Significance
The parathyroid gland (PTG), a tiny endocrine organ, is crucial in regulating calcium (Ca) homeostasis. Despite progress in tissue engineering in vitro for regenerative medicine, to make matured PTGs, sensitive to extracellular Ca concentration ([Ca]), from pluripotent stem cells remains difficult. Via single-step blastocyst complementation, we generated functional PTGs from mouse embryonic stem cells (mESCs) in parathyroid-deficient mice produced by CRISPR-Cas9-mediated zygote genome editing. The mESCs differentiated into mature PTGs that regulated PTH secretion in response to [Ca] and rescued aparathyroid hosts from neonatal death. These PTGs also on transplantation ameliorated postoperative hypoparathyroidism. Our results demonstrate a key concept in treatment of hypoparathyroidism.
Keywords: aparathyroid mouse, blastocyst complementation, embryonic stem cells, hypoparathyroid, parathyroid glands
Abstract
Patients with permanent hypoparathyroidism require lifelong replacement therapy to avoid life-threatening complications, The benefits of conventional treatment are limited, however. Transplanting a functional parathyroid gland (PTG) would yield better results. Parathyroid gland cells generated from pluripotent stem cells in vitro to date cannot mimic the physiological responses to extracellular calcium that are essential for calcium homeostasis. We thus hypothesized that blastocyst complementation (BC) could be a better strategy for generating functional PTG cells and compensating loss of parathyroid function. We here describe generation of fully functional PTGs from mouse embryonic stem cells (mESCs) with single-step BC. Using CRISPR-Cas9 knockout of Glial cells missing2 (Gcm2), we efficiently produced aparathyroid embryos for BC. In these embryos, mESCs differentiated into endocrinologically mature PTGs that rescued Gcm2−/− mice from neonatal death. The mESC-derived PTGs responded to extracellular calcium, restoring calcium homeostasis on transplantation into mice surgically rendered hypoparathyroid. We also successfully generated functional interspecies PTGs in Gcm2−/− rat neonates, an accomplishment with potential for future human PTG therapy using xenogeneic animal BC. Our results demonstrate that BC can produce functional endocrine organs and constitute a concept in treatment of hypoparathyroidism.
The parathyroid glands (PTGs) are small endocrine organs that regulate calcium (Ca) homeostasis. PTG chief cells express Ca-sensing receptor (CaSR), encoded by Casr, on their surface, permitting reaction to minute changes in extracellular Ca concentration ([Ca]). Responses include shifts in the secretion of parathyroid hormone (PTH); increases in PTH secretion yield rises in [Ca], and modulation of PTH secretion thus maintains plasma [Ca] within a very narrow range (Fig. 1A). Deficiency of PTG function can be acquired or, as with 22q11.2 deletion syndrome/“DiGeorge syndrome”, congenital. Neck surgery, the most common cause of acquired hypoparathyroidism, accounts for 75% of all instances of hypoparathyroidism (1). Surgical procedures with possible subsequent hypoparathyroidism include total thyroidectomy and radical neck dissection (1). The incidence of permanent hypoparathyroidism related to thyroidectomy ranges from 0 to 32%, although in most reports only a few percent (2). Patients with permanent hypoparathyroidism require lifelong replacement therapy and follow-up to avoid potentially life-threatening hypocalcemia.
Fig. 1.
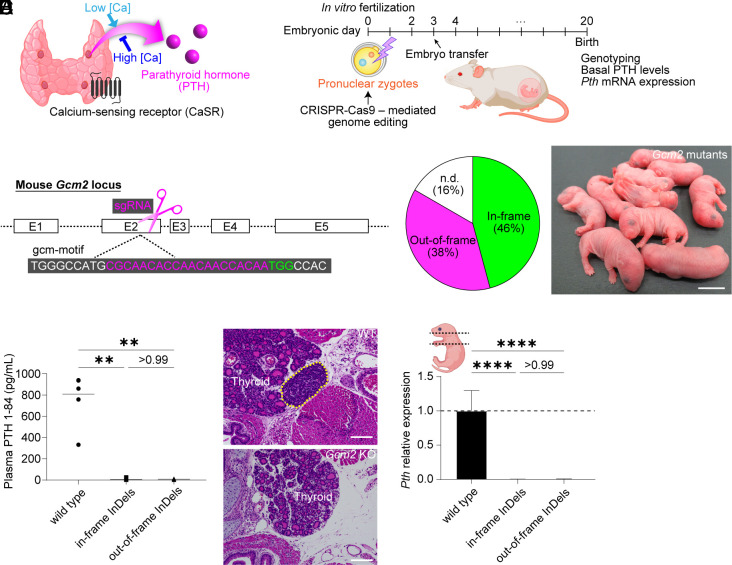
Generation and characterization of Gcm2 KO mice. (A) Schematic diagram of PTG function. (B) Zygote genome editing for generating PTG-deficient mice. (C) The target site of CRISPR-Cas9 to delete Gcm2 function. (D) Genotypes of Gcm2 mutant newborns. Those that could not be amplified by crude PCR with the mouse primer pairs were described as n.d. (E) The appearance of Gcm2 mutant pups just after birth. (Scale bar: 10 mm.) (F) Plasma PTH 1-84 levels in wild-type and Gcm2 mutant mice on B6D2F1 × C57BL/6N (BDF1B6) background at the neonatal stage. Wild type, n = 6. Gcm2 mutants with in-frame InDels, n = 11. Gcm2 mutants with out-of-frame InDels, n = 9. (G and H) Hematoxylin–eosin (HE) sections of wild-type (G) and Gcm2 mutant mice with in-frame deletion (H). The yellow dotted line indicates PTG. (Scale bars: 100 μm.) (I) Pth mRNA expression in wild-type and Gcm2 mutant neonates on BDF1B6 background. As shown in the illustration, all tissues from the neck to the mediastinum were sampled in each group. Wild type, n = 7. Gcm2 mutants with in-frame InDels, n = 5. Gcm2 mutants with out-of-frame InDels, n = 5. Graphed values, mean with s.d. **P ≤ 0.01 and ****P ≤ 0.0001, using the Kruskal–Wallis test with Dunn’s multiple comparisons test (F) and the one-way ANOVA test with Tukey’s multiple comparisons test (I).
Conventional management of hypoparathyroidism consists of Ca supplements and vitamin D analogs to keep plasma [Ca] within the low normal range. However, therapeutic benefits are limited. Even if normal [Ca] values are achieved with pharmaceutical supplementation, the quality of life of hypoparathyroidism patients is less than that of healthy persons (3). Why this is so is unknown; it is provisionally ascribed to deficiency of PTH itself. Furthermore, long-term treatment increases the risk of extraskeletal calcifications such as ureterolithiasis. Patients with permanent hypoparathyroidism are at high risk of chronic kidney disease because of hypercalciuria (4). Clinical use of full-length recombinant human PTH 1-84 (rhPTH 1-84) was recently approved in the United States and the European Union. It reduces the need for supplemental Ca and vitamin D analogs (5) and improves quality of life (6). However, whether rhPTH1-84 will replace conventional therapy is still unclear, as daily self-injection of rhPTH1-84 is inconvenient and its long-term safety is unknown (7).
The functional simplicity and independence of PTGs suit them well for transplantation. Subcutaneous grafting, an easy method widely used in clinical practice, confers further advantages. Accordingly, to generate functional PTGs from a patient’s own induced pluripotent stem cells (iPSCs) followed by PTG transplantation could be clinically significant. Recent PSC technology makes it possible to produce organoids or tissues in vitro and in vivo, and progress has been made toward induction of PTG-like cells from human PSCs in vitro (8–10), with increased concentrations of PTH in culture medium or expression of Pth at the mRNA level. In that work, however, response to extracellular [Ca] was not verified, and transplantation in animal models of hypoparathyroidism was not undertaken. Induction of endocrinologically mature PTG cells in vitro that regulate [Ca] in vivo thus remains unachieved.
We therefore attempted to generate functional PTGs from donor PSCs by blastocyst complementation (BC). In this method, an organ composed principally of PSC-derived cells is formed chimerically when PSCs are injected into preimplantation embryos of animals genetically engineered to lack ability to form an organ. After successfully generating rat pancreas in pancreatogenesis-disabled mice (11), in reciprocal experiments, we demonstrated that mouse PSC (mPSC)-derived islets generated in apancreatic rats normalized blood glucose in insulin-dependent diabetic mice for over one year without long-term immunosuppression, work constituting proof-of principle data for human transplantation medicine (12). Our group and others have since generated kidneys (13), lungs and bronchi (14), gametes (15), and vascular endothelial cells (16) using BC. Furthermore, BC-derived organ generation may be possible in pigs (17), a step fundamental to clinical application since pig and human organs are similar in size.
We here report generation via BC of functional PTGs, sensitive to [Ca], from mouse embryonic stem cells (mESCs). Our method, established in rodents, may yield approaches to the ultimate goal of human–animal BC-derived PTG generation.
Results
Efficient Generation of PTG-Deficient Mice Using CRISPR-Cas9.
During early embryogenesis, the PTG and thymic primordia emerge together from the third pharyngeal pouches (18). Glial cells missing2 (Gcm2), a mammalian homolog of the Drosophila gene Glial cells missing, is a master gene of early PTG organogenesis (19–21). Knockout (KO) of Gcm2 in mice leads to complete loss of PTGs (20, 21). Without Gcm2, differentiation of PTG precursors halts, followed by programmed cell death (22). Gcm2-KO PTG-deficient embryos thus offer a vacant developmental organ niche.
Embryos deficient in specific organs have generally been obtained by intercrossing heterozygous KO animals because homozygous KO animals do not survive to breeding age (17). Progress in genome editing has permitted generation of KO animals by directly targeting zygotes. We used CRISPR-Cas9-mediated genome editing to generate Gcm2 homozygous KO zygotes for BC (Fig. 1B).
Mouse Gcm2 has five exons. Exons 2 and 3 encode the entire DNA-binding domain (the gcm motif) (19). We considered this region the most critical portion of Gcm2. We therefore designed a single-guide RNA (sgRNA) on exon 2 of Gcm2, which encodes amino acid residues that are absolutely conserved among several species (19) (Fig. 1C). Hypoparathyroidism phenotypes of Gcm2 KO mice vary in severity depending on genetic background (21). Hence, we attempted to produce Gcm2 KO mice on three different genetic backgrounds, including C57BL/6N (hereafter B6), B6D2F1 × B6 (hereafter BDF1B6), and B6D2F1 (hereafter BDF1), intending to define their hypoparathyroidism phenotypes. The frequency of wild types and monoallelic Gcm2 mutants was approximately 5%, indicating that CRISPR-Cas9-mediated zygote genome editing was highly efficient (Fig. 1D, Table 1). While these Gcm2 mutant mice appeared normal at birth (Fig. 1E), they soon died, with mortality rates of >95% (SI Appendix, Table S1).
Table 1.
Gcm2 KO efficiency in mouse offspring after zygote genome editing using CRISPR-Cas9
| Strain | Embryos transferred | Living fetuses | Genotypes of Gcm2 mutants | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Monoallelic | Biallelic | Mosaic | n.d. | |||||||
| Out-of-frame | In-frame | Both | Out-of-frame | In-frame | Both | |||||
| B6 | 120 | 20 | 0 | 6 | 1 | 7 | 3 | 0 | 3 | 0 |
| BDF1B6 | 100 | 24 | 0 | 9 | 3 | 7 | 0 | 1 | 0 | 4 |
| BDF1 | 60 | 20 | 1 | 5 | 5 | 7 | 0 | 0 | 2 | 0 |
B6, BDF1B6, and BDF1, respectively, mean C57BL/6N, B6D2F1 × C57BL/6N, and B6D2F1. Those that could not be amplified by crude PCR with the mouse primer pairs were described as n.d.
We analyzed phenotypes of Gcm2 mutant pups to confirm that they lacked PTG cells. Having established that nearly 50% of the Gcm2-mutants harbored in-frame mutations, we compared phenotypes among wild-type pups, Gcm2 mutants with in-frame InDels, and Gcm2 mutants with out-of-frame InDels. Gcm2 mutants with both in-frame and out-of-frame InDels had lower levels of plasma PTH than wild-type pups (Fig. 1F and SI Appendix, Fig. S1 A and B). Histological examination of a Gcm2-mutant pup with an in-frame deletion found no PTGs in the neck (Fig. 1 G and H). Because in mice to identify PTGs is difficult, owing to their small size and variable anatomic distribution, we subjected DNA of tissues from neck to mediastinum to qPCR for Pth sequences. As predicted, Pth expression in Gcm2 mutants was below the lower limit of detection (Fig. 1I and SI Appendix, Fig. S1 C and D). In mice, PTGs can lie ectopically in thymus, resulting from incomplete separation during organogenesis (21). Levels of expression of Foxn1, associated with thymic epithelium, did not differ among pup cohorts, suggesting that the mediastinal tissues including the thymus were collected without individual differences (SI Appendix, Fig. S1 E–G). Unexpectedly, these phenotypes were the same in mutants, regardless of in-frame and out-of-frame InDel status. We infer that not only nonsense and frameshift mutations but also small in-frame mutations in the core domain of Gcm2 yielded a complete KO phenotype.
These data in sum demonstrate that CRISPR-Cas9-mediated zygote genome editing effectively generated Gcm2 KO PTG-deficient embryos suitable for BC.
Generation of mESC-Derived PTGs in Gcm2 KO Mice.
We next sought to generate PTGs in Gcm2 KO (Gcm2−/−) mice by single-step BC (Fig. 2A). We established mESCs with a knock-in of 2A-linked-tdTomato directly following the coding sequence of Pth in CAG-EGFP transgenic mESCs (12) [C57BL/6-Tg(CAG-EGFP)Pthtm1(P2A-tdTomato)ES1 (hereafter “Pth-tdTomato mESCs”)] (SI Appendix, Fig. S2A). Chimeric mice derived from Pth-tdTomato mESCs express GFP ubiquitously and tdTomato in PTG cells, permitting assessment of donor cell contribution throughout the body and visualization of PTGs, despite their small size, at various locations, orthotopic or ectopic. We chose Gcm2−/− embryos on an BDF1B6 background as recipients because of a high birth rate. Pth-tdTomato mESCs were microinjected directly into CRISPR-Cas9-treated embryos cultured till the blastocyst stage. The chimeric pups appeared normal at birth (Fig. 2 B and C). More than 90% of chimeric neonates survived into adulthood without any complications of hypoparathyroidism, such as tetany (Fig. 2D). Global donor cell chimerism of CD45-positive peripheral blood cells was assessed using flow cytometry. Extent of chimerism differed with genetic backgrounds of host embryos (Fig. 2E). Host embryos were genotyped by sorting GFP-negative blood cells.
Fig. 2.
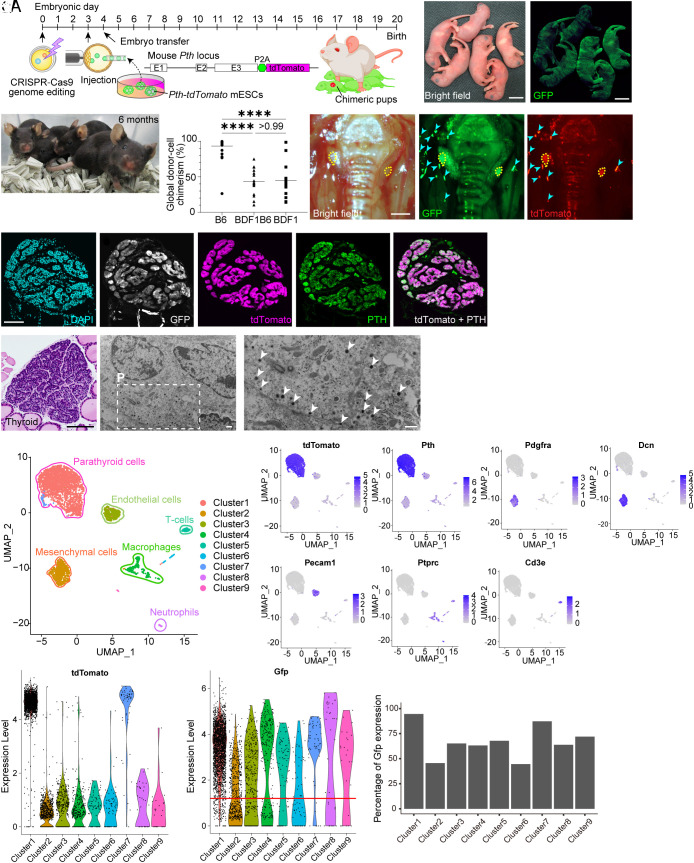
Generation of mESC-derived PTGs in Gcm2 KO mice by single-step BC. (A) Single-step BC for generating mESC-derived PTGs in Gcm2 KO mice. (B and C) Bright-field (B) and GFP-fluorescence (C) views of a Gcm2−/− mouse neonate complemented with mESCs. (Scale bars: 10 mm.) (D) Adult Gcm2−/− mice complemented with mESCs. (E) Global donor cell chimerism of CD45-positive peripheral blood cells in allogenic chimeras with three different genetic backgrounds. B6, n = 26. BDF1B6, n = 15. BDF1, n = 15. (F–H) Mouse ESC-derived PTGs generated in Gcm2−/− mice with small in-frame InDels. Yellow dotted lines indicate complemented PTGs. Note that cyan arrowheads are artifacts due to reflections. No tdTomato signaling was observed outside the PTGs in this field of view. The donor cell chimerism of peripheral blood cells was 30%. (Scale bars: 500 μm.) (I–M) Immunostaining of mESC-derived PTGs generated in Gcm2−/− mice with out-of-frame InDels. DAPI (cyan; I), GFP (white; J), tdTomato (magenta; K and M), PTH (green; L and M). (Scale bars: 100 μm.) (N) Hematoxylin/eosin–stained section of mESC-derived PTG generated in Gcm2−/− mouse with out-of-frame InDels. (Scale bar: 100 μm.) (O and P) Electron micrographs of mESC-derived PTGs. White arrowheads, secretory granules. (Scale bars: 500 nm.) (Q–AA) Single-cell transcriptome analysis. (Q) Uniform Manifold Approximation and Projection (UMAP) of mESC-derived PTGs. Each cluster was annotated as follows: Clusters 1 and 7; PTG cells. Cluster 2; mesenchymal cells. Cluster 3; endothelial cells. Cluster 4; macrophages. Cluster 5; T cells. Cluster 6; NK cells. Cluster 8; neutrophils. Cluster 9; vascular smooth muscle cells. (R–X) Feature plots on the UMAP showing tdTomato (R), Pth (S), Pdgfra (T), Dcn (U), Pecam1 (V), Ptprc (W), and Cd3e (X) gene expression. (Y) Violin plots displaying tdTomato expression. (Z) Violin plots displaying Gfp expression. Red line, threshold used as cutoff for Gfp expression. (AA) Percentage of Gfp expression in each cluster, based on threshold. ****P ≤ 0.0001, using the Kruskal–Wallis test with Dunn’s multiple comparisons test (E).
On fluorescence stereomicroscopy, PTGs generated in Gcm2−/− mice expressed both GFP and tdTomato (Fig. 2 F–H and SI Appendix, Fig. S2B). This suggests that the vacant developmental organ niche in Gcm2−/− mice was occupied by donor mESC-derived cells (11). Immunohistological analysis revealed that GFP+tdTomato+ PTGs diffusely expressed PTH (Fig. 2 I–M). Usual light microscopy of formalin-fixed, paraffin-embedded PTGs stained with hematoxylin and eosin showed that PTGs of chimeras consisted of clusters of chief cells as encountered in normal wild-type mice (23) (Fig. 2N). On ultrastructural study, these cells’ cytoplasm contained numerous secretory granules 150 to 180 nm in diameter, consistent with synthesis and storage of PTH in chief cells (Fig. 2 O and P).
Parathyroid glands contain blood vessels and supporting tissues in addition to PTH-releasing chief cells. We evaluated whether Gcm2 KO affects chimerism of these minor cell lineages by single-cell RNA-seq with 10× Genomics (Fig. 2 Q–AA and SI Appendix, Fig. S3 A–E). Based on differentially expressed genes (DEGs) specific to each cell lineage (Dataset S1), we identified and manually annotated nine major clusters (Fig. 2Q). Mouse ESC-derived PTGs are composed of tdTomato-positive Pth-positive parathyroid chief cells (Fig. 2 R and S); platelet-derived growth factor, alpha (Pdgfa)-positive and decorin (Dcn)-positive mesenchymal cells (Fig. 2 T and U); platelet/endothelial cell adhesion molecule 1 (Pecam1)-positive endothelial cells (Fig. 2V); peripheral blood cells including protein tyrosine phosphatase, receptor type, C (Ptprc)-positive macrophages (Fig. 2W); and CD3 antigen, epsilon polypeptide (Cd3e)-positive T cells (Fig. 2X). The Pth-positive and tdTomato-positive minor fraction was identified as cluster 7. DEGs seem to differ slightly between cluster 1 and cluster 7, although functional differences are unknown (SI Appendix, Fig. S3B). Parathyroid chief cells with high tdTomato expression (Fig. 2Y), which were clearly 100% donor mESC-derived on immunohistological analysis, expressed Gfp strongly at 94.6% (Fig. 2 Z and AA and SI Appendix, Fig. S3 C–E and Table S2). In contrast, percentages of Gfp expression by endothelial cells (65.2%) and mesenchymal cells (45.6%) were similar to percentage global donor cell chimerism of CD45-positive peripheral blood cells as determined by flow cytometry (50 to 70%).
These data in sum demonstrate that mESCs differentiated into histologically normal PTGs, rescuing PTG-deficient hosts from neonatal death, and that Gcm2-targeted single-step BC replaced only parathyroid chief cells, while other cell lineages were derived from both donor mESCs and host Gcm2 KO embryos.
In Vivo and Ex Vivo Functional Assessment of mESC-Derived PTGs.
We next examined endocrinologic function (PTH and [Ca] regulation) of mESC-derived PTGs generated in Gcm2−/− mice. We established a Pth-tdTomato reporter B6 mouse strain from Pth-tdTomato mESCs through classical germline-transmitted chimera formation (C57BL/6-Pthtm1(P2A-tdTomato)) to ensure uniformity of genetic background between cohorts. Control mice (C57BL/6-Pthtm1(P2A-tdTomato)/+) underwent parathyroidectomy, yielding a model of postsurgical hypoparathyroidism. As they did not undergo thymectomy, intrathymic ectopic PTG tissue may have been present. However, plasma [Ca] values in all mice that underwent parathyroidectomy indicated hypoparathyroidism (SI Appendix, Fig. S4A).
Plasma [Ca] values in Gcm2−/− mice complemented with mESCs were similar to those in control mice and significantly higher than those in postparathyroidectomy mice (P ≤ 0.001) (Fig. 3A). Basal PTH values were similar in chimeras and controls and were higher than in postparathyroidectomy mice, despite no statistically significant differences (P = 0.1137) (Fig. 3B). We speculate that no statistically significant difference was found because basal PTH levels in these mice were near the detection limit of the ELISA kit used for assessment. We next sought to examine the potential to secrete additional PTH in hypocalcemia. Sodium bicarbonate (NaHCO3) infusion tests are useful when estimating PTH-releasing capacity in clinical practice (24). Blood pH and bicarbonate ion concentrations increase following NaHCO3 injection, after which plasma [Ca] decreases. Healthy patients exhibit a marked and short-lasting rise in plasma PTH in response to the fall in plasma [Ca]. In contrast, plasma PTH rises slightly or not at all in patients with hypoparathyroidism. We found this approach to be an effective PTH stimulation test in mice (Fig. 3C and SI Appendix, Fig. S4 B and C). In wild-type B6 mice, intraperitoneal administration of NaHCO3 induced a rapid decrease in plasma [Ca] that was followed by a marked rise in plasma PTH levels maintained for >20 to 30 min. Postparathyroidectomy mice failed to respond to NaHCO3 infusion (SI Appendix, Fig. S4D), but Gcm2−/− mice complemented with mESCs responded well, secreting PTH in response to hypocalcemia (Fig. 3D). This indicates that mESC-derived PTGs were functional in vivo.
Fig. 3.
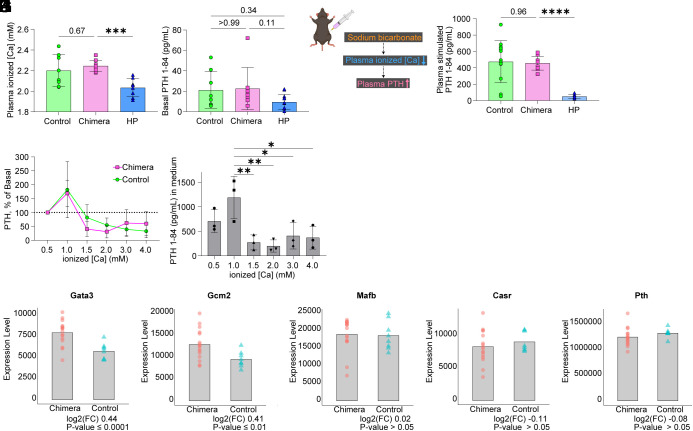
In vivo and ex vitro functional assessment of mESC-derived PTGs. (A) Plasma [Ca] in controls, Gcm2−/− mice complemented with mESCs (chimeras), and postparathyroidectomy mice. “HP” on the X axis means postparathyroidectomy mice with confirmed hypoparathyroidism. Controls, n = 8. Chimeras, n = 9. HP, n = 8. (B) Plasma basal PTH 1-84 levels in controls, Gcm2−/− mice complemented with mESCs, and postparathyroidectomy mice. Controls, n = 8. Chimeras, n = 8. HP, n = 12. (C) Schematic of sodium bicarbonate infusion test in mice. (D) Plasma stimulated PTH 1-84 levels in controls, chimeric mice, and postparathyroidectomy mice. Controls, n = 15. Chimeras, n = 9. HP, n = 8. (E) Ex vivo PTH release at different [Ca]. The PTGs were transferred from low to high calcium concentrations. Peak PTH secretion at 1.0 mM was set to 100%. Controls, n = 5. Chimeras, n = 3. (F) Ex vivo PTH 1-84 concentrations in the culture medium of PTGs generated in Gcm2−/− mice. n = 3. Same samples as (E). (G–K) Bulk RNA-seq of PTG cells. Differentially expressed gene (DEG) analysis using DESeq2. log2(FC) means fold changes of chimera gene counts against control (C57BL/6-Pthtm1(P2A-tdTomato)/+ mouse). *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001 using the Brown–Forsythe ANOVA test with Dunnett’s T3 multiple comparisons test (AD) and Kruskal–Wallis test with Dunn’s multiple comparisons test (B) and one-way ANOVA test with Šídák's multiple comparisons test (F).
We next performed detailed ex vivo assays of the regulation of PTH secretion in response to [Ca]. Excised PTGs secrete PTH in response to variation in external [Ca] ex vivo (25). The mESC-derived PTGs generated in Gcm2−/− mice, like PTGs in control mice, showed drastic changes in PTH secretion in response to [Ca] (Fig. 3E). Of note is that normal to high [Ca] suppressed PTH secretion (Fig. 3F). These findings demonstrate that mESC-derived PTGs regulate PTH release in response to external [Ca], in particular, that feedback mechanisms suppress PTH secretion when [Ca] is below normal.
To examine the gene expression profile essential for the development and maintenance of PTGs, we performed bulk RNA-seq (Fig. 3 G–K and SI Appendix, Fig. S4 E–I). We collected tdTomato-positive PTG cells under fluorescence microscopy, using micromanipulation to avoid contamination by neighboring cells such as thyroid or GFP-negative non-PTG cells. GATA-binding protein 3 (Gata3), Gcm2, and v-maf musculoaponeurotic fibrosarcoma oncogene homolog B (MafB) interact with each other and regulate development of PTGs during morphogenesis. They continue to be expressed in postnatal PTGs, where they are involved in transcriptional regulation of Pth (26, 27). Differential gene expression analysis between PTG cells generated in Gcm2−/− mice and PTG cells of control mice was performed using the DESeq2 algorithm (28). We found that mESC-derived PTG cells expressed Gata3, Gcm2, MafB, Casr, and Pth at levels comparable with or slightly higher than those in control mice (Fig. 3 G–K). Results for other genes expressed in PTGs, including Paired box 1 (Pax1), Pax9, T-box transcription factor 1 (Tbx1), and Sine oculis homeobox 1 (Six1) (18) were like those in controls (SI Appendix, Fig. S4 E–I). These data suggest that the mESC-derived PTGs were mature endocrine organs (adult PTGs) that could regulate PTH secretion in response to extracellular [Ca].
Therapeutic Potential of mESC-Derived PTGs in Transplantation and in Generation of mESC-Derived PTGs in Gcm2−/− Rat Neonates.
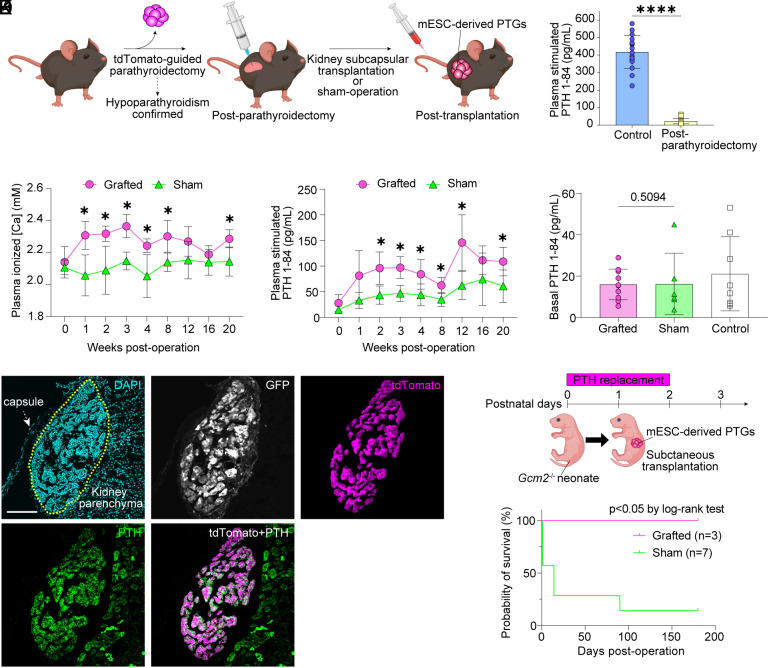
We assessed whether mESC-derived PTGs could serve as allografts. Neck surgery is the predominant cause of hypoparathyroidism in clinical practice, and to relieve iatrogenic hypoparathyroidism of this sort would be of value. We thus examined whether mESC-derived PTGs transplanted beneath the renal capsule could rescue postparathyroidectomy mice from hypoparathyroidism. After decreased PTH release on NaHCO3 stimulation confirmed hypoparathyroidism in postparathyroidectomy mice, we transplanted beneath the renal capsule mESC-derived PTGs from Gcm2−/− mice (Fig. 4 A and B). Blood samples were obtained before and after transplantation to assess Ca metabolism. Plasma [Ca] was significantly higher in the grafted group (Fig. 4C). NaHCO3 infusion testing showed that induction of PTH release was substantially greater in the grafted group than in a sham-operated group, indicating that the grafts responded to low plasma [Ca] levels induced by NaHCO3 injection (Fig. 4D). Basal PTH secretion did not differ significantly between grafted and sham-operated mice (Fig. 4E). Histologic findings demonstrated that GFP+ tdTomato+ grafts maintain expression of PTH (Fig. 4 F–J and SI Appendix, Fig. S5 A–D). Transcriptome analysis of tdTomato-positive cells in the grafts found that expression of genes involved in PTG maintenance and PTH secretion was the same as that in primary PTG cells (SI Appendix, Fig. S5 E–J), suggesting that grafted PTG cells retain their endocrinological function despite ectopic transplantation. All postparathyroidectomy mice had tdTomato-positive tissues in their thymus, indicating residual ectopic PTGs. Such mice thus might have not complete but partial hypoparathyroidism, as PTH secretion from PTG cells in the thymus is well established (21). We thus subjected Gcm2 KO neonates to subcutaneous transplantation of mESC-derived PTGs, examining how this affected complete hypoparathyroidism (Fig. 4K). Gcm2 B6-background KO neonates, a 100% lethal genotype, were kept alive by multiple daily injections of teriparatide, i.e., rhPTH (1-34). On the first postnatal day, mESC-derived PTGs were transplanted into dorsal subcutaneous tissue. After cessation of PTH replacement therapy, all mice in the grafted group survived, whereas all mice in the sham-operated group died but one (Fig. 4L). This demonstrated that in Gcm2 KO neonates, mESC-derived PTGs transplanted subcutaneously eliminated near-uniform mortality.
Fig. 4.
Transplantation of mESC-derived PTGs into mice modeling postsurgical hypoparathyroidism. (A) Schematic diagram of kidney subcapsular transplantation. (B) Plasma stimulated PTH 1-84 levels after sodium bicarbonate infusion in control mice (C57BL/6-Pthtm1(P2A-tdTomato) mice) and postparathyroidectomy mice used for this transplantation experiment. Control, n = 17. Postparathyroidectomy, n = 16. (C) Plasma [Ca] in grafted and sham-operated mice. Grafted, n = 7. Sham-operated, n = 5. (D) Plasma stimulated PTH 1-84 levels after sodium bicarbonate infusion in grafted and sham-operated mice. Grafted, n = 7. Sham-operated, n = 5. Same samples as in (C). (E) Basal blood levels of PTH 1-84 in grafted and sham-operated mice. Grafted, n = 10. Sham-operated, n = 6. Basal levels of PTH 1-84 in C57BL/6-Pthtm1(P2A-tdTomato) mice are provided as control (same samples as in Fig. 3B). (F–J) Immunostaining of grafted PTGs on posttransplantation day 14. Yellow dotted lines indicate the grafts. DAPI (cyan; F), GFP (white; G), tdTomato (magenta; H and J), and PTH (green; I and J). (Scale bars: 100 μm.) (K) Subcutaneous transplantation of mESC-derived PTGs into Gcm2 KO neonates on B6 backgrounds. (L) Survival curve of Gcm2 KO neonates. Grafted, n = 3. Sham-operated, n = 7. *P ≤ 0.05 and ****P ≤ 0.0001 using the Mann–Whitney U test (B and E), multiple t test (FDR 1%) (C and D), and log-rank test (L).
Last, to determine whether interspecific BC for clinical application holds for PTGs, we generated mESC-derived PTGs in PTG-deficient rats. The gcm motif encoded by Gcm2 is strongly conserved among species, including rodents and humans (19, 29). This suggested that to obtain PTG-deficient animals by deleting Gcm2 could succeed in animals other than mice. As we knew of no PTG-deficient rat lines, we generated PTG-deficient embryos of Wistar rats by CRISPR-Cas9 gene editing in zygotes (30) and complemented them with mESCs, using the single-step BC strategy that we had employed in mice (SI Appendix, Fig. S6 A and B and Table S3). The phenotypes of neonatal Gcm2−/− rats were evaluated. The Gcm2−/− rats had levels of plasma PTH significantly lower than those of wild-type pups (P ≤ 0.0001) (SI Appendix, Fig. S6C). Quantitative PCR revealed that Gcm2−/− rats expressed too little Pth to be detected, far less than wild-type pups (SI Appendix, Fig. S6D). Histological examination also detected no PTGs in Gcm2−/− rats (SI Appendix, Fig. S6 E and F). In sum, Gcm2−/− rats obtained by zygote genome editing lacked PTGs completely and were hypoparathyroid.
To generate xenogeneic PTGs in Gcm2−/− rats, we injected mESCs into Gcm2 KO embryos at the blastocyst stage. Unlike embryos of mice and humans, rat embryos cultured in vitro from one-cell to blastocyst stage die before birth when implanted into surrogate mothers (31, 32). We therefore transplanted gene-edited one- or two-cell stage rat embryos into pseudopregnant females for development in vivo. The embryos were then retrieved 72 h later, microinjected with mESCs, and transferred into different pseudopregnant females. In total, 100 embryos were transferred, resulting in three live-born interspecies chimeras. Immunofluorescence staining demonstrated that tdTomato-positive PTG cells expressed PTH (SI Appendix, Fig. S6 G–N). Furthermore, transcriptome analysis demonstrated that mESC-derived PTGs generated in Gcm2−/− rats expressed the transcription factors that regulate PTG development and Pth expression (SI Appendix, Fig. S6 O–V). However, all three Gcm2−/− rats complemented with mESC-derived PTGs had large umbilical hernias and died soon after birth. Since long-term survival was not achieved, further research is required for generation of PTGs by interspecies single-step BC.
Discussion
Despite many and various attempts, generation of functional PTGs from PSCs has not hitherto been achieved. We generated fully functional PTGs from mESCs. For animal-developed human PTG transplantation via BC to succeed, PTGs generated from human iPSCs must in vivo regulate PTH secretion in response to extracellular [Ca]. Thus PTG cells must 1) express CaSR, 2) maintain baseline PTH secretion, and 3) if extracellular [Ca] falls immediately secrete PTH in amounts adequate to restore normocalcemia. The PTG that we generated met all these criteria.
We genetically edited zygotes, disrupting Gcm2 via CRISPR-Cas9, to generate Gcm2 KO embryos for BC. The InDel mutations thus induced are unpredictable; nonhomologous end joining occasionally yields in-frame mutations with inadequate KO of the target gene. Although our work often produced in-frame mutations in Gcm2, with small InDels these generated severely PTH-deficient phenotypes like those generated by Gcm2 frame-shift mutations. That we targeted CRISPR-Cas9 to a core domain of Gcm2 may explain this. Overall efficiency in obtaining Gcm2−/− embryos via CRISPR-Cas9 was as high as 80%. Single-step BC technique and targeting of critical domains thus minimize animal use via more efficient KO embryo generation.
We further demonstrated that the generated PTGs were fully functional and mature endocrine organs in respect of immunohistological findings, electron microscopic findings, functional investigation results, and transcriptome analysis results. In vivo and ex vivo secretion studies revealed that mESC-derived PTGs regulate PTH secretion in response to environmental [Ca]. They do not secrete PTH autonomously but strictly maintain [Ca] homeostasis, crucial in mature PTGs. Our work demonstrated that mESC-derived PTGs transplanted under the renal capsule ameliorated postoperative hypoparathyroidism. Functional assessment of grafted PTGs is often challenging because PTH levels in the systemic circulation can be affected by residual normal parathyroid tissue (2). Because some PTG tissue may persist within the thymus of mice after parathyroidectomy, we backed demonstration of graft function by immunohistochemical studies and by transcriptome analysis as well as by hormone measurement in peripheral blood. The engrafted mESC-derived PTGs expressed CaSR and regulated plasma PTH levels despite ectopy. They also rescued Gcm2-deficient neonates from death, conferring prolonged survival. These data clearly establish that PSC-derived PTGs can function after ectopic transplantation.
One of the most critical concerns in the transplantation of organs established via BC is contamination of the chimeric tissues in the grafts. Our single-cell RNA-seq results show that blood vessels and supporting tissues derived from a mixture of donor mESCs and host embryos lie within mESC-derived PTGs. Our previous work with interspecies BC has suggested that contamination of such residual cells from a nonhost species is eliminated by the host immune system after transplantation and has no negative effect on the prognosis of grafts (12). In the present study, grafted mESC-derived PTGs functioned well without any immunosuppressants, suggesting that contamination of host embryo-derived cells is not a major problem.
Gcm2 is highly conserved and is involved in PTG development across species. Using interspecies BC, we generated Gcm2−/− rat neonates that had mESC-derived PTGs. Immunohistologic and transcriptome analysis data indicated that the mESC-derived PTGs generated in rat neonates were functional. Nonetheless, PTG-deficient rats were severely malformed and did not survive into adulthood. In addition, the birth rate of mouse–rat chimeras was extremely low. The causes of low birth rate and of survival failure in interspecies chimeras generated by single-step BC remain unclear. Success may yet be achieved using different rat embryo culture conditions and injection timing, as well as improved donor mESCs (e.g., different mESC lines or mESC culture conditions).
To realize the clinical application of human PTGs established by xenogeneic BC, generation of human PSC-derived interspecies chimeras is crucial. Low chimerism between injected human PSCs and animal embryos is at present a major issue for human–animal BC (33). We and others are attempting to solve this problem. The prevention of apoptosis in donor human PSCs might be one strategy to improve human–animal chimerism (34). We have proposed that to delete Insulin-like growth factor 1 (Igf1r) of host embryos may allow gradual increase of donor cell chimerism (35). This approach also has the potential to improve human PSC chimerism in interspecies chimeras. Ethical issues of human–animal BC to be fully discussed include issues of animal welfare and concerns about the possibility of “human-like” chimeric animals. We may need genetic modification of donor human PSCs to prevent the development of human-derived central nervous system and gonads within the chimeric animals.
In conclusion, our work generated functional PTGs responsive to peripheral [Ca] levels from mESCs. Our results add significantly to knowledge concerning single-step BC and suggest considerations for efficient organ complementation. Of importance, although improvement is needed, is that our findings in interspecies organ generation demonstrate substantial potential for clinical applications and represent an important demonstration that PSC-derived functional PTGs can be generated in a xenogeneic environment.
Materials and Methods
Animals.
C57BL/6NCrSlc, B6D2F1/Slc, DBA/2CrSlc, Slc:ICR, transgenic C57BL/6-Tg(CAG-EGFP) mice were purchased from SLC Japan. Crlj:WI rats were purchased from Charles River Laboratories Japan. All experiments were performed in accordance with the animal care and use committee guidelines of the Institute of Medical Science, University of Tokyo, and the University of Tsukuba. All mice and rats were housed in an environmentally controlled room with a 12-h dark/12-h light cycle at 23 ± 2 °C and humidity of 50 ± 10% and were given ad libitum access to a laboratory diet (CE-2; CLEA Japan, Tokyo, Japan) and sterilized water.
Cell Culture.
Mouse ESCs were cultured on mitomycin-C-treated mouse embryonic fibroblasts (MEFs) in six-well tissue culture plates (92006, TPP, Trasadingen, Switzerland). Mouse ESC maintenance medium was KnockOut™ Dulbecco’s modified Eagle medium [10829018, Thermo Fisher Scientific (Thermo)] supplemented with 20% HyClone™ fetal bovine plasma (SH30070.03, Cytiva), 1% 2-mercaptoethanol (ES-007-E, Merck), 1% nonessential amino acids (TMS-001-C, Merck), 1% nucleosides (ES-008-D, Merck), 1% L-glutamine (TMS-002-C, Merck), 100 U/mL penicillin-streptomycin (15140-122, Thermo), and 1,000 U/mL mouse leukemia inhibitory factor (ESG1107, Merck). Before use, 1 μM mirdametinib [PD0325901/162-25291, FUJIFILM Wako (Wako)] and 3 μM laduviglusib (CHIR-99021/TB4423-GMP, Bio-Techne) were added to mESC maintenance medium. Cells were passaged every 2 to 3 d.
For cryopreservation, CELLBANKER 1plus (CB023, Takara) was used. For thawing, 1 mL of phosphate-buffered saline (PBS, Wako, 048-29805) was added to a cryopreserved cell vial. Mouse ESCs were quickly dispersed by pipetting and transferred to 15-mL centrifuge tubes containing 9 mL of PBS. After centrifugation, the cells were suspended in mESC maintenance medium containing 1 μM PD0325901 and 3 μM CHIR-99021. The cell suspension was then seeded onto MEF feeder cells. The thawed mESCs were used for injection after more than one passage.
In Vitro Fertilization (IVF).
In mice, 8- to 10-wk-old female mice (C57BL/6N or B6D2F1) were superovulated by intraperitoneal injection with 7.5 IU of pregnant mare plasma gonadotropin (PMSG; ASKA Animal Health, Tokyo, Japan), followed by injection of 5 IU of human chorionic gonadotropin (hCG; ASKA Animal Health) 48 h later. The day before IVF, droplets of human tubal fluid medium (HTF; ARK Resource) were covered with paraffin oil (Nacalai Tesque) and placed in an incubator at 37 °C under 5% CO2 in air to allow equilibration. Cauda epididymis sperms from male mice (C57BL/6N or DBA/2) were placed into an HTF droplet and dispersed for capacitation for 1 h. Isolated oviduct ampullae from superovulated females were placed into paraffin oil in an insemination dish. Cumulus–oöcyte complexes (COCs) were pulled from cut oviduct ampullae with a 27-gauge hypodermic needle (SS-010F2713, TERUMO)—not using suction with an attached syringe—and placed into HTF droplets. Incubated sperms were then transferred into COCs droplets for insemination at a density of 1.0 to 2.0 × 105/mL. After incubation for about 5 h at 37 °C under 5% CO2 in air, oöcytes were washed and cultured in potassium-supplemented simplex optimized medium (KSOM) with amino acids (AAs) (KSOM-AA medium/MR020P, Merck) until electroporation. Electroporation was done 1 to 2 h after washing.
For rats, in vitro fertilization was conducted as described (30). In brief, 3-wk-old immature Crlj:WI females were injected intraperitoneally with 40 µg of [des-Gly10, D-Ala6]-luteinizing hormone-releasing hormone ethylamide acetate salt hydrate (L4513, Sigma-Aldrich) dissolved in 200 μL saline. After 52 to 53 h, the rats were injected with 300 IU/kg PMSG and 100 μL anti-inhibin plasma (Central Research, Tokyo, Japan), 48 to 50 h before injection of 300 IU/kg hCG. COCs were collected from oviducts 16 to 17 h after hCG injection (see below). The day before IVF, HTF droplets covered with paraffin oil were placed in an incubator at 37 °C under 5% CO2 in air to allow equilibration. Before COC collection, cauda epididymis sperms from Crlj:WI males were placed in a 1 mL-HTF droplet and dispersed for capacitation for 1 h. The incubated sperms in the drops were then employed for insemination at a density of 1.0 to 2.0 × 106/mL. The oviduct ampullae from superovulated females were isolated under anesthesia using intraperitoneally injected medetomidine (0.375 mg/kg, Zenoaq), midazolam (2 mg/kg, Sandoz), and butorphanol (2.5 mg/kg, Meiji Seika). COCs were pulled from cut oviduct ampullae with a 27-gauge hypodermic needle (SS-010F2713, TERUMO)—not using suction with an attached syringe—and placed into the droplets containing sperms. After incubation for 6 h at 37 °C under 5% CO2 in air, the inseminated oöcytes were washed and cultured in rat KSOM medium (ARK Resource) until electroporation. Electroporation was done 1 to 2 h after washing.
Generation of tdTomato–Knock-in mESCs at the Pth Locus (SI Appendix, Fig. S2A).
The original targeting vector was constructed from a OSDupDel. Neo Vector (MES3974, Open Biosystems), which contains the neomycin resistance cassette (neo) flanked by loxP, using the In-Fusion HD Cloning Kit (639650, Takara). The neomycin resistance cassette of OSDupDel. Neo Vector was replaced with an EF1-promoter-E2-crimson-T2A-neo cassette (hereinafter, OS_DupDel_EF1_E2-crimson_T2A_NeoR_rGpA vector). EF1-promoter and E2-crimson were from CSII-EF-MCS-IRES2-Venus (RDB04384, RIKEN BRC) and pE2-Crimson Vector (632553, Takara). The OS_DupDel_EF1_E2-crimson_T2A_NeoR_rGpA vector was digested with BamHI and NotI. The 1.2 kb 3’ homology arm of Pth (starting 23 bp downstream from the stop codon of the CDS) was then inserted by In-Fusion reaction. The resulting vector was digested with NheI and ClaI. The 1-kb 5′ homology arm of Pth (starting 908 bp upstream from the stop codon of the CDS) and P2A-tdTomato-rabbit globin pA were then inserted. P2A-tdTomato was constructed by adding P2A to tdTomato derived from ptdTomato-N1 (632532, Takara) with primer extension.
The final targeting vector was linearized with SwaI before electroporation (pulse voltage 1,400 V, pulse width 10 ms, pulse number 3). The targeting linearized vector (5 μg) was introduced into 3 × 106 GFP-expressing mESCs (SGE2) (12) using the Neon® Transfection System (Thermo). After 48 h, the electroporated mESCs underwent positive selection with 400 μg/mL G418 (108321-42-2, Merck). After a further 48 h, they underwent negative selection with 1 μM ganciclovir (21800AMX10277, Tanabe, Osaka, Japan). Positive clones were expanded, and the correct homologous recombination event at the Pth locus was confirmed by PCR. The junction region on the 5′ side was identified with the primers 5′-CTGAGTCATCCTTCTAGCCTAAGAC-3′ (primer 1, forward; upstream of the 5′ homology arm) and 5′-TCCTCCACGTCACCTGCTTG-3′ (primer 2, reverse; P2A-specific). The junction region on the 3′ side was identified with the primers 5′-CCTGCCGAGAAAGTATCCATCA-3′ (primer 3, forward; neo-specific) and 5′-AGTGGCTGGAAGTGAAAACCT-3′ (primer 4, reverse; downstream of the 3′ homology arm). The E2-crimson-T2A-neo cassette was removed by Cre-loxP treatment.
Generation of Chimeric Mice and Germline Transmission of a Targeted Mutation at the Pth Locus.
Pth-tdTomato reporter B6 mice (C57BL/6N-Pthtm1(P2A-tdTomato)) were established from Pth-tdTomato mESCs through classical germline-transmitted chimera formation. Recombinant mESCs were injected into blastocysts derived from C57BL/6N mice. The chimeric embryos were transferred into the uteri of pseudopregnant females. Chimeric males with >90% chimerism were mated to C57BL/6-Tg(CAG-EGFP) females. Germline transmission was identified by PCR with the P2A-specific primers 5′-CTGAGTCATCCTTCTAGCCTAAGAC-3′ (primer 1, forward) and 5′- TGCCCTGCACTGTCTAGAATA -3′ (primer 5, reverse). Homozygous male mice without GFP fluorescent signaling were mated to wild-type C57BL/6N females. Thus, all of the resulting offspring (C57BL/6-Pthtm1(P2A-tdTomato)/+) were GFP negative. They were used as controls and as recipients in PTG transplantation.
Generation of Gcm2 KO Mice by CRISPR-Cas9.
CRISPR-Cas9-mediated gene editing in mouse embryos was performed using Genome Editor™ (BEX). In brief, 20 to 30 pronuclear embryos were transferred into 5 μL Opti-MEM™ I medium (31985-062, Thermo) containing 100 ng/μL Alt-R® S.p. Cas9 Nuclease V3 (1081059, IDT, Skokie, IL) and 200 ng/μL sgRNA (custom, IDT) on a platinum electrode (LF501PT1-10, BEX). The sgRNA target was 5′-CGCAACACCAACAACCACAA-3′ (second exon, mouse Gcm2 locus). After electroporation (15 V, 3 ms ON, 97 ms OFF, Pd Alt 3), the embryos were cultured in KSOM-AA medium at 37 °C under 5% CO2 in air for 24 h until transfer to pseudopregnant females or microinjection. Recipient mice underwent cesarean section at 19.5 d post coitum (dpc).
Generation of Chimeric Mice by Single-Step BC.
We have confirmed that single-step BC is possible using SGE2, the origin of Pth-tdTomato mESCs (C57BL/6N-Tg(CAG-EGFP)Pthtm1(P2A-tdTomato)ES1). We established two clones of Pth-tdTomato mESCs and for subsequent detailed analysis used that with higher efficiency of chimera generation. Pth-tdTomato mESCs were dissociated (0.25% trypsin-EDTA; 25200-056, Thermo), yielding single cells, immediately before microinjection. From 2 to 5 mESCs were introduced into the blastocoel cavity of C57BL/6N, B6D2F1×C57BL/6N, and C57BL/6N×DBA/2 mouse blastocysts at E3.5 using piezo-actuated microinjection (Narishige Scientific Instrument Lab). Injected blastocysts were transferred into the uteri of 2.5 dpc pseudopregnant ICR females. Recipient mice underwent cesarean section at 19.5 dpc. The chimeric pups were fostered by surrogate ICR mothers.
Generation of Gcm2 KO Rats by CRISPR-Cas9.
Genome editing on Wistar rat pronuclear embryos was performed using the same procedure as for mice, but the conditions of electroporation differed (25 V, 3 ms ON, 97 ms OFF, Pd Alt 4 times). The sgRNA target site was 5′-CGCAACACCAACAACCACAA-3′ at the second exon of the rat Gcm2 locus. After electroporation, the embryos were transferred into the oviducts of pseudopregnant recipients (0.5 dpc) after overnight culture in rat KSOM medium at 37 °C under 5% CO2 in air. The recipient females underwent cesarean section at 22.5 dpc.
Generation of Interspecies Chimeras.
To generate Gcm2 KO rat embryos, pronuclear stage rat IVF embryos were treated with CRISPR-Cas9 under the same conditions as those used for mice and were transferred into oviducts of pseudopregnant recipients (0.5 dpc). At 3.5 dpc, blastocysts were retrieved from recipient oviducts by flushing with M2 medium (MR-015-D, Merck). The recovered blastocysts were cultured in paraffin oil–covered rat KSOM until microinjection. Pth-tdTomato mESCs were then injected into the blastocyst cavity at 3 to 5 cells/embryo. The injected blastocysts were transferred into the uteri of pseudopregnant Crlj:WI females (3.5 dpc). The recipient females underwent cesarean section at 22.5 dpc.
Genotyping and Analysis of Chimerism.
Genotyping of offspring was performed with crude lysate. In brief, tissue samples (ears, tails) were incubated in lysis buffer (all components, Sigma; 20 mM Tris–HCl, pH 8.0, [T1503], 100 mM NaCl [28-2270], 5 mM EDTA [09-1420], 0.1% SDS [28-3284], 200 μg/mL Proteinase K [P6556]) at 60 °C for 5 min to 24 h, followed by 98 °C Proteinase K heat-inactivation for 2 min. For chimeric mice, peripheral blood cells were stained with APC-labeled anti-mouse CD45 (17-0451-82, Invitrogen, 1:10,000) after lysis of red blood cells. To mark dead cells, 1 mg/mL DAPI solution (D1306, Invitrogen; 1:10,000) was used. Genomic DNA was extracted from fluorescent marker (GFP)-negative CD45-positive cells (2,000 to 3,000 cells) isolated from blood using an FACSAria™ III Cell Sorter (648282F1, BD Biosciences). The cells were incubated in lysis buffer at 60 °C for 5 min, followed by 98 °C Proteinase K heat inactivation for 2 min. For chimeric rats, rat embryonic fibroblasts (REFs) were isolated from E21.5 embryos. REFs on 6-well tissue culture plates (92006, TPP) were cultured in Dulbecco’s modified Eagle medium (Sigma, D5796) supplemented with 10% fetal bovine plasma (Gibco, 10437-028) and 100 U/mL penicillin–streptomycin. Genomic DNA was extracted from GFP-negative cells (2,000 to 3,000 cells) isolated from REFs using an FACSAria™ III Cell Sorter. DAPI (D1306, Invitrogen; 1:10,000) was used to mark dead cells.
Crude PCR was performed with KOD Multi&Epi DNA Polymerase (KME-101, TOYOBO) and mouse or rat primer pairs (mice, forward 5′-AGATGTTCGCCAGTTACCCTG-3′, reverse 5′-CCCGTCTGTCTGTCCGTTG-3′; rats, forward 5′-CACACATAGCGGAAGGGCTA-3′, reverse 5′-CAACCTGTCCGGTGATTCCT-3′. The PCR products were purified using QIAquick® PCR purification kits (28106, Qiagen) and micro-elute columns (FAMGC-C50, Favorgen). To determine small In-Dels in the Gcm2 locus, purified PCR products underwent Sanger sequencing (FASMAC). These purified PCR products were used for the In-Fusion reaction described below. Newborns were genotyped by Sanger sequencing, using the Inference of CRISPR Edits (ICE) algorithm (36) (Synthego). We defined biallelic Gcm2 mutants with out-of-frame InDels and mosaics of out-of-frame mutant alleles as “Gcm2 mutants with out-of-frame InDels” and biallelic Gcm2 mutants with in-frame InDels, biallelic Gcm2 mutants with out-of-frame and in-frame InDels, mosaics of in-frame mutant alleles, and mosaics of in-frame and out-of-frame mutant alleles as “Gcm2 mutants with in-frame InDels”.
For analysis of large deletions in mice (>100 bases), In-Fusion cloning was done. The backbone vector pUC19 was amplified by inverse PCR with primers that allow the addition of homology arms (forward 5′-caacggacagacagacgggAAGCTTGGCGTAATCATGG-3′, reverse 5′-cagggtaactggcgaacatctGAATTCACTGGCCGTCGTT-3′). For In-Fusion cloning reaction, a mixture of 50 ng purified crude PCR product, 100 ng purified inverse PCR products, and 5× In-Fusion HD Enzyme Premix 1 µL (639650, Takara) was incubated for 15 min at 50 °C. For transformation, 2.5 µL of the In-Fusion reaction mixture was added to 50 µL of E. coli competent cells DH5α (DNA-913, TOYOBO) and incubated for 20 min on ice. A heat shock of 35 s at 42 °C was applied. After 30 min of recovery with 450 µL of SOC medium, each transformation reaction was seeded onto ampicillin-resistant Luria Bertani agar plates. The next day, individual isolated colonies were picked from each LB plate. Plasmid DNAs were isolated using NucleoSpin® Plasmid QuickPure (740615.250, Takara).
Immunohistochemical Studies.
Tissues from mice and rats were fixed in 4% paraformaldehyde (163-20145, Wako) overnight at 4°C. For cryosections, the fixed tissues were infiltrated with 20% sucrose (196-00015, Wako), mounted in OCT embedding compound (45833, Sakura Finetek), and frozen at −80 °C. Frozen tissues were sectioned at 10 to 20 µm (20-µm thickness for SI Appendix, Fig. S2B, 10-µm thickness for others) using a cryostat (CM3050S, Leica). The cryosections were picked up on glass slides, washed with PBS, and blocked with blocking medium (15252, Active Motif) for 30 min at room temperature. Primary antibodies diluted in 0.02% Triton/PBS (04605-250, Wako) were applied to the sections, which were incubated at 4 °C overnight. The next day, after washing with PBS, secondary antibodies diluted with 0.02% Triton/PBS were applied, and the sections were incubated at room temperature for 60 min. Coverslips (C024601, Matsunami) were placed using Fluorescent Mounting Medium (S3023, Dako). Primary antibodies employed, with animal source and dilution, were chicken anti-GFP (A10262, Invitrogen, 1:500), mouse anti-DsRed (Z2392N, Clontech, 1:100), and rabbit anti-PTH (ab213557, Abcam, 1:100). Secondary antibodies used were Alexa Fluor 488 goat anti-chicken IgY (A11039, Invitrogen, 1:1,000), Alexa Fluor 546 donkey anti-mouse IgG (A10036, Invitrogen, 1:1,000), and Alexa Fluor 647 donkey anti-rabbit IgG (A10036, Invitrogen, 1:1,000). Nuclear counterstaining was performed with 1 mg/mL DAPI solution (D1306, Invitrogen; 1:1,000). Immunostained cryosections were observed by confocal laser scanning microscopy (FV3000, Olympus, Tokyo, Japan).
qPCR.
All tissues from the neck to the mediastinum of neonatal Gcm2 mutant mice and rats were excised en bloc and immersed overnight in RNAlater™ Stabilization Solution (AM7021, Thermo) at 4 °C before storage at −80 °C until RNA extraction. Total RNA was isolated from the tissue using TRIzol® Reagent (15596-018, Thermo). The tissues were subdivided into 100-mg aliquots, and 1 mL of TRIzol® Reagent was added per 100 mg tissue. The tissues were homogenized using a homogenizer (TissueLyserLT, Qiagen), and lysates from the same individual were combined with TRIzol® Reagent to yield a volume of 1 mL. Chloroform, 200 µL (033-15721, Wako), was added. After centrifugation, the upper aqueous phase containing RNA was collected, and RNA purification was performed using the RNeasy Plus Mini Kit (74134, Qiagen). The RNA yield was determined using Nanodrop™ 2000c (Thermo). Reverse transcription from total RNA was performed using the ReverTra Ace™ qPCR RT Master Mix (FSQ-201, TOYOBO). In all individuals, 2 µg each of RNA template was used for reverse transcription. For real-time PCR, TaqMan® Fast Advanced Master Mix (4444557, Thermo) was used. Primers and probes used were mouse Pth, forward 5′-ccagttcatcagctgtctggt-3′, reverse 5′-gtgtttgcagacatcatctttacat-3′, Universal Probe Library #78 (Roche); mouse Foxn1, forward 5′-tgacggagcacttcccttac-3′, reverse 5′-gacaggttatggcgaaca-3′, Universal Probe Library #68 (Roche); and rat Pth, forward 5′-gtctggcttactccagcttaat-3′, reverse 5′-catgaggatcatcaccttagcc-3′, probe 5′-acagggtcactcctgaaggatcct-3′. For Gapdh, TaqMan® Rodent GAPDH Control Reagents (4308313, Thermo) were used. Quantitative PCR was performed using the QuantStudio 7 Flex real-time qPCR system (4485701, Thermo). The data were normalized to Gapdh mRNA expression.
Electron Microscopy.
To prepare specimens for electron microscopy, PTGs were fixed in 2.5% glutaraldehyde (G004, TAAB Laboratories Equipment) for 3 d at 4°C and postfixed in 1% osmium tetraoxide (O018, TAAB Laboratories Equipment) for 1 h. The tissue block was dehydrated in a graded series of alcohol solutions and embedded in Quetol-812 (340-H, Nissin EM). Ultrathin sections were then prepared, stained with 2% uranyl acetate and 0.2% lead citrate, and examined under a transmission electron microscope (H-7500, Hitachi).
Statistical Analyses.
All data were analyzed using Prism 9.5.1 software (https://www.graphpad.com) (GraphPad).
Biochemical Analyses of Blood Samples.
[Ca] in heparinized plasma was colorimetrically determined (MAK022, Sigma). PTH concentrations in EDTA plasma were determined by mouse PTH1-84 enzyme-linked immunosorbent assay (ELISA; 60-2305, Quidel). NaHCO3 infusion testing was performed in mice anesthetized with isoflurane injected intraperitoneally with 25 µL/g of 8.4% (w/v) NaHCO3 (Otsuka Pharmaceutical). Blood was sampled in EDTA before injection and 30 min thereafter.
Ex Vivo Analysis of PTG Function.
Ex vivo analysis of PTG function was performed as described in ref. 25, with minor modifications. Two PTGs from each chimeric mouse were isolated. They were submerged in basal medium consisting of Ca-free Dulbecco’s modified Eagle medium (16972-45, Nacalai Tesque) supplemented with 0.2% bovine serum albumin (15260-037, Thermo), 20 mM HEPES (15630-080, Thermo), and 1 % penicillin–streptomycin (15140-122, Thermo) containing 3.0 mM [Ca] at room air. Thirty minutes later, each pair of PTGs was transferred into a 96-well flat-bottom tissue culture plate (92096, TPP) and exposed for 30-min intervals to media containing increasing [Ca] (from 0.5 to 4.0 mM), ending with a drop to medium containing 0.5 mM [Ca] (two successive exposures).
PTG Isolation and Transplantation.
A hypoparathyroidism mouse model was generated in 8- to 10-wk-old Pth-tdTomato reporter mice (C57BL/6N-Pthtm1(P2A-tdTomato)) anesthetized by intraperitoneal injection (medetomidine 0.75 mg/kg, midazolam 4 mg/kg, and butorphanol 5 mg/kg). Under fluorescence stereomicroscopy (SMZ18 microscope, Nikon), tdTomato-guided parathyroidectomy was performed using Vannas scissors (14003-G, WPI)/tapered tweezers (7-562-05, Dumont). Although tdTomato-expressing tissue, interpreted as PTG, was also found in the thymus, thymectomy was not performed. Postsurgical hypoparathyroidism was confirmed by analyses of PTH and Ca values as stimulated by NaHCO3 infusion. Isolated mESC-derived PTG tissue was injected beneath the right renal capsule of postparathyroidectomy mice using tapered tweezers (7-562-05, Dumont).
Gcm2 KO neonates on C57BL/6N × C57BL/6-Tg(CAG-EGFP) background were kept alive by injections (1.25 µg, 3×/d) of teriparatide (224-08739-9, Mochida Pharmaceutical). On the first postnatal day, a pair of mESC-derived PTGs was implanted into the dorsal subcutaneous tissue of Gcm2 KO neonates with Vannas scissors (14003-G, WPI)/tapered tweezers (7-562-05, Dumont). Three Gcm2 KO neonates were transplanted and seven were sham operated. Teriparatide therapy was discontinued on posttransplantation day 1.
Transcriptome Analysis.
Bulk RNA-seq.
The SMART-Seq® HT Kit (634438, Takara) was used according to the manufacturer’s instructions, minimally modified, to synthesize cDNA from total RNA derived from PTG cells. Under fluorescent light illumination, 10 to 20 tdTomato-expressing PTG cells were manually collected using a micromanipulator (IX73, Olympus). The cells were added to PCR tubes prefilled with nuclease-free phosphate-buffered saline solution (4 µL). A mixture (6 µL) of 0.76 µL 10× lysis buffer, 0.04 µL RNase inhibitor, 0.8 µL 3′ SMART-Seq CDS primer IIA, and 4.4 µL nuclease-free water was added, pipetted several times to mix, snap-frozen using dry ice, and held at −80 °C until cDNA synthesis was undertaken. For one-step first-strand cDNA synthesis and double-stranded cDNA amplification, an equal volume of one-step master mix composed of 0.56 µL nuclease-free water, 6.4 µL one-step buffer, 0.8 µL SMART-Seq HT oligonucleotide, 0.4 µL RNase inhibitor, 0.24 µL SeqAmp DNA polymerase, and 1.6 µL SMARTScribe reverse transcriptase was added to 10 µL of lysis solution and subjected to 16 cycles in a thermal cycler (TP350, Takara) according to the manufacturer’s instructions. PCR-amplified cDNA was purified using the AMPure XP Kit (A63881, Beckman Coulter). The ratio of beads to sample was 1:1 (20 µL of each). The Nextera XT DNA Library Preparation Kit (FC-131-1024, Illumina) was used according to the manufacturer’s instructions, minimally modified, to prepare cDNA libraries. To tagment cDNA, a mixture of 2 µL tagment DNA buffer, 1 µL amplification tagment mix, and 200 pg of cDNA (in a volume of 1 µL) was prepared. Immediately after the tagmentation program in a thermal cycler according to the manufacturer’s instructions ended, 1 µL of NT buffer was added. A total of 10 µL of PCR reaction solution was prepared by adding 3 µL of Nextera PCR master mix, 1 µL of index 1 primer, and 1 µL of index 2 primer to 5 µL of the tagmented sample. PCR amplification was performed as described in the manufacturer’s instructions. PCR-amplified, tagmented cDNA was purified using the AMPure XP Kit. The ratio of beads to sample was 0.65:1. The Qubit dsDNA HS Assay (Q32851, Invitrogen) was used to quantify cDNA. TapeStation (4150, Agilent, Santa Clara, CA) and High Sensitivity DNA Kits (5067-5584 and 5067-5585, all Agilent) were used to evaluate the profiles of cDNA libraries. Next-generation sequencing was performed in a GENEWIZ laboratory (South Plainfield, NJ) on HiSeq X ten (Illumina, 2 × 150 bp). RNA-seq datasets were analyzed using Galaxy software (https://usegalaxy.org/). Sequence data were annotated by the reference genome (GRCm39_version104) using the Salmon quant in Galaxy. Normalization and DEG analysis were performed using DESeq2 in Galaxy and visualized using ggplot2 in R v4.1.2. Volcano plots were generated using ggplot2 and dplyr in R v4.1.2.
Single-cell RNA-seq.
We performed a series of experiments twice (each experiment was titled para10_215 and para10_222). Adult Gcm2−/− mice complemented with mESCs showing 50 to 70% chimerism of peripheral blood cells were used (three for para10_215 and five for para10_222). A pair of mESC-derived PTGs was removed as described above and placed in a 20-µL PBS drop. Isolated mESC-derived PTGs were cut into small pieces using Vannas scissors. Minced mESC-derived PTGs were transferred into a 1.5 mL siliconized tube (131-615CH, Watson). They were incubated in 25 µl of papain solution (LK003150, Worthington) for 15 min at 37 °C under 5% CO2 in air. Subsequently, 25 µL of 10 mg/mL collagenase XI (C7657, Sigma)/HBSS (084-08965, Wako) supplemented with 120 µL of 170 mM CaCl2 was added into the same tube, which was incubated for 15 min at 37 °C under 5% CO2 in air. Finally, 10 µL of 0.25% trypsin-EDTA was added. After 1 min, pipetting was done several times, yielding single cells that were suspended in 140 µL of medium containing equal volumes of 10% FBS/DMEM and ovomucoid protease inhibitor (LK003150, Worthington) and placed on ice. Doublets were discriminated using the FACSAria™ Fusion Flow Cytometer (BD Biosciences). Sorted single cells were collected in 500 µL 0.1% BSA (126615, Merck)/PBS in a 1.5-mL siliconized tube. The cell suspension was centrifuged at 300g at room temperature for 5 min using an Allegra X-15R centrifuge (Beckman Coulter) and swing bucket rotors (339175, Beckman Coulter). Cell concentration was determined using CellDrop™ automated cell counters (DeNovix, Wilmington, DE). Single-cell gel beads in emulsion were generated using the Chromium Controller (10× Genomics). Libraries were generated using the Single Cell 3′ Reagent Kit v.3.1 for dual index (1000269 and 1000215, 10× Genomics) according to the manufacturer’s instructions. In GEM generation, cell stock concentration and volume of cell suspension stock per reaction were 8 × 104/mL, 30 µL (para10_215) and 1.42 × 105/mL, 41.3 µL (para10_222). PCR was done with the Applied Biosystems™ VeritiPro™ thermal cycler (A48141, Thermo). Total cycles of sample index PCR were 13 for para10_215 and 12 for para10_222. We performed cDNA and postlibrary QC using TapeStation (2200, Agilent) and High Sensitivity D5000 screen tape (5067-5592, Agilent). Next-generation sequencing was performed in a GENEWIZ laboratory on HiSeq X ten (Illumina). Sequence data were mapped to the reference genome (mm10) with Gfp and tdTomato using Cell Ranger v.7.0.0. Subsequent analysis was performed using Seurat v.4.2.1 in R. We filtered out cells that had unique features of over 7,500 or less than 1,000 and mitochondrial gene counts higher than 7.5%. Filtered data were normalized with SCTransform function. To integrate data, we used SelectIntegrationFeatures, PrepSCTIntegration, FindIntegrationAnchors, and IntegrateData functions. Principal component analysis (PCA) was performed using the RunPCA function; uniform manifold approximation and projection (UMAP) was then performed using the RunUMAP function. FindNeighbors function was used to construct a shared nearest neighbor graph, and FindClusters function was used to cluster cells (resolution = 0.1). Cell cycle status was scored with CellCycleScoring function and then regressed out using SCTransform function with vars.to.regress = c(“S.Score”, “G2M.Score”). Results were visualized using FeaturePlot and Vlnplot function in Seurat v.4.2.1 and ggplot2 in R.
In 10× Genomics, read counts show wide dispersion, and Gfp false-positive cells may occur due to ambient RNA contamination. In addition, Gfp expression varies even within the same cell lineage. Clear division of Gfp-positive from Gfp-negative cells thus is difficult. To estimate donor cell contribution to each cell lineage, we displayed the normalized Gfp count data of all cells in a histogram. The inflection point of the density curve was set as the cusp between Gfp-high expression and Gfp-low expression, serving as the cutoff for Gfp expression.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (CSV)
Acknowledgments
We are grateful to Dr. Motoo Watanabe (Tokyo Medical and Dental University), Dr. Toshihiro Kobayashi (Institute of Medical Science, University of Tokyo), and Dr. Satoshi Yamazaki (University of Tsukuba) for invaluable advice in preparing the manuscript, Ms. Kyoko Okada (Tokyo Medical and Dental University) for secretarial support, and to all members of the Nakauchi lab for discussions. We thank Ms. Tomoko Ando (Institute of Medical Science, University of Tokyo) for supporting histological analysis. We thank Mr. Yuji Yamazaki (University of Tsukuba) for supporting cell sorting. We thank Dr. Ryusaku Matsumoto (Center for iPS Cell Research and Application, Kyoto University) for supporting transcriptome analysis. We thank Dr. Alex Knisely for invaluable help in proofreading and editing this manuscript. We used BioRender (https://biorender.com) in creating the figures. This work was supported by grants from the Centers for Clinical Application Research on Specific Disease/Organ (to H. Nakauchi) of the Research Center Network for Realization of Regenerative Medicine, funded by the Japan Agency for Medical Research and Development under Grant Number JP22bm1004002; Grant-in-Aid for Scientific Research (B) (to E.M.), funded by the Japan Society for the Promotion of Science (JSPS) KAKENHI under Grant Number JP20H03638; and Grant-in-Aid for Early-Career Scientists (to M. Kano), funded by JSPS KAKENHI under Grant Number JP21K16337.
Author contributions
M. Kano, N.M., Y.I., T.Y., H. Suga, E.M., and H. Nakauchi designed research; M. Kano, N.M., H. Sato, R.H., N.I., H. Nagano, M. Kasai, H.Y., H. Suga, and E.M. performed research; M. Kano, N.M., T.K., H. Suga, H.M., E.M., and H. Nakauchi analyzed data; and M. Kano, N.M., T.K., H. Suga, E.M., and H. Nakauchi wrote the paper.
Competing interests
H. Nakauchi is a co-founder and shareholder in ReproCELL, Megakaryon, and Century Therapeutics.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Eiji Mizutani, Email: emizutani@md.tsukuba.ac.jp.
Hiromitsu Nakauchi, Email: nakauchi@stanford.edu.
Data, Materials, and Software Availability
All RNA-seq data have been deposited in the Gene Expression Omnibus under the accession code GSE232602 (37). All other data are included in the manuscript and/or supporting information.
Supporting Information
References
- 1.Mannstadt M., et al. , Hypoparathyroidism. Nat. Rev. Dis. Primers 3, 17055 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Lo C. Y., Parathyroid autotransplantation during thyroidectomy. ANZ J. Surg. 72, 902–907 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Buttner M., Musholt T. J., Singer S., Quality of life in patients with hypoparathyroidism receiving standard treatment: A systematic review. Endocrine 58, 14–20 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Mitchell D. M., et al. , Long-term follow-up of patients with hypoparathyroidism. J. Clin. Endocrinol. Metab. 97, 4507–4514 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tay Y.-K.D., et al. , Therapy of hypoparathyroidism with rhPTH(1–84): A prospective, 8-year investigation of efficacy and safety. J. Clin. Endocrinol. Metab. 104, 5601–5610 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tabacco G., et al. , Quality of life in hypoparathyroidism improves with rhPTH(1–84) throughout 8 years of therapy. J. Clin. Endocrinol. Metab. 104, 2748–2756 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bilezikian J. P., Hypoparathyroidism. J. Clin. Endocrinol. Metab. 105, 1722–1736 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bingham E. L., Cheng S. P., Woods Ignatoski K. M., Doherty G. M., Differentiation of human embryonic stem cells to a parathyroid-like phenotype. Stem Cells Dev. 18, 1071–1080 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Lawton B. R., et al. , Differentiation of PTH-expressing cells from human pluripotent stem cells. Endocrinology 161, bqaa141 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woods Ignatoski K. M., Bingham E. L., Frome L. K., Doherty G. M., Differentiation of precursors into parathyroid-like cells for treatment of hypoparathyroidism. Surgery 148, 1186–1189; discussion 1189–1190 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Kobayashi T., et al. , Generation of rat pancreas in mouse by interspecific blastocyst injection of pluripotent stem cells. Cell 142, 787–799 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Yamaguchi T., et al. , Interspecies organogenesis generates autologous functional islets. Nature 542, 191–196 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Goto T., et al. , Generation of pluripotent stem cell-derived mouse kidneys in Sall1-targeted anephric rats. Nat. Commun. 10, 451 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mori M., et al. , Generation of functional lungs via conditional blastocyst complementation using pluripotent stem cells. Nat. Med. 25, 1691–1698 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobayashi T., et al. , Blastocyst complementation using Prdm14-deficient rats enables efficient germline transmission and generation of functional mouse spermatids in rats. Nat. Commun. 12, 1328 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamanaka S., et al. , Generation of vascular endothelial cells and hematopoietic cells by blastocyst complementation. Stem Cell Rep. 11, 988–997 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsunari H., et al. , Blastocyst complementation generates exogenic pancreas in vivo in apancreatic cloned pigs. Proc. Natl. Acad. Sci. U.S.A. 110, 4557–4562 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gordon J., Manley N. R., Mechanisms of thymus organogenesis and morphogenesis. Development 138, 3865–3878 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akiyama Y., Hosoya T., Poole A. M., Hotta Y., The gcm-motif: A novel DNA-binding motif conserved in Drosophila and mammals. Proc. Natl. Acad. Sci. U.S.A. 93, 14912–14916 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gunther T., et al. , Genetic ablation of parathyroid glands reveals another source of parathyroid hormone. Nature 406, 199–203 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Liu Z., et al. , Thymus-associated parathyroid hormone has two cellular origins with distinct endocrine and immunological functions. PLoS. Genet. 6, e1001251 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Z., Yu S., Manley N. R., Gcm2 is required for the differentiation and survival of parathyroid precursor cells in the parathyroid/thymus primordia. Dev. Biol. 305, 333–346 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kittel B., Ernst H., Kamino K., “Anatomy, histology, and ultrastructure, parathyroid, mouse” in Endocrine System, Jones T. C., Capen C. C., Mohr U., Eds. (Springer, Berlin Heidelberg, Berlin, Heidelberg, 1996), pp. 328–330, 10.1007/978-3-642-60996-1_35. [DOI] [Google Scholar]
- 24.Iwasaki Y., et al. , Sodium bicarbonate infusion test: A new method for evaluating parathyroid function. Endocr. J. 50, 545–551 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Cheng Z., et al. , Sex and age modify biochemical and skeletal manifestations of chronic hyperparathyroidism by altering target organ responses to Ca2+ and parathyroid hormone in mice. J. Bone. Miner. Res. 28, 1087–1100 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kamitani-Kawamoto A., et al. , MafB interacts with Gcm2 and regulates parathyroid hormone expression and parathyroid development. J. Bone. Miner. Res. 26, 2463–2472 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Han S. I., Tsunekage Y., Kataoka K., Gata3 cooperates with Gcm2 and MafB to activate parathyroid hormone gene expression by interacting with SP1. Mol. Cell. Endocrinol. 411, 113–120 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okabe M., Graham A., The origin of the parathyroid gland. Proc. Natl. Acad. Sci. U.S.A. 101, 17716–17719 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Honda A., et al. , Efficient derivation of knock-out and knock-in rats using embryos obtained by in vitro fertilization. Sci. Rep. 9, 11571 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miyoshi K., Abeydeera L. R., Okuda K., Niwa K., Effects of osmolarity and amino acids in a chemically defined medium on development of rat one-cell embryos. J. Reprod. Fertil. 103, 27–32 (1995). [DOI] [PubMed] [Google Scholar]
- 32.Nakamura K., et al. , The effect of supplementation of amino acids and taurine to modified KSOM culture medium on rat embryo development. Theriogenology 86, 2083–2090 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Kano M., Mizutani E., Homma S., Masaki H., Nakauchi H., Xenotransplantation and interspecies organogenesis: Current status and issues. Front. Endocrinol. (Lausanne) 13, 963282 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masaki H., et al. , Inhibition of apoptosis overcomes stage-related compatibility barriers to chimera formation in mouse embryos. Cell Stem Cell 19, 587–592 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Nishimura T., et al. , Generation of functional organs using a cell-competitive niche in intra- and inter-species rodent chimeras. Cell Stem Cell 28, 141–149.e143 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Conant D., et al. , Inference of CRISPR edits from Sanger trace data. CRISPR J. 5, 123–130 (2022). [DOI] [PubMed] [Google Scholar]
- 37.Kano M., Mizuno N., Kimura T., Mizutani E., Bulk RNA-seq and Single-cell RNA-seq data of parathyroid gland (PTG) cells. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE232602. Deposited 16 May 2023. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (CSV)
Data Availability Statement
All RNA-seq data have been deposited in the Gene Expression Omnibus under the accession code GSE232602 (37). All other data are included in the manuscript and/or supporting information.