Significance
Energy transport over long distances with high efficiency is a critical design parameter of photosynthetic light harvesting, where photonic energy travels across multiple antenna proteins to reach the reaction center. Understanding transport requires that energy transfer between proteins be resolved, but this crucial step has been experimentally inaccessible. Here, we used near-native membrane nanodiscs to isolate two antenna proteins from purple bacteria, one of the most ancient and most efficient photosynthetic organisms. By measuring the structural organization and dynamics, we characterized interprotein energy transfer across the physiological range of distances. Simulations revealed that the presence of fast energy transfer steps enhances long-distance transport. These results point to pairwise interactions as a key mediator of energy transport.
Keywords: photosynthesis, purple bacteria, light harvesting, ultrafast spectroscopy, cryogenic electron microscopy
Abstract
In photosynthesis, absorbed light energy transfers through a network of antenna proteins with near-unity quantum efficiency to reach the reaction center, which initiates the downstream biochemical reactions. While the energy transfer dynamics within individual antenna proteins have been extensively studied over the past decades, the dynamics between the proteins are poorly understood due to the heterogeneous organization of the network. Previously reported timescales averaged over such heterogeneity, obscuring individual interprotein energy transfer steps. Here, we isolated and interrogated interprotein energy transfer by embedding two variants of the primary antenna protein from purple bacteria, light-harvesting complex 2 (LH2), together into a near-native membrane disc, known as a nanodisc. We integrated ultrafast transient absorption spectroscopy, quantum dynamics simulations, and cryogenic electron microscopy to determine interprotein energy transfer timescales. By varying the diameter of the nanodiscs, we replicated a range of distances between the proteins. The closest distance possible between neighboring LH2, which is the most common in native membranes, is 25 Å and resulted in a timescale of 5.7 ps. Larger distances of 28 to 31 Å resulted in timescales of 10 to 14 ps. Corresponding simulations showed that the fast energy transfer steps between closely spaced LH2 increase transport distances by ∼15%. Overall, our results introduce a framework for well-controlled studies of interprotein energy transfer dynamics and suggest that protein pairs serve as the primary pathway for the efficient transport of solar energy.
Photosynthesis supports nearly all life on Earth by converting light energy to chemical energy with a remarkable near-unity quantum efficiency (1–5). The quantum efficiency is the yield of charge separation per absorbed photon rather than the overall power conversion efficiency, which is much lower at ∼5% (6–8). The high quantum efficiency in these first steps is achieved through a network of antenna proteins that absorb sunlight and then rapidly transport the resultant photoenergy over distances of 25 to 200 nm to reach reaction centers for charge separation. While nanometer-scale energy transfer within individual antenna proteins has been extensively characterized (3, 9–25), energy transport on the tens to hundreds of nanometer distances relies on interprotein energy transfer. However, these energy transfer steps have been more challenging to probe due to the number of proteins involved, their heterogeneous organization, and their overlapping spectral features (26–31). As a result, many open questions remain about interprotein energy transfer and how it gives rise to efficient energy transport over long distances.
Purple bacteria serve as model organisms for studies of photosynthetic light harvesting due to their high efficiency and well-separated spectral features (7). The primary antenna protein, light-harvesting complex 2 (LH2) or the B800–850 complex, is formed from subunits consisting of a protein heterodimer that bind one carotenoid and three bacteriochlorophyll a (BChla). The subunits further assemble into an octameric, nonameric, or decameric cylindrical structure in a species-dependent manner (28, 32–34). Upon assembly, two concentric rings of BChla are generated. One of the rings, denoted B800, consists of one BChla from each subunit and absorbs at 800 nm, whereas the other, denoted B850, consists of two BChla from each subunit and usually absorbs at 850 nm (35–38). While these are the most common absorption features, in some species, the blue-shift of B850 ring to 820 nm under stress conditions (primarily low-light irradiation) is attributed to a single amino acid substitution that restructures the hydrogen bonding network between the protein and the BChlas (35, 39, 40). This variant of LH2 is known as LH3 or the B800–820 complex (36, 41). For all variants, energy rapidly transfers from the B800 to the B820/B850 ring and then around the B820/B850 ring (3, 10–24, 35, 42–47).
After absorption and relaxation within LH2, energy transfers between the LH2 to reach light-harvesting complex 1, which encircles the bacterial reaction center (LH1–RC) (3, 35, 42, 43, 48–50). The LH2 and LH1–RC are organized in the photosynthetic membrane in heterogeneous arrangements and protein ratios, which further vary with species and growth conditions (5, 26, 29, 39, 51–54). To understand the energy transfer dynamics within the protein network, energy transfer timescales from LH2 to LH1 and among LH2 have been measured. Timescales of 2 to 10 ps were determined for LH2-to-LH1 energy transfer (15, 35, 42, 55–57) and 3 to 5 ps for LH2-to-LH2 energy transfer (58, 59). However, because LH2-to-LH2 energy transfer does not have any associated spectral changes, the timescale of this step was only indirectly resolved. Furthermore, measurements of interprotein energy transfer were all performed in vivo or on isolated membrane segments, which exhibit significant heterogeneity in the interprotein distances. Energy transfer is highly dependent on the interprotein distance, and so the reported values likely represent averages over a range of underlying timescales. The heterogeneous arrangements may also contribute to the heterogeneity in reported values. Due to this complexity, the specific timescales for protein-to-protein energy transfer have not yet been isolated, despite their key role in long-distance energy transport.
In this work, we isolate and interrogate energy transfer between LH2 variants by reconstituting LH2 and LH3 from Phaeospirillum (Ph.) molischianum together within a model membrane, known as a nanodisc (Fig. 1A). Nanodiscs are discoidal lipid bilayers with well-controlled diameters and protein composition and thereby provide a near-physiological yet homogeneous membrane-protein system (60–62).
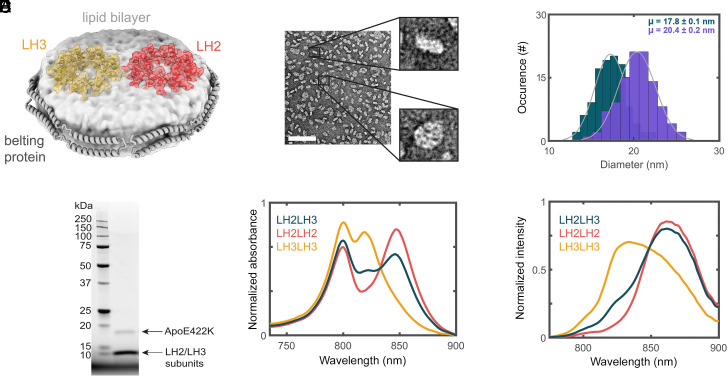
Fig. 1.
Successful production of LH2–LH3 DLDs. (A) Schematic of LH2 (red) and LH3 (orange) embedded in a nanodisc. The discoidal lipid bilayer (light gray) is encircled by the ApoE422K belting protein (gray ribbons). (B) nsTEM image of nanodiscs. The scale bar (white line) is 100 nm. (C) Histogram of nanodisc diameters for small (teal, 17.8 ± 0.1 nm) and large (purple, 20.4 ± 0.2 nm) DLDs. (D) SDS-PAGE of the LH2–LH3 DLDs showing the presence of belting protein along with LH2 and LH3 subunits. (E) Linear absorption and (F) fluorescence spectra of LH2–LH2 DLDs (red), LH3–LH3 DLDs (orange), and LH2–LH3 DLDs (teal). All spectra are normalized by area. The excitation wavelength for the fluorescence data is 532 nm.
By changing the diameter of the nanodisc, we produced interprotein distances across the physiological range (26, 63–65). Cryogenic electronic microscopy (cryoEM) revealed interprotein distances of ∼25, ∼28, and ∼31 Å. Transient absorption (TA) spectroscopy revealed corresponding LH3-to-LH2 energy transfer timescales of 5.7, 9.8, and 14.7 ps. Supporting simulations emphasize that the closely packed protein pairs improve long-distance energy transport, effectively creating a superhighway for energy migration that may underlie the high quantum efficiency. Overall, these results identify the timescales of this critical energy transfer step and establish the utility of nanodiscs as a platform for bottom–up investigation of the photosynthetic antenna network.
Results and Discussion
LH2 Variants in Doubly Loaded Membrane Discs.
To isolate energy transfer between LH2 proteins, we constructed doubly loaded nanodiscs (DLDs) with both LH2 and LH3 incorporated as illustrated in Fig. 1A. Nanodiscs self-assemble into a discoidal membrane encircled by a stabilizing belting protein. The membrane composition and size depend on the ratios of the belting proteins, constituent lipids, and target membrane proteins in the initial mixture. By varying the stoichiometry of the mixture, the nanodiscs were optimized for two LH2 per disc with different diameters. Negative-stain transmission electron microscopy (nsTEM) and cryoEM analysis of the DLDs revealed discoidal structures, confirming nanodisc formation. The structures exhibited apparent diameters of 17.8 ± 0.1 nm and 20.4 ± 0.2 nm for the small and large nanodiscs, respectively (Fig. 1 B and C). Successful incorporation of LH2 into nanodiscs was suggested by SDS-PAGE of the purified DLDs samples showing both LH monomer subunits and the belting protein, ApoE422K (Fig. 1D).
The integrity of the LH2 variants within the nanodiscs was established with steady-state linear absorption and fluorescence spectra (Fig. 1 E and F) for three DLDs samples: LH2 only (LH2–LH2 DLDs), LH3 only (LH3–LH3 DLDs), and the LH2–LH3 mixture (LH2–LH3 DLDs). The overall peak profiles are similar to the proteins in detergent. For LH2–LH2 DLDs, the maximum of the B850 band red-shifted from 846 to 849 nm upon incorporation into the lipid bilayer, consistent with previous reports (13, 14). Similarly, for LH3–LH3 DLDs, the maximum of the B820 band red-shifted from 817 nm to 819 nm upon incorporation into the bilayer. The red tail of the B820 band for LH3–LH3 DLDs is due to a contribution of 5 to 10% LH2 (SI Appendix, Fig. S6), as full conversion has not been achieved in this species (14, 40). For the LH2–LH3 DLDs, incorporation of both LH2 and LH3 into the nanodiscs was confirmed through the linear absorption spectrum of the purified sample, which was decomposed into a ∼50%/50% combination of the spectra of the two variants (Fig. 1E and SI Appendix, Fig. S8).
Finally, commixture of LH2 and LH3 within nanodiscs was established through the fluorescence spectrum of the LH2–LH3 DLDs sample (Fig. 1F). The LH2–LH3 DLDs sample contained a mixture of LH2–LH2 DLDs, LH3–LH3 DLDs, and LH2–LH3 DLDs owing to the stochastic nature of the nanodisc formation reaction. Decomposition of the fluorescence spectrum into those of the two individual variants showed a ∼35 to 40% increase in LH2 content (SI Appendix, Fig. S8), consistent with a contribution from LH3-to-LH2 energy transfer at the level expected for stochastic insertion. At the 0.1 μM concentration of the fluorescence measurement, the presence of interprotein energy transfer strongly implies incorporation of LH2 and LH3 into the same nanodisc.
Cryogenic Electron Microscopy of Doubly Loaded Nanodiscs.
To determine the architecture of the proteins within the DLDs, we used cryoEM. Single-particle reconstruction of the LH2–LH3 DLDs revealed how the proteins arrange in the nanodisc by resolving the structures to a resolution of up to 6.5 Å (SI Appendix, Figs. S3 and S4). As shown in Fig. 2, the formation of the DLDs was clearly observed in the electron density, with the encircling belting protein and two embedded membrane proteins all distinctly visible. Each of the embedded proteins had an apparent diameter of 7.1 nm and an apparent octameric symmetry, consistent with both the crystal structure of LH2 (PDB Code 1LGH) and the homology model of LH3 from Ph. molischianum. These features were used to dock one LH2 and one LH3 into the densities.
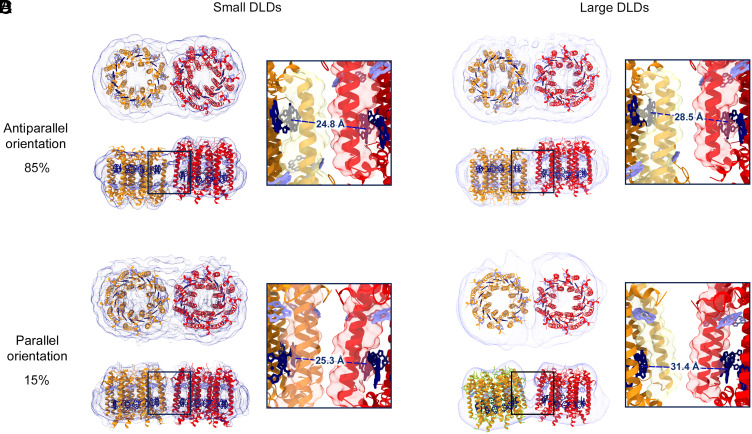
Fig. 2.
Structural organization of LH2 variants in nanodiscs. CryoEM analysis of small (A and B) and large (C and D) LH2–LH3 DLDs showing density maps (dark blue for small nanodiscs and light blue for large nanodiscs) with docked crystal structures (LH3 helices, orange; LH2 helices, red; B800 BChla, light blue; B820/B850 BChla, dark blue). In both small and large DLDs, the antiparallel and the parallel orientations are 85% and 15% of the total population, respectively. The composition of LH2 and LH3 in the DLDs was determined through analysis of the steady-state spectra (SI Appendix, section 1.7 and Table S4). Top views of all nanodisc types illustrate the DLDs assembly with the electron densities of belting proteins encircling the two LH proteins. Side views illustrate that the two LH proteins are at the same approximate vertical position within the lipid bilayer for the parallel orientation (B and D) and are displaced vertically for the antiparallel orientation (A and C). For the small nanodisc, both orientations are closely packed with ∼25 Å between the BChla of the B820 and B850 rings (A and B; black squares). For the large nanodisc, the orientations are less packed, and the antiparallel shows association with ∼28.5 Å between the BChla of the B820 and B850 rings, while in the parallel case, the association brings the two bands ∼31.4 Å apart (C and D; black squares).
Two types of structures emerged from the analysis. One is similar to the native membrane arrangement with both LH complexes in a parallel orientation (with respect to facing the cytosol/periplasm) within their surrounding nanodisc (Fig. 2 B and D). The other showed the two LH proteins in an antiparallel orientation (with respect to facing the cytosol/periplasm) within their surrounding nanodisc (Fig. 2 A and C). Reconstituted systems tend to adopt this antiparallel orientation (30, 31). Despite containing some nonphysiological orientations, these systems have consistently exhibited spectroscopic signatures similar to those observed in the native system and provided many insights into protein–protein interactions (66–68). The antiparallel orientation dominates our samples, accounting for ∼85% of total particles analyzed. The antiparallel orientation also shows a vertical displacement between neighboring proteins of ∼12 Å along the perpendicular axis to the membrane plane (Fig. 2 A and C). As a result of this displacement, the two low-energy chlorophyll rings in the antiparallel orientation remain at nearly identical vertical height as in the parallel orientation. The geometry of the B850 band also means that the relative angles of the transition dipole moments are similar in the two orientations (SI Appendix, Fig. S21A). For the small DLDs, the antiparallel and parallel orientations have similar distance values of 24.8 and 25.3 Å, respectively. The similar values are due to the confinement of the nanodisc, which positions the proteins in van der Waals contact as illustrated by the protein density in Fig. 2 A and B, Inset. For the large DLDs, the antiparallel and parallel orientations have distance values of 28.5 and 31.4 Å, respectively (Fig. 2 C and D). All distance values are consistent with typical LH2–LH2 distances as measured by atomic force microscopy (AFM) (28, 65, 69).
The tendency to adopt the antiparallel orientation suggests that it is energetically favored. Perhaps, the same surface charge distribution of the two proteins disfavors parallel orientation in the membrane due to symmetry-derived repulsion. Consistently, the proteins adopted an angled interaction in the parallel orientation with a tilt of 5° and 15° between the proteins for the small and large DLDs, respectively, both within the range of tilt angles observed for these proteins through other methods (64, 70). The repulsion in the native orientation may be required in vivo for the disassembly and reorganization of LH2 into less densely packed organizations under high-light conditions or to comply with the curvature of the membrane in chromatophores from purple bacteria (70).
Interprotein Energy Transfer between LH2 Variants.
To probe the dynamics of LH3-to-LH2 energy transfer, spectrally resolved TA spectroscopy was performed on the DLDs as shown in Fig. 3A and SI Appendix, Fig. S15. In order to accurately extract the interprotein energy transfer timescale, LH2–LH2 DLDs and LH3–LH3 DLDs were also measured separately (SI Appendix, Figs. S13 and S14). By analysis of the spectral and temporal evolution of the TA data in a self-consistent manner, we resolved the dynamics of each sample and isolated the LH3-to-LH2 energy transfer timescales.
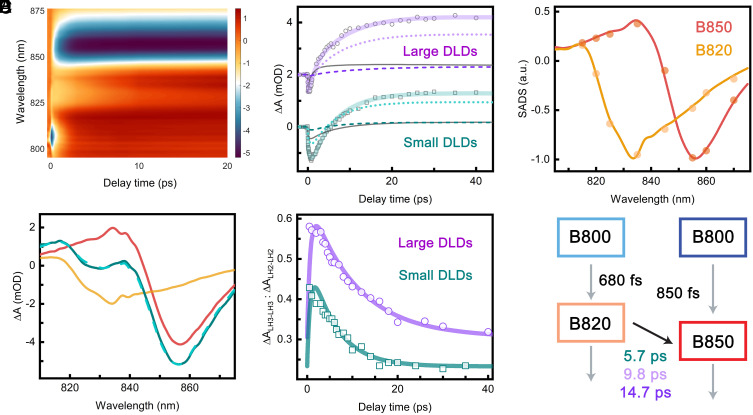
Fig. 3.
LH3-to-LH2 energy transfer dynamics. (A) 2D representation of TA spectra of small LH2–LH3 DLDs. The color bar describes signal intensity (ΔmOD). (B) Absorption transients of small (teal) and large (purple) LH2–LH3 DLDs at 840 nm. Experimental values are shown as open symbols, and the fitted kinetics are shown as solid lines. The large DLDs transient was deconvoluted into antiparallel (light purple dotted line) and parallel (purple dashed line) LH2–LH3 and combined LH2–LH2 and LH3–LH3 (dark gray line) contributions. The transient of small DLDs was deconvoluted into antiparallel (turquoise dotted line) and parallel (dark cyan dashed line) LH2–LH3 and combined LH2–LH2 and LH3–LH3 (dark gray line) contributions. The large DLDs transient is offset by 2 mOD. (C) Normalized SADS of B820 (orange) and B850 (red) components from the TA data of small LH3–LH3 DLDs and LH2–LH2 DLDs, respectively. Results are shown from both simultaneous fitting of the single-wavelength kinetics (dots) and global analysis (lines). (D) Transient absorption spectra of small LH2–LH3 DLDs (teal), LH3–LH3 DLDs (orange), and LH2–LH2 DLDs (red) at 5-ps time delay. The turquoise dashed line is the LH2–LH3 DLDs spectrum constructed from the linear combination of the spectra from LH3 and LH2 DLDs. (E) The population dynamics of excited B820 from the model fitting (solid lines) and the results of fitting the TA spectra (open symbols). The relative contributions of the LH3–LH3 and LH2–LH2 DLDs spectra to the small (teal) and large (purple) LH2–LH3 DLDs spectra are plotted as a function of time. The correspondence between the two indicates that the fitting successfully extracted the kinetics of LH3-to-LH2 energy transfer. (F) Schematic of the model used to fit the LH2–LH3 DLDs with corresponding timescales. The values held fixed from LH2–LH2 DLDs and LH3–LH3 DLDs are shown as gray arrows, and the extracted LH3-to-LH2 energy transfer timescale is shown as a black arrow.
Both individual protein samples, the LH2–LH2 DLDs and LH3–LH3 DLDs, were fit using global analysis, which identifies the characteristic time constants and their associated spectral profiles, known as species-associated-decay-spectra (SADS) (71). The data were best fit with a sequential model in which the system evolves through three components (SI Appendix, Fig. S11A). The first component decayed on a subpicosecond timescale (850 fs for LH2–LH2 DLDs; 680 fs for LH3–LH3 DLDs), and the SADS was dominated by a negative peak at ∼805 nm from the ground state bleach (GSB)/stimulated emission (SE) of the B800 band. Therefore, this component was assigned to B800 to B820/B850 energy transfer. Consistent with this assignment, the SADS associated with the second component were dominated by a negative peak at ∼856 nm for LH2 and ∼836 nm for LH3 from the GSB/SE of the B850 and B820 bands, respectively, as shown in Fig. 3C. A broad positive feature at lower wavelengths (< 842 nm for LH2 and < 818 nm for LH3) was also present due to excited-state absorption (ESA) primarily from the B850/B820 bands. The timescale of the second component was 6.6 to 14 ps for LH2–LH2 DLDs and 8.1 ps for LH3–LH3 DLDs and was assigned to vibrational relaxation. The final component exhibited a similar, but slightly red-shifted, spectral profile and decayed on a ∼1.3-ns timescale, which is the approximate fluorescence lifetime of the LH2 variants. The time constants and their associated spectra are consistent with previous work (13, 14, 23, 44).
The spectra were also fit using a three-step kinetic model in which, after photoexcitation of B800, i) energy transfers from B800 to B850, ii) B850 undergoes vibrational relaxation, and iii) B850 relaxes to the ground state (SI Appendix, Fig. S11). Single wavelength transients were taken at 2.5-nm intervals from 790 to 880 nm and globally fit to the kinetic model. The amplitudes associated with step (ii) of the kinetic model are overlaid on the SADS from global analysis in Fig. 3C. As illustrated, the extracted time constants and spectral profiles were consistent with the components from the global analysis.
For the LH2–LH3 DLDs, LH3-to-LH2 energy transfer is present, and so energy transfer can also be observed in the spectra. As shown in Fig. 3A, the GSB/SE signal centered at ∼835 nm from the B820 band decays with a corresponding rise in the GSB/SE signal centered at ∼855 nm from the B850 band. In order to extract the energy transfer time, the TA data were fit through the single-wavelength kinetics, as the complexity of the data precluded direct fitting via global analysis. Owing to the stochastic nature of the nanodisc self-assembly process, the LH2–LH3 DLDs samples contain a mixture of LH2–LH2 DLDs, LH3–LH3 DLDs, and LH2–LH3 DLDs. Therefore, the signal was fit to a sum of the evolution of the three samples described by kinetic models (SI Appendix, Fig. S11). The intra-LH2 and intra-LH3 time constants and the SADS for all species were held fixed to the values determined through global fitting of the LH2–LH2 DLDs and the LH3–LH3 DLDs, leaving only the LH3-to-LH2 energy transfer time and the relative amplitudes for the samples as free parameters. LH3-to-LH2 energy transfer is most prominent within the spectral regions from 825 to 850 nm where the SADS of B820 and B850 have opposite signs (Fig. 3C). In this region, the negative signal becomes positive due to the decay of the negative B820 and the rise of the positive B850 as illustrated in Fig. 3B. Therefore, transients within this spectral range were simultaneously fit in order to determine the LH3-to-LH2 energy transfer timescales. For the small LH2–LH3 DLDs, a timescale of 5.7 ps was extracted. Applying two LH3-to-LH2 timescales to capture the two orientations gave comparable fitting quality and nearly coincident values, consistent with the similar distances in the cryoEM structures (SI Appendix, Fig. S16). For the large LH2–LH3 DLDs, the two orientations give rise to different distances (Fig. 2 C and D). Consistent with this model, two timescales of 9.8 ps and 14.7 ps were extracted, likely corresponding to the two orientations. The deconvoluted amplitudes of the components are 70%, 13%, and 17% for the antiparallel LH2–LH3, the parallel LH2–LH3, and the LH2–LH2 and LH3–LH3 combined signal in the key wavelength for our analysis (840 nm).
To confirm the LH3-to-LH2 energy transfer timescale was successfully described by fitting to the kinetic model, the TA spectra from the LH2–LH3 DLDs were also decomposed into a linear combination of the TA spectra from the LH2–LH2 and LH3–LH3 DLDs as illustrated in Fig. 3D. The ratios of the contributions of LH3 to the contributions of LH2 were calculated as shown as teal dots in Fig. 3E. The ratio decreased with delay time in the LH2–LH3 DLDs, consistent with the presence of LH3-to-LH2 energy transfer. In contrast, the ratio was nearly constant with delay time in a control sample of a simple mixture of LH2 and LH3 (SI Appendix, Figs. S17–S19). The ratio from the LH2–LH3 DLDs was overlaid with the population dynamics of B820 from the fitting results for the TA spectra of LH2–LH3 DLDs. The evolution from both approaches showed good agreement, supporting the extracted timescales for energy transfer within the LH2–LH3 DLDs.
Calculations of Energy Transfer Timescale.
The timescales of LH3-to-LH2 energy transfer were also calculated using generalized Förster theory as shown in Fig. 4A. Typically, B850-to-B850 energy transfer dominates in vivo, and so we focus on energy transfer between the low-energy rings (B820 to B850). For the small DLDs, the ∼25 Å distance extracted from the cryoEM structures had a theoretical timescale of 6.5 ps (SI Appendix), similar to the experimental value of 5.7 ps. For the large DLDs, the ∼28.5 and ∼31.4 Å distances extracted from the cryoEM structures had theoretical timescales of 11.3 and 18.1 ps, respectively (SI Appendix, Fig. S24). These were again similar to the experimental values of 9.8 and 14.7 ps, respectively. Generalized Förster theory describes the intermediate regime appropriate for photosynthetic light-harvesting apparatuses, i.e., the interbacteriochlorophyll distances give rise to interactions between the far-field and near-field limits (SI Appendix) (72–75). The good agreement between experiment and theory also illustrates that, even for interprotein energy transfer, the far-field regime breaks down due to the nanoscale distances involved.
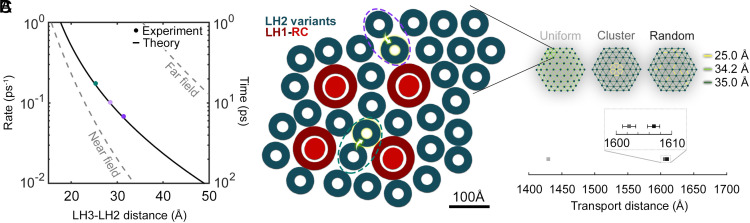
Fig. 4.
LH3-to-LH2 energy transfer within the photosynthetic membrane. (A) The rate (left axis) and timescale (right axis) for interprotein energy transfer across the biologically relevant range as a function of the LH3-to-LH2 distance, which was defined as the separation between the nearest B820 and B850 BChla. Energy transfer rates in the far-field (dashed gray line, Top) and near-field (dashed gray line, Bottom) regimes are also plotted. The experimentally measured LH3-to-LH2 distances and energy transfer timescales for the small and large DLDs are indicated in teal and two shades of purple dots, respectively. The theoretical generalized FRET rate is indicated by the solid line (65, 69). (B) Schematic representation of the Ph. molischianum photosynthetic membrane (26). Dotted ovals highlight LH2 pairs expected to exhibit faster and slower energy transfer (teal and purple, respectively). (C) The energy transport distance was simulated using an LH2 network with hexagonal packing for 156 interprotein distances. Three organizations with the same average spacings (Top) were compared: uniform spacing (Left), tightly interacting cluster (Center), and random tightly interacting pairs (Right). Total transport distances (Bottom) were calculated for all three organizations as the distance traveled after 1 ns, which is approximately the lifetime of LH2.
The theoretical energy transfer timescales were also calculated for LH3-to-LH3 and LH2-to-LH2 energy transfer (SI Appendix, Fig. S24). Overall, similar timescales and distance scaling were observed for the three pairs of variants, confirming that LH3-to-LH2 energy transfer captures the behaviors associated with energy transfer among LH2 variants in the membrane. The similar timescales also suggest that LH3-to-LH2 conversion does not interfere with robust energy transfer, enabling spectral broadening without a corresponding cost in energy transport efficiency. That is, these timescales prevent LH2 traps for transport between LH3 or LH3 hills for transport between LH2. In fact, the introduction of LH3 actually leads to slightly faster interprotein energy transfer in vivo. At thermal equilibrium, the B800 population is larger for LH3 than for LH2 due to the smaller energy gap between the two bands, increasing the interprotein energy transfer via the B800 band from 10% to 14% (SI Appendix, Fig. S21). Overall, for all protein pairs, the predominant energy transfer pathway is via the lower energy bands, leading to similar behavior (SI Appendix, Table S8). Thus, the conversion to LH3 under low light not only helps with light harvesting through spectral broadening but also maintains efficient energy transport.
Energy Transfer in the Heterogeneous Membrane.
AFM images of native membranes from purple bacteria revealed a range of distances within the heterogeneous protein network (26, 51–54). The most common, and closest possible, distance between LH2 was 25 Å, which was replicated in the small DLDs and gave rise to a 5.7-ps timescale of LH3-to-LH2 energy transfer. Distances up to 50 Å were also present with decreasing probability. Other fairly common distances were ∼30 Å, which were replicated in the large DLDs and gave rise to 10- to 14-ps timescales of LH3-to-LH2 energy transfer.
Previous indirect measurements of LH2-to-LH2 energy transfer in these native membranes found 3–5-ps timescales. Consistently, measurements of LH2-to-LH1 energy transfer, which has a similar energy gap as LH3-to-LH2 transfer, found 5- to 20-ps timescales (15, 56–59, 76). Remarkably, the measurements on native membranes recovered values similar to the closely spaced protein pairs despite averaging over the full distributions of distances. The similarity may in part arise from the ability of the fastest component to dictate the rise of a spectroscopic signal, obfuscating slower dynamics. Furthermore, energy transfer will occur preferentially to the closest of multiple neighboring LH2, and so energy transfer between these proteins can also dominate the signal. These energy transfer steps between closely spaced proteins, however, describe local equilibration, and such local effects do not capture the dynamics of long-distance transport (77). Instead, energy transfer timescales between proteins separated by multiple distances are needed for such descriptions.
To investigate the role of the different energy transfer timescales in long-distance energy transport, total energy transport distances were simulated. The LH2 variants were positioned in a hexagonal organization, which is a well-established model for the arrangement of LH2 in the bacterial membrane (Fig. 4 B and C) (78–80). The total transport distance within the one nanosecond lifetime of LH2 was calculated for three different membrane arrangements, as shown in Fig. 4C: i) uniform organization with equivalent interprotein distances; ii) protein cluster with 12 short distances and 144 long distances within and without the cluster, respectively; and iii) random short distances within a lattice of long distances. Of note, 25 and 35 Å were selected as the short and long distances, respectively, because both correspond to the distances found experimentally in the DLDs and because they are both observed in native membranes, with 25 Å the most common and closest possible distance and 35 Å a longer distance (65, 69). The distance in the uniform organization is the weighted average of the short and long distances in the other two organizations.
As shown in Fig. 4C and SI Appendix, Table S9, the simulations revealed a ∼15% increase in transport distances for the heterogeneous arrangements (ii, iii) over the uniform arrangement (i). The overall similarity in transport distance between the cluster arrangement (ii) and the random arrangement (iii) suggests that enhancement of energy migration occurs without ordered positioning of membrane complexes. Fundamentally, the increase in transport distance comes from the nonlinear relationship between energy transfer time and distance. That is, a small decrease in spacing can lead to a many-fold speed-up, and so it is better to have a few short distances than a uniform spread of longer distances. Based on this enhancement, the closely spaced protein pairs facilitate long-distance migration by extending the transport distances through the creation of a “superhighway” for photonic energy through the protein network.
Conclusion
In summary, we report the reconstitution of nanoconfined pairs of LH2 and the low-light variant LH3 from Ph. molischianum and their structural and spectroscopic characterization. Our results establish a method to reconstitute heterogeneous antennae, a long-standing challenge in the field. Overall, nanodiscs, ultrafast TA spectroscopy, and cryoEM together enable investigations of interprotein energy transfer in photosynthetic light harvesting.
Through this combination, we measured distance-dependent interprotein energy transfer timescales from ∼5 to 15 ps between protein pairs. Simulations and comparison to previous measurements suggest that interprotein energy transfer is dominated by these closely spaced protein pairs, and their presence may be required for long-distance energy transport. The ability of these protein pairs to dominate interprotein energy transfer may also be a mechanism by which similar timescales are maintained despite heterogeneous protein organizations, allowing the light-harvesting dynamics to be robust to the variations in antenna protein expression and organization by which the light-harvesting machinery responds to the fluctuating conditions of natural environments.
Materials and Methods
Sample Preparation.
The LH2 and LH3 proteins were isolated from the purple nonsulfur bacteria Phaeospirillum molischianum and were stored in a Tris buffer containing 20 mM Tris HCl and 0.1% n-dodecyl-β-D-maltoside (DDM) with pH 8.0. The belting protein ApoE422K used for double-loaded nanodiscs was expressed and purified with the standard protocols as described in SI Appendix. To produce double-loaded nanodiscs, ApoE422K, LH2+LH3, and DMPC (Avanti Polar Lipids) were mixed together in the molar ratio of 1:0.25:22.2 for small DLDs and 1:0.25:30 for large DLDs. Bio-Beads SM-2 Resin (Bio-Rad Laboratories) was added to remove detergent. The nanodiscs were purified by size-exclusion chromatography with a Superdex 200 Increase 10/300 GL column (Cytiva).
CryoEM Procedures and Structural Determination.
To glow-discharged, gold standard R 1.2/1.3 holey carbon grids 300 mesh (Quantifoil), 3 μL drops of LH2–LH3 DLDs at a concentration range of 0.75 to 1 mg/mL were applied. Vitrification was performed using a Vitrobot Mark IV (Thermo Fisher/FEI). Data were collected on a Titan Krios transmission electron microscope (Thermo Fisher/FEI). Electron images were recorded using a Falcon III direct electron detect camera (FEI). Data processing was carried out using Relion 3.0 suite (81). Alignment of frames was done using Relion implementation of MotionCorr. Particle picking was done using LoG, which led to 2D/3D classifications followed by 3D refinements. Final models were docked into cryoEM maps as rigid bodies in ChimeraX (82). More experimental details can be found in SI Appendix.
TA Spectroscopy.
Details of the TA apparatus are provided in SI Appendix. In brief, all the protein samples were pumped at 800 nm with a femtosecond Ti:Sapphire laser (Coherent Libra). The pump pulse energy was attenuated to 10 nJ/pulse before being focused on the DLDs samples in a flow cuvette. The sample was flowed using a peristaltic pump and stored on ice during data acquisition. The white-light continuum probe pulse was generated by focusing the 800-nm pulses through a tube of argon gas. The polarization between the pump and the probe pulses was set to be the magic angle (54.7°) with ∼110 fs instrument response. The probe was detected with a CCD array (AViiVA EM2) on a shot-to-shot basis for data acquisition and analysis.
Theoretical Calculations.
Frenkel exciton models were employed to describe the excitonic structure of LH2/LH3. On-site energies and short-range couplings were based on ref. 83 and adjusted to better fit the absorption and emission spectra. Long-range (second neighbors and farther) couplings were calculated using transition monopole approximation (84, 85). Transition monopoles (charges) were calculated using the ZINDO method implemented in Gaussian (86). Near-field effect in energy transfer was accounted for using Generalized FRET formalism (87, 88). Details of the theoretical calculation are described in SI Appendix.
Energy Transport Simulations.
A hexagonal lattice model was used to simulate excitation energy transfer dynamics between LH2 complexes. The hopping probability was proportional to the energy transfer rate (ps-1) calculated with Förster theory. The exciton transport distance was calculated as the distance within a 1-ns lifetime. The exciton transport distance was averaged for 30,000 simulation trajectories for each arrangement.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This work was primarily supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, Division of Chemical Sciences, Geosciences, and Biosciences under Award DE-SC0018097 to G.S.S.-C. Y.M. also acknowledges the US Department of Energy, Office of Science, Office of Basic Energy Sciences, Division of Chemical Sciences, Geosciences, and Biosciences under Award DE-SC0022956. J.C. would like to acknowledge the NSF (CHE 1800301 and CHE 1836913). J.N.S. has received funding for this research from Excellence Initiative of Aix-Marseille University—A*MIDEX, a French “Investissements d’Avenir” program. O.C.F. gratefully acknowledges support from an NSF Graduate Research Fellowship. D.H. would like to acknowledge the Yad Hanadiv (Rothschild) Foundation, the Zuckerman STEM Leadership Program, and the Israel Council for Higher Education (CHE) for their generous financial support. We thank Moungi Bawendi for help with the near-infrared fluorescence measurements. This work made use of the MRSEC Shared Experimental Facilities at MIT, supported by the NSF under award number DMR-1419807. This work was carried out in part through the use of the cryoEM facility at MIT.nano. A previous version of this work originally appeared in the Ph.D. thesis of O.C.F.
Author contributions
D.W., O.C.F., D.H., A.L.T., J.I.O., J.C., J.N.S., Y.M., and G.S.S.-C. designed research; D.W., O.C.F., D.H., H.T., Y.J., C.C., M.N., A.L.T., J.I.O., S.M.H., and Y.M. performed research; Y.J., M.N., and J.N.S. contributed new reagents/analytic tools; D.W., O.C.F., D.H., H.T., M.N., and Y.M. analyzed data; and D.W., O.C.F., D.H., and G.S.S.-C. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission. G.E. is a guest editor invited by the Editorial Board.
Data, Materials, and Software Availability
Atomic coordinates and the cryoEM density maps have been deposited in the Protein Data Bank (https://www.rcsb.org) and the Electron Microscopy Data Bank (https://www.ebi.ac.uk/emdb/) under the following access codes: 7TV3 and EMD-2613 (small DLDs, parallel orientation), 7TUW and EMD-26134 (small DLDs, antiparallel orientation), 8FBB and EMD-28963 (large DLDs, parallel orientation), and 8FB9 and EMD-28962 (large DLDs, antiparallel orientation). All other data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.van Grondelle R., Dekker J. P., Gillbro T., Sundström V., Energy transfer and trapping in photosynthesis. Biochim. Biophys. Acta 1187, 1–65 (1994). [Google Scholar]
- 2.Pullerits T., Sündstrom V., Photosynthetic light-harvesting pigment–protein complexes: Toward understanding how and why. Acc. Chem. Res. 29, 381–389 (1996). [Google Scholar]
- 3.Fleming G. R., van Grondelle R., Femtosecond spectroscopy of photosynthetic light-harvesting systems. Curr. Opin. Struct. Biol 7, 738–748 (1997). [DOI] [PubMed] [Google Scholar]
- 4.Mirkovic T., et al. , Light absorption and energy transfer in the antenna complexes of photosynthetic organisms. Chem. Rev. 117, 249–293 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Hu X., Ritz T., Damjanović A., Schulten K., Pigment organization and transfer of electronic excitation in the photosynthetic unit of purple bacteria. J. Phys. Chem. B 101, 3854–3871 (1997). [Google Scholar]
- 6.Melis A., Solar energy conversion efficiencies in photosynthesis: Minimizing the chlorophyll antennae to maximize efficiency. Plant Sci. 117, 272–280 (2009). [Google Scholar]
- 7.Blankenship R. E., et al. , Comparing photosynthetic and photovoltaic efficiencies and recognizing the potential for improvement. Science 332, 805–809 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Perrine Z., Negi S., Sayre R., Optimization of photosynthetic light energy utilization by microalgae. Algal Res. 1, 134–142 (2012). [Google Scholar]
- 9.Novoderezhkin V., Wendling M., van Grondelle R., Intra- and interband transfers in the B800–B850 antenna of Rhodospirillum molischianum: Redfield theory modeling of polarized pump-probe kinetics. J. Phys. Chem. B 107, 11534–11548 (2003). [Google Scholar]
- 10.Sundström V., Pullerits T., van Grondelle R., Photosynthetic light-harvesting: Reconciling dynamics and structure of purple bacterial LH2 reveals function of photosynthetic unit. J. Phys. Chem. B 103, 2327–2346 (1999). [Google Scholar]
- 11.Monshouwer R., van Grondelle R., Excitations and excitons in bacterial light-harvesting complexes. Biochim. Biophys. Acta 1275, 70–75 (1996). [DOI] [PubMed] [Google Scholar]
- 12.Hess S., Akesson E., Cogdell R. J., Pullerits T., Sundström V., Energy transfer in spectrally inhomogeneous light-harvesting pigment–protein complexes of purple bacteria. Biophys. J. 69, 2211 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ogren J. I., et al. , Impact of the lipid bilayer on energy transfer kinetics in the photosynthetic protein LH2. Chem. Sci. 9, 3095–3104 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tong A. L., et al. , Comparison of the energy transfer rates in structural and spectral variants of the B800–850 complex from purple bacteria. J. Phys. Chem. B 124, 1460–1469 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Hess S., et al. , Temporally and spectrally resolved subpicosecond energy transfer within the peripheral antenna complex (LH2) and from LH2 to the core antenna complex in photosynthetic purple bacteria. Proc. Natl. Acad. Sci. U.S.A. 92, 12333–12337 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hess S., et al. , Femtosecond energy transfer within the LH2 peripheral antenna of the photosynthetic purple bacteria Rhodobacter sphaeroides and Rhodopseudomonas palustris LL. Chem. Phys. Lett. 216, 247–257 (1993). [Google Scholar]
- 17.Fidler A. F., Singh V. P., Long P. D., Dahlberg P. D., Engel G. S., Probing energy transfer events in the light harvesting complex 2 (LH2) of Rhodobacter sphaeroides with two-dimensional spectroscopy. J. Chem. Phys. 139, 155101 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jimenez R., Dikshit S. N., Bradforth S. E., Fleming G. R., Electronic excitation transfer in the LH2 complex of Rhodobacter sphaeroides. J. Phys. Chem. 100, 6825–6834 (1996). [Google Scholar]
- 19.Herek J. L., et al. , B800→B850 energy transfer mechanism in bacterial LH2 complexes investigated by B800 pigment exchange. Biophys. J. 78, 2590–2596 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pullerits T., Hess S., Herek S. J. L., Sundström V., Temperature dependence of excitation transfer in LH2 of Rhodobacter sphaeroides. J. Phys. Chem. B 101, 10560–10567 (1997). [Google Scholar]
- 21.Joo T., Jia Y., Yu J.-Y., Jonas D. M., Fleming G. R., Dynamics in isolated bacterial light harvesting antenna (LH2) of Rhodobacter sphaeroides at room temperature. J. Phys. Chem. 100, 2399–2409 (1996). [Google Scholar]
- 22.Kennis J. T. M., et al. , Femtosecond dynamics in isolated LH2 complexes of various species of purple bacteria. J. Phys. Chem. B 101, 7827–7834 (1997). [Google Scholar]
- 23.Ihalainen J. A., et al. , Energy transfer in LH2 of Rhodospirillum molischianum, studied by subpicosecond spectroscopy and configuration interaction exciton calculations. J. Phys. Chem. B 105, 9849–9856 (2001). [Google Scholar]
- 24.Pullerits T., Chachisvilis M., Sundström V., Exciton delocalization length in the B850 antenna of Rhodobacter sphaeroides. J. Phys. Chem. 100, 10787–10792 (1996). [Google Scholar]
- 25.Schlau-Cohen G. S., Wang Q., Southall J., Cogdell R. J., Moerner W. E., Single-molecule spectroscopy reveals photosynthetic LH2 complexes switch between emissive states. Proc. Natl. Acad. Sci. U.S.A. 110, 10899–10903 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scheuring S., Sturgis J. N., Chromatic adaptation of photosynthetic membranes. Science 309, 484–487 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Scheuring S., et al. , Nanodissection and high-resolution imaging of the Rhodopseudomonas viridis photosynthetic core complex in native membranes by AFM. Proc. Natl. Acad. Sci. U.S.A. 100, 1690–1693 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scheuring S., Rigaud J.-L., Sturgis J. N., Variable LH2 stoichiometry and core clustering in native membranes of Rhodospirillum photometricum. EMBO J. 23, 4127–4133 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frese R. N., et al. , The long-range organization of a native photosynthetic membrane. Proc. Natl. Acad. Sci. U.S.A. 101, 17994–17999 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gonçalves R. P., Busselez J., Lévy D., Seguin J., Scheuring S., Membrane insertion of Rhodopseudomonas acidophila light harvesting complex 2 investigated by high resolution AFM. J. Struct. Biol. 149, 79–86 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Stamouli A., et al. , The ring structure and organization of light harvesting 2 complexes in a reconstituted lipid bilayer, resolved by atomic force microscopy. Biophys. J. 84, 2483–2491 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cogdell R. J., et al. , The structural basis of light-harvesting in purple bacteria. FEBS Lett. 555, 35–39 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Bahatyrova S., et al. , The native architecture of a photosynthetic membrane. Nature 430, 1058–1062 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Cogdell R. J., et al. , The structure and function of the LH2 (B800–850) complex from the purple photosynthetic bacterium Rhodopseudomonas acidophila strain 10050. Prog. Biophys. Mol. Biol. 68, 1–27 (1997). [DOI] [PubMed] [Google Scholar]
- 35.Cogdell R. J., Gall A., Köhler J., The architecture and function of the light-harvesting apparatus of purple bacteria: From single molecules to in vivo membranes. Q. Rev. Biophys. 39, 227–324 (2006). [DOI] [PubMed] [Google Scholar]
- 36.McLuskey K., Prince S. M., Cogdell R. J., Isaacs N. W., The crystallographic structure of the B800–820 LH3 light-harvesting complex from the purple bacteria Rhodopseudomonas acidophila strain 7050. Biochemistry 40, 8783–8789 (2001). [DOI] [PubMed] [Google Scholar]
- 37.Koepke J., Hu X., Muenke C., Schulten K., Michel H., The crystal structure of the light-harvesting complex II (B800–850) from Rhodospirillum molischianum. Structure 4, 581–597 (1996). [DOI] [PubMed] [Google Scholar]
- 38.Papiz M. Z., Prince S. M., Howard T., Cogdell R. J., Isaacs N. W., The structure and thermal motion of the B800–850 LH2 complex from Rps. acidophila at 2.0 Å resolution and 100 K: New structural features and functionally relevant motions. J. Mol. Biol. 326, 1523–1538 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Gardiner A. T., Cogdell R. J., Takaichi S., The effect of growth conditions on the light-harvesting apparatus in Rhodopseudomonas acidophila. Photosynth. Res. 38, 159–167 (1993). [DOI] [PubMed] [Google Scholar]
- 40.Mascle-Allemand C., Duquesne K., Lebrun R., Scheuring S., Sturgis J. N., Antenna mixing in photosynthetic membranes from Phaeospirillum molischianum. Proc. Natl. Acad. Sci. U.S.A. 107, 5357–5362 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chmeliov J., et al. , Excitons in the LH3 complexes from purple bacteria. J. Phys. Chem. B 117, 11058–11068 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Yoneda Y., et al. , Ultrafast photodynamics and quantitative evaluation of biohybrid photosynthetic antenna and reaction center complexes generating photocurrent. J. Phys. Chem. C 124, 8605–8615 (2020). [Google Scholar]
- 43.Ma F., Yu L.-J., Wang-Otomo Z.-Y., van Grondelle R., Temperature dependent LH1→RC energy transfer in purple bacteria Tch. tepidum with shiftable LH1-Qy band: A natural system to investigate thermally activated energy transfer in photosynthesis. Biochim. Biophys. Acta 1857, 408–414 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Markus M., et al. , Low-intensity pump-probe measurements on the B800 band of Rhodospirillum molischianum. Biophys. J. 84, 440–449 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scholes G. D., Fleming G. R., On the mechanism of light harvesting in photosynthetic purple bacteria: B800 to B850 energy transfer. J. Phys. Chem. B 104, 1854–1868 (2000). [Google Scholar]
- 46.Saga Y., et al. , Excitation energy transfer from bacteriochlorophyll b in the B800 site to B850 bacteriochlorophyll a in light-harvesting complex 2. J. Phys. Chem. B 125, 2009–2017 (2021). [DOI] [PubMed] [Google Scholar]
- 47.Zigmantas D., et al. , Two-dimensional electronic spectroscopy of the B800–B820 light-harvesting complex. Proc. Natl. Acad. Sci. U.S.A. 103, 12672–12677 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim J., et al. , Vibrational modes promoting exciton relaxation in the B850 band of LH2. J. Phys. Chem. Lett. 13, 1099–1106 (2022). [DOI] [PubMed] [Google Scholar]
- 49.Ma F., et al. , Excitation dynamics of two spectral forms of the core complexes from photosynthetic bacterium Thermochromatium tepidum. Biophys. J. 95, 3349–3357 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schlau-Cohen G. S., De Re E., Cogdell R. J., Fleming G. R., Determination of excited-state energies and dynamics in the B band of the bacterial reaction center with 2D electronic spectroscopy. J. Phys. Chem. Lett. 3, 2487–2492 (2012). [DOI] [PubMed] [Google Scholar]
- 51.Sturgis J. N., Niederman R. A., Atomic force microscopy reveals multiple patterns of antenna organization in purple bacteria: Implications for energy transduction mechanisms and membrane modeling. Photosynth. Res. 95, 269–278 (2008). [DOI] [PubMed] [Google Scholar]
- 52.Liu L. N., Sturgis J. N., Scheuring S., Native architecture of the photosynthetic membrane from Rhodobacter veldkampii. J. Struct. Biol. 173, 138–145 (2011). [DOI] [PubMed] [Google Scholar]
- 53.Adams P. G., Hunter C. N., Adaptation of intracytoplasmic membranes to altered light intensity in Rhodobacter sphaeroides. Biochim. Biophys. Acta 1817, 1616–1627 (2012). [DOI] [PubMed] [Google Scholar]
- 54.Mullineaux C. W., Liu L. N., Membrane dynamics in phototrophic bacteria. Annu. Rev. Microbiol. 74, 633–654 (2020). [DOI] [PubMed] [Google Scholar]
- 55.Zhang F. G., van Grondelle R., Sundström V., Pathways of energy flow through the light-harvesting antenna of the photosynthetic purple bacterium Rhodobacter sphaeroides. Biophys. J. 61, 911–920 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nagarajan V., Parson W. W., Excitation energy transfer between the B850 and B875 antenna complexes of Rhodobacter sphaeroides. Biochemistry 36, 2300–2306 (1997). [DOI] [PubMed] [Google Scholar]
- 57.Lüer L., et al. , Tracking energy transfer between light harvesting complex 2 and 1 in photosynthetic membranes grown under high and low illumination. Proc. Natl. Acad. Sci. U.S.A. 109, 1473–1478 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Agarwal R., et al. , Nature of disorder and inter-complex energy transfer in LH2 at room temperature: A three pulse photon echo peak shift study. J. Phys. Chem. A 106, 7573–7578 (2002). [Google Scholar]
- 59.Dahlberg P. D., et al. , Mapping the ultrafast flow of harvested solar energy in living photosynthetic cells. Nat. Commun. 8, 1–7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Denisov I. G., Sligar S. G., Nanodiscs for structural and functional studies of membrane proteins. Nat. Struct. Mol. Biol. 23, 481–486 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Denisov I. G., Sligar S. G., Nanodiscs in membrane biochemistry and biophysics. Chem. Rev. 117, 4669–4713 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hartley M. D., Schneggenburger P. E., Imperiali B., Lipid bilayer nanodisc platform for investigating polyprenol-dependent enzyme interactions and activities. Proc. Natl. Acad. Sci. U.S.A. 110, 20863–20870 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sturgis J. N., Tucker J. D., Olsen J. D., Hunter C. N., Niederman R. A., Atomic force microscopy studies of native photosynthetic membranes. Biochemistry 48, 3679–3698 (2009). [DOI] [PubMed] [Google Scholar]
- 64.Olsen J. D., et al. , The organization of LH2 complexes in membranes from Rhodobacter sphaeroides. J. Biol. Chem. 283, 30772–30779 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gonçalves R. P., Bernadac A., Sturgis J. N., Scheuring S., Architecture of the native photosynthetic apparatus of Phaeospirillum molischianum. J. Struct. Biol. 152, 221–228 (2005). [DOI] [PubMed] [Google Scholar]
- 66.Ruban A., et al. , Identification of a mechanism of photoprotective energy dissipation in higher plants. Nature 450, 575–578 (2007). [DOI] [PubMed] [Google Scholar]
- 67.Wilk L., Grunwald M., Liao P. N., Walla P. J., Kühlbrandt W., Direct interaction of the major light-harvesting complex II and PsbS in nonphotochemical quenching. Proc. Natl. Acad. Sci. U.S.A. 110, 5452–5456 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Son M., Moya R., Pinnola A., Bassi R., Schlau-Cohen G. S., Protein–protein interactions induce pH-dependent and zeaxanthin-independent photoprotection in the plant light-harvesting complex, LHCII. J. Am. Chem. Soc. 143, 17577–17586 (2021). [DOI] [PubMed] [Google Scholar]
- 69.Scheuring S., et al. , Watching the photosynthetic apparatus in native membranes. Proc. Natl. Acad. Sci. U.S.A. 101, 11293–11297 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chandler D., Hsin J., Harrison C., Gumbart J., Schulten K., Intrinsic curvature properties of photosynthetic proteins in chromatophores. Biophys. J. 95, 2822–2836 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van Stokkum I. H., Larsen D. S., van Grondelle R., Global and target analysis of time-resolved spectra. Biochim. Biophys. Acta 1657, 82–104 (2004). [DOI] [PubMed] [Google Scholar]
- 72.Mukai K., Abe S., Sumi H., Theory of rapid excitation-energy transfer from B800 to optically-forbidden exciton states of B850 in the antenna system LH2 of photosynthetic purple bacteria. J. Phys. Chem. B 103, 6096–6102 (1999). [Google Scholar]
- 73.Sumi H., Theory on rates of excitation-energy transfer between molecular aggregates through distributed transition dipoles with application to the antenna system in bacterial photosynthesis. J. Phys. Chem. B 103, 252–260 (1999). [Google Scholar]
- 74.Cleary L., Cao J., Optimal thermal bath for robust excitation energy transfer in disordered light-harvesting complex 2 of purple bacteria. New J. Phys. 15, 125030 (2013). [Google Scholar]
- 75.Ma J., Cao J., Förster resonance energy transfer, absorption and emission spectra in multichromophoric systems. I. Full cumulant expansions and system-bath entanglement. J. Chem. Phys. 142, 094106 (2015). [DOI] [PubMed] [Google Scholar]
- 76.Niedzwiedzki D. M., Gardiner A. T., Blankenship R. E., Cogdell R. J., Energy transfer in purple bacterial photosynthetic units from cells grown in various light intensities. Photosynth. Res. 137, 389–402 (2018). [DOI] [PubMed] [Google Scholar]
- 77.Akselrod G. M., et al. , Subdiffusive exciton transport in quantum dot solids. Nano Lett. 14, 3556–3562 (2014). [DOI] [PubMed] [Google Scholar]
- 78.Pflock T. J., et al. , The electronically excited states of LH2 complexes from Rhodopseudomonas acidophila strain 10050 studied by time-resolved spectroscopy and dynamic Monte Carlo simulations. II. Homo-arrays of LH2 complexes reconstituted into phospholipid model membranes. J. Phys. Chem. B 115, 8821–8831 (2011). [DOI] [PubMed] [Google Scholar]
- 79.Onizhuk M., Sohoni S., Galli G., Engel G. S., Spatial patterns of light-harvesting antenna complex arrangements tune the transfer-to-trap efficiency of excitons in purple bacteria. J. Phys. Chem. Lett. 12, 6967–6973 (2021). [DOI] [PubMed] [Google Scholar]
- 80.Cleary L., Chen H., Chern C., Silbey R. J., Cao J., Optimal fold symmetry of LH2 rings on a photosynthetic membrane. Proc. Natl. Acad. Sci. U.S.A. 110, 8537–8542 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zivanov J., et al. , New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Goddard T. D., et al. , UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chmeliov J., et al. , Excitons in the LH3 complexes from purple bacteria. J. Phys. Chem. B 117, 11058–11068 (2013). [DOI] [PubMed] [Google Scholar]
- 84.Sauer K., Philipson K. D., Tsai S. C., Circular dichroism of chlorophyll and related molecules calculated using a point monopole model for the electronic transitions. J. Phys. Chem. 75, 1440–1445 (1971). [Google Scholar]
- 85.Chang J. C., Monopole effects on electronic excitation interactions between large molecules. I. Application to energy transfer in chlorophylls. J. Chem. Phys. 67, 3901–3909 (1977). [Google Scholar]
- 86.Frisch M. J., et al. , Gaussian∼16 Revision C.01 (Gaussian Inc., Wallingford, CT, 2016). [Google Scholar]
- 87.Cleary L., Cao J., Optimal thermal bath for robust excitation energy transfer in disordered light-harvesting complex 2 of purple bacteria. New J. Phys. 15, 125030 (2013). [Google Scholar]
- 88.Ma J., Cao J., Förster resonance energy transfer, absorption and emission spectra in multichromophoric systems. I. Full cumulant expansions and system-bath entanglement. J. Chem. Phys. 142, 094106 (2015). [DOI] [PubMed] [Google Scholar]
- 89.O. C. Fiebig, “Understanding ultrafast energy transfer across the photosynthetic membrane of purple bacteria with near-native systems,” PhD thesis, Massachusetts Institute of Technology, Cambridge, MA (2022).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
Atomic coordinates and the cryoEM density maps have been deposited in the Protein Data Bank (https://www.rcsb.org) and the Electron Microscopy Data Bank (https://www.ebi.ac.uk/emdb/) under the following access codes: 7TV3 and EMD-2613 (small DLDs, parallel orientation), 7TUW and EMD-26134 (small DLDs, antiparallel orientation), 8FBB and EMD-28963 (large DLDs, parallel orientation), and 8FB9 and EMD-28962 (large DLDs, antiparallel orientation). All other data are included in the article and/or SI Appendix.