Significance
Jasmonic acid (JA) plays an important role in plant defense against insects, pathogenic fungi and viruses mediated by cell-surface pattern recognition receptors, and intracellular nucleotide-binding leucine-rich repeat (NLR) class immune receptors. In this study, we uncovered a previously unknown role for MED10b and MED7 of the Mediator complex in the jasmonate-dependent transcription response, and coiled-coil NLRs (CNLs) in Solanaceae modulate MED10b/MED7 to activate immunity. MED10b, MED7, and transcription repressor JAZs interact with each other to repress the expression of jasmonate-dependent defense genes. Whereas, CC domains of Sw-5b and various other CNLs from Solanaceae interfere with the interaction between MED10b and MED7 thereby derepressing the repressor activity of MED10b–MED7–JAZ to activate immunity.
Keywords: Mediator complex, NLRs, Sw-5b, effector-triggered immunity
Abstract
Plant intracellular nucleotide-binding domain, leucine-rich repeat-containing receptors (NLRs) activate a robust immune response upon detection of pathogen effectors. How NLRs induce downstream immune defense genes remains poorly understood. The Mediator complex plays a central role in transducing signals from gene-specific transcription factors to the transcription machinery for gene transcription/activation. In this study, we demonstrate that MED10b and MED7 of the Mediator complex mediate jasmonate-dependent transcription repression, and coiled-coil NLRs (CNLs) in Solanaceae modulate MED10b/MED7 to activate immunity. Using the tomato CNL Sw-5b, which confers resistance to tospovirus, as a model, we found that the CC domain of Sw-5b directly interacts with MED10b. Knockout/down of MED10b and other subunits including MED7 of the middle module of Mediator activates plant defense against tospovirus. MED10b was found to directly interact with MED7, and MED7 directly interacts with JAZ proteins, which function as transcriptional repressors of jasmonic acid (JA) signaling. MED10b–MED7–JAZ together can strongly repress the expression of JA-responsive genes. The activated Sw-5b CC interferes with the interaction between MED10b and MED7, leading to the activation of JA-dependent defense signaling against tospovirus. Furthermore, we found that CC domains of various other CNLs including helper NLR NRCs from Solanaceae modulate MED10b/MED7 to activate defense against different pathogens. Together, our findings reveal that MED10b/MED7 serve as a previously unknown repressor of jasmonate-dependent transcription repression and are modulated by diverse CNLs in Solanaceae to activate the JA-specific defense pathways.
Plants use two tiers of innate immune system known as the pattern-triggered immunity (PTI) and the effector-triggered immunity (ETI) to defend against microbial pathogens (1, 2). The PTI is initiated by cell-surface pattern recognition receptors upon recognition of microbe-associated molecular patterns (3–5), whereas the ETI is initiated by intracellular nucleotide-binding leucine-rich repeat-containing receptors (NLRs) upon recognition of pathogen effectors (1, 2, 6, 7). The PTI-induced plant defense is often mild and transient, whereas the defense by ETI is robust and persistent (6). The downstream defense outputs of PTI and ETI largely overlap (8, 9), and the PTI can boost the ETI defense signaling (10, 11).
NLRs contain an N-terminal coiled-coil (CC) or a Toll/interleukin-1 receptor (TIR) homology domain, a nucleotide-binding adaptor shared by Apaf-1, certain resistance proteins, CED-4 domain (NB-ARC), and a leucine-rich repeat domain (2, 12). Upon recognition of the pathogen effectors directly or indirectly, the NLRs switch from the ADP-bound (inactive) state to the ATP-bound (active) state (13, 14). The activated NLRs then induce robust downstream defense response, which is typically associated with ROS production, influx of calcium, and other responses that culminate into hypersensitive response (HR) cell death (6). Recently, it was shown that activated Arabidopsis CC–type NLR (CNL) ZAR1 assembles into a pentamer structure called resistosome and associate with the plasma membrane to function as a calcium-permeable cation channel to trigger cell death and immunity (15, 16). It was also reported recently that the TIR-containing NLR (TNL) proteins can catalyze NAD+ and produce two types of signaling molecules that selectively activate EDS1–SAG101 and EDS1–PAD4 modules. The two EDS1 signaling modules then activate the helper NLRs AtADR1 and AtNRG1, respectively (17, 18). Recently, the AtADR1 and AtNRG1 were also found to function as calcium-permeable cation channels in triggering cell death (19). Despite this, cell death is not sufficient to restrict the spread of pathogen (20, 21). Even when the plant induced cell death, the plant still get infected by pathogen. In addition to HR cell death, NLRs can also induce major transcriptome reprogramming of nuclear genes that play a role in defense (6). However, how NLRs upon pathogen recognition induce the expression of defense-related genes remains largely unknown.
Mediator is a multiprotein complex that plays a central role in gene transcription/activation in eukaryotes. The Mediator serves as a molecular bridge to link transcription factors to RNA polymerase II (Pol II) (22, 23). The Mediator complex transduces signals from gene-specific transcription factors to the transcription machinery to activate target gene expression (22, 23). During transcription initiation, transcription factors bind to the promoters of specific genes and recruit the Mediator complex. The Mediator complex then recruits Pol II to the promoters to form the transcriptional preinitiation complexes (24, 25). Yeast and animal Mediator complexes are known to contain about 25 and 31 subunits, respectively. Plant Mediator complex includes approximately 34 subunits (26). The core Mediator complex and the head, middle, and tail modules are well conserved in yeast, animals, and plants. As the master coordinators of gene transcription, the Mediator complex participates in many signaling pathways, including those involved in cell division, cell fate, organogenesis, hormone-associated responses, and plant immunity (27, 28).
Jasmonic acid (JA) plays a crucial role in regulating various plant stress responses including insect defense and pathogen resistance (29, 30). MYELOCYTOMATOSIS 2/3/4 (MYC2/3/4) transcription factors, which belong to bHLH transcription factor family, are the core transcription activators of JA response genes. The transcriptional repressor JASMONATE ZIM DOMAIN (JAZ) proteins physically associate with MYC2/3/4 and inhibit the activation of these transcription factors. At the same time, JAZ proteins recruit the transcriptional corepressor TOPLESS (TPL) directly or through interacting with EAR motif–containing adapter protein NINJA, another corepressor, to further inhibit the activation of MYC2/3/4 (31, 32). It remains unknown yet whether any other transcriptional corepressor works together with JAZ proteins to repress the transcription of JA-dependent defense genes.
Not only does JA play an important role in regulating plant defense responses against necrotrophic, biotrophic, and hemi-biotrophic pathogens (33–35), several recent studies also showed that JA signaling plays an important role in plant defense against viruses. Silencing the JA biosynthesis gene allene oxide cyclase (AOC) increases plant susceptibility to Turnip mosaic virus (TuMV) in Nicotiana benthamiana (36). Application of methyl jasmonate (MeJA) significantly reduces the infection of Rice black–streaked dwarf virus (RBSDV) in rice, while a Rice OsCOI1 RNAi line (coi1-13) is more susceptible to RBSDV (37). Overexpression of OsMYC2 or OsMYC3 increases plant resistance to Rice stripe virus (RSV) or Southern rice black–streaked dwarf virus (SRBSDV), whereas osmyc2 or osmyc3 mutant shows increased plant susceptibility to RSV or SRBSDV (38, 39).
Tomato spotted wilt orthotospovirus (TSWV) is among the most destructive plant viruses (40) and causes significant economic losses annually, posing serious threats to global food security (41, 42). Tomato resistance gene Sw-5b is the most effective resistance gene to control TSWV infection (43, 44) and has been widely used in tomato-breeding projects (45, 46). The Sw-5b belongs to CNL, and many CNLs including R8, Mi-1.2, Rpi-blb2, and Rx have been characterized in solanaceous plants. However, little is known about how these solanaceae NLRs induce downstream immunity. N. benthamiana is an ideal model plant for plant–microbe interaction studies (47). Many NLRs, including Sw-5b in the family of Solanaceae, can provide strong immunity to their cognate pathogens when those NLRs were expressed in N. benthamiana (48). This indicates that the downstream immune signaling controlled by those NLRs is conserved in N. benthamiana and other solanaceous plants.
In this study, we used tomato Sw-5b and various solanaceous CNLs and N. benthamiana as our assay model. We demonstrated that the CC domains of Sw-5b and other CNLs can interact directly with the Mediator 10b (MED10b), a subunit in the middle module of the Mediator complex. We found that MED10b–MED7–JAZ proteins interact with each other and corepress jasmonate-specific defense gene expressions. The CC domains of Sw-5b and other CNLs including NRC helper NLRs can interfere with the interaction between MED10b and MED7, therefore derepressing the repressor activity of MED10b–MED7–JAZ proteins on JA-dependent defense gene expressions. Our findings uncovered a previously unknown role for MED10b/MED7 in jasmonate-dependent transcription response, and CNLs in Solanaceae modulate MED10b/MED7 to activate immunity.
Results
Sw-5b CC Domain Directly Interacts with MED10b.
The CC domain of several CNLs has been shown to play critical role in the activation of downstream immune signaling (49–53). To test whether the CC domain of Sw-5b NLR can activate immune defense against tospovirus, we coexpressed the Sw-5b CC domain with the previously reported TSWV infectious replicons (L(+)opt+M(−)opt+SR(+)eGFP) (54) in N. benthamiana leaves through agro-infiltration. The expression of Sw-5b CC domain alone in N. benthamiana leaves did not cause cell death (SI Appendix, Fig. S1A). However, coexpression of FLAG-Sw-5b-CC significantly inhibited eGFP expression from L(+)opt+M(−)opt+SR(+)eGFP viral replicon compared to the leaves coexpressing L(+)opt+M(−)opt+SR(+)eGFP and the empty vector (EV) (SI Appendix, Fig. S1 B and C). These results indicate that CC domain can induce a defense response against TSWV.
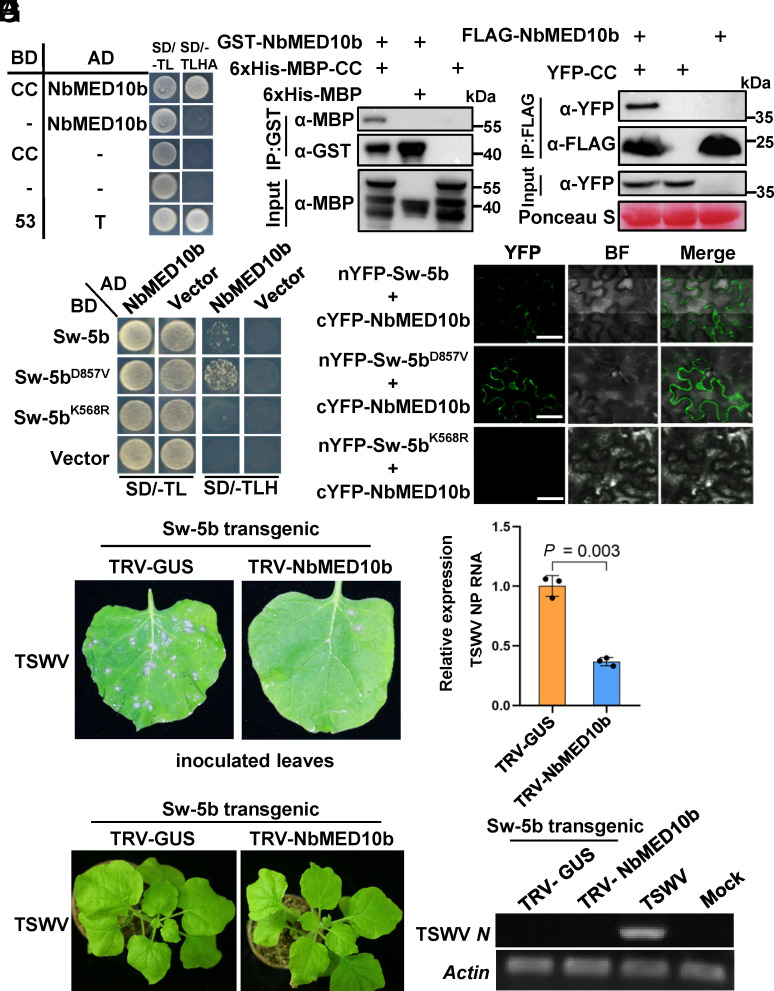
To elucidate the downstream components involved in the Sw-5b CC domain–induced immune signaling, we used the CC domain as a bait to screen its interacting proteins through a yeast two-hybrid (Y2H) screening using cDNA library from N. benthamiana. The Mediator 10b (NbMED10b), a subunit in the middle module of Mediator complex (55), was identified as the CC-interacting protein (Fig. 1A and SI Appendix, Fig. S2A). The GST pull-down, coimmunoprecipitation (Co-IP), and bimolecular fluorescence complementation (BiFC) assay results confirmed that CC domain interacts with NbMED10b in vitro and in planta (Fig. 1 B and C and SI Appendix, Fig. S2B). The Y2H result also showed that the autoactive mutant of full-length Sw-5bD857V (21) interacted strongly with NbMED10b (Fig. 1D). In contrast, the Sw-5bK568R, a P-loop defective mutant that does not induce HR in plants (21), failed to interact with NbMED10b (Fig. 1D). These interactions were confirmed by the result from BiFC assays (Fig. 1E).
Fig. 1.
Sw-5b CC domain interacts with NbMED10b, a negative regulator of the Sw-5b-mediated defense against TSWV infection. (A) A yeast two-hybrid (Y2H) assay result showing an interaction between NbMED10b and Sw-5b CC domain. The transformed yeast cultures were grown on the solid SD dropout medium lacking Trp and Leu (SD/-TL) and the dropout medium lacking Trp, Leu, His, and Ade (SD/-TLHA), respectively. (B) A GST pull-down assay result showing the interaction between the GST-NbMED10b and 6×His-MBP–Sw-5b-CC. 6×His-MBP–Sw-5b-CC, 6×His-MBP, and GST-NbMED10b were expressed individually in Escherichia coli and then purified. The purified 6×His-MBP–Sw-5b-CC or 6×His-MBP was incubated with GST-NbMED10b followed by the pull-down assay using glutathione-sepharose beads. The blots were probed with an anti-GST- or anti-MBP-specific antibody. (C) A coimmunoprecipitation (Co-IP) assay for the interaction between NbMED10b and the Sw-5b CC domain. FLAG-NbMED10b was coimmunoprecipitated with YFP-CC from N. benthamiana leaf extracts. The blots were then probed using an anti-FLAG or an anti-YFP antibody. (D) A Y2H assay result showing the interaction between NbMED10b and Sw-5b, Sw-5bD857V, or Sw-5bK568R mutant. The transformed yeast cultures were grown on the SD/-TL and the SD/-TLH dropout plates, respectively. (E) Bimolecular fluorescence complementation (BiFC) assay results showing the interaction between NbMED10b and Sw-5b, Sw-5bD857V, or Sw-5bK568R mutant. The nYFP- or cYFP-tagged proteins were transiently coexpressed in N. benthamiana leaf cells and then imaged under confocal microscopy. The YFP signal is shown in green (Scale bars, 20 μm.). (F) The NbMED10b-silenced or nonsilenced Sw-5b transgenic N. benthamiana plant leaves were inoculated with TSWV-infected crude leaf extracts. The TSWV-inoculated leaves were photographed at 3 d post TSWV inoculation (dpi) to show the number of HR loci. (G) The accumulation level of TSWV RNA in the inoculated leaves shown in (F) was determined through qRT-PCR using TSWV N gene–specific primers. All the inoculated leaves were harvested at 3 dpi. The expression levels of NbActin in these assayed leaf samples were used as the internal control. Data are presented as the means ± SE (three biological samples per treatment). (H) Phenotype of systemic leaves of the NbMED10b-silenced and nonsilenced Sw-5b transgenic N. benthamiana plants inoculated with TSWV. The plants were photographed at 14 dpi. (I) RT-PCR detection of TSWV RNA in the systemic leaves of the plants shown in the panel (H) at 14 dpi using TSWV N gene–specific primers. The crude extract from a TSWV-infected N. benthamiana leaf sample was used as the positive control. The leaf sample from a mock-inoculated Sw-5b transgenic N. benthamiana plant was used as the negative control. The expression levels of NbActin in the assayed samples were used as the internal controls.
MED10b Negatively Regulates Sw-5b-Mediated Defense.
To elucidate the function of NbMED10b in Sw-5b-mediated resistance, we silenced NbMED10b expression in the Sw-5b transgenic N. benthamiana plants (56) through virus-induced gene silencing (VIGS) using a Tobacco rattle virus (TRV) (SI Appendix, Figs. S3 and S4). Three weeks post-TRV treatment, the TRV-NbMED10b-treated (referred to as NbMED10b-silenced) plants or the TRV-GUS (nonsilenced) control plants were inoculated with the TSWV L(+)opt+M(−)opt+SR(+)eGFP clones via agro-infiltration. The results showed that, compared to the nonsilenced control plants, the NbMED10b-silenced Sw-5b transgenic N. benthamiana plants accumulated much less eGFP expressed from TSWV (SI Appendix, Fig. S4 A and B). When the NbMED10b-silenced or nonsilenced plants were rub-inoculated with TSWV-infected leaf extracts, it induced numerous HR loci in the nonsilenced Sw-5b transgenic plant leaves (Fig. 1F). In contrast, TSWV inoculation caused very few HR loci in the NbMED10b-silenced Sw-5b transgenic plant leaves (Fig. 1F). Generally, even with HR, certain level of viral accumulation including replication and intercellular movement of the virus occurs. Complete absence of HR cell death in NbMED10b-silenced Sw-5b transgenic plant leaves resembles the phenotype of extreme resistance observed in the case of some R genes which no longer allow viral accumulation during defense response (57, 58). Analysis of TSWV-inoculated leaves through qRT-PCR showed that the accumulation level of viral RNA in the TSWV-inoculated NbMED10b-silenced leaves was significantly reduced compared to that in the TSWV-inoculated nonsilenced leaves (Fig. 1G). In addition, we also examined whether silencing NbMED10b has any effect on TSWV systemic infection in Sw-5b transgenic plant. No TSWV was detected in the newly emerged systemic leaves of the NbMED10b-silenced plants (Fig. 1 H and I).
We then examined the interaction between Sw-5b CC and SlMED10b from tomato. The results of Y2H and Co-IP assays showed that the Sw-5b CC domain interacted with SlMED10b in vitro and in planta (SI Appendix, Fig. S5 A and B). We silenced SlMED10b expression in tomato cv. IVF3545 plants (with Sw-5b gene) followed by inoculation with TSWV (SI Appendix, Fig. S5 C and D). Similar to transgenic N. benthamiana with Sw-5b, the numbers of HR loci and the accumulation level of TSWV RNA in the TSWV-inoculated leaves of the SlMED10b-silenced tomato plants were significantly reduced compared to the control plants (SI Appendix, Fig. S5 E and F). In addition, no systemic TSWV infection was detected in the newly emerged systemic leaves of the TSWV-inoculated SlMED10b-silenced tomato plants (SI Appendix, Fig. S5 G and H). These results indicate that MED10b functions as a negative regulator of Sw-5b-mediated immune response to TSWV.
Knockout/down of NbMED10b and Other Subunits in the Middle Module of Mediator Complex Activate Defense against TSWV Infection.
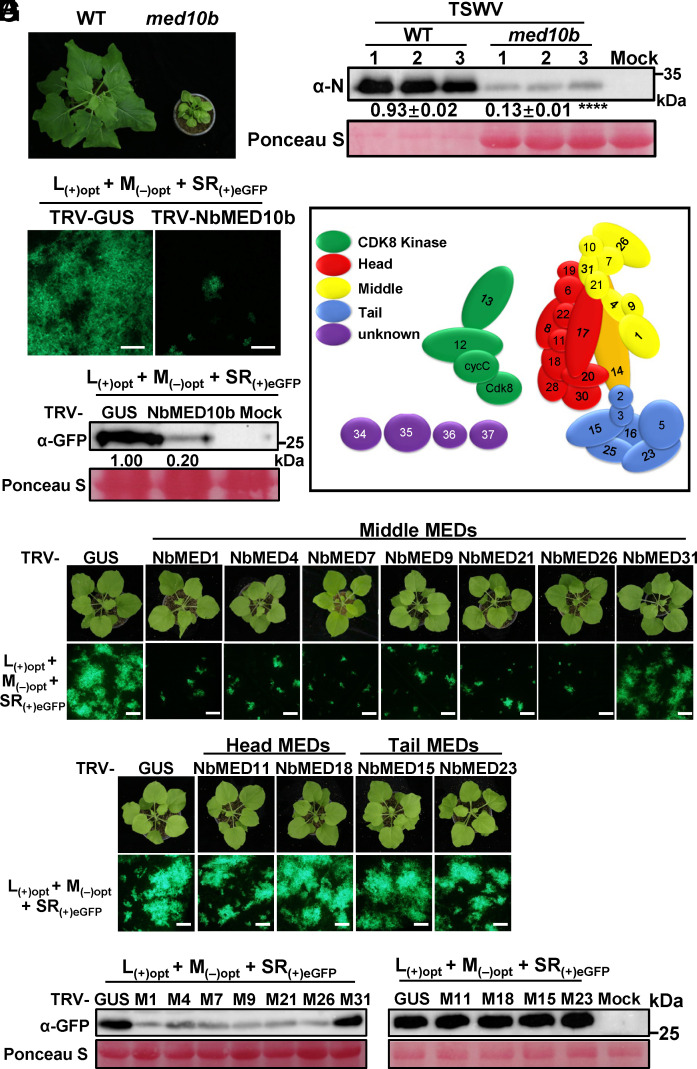
To further investigate how MED10b negatively regulates resistance, we generated med10b knockout mutant lines in wild-type N. benthamiana using CRISPR/Cas9-based technology (SI Appendix, Fig. S6 A–C). Compared to the wild-type plants, the Nbmed10b knockout mutant plants showed a strong dwarfing phenotype (Fig. 2A and SI Appendix, Fig. S6D). The Nbmed10b knockout plants were rub-inoculated with TSWV-infected leaf extracts. The accumulation level of TSWV RNA in the leaves of the Nbmed10b knockout plants was significantly reduced (Fig. 2B), suggesting that knockout of NbMED10b activated plant defense against TSWV infection. As Nbmed10b knockout caused strong dwarfing phenotype, we also silenced NbMED10b expression in wild-type N. benthamiana plants through TRV-mediated gene silencing followed by inoculation of TSWV L(+)opt+M(−)opt+SR(+)eGFP infectious clone or TSWV-containing sap. Similar to the above results, the accumulation of TSWV was significantly lower in the leaves of the NbMED10b-silenced plants compared to the control plants (Fig. 2 C and D and SI Appendix, Fig. S7). Next, we silenced SlMED10b expression in tomato cv. Moneymaker (without Sw-5b) through VIGS followed by TSWV inoculation (SI Appendix, Fig. S8 A and B). The results showed that the accumulation level of TSWV RNA was also significantly reduced in leaves of the SlMED10b-silenced tomato plants (SI Appendix, Fig. S8 C and D).
Fig. 2.
Knockout/down of MED10b and other subunits in the middle module of Mediator complex in wild-type N. benthamiana activate plant defense against TSWV infection. (A) A photograph showing a 12-wk-old N. benthamiana plant and an Nbmed10b knockout mutant plant. (B) Western blot analysis of TSWV N protein accumulation in the TSWV-inoculated leaves of the wild-type N. benthamiana plant or Nbmed10b mutant plant at 5 dpi using an N protein–specific antibody. (C) NbMED10b-silenced (TRV-NbMED10b) or nonsilenced (TRV-GUS control) N. benthamiana plants were inoculated with TSWV infectious clone [L(+)opt+M(−)opt+SR(+)eGFP] via agro-infiltration. The infiltrated N. benthamiana plant leaves were harvested at 60 h post-agro infiltration (hpai) and imaged for eGFP fluorescence under an inverted fluorescence microscope (Scale bars, 800 μm.). (D) Western blot results showing the accumulation level of eGFP in the infiltrated leaves shown in (C) using a GFP-specific antibody at 60 hpai. (E) A diagram showing the subunits in the head, middle, and tail modules of Mediator complex in N. tabacum. (F) Subunits NbMED1, NbMED4, NbMED7, NbMED9, NbMED21, NbMED26, and NbMED31 in the middle module; subunits NbMED11 and NbMED18 in the head module; and subunits NbMED15 and NbMED23 in the tail module of Mediator complexes were individually silenced through VIGS using a TRV-based VIGS vector. These N. benthamiana plants were then inoculated individually with the TSWV infectious clone L(+)opt+M(–)opt+SR(+)eGFP via agro-infiltration. The eGFP fluorescence in the inoculated leaves was imaged at 60 h post TSWV inoculation (hpi) under an inverted fluorescence microscope (Scale bars, 800 μm). (G) Western blot analysis results showing eGFP accumulations in various assayed leaf samples shown in (F) using GFP antibody.
As knockout/down MED10b lead to immune activation, we then analyzed the role of other subunits in the middle module of the Mediator complex, i.e., NbMED1, NbMED4, NbMED7, NbMED9, NbMED21, NbMED26, and NbMED31 through VIGS (Fig. 2E). The expressions of NbMED1, NbMED4, NbMED7, NbMED9, NbMED21, NbMED26, and NbMED31 in N. benthamiana plants were silenced individually using the TRV-based VIGS vectors (SI Appendix, Fig. S9 A and B) followed by inoculation of TSWV replicon via agro-infiltration. Strikingly, silencing the expressions of NbMED1, NbMED4, NbMED7, NbMED9, NbMED21, and NbMED26 also suppressed the accumulation of TSWV replicon (Fig. 2 F and G), indicating that knockdown of expression of these subunits in the middle module of Mediator complex can also activate plant defense against TSWV infection.
We also analyzed whether subunits in the head and the tail module of Mediator complex have any effect against TSWV. The expressions of NbMED11 and NbMED18 in the head module and those of NbMED15 and NbMED23 in the tail module were silenced through VIGS (SI Appendix, Fig. S9 A and B). The results showed that, unlike NbMED10b or other subunits in the middle module, silencing of NbMED11, NbMED18, NbMED15, or NbMED23 had no effect on TSWV accumulation (Fig. 2 F and G).
MED10b Directly Interacts with MED7, Which Also Negatively Regulates Sw-5b-Mediated Defense.
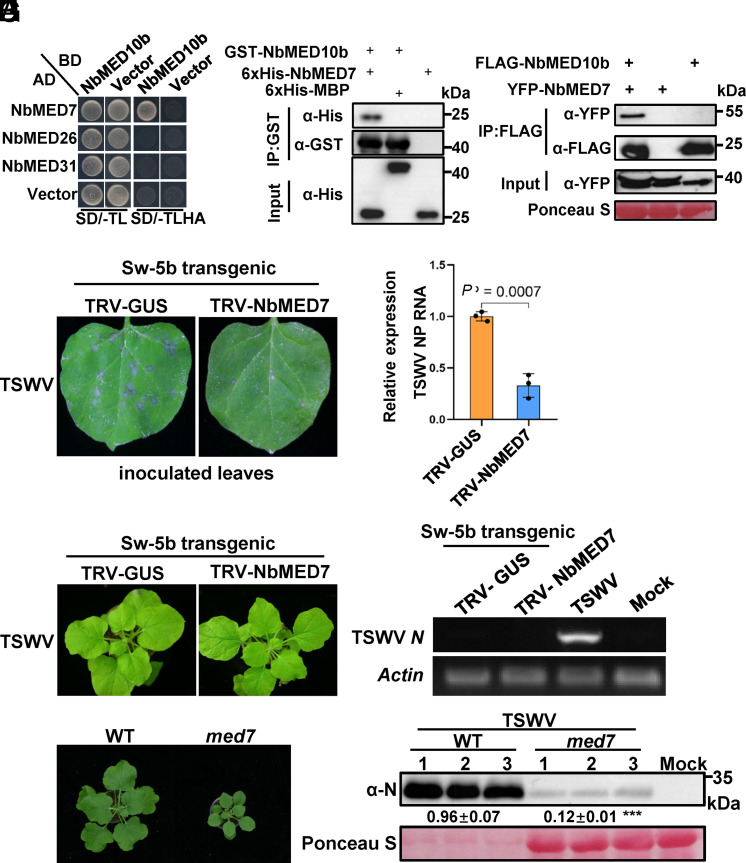
Based on the above results, we hypothesized that MED10b may interact with other subunit(s) of the middle module of Mediator complex to function as repressors of plant defense against TSWV. Whereas, binding of Sw-5b CC domain to MED10b might disrupt the interaction between MED10b with its adjacent subunit(s) in the middle module of Mediator complex and consequently derepresses the repressor activity of those subunits, leading to an activation of host defense against TSWV infection. To test this hypothesis, we examined the interactions between NbMED10b and other three subunits, NbMED7, NbMED26, and NbMED31, that were predicted to be in close proximity to NbMED10b in the middle module (59) (Fig. 2E). The results of BiFC assay showed that NbMED10b could interact with NbMED7, NbMED26, or NbMED31 (SI Appendix, Fig. S10A). However, the result of Y2H assay showed that NbMED10b could directly interact with NbMED7, but not with the other two subunits (Fig. 3A). The interaction between NbMED10b and NbMED7 was further confirmed through GST-pull down, Co-IP, and split-luciferase (SLC) assays (Fig. 3 B and C and SI Appendix, Fig. S10B).
Fig. 3.
MED10b interacts with MED7, which also negatively regulates Sw-5b-mediated defense. (A) A Y2H assay result showing the interaction between NbMED10b and NbMED7, NbMED26, or NbMED31. The transformed yeast cells were grown on the SD/-TL and the SD/-TLHA dropout plates, respectively. (B) GST pull-down assay results showing the interaction between the GST-NbMED10b and 6×His-NbMED7. 6×His-NbMED7, 6×His-MBP, and GST-NbMED10b were expressed individually in E. coli and then purified. The purified 6×His-NbMED7 or 6×His-MBP was incubated with GST-NbMED10b followed by the pull-down assay using glutathione-sepharose beads. The blots were probed with an anti-GST- or anti-His-specific antibody. (C) Co-IP assay results showing the interaction between the FLAG-NbMED10b and YFP-NbMED7. The FLAG-NbMED10b was used to coimmunoprecipitate YFP-NbMED7 from N. benthamiana leaf extracts. The blots were probed with an anti-FLAG or an anti-YFP antibody. (D) The NbMED7-silenced or nonsilenced Sw-5b transgenic N. benthamiana plants were inoculated with TSWV-infected crude leaf extracts. The inoculated leaves were photographed at 3 dpi. The number and size of HR loci in these inoculated leaves were determined. (E) The accumulation level of TSWV RNA in the inoculated leaves shown in (D) was determined through qRT-PCR using the TSWV N gene–specific primers at 3 d post TSWV inoculation. The expression levels of NbActin in the assayed samples were used as the internal controls. Data are presented as the means ± SE (three biological samples per treatment). (F) The TSWV-inoculated NbMED7-silenced or nonsilenced Sw-5b transgenic N. benthamiana plants were photographed at 14 d post TSWV inoculation. (G) Detection of TSWV RNA in the systemic leaves of the TSWV-inoculated NbMED7-silenced or nonsilenced Sw-5b transgenic N. benthamiana plants through RT-PCR using TSWV N gene–specific primers. The systemic leaves of the TSWV-infected N. benthamiana plants were used as the positive control. The systemic leaves of the mock-inoculated plants were used as the negative controls. The expression levels of NbActin in the assayed samples were used as the internal controls. (H) A photograph showing 12-wk-old wild-type N. benthamiana plant and an Nbmed7 knockout mutant plant. (I) Western blot analysis of TSWV N protein accumulation in the TSWV-inoculated leaves of the wild-type N. benthamiana plant or Nbmed7 mutant plant at 5 dpi using an N protein–specific antibody.
To determine the role of NbMED7 in Sw-5b-mediated resistance to TSWV infection, we silenced NbMED7 expression in Sw-5b transgenic N. benthamiana plants through VIGS (SI Appendix, Fig. S11) followed with rub inoculation of TSWV-infected leaf extracts at 21 d post TRV treatment. By 3 d post TSWV-inoculation, the numbers of HR loci and the accumulation level of TSWV RNA in the TSWV-inoculated leaves of the NbMED7-silenced plants were significantly lower than those in the TSWV-inoculated leaves of the control plants (Fig. 3 D and E). In addition, we examined whether silencing NbMED7 has any effect on TSWV systemic infection in Sw-5b transgenic plant. No TSWV systemic infection was detected in the newly merged leaves of the NbMED7-silenced plants (Fig. 3 F and G), indicating that NbMED7 is also an important negative regulator in the Sw-5b-mediated resistance to TSWV infection. We generated Nbmed7 knockout N. benthamiana mutant lines. The Nbmed7 knockout mutant also showed a strong dwarfing phenotype (Fig. 3H and SI Appendix, Fig. S12). Similar to Nbmed10b, the accumulation of TSWV was significantly reduced in Nbmed7 knockout mutant (Fig. 3I).
CC Domain of the Activated Sw-5b Interferes with the Interaction between MED10b and MED7.
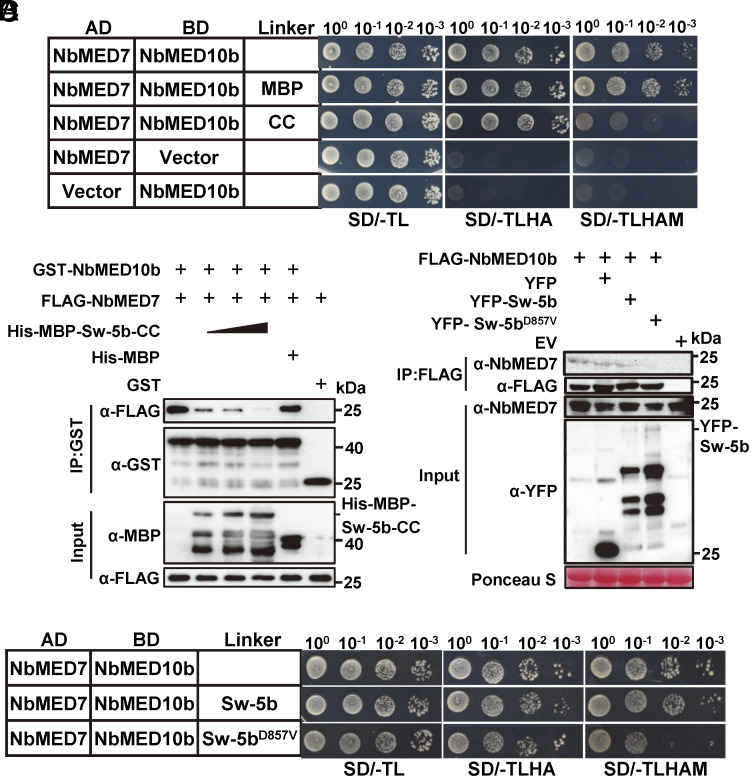
We conducted yeast three hybrid (Y3H) assay to examine whether Sw-5b CC can interfere with the interaction between NbMED10b and NbMED7 in yeast cells. The result showed that in the presence of Sw-5b CC, but not MBP, the interaction between NbMED10b and NbMED7 was strongly inhibited (Fig. 4A). The GST pull-down assay result also showed that when the amount of MBP–Sw-5b-CC was increased, the amount of FLAG-NbMED7 pulled down by GST-NbMED10b was decreased (Fig. 4B). In the same assay, the addition of His-MBP had no clear effect on the interaction between GST-NbMED10b and FLAG-NbMED7 (Fig. 4B). Co-IP assay confirmed that the interaction between NbMED10b and NbMED7 was significantly reduced in the presence of YFP-Sw-5b-CC but not YFP in vivo (SI Appendix, Fig. S13). The Y3H assays and GST pull-down result also showed that Sw-5b CC could interfere with the interaction between SlMED10b and SlMED7 from tomato (SI Appendix, Fig. S14 A and B).
Fig. 4.
CC domain of the activated Sw-5b interferes with the interaction between MED10b and MED7. (A) Y3H assay results showing the effects of the CC domain on the interaction between NbMED10b and NbMED7. Yeast cells were cotransformed with pGAD-NbMED7 and pBridge-NbMED10b, pGAD-NbMED7 and pBridge-NbMED10b+MBP, or pGAD-NbMED7 and pBridge-NbMED10b + Sw-5b CC domain. The cotransformed yeast cells were grown on the SD/-TL, SD/-TLHA, or SD/-TLHAM (lacking Trp, Leu, His, Ade, and Met) plates, respectively, for 5 d. (B) GST pull-down assay results showing the effect of Sw-5b CC domain on the interaction between NbMED10b and NbMED7. Fixed amount of GST-MED10b and FLAG-MED7 was incubated with increasing amounts of purified His-MBP–Sw-5b-CC or purified His-MBP protein (control). Proteins in the samples were then pulled down using glutathione-sepharose beads followed by western blot assays with an anti-GST-, anti-MBP-, or anti-FLAG-specific antibody. (C) Y3H assay results showing that the full-length Sw-5bD857V, but not Sw-5b, interrupted the interaction between NbMED10b and NbMED7. Yeast cells were cotransformed with constructs expressing AD-NbMED7 + BD-NbMED10b + empty vector, AD-NbMED7 + BD-NbMED10b + Sw-5b, or AD-NbMED7 + BD-NbMED10b + Sw-5bD857V. The transformed cells were grown on the SD/-TL, SD/-TLHA, and SD/-TLHAM plates, respectively, to assess the NbMED7 and NbMED10b interaction. (D) FLAG-NbMED10b was used to immunoprecipitate the endogenous NbMED7 in N. benthamiana leaves in the presence of YFP, YFP-Sw-5b, and YFP-Sw-5bD857V, respectively. Western blots were probed using an anti-FLAG, anti-YFP, or anti-NbMED7 antibody.
In the absence of TSWV NSm effector, Sw-5b stays in an inactive state and upon recognition of NSm, Sw-5b NLR converts to an activated state to trigger defense response (43, 60). To determine which form of Sw-5b could interfere the interaction between NbMED10b and NbMED7, we first performed Y3H assay. The results showed that the full-length autoactive Sw-5bD857V, but not the inactive Sw-5b, reduced the interaction between NbMED10b and NbMED7 (Fig. 4C). The activated form of Sw-5b (Sw-5bD857V) also interfered with the interaction between NbMED10b and NbMED7 in vivo (Fig. 4D). Western blot analyses showed that the protein accumulations of endogenous NbMED10b and NbMED7 were not affected by the Sw-5bD857V (SI Appendix, Fig. S15).
MED10b/MED7 Repress the Expression of Jasmonate-Dependent Defense Response Genes.
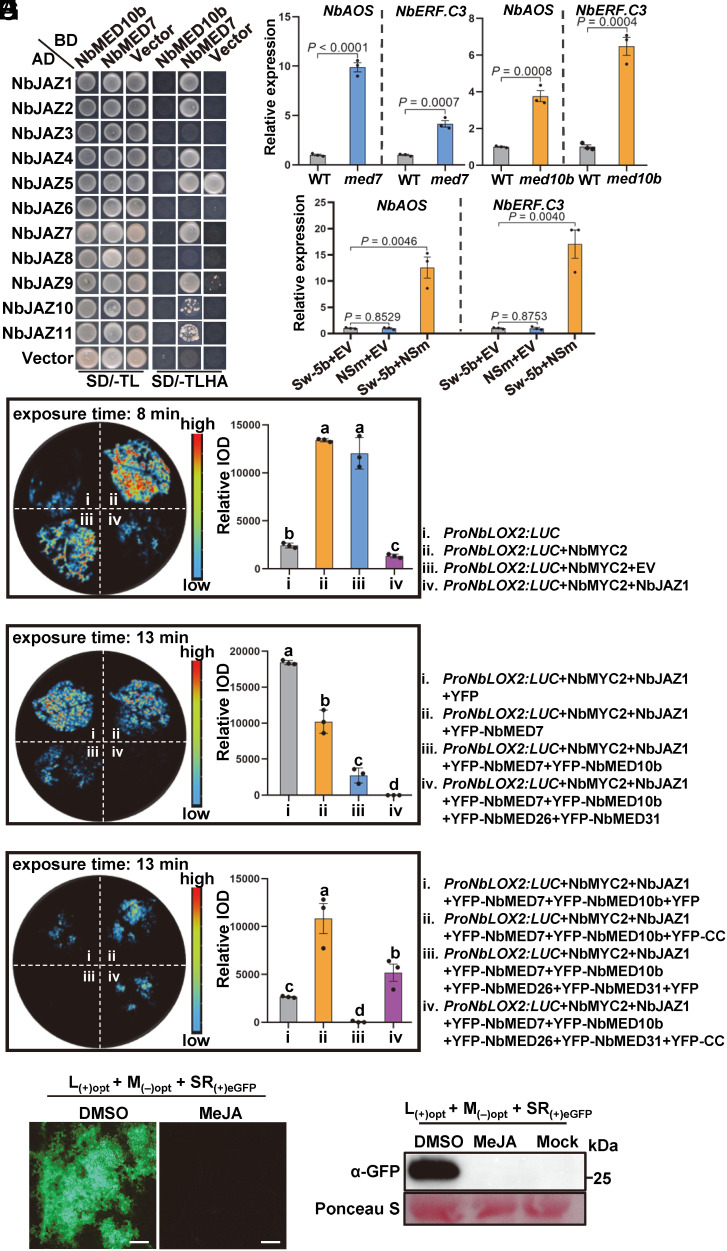
To further elucidate the downstream components in the CC-induced immune signaling, we used the MED10b or MED7 as the bait to screen its interacting proteins through a Y2H screening. The NbJAZ7, a transcription repressor of JA signaling pathway, was identified to interact with NbMED7 (SI Appendix, Fig. S16A). We then examined the interaction of MED7 and MED10b with all 11 members of JAZ proteins from N. benthamiana or tomato. NbMED10b does not interact with any of the other JAZ proteins. In addition to NbJAZ7, NbMED7 also interacted with NbJAZ1, NbJAZ2, NbJAZ4, and NbJAZ9–NbJAZ11 in Y2H assay (Fig. 5A). SlMED7 but not SlMED10b also interacts with many SlJAZs (SI Appendix, Fig. S16B). TPL and NINJA are corepressors of JAZs in JA signaling pathway. Knockout or downregulation of TPL and NINJA can cause the activation of JA signaling (31, 32). We then examined whether the NbMED10b and NbMED7 could interact with NbTPL and NbNINJA. Y2H results showed that neither NbMED10b nor NbMED7 interacts with NbTPL and NbNINJA (SI Appendix, Fig. S17).
Fig. 5.
MED10b/MED7 mediate jasmonate-dependent transcription repression. (A) A Y2H assay result showing the interaction between NbJAZs and NbMED10b or between NbJAZs and NbMED7. The transformed yeast cells were grown on the SD/-TL and the SD/-TLHA plates, respectively. (B) Quantitative RT-PCR analysis results showing the expressions of representative marker genes involved in the JA signaling pathways in wild-type (WT) N. benthamiana Nbmed7 knockout, and the Nbmed10b knockout plants. The expression levels of NbActin in these samples were used as the internal controls. Data are presented as the means ± SE (three independent biological sample per treatment). (C) Quantitative RT-PCR analysis results showing the expressions of representative marker genes involved in the JA signaling pathways in N. benthamiana coexpressing Sw-5b with EV, NSm with EV, or Sw-5b with NSm. The expression levels of NbActin in these samples were used as the internal controls. Data are presented as the means ± SE (three independent biological samples per treatment). (D) Transient overexpression of NbMYC2 transcription factor activated the expression of luciferase (LUC) driven by the NbLOX2 promoter, and addition of NbJAZ1 into NbMYC2 reduced the expression of LUC. The luciferase activity (integrated optical density, IOD) in the treated leaves was quantified and shown in the right. Data are presented as the means ± SE (three independent biological samples per treatment). (E) Coexpression of NbJAZ1, or NbMED7 and NbJAZ1, or NbMED7, NbMED10b, and NbJAZ1, or NbMED7, NbMED10b, NbMED26, NbMED31, and NbJAZ1 with NbMYC2 reduced the expression of LUC. The luciferase activity (integrated optical density, IOD) in the treated leaves was quantified and shown in the right. Data are presented as the means ± SE (three independent biological samples per treatment). (F) Coexpression of Sw-5b CC with NbMYC2, NbJAZ1, NbMED7 and NbMED10b, or NbMYC2, NbJAZ1, NbMED7, NbMED10b, NbMED26, and NbMED31 activated the expression of LUC. The luciferase activity (integrated optical density, IOD) in the treated leaves was quantified and shown in the right. Data are presented as the means ± SE (three independent biological samples per treatment). (G) N. benthamiana plants were sprayed with DMSO or 100 µM MeJA. At 2 d post treatment, phytohormone-treated leaves were inoculated again with TSWV infectious clone [L(+)opt+M(–)opt+SR(+)eGFP] via agro-infiltration. The infiltrated N. benthamiana plant leaves were harvested at 60 hpai and imaged for eGFP fluorescence loci under an inverted fluorescence microscope (Scale bars, 800 μm.). (H) Western blot assay results showing the accumulation level of eGFP at 60 hpai in the infiltrated leaves shown in (G), using GFP antibody.
Next, we examined the JA response genes in med7 and med10b knockout N. benthamiana plants. The expression levels of JA response genes NbAOS, NbERF.C3, and NbPR-STH2 were significantly up-regulated in both med7 and med10b knockout plants (Fig. 5B and SI Appendix, Fig. S18A). We also examined whether Sw-5b can activate JA signaling pathway upon the recognition of TSWV NSm effector. Coexpression of Sw-5b with NSm significantly up-regulated the expression levels of these JA response genes but not by coexpression of Sw-5b with EV or NSm with EV (43) (Fig. 5C and SI Appendix, Fig. S18B). Knockdown of a component of the middle Mediator module NbMED26, but not NbMED18 component of the head module or NbMED23 in the tail module, also up-regulated the expressions of JA response genes (SI Appendix, Fig. S18 C–E). Knockdown of SlMED10b or SlMED7 also up-regulated the expression levels of JA response genes SlAOS, SlERF.C3, and SlPR-STH2 in tomato (SI Appendix, Fig. S19 A and B). Inoculation of TSWV onto tomato cv. IVF3545 (with Sw-5b) also activated these JA marker genes (SI Appendix, Fig. S19C). Transcription factor MYC2 can directly bind to the promoter and activate LOX2, a representative JA response gene (61). The addition of NbMYC2 transcription factor strongly activated the luciferase (LUC) reporter gene driven by the NbLOX2 promoter. However, the addition of NbJAZ1 into NbMYC2 reduced the expression of LUC (Fig. 5D). The coaddition of NbMED7 and NbJAZ1 to NbMYC2 further inhibited the expression of LUC. The coexpression of NbMED7, NbMED10b, and NbJAZ1 inhibited the expression of LUC further more (Fig. 5E), while the coexpression of NbMED10b and NbJAZ1 without NbMED7 did not further reduce the expression of LUC (SI Appendix, Fig. S20A). Moreover, the coexpression of NbMED7, NbMED10b, NbMED26, NbMED31, and NbJAZ1 with NbMYC2 completely suppressed the expression of LUC (Fig. 5E). These effects on LOX2 promoter were not observed in the absence of JAZ1 (SI Appendix, Fig. S20B), suggesting that MED7/MED10b work together with JAZ1 to suppress the transcription of JA response genes. Next, we tested whether Sw-5b can derepress the inhibitory effect of JAZ1–MED7–MED10b. The results showed that coexpression of Sw-5b and NSm caused fluorescence quenching and no LUC expression was detected (SI Appendix, Fig. S20C). However, the addition of Sw-5b CC activated the expression of LUC, suggesting that Sw-5b CC can derepress JAZ1–MED7–MED10b-mediated repression of JA-dependent transcription (Fig. 5F). Furthermore, we investigated the role of MED7/MED10b repression and Sw-5b derepression on plant resistance to TSWV (SI Appendix, Fig. S21 A and B). The results showed that coexpression of the transcription factor NbMYC2 with TSWV infectious clones L(+)opt+M(−)opt+SR(+)eGFP reduced the expression of the TSWV eGFP reporter. Coexpression of NbMED7, NbMED10b, and NbJAZ1 attenuated the inhibitory effect of MYC2 on TSWV reporter accumulation. However, coexpression of Sw-5b or Sw-5b CC with NbMYC2, NbJAZ1, NbMED7, and NbMED10b strongly inhibited TSWV accumulation in N. benthamiana leaves. We have shown recently that exogenous application of JA induces defense against TSWV (62). Here, we further treated the wild-type N. benthamiana with MeJA followed by inoculation of TSWV L(+)opt+M(−)opt+SR(+)eGFP infectious clones via agro-infiltration. The results further showed that the JA treatment significantly inhibited TSWV infection in N. benthamiana (Fig. 5 G and H).
CC Domain of Various Other CNLs from Solanaceae Modulates MED10b/MED7 Interaction to Activate Jasmonate-Dependent Immune Pathway.
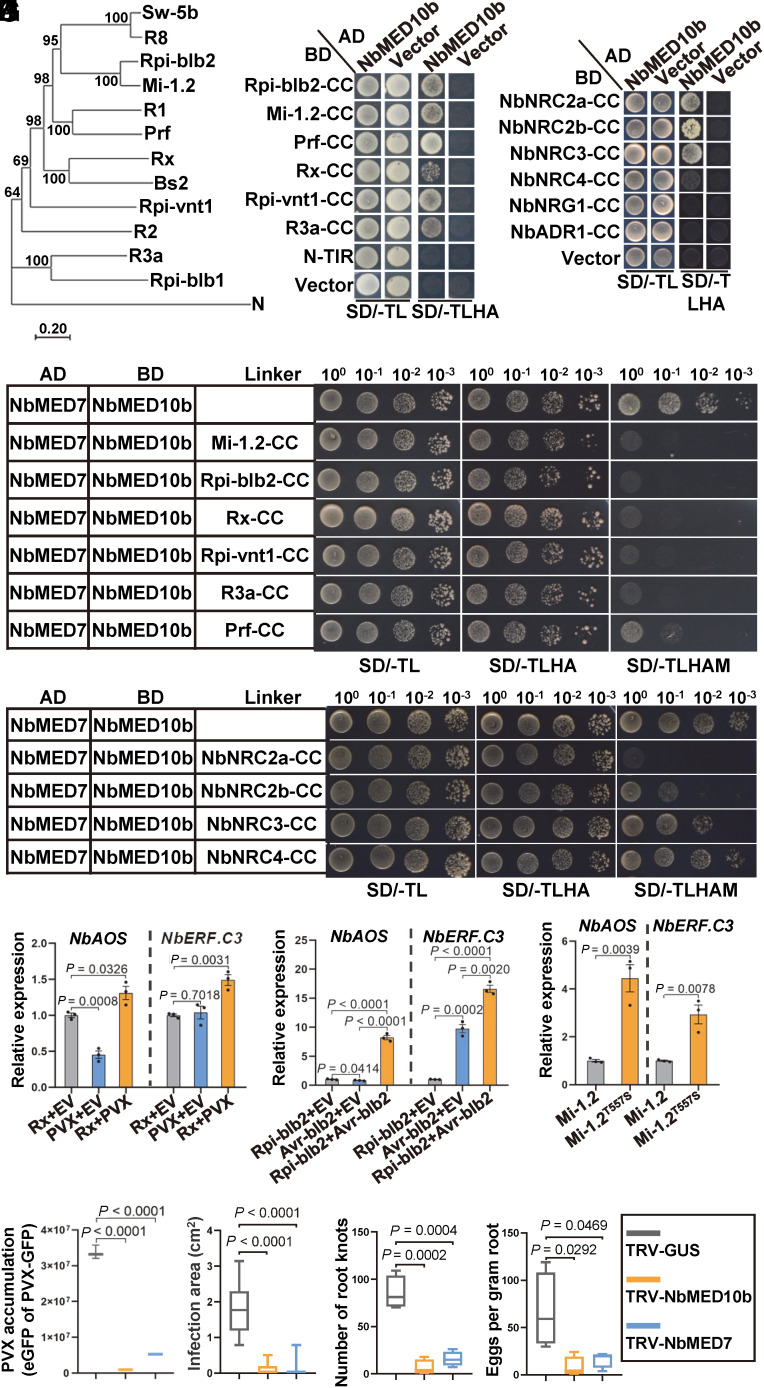
We next investigated whether CC domain of other CNLs from Solanaceae (Fig. 6A) has effect on MED10b and MED7 interaction. Y2H and BiFC assay results showed that the CC domains of Mi-1.2, Rpi-blb2, Rpi-vnt1, R3a, Prf, and Rx were all capable of interacting with NbMED10b (Fig. 6B and SI Appendix, Fig. S22). The TIR domain of tobacco NLR N was, however, incapable of interacting with NbMED10b in yeast (Fig. 6B). We then cloned SlMED10b from tomato and StMED10b from potato and tested their abilities to interact with the CC domains described above. As shown in SI Appendix, Fig. S23 A and B, SlMED10b did interact with the CC domains of Mi-1.2 and Prf, whereas StMED10b interacted with the CC domains of Rpi-blb2, Rpi-vnt1, R3a, and Rx. Based on these findings, we conclude that MED10bs in various solanaceous species can interact with the CC domains of NLR receptors. Besides the CC domains of sensor NLRs, NbMED10b also interacted strongly with the CC domains of helper NLRs NbNRC2a, NbNRC2b, and NbNRC3 and weakly with the CC domain of NbNRC4 (Fig. 6C). NbMED10b, however, did not interact with the CC domains of helper NLRs NbNRG1 and NbADR1 (Fig. 6C).
Fig. 6.
CC domain of CNLs in Solanaceae modulates MED10b/MED7 interaction to activate jasmonate-dependent immune pathway. (A) A phylogenetic tree constructed using CNLs from various solanaceous species. Sequences of these NLRs were retrieved from the GenBank, and the phylogenetic tree was conducted using the neighbor-joining method in the MEGA7 package. (B) Y2H assay results showing the interactions between NbMED10b and various CC domains of CNLs from different solanaceous species. The transformed yeast cells were grown on the SD/-TL and the SD/-TLHA plates, respectively. (C) Y2H assay results showing the interactions between NbMED10b and the CC domains in the helper NLRs NRC2/3/4. The transformed yeast cells were grown on the SD/-TL and the SD/-TLHA plates, respectively. (D) Y3H assay results showing the effects of CC domains from different CC-type NLRs on the interaction between NbMED10b and NbMED7. Yeast cells coexpressing AD-NbMED7, BD-NbMED10b, Mi-1.2-CC, Rip-blb2-CC, Rx-CC, Rpi-vnt1-CC, R3a-CC, or Prf-CC were grown, respectively, on the SD/-TL, SD/-TLHA, or SD/-TLHAM medium plates to determine the interaction between NbMED7 and NbMED10b. (E) Y3H assay results showing the effects of the CC domains of NRC2a, NRC2b, NRC3, and NRC4 on the interaction between NbMED10b and NbMED7. Yeast cells coexpressing AD-NbMED7, BD-NbMED10b, Vector, NbNRC2a-CC, NbNRC2b-CC, NbNRC3-CC, or NbNRC4-CC were grown on the SD/-TL, SD/-TLHA, or SD/-TLHAM medium plates to determine the interaction between NbMED7 and NbMED10b. (F) Quantitative RT-PCR analysis results showing the expressions of representative marker genes involved in the JA signaling pathways in N. benthamiana coexpressing Rx with EV, PVX with EV, or Rx with PVX. The expression levels of NbActin in these samples were used as the internal controls. Data are presented as the means ± SE (three independent biological samples per treatment). (G) Quantitative RT-PCR analysis results showing the expression of representative marker genes involved in the JA signaling pathways in N. benthamiana coexpressing Rpi-blb2 with EV, Avr-blb2 with EV, or Rpi-blb2 with Avr-blb2. The expression levels of NbActin in these samples were used as the internal controls. Data are presented as the means ± SE (three independent biological samples per treatment). (H) Quantitative RT-PCR analysis results showing the expressions of representative marker genes involved in the JA signaling pathways in N. benthamiana expression of Mi-1.2 or Mi-1.2T557S. The expression levels of NbActin in these samples were used as the internal controls. Data are presented as the means ± SE (three independent biological samples per treatment). (I) The accumulation level of eGFP expressing from PVX in NbMED10b-silenced, NbMED7-silenced, or nonsilenced control (TRV-GUS) leaves of N. benthamiana in the infiltrated leaves was measured and quantified using the Image J software. Statistical analyses were done using the two-tailed Student’s t test. Data are presented as the means ± SE (three biological samples per treatment). (J) The size of P. infestans lesions in NbMED10b-silenced, NbMED7-silenced, or nonsilenced control leaves of N. benthamiana was measured and quantified using the Image J software. Statistical analyses were done using the two-tailed Student’s t test. Data are presented as the means ± SE (10 biological samples per treatment). (K) The numbers of root knots on NbMED10b-silenced, NbMED7-silenced, or nonsilenced control (TRV-GUS) roots of N. benthamiana plants and the statistical differences between the treatments were determined using the two-tailed Student’s t test. Data are presented as the means ± SE (four biological samples per treatment). (L) The numbers of nematode egg masses on the assayed plant roots. The statistical differences between the treatments were determined using the two-tailed Student’s t test. Data are presented as the means ± SE (four biological samples per treatment).
Next, we used Y3H assay to test the abilities of different CC domains to disrupt the interaction between MED7 and MED10b. The results showed that the CC domains of sensor (Fig. 6D) and helper NLRs (Fig. 6E) that interact with NbMED10b could all reduce the NbMED7 and NbMED10b interaction. In addition, the Y3H assay results showed that the CC domains of these solanaceous NLRs could disrupt the interaction between SlMED7 and SlMED10b or the interaction between StMED7 and StMED10b (SI Appendix, Fig. S23 C and D). The GST pull-down assay results confirmed that MBP–Mi-1.2-CC or MBP–Rpi-blb2-CC interfered with the interactions between NbMED7 and NbMED10b, SlMED7 and SlMED10b, or StMED7 and StMED10b (SI Appendix, Fig. S24 A–D). GST pull-down assays also showed that the addition of His-MBP–NbNRC2a-CC or His-MBP–NbNRC3-CC reduced the interaction between NbMED7 and NbMED10b (SI Appendix, Fig. S25 A and B).
Next, we investigated whether Rx, Rpi-blb2, or Mi-1.2 can activate JA signaling pathway upon the recognition of corresponding effector protein or when NLR switched into an active form. Coexpression of Rx and PVX or Rpi-blb2 and Avr-blb2 significantly up-regulated the expression levels of JA response genes NbAOS, NbERF.C3, and NbPR-STH2 but not by the expression of Rx or Rpi-blb2 alone (Fig. 6 F and G and SI Appendix, Fig. S26 A and B). Similarly, the autoactive Mi-1.2T557S mutant (63) also up-regulated the expression levels of these JA response genes (Fig. 6H and SI Appendix, Fig. S26C).
Because the CC domains of various NLRs in Solanaceae could interfere with the interaction between MED7 and MED10b, we also investigated whether downregulation of MED10b and MED7 activates the host resistance against different pathogens. As med10b and med7 knockout plant grow too small to perform these experiments, we silenced the expression of NbMED10b and NbMED7 in N. benthamiana plants through VIGS using TRV-NbMED10b and TRV-NbMED7, respectively (SI Appendix, Fig. S27 A and B). The gene silenced or nonsilenced control plants were inoculated with PVX-GFP, P. infestans, or M. incognita. Compared to the control plants (TRV-GUS), silencing of NbMED10b or NbMED7 expression in N. benthamiana plants strongly inhibited PVX-GFP infection (Fig. 6I and SI Appendix, Fig. S27 C and D) and P. infestans infection (Fig. 6J and SI Appendix, Fig. S27E). The NbMED10b- or NbMED7-silenced N. benthamiana plants also showed a strong resistance to M. incognita infection. The numbers of M. incognita-induced root knots or egg masses in the roots of the NbMED10b- or NbMED7-silenced plants were significantly reduced compared to the nonsilenced control plants (Fig. 6 K and L and SI Appendix, Fig. S27F). These results show that NbMED10b and NbMED7 function as negative regulators of CNL-mediated immunity in Solanaceae.
Discussion
In this study, we demonstrate that MED10b/MED7 mediate jasmonate-dependent transcription repression, and CC domains of CNLs from Solanaceae derepress the repressor activity of MED10b–MED7–JAZ and activate immunity. Using Sw-5b NLR as a model, we show that Sw-5b CC domain directly interacts with MED10b. Knockout/down subunits including MED10b and MED7 cause the activation of defense against TSWV. MED10b was found to directly interact with MED7, whereas MED7 directly interacts with JAZ transcription repressor proteins. MED10b–MED7–JAZ proteins together can strongly corepress the expression of JA defense response genes. However, Sw-5b CC interferes with the interaction between MED10b and MED7, and consequently derepresses the corepressor activity of MED10b–MED7–JAZ, leading to the activation of JA-dependent defense response against TSWV infection. Furthermore, using various other CNLs from Solanaceae, we show that the CC domains of those CNLs including helper NLR NRCs can also modulate MED10b/MED7 to activate defense against different pathogens.
NINJA (NOVEL INTERACTOR OF JAZ) and TPL (TOPLESS) were previously shown to serve as corepressors of JAZ proteins in JA pathway (31, 32). Our findings reveal that MED10b and MED7 proteins serve as previously unknown repressors of jasmonate-dependent transcription through interaction with JAZ proteins. MED7 was found to directly interact with JAZ proteins. Coexpression of MED10b, MED7, JAZ, and other middle Mediator module components together can strongly cosuppress the expression of JA response genes. This corepression is dependent on JAZ proteins. It was previously shown that knockout of either NINJA or TPL activates JA signaling pathway (31). It was also reported that transcription corepressor TPL interacts with AtMED21 and AtMED10a in Arabidopsis, and this interaction is necessary for TPL to function as a corepressor in Auxin signaling (64). In this study, knockout/down of MED10b, MED7, and other subunits can activate the JA pathway, and it mediates defense response against TSWV. Furthermore, knockdown subunits in the head and tail modules of Mediator do not lead to the activation of JA response genes, suggesting that subunits in the middle module mainly participate in the repression function. Together, we propose that MED10b–MED7–JAZ and other subunits of the middle module may form a complex, and this entire supercomplex functions as a repressor of JA response genes. Knockout/down of either MED10b, MED7, or TPL in this supercomplex repressor possibly disrupt the complex formation and hence lead to activation of JA-specific defense response.
The Mediator complex in eukaryotes serves as a molecular bridge to link specific enhancer-bound transcription factors to RNA polymerase II to engage in the initiation of gene transcription/activation (22, 23). Our results suggest that the entire middle module of the Mediator complex may function as a corepressor of JAZ proteins in JA pathway. How can the Mediator complex act both as a corepressor and a molecular linker to activate downstream JA defense genes? It has been reported previously that the transcriptional corepressor TPL forms a bridge between IAA14 and the CKM component MED13 through the physical interaction. Auxin induces the dissociation of MED13 from the ARF7-binding region upstream of its target gene (65). These findings indicate that auxin induced compositional change in the Mediator in ARF7- and ARF19-mediated transcription. Chen and colleagues recently reported that the mammalian Mediator complex has several different conformations (66). The biological significance of these conformations remains unclear. We speculate that these different conformations may have distinct functions in gene transcription regulations. For instance, one form of Mediator complex acts as a corepressor to inhibit JA defense gene expressions through physical interaction of subunits in the middle module with transcription repressor JAZs and corepressor TPL. When its conformation changes to another form, it functions as a molecular bridge to connect specific transcription factor(s) to Pol II and consequently activates JA-dependent defense gene expressions. These conformation changes are perhaps the most economical way to recruit and bridge the Mediator complexes with Pol II during host defense responses. When JA pathway was not activated, the subunits in the middle module physically interact with JAZs, TPL, and Mediator, which maybe already on the transcription factors but act as a corepressor. Upon triggering the activation of JA pathway, the JAZs are degraded or the MED10b/MED7 interaction is disturbed, either of which may change the conformation of the Mediator from a repressor into a molecular bridge to connect between MYC2/3/4 transcription factors and Pol II. This role of the Mediator may specifically act in stress response, as it needs to respond quickly to environmental clues. Further studies are needed to elucidate the substantial role and mechanism underlying Mediator complex conformational changes in regulating defense gene repression and activation.
Recent studies have demonstrated an important role of JA signaling in plant defense against various pathogens including viruses (34). In this study, we showed that coexpression of Sw-5b NLR and NSm elicitor activated the JA signaling. Consistent with this, the application of JA significantly reduced TSWV infection in N. benthamiana and in pepper plants (62). Activation of the JA signaling pathway was also found in another tomato NLR gene, Sl5R-1, which also confers resistance to TSWV and was map cloned recently (67). The expression levels of JA response genes SlMYC2, SlJAZ, SlAOC, and SlAOS were significantly up-regulated in TSWV-inoculated Sl5R-1 resistant plants compared to noninoculated resistant plants. In contrast, these genes were down-regulated in susceptible plants. The NLRs that confer the resistance to bacteria typically activate salicylic acid (SA) signaling rather than JA signaling. Consistent with this, SA, but not JA, is critical for regulating plant defense against bacterial infection. Interestingly, we found here that coexpression of potato NLR Rpi-blb2 and Avr-blb2 from Phytophthora up-regulated the JA response genes. Recently, exogenous application of JA has been shown to improve plant resistance to Phytophthora (68). The tomato NLR Mi-1.2 confers the resistance to nematode and the constitutive mutant Mi-1.2T557S can also activate JA signaling. It has been previously reported that JA application reduces nematode reproduction on tomato plants (69).
Previous studies have identified some proteins that interact with NLRs to activate downstream immune signaling (49–53, 70–72). Interestingly, the CC domain of barley CNL MLA has been shown to interact with HvWRKY1 or HvWRKY2, and this interaction derepresses the inhibitory effect of HvWRKY1 or HvWRKY2 on plant defense (49). However, those protein-mediated immune signaling pathways are usually used by specific NLR. In this study, we show that different CNLs in the family of Solanaceae can interact with MED10b and consequently interfere with the interaction between MED10b and MED7. This suggests that derepression of repressor activities of MED10–MED7 in the middle module of Mediator complex might be a conserved mechanism used by diverse CNLs in the family of Solanaceae to activate immune pathway. The helper NLR NRCs can also interfere with MED10b and MED7 interaction; therefore, the helper NLRs might further amplify the expressions of JA signaling–dependent defense genes through derepressing the corepressor activities of MED10b/MED7.
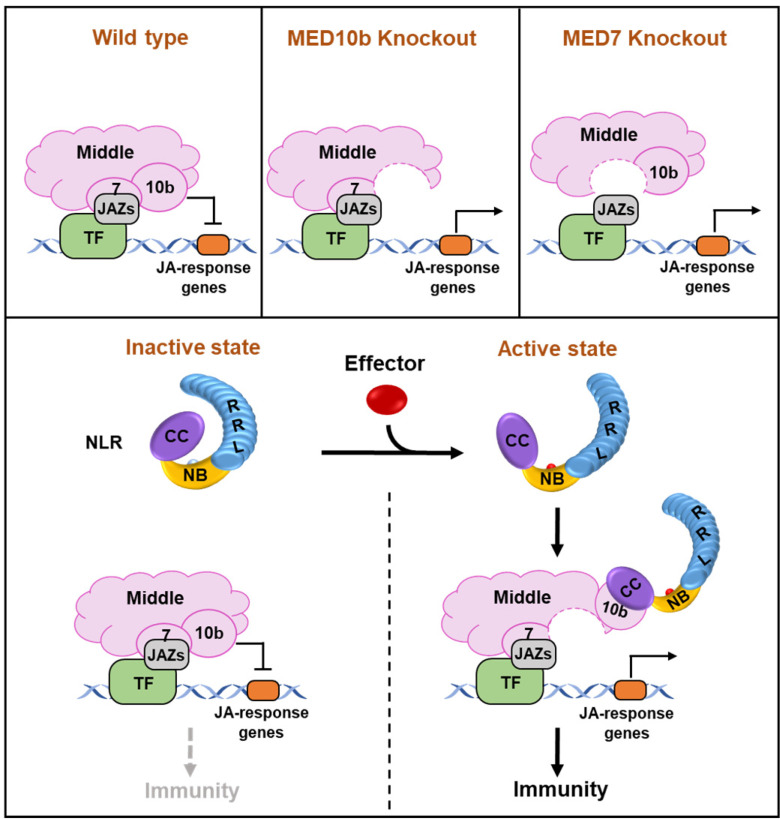
Based on these findings, we propose that the MED10b, MED7, and other subunits in the middle module of Mediator complex function as corepressors for JA defense signaling through MED7 and JAZ interaction (Fig. 7). Knockout of MED10b or MED7 derepresses the corepressor activity of the middle module on JA defense gene expression. Upon recognition of pathogen effectors, the CNLs switch from an inactive state into an active state and the CC domain of the active CNLs binds to MED10b. This binding interferes with the MED10b and MED7 association and derepresses the MED7–MED10b–JAZ-mediated repression on JA-dependent gene transcription, thus leading to the activation of plant immunity to different pathogens (Fig. 7).
Fig. 7.
A model describing the role of MED10b/MED7 in the middle module of Mediator complex as corepressors of JA defense signaling. Knockout MED10b or MED7 derepresses the corepressor activity of the middle module on JA defense gene expression. Upon recognition of pathogen effectors, the CC containing NLRs switch from an inactive state into an activate state and the CC domain of the activated NLRs binds to MED10b. This binding interferes with MED10b and MED7 association and derepresses the MED7–MED10b–JAZ-mediated repression on JA-dependent gene transcription, thus leading to the activation of plant immunity to different pathogens.
Materials and Methods
Details of the methodology used are provided in SI Appendix, Materials and Methods, and include plant material and growth conditions, plasmid construction, HR assay, Y2H assay, Y3H assay, VIGS assay, virus inoculation, total RNA extraction, RT-PCR, qRT-PCR, antibody production, coimmunoprecipitation assay, protein expression, protein purification, GST pull-down assay, BiFC assay, SLC complementation assay, CRISPR/Cas9 technology, inoculation of P. infestans assay, trypan blue staining assay, nematode inoculation assay, quantification, and statistical analysis. Primers used in this study are listed in SI Appendix, Table S1.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We sincerely thank Professor Jane E. Parker at Max Planck Institute for Plant Breeding Research, Germany, and Richard Kormelink at the Wageningen University & Research, the Netherlands, for their constructive comments and suggestions. We are also grateful to Professors David Baulcombe, Sophien Kamoun, and Jianming Zhou and Drs. Fan Zhang, Chenjie Fang, Hongyu Chen, and Xiaojiao Chen for providing resources. We thank Prof. Xin Shun Ding for critical reading of the manuscript and also thank our laboratory members for helpful discussions. This work is supported by the National Natural Science Foundation of China (31925032, 32220103008, and 31972241), the Funds from the Independent Innovation of Agricultural Science and Technology of Jiangsu Province [CX(22)2039], the Key Science and Technology Program of Hainan Province (ZDKJ2021007), and the “111” project.
Author contributions
Q.W. and X.T. designed research; Q.W., C.T., Z.C., S.H., X.Z., H.H., M.F., H.W., M.X., Y.Y., and H.C. performed research; J.L., D.S., G.A., J.L., H.Z., C.H., Z.Z., S.D., X.W., M.Z., and S.P.D.-K. contributed new reagents/analytic tools; Q.W., Y.X., and X.T. analyzed data; and Q.W., S.P.D.-K., and X.T. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Jones J. D., Dangl J. L., The plant immune system. Nature 444, 323–329 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Jones J. D., Vance R. E., Dangl J. L., Intracellular innate immune surveillance devices in plants and animals. Science 354, aaf6395 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Couto D., Zipfel C., Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 16, 537–552 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Zhou J. M., Zhang Y., Plant immunity: Danger perception and signaling. Cell 181, 978–989 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Yu X., Feng B. M., He P., Shan L. B., From chaos to harmony: Responses and signaling upon microbial pattern recognition. Annu. Rev. Phytopathol. 55, 109–137 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cui H., Tsuda K., Parker J. E., Effector-triggered immunity: From pathogen perception to robust defense. Annu. Rev. Plant Biol. 66, 487–511 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Caplan J., Padmanabhan M., Dinesh-Kumar S. P., Mant NB-LRR immune receptors: From recognition to transcriptional reprogramming. Cell Host Microbe 3, 126–135 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Peng Y., van Wersch R., Zhang Y., Convergent and divergent signaling in PAMP-triggered immunity and effector-triggered immunity. Mol. Plant Microbe Interact. 31, 403–409 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Tsuda K., Katagiri F., Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr. Opin. Plant Biol. 13, 459–465 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Ngou B. P. M., Ahn H. K., Ding P., Jones J. D. G., Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature 592, 110–115 (2021). [DOI] [PubMed] [Google Scholar]
- 11.Yuan M., et al. , Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 592, 105–109 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang X., Dodds P. N., Bernoux M., What do we know about NOD-like receptors in plant immunity? Annu. Rev. Phytopathol. 55, 205–229 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Takken F. L., Tameling W. I., To nibble at plant resistance proteins. Science 324, 744–746 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Monteiro F., Nishimura M. T., Structural, functional, and genomic diversity of plant NLR proteins: An evolved resource for rational engineering of plant immunity. Annu. Rev. Phytopathol. 56, 243–267 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Bi G., et al. , The ZAR1 resistosome is a calcium-permeable channel triggering plant immune signaling. Cell 184, 3528–3541.e3512 (2021). [DOI] [PubMed] [Google Scholar]
- 16.Wang J., et al. , Reconstitution and structure of a plant NLR resistosome conferring immunity. Science 364, 5868–5870 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Huang S. J., et al. , Identification and receptor mechanism of TIR-catalyzed small molecules in plant immunity. Science 377, 487 (2022). [DOI] [PubMed] [Google Scholar]
- 18.Jia A. L., et al. , TIR-catalyzed ADP-ribosylation reactions produce signaling molecules for plant immunity. Science 377, 488 (2022). [DOI] [PubMed] [Google Scholar]
- 19.Jacob P., et al. , Plant “helper” immune receptors are Ca(2+)-permeable nonselective cation channels. Science 373, 420–425 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kunstler A., Bacso R., Gullner G., Hafez Y. M., Kiraly L., Staying alive—Is cell death dispensable for plant disease resistance during the hypersensitive response? Physiol. Mol. Plant Pathol. 93, 75–84 (2016). [Google Scholar]
- 21.Chen X., et al. , A multilayered regulatory mechanism for the autoinhibition and activation of a plant CC-NB-LRR resistance protein with an extra N-terminal domain. New Phytol. 212, 161–175 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Kim Y. J., Bjorklund S., Li Y., Sayre M. H., Kornberg R. D., A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell 77, 599–608 (1994). [DOI] [PubMed] [Google Scholar]
- 23.Kornberg R. D., Mediator and the mechanism of transcriptional activation. Trends Biochem. Sci. 30, 235–239 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Davis J. A., Takagi Y., Kornberg R. D., Asturias F. A., Structure of the yeast RNA polymerase II holoenzyme: Mediator conformation and polymerase interaction. Mol. Cell 10, 409–415 (2002). [DOI] [PubMed] [Google Scholar]
- 25.Holstege F. C., et al. , Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95, 717–728 (1998). [DOI] [PubMed] [Google Scholar]
- 26.Allen B. L., Taatjes D. J., The mediator complex: A central integrator of transcription. Nat. Rev. Mol. Cell Biol. 16, 155–166 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kidd B. N., Cahill D. M., Manners J. M., Schenk P. M., Kazan K., Diverse roles of the mediator complex in plants. Semin. Cell Dev. Biol. 22, 741–748 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Buendia-Monreal M., Gillmor C. S., Mediator: A key regulator of plant development. Dev. Biol. 419, 7–18 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Browse J., Jasmonate passes muster: A receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 60, 183–205 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Wasternack C., Hause B., Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in annals of botany. Ann. Bot. 111, 1021–1058 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pauwels L., et al. , NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464, 788–791 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang F., et al. , Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling. Nature 525, 269–273 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kloek A. P., et al. , Resistance to pseudomonas syringae conferred by an arabidopsis thaliana coronatine-insensitive (coi1) mutation occurs through two distinct mechanisms. Plant J. 26, 509–522 (2001). [DOI] [PubMed] [Google Scholar]
- 34.Li R., et al. , Jasmonate-based warfare between the pathogenic intruder and host plant: Who wins?. J. Exp. Bot. 74, 1244–1257 (2023). [DOI] [PubMed] [Google Scholar]
- 35.Qiu J. H., et al. , Warm temperature compromises JA-regulated basal resistance to enhance magnaporthe oryzae infection in rice. Mol. Plant 15, 723–739 (2022). [DOI] [PubMed] [Google Scholar]
- 36.Ji M. F., et al. , Turnip mosaic virus P1 suppresses JA biosynthesis by degrading cpSRP54 that delivers AOCs onto the thylakoid membrane to facilitate viral infection. Plos Pathog. 17, e1010108 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He Y. Q., et al. , Jasmonic acid-mediated defense suppresses brassinosteroid-mediated susceptibility to rice black streaked dwarf virus infection in rice. New Phytol. 214, 388–399 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Hu J. L., et al. , Rice stripe virus suppresses jasmonic acid-mediated resistance by hijacking brassinosteroid signaling pathway in rice. Plos Pathog. 16, e1008801 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li L. L., et al. , A class of independently evolved transcriptional repressors in plant RNA viruses facilitates viral infection and vector feeding. Proc. Natl. Acad. Sci. U.S.A. 118, e2016673118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scholthof K. B., et al. , Top 10 plant viruses in molecular plant pathology. Mol. Plant Pathol. 12, 938–954 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pappu H. R., Jones R. A., Jain R. K., Global status of tospovirus epidemics in diverse cropping systems: Successes achieved and challenges ahead. Virus Res. 141, 219–236 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Oliver J. E., Whitfield A. E., The genus tospovirus: Emerging bunyaviruses that threaten food security. Annu. Rev. Virol. 3, 101–124 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Zhu M., et al. , The intracellular immune receptor Sw-5b confers broad-spectrum resistance to tospoviruses through recognition of a conserved 21-amino acid viral effector epitope. Plant Cell 29, 2214–2232 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu M., van Grinsven I. L., Kormelink R., Tao X. R., Paving the way to tospovirus infection: Multilined interplays with plant innate immunity. Annu. Rev. Phytopathol. 57, 41–62 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Brommonschenkel S. H., Frary A., Frary A., Tanksley S. D., The broad-spectrum tospovirus resistance gene Sw-5 of tomato is a homolog of the root-knot nematode resistance gene Mi. Mol. Plant Microbe Interact. 13, 1130–1138 (2000). [DOI] [PubMed] [Google Scholar]
- 46.Spassova M. I., et al. , The tomato gene Sw5 is a member of the coiled coil, nucleotide binding, leucine-rich repeat class of plant resistance genes and confers resistance to TSWV in tobacco. Mol. Breeding 7, 151–161 (2001). [Google Scholar]
- 47.Bombarely A., et al. , A draft genome sequence of nicotiana benthamiana to enhance molecular plant-microbe biology research. Mol. Plant Microbe Interact. 25, 1523–1530 (2012). [DOI] [PubMed] [Google Scholar]
- 48.Wu C. H., et al. , NLR network mediates immunity to diverse plant pathogens. Proc. Natl. Acad. Sci. U.S.A. 114, 8113–8118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shen Q. H., et al. , Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315, 1098–1103 (2007). [DOI] [PubMed] [Google Scholar]
- 50.Chang C., et al. , Barley MLA immune receptors directly interfere with antagonistically acting transcription factors to initiate disease resistance signaling. Plant Cell 25, 1158–1173 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu L., et al. , The coiled-coil and nucleotide binding domains of brown planthopper resistance 14 function in signaling and resistance against planthopper in rice. Plant Cell 29, 3157–3185 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhai K., et al. , RRM transcription factors interact with NLRs and regulate broad-spectrum blast resistance in rice. Mol. Cell 74, 996–1009.e1007 (2019). [DOI] [PubMed] [Google Scholar]
- 53.Inoue H., et al. , Blast resistance of CC-NB-LRR protein Pb1 is mediated by WRKY45 through protein-protein interaction. Proc. Natl. Acad. Sci. U.S.A. 110, 9577–9582 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Feng M., et al. , Rescue of tomato spotted wilt virus entirely from complementary DNA clones. Proc. Natl. Acad. Sci. U.S.A. 117, 1181–1190 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koschubs T., et al. , Preparation and topology of the mediator middle module. Nucleic Acids Res. 38, 3186–3195 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen H., et al. , Cytoplasmic and nuclear Sw-5b NLR act both independently and synergistically to confer full host defense against tospovirus infection. New Phytol. 231, 2262–2281 (2021). [DOI] [PubMed] [Google Scholar]
- 57.Bendahmane A., Kanyuka K., Baulcombe D. C., The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell 11, 781–792 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hajimorad M. R., Hill J. H., Rsv1-mediated resistance against soybean mosaic virus-N is hypersensitive response-independent at inoculation site, but has the potential to initiate a hypersensitive response-like mechanism. Mol. Plant Microbe Interact. 14, 587–598 (2001). [DOI] [PubMed] [Google Scholar]
- 59.Zhai Q., Li C., The plant mediator complex and its role in jasmonate signaling. J. Exp. Bot. 70, 3415–3424 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li J., et al. , A plant immune receptor adopts a two-step recognition mechanism to enhance viral effector perception. Mol. Plant 12, 248–262 (2019). [DOI] [PubMed] [Google Scholar]
- 61.You Y., Zhai Q., An C., Li C., LEUNIG_HOMOLOG mediates MYC2-dependent transcriptional activation in cooperation with the coactivators HAC1 and MED25. Plant Cell 31, 2187–2205 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen J., et al. , NLR surveillance of pathogen interference with hormone receptors induces immunity. Nature 613, 145–152 (2023). [DOI] [PubMed] [Google Scholar]
- 63.Gabriels S. H., et al. , An NB-LRR protein required for HR signalling mediated by both extra- and intracellular resistance proteins. Plant J. 50, 14–28 (2007). [DOI] [PubMed] [Google Scholar]
- 64.Leydon A. R., et al. , Repression by the arabidopsis TOPLESS corepressor requires association with the core mediator complex. Elife 10, e66739 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ito J., et al. , Auxin-dependent compositional change in mediator in ARF7- and ARF19-mediated transcription. Proc. Natl. Acad. Sci. U.S.A. 113, 6562–6567 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen X., et al. , Structures of the human mediator and mediator-bound preinitiation complex. Science 372, abg0635 (2021). [DOI] [PubMed] [Google Scholar]
- 67.Qi S., et al. , A new NLR gene for resistance to tomato spotted wilt virus in tomato (Solanum lycopersicum). Theor. Appl. Genet. 135, 1493–1509 (2022). [DOI] [PubMed] [Google Scholar]
- 68.Zhao Y., et al. , The phytophthora effector Avh94 manipulates host jasmonic acid signaling to promote infection. J. Integr. Plant Biol. 64, 2199–2210 (2022). [DOI] [PubMed] [Google Scholar]
- 69.Cooper W. R., Jia L., Goggin L., Effects of jasmonate-induced defenses on root-knot nematode infection of resistant and susceptible tomato cultivars. J. Chem. Ecol. 31, 1953–1967 (2005). [DOI] [PubMed] [Google Scholar]
- 70.Padmanabhan M. S., et al. , Novel positive regulatory role for the SPL6 transcription factor in the N TIR-NB-LRR receptor-mediated plant innate immunity. PLoS Pathog. 9, e1003235 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhu Z., et al. , Arabidopsis resistance protein SNC1 activates immune responses through association with a transcriptional corepressor. Proc. Natl. Acad. Sci. U.S.A. 107, 13960–13965 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.El Kasmi F., How activated NLRs induce anti-microbial defenses in plants. Biochem. Soc. Trans. 49, 2177–2188 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix.