Significance
Generalization from the training data is a test of a true AI. Methods for computing protein structure using AI have been criticized as merely using pattern recognition rather than knowledge of the rules of physics and chemistry. Here, we show that contrary to this view, AI methods can accurately identify rarely observed features in crystal structures, provided these are not induced by the crystal environment. In contrast, pattern recognition would usually select the more commonly observed structural feature. We propose that in this application, the AI network has learned the principles of protein structure such that it can successfully apply those to previously unobserved situations.
Keywords: CASP14, alphaFold2, AI, structure analysis
Abstract
The CASP14 experiment demonstrated the extraordinary structure modeling capabilities of artificial intelligence (AI) methods. That result has ignited a fierce debate about what these methods are actually doing. One of the criticisms has been that the AI does not have any sense of the underlying physics but is merely performing pattern recognition. Here, we address that issue by analyzing the extent to which the methods identify rare structural motifs. The rationale underlying the approach is that a pattern recognition machine tends to choose the more frequently occurring motifs, whereas some sense of subtle energetic factors is required to choose infrequently occurring ones. To reduce the possibility of bias from related experimental structures and to minimize the effect of experimental errors, we examined only CASP14 target protein crystal structures determined to a resolution limit better than 2 Å, which lacked significant amino acid sequence homology to proteins of known structure. In those experimental structures and in the corresponding models, we track cis peptides, π-helices, 310-helices, and other small 3D motifs that occur in the PDB database at a frequency of lower than 1% of total amino acid residues. The best-performing AI method, AlphaFold2, captured these uncommon structural elements exquisitely well. All discrepancies appeared to be a consequence of crystal environment effects. We propose that the neural network learned a protein structure potential of mean force, enabling it to correctly identify situations where unusual structural features represent the lowest local free energy because of subtle influences from the atomic environment.
Protein structure prediction using artificial intelligence (AI) techniques, specifically the AlphaFold2 (AF2) deep learning network, developed by the DeepMind team (1, 2), performed spectacularly well during the fourteenth season of the Critical Assessment of Structure Prediction experiment (CASP14) (3). Analysis of the results showed that AF2 generated structures in many cases rival the accuracy of structures determined by high-resolution X-ray crystallographic methods (3). Indeed, in some cases, the calculated structures may represent the biologically relevant ones better than the crystal structures because the latter are influenced by crystal packing forces absent in biological systems (4). Yet, the AI methods have also been criticized. Here, we address the criticism that current deep learning AI methods are only capable of recognizing structural patterns present in the training data and have no sense of the energetic subtleties that determine the details of protein structures. As Moore and colleagues put it, the predictions might suffer from “bias toward structural patterns observed in repositories” (5). More concretely, Skolnick and colleagues (6) argued that the neural networks have so many parameters [10 s of millions (2)] that they simply store known protein structures in detail and regurgitate parts of them where appropriate. If that is the case, these methods do not solve the protein folding problem in a meaningful sense, rather we have just collected enough experimental structures to allow the equivalent of an effective database lookup to provide accurate answers. More broadly, a key test of any AI is the extent to which it can generalize from the training data to effectively deal with new situations. Does AF2 pass that test?
One argument that substantial generalization is achieved is based on rough estimates of the number of possible local atomic configurations at the 1 Å accuracy often achieved. That number appears to be astronomically larger than the number of such configurations represented in the PDB. But, it is difficult to make such a back-of-the-envelope approach numerically robust. Here, we use a different approach. Many structural features in proteins occur frequently: for example, α-helices, β-strands, and a small number of common turns (7). A machine that is effectively consulting a database of observed structures could identify these features fairly easily. But, there are also less common features, for example 310- or π-helices; β-bulges; and unusual turns. By and large, these are less energetically favorable when considered in isolation, and they occur when some other structural features directly or indirectly compensate for the local-in-sequence relative energetic strain. For example, about 1 in 20 proline residues in the PDB occurs with a cis peptide (8) and calculations suggest that this rarity is a consequence of the local steric strain (9). Thus, consistent reproduction of rare features requires a computational method that in some sense has an appreciation of the subtle balance of the interaction energies involved.
The goal of the current study was to investigate and as far as possible quantitate the extent to which the AF2 method, as deployed during the CASP14 experiment (1), reproduced rare structural features. CASP conducts biannual community experiments with the aim of determining the state-of-the-art computing protein structure from amino acid sequence. Participants are blind to the corresponding experimental structures (3).
We focused on the subset of the CASP14 structure prediction targets classified as template free (10), so that previously obtained experimental structure information could not be directly utilized by the machine. We also examined only structures determined at high resolution because analysis of the CASP results showed that agreement between AF2 models and experimental structures decreased with experimental data quality, suggesting an increasing impact of experimental error with lower resolution (3). We searched the selected experimental structures and their AF2 models for uncommon structure features. Where there are differences between the computed and experimental structures, we also examined whether protein–protein interactions or crystal contacts were involved and whether bound ligands could influence the local conformation because the AF2 models were calculated as stand-alone free molecules.
The results show that AF2 structures include all the unusual features in the set of proteins not affected by the crystal environment. We therefore conclude that this class of deep learning AI method does generalize from its training data so as to correctly determine seldom seen subtle and energetically complex structural features. Given the nature of the method, the likely reason for this is that the machine learns a potential of mean force between the different atom types (11) that can be used to evaluate the relative free energy of any considered constellation of atoms.
Results
Table 1 lists the six selected CASP14 targets. Residue numbering is that of the target sequences provided to CASP participants and sometimes differs from that of the final experimental structures deposited in the PDB. The structures of targets T1046s1 and T1065s2 were determined in complex with partner proteins, while the remaining proteins were reported by the experimentalists to function as monomers, and examination of the corresponding crystal contacts also supports the monomeric state.
Table 1.
CASP14 targets selected for analysis
| Target | Protein | Number of residues* | PDB code | Resolution (Å) | Rwork/Rfree | Reference | AF2 GDT-TS |
|---|---|---|---|---|---|---|---|
| T1046s1† | Antiholin | 157 | 6px4 | 1.65 | 0.195/0.225 | (12) | 97.22 |
| T1049 | Receptor-binding domain of adhesin MrpH | 141 | 6y4f6y4e | 1.75 1.02 | 0.153/0.183 0.107/0.128 | (13) | 93.10 |
| T1065s2† | Immunity protein CDII | 98 | 7m5f | 1.59 | 0.140/0.167 | Michlska et al, unpublished; | 98.47 |
| T1074 | Bdellovibrio bacteriovorus Bd0675 | 202 | 7oc9 | 1.50 | 0.197/0.213 | (14) | 89.77 |
| T1082 | Bacteriophage T4 spackle protein | 97 | 7cn67cn7‡ | 1.60 1.15 | 0.179/0.228 0.107/0.121 | (15) | 95.33 |
| T1090 | N-terminal domain of chromatin remodeling protein Ssr4 | 193 | 7k7w | 1.77 | 0.177/0.206 | (16) | 89.02 |
*The number of residues corresponds to the sequences provided to the CASP14 predictors.
†T1046s1 was determined in complex with holin and T1065s2 was determined in complex with CdiA.
‡PDB entry code 7cn7 includes the T1082 target in complex with the lysozyme domain of GP5 tail lysozyme.
Cis Peptides.
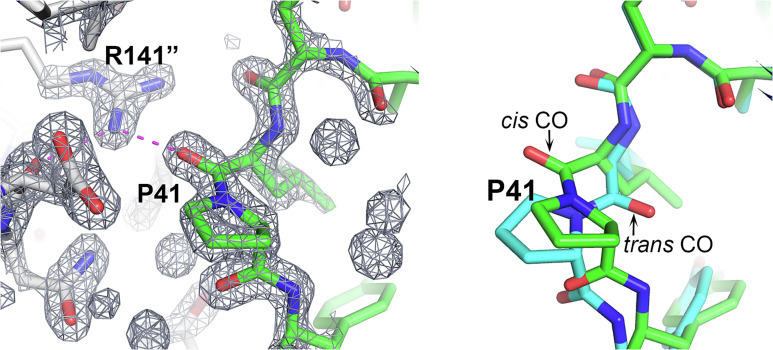
Cis peptides other than those involving proline residues in the second position are energetically strained and rare [less than 1 in 1,000 (17, 18)]. For prolines, the cis/trans energy difference is smaller, but still significant (9), and about 1 in 20 prolines in the PDB is reported as cis (8). In the CASP analysis set, there are no nonproline cis peptides and of the 46 prolines, four are reported as cis (Table S2). Surprisingly, 21 prolines are in one target, T1090 (16), which is only 191 residues long. Of these 21 prolines, three are involved in cis peptides. The AF2 best models agree with experiment for all 42 trans prolines and three of the four cis prolines. The cis proline peptide (Leu40–Pro41) in the T1090 experimental structure is predicted by all five AF2 models as trans, with an estimated average coordinate error of 0.5 Å. Examination of the experimental electron density in this region unambiguously supports the assignment of the cis peptide (Fig. 1). The residue preceding cis peptide, Leu40, adopts α-helical backbone dihedral angles, which is rarely observed in crystal structures because of the resulting sterically strained interaction between the Cβ atom and the proline backbone Cα and C atoms (8). Here, this local conformation allows the carbonyl oxygen of Leu40 to form favorable electrostatic interactions with the Arg141 guanidinium group of a symmetry-related molecule and the side chain is buried in the interface. In contrast, the trans proline in the AF2 structures, in the absence of crystal contacts, results in a favored Leu40 conformation with no steric strain between the Cβ atom and Pro41 atoms. Consequently, the Leu40 CO group in the AF2 calculated structure is oriented so that the interprotein electrostatic interaction could not be made (Fig. 1). Notably, a substantial contribution to this crystal contact interface is provided by a nonnative 18-residue N-terminal tag that forms an α-helix (16). These observations suggest that AF2’s trans peptide is not an error, rather it is likely the preferred conformation in solution and the experimental cis conformation is a crystallographic artifact (Fig. 1). Thus, overall, on this small sample, AF2 correctly assigned cis and trans proline states in all cases, an assignment that is sometimes difficult to make experimentally in structures determined at low or moderate resolution.
Fig. 1.
A cis-trans proline change induced by crystal packing. In the CASP target T1090 (chromatin remodeling protein Ssr4 N-terminal domain) crystal structure, Pro41 has a cis peptide, but it is trans in the AF2-calculated structure. (Left) Pro41 and its crystal environment together with the associated electron density map. The Leu40 backbone CO group interacts with the solvent inaccessible Arg141 guanidinium group of a neighboring molecule. Interaction is shown by magenta dashed lines, neighboring molecule carbon atoms are colored gray, and Arg141 is superscripted with “. (Right) Pro41 superposition of the crystal (green) and AF2 (sky blue) structures highlighting the difference between the cis and trans peptides. Trans does not allow formation of the intermolecular interaction. It appears that the AF2 structure’s trans conformation likely represents the in vivo solution state.
π- and 310-Helices.
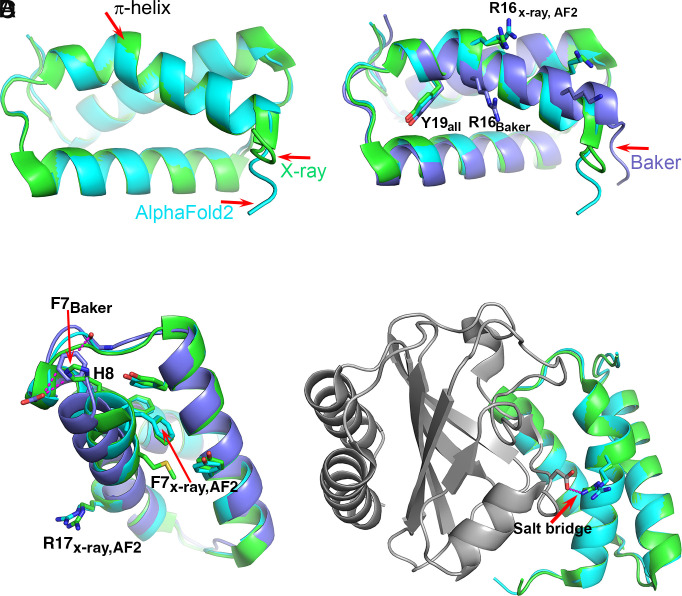
A survey of 5,620 protein structures identified a total of 1,010 π-helices (19). Of these, 80% are comprised of a single residue insertion into α-helices, forming a single π-helix turn, also termed π-bulge or α-bulge, with two i to i+5 backbone hydrogen bonds. The exact per residue frequency is not provided in the study, but assuming an average of 200-residue long chains, it is <0.1%. An earlier survey of 936 proteins containing a total of 224,046 amino acids identified a total of 728 residues forming π-helices, an overall frequency of 0.3% (20). In agreement with this low frequency, only one of the six selected CASP14 experimental structures contains a π-helix (Table 2), and there are no other π-helices in the calculated structures, consistent with the experimental ones. The experimental π-helix occurs in target T1046S1, an antiholin, which forms a complex with holin. The complex regulates T4 phage lysis of an infected host cell. Holin forms lesions in the host membrane and antiholin inhibits this activity (12). Antiholin folds into a three-helical bundle, with the first α-helix distorted at His17 to form a single α-helix turn spanning Gly14–Met20. The AF2 best model has a very high overall level of agreement with the crystal structure, as reflected in a 97.6 GDT-TS score (21). The model also contains the π-helix turn, with estimated atomic errors of ~0.4 Å. Consistent with that and the high quality of the crystal structure (1.65 Å resolution), the superposition of the model and the experimental structure show deviation in backbone atomic positions of 0.2 to 0.4 Å (Fig. 2A). The prediction of the π-helix is not a trivial outcome of the core packing, as demonstrated by the second-best predicted model (Baker Group), where there is an unbroken α-helix, displaced relative to the experimental structure (Fig. 2B). The motif is also not retrievable from the PDB by sequence similarity: The closest match of the 11 residues around the π-helix turn has 55% identity and forms a β-strand (PDB entry 8A3T). Neither is it straightforwardly predicted by coevolution information: Replacement of the three turn residues by gap characters in the multiple sequence alignment does not affect the local conformation.
Table 2.
Statistics of uncommon features in CASP14 template-free high-resolution structures
| Total number in crystal structures | 46 | |
|---|---|---|
| Prolines | Number of cis Pro in crystal structures | 4 |
| Cis Pro present in AF2 models | 3 | |
| Cis Pro not predicted, affected by crystal contacts | 1 | |
| p-helices | Crystal structures | 1 |
| AF2 models | 1 | |
| 310-helices (three-residues) | Crystal structures | 13 |
| AF2 models | 11 | |
| Predicted but contains four residues | 1 | |
| Not predicted, affected by crystal contacts | 1 | |
| 310-helices (six-residues) | Crystal structures | 1 |
| AF2 models | 1 | |
| Small 3D structure motifs | Total number in crystal structures | 18 |
| Present in AF2 structures | 13 | |
| Not in AF2 structures, affected by crystal contacts/environment | 5 | |
| Small 3D structure motifs | Found in AF2 structures, not observed experimentally, crystal contacts | 1 |
Fig. 2.
The T1046S1 (antiholin) π-helix. (A) Superposition of the crystal (green) and the AF2 (sky blue) structures. Both contain a short π-helix. (B) Superposition of the crystal (green), AF2 (sky blue), and Baker’s group (lavender) structures. The Baker group structure has a continuous α-helix with no π interruption, resulting in a different position of Arg16. (C) The environment of Phe7 in the three structures. In the crystal and AF2 structures, Phe7 is located within a hydrophobic core and His8 is exposed to solvent, whereas in the Baker’s group structure, Phe7 is located in approximately the same position as His8 in the crystal structure and interacts unfavorably with a backbone CO and a carboxyl group. The location of Arg16 on the experimental and AF2 structures is indicated. (D) Superposition of the crystal (green) and AF2 (sky blue) structures in the context of the complex with holin (gray). Arg16 forms a salt bridge with an aspartic acid on the partner holin protein. Without the π-helix segment, that key intermolecular interaction cannot be made. In this case, the AF2 structure includes a rare motif that is stabilized by interactions nine residues away, and that is critical to function.
Careful inspection of the environment for the residues on the helix revealed that in both the X-ray structure and the AF2 model, the side chain of Phe7, seven residues away, forms favorable hydrophobic interactions with neighboring residues including two edge-to-face interactions with the side chains of Tyr4 and Tyr34 on the second α-helix. In contrast, the Phe7 side chain in the Baker’s group model is tucked close to the loop between the second and third α-helices, forming two unfavorable interactions, one with the carboxylic group of Asp49 (3.1 Å) and the second with the backbone carbonyl group of Cys46 (3.2 Å). Conversely, in the X-ray structure and AF2 model, this position is occupied by the side chain of His6, which provides a more favorable electrostatic environment than that of the Phe7 benzyl group (Fig. 2C). The energy cost of inserting a residue to form a π-helix turn has been estimated to be ~3 to 6 kcal/mol (22, 23). It appears that in this case, that strain is compensated by improved interactions seven residues away.
It has been proposed that the protein destabilization due to insertion of a residue to form a π-helix would tend to be selected against unless it was associated with a functional advantage (19, 24, 25). The example of antiholin supports this proposal. In the experimental holin:antiholin complex, Arg16 of antiholin forms a salt bridge with an aspartic acid on holin (Fig. 2D), an apparently important feature of the protein–protein interface. The Baker group model without the π-bulge has Arg16 and the preceding helical amino acids out of register by one residue, so that the interprotein salt bridge cannot form. That is, the π-bulge is critical to function.
310-helices are more abundant than π-helices, accounting for approximately 4% of amino acids (26, 27). However, most are short, with only three amino acids. Four-, five-, and six-residue 310-helices are uncommon, occurring in 0.8%, 0.4%, and 0.2% of total number of amino acids, respectively (27). In line with those observations, the selected crystal structures contain thirteen three-residue 310-helices, of which eleven are present in both the crystal structure and the AF2 model (Table 2), and there are no 310-helices in the models that are absent in the experimental structure.
One of the two 310-helices absent in the AF2 model comprises residues 100 to 103 of T1049, the tip adhesin MrpH (13). The five AF2 models have a range of conformations here with an estimated average coordinate error of 0.9 Å. This experimental 310-helix is associated with crystal contacts. Glu103 in the crystal structure is involved in a salt bridge with Arg119 of a crystal symmetry–related molecule. Moreover, the C-terminal region of the AF2 model, which includes a 6xHis tag, extends the C-terminal β-strand. That polypeptide would clash with the 310-helix of the crystal structure and the 6xHis tag would clash with a second neighboring molecule. Instead, the crystal structure’s C-terminal region reverses direction to provide space for the 6xHis tag in a solvent channel. We also note that Glu103 may have a functional role, which is indicated by the AF2 model but not by the crystal structure. In the calculated structure, it forms a hydrogen bond with Tyr18 and both residues are placed in the vicinity of a zinc ion that coordinates three histidine residues and a carboxylic group of a ligand (two crystal structures are available, one with a bound glutamic acid and the second with a bound tartrate). The zinc ion is critical for the protein function in biofilm formation (13). However, Glu103 in the crystal structure is positioned further from the zinc center and from Tyr18, and instead forms the salt bridge with the neighboring molecule. Hence, the AF2 model likely better represents the biologically relevant structure. A sequence search of the 310-helix and the flanking three residues on each end identified no identical sequence. The closest homolog had 70% sequence identity and an entirely different fold in this region. Yet again, AF2 had no template peptide that could guide the prediction.
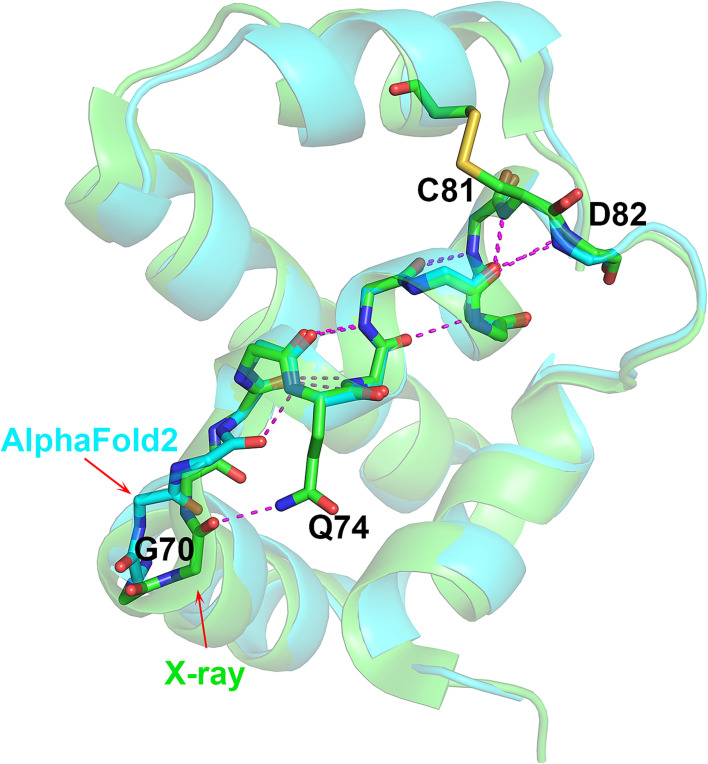
The second 310-helix discrepancy occurs in T1082 (bacteriophage T4 Spackle protein). The crystal structure has a three-residue 310-helix at Val72–Tyr75. The AF2 model has this feature, but with an extra 310 residue at Gly71, making a rarer four-residue 310-helix (Gly71–Tyr75). The Ser69–Gly70–Gly71 tripeptide preceding Val72 adopts a different conformation in the experimental and AF2 structures. Gly70 CO group forms a hydrogen bond with Gln74 side chain in the crystal structure, a feature not present in the AF2 structure. Instead, all five AF2 models have an i to i+3 backbone hydrogen bond between the backbone Gly71 CO and Gln74 NH, so extending the length of the 310-helix by one residue (Fig. 3). No clear evidence exists that the Ser69–Gly70–Gly71 conformation is affected by crystal contacts or ion binding even though this region is quite close to a neighboring molecule. The crystallographic temperature factors of these residues are low and the AF2-estimated atomic position errors are also low (~0.6 Å). Thus, the extra 310 turn may be a calculation error. Nevertheless, the three-residue 310-helix determined in the crystal structure is contained within the four-residues of the AF2-calculated structure and in that sense, the prediction is correct.
Fig. 3.
Different-length 310 helices in AF2 and experimental structures. Superposition of T1082 (bacteriophage T4 spackle protein) crystal (green) and AF2 (sky blue) structures highlighting the difference between the three- (experimental) and four-residue (calculated) 310 helices, and the similar ensuing complex helical architecture Gly70 CO in the crystal structure forms a hydrogen bond with Gln74 side chain, and Gly71 CO in the AF2 structure forms an additional 310-helix hydrogen bond. This one hydrogen bond difference may be a computational error, the only one involving a rare motif found in these protein structures. The error introduces a rarer structural feature than the one found by experiment.
The selected CASP14 experimental structures contain one rare six-residue 310-helix, at residues Val28–Gln33 of the T1090 structure, connecting the native N-terminal α-helix (which follows an engineered affinity tag α-helical segment) to the first β-strand. AF2 captured this rare long 310-helix precisely.
Small 3D Structure Motifs.
A number of classifications for small 3D structure motifs have been published, based on geometric criteria and hydrogen bonding patterns and independent of amino acid sequence (7, 28–32). Analysis of ~50,000 PDB structures has been reported (33). The statistics of the small 3D structure motifs and their subtype in this set are listed in SI Appendix, Table S1. The motifs included are alpha–beta-motif, asx-motif, asx-turn, beta-bulge, beta-bulge-loop, beta-turn, gamma-turn, nest, niche, Schellman-loop, st-motif, st-staple, and st-turn. We use the same terminology for these motifs as employed by Golovin and Herick and listed in PDBeMotif. The survey found that 16 of the motifs occur in less than 1% of the total number of PDB amino acids. We extracted data on the occurrence of these uncommon motifs in the selected CASP14 targets for both the experimental structures and the AF2 models using the PDBeMotifs server. Nine out of the sixteen motif types were found, involving four of the six structures. A total of eighteen motifs were identified in the experimental structures. Statistics of all the 3D structure motifs considered are provided in Table 2, and the details are provided in SI Appendix, Table S3. All the motifs not present in the calculated structures are associated with crystal environment effects. Where the calculation does not agree with the crystal structure, we examined all five AF2 models to confirm that none predicted the rare motif. Below, we describe interesting cases.
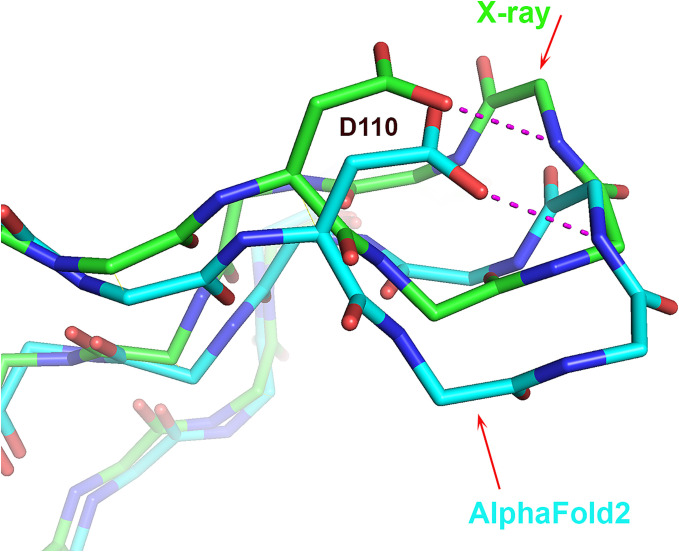
In one case, an experimentally observed rare motif is also found in the calculated structure, though the local structure is displaced relative to experiment. T1090 contains a β-turn il conformation (0.6% frequency) comprising Val111 to Lys114 at the tip of a solvent-exposed β-hairpin, with an estimated average coordinate error of 0.5 Å (Fig. 4). There is an orientation difference of the β-turn between the experimental and calculated structures of up to 3 Å, yet the unusual β-turn il conformation is present in both. Coevolution appears not to determine this feature, since an AF2 calculation in which residues in positions 111 to 114 in the multiple sequence are replaced by gap characters still produced models containing the same β-turn il conformation.
Fig. 4.
Agreement between an experimental and calculated rare 3D structure motif despite local rigid body displacement of a loop. Superposition of part of the T1090 (chromatin remodeling protein Ssr4 N-terminal domain) crystal (green) and AF2 (sky blue) structures, showing an uncommon β-turn il (residues 111 to 114) that is present in both the crystal and calculated structures despite the ~3 Å hinge-like shift of the β-hairpin in the model relative to the crystal structure. The superposition included the entire molecule. There are no crystal contacts in the vicinity of the loop, and its lack of contacts with the rest of the protein suggests a shallow and broad energy minimum.
The crystallographic temperature factors in this region are somewhat elevated compared with those in the protein core, as often observed in solvent-exposed regions within crystals. There are no crystal effects that appear to affect the loop conformation and there is no obvious factor in the protein environment that appears to influence one loop orientation over another. That is, the local energy surface appears fairly flat. Thus, in a structural sense, the calculated loop structure appears to be in error, but the energy discrepancy is likely small. Most importantly, the rare motif is modeled.
Strikingly, thirteen of the eighteen small 3D structure motifs occur in T1074, the BD0675 protein from Bdellovibrio bacteriovorus, a 202-residue protein of unknown function that folds into a distorted β-roll-like structure (Table 1). The AF2 model agrees with eight of these thirteen uncommon motifs. Three of the discordant motifs occur within the loop encompassing residues 88 to 98. All five AF2 structures have loop conformations with no uncommon motifs. The loop is involved in crystal contacts, and superposition of the AF2 structures on the crystal structure shows that the AF2 loop conformations cannot be accommodated in the crystal context because of clashes with a neighboring molecule. Thus, these three experimental rare motifs appear to be an artifact of the crystal packing.
Two more uncommon small 3D motifs in T1074 are not present in the AF2 models. One comprises residues 174 to 177, which adopt a β-turn il conformation in the crystal structure, and the more common type ir in the five AF2 models (estimated model average coordinate error of 0.6 Å). The difference between the two motifs is a flipped peptide bond with little difference in the other atomic positions. In the experimental structure, this β-turn is also intimately involved in a crystal contact and the common ir turn motif in the calculated structure cannot be accommodated because of clashes with the neighboring molecule. In contrast, the experimentally observed peptide enables hydrogen bonding between the peptide CO group and the side chain of Gln201 on the adjacent β-strand, which in turn is also hydrogen bonded to the side chain of Gln177. So here too, the difference appears to be due to crystal packing interactions.
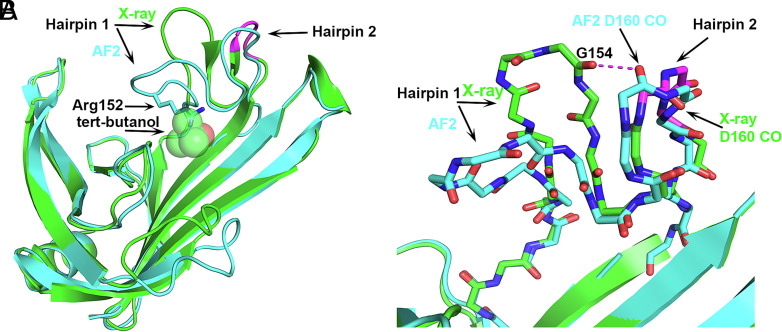
The fifth disagreement between the X-ray and AF2 structures involves a hairpin loop on T1074 that is placed differently in the two structures (labeled Hairpin 1 in Fig. 5). The AF2 placement of Hairpin 1 cannot occur in the crystal structure because it would overlap with a bound tert-butanol molecule, in particular clashing with the side chain of Arg152 on this loop (Fig. 5A). The tert-butanol was included in the crystallization solution but not included in the AF2 calculations. A clash with the tert-butanol is avoided by Hairpin 1 of the crystal structure adopting a more open conformation, i.e., moving further away to enlarge the cleft where the tert-butanol binds. In turn, the open Hairpin 1 conformation requires that an adjacent hairpin loop (Hairpin 2 in Fig. 5) adopts an uncommon 3D motif, niche3l (0.1% frequency), so as to prevent an unfavorable electrostatic interaction (2.9 Å) between two backbone CO groups, one of Gly154 on Hairpin 1, and the second of Asp160 on Hairpin 2 (Fig. 5B). In contrast, Gly154 in the closed Hairpin 1 conformation is placed remotely from Hairpin 2 and Asp160 CO flips over (Fig. 5B). Hairpins 1 and 2 flank a cleft that was proposed to contain a potential ligand binding site (14). Therefore, their mobility may be relevant to the function of the protein, which is currently unknown. That is, a closed Hairpin 1 conformation similar to the one in the AF2 structure may be preferred at the apo-state and the more open crystal structure conformation may be similar to that when the postulated ligand binds. Elevated crystallographic temperature factors in this region (average backbone atoms’ B = 64) and higher predicted coordinate errors by AF2 (1.9 Å) also suggest loop flexibility. Confirmation of this proposal awaits discovery of the protein function and its ligand.
Fig. 5.
Bound ligand (tert-butanol) present in the crystallization solution affecting a hairpin loop location and formation of an uncommon small 3D motif in the crystal structure. Superposition of T1074 (Bdellovibrio bacteriovorus Bd0675) crystal structure (green) and the AF2 model (sky blue). (A) Hairpin 1 open (crystal structure) and closed (AF2 model) conformations. The uncommon niche3l motif on the crystal hairpin 2 of the crystal structure is colored magenta. A tert-butanol is bound in the experimental structure but was not included in the calculated structure. The side chains of Arg152 in the AF2 structure overlap with the tert-butanol molecule. (B) Atomic detail of hairpins 1 and 2 showing that the Asp160 CO group in the uncommon nich3l conformation avoids the short contact with Gly154 CO, which would occur if the Hairpin 2 conformation resembles that in the AF2 calculated structure. Here, the AF2 Hairpin 2 conformation is likely more representative of the in vivo structure in a postulated ligand-free state.
Uncommon Structural Features in AF2 Models Absent in the Experimental Structures.
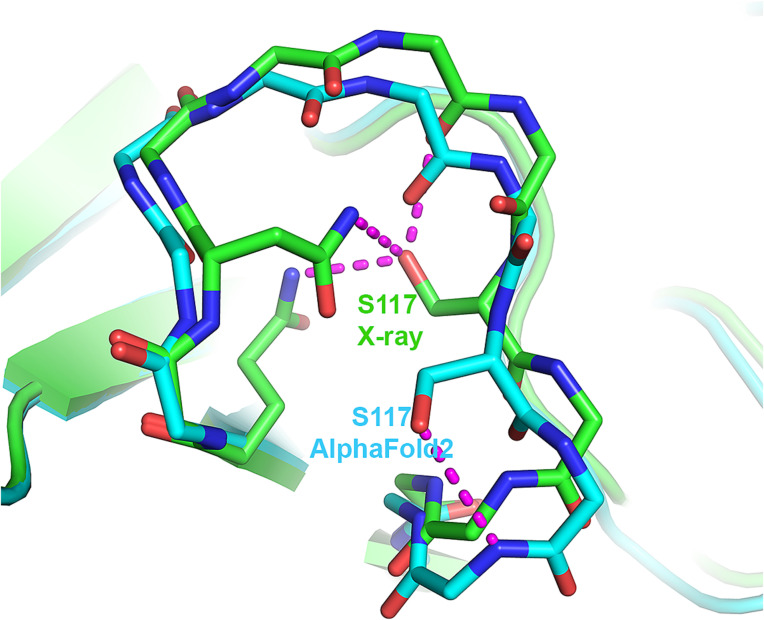
None of the AF2 structures contain π- or 310-helices that are not found in the experimental structures. However, all five AF2 structures of T1049 contain a single uncommon 3D structure motif that is absent in the experimental structure, a st-turn iil (0.8% frequency) (Table 1 and SI Appendix, Table S3). This motif comprises residues Ser117–Pro118–Arg119 in an omega loop spanning residues 110 to 120 where the estimated average coordinate accuracy is 0.8 Å (Fig. 6). Ser117 is buried in the experimental structure, and the side chain hydroxyl group is well compensated by electrostatic interactions with surrounding hydrophilic groups. In contrast, the Ser117 hydroxyl group of the AF2 structure is on the protein surface and interacts with the backbone NH group Arg119 (Fig. 6). Examination of crystal contacts shows that the AF2 structure in this region cannot be accommodated in the crystal environment, suggesting that the absence of this rare feature in the experimental structure is a consequence of the crystal environment. A BLAST sequence search against the PDB for the Ser–Pro–Arg motif together with the three flanking residues on each side finds ten proteins containing Ser–Pro–Xaa sequences. Of these, five exhibit the rare ST-turn iil conformation, suggesting that a proline in the second position enriches the occurrence of the ST-turn iil compared with other residues. Whichever energetic consideration is involved, for a proline in the second position, the occurrence of the ST-turn iil increases substantially.
Fig. 6.
The single instance where an AF2 model contains an uncommon feature, not present in the corresponding crystal structure. Local superposition of T1049 (receptor-binding domain of adhesin MrpH) crystal structure (green) and AF2 model (sky blue) ω-loop showing the uncommon Ser116 st-turn iil in the AF2 model that is absent in the experimental structure. Instead, Ser116 in the crystal structure is buried and forms three favorable electrostatic interactions. Ser116 in the modeled structure is exposed to the solvent. The ω-loop is involved in crystal contacts, and superposing the entire AF2 model on the experimental structure reveals that the predicted loop conformation would clash badly with a symmetry-related molecule.
Discussion
This study shows that an AI deep learning network is very successful in capturing structural features that occur infrequently in protein structures. Overall, of the 37 rare structural features found in the experimental structures, 30 are found in the corresponding calculated structures. Of the seven not found, six appear to be a consequence of crystal contacts and one a consequence of the presence of a bound organic molecule included in the crystallization solution. Thus, the calculated structures appear to contain all the experimentally observed rare motifs that are not artifacts of the crystal environment. There are two instances of rare motifs found in the calculated structures that are not present in the corresponding experimental ones: an extra residue in a 310-helix which may be a computational error, and a rare turn that the crystal environment could not accommodate. That is, its absence in the experimental structure appears to be a crystallographic artifact.
The success in the identification of rare features provides support for the view that the machine is generalizing from the training data such that it can determine when unusual structural features are the energetically optimal solution. How the machine does this is less obvious. Given the type of data used for training, the implication is that it does something akin to learning a knowledge potential. That is, it learns the probability distributions for the interatomic distances between each pair of atom types in the PDB. These distributions are used to determine the relative probability of the set of interatomic distances present in any conformation. In this sense, the machine likely does something equivalent to learning a potential of mean force (11). Some aspects of the potential, especially packing, are incorporated in the training loss function (2). Most, including electrostatics, are not. The information representing the potential is distributed over the trained network, and thus over a very large number of parameter values. As a result, it may be a more nuanced force field than those traditionally used to represent potentials and may also effectively include higher-than-pairwise terms not usually part of physics-based potentials. We note that this conclusion is not based on direct evidence but is a likely explanation for the machine’s capabilities. A possible confounding factor is the role of coevolution information in determining conformational details. In the limited tests we have done, removing local coevolution signals does not appear to be key. Another study of AF2 properties (34) provides some support for these points of view. That work shows that sets of conformational decoys can be ranked with AF2 in the absence of coevolution information (although coevolution is often important for finding the approximate global minimum—see below).
An algorithm that can compute protein structure from sequence must solve two problems. One is to be able to recognize the lowest free energy conformation among the set of sampled conformations, and there are data showing that classical physics-inspired potentials are able to do this, although the AF2 potential achieves a higher level of agreement with experimental coordinates. The second problem is finding the global free energy conformation, which classical methods have generally not achieved yet, even when starting from a conformation close to the experimental one (4). The examples here demonstrate how difficult this is. For instance, for the π-helix case, creating that feature allows better interactions for an amino acid nine residues away from the inserted residue (Fig. 2), and inspection suggests that there is no physical space smooth energy gradient between the simpler continuous α-helical conformation and the observed arrangement. How AF2 and related machines find the minimum is unclear. One view is that the large neural network transforms the data into a space where there is a monotonic relationship between free energy and conformation (35). A second explanation is that the sequence coevolution restricts the conformational options sufficiently to provide an approximate solution, and the Rooney and Ovchinnikov study (34) takes that view. Note that because of detailed conformational differences within protein families, coevolution is likely inherently not atomic resolution.
It is not a goal of this study to analyze other sorts of discrepancy between the experimental and calculated structures, though we do report a minor one that is possibly a computational error. A recent paper by Terwilliger and colleagues (36) asserts that detailed computational errors are common. In the CASP14 assessment, discrepancies are also fairly common, but a large fraction of those are likely attributable to crystal artifacts (4). Similarly, in this analysis, about 20% of experimental rare features are not found in the calculated structures, but all appear to be crystallographic artifacts. The Terwilliger et al. study uses structure comparisons of pairs of proteins with identical sequences, each crystallized in different lattices to provide an estimate of experimental uncertainty caused by crystal effects. They make an interesting observation that differences are very small at short or moderate distances between residues (at 15 Å Cα atom separation, about 0.1 Å median discrepancy between crystal structure pairs and about 0.25 Å between calculated/experimental pairs). Taken at face value, these data suggest that at this Cα atom separation, the median computed structure inter-Cα error is ~0.15 Å, or 1%. However, that is likely an overestimate, since the comparison of crystal structures in this way is not bias free. First, experimental procedures may tend to make the structures too similar, since one structure is often solved based on the other. Second, different crystal forms of the same protein tend to employ the same regions on the protein surface for crystal contacts, inducing the same crystallographic artifacts. A detailed analysis of this phenomenon has been reported for 25 nonisomorphous crystal forms of T4 lysozyme (37). The protein molecules in this set of structures showed quasi-equivalent association due to utilization of common crystallographic symmetry axes.
There are limitations to our study. Because there are no automated methods for relating structural differences to crystal environmental effects, the number of features analyzed is small. While this is an insufficient number to provide precise statistics on the fraction of rare features produced in calculated structures, it is sufficient to show that fraction is large. Because human interpretation of structural features is required, there is potential for bias. We endeavored to minimize that by scrutinizing each case from the point of view of a champion of computational methods (JM) and an experimentalist (OH) who if anything is biased toward the superiority of experiments. We also decided in advance which rare feature criteria and which structures to include and did not alter these during the work.
Methods
CASP14 single protein or domain targets were considered. Targets were classified as template based (easy and difficult) or template free by the CASP14 organizers based on hidden Markov model analyses and structure similarity to PDB entries (Kinch et al, Proteins 89:1618, 2021). For the purpose of the current study, 23 free modeling proteins were examined. Of these, six structures determined to a resolution limit of 2 Å or better were selected for detailed evaluation. The analyses utilized the computer programs DSSP (38) for secondary structure classification, PDBeMotif (33) for identifying rare motifs, and PyMol (39) for visual inspection and for local and global structure alignment. The features examined are cis peptides, 310-helices, π-helices, and small 3D motifs that occur in the PDB at frequency lower than 1% of the total number of residues, as defined previously (33). The statistics are provided in SI Appendix, Table S1.
CASP allows participants to submit up to five models per target. For each target, we first analyzed both the experimental structure and the AF2 “Best Model” [the model with the smallest overall backbone difference to the experimental structure based on the GDT-TS criterion used by CASP (40)]. Where there was a disagreement between the two structures, we examined all five AF2 predictions to see whether any of the models agreed with the crystal structure and confirmed that no such cases occurred. When a discrepancy between the calculated and experimental structures was identified, we checked for a possible influence of crystal or partner protein contacts on the crystal structure. A previous analysis by a CASP14 assessor showed that these are a very common cause of such discrepancies (4).
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
Author contributions
O.H. and J.M. designed research; performed research; analyzed data; and wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Osnat Herzberg, Email: osnat@umd.edu.
John Moult, Email: jmoult@umd.edu.
Data, Materials, and Software Availability
All study data are included in the main text and/or SI Appendix.
Supporting Information
References
- 1.Jumper J., et al. , Applying and improving AlphaFold at CASP14. Proteins 89, 1711–1721 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jumper J., et al. , Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kryshtafovych A., Schwede T., Topf M., Fidelis K., Moult J., Critical assessment of methods of protein structure prediction (CASP)-Round XIV. Proteins 89, 1607–1617 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simpkin A. J., Sanchez Rodriguez F., Mesdaghi S., Kryshtafovych A., Rigden D. J., Evaluation of model refinement in CASP14. Proteins 89, 1852–1869 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moore P. B., Hendrickson W. A., Henderson R., Brunger A. T., The protein-folding problem: Not yet solved. Science 375, 507 (2022). [DOI] [PubMed] [Google Scholar]
- 6.Skolnick J., Gao M., Zhou H., Singh S., AlphaFold 2: Why it works and its implications for understanding the relationships of protein sequence, structure, and function. J. Chem. Inf. Model. 61, 4827–4831 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richardson J. S., The anatomy and taxonomy of protein structure. Adv. Protein Chem. 34, 167–339 (1981). [DOI] [PubMed] [Google Scholar]
- 8.MacArthur M. W., Thornton J. M., Influence of proline residues on protein conformation. J. Mol. Biol. 218, 397–412 (1991). [DOI] [PubMed] [Google Scholar]
- 9.Wuthrich K., Grathwohl C., A novel approach for studies of the molecular conformations in flexible polypeptides. FEBS Lett. 43, 337–340 (1974). [DOI] [PubMed] [Google Scholar]
- 10.Kinch L. N., Schaeffer R. D., Kryshtafovych A., Grishin N. V., Target classification in the 14th round of the critical assessment of protein structure prediction (CASP14). Proteins 89, 1618–1632 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sippl M. J., Boltzmann’s principle, knowledge-based mean fields and protein folding. An approach to the computational determination of protein structures. J. Comput. Aided. Mol. Des. 7, 473–501 (1993). [DOI] [PubMed] [Google Scholar]
- 12.Krieger I. V., et al. , The structural basis of T4 phage lysis control: DNA as the signal for lysis inhibition. J. Mol. Biol. 432, 4623–4636 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang W., et al. , MrpH, a new class of metal-binding adhesin, requires zinc to mediate biofilm formation. PLoS Pathog. 16, e1008707 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alexander L. T., et al. , Target highlights in CASP14: Analysis of models by structure providers. Proteins 89, 1647–1672 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanamaru S., et al. , Structure and function of the T4 spackle protein Gp61.3. Viruses 12, 1070 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newman J., Nebl T., Van H., Peat T. S., The X-ray crystal structure of the N-terminal domain of Ssr4, a Schizosaccharomyces pombe chromatin-remodelling protein. Acta Crystallogr. F Struct. Biol. Commun. 76, 583–589 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herzberg O., Moult J., Analysis of the steric strain in the polypeptide backbone of protein molecules. Proteins 11, 223–229 (1991). [DOI] [PubMed] [Google Scholar]
- 18.Jabs A., Weiss M. S., Hilgenfeld R., Non-proline cis peptide bonds in proteins. J. Mol. Biol. 286, 291–304 (1999). [DOI] [PubMed] [Google Scholar]
- 19.Cooley R. B., Arp D. J., Karplus P. A., Evolutionary origin of a secondary structure: Pi-helices as cryptic but widespread insertional variations of alpha-helices that enhance protein functionality. J. Mol. Biol. 404, 232–246 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fodje M. N., Al-Karadaghi S., Occurrence, conformational features and amino acid propensities for the pi-helix. Protein Eng. 15, 353–358 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Zemla A., LGA: A method for finding 3D similarities in protein structures. Nucleic Acids Res. 31, 3370–3374 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heinz D. W., Baase W. A., Dahlquist F. W., Matthews B. W., How amino-acid insertions are allowed in an alpha-helix of T4 lysozyme. Nature 361, 561–564 (1993). [DOI] [PubMed] [Google Scholar]
- 23.Keefe L. J., Sondek J., Shortle D., Lattman E. E., The alpha aneurism: A structural motif revealed in an insertion mutant of staphylococcal nuclease. Proc. Natl. Acad. Sci. U.S.A. 90, 3275–3279 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cartailler J. P., Luecke H., Structural and functional characterization of pi bulges and other short intrahelical deformations. Structure 12, 133–144 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Weaver T. M., The pi-helix translates structure into function. Protein Sci. 9, 201–206 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barlow D. J., Thornton J. M., Helix geometry in proteins. J. Mol. Biol. 201, 601–619 (1988). [DOI] [PubMed] [Google Scholar]
- 27.Karpen M. E., de Haseth P. L., Neet K. E., Differences in the amino acid distributions of 3(10)-helices and alpha-helices. Protein Sci. 1, 1333–1342 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abdel-Rahman S. M., Nahata M. C., Stability of fumagillin in an extemporaneously prepared ophthalmic solution. Am. J. Health Syst. Pharm. 56, 547–550 (1999). [PubMed] [Google Scholar]
- 29.Duddy W. J., Nissink J. W., Allen F. H., Milner-White E. J., Mimicry by asx- and ST-turns of the four main types of beta-turn in proteins. Protein Sci. 13, 3051–3055 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Milner-White E. J., Beta-bulges within loops as recurring features of protein structure. Biochim. Biophys. Acta 911, 261–265 (1987). [DOI] [PubMed] [Google Scholar]
- 31.Sibanda B. L., Blundell T. L., Thornton J. M., Conformation of beta-hairpins in protein structures. A systematic classification with applications to modelling by homology, electron density fitting and protein engineering. J. Mol. Biol. 206, 759–777 (1989). [DOI] [PubMed] [Google Scholar]
- 32.Watson J. D., Milner-White E. J., The conformations of polypeptide chains where the main-chain parts of successive residues are enantiomeric. Their occurrence in cation and anion-binding regions of proteins. J. Mol. Biol. 315, 183–191 (2002). [DOI] [PubMed] [Google Scholar]
- 33.Golovin A., Henrick K., MSDmotif: Exploring protein sites and motifs. BMC Bioinformatics 9, 312 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roney J. P., Ovchinnikov S., State-of-the-art estimation of protein model accuracy using alphafold. Phys. Rev. Lett. 129, 238101 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang H. R., Shao J., Salakhutdinov R., "Deep neural networks with multi-branch architectures are intrinsically less non-convex" in Proceedings of the Twenty-Second International Conference on Artificial Intelligence and Statistics, Kamalika C., Masashi S., Eds. (PMLR, Naha, Okinawa, Japan, 2019), pp. 1099–1109. [Google Scholar]
- 36.Terwilliger T. C., et al. , Improved alphafold modeling with implicit experimental information. Nat. Methods 19, 1376–1382 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang X. J., Wozniak J. A., Matthews B. W., Protein flexibility and adaptability seen in 25 crystal forms of T4 lysozyme. J. Mol. Biol. 250, 527–552 (1995). [DOI] [PubMed] [Google Scholar]
- 38.Kabsch W., Sander C., Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22, 2577–2637 (1983). [DOI] [PubMed] [Google Scholar]
- 39.DeLano W. L., The PyMOL Molecular Graphics System (DeLano Scientific, Palo Alto, CA, USA, 2002). [Google Scholar]
- 40.Zemla A., Venclovas J., Moult K. Fidelis., Processing and evaluation of predictions in CASP4. Proteins 5, 13–21 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the main text and/or SI Appendix.