C) were found to have no evident impact on DNA nicking or linearization in vitro and were not pursued further. Efforts to directly purify hTOP2βD759G and hTOP2βR761C unfortunately proved unsuccessful due to poor expression of the mutant enzymes, suggesting that they might be toxic; this outcome prompted us to conduct genetic studies of these two variants along with the two remaining mutants in the pool, hTOP2βV111I and hTOP2βK646N. The behavior of hTOP2βK646N in cells proved to be erratic and irreproducible, so this mutant was not genetically analyzed further (SI Appendix, Supplemental Methods). However, the remaining three mutants were able to successfully complement growth of the temperature-sensitive top2-4 allele (SI Appendix, Fig. S6, left set of plates). Moreover, both hTOP2βV111I and hTOP2βD759G proved lethal when expressed in the top2-4-/rad52-deficient yeast strain (SI Appendix, Fig. S6, right set of plates), and the two mutants also conferred hypersensitivity to etoposide (SI Appendix, Fig. S7). Hence, two of the mutants from our panel exhibited clear DNA-damaging potential in cells. For in vitro studies, supercoil relaxation assays using purified preparations of the two remaining enzymes we could express—hTOP2βV111I and hTOP2βK646N—revealed that both proteins possessed essentially wild-type catalytic activity (Fig. 5A, first panel, compare the relative ratios of supercoiled torelaxed species across the enzyme titration). However, while the two mutations had little discernible effect on nicking (Fig. 5 D and E), they did display significantly elevated levels of both reversible and irreversible linearization (Fig. 5 B and C). Thus, combined with our in vivo results, three of the ten mutations highlighted from our MD-DNA analysis of cancer genome alterations in TOP2B were found to exhibit intrinsic DNA-damaging behavior.
Fig. 5.
Elevated reversible and irreversible cleavage complex formation by hTOP2β mutations present in cancer genome datasets. (A) Innate (drug free) nicking and linearization of DNA during the relaxation of negatively supercoiled plasmid DNA by two hTOP2β mutants are compared with wild-type hTOP2β. Representative agarose gels of supercoil relaxation and cleavage reactions conducted in the absence of etoposide are shown for purified mutant proteins identified from cancer genome databases. Enzyme titrations are denoted as the molar ratio of hTOP2β dimer to plasmid DNA. Red dotted boxes indicate enzyme concentrations necessary to achieve levels of supercoil relaxation comparable to wild type (WT). The middle panel of (A) was quenched to reveal reversible cleavage, whereas the bottom panel was quenched to reveal irreversible cleavage. (B–E) Reactions were stopped by (B and D) 1% SDS alone to trap reversible cleavage complexes, or by (C and E) 1% SDS and 50 mM EDTA to isolate irreversible cleavage complexes. Plots show a summary of DNA linearization and nicking activity by hTOP2βK646N and hTOP2βV111I, as quantified by densitometry of agarose-gel-resolved cleavage reactions. Error bars represent the SD of three experimental replicates.
Discussion
Type II topoisomerases create transient double-strand breaks in DNA to facilitate the physical passage of DNA strands through one another. This activity is essential both for enabling chromosomal segregation and to counteract the topological consequences of replication and transcription. Due to the potential cytotoxicity of DNA breaks, the equilibrium between topoisomerase-dependent DNA cleavage and religation is critical and must be precisely controlled. Both therapeutic antagonists (12, 19, 37) and mutations (20, 38) that impact type II topoisomerases can corrupt this equilibrium, resulting in DNA damage and genetic instabilities.
Given the importance of topoisomerase poisons to cancer therapy (3) and the role of hTOP2β in promoting therapy-related secondary malignancies (12, 19, 37), we set out to characterize factors that modulate the sensitivity of hTOP2β to etoposide, a commonly used chemotherapeutic drug. As seen with mutations that impart resistance to this agent (24), etoposide-sensitizing mutations were distributed throughout the enzyme and frequently did not localize near the drug-binding site, which resides in the DNA-gate region of the enzyme (SI Appendix, Fig. S2). We observed that several mutants with strong sensitivity phenotypes tended to be substitutions that localize near enzyme subunit interfaces and introduce steric clashes or disrupt salt-bridge interactions (e.g., hTOP2βS396F, hTOP2βK600T, hTOP2βK646E, and hTOP2βR757W). Notably, these variants led the enzyme to generate elevated levels of drug-free reversible and/or irreversible DNA damage in vitro (both nicking and double-strand break formation, Fig. 2). It is intriguing that a screen for etoposide hypersensitivity inadvertently yielded a large number of innately DNA-damaging alleles of hTOP2β. Whether this adaptation is unique to hTOP2β will require additional studies, although it has been suggested that topoisomerase II mutants which are relatively deficient in covalent complex religation might also frequently manifest hypersensitivity to antitopoisomerase poisons (39).
To better understand some of the structural mechanisms that allow mutations to convert hTOP2β into a DNA-damaging agent, we used MD simulations to construct an interaction network model for a ternary complex between hTOP2β, ATP, and DNA. Dynamics-based models have several advantages over models built from static structural data, the most significant being their ability to represent conformational ensembles (40, 41). Most structural data are derived from a single or few physical configurations (often frozen at some local minimum) that can at times also be influenced by nonbiological factors used to facilitate imaging (e.g., buffer composition, detergents, crystal packing, and cryo-preservation). By comparison, MD simulations sample a region of conformational space to highlight important contacts that may not be immediately evident from a single structure. The visualization of persistent local interactions during dynamical modeling can aid in tracing allosteric signal propagation paths within the network. Further analysis of such networks can be used to characterize features like community structures (31), which partition multidomain proteins into groups of interconnected nodes. The community structure for an hTOP2β•ATP•DNA ternary complex revealed an interesting trend in which etoposide-sensitizing mutations—which were found to frequently make topoisomerase II more innately cleavage-prone—also tended to reside at the interfaces between motion-coupled communities (Fig. 4 and SI Appendix, Fig. S2).
While analyzing our mutational data, we discovered that one of our identified mutants (S396F) is present in the cBioPortal cancer genome database (35) and another (K600T) is listed in the COSMIC database (36). Interestingly, both of these mutations give rise to enzymes with aberrant functional properties. For example, the S396F substitution severely compromises strand passage activity (Fig. 2 B, Upper), which potentially accounts for its relatively weak EtopHS phenotype. This low level of activity also suggests that hTOP2βS396F may struggle in supporting supercoil removal during times of increased demand. We speculate that such a deficiency could lead to a buildup of superhelical strain capable of disrupting gene expression programs and/or contributing to genetic instabilities in certain contexts [e.g., by increasing the frequency of RNA polymerase stalling and R-loop formation (42)]. DNA nicking and linearization by the S396F allele was also strongly sensitized to etoposide (Fig. 1 C and D), indicating that the alteration compromises the fidelity of the DNA religation reaction by either enhancing drug binding or diminishing drug dissociation. In yeast cells, these phenotypes resulted in an incompatibility of the S396F mutant with a repair-deficient rad52− background (SI Appendix, Fig. S3), indicating that the allele induces genetic damage in cells that is not detectable by the simple cleavage assays used here. In comparison, hTOP2βK600T is highly etoposide sensitive in vivo and also one of the most innately cleavage-prone enzymes tested in vitro (both reversible and irreversible) (Fig. 2). Concordantly, the expression of hTOP2βK600T in rad52− yeast cells proved lethal (Fig. 3), indicating that it also generates elevated levels of DNA damage in cells.
The realization that several of our etoposide-sensitizing mutations induce DNA damage and frequently share a similar dynamical signature (i.e., residing on a community interface), yet also are found in human cancer cells, prompted us to scan cancer genome databases for mutations with a similar profile. Mutational data were filtered for residues that were either directly mapped to or formed a direct contact with a critical node, producing a set of 10 previously uncharacterized mutations that were selected for follow-up biochemical and genetic studies. Of these, two (hTOP2βK646N and hTOP2βV111I) showed significantly elevated levels of DNA damage propensity in vitro (Fig. 5). Upon examination in yeast cells, two of the mutants also exhibited lethality when expressed in repair-deficient yeast cells (hTOP2βV111I and hTOP2βD759G), and both mutants conferred etoposide hypersensitivity (SI Appendix, Figs. S6 and S7). In terms of placement, hTOP2βV111I (identified in a stomach cancer sample) sits at the boundary between the GHKL and transducer elements and resides near the site of ATP binding and hydrolysis (SI Appendix, Fig. S2G). hTOP2βK646N (also identified in a stomach cancer sample) occurs at a critical node that participates in an intersubunit electrostatic interaction with Asp984 (SI Appendix, Fig. S2D); like K600T and S396F, this particular amino acid was also found to be altered in our etoposide screen, but to Glu (a more severe change) as opposed to Asn. The D759G mutation (which has been identified in mixed cancer types) appears to alter electrostatic interactions with Arg757 (which itself was identified as a mutation site in our etoposide hypersensitivity screen) and maps to a critical node that forms part of the subunit interface that cleaves and opens bound DNA (SI Appendix, Fig. S2F). Overall, our findings indicate that the cancer genome data for hTOP2β appear to contain numerous mutations that impact critical dynamical pathways in the enzyme, the disruption of which can lead to aberrant cleavage activity. We note that an analogous enrichment of cancer genome mutations at critical nodes has been reported in the literature for other systems such as proteins involved in regulating kinase activities (43).
Why should mutations at critical nodes in hTOP2β correlate with an abundance of DNA-damaging alleles? We hypothesize that the induction of DNA damage (and thereby genomic instability) could provide a selective advantage in certain cancer cell backgrounds. Although only three of the 10 cancer mutations tested here had a discernible functional impact, the absence of a phenotype in the select number of assays used here does not necessarily demonstrate that the changes have no functional impact on cells. For example, critical node mutations that impact a particular biological function of hTOP2β (e.g., the support of transcriptional bursting or chromosomal segregation) may not exhibit a DNA hypercleavage signature but instead could impair overall activities such as supercoil removal (S396F may represent such an allele). The observation that five damage-prone critical node alleles were revealed in the cancer genome data from just two screens (etoposide hypersensitivity, MD) indicates that it may be relatively straightforward for hTOP2β to acquire alterations capable of promoting DNA damage.
The finding that hTOP2β can naturally acquire damage-promoting mutations also raises the possibility that some enzyme variants found in cancer cells may serve as clastogens capable of driving cellular transformation. To this end, a mutational signature has recently been reported for gastric cancer cells carrying a K743N mutation in hTOP2α that is consistent with the repair of type II topoisomerase cleavage complexes (20). Interestingly, although a corresponding mutation in hTOP2β (K764N) has not been identified in any cancer samples sequenced thus far, Lys764 does share the same dynamical network signature in hTOP2β (proximal to a critical node, disrupts a conserved salt bridge interaction) as the other mutations reported here. This correlation suggests that some of the mutations we have investigated may in fact be driver mutations and further demonstrates that both drug sensitivity screening and dynamical network analyses can be useful approaches for identifying mutations of potential functional impact and sorting them from benign passenger mutations that are frequently present in cancer genome samples. Examining the downstream effects of the mutations studied here in relevant cancer model systems will be an important future avenue of inquiry for defining the prospects and limits of this combined approach.
Methods
Yeast Strains, Plasmids, and Growth Conditions.
Yeast strains and growth conditions are presented in SI Appendix, Supplemental Methods.
Source of Proteins.
PCR-amplified full-length topoisomerase genes were inserted by LIC, ligation-independent cloning (44), into 12UraB (Addgene #48304), a modified version of pRS426 (45). The resulting plasmid (hTOP2β-12UraC) contained galactose-inducible fusions with an N-terminal tobacco etch virus protease-cleavable hexahistidine tag. Mutant hTOP2β constructs were generated by site-directed mutagenesis of the hTOP2β-12UraC construct. Purification details are outlined in SI Appendix, Supplemental Methods.
Topoisomerase DNA Cleavage and Supercoil Relaxation Assays.
Reaction mixtures contained varying amounts of full-length hTOP2β dimer (0 to 250 nM, 31.25 nM in assays with etoposide), 12.5 nM supercoiled pSG483, variable drug content (or solvent), 1 mM ATP, 20 mM Tris [pH 7.5], 100 mM KOAc, 10 mM MgOAc, 1.2 mM TCEP, 35 µg mL−1 BSA, and 10% [vol vol−1] glycerol. Following incubation, reactions were first quenched with 2 µL of stopping buffer containing either 5% (wt vol−1) SDS (for etoposide-containing reactions, reversible cleavage measurements, and analysis of supercoil relaxation activity) or 5% (wt vol−1) SDS and 125 mM EDTA (for irreversible cleavage measurements only). Stopped reactions were subsequently treated with 1 µL of 12 mg mL−1 proteinase K and a further incubation at 37 °C for 30 min. Samples were stored on ice until gel loading, whereupon a 6× agarose gel–loading dye was added and the samples were warmed to 37 °C for 5 min. The samples were separated by electrophoresis in 1.4% (wt vol−1) TAE agarose gels (50 mM Tris-HCl [pH 7.9], 40 mM NaOAc, and 1 mM EDTA [pH 8.0] running buffer), for 18 to 22 h at 2 to 2.5 V cm−1. To visualize the DNA, gels were poststained with 0.5 μg mL−1 ethidium bromide in TAE buffer for 30 min, destained in TAE buffer for a further 30 min, and exposed to UV illumination. Gel images were analyzed using ImageJ (46), and data were plotted using Prism (GraphPad Software). Specific activity for DNA supercoil relaxation was measured by comparing enzyme concentrations required to produce fully relaxed plasmid in 30 min.
MD Simulations.
An hTOP2β–DNA–ATP ternary complex model was built using iTASSER (47) based on previous structures of full-length scTOP2 (PDB: 4GFH) and the hTOP2β nucleolytic core (PDB: 3QX3). The fully solvated system contained a total of 465,199 atoms. The initial protein configuration was equilibrated for 200 ns in preparation for long-timescale simulations. The preproduction and equilibration runs were performed using NAMD 2.9 (48) on the Anton2 supercomputer at the Pittsburgh Supercomputing Center (PSC). Trajectories were used to construct allosteric network models as outlined in ref. 31. The complete simulation setup is outlined in SI Appendix, Supplemental Methods.
Statistical Analysis.
Monte Carlo simulations were used to generate 10,000 sets of mutations distributed randomly throughout the catalytic core of hTOP2β. For each mutation in a given set, an array of Cα–Cα distances was computed for each mutation/critical node pair. The shortest distance in the array was then recorded as the corresponding residue’s proximity to its nearest critical node. This process was iteratively repeated for each mutation in a set, with a final median nearest distance recorded for each set. The set of 10,000 median values was then fit to a random distribution, and the P-value for the actual cancer mutant dataset was computed using a simple t test.
Construction of Top2β Mutants Derived from MD.
Mutants identified as having elevated DNA cleavage when expressed from hTOP2β-12UraC were introduced into pKN27 using standard subcloning techniques. Mutations were verified by Sanger sequencing. The plasmids were then introduced into JN362at2-4 or JN394t2-4 as described above. We observed that some isolates, particularly those carrying K646N, gave variable results following transformation into JN362at2-4 or JN394t2-4 and occasionally failed to complement the top2-4 allele. Since the lack of complementation was not reproducible, the pKN27:K646N mutant was not subjected to further cell-based analyses.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (PDF)
Dataset S02 (CSV)
Dataset S03 (TXT)
MD simulation trajectory for hTOP2βߦATPߦDNA complex (500 ns).
Full view of community representation for hTOP2βߦATPߦDNA complex shown in Fig. 4.
Acknowledgments
We thank Alagar Boopathy, Natalie Bitar, Gabriela Swan, and MBT530 students for assistance with the yeast experiments. This work was supported by the NIH [T32-GM008403-28 to A.F.B., R01-CA077373 and R35-CA263778 to J.M.B.]; the National Cancer Institute [CA 216010 to J.L.N.]; and European Molecular Biology Organization [Long-Term Fellowship to T.R.B.]. Anton 2 computer time (MCB190063P) was provided by the PSC through NIH grant R01GM116961 (to J.M.B.); the Anton 2 machine at PSC was generously made available by D.E. Shaw Research. We also used resources provided by the Maryland Advanced Research Computing Center at Johns Hopkins University.
Author contributions
A.F.B., T.R.B., K.C.N., A.Y.L., J.L.N., and J.M.B. designed research; A.F.B., K.C.N., R. Gupta, and R. Guha performed research; A.F.B. and T.R.B. contributed new reagents/analytic tools; A.F.B., K.C.N., A.Y.L., J.L.N., and J.M.B. analyzed data; and A.F.B., T.R.B., J.L.N., and J.M.B. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
Reviewers: R.M., The University of Texas Southwestern Medical Center; and K.C.N., Intramural Research Program (NIH).
Contributor Information
John L. Nitiss, Email: jlnitiss@uic.edu.
James M. Berger, Email: jmberger@jhmi.edu.
Data, Materials, and Software Availability
All study data are included in the article and/or supporting information.
Supporting Information
References
- 1.Brown P. O., Cozzarelli N. R., A sign inversion mechanism for enzymatic supercoiling of DNA. Science 206, 1081–1083 (1979). [DOI] [PubMed] [Google Scholar]
- 2.Roca J., Wang J. C., DNA transport by a type II DNA topoisomerase: Evidence in favor of a two-gate mechanism. Cell 77, 609–616 (1994). [DOI] [PubMed] [Google Scholar]
- 3.Nitiss J. L., Targeting DNA topoisomerase II in cancer chemotherapy. Nat. Rev. Cancer 9, 338–350 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pommier Y., Sun Y., Huang S. N., Nitiss J. L., Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat. Rev. Mol. Cell Biol. 17, 703–721 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pommier Y., Leo E., Zhang H., Marchand C., DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem. Biol. 17, 421–433 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collin F., Karkare S., Maxwell A., Exploiting bacterial DNA gyrase as a drug target: Current state and perspectives. Appl. Microbiol. Biotechnol. 92, 479–497 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deweese J. E., Osheroff N., The DNA cleavage reaction of topoisomerase II: Wolf in sheep’s clothing. Nucleic Acids Res. 37, 738–748 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nitiss K. C., Nitiss J. L., Hanakahi L. A., DNA damage by an essential enzyme: A delicate balance act on the tightrope. DNA Repair (Amst). 82, 102639 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Toyoda E., et al. , NK314, a topoisomerase II inhibitor that specifically targets the α isoform. J. Biol. Chem. 283, 23711–23720 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans C. D., Mirski S. E., Danks M. K., Cole S. P., Reduced levels of topoisomerase II alpha and II beta in a multidrug-resistant lung-cancer cell line. Cancer Chemother. Pharmacol. 34, 242–248 (1994). [DOI] [PubMed] [Google Scholar]
- 11.Chen M., Beck W. T., DNA topoisomerase II expression, stability, and phosphorylation in two VM-26-resistant human leukemic CEM sublines. Oncol. Res. 7, 103–111 (1995). [PubMed] [Google Scholar]
- 12.Azarova A. M., et al. , Roles of DNA topoisomerase II isozymes in chemotherapy and secondary malignancies. Proc. Natl. Acad. Sci. U.S.A. 104, 11014–11019 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Willmore E., Frank A. J., Padget K., Tilby M. J., Austin C. A., Etoposide targets topoisomerase IIα and IIβ in leukemic cells: Isoform-specific cleavable complexes visualized and quantified in situ by a novel immunofluorescence technique. Mol. Pharmacol. 54, 78–85 (1998). [DOI] [PubMed] [Google Scholar]
- 14.Pendleton M., Lindsey R. H., Felix C. A., Grimwade D., Osheroff N., Topoisomerase II and leukemia. Ann. N. Y. Acad. Sci. 1310, 98–110 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haffner M. C., De Marzo A. M., Meeker A. K., Nelson W. G., Yegnasubramanian S., Transcription-induced DNA double strand breaks: Both oncogenic force and potential therapeutic target? Clin. Cancer Res. 17, 3858–3864 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zagnoli-Vieira G., Caldecott K. W., Untangling trapped topoisomerases with tyrosyl-DNA phosphodiesterases. DNA Repair (Amst). 94, 102900 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Hinds M., et al. , Identification of a point mutation in the topoisomerase II gene from a human leukemia cell line containing an amsacrine-resistant form of topoisomerase II. Cancer Res. 51, 4729–4731 (1991). [PubMed] [Google Scholar]
- 18.Bugg B. Y., Danks M. K., Beck W. T., Suttle D. P., Expression of a mutant DNA topoisomerase II in CCRF-CEM human leukemic cells selected for resistance to teniposide. Proc. Natl. Acad. Sci. U.S.A. 88, 7654–7658 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cowell I. G., Austin C. A., Mechanism of generation of therapy related leukemia in response to anti-topoisomerase II agents. Int. J. Environ. Res. Public Health 9, 2075–2091 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boot A., et al. , Recurrent mutations in topoisomerase IIα cause a previously undescribed mutator phenotype in human cancers. Proc. Natl. Acad. Sci. U.S.A. 119, e2114024119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilroy K. L., Leontiou C., Padget K., Lakey J. H., Austin C. A., mAMSA resistant human topoisomerase II mutation G465D has reduced ATP hydrolysis activity. Nucleic Acids Res. 34, 1597–1607 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen S. H., Chan N.-L., Hsieh T., New mechanistic and functional insights into DNA topoisomerases. Annu. Rev. Biochem. 82, 139–170 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Elsea S. H., Hsiung Y., Nitiss J. L., Osheroff N., A yeast type II topoisomerase selected for resistance to quinolones. J. Biol. Chem. 270, 1913–1920 (1995). [DOI] [PubMed] [Google Scholar]
- 24.Blower T. R., et al. , A complex suite of loci and elements in eukaryotic type II topoisomerases determine selective sensitivity to distinct poisoning agents. Nucleic Acids Res. 47, 8163–8179 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jensen S., Redwood C. S., Jenkins J. R., Andersen A. H., Hickson I. D., Human DNA topoisomerases II alpha and II beta can functionally substitute for yeast TOP2 in chromosome segregation and recombination. Mol. Genet. Genom. 252, 79–86 (1996). [PubMed] [Google Scholar]
- 26.Meczes E. L., Marsh K. L., Fisher L. M., Rogers M. P., Austin C. A., Complementation of temperature-sensitive topoisomerase II mutations in Saccharomyces cerevisiae by a human TOP2? construct allows the study of topoisomerase II? inhibitors in yeast Cancer Chemother. Pharmacol. 39, 367–375 (1997). [DOI] [PubMed] [Google Scholar]
- 27.Nelson E. M., Tewey K. M., Liu L. F., Mechanism of antitumor drug action: Poisoning of mammalian DNA topoisomerase II on DNA by 4’-(9-acridinylamino)-methanesulfon-m-anisidide. Proc. Natl. Acad. Sci. U.S.A. 81, 1361–1365 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li T.-K., Liu L. F., Tumor cell death induced by topoisomerase-targeting drugs. Annu. Rev. Pharmacol. Toxicol. 41, 53–77 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Hsiung Y., Elsea S. H., Osheroff N., Nitiss J. L., A mutation in yeast TOP2 homologous to a quinolone-resistant mutation in bacteria. J. Biol. Chem. 270, 20359–20364 (1995). [DOI] [PubMed] [Google Scholar]
- 30.Mortensen U. H., Lisby M., Rothstein R., Rad52. Curr. Biol. 19, R676–R677 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Sethi A., Eargle J., Black A. A., Luthey-Schulten Z., Dynamical networks in tRNA:protein complexes. Proc. Natl. Acad. Sci. U.S.A. 106, 6620–6625 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmidt B. H., Osheroff N., Berger J. M., Structure of a topoisomerase II–DNA–nucleotide complex reveals a new control mechanism for ATPase activity. Nat. Struct. Mol. Biol. 19, 1147–1154 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Girvan M., Newman M. E. J., Community structure in social and biological networks. Proc. Natl. Acad. Sci. U.S.A. 99, 7821–7826 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bax B. D., Murshudov G., Maxwell A., Germe T., DNA topoisomerase inhibitors: Trapping a DNA-cleaving machine in motion. J. Mol. Biol. 431, 3427–3449 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cerami E., et al. , The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bamford S., et al. , The COSMIC (Catalogue of somatic mutations in cancer) database and website. Br. J. Cancer 91, 355–358 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith K. A., Cowell I. G., Zhang Y., Sondka Z., Austin C. A., The role of topoisomerase II beta on breakage and proximity of RUNX1 to partner alleles RUNX1T1 and EVI1. Genes Chromosomes Cancer 53, 117–128 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Stantial N., et al. , Trapped topoisomerase II initiates formation of de novo duplications via the nonhomologous end-joining pathway in yeast. Proc. Natl. Acad. Sci. U.S.A. 117, 26876–26884 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rogojina A. T., Nitiss J. L., Isolation and characterization of mAMSA-hypersensitive mutants. J. Biol. Chem. 283, 29239–29250 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frauenfelder H., Sligar S. G., Wolynes P. G., The energy landscapes and motions of proteins. Science 254, 1598–1603 (1991). [DOI] [PubMed] [Google Scholar]
- 41.Nussinov R., Tsai C.-J., Allostery in disease and in drug discovery. Cell 153, 293–305 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Christman M. F., Dietrich F. S., Fink G. R., Mitotic recombination in the rDNA of S. cerevisiae is suppressed by the combined action of DNA topoisomerases I and II. Cell 55, 413–425 (1988). [DOI] [PubMed] [Google Scholar]
- 43.Kumar S., Clarke D., Gerstein M. B., Leveraging protein dynamics to identify cancer mutational hotspots using 3D structures. Proc. Natl. Acad. Sci. U.S.A. 116, 18962–18970 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aslanidis C., de Jong P. J., Ligation-independent cloning of PCR products (LIC-PCR). Nucleic Acids Res. 18, 6069–6074 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Christianson T. W., Sikorski R. S., Dante M., Shero J. H., Hieter P., Multifunctional yeast high-copy-number shuttle vectors. Gene 110, 119–122 (1992). [DOI] [PubMed] [Google Scholar]
- 46.Schneider C. A., Rasband W. S., Eliceiri K. W., NIH image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y., I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9, 40 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Phillips J. C., et al. , Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (PDF)
Dataset S02 (CSV)
Dataset S03 (TXT)
MD simulation trajectory for hTOP2βߦATPߦDNA complex (500 ns).
Full view of community representation for hTOP2βߦATPߦDNA complex shown in Fig. 4.
Data Availability Statement
All study data are included in the article and/or supporting information.