Significance
As Dobzhansky stated, nothing in biology makes sense except in the light of evolution. But, creationists have used the bacterial flagellar system as an example of “irreducible complexity,” arguing that it could never have evolved from a simpler system. In archaea, a flagellar motility system arose independently from that in bacteria and is thus an example of convergent evolution. However, little has been known about the evolution of either the bacterial or archaeal flagellar system. We show how the archaeal flagellum likely evolved from simpler archaeal type IV pili, taking advantage of the structural diversity among the archaeal type IV pili to build a flagellar filament that supercoils. It is the supercoiled filament that generates thrust when rotated.
Keywords: motility, helical polymers, cryo-EM
Abstract
Flagellar motility has independently arisen three times during evolution: in bacteria, archaea, and eukaryotes. In prokaryotes, the supercoiled flagellar filaments are composed largely of a single protein, bacterial or archaeal flagellin, although these two proteins are not homologous, while in eukaryotes, the flagellum contains hundreds of proteins. Archaeal flagellin and archaeal type IV pilin are homologous, but how archaeal flagellar filaments (AFFs) and archaeal type IV pili (AT4Ps) diverged is not understood, in part, due to the paucity of structures for AFFs and AT4Ps. Despite having similar structures, AFFs supercoil, while AT4Ps do not, and supercoiling is essential for the function of AFFs. We used cryo-electron microscopy to determine the atomic structure of two additional AT4Ps and reanalyzed previous structures. We find that all AFFs have a prominent 10-strand packing, while AT4Ps show a striking structural diversity in their subunit packing. A clear distinction between all AFF and all AT4P structures involves the extension of the N-terminal α-helix with polar residues in the AFFs. Additionally, we characterize a flagellar-like AT4P from Pyrobaculum calidifontis with filament and subunit structure similar to that of AFFs which can be viewed as an evolutionary link, showing how the structural diversity of AT4Ps likely allowed for an AT4P to evolve into a supercoiling AFF.
Across the three domains of life, single-celled organisms have evolved various means of motility, such as the ability to swim in liquids or to crawl or glide across surfaces. The motility structures organisms use for swimming motility have been collectively called flagella (1–3). However, just as bats and bees both have wings for flight, which are analogous but not homologous structures that have evolved to fulfill the same function, bacteria, archaea, and eukaryotes have flagella that are analogous but not homologous. Thus, these three flagellar systems are an example of convergent evolution as they have evolved independently to support swimming motility. The eukaryotic flagellum, which contains ~600 different proteins (4), is based upon microtubules and the motor protein dynein distributed throughout the flagellum, producing a beating motion rather than a rotation. The prokaryotes, bacteria and archaea, have flagellar systems which are analogous to each other in that both involve the motor-driven rotation of a flagellar filament. These filaments are typically homopolymers of a single protein and supercoil to form rather rigid corkscrew-like structures.
In both prokaryotic flagellar systems, extracellular flagellar filaments behave as Archimedean screws when rotated by a motor at the base of their filaments, generating thrust. While supercoiling is essential to function, much of the early structural studies of the bacterial flagellar filament (BFF) were based upon finding mutant flagellins that formed nonmotile straight filaments that had the helical symmetry needed in those times for structure determination (5). Recently, it was shown with high-resolution cryo-EM structures that despite having structurally distinct subunits, the supercoiling of both BFFs and archaeal flagellar filaments (AFFs) is achieved through a similar mechanism which has convergently emerged in the two structures (6). For BFFs, the supercoiled filaments consist of 11 protofilaments, each of which adopt a unique conformation with adjacent protofilaments adopting more similar states. The supercoiled AFF structure revealed 10 protofilaments each of which also had a unique state. A striking similarity between supercoiling in BFFs and AFFs was that subunits on the same protofilament displayed a remarkable cooperativity extending over microns and were in the same conformation. Thus, all one would need to generate a reliable model for a supercoiled AFF or BFF several microns in length would be a structure of the supercoiled filament only several hundred Ångstroms in length. In contrast to the bacterial filaments, in which the most distinct conformations are on opposite ends of the filament, the most different conformations of AFF subunits are on adjacent protofilaments on the concave surface of the structure. While 9 of the 10 interfaces between the AFF protofilaments were virtually identical, the 10th protofilament on the concave surface was different and interactions between N-terminal helices were shifted downward by 4 to 5 Å, approximately one turn of an α-helix. The singular interface which was shifted downward relative to the other ones was termed a “seam interface,” while the other nine were called “nonseam interfaces.”
While bacterial swimming motility and the bacterial flagellum have been studied intensively for over 70 y (3, 7), with some studies dating back even much further (8), the earliest studies regarding the archaeal flagellum are much more recent (1, 9). Almost 30 y ago, it was suggested that archaeal flagella are homologs of bacterial type IV pili (BT4Ps) (10, 11). More recently, it has been suggested that BT4Ps are a result of a gene fusion between an N-terminal α-helical domain and a C-terminal globular domain (12), and that the homology between archaeal flagellins and BT4Ps only involves the common N-terminal domain. Structural studies have provided support for this hypothesis, showing that BT4Ps and AFFs share the N-terminal α-helical domain and are processed in a similar way (6, 13–18). It has been shown that archaeal flagellins share a more recent common ancestor with AT4Ps and, besides the N-terminal α-helical domain, both have a C-terminal immunoglobulin (Ig)–like domain, which has no homology with the C-terminal domain present in BT4P (19–22).
Comprehensive comparative genomic analysis has uncovered a striking diversity of archaeal type IV pili–like systems, including both pili and flagella (23). The corresponding AT4P operons vary greatly in terms of sequence similarity between the core components and complexity of gene complements and are particularly abundant in hyperthermophilic archaea of the order Thermoproteales, where they were judged to reach extreme diversity far exceeding that in other archaeal lineages (23). The core of AT4P consists of four main structural and functional components: pilins, an assembly ATPase, a TadC-like membrane platform, and a prepilin peptidase responsible for the proteolytic processing of the class III signal peptide (24). Flagellar operons contain additional conserved components, such as the stator proteins FlaF/ArlF and FlaG/ArlG (25), and the ATP-binding protein FlaH/ArlH (26). FlaF/ArlF and FlaG/ArlG have apparently evolved by duplication from the main pilin protein FlaB/ArlB, whereas FlaH/ArlH is homologous to KaiC-like proteins capable of autophosphorylation and dephosphorylation, an activity important for FlaH/ArlH hexamerization and archaeal motility (26). However, the exact evolutionary relationship between AT4Ps and AFFs has remained unclear, presumably due in part to the relatively few atomic structures that exist for each. Further, the structural basis for AFF supercoiling remained unsolved until very recently (6). Importantly, despite structural similarity between AFF and AT4P subunits, there is a major difference between the two types of filaments which underlies their respective properties and functions, namely, whereas AFFs supercoil (red arrows Fig. 1A), AT4Ps do not (blue arrows Fig. 1A). It is the supercoiled filament that generates thrust when rotated, whereas rotation of a straight AT4P would not result in movement.
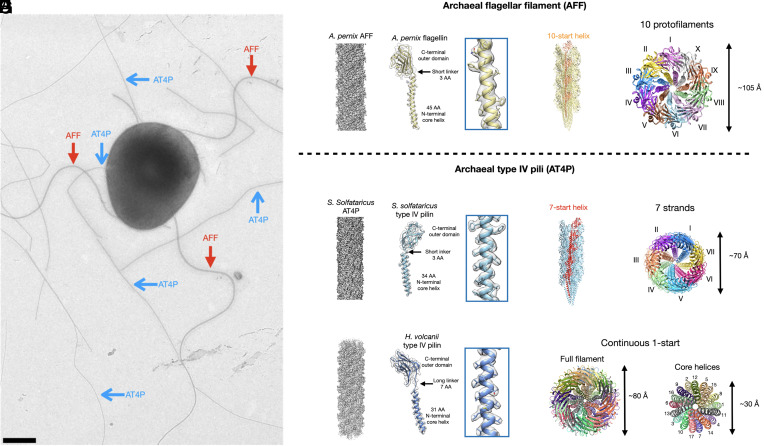
Fig. 1.
Cryo-EM structures of an AFF and two archaeal type IV pili. (A) Electron micrograph of a Saccharolobus islandicus REY15A cell showing supercoiled AFF (red arrows) and nonsupercoiled archaeal type IV pili (AT4P, blue arrows). The sample has been positively stained with uranyl acetate (2%). The scale bar is 500 nm. (B) Cryo-EM density map of the filament (Left) and single flagellin model fit into its corresponding map (Right) of the Aeropyrum pernix AFF. (C) The full filament model of the A. pernix AFF is shown, with subunits along a single 10-start helix colored orange. (D) Top-down view of the A. pernix AFF, with each of its 10 protofilaments colored differently. (E) Cryo-EM density map of the Saccharolobus solfataricus AT4P filament (Left) and single pilin model fit into its corresponding map (Right). (F) The full filament model of the S. solfataricus T4P is shown, with subunits along a single 7-start helix colored red. (G) Top-down view of the S. solfataricus T4P, with each of the 7-start strands colored uniquely. (H) Cryo-EM density map of the Haloferax volcanii AT4P filament (Left) and single pilin model fit into its corresponding map (Right). (I) Top-down views of the H. volcanii filament showing the full filament (Left) and just the core N-terminal helices (Right). Each subunit in the filament model is colored distinctly.
Here, we describe further cryo-electron microscopy (cryo-EM) structural details for the previously reported Aeropyrum pernix AFF (27) and Saccharolobus solfataricus AT4P (20). We find that the Aeropyrum AFF has the expected 10 protofilaments previously reported for an AFF from Saccharolobus islandicus REY15A (6), whereas the S. solfataricus AT4P has seven prominent helical strands. We then solve the cryo-EM structures of the H. volcanii and Pyrobaculum calidifontis AT4Ps. We classify the P. calidifontis structure as a flagellar-like AT4P due to its 10 helical strands but lack of supercoiling. We show that AT4Ps, much like their bacterial counterparts (BT4Ps), have a great diversity in their subunit packing which likely allowed an AT4P to evolve into an AFF. In contrast, both the BFFs and the AFFs, which have no homology to each other, show little structural diversity due to the exceedingly strong constraints needed to maintain supercoiling.
Results
Structure of an AFF from Aeropyrum.
To gain deeper understanding into the structural diversity of AFFs and AT4Ps, we investigated four structures from archaeal species representing four different taxonomic orders using cryo-EM (Figs. 1 and 2 and SI Appendix, Figs. S1–S5). SI Appendix, Table S1 shows the image processing and model refinement statistics for each structure. We started by reanalyzing a recently published (27) 3.5 Å resolution AFF structure belonging to a species of the genus Aeropyrum (order Desulfurococcales) in further detail (Fig. 1B). As expected for an AFF (6), we found the 10-start helix (Fig. 1C) results in ten protofilaments (Fig. 1D). The helical parameters of this filament were a rise of 5.5 Å and a twist of 108.0°. For any helical filament, a 1-start helix passes through subunits Sn and Sn+1 where the two subunits are related by the rise and twist parameters. The 10-start helix in the A. pernix AFF structure relates subunit Sn and another subunit which is 10 subunits away along the filament, Sn+10. Since the concept of a helix here is a mathematical line, there are an infinite number of helices that can occur in a filament. However, the ones that are most meaningful physically are those along which subunits make contacts with each other. For the A. pernix AFF, each subunit, Sn, makes contacts with subunits that are 1 (Sn+1), 3 (Sn+3), 4 (Sn+4), 7 (Sn+7), and 10 (Sn+10) subunits away (SI Appendix, Fig. S2A). Thus, the most important helices in the A. pernix AFF are the 1-, 3-, 4-, 7-, and 10-start helices, which can be depicted using a helical net (SI Appendix, Fig. S2B). The 10 protofilaments of the A. pernix AFF occur because the N-terminal helices are oriented along the 10-start helix.
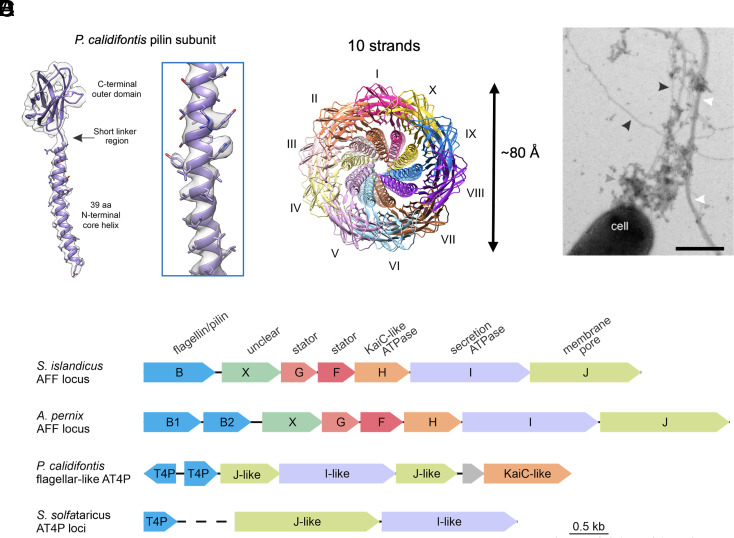
Fig. 2.
Cryo-EM structure of the P. calidifontis flagellar–like type IV pilus. (A) The model (purple) and map (light gray) of a single subunit of the P. calidifontis filament. (B) The top-down view of the P. calidifontis filament reveals 10 prominent strands similar to the 10 protofilaments found in AFFs. (C) Electron micrograph of P. calidifontis cells expressing various types of filaments positively stained with uranyl acetate (2%). Flagellar-like AT4P, bundling pili, and presumed exopolysaccharide are indicated with black, white, and gray arrowheads, respectively. (Scale bar, 500 nm.) Note that the AT4P filament does not supercoil. (D) Genomic loci encoding archaeal flagella and type IV pili. The genes in the depicted loci are shown as arrows, with homologous genes colored the same way. Dashed line represents discontinuity between the two loci encoding the adhesive pilus system in S. solfataricus. Depicted loci encompass the following genomic regions: S. islandicus REY15A (GenBank accession NC_017276, region 104950-111297); A. pernix K1 (NC_000854, region 1201618-1209093); P. calidifontis JCM 11548 (NC_009073, region 1259287-1264747); S. solfataricus POZ149 (CP050869, regions 1531910-1532347 and 1388946-1392553).
When we reanalyzed the deposited model (PDB 7TXI) and map (EMD 26158), we found that there was apparent protein density which was left unmodeled (SI Appendix, Fig. S3), originally assumed to be glycosylation. The A. pernix genome encodes two flagellins, FlaB1 and FlaB2 (SI Appendix, Fig. S3B), and the original model (7TXI) was built from FlaB1. After further analysis, we concluded that the most reasonable explanation for this additional protein density was heterogeneity in the filaments arising from segments being either FlaB1, FlaB2, or both flagellins organized into a hexameric asymmetric unit (SI Appendix, Figs. S3 and S4 and Methods) similar to the M. villosus flagellar filament (16).
Structural Examination of Several Archaeal T4Ps with Different Subunit Packing.
We improved the resolution of the previously published structure (20) of the AT4P from a hyperthermophilic and acidophilic archaeon S. solfataricus (order Sulfolobales) from 3.4 Å to 3.0 Å (Fig. 1E). While this seems like an incremental improvement, this is actually an increase in the information content by ~50% [since it scales as (3.4/3.0)3]. We found that the S. solfataricus AT4P has contacts along the 1-, 3-, 4-, and 7-start helices but not along the 10-start helices (SI Appendix, Fig. S2 C and D). The N-terminal helix of the S. solfataricus type IV pilin is 11 amino acid residues shorter in length than that of the A. pernix archaeal flagellin. These pilin subunits are oriented along the 7-start helices resulting in seven prominent right-handed strands (Fig. 1 F and G) which are easily viewed when looking at the filament model from top to down. We termed these “strands” to avoid confusing them with the protofilaments of AFFs.
We then determined to 3.0 Å the structure of an AT4P composed of PilA2 (28–30), one of the two major pilins of a hyperhalophilic archaeon H. volcanii (order Haloferacales) (Fig. 1H). The subunit architecture of the H. volcanii AT4Ps was quite unique with a short N-terminal helix similar to the S. solfataricus AT4P, but the linker region connecting the N-terminal helix to the C-terminal globular domain was much longer than the H. volcanii type IV pilin or A. pernix archaeal flagellin linker regions. The symmetry of this AT4P was quite different as axial views of the filament did not reveal either 7 or 10 strands (Fig. 1I). Instead, in a top view of a 17-subunit model, each subunit was distinctly separated, especially when viewing just the N-terminal core helices (right model Fig. 1I). As such, we termed this packing “continuous 1-start” as the features of the 1-start helix are prominently displayed in top-down views of this filament. Each H. volcanii type IV pilin subunit makes contacts along the 1-, 3-, 4-, 7-, and 10-start helices (SI Appendix, Fig. S2 E and F).
On the surface of the H. volcanii AT4P, we found N-linked glycosylation at three asparagine residues (Asn59, Asn63, and Asn113; SI Appendix, Fig. S5 A and B), all consistent with the previous results from mass spectrometry analysis (28). Unlike in other AT4Ps, the surface of the H. volcanii AT4P was highly negatively charged (SI Appendix, Fig. S5C), a well-known adaptation to high salt environments, compensating for the extreme ionic conditions (31).
Structure of a Flagellar-Like AT4P.
We solved the structure of a type IV pilus from a hyperthermophilic archaeon P. calidifontis (order Thermoproteales) to 4.3 Å (Fig. 2). Each P. calidifontis subunit makes contacts along the 1-, 3-, 4-, 7-, and 10-start helices (SI Appendix, Fig. S2 G and H). On the surface of the P. calidifontis filament, we found one clearly glycosylated residue, Asn85 (SI Appendix, Fig. S6 A and B). This glycosylation occurred in a groove between strands along the 3-start helix (SI Appendix, Fig. S6C). Additionally, the P. calidifontis pilin subunit contained an intramolecular disulfide bond (SI Appendix, Fig. S6D). Both glycosylation and the disulfide bond likely stabilize the T4P at the extreme temperatures (90 °C) under which P. calidifontis thrives. The P. calidifontis structure was particularly unique because while its N-terminal sequence length more closely resembles that of archaeal type IV pilins, its symmetry and subunit packing was much more similar to those of AFFs, as it has 10 prominent strands (Fig. 2B) and a twist of 108.9° resembling the 10 protofilaments of AFFs but with a rather large right-handed tilt. To determine whether the P. calidifontis filament was an AFF or an AT4P, P. calidifontis cells were negatively stained and observed under TEM (Fig. 2C). After analyzing ~300 cells, we found no evidence of supercoiling of the P. calidifontis filament, which instead had an appearance typical of T4P (black arrowheads in Fig. 2C). Notably, similar to other members of the genus Pyrobaculum (32), P. calidifontis genome lacks a canonical flagellar operon (23). Thus, we classified the P. calidifontis filament as a flagellar-like AT4P. However, unlike most other AT4P systems, the P. calidifontis T4P locus contains a gene for an ATPase of the KaiC family (Fig. 2D), which also includes the FlaH ATPase, a conserved component of the flagellar operons (23).
Classification of AT4Ps and AFFs on the Basis of Symmetry and Structure.
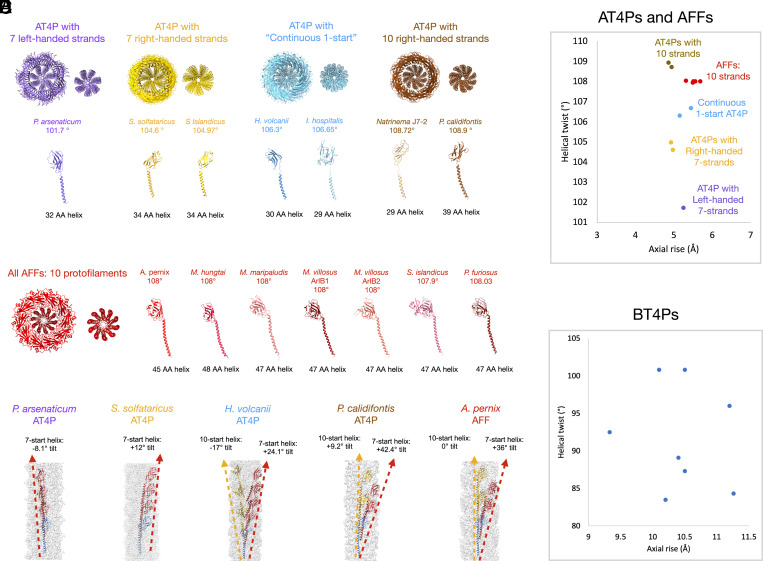
We next wanted to compare the properties of known AFF and AT4P structures reported here or in previous studies (6, 13, 15–18, 20, 21). We had very little success using sequence-based alignments to compare the subunits from each structure. We performed a detailed analysis of the subunit and quaternary filament structures of the AFFs and AT4Ps in the study as well as all prior ones (Fig. 3 and SI Appendix, Fig. S6). All AT4Ps and AFFs made similar contacts along their 1-start helix which were not distinguishable in the different classes of filaments. However, the AT4Ps could be classified into four groups (Fig. 3A) based on the subunit packing along the 7- and 10-start helices and whether seven or 10 strands are prominent in axial views of the structure. The P. arsenaticum AT4P has seven prominent strands which are left handed (purple model and text in Fig. 3). The S. solfataricus and S. islandicus LAL14 AT4Ps have seven strands as well but they are right handed (gold model and text in Fig. 3). These seven-stranded structures all had pilins with short N-terminal helices (32 to 34 amino acids) and short linker regions (~2 amino acids). The axial views of AT4Ps from H. volcanii and I. hospitalis show neither seven nor 10 prominent strands (cyan model in Fig. 3A). Both of these “continuous 1-start” AT4Ps had similar subunit architectures with short N-terminal helices (~30 amino acids) and longer linker regions (~7 to 8 amino acids). The last class of AT4P structures had 10 prominent strands (brown models in Fig. 3A) and included the flagellar-like AT4P P. calidifontis with its longer N-terminal helix as well as the Natrinema J7-2 AT4P which has a similar subunit to the H. volcanii and I. hospitalis AT4Ps. In contrast to the great structural diversity of AT4Ps, we found that all AFFs have N-terminal helices of similar size and all assemble to form a 10-stranded structure (Fig. 3B). Contacts along the 7-start and 10-start helices (or lack thereof) are key to the packing differences between the AT4P classes and AFFs. Subunits with no 10-start contacts assemble into the 7-stranded structures with either left-handed (Fig. 3C P. arsenaticum) or right-handed 7-start helices (Fig. 3C S. solfataricus). The “continuous 1-start” AT4Ps were unique in that they had 10-start and 7-start helices tilted at similar angles (~20°) but with opposite hands (Fig. 3C H. volcanii), where the angle is measured by the rotation between adjacent subunits along the same 10-start or 7-start helix. The flagellar-like P. calidifontis AT4P had a right-handed 10-start helix with a tilt of +9.2° and a right-handed 7-start with a tilt of 42.4°. In contrast, all AFFs had 10 protofilaments which were nearly vertical with respect to the filament axis.
Fig. 3.
Comparison of the filament symmetry and subunit features of AT4P and AFF. (A) Structural classification of AT4P structures from this and previous studies. Top: Axial view of AT4P models with approximately 20 subunits shown for each. For each class, the left image shows the full filament and the right image shows the only N-terminal core helices. From left to right, there are four broad classes of AT4P structures starting with 7 prominent left-handed strands (purple), AT4Ps with 7 prominent right-handed strands (gold), AT4P structures where the subunits form neither 7- nor 10-start strands (“continuous 1-start,” blue), and AT4P with prominent right-handed 10-start strands. Below each structure are the subunit models for each archaeal pilin that assembles into that structure. Pilin subunits are colored the same as their corresponding structural class. (B) Structural classification of AFF structures. All archaeal flagellins assemble into AFFs which have 10 protofilaments which are vertical with 0° of tilt with the exception of the S. islandicus REY15A AFF which has protofilaments with a slight left-handed tilt. (C) Filament density maps (light gray) with models showing a subunit Sn (light blue) and subsequent subunits along 7-start (red) and 10-start (gold) helices for each of the AT4P and AFF structural classes. The dashed lines with arrows indicate the helices. (D) Plot of helical twist versus axial rise for each AFF and AT4P structure. The colors and text indicate which symmetry group to which each filament belongs. (E) Plot of helical twist versus axial rise for all deposited bacterial T4P structures.
We quantified the structural diversity of AT4Ps versus AFFs by plotting helical twist (y axis) versus helical rise (x axis) for all known AT4P and AFF structures (Fig. 3D). This plot explicitly shows that the symmetries of AT4Ps vary to a much greater extent than the symmetries of AFFs, which is also evident from the analysis of the models. This large variation in AT4P symmetry is quite similar to that of the unrelated BT4P symmetries (Fig. 3E).
N-Terminal Core Helix Length Determines the Packing of AT4Ps versus Flagellar Filaments.
The long linker region in the H. volcanii and I. hospitalis structures results in 10-start contacts between the loop and outer domains of subunit Sn and the N-terminal helix of subunit Sn+10 (SI Appendix, Fig. S7 A and B). In contrast, the subunits in seven stranded AT4Ps, such as from S. solfataricus, make no 10-start contacts (SI Appendix, Fig. S7C), while those in the P. calidifontis flagellar–like AT4P and AFFs, such as the one from A. pernix, form contacts between the core domains of two subunits along the 10-start helix (SI Appendix, Fig. S7 D and E).
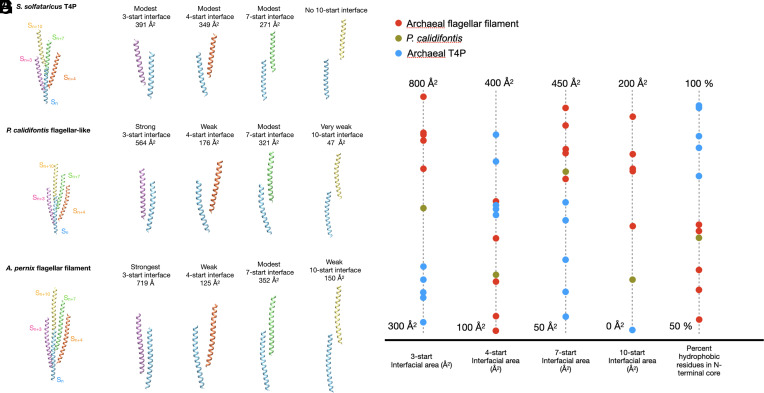
What appears to distinguish the P. calidifontis pilin from all other AT4Ps is that the N-terminal core helix is 5 to 11 amino acids longer than that in all other AT4Ps but is still 6 to 9 amino acids shorter than that in all bona fide AFFs. Thus, the P. calidifontis helix is intermediate in size between AT4Ps and AFFs. To determine the extent to which the N-terminal helix length affects the packing of the P. calidifontis flagellar–like AT4P compared to the other AT4Ps and AFFs, we examined the packing of the core domains of each structure (Fig. 4). Although the AT4P of S. solfataricus is seven stranded, there are only modest contacts along these 7-start helices (Fig. 4 A and B) as was true for all other AT4Ps (Fig. 4G). The P. calidifontis flagellar–like AT4P structure (Fig. 4 C and D) had stronger contacts along its 3- and 7-start helices than those of all other AT4P structures and in addition displayed 10-start core contacts, albeit very weak ones. None of the AT4Ps other than the flagellar-like P. calidifontis had core contacts along the 10-start helix. All AFF structures had more extensive 3-start interactions than those of the AT4Ps in addition to the 10-start contacts (Fig. 4 E–G). These results suggest that the elongated N-terminal helix of the P. calidifontis filament causes it to assemble into a flagellar-like structure.
Fig. 4.
Comparison of N-terminal core interactions in S. solfataricus T4P, P. calidifontis flagellar–like filament, and the A. pernix flagellar filament. (A) View of just the core domains from adjacent subunits related by the 3-start (pink), 4-start (orange), 7-start (green), and 10-start (yellow) helices relative to a subunit Sn (blue) in the S. solfataricus T4P. (B) From left to right, the 3-, 4-, 7-, and 10-start interfaces or lack thereof are shown for the S. solfataricus T4P, with the buried surface area for each interface shown. (C) View of the P. calidifontis subunits along the 3-start, 4-start, 7-start, and 10-start helices relative to subunit Sn showing just the core domains. (D) From left to right, the 3-, 4-, 7-, and 10-start interfaces are shown for the P. calidifontis flagellar–like filament. (E) View of the A. pernix core domain subunits along the 3-start, 4-start, 7-start, and 10-start helices relative to subunit Sn. (F) From left to right, the 3-, 4-, 7-, and 10-start interfaces are shown for the A. pernix flagellar filament. (G) Plot of the interfacial surface area of the main interfacial contact and percent of hydrophobic residues for the core domain helices of each archaeal T4P and AFF structure.
N-Terminal Core Helix Properties of Archaeal T4P and AFFs.
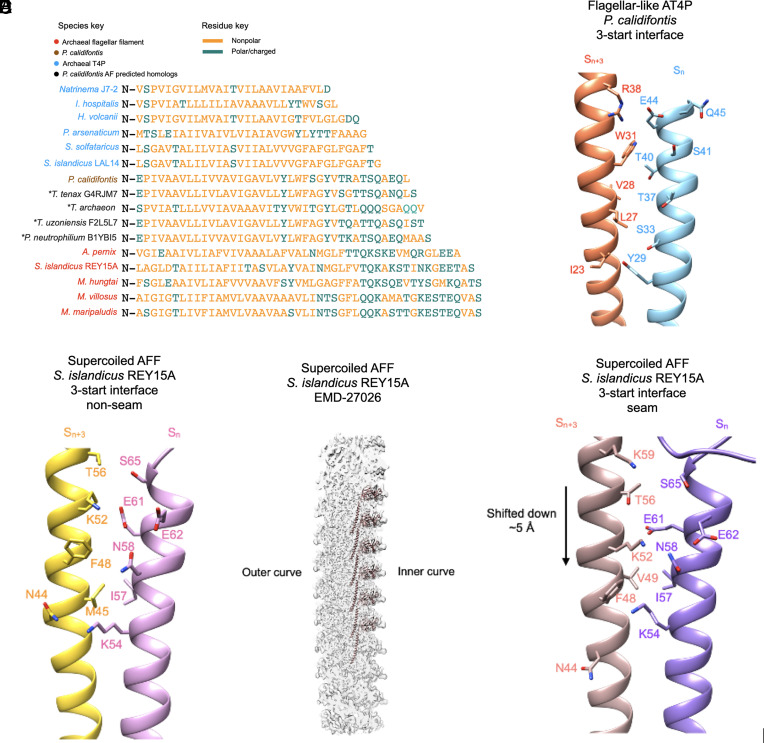
In addition to analyzing the interfacial surface area of each AT4P, we also examined the percent hydrophobicity of the core helix residues for all known AT4P and AFF structures (right-most column in Fig. 4G). We found that most archaeal type IV pilins had 80 to 90% hydrophobic or nonpolar residues in their N-terminal helix. By contrast, only 72% of the P. calidifontis core helix residues were nonpolar, which falls close to the range of the archaeal flagellins which had 57 to 71% of their residues being nonpolar. As mentioned above, in terms of length of the N-terminal helix, P. calidifontis AT4P falls in between other AT4P and AFF. To explore whether other flagellar-like T4Ps might exist and display similar sequence features, we searched for homologs in sequence databases using BLASTP. Close homologs of P. calidifontis T4P were found in other members of the order Thermoproteales. The structural models of the identified homologs were obtained from the AlphaFold database (33) and aligned with the available AT4P and AFF structures. Notably, some of the identified Thermoproteales AT4P contained even longer N-terminal helices compared to P. calidifontis AT4P, further closing the length gap between AT4P and AFF (Fig. 5A). The overall alignments show a clear pattern where the N-terminal helix residues for most pilins are largely nonpolar. The larger, more flagellar-like pilins (P. calidifontis and the AlphaFold predictions), on the one hand, share hydrophobic residues with the other pilins, but on the other hand, in the extended C-terminal region of the helix, contain charged/polar residues not present in the shorter pilins but conserved in flagellins. We expect that the shorter N-terminal helices will be overwhelmingly hydrophobic in sequence due to being transmembrane helices prior to assembly, and the typical length for transmembrane helices is roughly ~20 to 30 residues. It is likely that as the N-terminal α-helical sequences get longer, they will become more hydrophilic at the C-terminal end due to the lack of pressure for these residues to be hydrophobic.
Fig. 5.
Interfacial interactions and sequence of N-terminal core helices of archaeal type IV pili and AFFs. (A) Structural alignment of the core domain helices of the archaeal flagellins examined in this study in addition to several homologs whose structure was predicted by AlphaFold which are closely related to the P. calidifontis pilin. The “*” symbol next to the predicted homologs indicates that they are not from experimental filament structures. The names of the species are colored in sky blue for AT4P, light brown for the P. calidifontis AT4P, black for the AlphaFold-predicted P. calidifontis homologs, and red for AFFs. Amino acids colored gold are nonpolar residues, while amino acids colored green are polar/charged residues. (B) Charged/polar contacts along the 3-start interface of the P. calidifontis flagellar–like T4P. The two subunits, Sn and Sn+3, are on adjacent 10-start strands. (C) The charged/polar contacts along the core helix 3-start interface between nine of the ten protofilaments in the S. islandicus REY15A AFF (PDB:8CWM, EMD-27026). The two subunits are on adjacent protofilaments. (D) The density map of the supercoiled REY15A AFF with the model of the inner curve protofilament shown (light brown). (E) The charged/polar contacts along the inner curve seam 3-start interface of the REY15A AFF. The light brown subunit is from the 10-start protofilament in D.
We examined the location of the core helix polar/charged residues in P. calidifontis flagellar–like AT4P and found that many of these residues are near the long 3-start interface (Fig. 5B). We then examined the charged/polar N-terminal helix residues in the supercoiled S. islandicus REY15A AFF (PDB 8WCM) and found that those also made contacts along the 3-start interface (Fig. 5C). It is important to note that the REY15A supercoiled flagellar filament at the innermost region of the curve (Fig. 5D) has a 3-start interface that is shifted downward by a complete α-helical turn (6). While the interface is shifted downward by ~5 Å, the charged/polar residues make contacts with complementary residues in both the nonseam (Fig. 5C) and seam interfaces (Fig. 5E). Other AFF models also had charged/polar contacts along their 3-start interfaces, but since all of these had helical symmetry imposed on filaments that were most probably supercoiled, the details of these interfaces are questionable. The contacts along the 3-start interface for both the P. calidifontis flagellar–like AT4P and all AFFs represent the most extensive contacts between subunits on adjacent 10-start strands or protofilaments.
Flexibility of the Linker Region between N- and C-Terminal Domains in Archaeal T4P.
It has been suggested that the key difference between AT4P and AFFs is increased flexibility in the AFFs in the small linker region connecting the N-terminal helix to the C-terminal globular domain (16). A 3D variability analysis of an AFF was done to show flexibility in this region, but no comparative analysis was done for an AT4P. We have therefore made precisely such a comparison, between the S. solfataricus T4P and the S. islandicus AFF (SI Appendix, Fig. S8). Like AFFs (6, 16), the AT4P structure we examined had both curved and straight states as observed by 3D variability analysis using cryoSPARC (34) with output in simple mode. We then selected a cluster of particles and their curved volume from the 3D variability analysis as the starting input for an asymmetric reconstruction using cryoSPARC’s local refinement. The final map from this local refinement job showed a noticeable degree of curvature (SI Appendix, Fig. S8A) and from the models built for this structure, we found that subunits built on opposite ends of the filament had linker regions (SI Appendix, Fig. S8B) with about the same degree of flexibility as that of the supercoiled AFF structure (SI Appendix, Fig. S8 C and D), with displacements of ~2 Å for both structures’ linker regions, suggesting that the flexibility of the linker region is not a significant factor distinguishing AT4Ps from AFFs.
Discussion
BT4Ps have been characterized, from both sequence analysis and structural studies, as having an N-terminal domain that is largely a single extended α-helix, and a C-terminal globular domain that is in most cases largely β-stranded (35, 36). The sequence of the N-terminal domain is highly conserved, while the C-terminal domain is highly variable. A BT4P associated with competence in gram-positive (monoderm) bacteria of the class Bacilli has now been shown to share the same highly conserved N-terminal domain, but to have a C-terminal domain that is all α-helical (37). This suggests that the two domains evolved independently, and that the N-terminal T4P signature domain can be coupled with structurally distinct C-terminal domains (12). Consistent with this notion, a BT4P has now been found in Geobacter sulfurreducens that is encoded by two genes, one for the N-terminal domain and one for the C-terminal domain (38, 39). The subunits in this pilus thus contain two chains, encoded by each of these two genes.
The homology between BT4Ps and AT4Ps does not extend beyond the N-terminal domain. In contrast, the C-terminal domains in both the AFFs and AT4P have an Ig-domain fold, characterized by a β-sandwich. We can imagine three possibilities for the evolution of AT4P and flagellar filaments, starting with the inheritance of the N-terminal domain from a common ancestor with bacteria: i) AFF evolved from an early AT4P, ii) AT4P evolved from an early AFF, or iii) both evolved independently from the conserved N-terminal domain through parallel acquisition of homologous Ig-like domains. Given the obvious homology of Ig-like domains of AT4P and flagellins and their close structural similarity to each other, we do not think that independent acquisition of the same Ig domain and hence independent origin from a common ancestor is likely, and we can thus dismiss the third possibility. Since BT4P and the widespread AT4P all function as pili, it is highly likely that pilus formation was the ancestral function of this protein in archaea. This suggests that archaeal flagella have evolved from a subset of T4P which could support supercoiling, rather than the pili having evolved from the flagella with the loss of supercoiling.
Bacterial T4Ps display a high degree of structural diversity which is reflective in the diversity of their symmetries (Fig. 3E). The diversity in BT4P structure can reflect differences in function as has been shown with the two types of BT4Ps produced by Thermus thermophilus (wide and narrow) which are tied to separate functions of twitching motility and natural transformation (40). The similar diversity of AT4P structure could also reflect differences in functions which have yet to be elucidated.
This structural diversity in AT4P packing has not been previously discussed at length, mostly due to the paucity of AT4P structures. Additionally, the I. hospitalis AT4P was previously denoted as flagellar like (13). We have proposed that the P. calidifontis T4P can be viewed as an evolutionary intermediate between AT4Ps and AFFs. It is also possible that the continuous 1-start packing filaments (from H. volcanii and I. hospitalis) as well as the Natrinema sp. J7-2 AT4P with its 10-strand packing are also evolutionary links between the 7-stranded AT4Ps and 10-stranded AFFs. The extended linker regions in these structures produce contacts along the 10-start helix with the linker and outer domain of subunit Sn contacting the N-terminal core helix of subunit Sn+10. These 10-start contacts change how the pilin N-terminal helices pack into the filament when comparing the 7-start interfaces of the S. solfataricus and H. volcanii AT4Ps (SI Appendix, Fig. S9). A feature present in all the AT4P and AFF subunits is that the N-terminal core helix is curved. The curvature is especially noticeable in the P. calidifontis pilin and archaeal flagellins where the N-terminal helices are banana shaped (Fig. 3 A and B).
We describe AFFs as having 10 protofilaments, while the P. calidifontis flagellar–like AT4P has 10 strands. In AFFs, there is a cooperativity in subunit conformation along each protofilament, and a similar cooperativity was observed for BFF protofilaments. This cooperativity is an essential property of these supercoiled filaments and would not be maintained for AT4Ps which are flexible but do not supercoil.
The results of our structural analyses suggest a critical relationship between the N-terminal helix length and AFF structure. The P. calidifontis pilus has 10-strand packing like all AFFs; however, we did not observe any supercoiling and its genomic locus (Fig. 2D) lacks the genes for the stator proteins FlaF/ArlF and FlaG/ArlG (25) typical of flagellar operons. As a result, we have classified it as a flagellar-like AT4P. The longer P. calidifontis N-terminal helix likely favors the assembly of the subunits into a 10-strand architecture. The presence of 10-start contacts as well as increased 3-start contacts compared to other AT4Ps provides the greater rigidity needed to maintain a regular supercoil. It may be premature to draw firm conclusions about the evolutionary relationship between the P. calidifontis pilus and AFFs given that we have only one example of such a putative intermediate. However, we note that computational analyses suggest that structurally similar type IV pilins, some of which are even closer to archaeal flagellins than those of the P. calidifontis pilin, are encoded by diverse Thermoproteales species (Fig. 5A). Additionally, the 10-start protofilaments in all AFF structures are either vertical with no tilt such as in A. pernix (SI Appendix, Fig. S6B) and most other AFFs or very slightly tilted as in the S. islandicus REY15A AFF (6). The role of these nearly vertical 10-start protofilaments may be critical to the supercoiling process and is only beginning to be understood (6). Although we found no evidence for the P. calidifontis filament being able to supercoil, motility has been reported for this organism (41) and other Pyrobaculum species (42), despite the absence of a bona fide flagellar operon (32). We suspect that the motility observed may be due not to swimming, but to twitching or gliding, both of which require a surface, but this remains to be determined.
We also showed the relationship between the sequence properties of the N-terminal core helices of archaeal type IV pilins and flagellins. While most of the solved AT4P structures have pilins whose core helix domains are predominantly hydrophobic, flagellar-like AT4Ps from Thermoproteales, including P. calidifontis, have additional polar and charged residues at the C-terminal end of this helix. Archaeal flagellins also have these additional polar residues in their core helices. The interactions between these residues may provide an additional stability to the filaments, which cannot be provided by relatively nonspecific hydrophobic interactions. It has been shown for an archaeal virus that purely hydrophobic interactions can allow for helices to slide past each other relatively easily (43). Many of the P. calidifontis polar/charged residues such as serine, threonine, tyrosine, and glutamine are able to interface with both polar and hydrophobic residues. The polar/charged region of the core helix in archaeal flagellins contains more polar/charged residues such as lysine, arginine, glutamate, aspartate, glutamine, and asparagine, which are less likely to be found in hydrophobic interfaces.
It has been suggested (16) that flexibility is essential for the AFF function, and that AFFs are more flexible than AT4Ps. However, we find a similar degree of flexibility (SI Appendix, Fig. S7) based upon the same type of analysis carried out by Gambelli et al. and conclude instead that a rather rigid supercoil that is resistant to torsional stress is essential for flagellar function and what will distinguish AFFs from AT4Ps. Rather than flexibility, it is the capacity of flagellar subunits, whether from bacteria or archaea, to adopt multiple structural states within the same filament to form rather fixed supercoils that underlies the motility (6). This implies, of course, that a flagellin subunit has the ability to exist in multiple conformations, but these multiple states do not occur randomly in the supercoiled filaments. While AT4Ps are flexible, so are many other filamentous structures like F-actin (44) or filamentous plant and archaeal viruses (45, 46), yet none of these supercoil.
Structural alignments were previously used to compare the evolutionary relationships between AT4Ps and AFFs (20, 21). Performing an “all against all” analysis using the Dali server (47), we find that the P. calidifontis structure clusters with AT4Ps but also aligns well with flagellar stator proteins FlaF and FlaG (SI Appendix, Fig. S10). This further suggests that P. calidifontis is indeed a flagellar-like AT4P. Could the P. calidifontis T4P have evolved from an AFF rather than the opposite path that we propose? This seems unlikely, as phylogenetic analysis of the FlaI-like secretion ATPases places the ATPase of P. calidifontis and other Thermoproteales T4P operons at the base of the clade including all flagellar FlaI orthologs (23).
The C-terminal globular domain size appears quite variable across all AT4Ps and AFFs. However, in archaea, all of these C-terminal domains, despite their variable size, appear to have an Ig-like fold, while in bacteria, the overall folds of the C-terminal domain can diverge greatly. It has previously been shown that the outer domains in at least one bacterial flagellin can be largely removed without affecting motility (48, 49), and in bacteria, such as Bacillus subtilis (50), Kurthia (51), or Caulobacter (52), the outer domains are almost entirely absent. Thus, the C-terminal domains of AFFs and BFFs likely play little to no role determining the packing of the filament. However, it has also been shown for BFFs that the outer domains can modulate motility (53), enhance motility in high-viscosity conditions (54), or stabilize a structure that has weakened innerdomain interactions (54, 55). We therefore think that variations in the C-terminal domains of both AFFs and AT4Ps may be the result of species-specific adaptations.
Ever since Darwin (56), a cornerstone of evolutionary theory has been that randomly occurring variations are selected, either positively or negatively, based upon the fitness advantage or disadvantage they convey to an organism. We think that this simple paradigm can provide insights into the evolution of AFFs from AT4P. Given the possible functions of bacterial and archaeal T4Ps in adhesion, surface sensing, biofilm formation, natural transformation, and twitching motility, few constraints have been placed on the details of the quaternary structure. This is why such divergence has been seen in the helical symmetries for both archaeal (Fig. 3A) and bacterial (Fig. 3B) T4Ps. An analogy might be made with bacterial actins, where an even greater diversity of filament symmetries has now been found (57, 58). In contrast, the requirement of rigid supercoils for both BFF and AFFs has placed severe constraints on the helical symmetry and structure of these flagellar filaments. While BFFs have been found with twist values from 65.3° to 65.8° (59), recent observations suggest that the actual range in wild-type flagellar filaments is from 65.4° to 65.54°, and the previously described larger range comes from nonmotile mutants (6, 50, 59, 60). We thus consider it plausible that the naturally occurring variation in AT4Ps (Fig. 3A) has allowed for the selection of a flagellar-like filament, such as that from P. calidifontis. In this context, it is notable that the T4P locus of P. calidifontis includes a gene encoding a KaiC-like ATPase, which is homologous to the FlaH component of archaeal flagella. Thus, the only major components which are missing in the P. calidifontis AT4P system compared to AFF are the two proteins, FlaF and FlaG, which form the stator complex (61). Structural comparisons of the two proteins have strongly suggested that both have evolved by tandem duplication and subfunctionalization of the gene encoding the flagellin protein FlaB (21). Collectively, the comparisons of the AFF and AT4P structures and their genomic loci are both pointing to the AT4P systems of Thermoproteales as the likely ancestors of the archaeal motility apparatus.
Methods
Sample Preparation.
P. calidifontis DSM 21063 (41) cells were purchased from the DSMZ culture collection and grown in 1,090 medium (1.0% tryptone, 0.1% yeast extract, 0.3% sodium thiosulfate, pH 7) at 90 °C without agitation. Preculture (30 mL) was started from a 200-μL cryo-stock, grown for 2 d, and then diluted into 200 mL of fresh medium. When OD600 reached ∼0.2, the cells were collected by centrifugation (Sorval SLA1500 rotor, 7,000 rpm, 10 min, 20 °C) and resuspended in 10 mL phosphate-buffered saline (PBS) buffer. The extracellular filaments were collected as described previously (62, 63). Briefly, the cell suspension was vortexed for 15 min to shear off the extracellular filaments. The cells were removed by centrifugation (Eppendorf F-35-6-30 rotor, 7,830 rpm, 20 min, 20 °C), whereas extracellular filaments were pelleted by ultracentrifugation (Beckman SW60Ti rotor, 38,000 rpm, 2 h, 15 °C). After the run, the supernatant was removed, and the pellet was resuspended in 200 μL PBS.
H. volcanii H53 cells (64) were kindly provided by Thorsten Allers (University of Nottingham) and grown in Hv-YPC medium (64) containing (per liter) 144 g NaCl, 21 g MgSO4∙7H2O, 18 g MgCl2∙6H2O, 4.2 g KCl, 12 mM Tris HCl (pH 7.5), 0.5% yeast extract (Difco), 0.1% peptone (Oxoid), 0.1% casamino acids (Difco), and 3 mM CaCl2. A preculture of 5 mL was grown to stationary phase at 45 °C under agitation, then diluted into 500 mL culture and grown to exponential phase (OD600 ≈ 0.5). Cells were collected by centrifugation (Sorval SLA1500 rotor, 8,000 rpm, 10 min, 20 °C). The cell pellet was resuspended in 20 mL of 18% SW buffer (Hv-YPC medium in the absence of yeast extract, peptone, and casamino acids), and the cell suspension was vortexed for 30 min to shear off the pili. The cells were removed by centrifugation (Sorval SLA1500 rotor, 8,000 rpm, 30 min, 20 °C). The supernatant was collected, and the pili were pelleted by ultracentrifugation (Beckman 60 Ti rotor, 35,000 rpm, 2 h, 4 °C). The supernatant was removed, and the pellet was resuspended in 500 μL of 18% SW buffer.
Aeropyrum pernix K1 DSM 11879 (65) cells were purchased from the DSMZ culture collection and grown in 3ST medium [35 g/L Sea salts (Sigma), 0.1 % tryptone, 0.1 % yeast extract, 0.1 % sodium thiosulfate, pH 7] at 90 °C without agitation. Preculture (10 mL) was started from a 1 mL cryo-stock, grown for 3 d, and then diluted into 60 mL of fresh 3ST medium. After 3 d of growth, 100 mL of the fresh medium was added to the culture and the growth was continued for another 3 d. Then, the cells were removed by centrifugation (Sorval SLA1500 rotor, 7,000 rpm, 10 min, 20 °C), whereas extracellular filaments were collected by ultracentrifugation (Beckman SW60Ti rotor, 38,000 rpm, 2 h, 15 °C) and resuspended in 200 μL PBS.
Negative-Stain TEM of P. calidifontis Cells.
For negative-staining transmission electron microscopy, 10 μL of cell suspension was adsorbed onto glow-discharged copper grids with carbon-coated Formvar film and negatively stained with 2.0% (wt/vol) uranyl acetate. The samples were observed under FEI Tecnai Spirit BioTwin 120 microscope operated at 120 kV. To assess the supercoiling of flagellar-like AT4P of P. calidifontis, ~300 individual cells were observed at 18,000× magnification.
Cryo-EM Grid Preparation.
For each archaeal filament sample, a 3 to 4 μL aliquot was placed onto a lacey carbon grid (300 mesh, Cu) which was then blotted and plunge frozen into liquid ethane using a Leica EM GP plunge freezer.
Cryo-EM Imaging and Image Processing.
All cryo-EM movies were collected on a Titan Krios equipped with a K3 direct electron detector at a pixel size of 1.08 Å/pixel. Subsequent steps from movie processing up to the final reconstruction were performed in cryoSPARC (34). The basic movie processing workflow started with Patch Motion Correction, followed by Patch CTF Estimation and then Curate Exposures. Initial subsets of particles were then manually picked and then subject to 2D classification. The best classes were then selected and used as inputs for particle selection using either Filament Tracer or Template Picker, both of which adequately selected filament particles. The larger particle sets from automated particle picking were subjected to at least one round of 2D classification. More heterogeneous datasets were subjected to several rounds of 2D classification. For each filament dataset, helical reconstruction was performed using a starting cylindrical volume with a diameter slightly bigger than that measured for the filaments in 2D images. After successful helical reconstruction, CTF refinement and a final helical reconstruction were performed for each dataset. The local resolution was estimated for each volume and finally the maps were subject to local filtering.
Model Building and Refinement.
Initial models were built using either the DeepTracer-ID method (27), or built by refining an initial structure already deposited into the new map (S. solfataricus). These initial models were then subject to refinement in Coot (66) and using Phenix real-space refinement (67). Models were subsequently validated in Phenix (68, 69).
Structural Alignment of Archaeal Type IV Pilin and Flagellin N-Terminal Helices.
For the comparison of the properties of archaeal T4P and AFF N-terminal helix sequences (Fig. 5A), the model for each structure’s N-terminal helix was displayed using UCSF Chimera (70) and the start point of each species sequence in the alignment was taken to be the beginning of the model and the end point was taken to be the last residue in the alpha helix. The starting position of the archaeal flagellar–like type IV pilins and flagellins was moved by a residue of two, depending on the sequence, to align all helices as best as possible.
Analysis of the A. pernix AFF.
Due to the additional density found in the previously deposited A. pernix AFF map not explained by the flagellin (FlaB1) model (SI Appendix, Fig. S2A), we generated a sequence alignment of the two A. pernix flagellins (SI Appendix, Fig. S2B). For the reassessment of the Aeropyrum filament, a preliminary model for the A. pernix flagellin FlaB2 was generated using AlphaFold2 (71). This model was then fit into the A. pernix AFF density map and refined using Coot.
The deposited A. pernix FlaB1 model was unable to explain density in two loop regions of the map (SI Appendix, Fig. S2C), while the A. pernix FlaB2 model (SI Appendix, Fig. S2D) could explain this density. There was a region in the map which was fit by the A. pernix FlaB1 quite nicely (SI Appendix, Fig. S2C turn region) in comparison to the A. pernix FlaB2 model which had 8 unmodeled side chains (SI Appendix, Fig. S2D turn region).
Due to the recent discovery of a heterodimeric AFF from Methanococcus villosus with an asymmetric unit containing six flagellin subunits (16), we analyzed the power spectrum of the A. pernix AFF (SI Appendix, Fig. S3A) which showed the expected meridional layer line at 1/5.5 Å as well as an unexpected meridional layer line at 1/33 Å, which is 6 × 5.5 Å. This would be consistent with an asymmetric unit containing six flagellins, as observed for the heterodimeric AFF (16). The M. villosus heterodimeric filament was composed of two different flagellin subunits, and we suspected that the same could be the case for our A. pernix filament, due to the presence of two flagellins in the A. pernix genome. We then did a helical reconstruction with AFF symmetry parameters six times (a rise of 6 × 5.5 Å = 33 Å and a twist of 6 × 108.0° = −72.0°; SI Appendix, Fig. S3B) that have been used for the initial structure (SI Appendix, Fig. S2A). For some of the six subunits in the asymmetric unit, there was clear density in the previously unmodeled loop region at moderate thresholds (top images in SI Appendix, Fig. S3B), while in others, the density was absent or broken (bottom images in SI Appendix, Fig. S3B). However, all subunits showed loop density at lower thresholds. Some of the inconsistencies in the density map may best be explained by heterogeneity, with only some of the flagellar filament segments being of a heterodimeric assembly similar to the M. villosus AFF. Alternatively, or in addition, the register of the hexamers may change throughout the filaments, leading to an averaging of the two different flagellins.
To test the viability of the A. pernix flagellar filament being composed of a hexamer, we generated an asymmetric unit consisting of six subunits, containing three FlaB1 and three FlaB2 copies in the same manner as the M. villosus AFF (SI Appendix, Fig. S3C). We then imposed a helical symmetry with a rise of 33 Å and a twist of −72° corresponding to a hexameric ASU and generated a long filament mode (SI Appendix, Fig. S3D). We then simulated a density map to 3.5 Å (SI Appendix, Fig. S3E) using the EMAN2 program e2pdb2mrc (72). We converted this density map from mrc to spider format and then using the spider software processing suite (73) generated a 2D projection of the simulated volume and then a power spectrum. This power spectrum had meridional layer lines at 1/33 Å and 1/5.5 Å (SI Appendix, Fig. S3F), consistent with the averaged power spectrum from image segments. The averaged power spectrum from image segments (SI Appendix, Fig. S3A) had a weaker meridional layer line at 1/33 Å than that of the power spectrum from the simulated hexameric symmetry map. This is consistent with our hypothesis that the A. pernix filament image segments contain some degree of heterogeneity, with only some of the segments being from hexameric filaments.
3D Variability Analysis and Asymmetric Reconstruction and Modeling of the S. solfataricus aT4P.
As done previously for archaeal and BFFs (6), the helically symmetric S. solfataricus AT4P volume was used as input for 3D variability analysis in cryoSPARC. From this, several reconstructions were generated using the 3D variability display function with outputs in either simple mode or cluster mode. The most curved volume output from cluster mode and its associated particles were then used as the input for a local refinement job, and a final map at 4.0 Å resolution was generated. From this, a model for the curved AT4P structure was created using the same modeling techniques as described above.
Comparisons between Archaeal Flagellins and Pilins.
For the comparison of symmetries and subunits shown in Figs. 3 and 4, we examined the N-terminal helices of all AT4P and AFF structures. The end of each N-terminal helix was defined as the beginning of the linker region between the N-terminal helix and the C-terminal outer domain. The linker region lengths were somewhat ambiguous to determine for some structures, but in general, the beginning of the C-terminal outer domain was taken to be the start of beta-sheet- or near-beta-sheet-forming interactions. The interfacial surface areas between contacting N-terminal core helices were calculated using PDBePISA (74).
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
The cryo-EM imaging was done at the Molecular Electron Microscopy Core Facility at the University of Virginia, which is supported by the School of Medicine. This research was, in part, supported by the National Cancer Institute’s National Cryo-EM Facility at the Frederick National Laboratory for Cancer Research under contract 75N91019D00024. This work was supported by NIH Grant GM122510 (E.H.E.) and GM138756 (F.W.). The work in the M.K. laboratory was supported by grants from l'Agence Nationale de la Recherche (ANR-20-CE20-009-02 and ANR-21-CE11-0001-01) and Ville de Paris Emergence(s) project. We would like to acknowledge B.F. Luisi at the University of Cambridge and J.V. Pratap at the Central Drug Research Institute in Lucknow, India, for helpful discussions about the AFF and AT4P structures.
Author contributions
M.A.B.K., M.K., and E.H.E. designed research; M.A.B.K., V.C.-K., Y.L., D.P.B., J.L., R.R.S., and F.W. performed research; V.C.-K., Y.L., D.P.B., and J.L. contributed new reagents/analytic tools; M.A.B.K., R.R.S., C.R.C., F.W., M.K., and E.H.E. analyzed data; and M.A.B.K., M.K., and E.H.E. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
Reviewers: L.L.B., McMaster University; and M.W., Okinawa Institute of Science and Technology Graduate University.
Contributor Information
Fengbin Wang, Email: jerrywang@uab.edu.
Mart Krupovic, Email: mart.krupovic@pasteur.fr.
Edward H. Egelman, Email: egelman@virginia.edu.
Data, Materials, and Software Availability
Atomic model data have been deposited in the Protein Data Bank: 8FJS (75), 8FK7 (76), 8FJ5 (77), 7TXI (78), and 8FK0 (79). Cryo-EM maps have been deposited in the Electron Microscopy Data Bank: EMD-29246 (80), EMD-29249 (81), EMD-29215 (82), EMD-26158 (83), and EMD-29247 (84).
Supporting Information
References
- 1.Alam M., Oesterhelt D., Morphology, function and isolation of halobacterial flagella. J. Mol. Biol. 176, 459–475 (1984). [DOI] [PubMed] [Google Scholar]
- 2.Brown H. P., On the structure and mechanics of the protozoan flagellum. Ohio J. Sci. 45, 247–301 (1945). [PubMed] [Google Scholar]
- 3.Pijper A., Abraham G., Wavelengths of bacterial flagella. J. Gen. Microbiol. 10, 452–456 (1954). [DOI] [PubMed] [Google Scholar]
- 4.Pazour G. J., Agrin N., Leszyk J., Witman G. B., Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 170, 103–113 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hyman H. C., Trachtenberg S., Point mutations that lock Salmonella typhimurium flagellar filaments in the straight right-handed and left-handed forms and their relation to filament superhelicity. J. Mol. Biol. 220, 79–88 (1991). [DOI] [PubMed] [Google Scholar]
- 6.Kreutzberger M. A. B., et al. , Convergent evolution in the supercoiling of prokaryotic flagellar filaments. Cell 185, 3487–3500.e14 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strick J., Swimming against the tide: Adrianus Pijper and the debate over bacterial flagella, 1946–1956. Isis 87, 274–305 (1996). [Google Scholar]
- 8.Reichert K. B., Über die Sichtbarmachung der Geisseln und die Geisselbewegung der Bakterien (Gustav Fischer, 1909). [Google Scholar]
- 9.Schimz A., Hildebrand E., Chemosensory responses of Halobacterium halobium. J. Bacteriol. 140, 749–753 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bayley D. P., Jarrell K. F., Further evidence to suggest that archaeal flagella are related to bacterial type IV pili. J. Mol. Evol. 46, 370–373 (1998). [PubMed] [Google Scholar]
- 11.Faguy D. M., Jarrell K. F., Kuzio J., Kalmokoff M. L., Molecular analysis of archael flagellins: Similarity to the type IV pilin-transport superfamily widespread in bacteria. Can J. Microbiol. 40, 67–71 (1994). [DOI] [PubMed] [Google Scholar]
- 12.Wang F., Craig L., Liu X., Rensing C., Egelman E. H., Microbial nanowires: Type IV pili or cytochrome filaments? Trends Microbiol. 31, 384–392 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Braun T., et al. , Archaeal flagellin combines a bacterial type IV pilin domain with an Ig-like domain. Proc. Natl. Acad. Sci. U.S.A. 113, 10352–10357 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohen-Krausz S., Trachtenberg S., The structure of the archeabacterial flagellar filament of the extreme halophile Halobacterium salinarum R1M1 and its relation to eubacterial flagellar filaments and type IV pili. J. Mol. Biol. 321, 383–395 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Daum B., et al. , Structure and in situ organisation of the Pyrococcus furiosus archaellum machinery. Elife 6, e27470 (2017), 10.7554/eLife.27470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gambelli L., et al. , An archaellum filament composed of two alternating subunits. Nat. Commun. 13, 710 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meshcheryakov V. A., et al. , High-resolution archaellum structure reveals a conserved metal-binding site. EMBO Rep. 20, e46340 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poweleit N., et al. , CryoEM structure of the Methanospirillum hungatei archaellum reveals structural features distinct from the bacterial flagellum and type IV pilus. Nat. Microbiol. 2, 16222 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jarrell K. F., Ding Y., Nair D. B., Siu S., Surface appendages of archaea: Structure, function, genetics and assembly. Life (Basel) 3, 86–117 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang F., et al. , The structures of two archaeal type IV pili illuminate evolutionary relationships. Nat. Commun. 11, 3424 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang F., et al. , An extensively glycosylated archaeal pilus survives extreme conditions. Nat. Microbiol. 4, 1401–1410 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pohlschroder M., Pfeiffer F., Schulze S., Abdul Halim M. F., Archaeal cell surface biogenesis. FEMS Microbiol. Rev. 42, 694–717 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Makarova K. S., Koonin E. V., Albers S. V., Diversity and evolution of type IV pili systems in archaea. Front Microbiol. 7, 667 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chaudhury P., Quax T. E. F., Albers S. V., Versatile cell surface structures of archaea. Mol. Microbiol. 107, 298–311 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Umrekar T. R., et al. , Evolution of archaellum rotation involved invention of a stator complex by duplicating and modifying a core component. Front Microbiol. 12, 773386 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Sousa Machado J. N., et al. , Autophosphorylation of the KaiC-like protein ArlH inhibits oligomerization and interaction with ArlI, the motor ATPase of the archaellum. Mol. Microbiol. 116, 943–956 (2021). [DOI] [PubMed] [Google Scholar]
- 27.Chang L., et al. , DeepTracer-ID: De novo protein identification from cryo-EM maps. Biophys. J. 121, 2840–2848 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Esquivel R. N., Schulze S., Xu R., Hippler M., Pohlschroder M., Identification of Haloferax volcanii pilin N-glycans with diverse roles in pilus biosynthesis, adhesion, and microcolony formation. J. Biol. Chem. 291, 10602–10614 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pohlschroder M., Esquivel R. N., Archaeal type IV pili and their involvement in biofilm formation. Front. Microbiol. 6, 190 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Esquivel R. N., Xu R., Pohlschroder M., Novel archaeal adhesion pilins with a conserved N terminus. J. Bacteriol. 195, 3808–3818 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reed C. J., Lewis H., Trejo E., Winston V., Evilia C., Protein adaptations in archaeal extremophiles. Archaea. 2013, 373275 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Desmond E., Brochier-Armanet C., Gribaldo S., Phylogenomics of the archaeal flagellum: Rare horizontal gene transfer in a unique motility structure. BMC Evol. Biol. 7, 106 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Varadi M., et al. , AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50, D439–d444 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Craig L., Forest K. T., Maier B., Type IV pili: Dynamics, biophysics and functional consequences. Nat. Rev. Microbiol. 17, 429–440 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Giltner C. L., Nguyen Y., Burrows L. L., Type IV pilin proteins: Versatile molecular modules. Microbiol. Mol. Biol. Rev. 76, 740–772 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sheppard D., et al. , The major subunit of widespread competence pili exhibits a novel and conserved type IV pilin fold. J. Biol. Chem. 295, 6594–6604 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gu Y., et al. , Structure of Geobacter pili reveals secretory rather than nanowire behaviour. Nature 597, 430–434 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang F., et al. , Cryo-EM structure of an extracellular Geobacter OmcE cytochrome filament reveals tetrahaem packing. Nat. Microbiol. 7, 1291–1300 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neuhaus A., et al. , Cryo-electron microscopy reveals two distinct type IV pili assembled by the same bacterium. Nat. Commun. 11, 2231 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Amo T., et al. , Pyrobaculum calidifontis sp. nov., a novel hyperthermophilic archaeon that grows in atmospheric air. Archaea 1, 113–121 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Volkl P., et al. , Pyrobaculum aerophilum sp. nov., a novel nitrate-reducing hyperthermophilic archaeum. Appl. Environ. Microbiol. 59, 2918–2926 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang F., et al. , Spindle-shaped archaeal viruses evolved from rod-shaped ancestors to package a larger genome. Cell 185, 1297–1307.e11 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reynolds M. J., Hachicho C., Carl A. G., Gong R., Alushin G. M., Bending forces and nucleotide state jointly regulate F-actin structure. Nature 611, 380–386 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DiMaio F., et al. , The molecular basis for flexibility in the flexible filamentous plant viruses. Nat. Struct. Mol. Biol. 22, 642–644 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang F., et al. , Structures of filamentous viruses infecting hyperthermophilic archaea explain DNA stabilization in extreme environments. Proc. Natl. Acad. Sci. U.S.A. 117, 19643–19652 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holm L., Dali server: Structural unification of protein families. Nucleic Acids Res. 50, W210–W215 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Malapaka R. R., Adebayo L. O., Tripp B. C., A deletion variant study of the functional role of the Salmonella flagellin hypervariable domain region in motility. J. Mol. Biol. 365, 1102–1116 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Yoshioka K., Aizawa S., Yamaguchi S., Flagellar filament structure and cell motility of Salmonella typhimurium mutants lacking part of the outer domain of flagellin. J. Bacteriol. 177, 1090–1093 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang F., et al. , A structural model of flagellar filament switching across multiple bacterial species. Nat. Commun. 8, 960 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blum T. B., Filippidou S., Fatton M., Junier P., Abrahams J. P., The wild-type flagellar filament of the Firmicute Kurthia at 2.8 Å resolution in vivo. Sci. Rep. 9, 14948 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Montemayor E. J., et al. , Flagellar structures from the bacterium Caulobacter crescentus and implications for Phage varphi CbK predation of multiflagellin bacteria. J. Bacteriol. 203, e00399-20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kreutzberger M. A. B., et al. , Flagellin outer domain dimerization modulates motility in pathogenic and soil bacteria from viscous environments. Nat. Commun. 13, 1422 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nedeljković M., et al. , An unbroken network of interactions connecting flagellin domains is required for motility in viscous environments. PLoS Pathog. 19, e1010979 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kreutzberger M. A. B., Ewing C., Poly F., Wang F., Egelman E. H., Atomic structure of the Campylobacter jejuni flagellar filament reveals how epsilon Proteobacteria escaped Toll-like receptor 5 surveillance. Proc. Natl. Acad. Sci. U.S.A. 117, 16985–16991 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gould S. J., Ever Since Darwin: Reflections in Natural History (Norton, New York, 1977), p. 285. [Google Scholar]
- 57.Orlova A., et al. , The structure of bacterial ParM filaments. Nat. Struct. Mol. Biol. 14, 921–926 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Usluer G. D., et al. , Cryo-EM structure of the bacterial actin AlfA reveals unique assembly and ATP-binding interactions and the absence of a conserved subdomain. Proc. Natl. Acad. Sci. U.S.A. 115, 3356–3361 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maki-Yonekura S., Yonekura K., Namba K., Conformational change of flagellin for polymorphic supercoiling of the flagellar filament. Nat. Struct. Mol. Biol. 17, 417–422 (2010). [DOI] [PubMed] [Google Scholar]
- 60.Yonekura K., Maki-Yonekura S., Namba K., Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature 424, 643–650 (2003). [DOI] [PubMed] [Google Scholar]
- 61.Tsai C. L., et al. , The structure of the periplasmic FlaG-FlaF complex and its essential role for archaellar swimming motility. Nat. Microbiol. 5, 216–225 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang F., Cvirkaite-Krupovic V., Krupovic M., Egelman E. H., Archaeal bundling pili of Pyrobaculum calidifontis reveal similarities between archaeal and bacterial biofilms. Proc. Natl. Acad. Sci. U.S.A. 119, e2207037119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Beltran L. C., et al. , Archaeal DNA-import apparatus is homologous to bacterial conjugation machinery. Nat. Commun. 14, 666 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Allers T., Ngo H. P., Mevarech M., Lloyd R. G., Development of additional selectable markers for the halophilic archaeon Haloferax volcanii based on the leuB and trpA genes. Appl. Environ. Microbiol. 70, 943–953 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sako Y., et al. , Aeropyrum pernix gen. nov., sp. nov., a novel aerobic hyperthermophilic archaeon growing at temperatures up to 100 degrees C. Int. J. Syst. Bacteriol. 46, 1070–1077 (1996). [DOI] [PubMed] [Google Scholar]
- 66.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Afonine P. V., et al. , Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D. Struct. Biol. 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Afonine P. V., et al. , New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallogr. D. Struct. Biol. 74, 814–840 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Williams C. J., et al. , MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pettersen E. F., et al. , UCSF Chimera–A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 71.Jumper J., et al. , Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tang G., et al. , EMAN2: An extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007). [DOI] [PubMed] [Google Scholar]
- 73.Frank J., et al. , SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116, 190–199 (1996). [DOI] [PubMed] [Google Scholar]
- 74.Krissinel E., Henrick K., Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007). [DOI] [PubMed] [Google Scholar]
- 75.Kreutzberger M. A. B., Wang F., Krupovic M., Egelman E. H., 8FJS, Structure of the Saccharolobus solfataricus archaeal type IV pilus at 3 Angstrom resolution. Protein Data Bank. https://www.rcsb.org/structure/8FJS. Deposited 20 December 2022.
- 76.Wang F., Kreutzberger M. A. B., Cvirkaite-Krupovic V., Krupovic M., Egelman E. H., 8FK7, Structure of the Pyrobaculum calidifontis flagellar-like archaeal type IV pilus. Protein Data Bank. https://www.rcsb.org/structure/8FK7. Deposited 20 December 2022.
- 77.Wang F., Kreutzberger M. A. B., Baquero D. P., Krupovic M., Egelman E. H., 8FJ5, Structure of the Haloferax volcanii archaeal type IV pilus. Protein Data Bank. https://www.rcsb.org/structure/8FJ5. Deposited 19 December 2022.
- 78.Wang F., Cvirkaite-Krupovic V., Baquero D. P., Krupovic M., Egelman E. H., 7TXI, Cryo-EM of A. pernix flagellum. Protein Data Bank. https://www.rcsb.org/structure/7TXI. Deposited 9 February 2022.
- 79.Kreutzberger M. A. B., Wang F., Krupovic M., Egelman E. H., 8FK0, Asymmetric cryo-EM structure of a curved Saccharolobus solfataricus type IV pilus. Protein Data Bank. https://www.rcsb.org/structure/8FK0. Deposited 20 December 2022.
- 80.Kreutzberger M. A. B., Wang F., Krupovic M., Egelman E. H., EMD-29246. Electron Microscopy Database. https://www.ebi.ac.uk/emdb/EMD-29246. Deposited 20 December 2022.
- 81.Wang F., Kreutzberger M. A. B., Cvirkaite-Krupovic V., Krupovic M., Egelman E. H., EMD-29249. Electron Microscopy Database. https://www.ebi.ac.uk/emdb/EMD-29249. Deposited 20 December 2022.
- 82.Wang F., Kreutzberger M. A. B., Baquero D. P., Krupovic M., Egelman E. H., EMD-29215, Electron Microscopy Database. https://www.ebi.ac.uk/emdb/EMD-29215. Deposited 19 December 2022.
- 83.Wang F., Cvirkaite-Krupovic V., Baquero D. P., Krupovic M., Egelman E. H., EMD-26158, Electron Microscopy Database. https://www.ebi.ac.uk/emdb/EMD-26158. Deposited 9 February 2022.
- 84.Kreutzberger M. A. B., Wang F., Krupovic M., Egelman E. H., EMD-29247, Electron Microscopy Database. https://www.ebi.ac.uk/emdb/EMD-29247. Deposited 20 December 2022.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
Atomic model data have been deposited in the Protein Data Bank: 8FJS (75), 8FK7 (76), 8FJ5 (77), 7TXI (78), and 8FK0 (79). Cryo-EM maps have been deposited in the Electron Microscopy Data Bank: EMD-29246 (80), EMD-29249 (81), EMD-29215 (82), EMD-26158 (83), and EMD-29247 (84).