Significance
During human pregnancy, the placenta is crucial for normal fetal growth and development. Defects in early placenta development can result in severe pregnancy complications. To better understand the molecular mechanisms underlying placenta development, we investigated the role of the paralogous proteins E1A-binding protein p300 (EP300) and CREB-binding protein (CREBBP) in trophoblast differentiation using human first-trimester placenta cell models. We found that EP300, but not CREBBP, is required for differentiation into the two major trophoblast cell types. This may explain the increased incidence of severe pregnancy complications associated with mutations in EP300, and not CREBBP, in Rubinstein–Taybi syndrome pregnancies.
Keywords: placenta development, Rubinstein–Taybi syndrome, E1A-binding protein p300, CREB-binding protein, organoids
Abstract
Early placenta development involves cytotrophoblast differentiation into extravillous trophoblast (EVT) and syncytiotrophoblast (STB). Defective trophoblast development and function may result in severe pregnancy complications, including fetal growth restriction and pre-eclampsia. The incidence of these complications is increased in pregnancies of fetuses affected by Rubinstein–Taybi syndrome, a developmental disorder predominantly caused by heterozygous mutations in CREB-binding protein (CREBBP) or E1A-binding protein p300 (EP300). Although the acetyltransferases CREBBP and EP300 are paralogs with many overlapping functions, the increased incidence of pregnancy complications is specific for EP300 mutations. We hypothesized that these complications have their origin in early placentation and that EP300 is involved in that process. Therefore, we investigated the role of EP300 and CREBBP in trophoblast differentiation, using human trophoblast stem cells (TSCs) and trophoblast organoids. We found that pharmacological CREBBP/EP300 inhibition blocks differentiation of TSCs into both EVT and STB lineages, and results in an expansion of TSC-like cells under differentiation-inducing conditions. Specific targeting by RNA interference or CRISPR/Cas9-mediated mutagenesis demonstrated that knockdown of EP300 but not CREBBP, inhibits trophoblast differentiation, consistent with the complications seen in Rubinstein–Taybi syndrome pregnancies. By transcriptome sequencing, we identified transforming growth factor alpha (TGFA, encoding TGF-α) as being strongly upregulated upon EP300 knockdown. Moreover, supplementing differentiation medium with TGF-α, which is a ligand for the epidermal growth factor receptor (EGFR), likewise affected trophoblast differentiation and resulted in increased TSC-like cell proliferation. These findings suggest that EP300 facilitates trophoblast differentiation by interfering with at least EGFR signaling, pointing towards a crucial role for EP300 in early human placentation.
The human placenta is an extra-embryonic organ essential for proper growth, development and survival of the embryo and fetus. During the first trimester of human pregnancy, proliferative cytotrophoblasts (CTBs) differentiate into syncytiotrophoblast (STB) and extravillous trophoblast (EVT) (1, 2). The multinucleated STB layer facilitates maternal–fetal exchange of nutrients and gases, functions as a protective immunological barrier and secretes hormones into the maternal circulation to maintain pregnancy. On the other hand, EVTs undergo epithelial-mesenchymal transition (EMT) and invade the maternal decidua and spiral arteries (3). Subsequently, EVTs remodel the spiral arteries, thereby allowing an appropriate blood supply towards the fetus. Defective trophoblast development and function are associated with severe pregnancy complications, including early-onset fetal growth restriction (FGR) and pre-eclampsia (1, 2). Currently, these pregnancy complications cannot be cured and account for a large proportion of neonatal and maternal morbidity (1).
The lack of proper model systems to study early human placentation has been a major issue in placenta research. However, derivation of human trophoblast stem cells (TSCs) from first-trimester placental tissue has recently been reported (4). These TSCs can be cultured long term, induced to differentiate into STB and EVT lineages, and they fulfill the criteria for first-trimester trophoblast cells as defined by Lee et al. (5). In addition, trophoblast organoids have been established from first-trimester placentas, which can also undergo EVT differentiation (6, 7). These model systems have considerably increased our possibilities to study human first-trimester placenta development in vitro. Recent studies have already reported some important regulatory pathways that control trophoblast proliferation and differentiation (8–17), yet our understanding of these processes remains limited.
A meta-analysis of human term placental transcriptome data has indicated a role for reduced levels of CREB-binding protein (CREBBP) and E1A-binding protein p300 (EP300) in pre-eclamptic pregnancies compared to healthy pregnancies (18). The paralogs CREBBP and EP300 are ubiquitously expressed and involved in many fundamental biological processes, including cell growth, proliferation, differentiation and apoptosis (19). They function as lysine acetyltransferases and transcriptional coactivators, and interact with a large number of proteins (20–22). Heterozygous mutations in CREBBP or EP300 are also known to cause Rubinstein–Taybi syndrome, a congenital developmental disorder characterized by intellectual disability, postnatal growth retardation, skeletal abnormalities and facial dysmorphisms (23). Remarkably, an increased incidence of FGR and pre-eclampsia have been reported in pregnancies of Rubinstein–Taybi syndrome-affected fetuses with EP300 mutations, but not CREBBP mutations (24, 25). We hypothesized that these pregnancy complications have their origin in early placenta development, and that particularly EP300 plays an important role in this process by regulating trophoblast differentiation and/or function.
In this study, we investigated the role of CREBBP and EP300 in trophoblast differentiation. We generated human TSCs from first-trimester placental tissue and cultured them as monolayer cells and trophoblast organoids. By inhibition of CREBBP/EP300 in these models using pharmacological inhibitors, RNA interference and CRISPR/Cas9-mediated mutagenesis, we demonstrated that EP300, but not CREBBP, is required for differentiation of TSCs into both EVT and STB lineages. These data indicate that EP300 is an important regulator of trophoblast differentiation, offering insights into the molecular mechanisms driving normal and pathological placenta development.
Results
Generation of Human TSCs and Trophoblast Organoids.
We generated three independent TSC lines (TSC_1, TSC_2 and TSC_3) from first-trimester placentas using methods described by Okae et al. (4) (SI Appendix, Fig. S1A). The three TSC lines slightly differed in proliferation rate (SI Appendix, Fig. S1B) and differentiation characteristics (SI Appendix, Fig. S1 C and D). However, all TSC lines continued to proliferate for at least 28 passages and displayed characteristic cobblestone-shaped morphology and TEA domain transcription factor 4 (TEAD4)/tumor protein p63 (TP63) expression. By changing components in the culture medium, the TSCs could differentiate into chorionic gonadotropin subunit beta (CGB, encoding hCGβ)/syndecan 1 (SDC1)-positive, multinucleated STBs, and major histocompatibility complex, class I, G(HLA-G)/matrix metallopeptidase 2(MMP2)-positive EVTs with mesenchymal morphology. In addition, we generated trophoblast organoids from the TSC lines by seeding them in Matrigel droplets. Organoids from all TSC lines could also give rise to HLA-G/MMP2-positive EVTs (SI Appendix, Fig. S1 C and D). Unless otherwise stated, we used TSC_1 for further experiments.
Organoids were further characterized by immunofluorescence staining and confocal imaging. The trophoblast organoids consisted of an outer layer of TP63/E-cadherin-positive CTBs and an inner layer of hCG-positive STBs (SI Appendix, Fig. S2A). Under EVT-inducing conditions, the organoids formed protrusions of HLA-G-positive EVTs that invaded the Matrigel radially (SI Appendix, Fig. S2B).
Pharmacological CREBBP/EP300 Inhibition Blocks EVT and STB Differentiation.
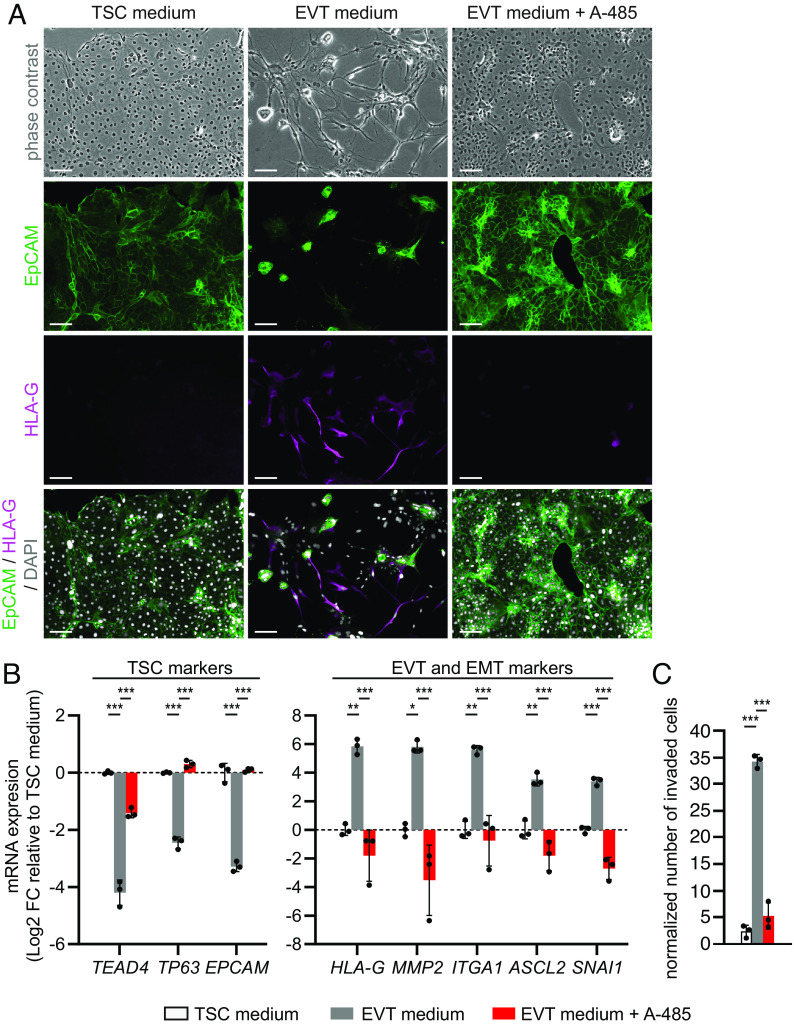
To investigate the effect of CREBBP/EP300 inhibition on trophoblast differentiation, the CREBBP/EP300 lysine acetyltransferase domain inhibitor A-485 (26) was added to EVT or STB medium throughout the differentiation protocols. Under EVT-inducing conditions, A-485-treated cells did not acquire the mesenchymal morphology characteristic of EVT cells, but rather looked like cobblestone-shaped TSC-like cells forming patches (Fig. 1A). Immunostaining showed that the A-485-treated cells were negative for the EVT marker HLA-G and positive for the TSC marker epithelial cell adhesion molecule (EpCAM) (27). In line with this, the mRNA levels of TSC markers (TEAD4, TP63, EPCAM) were increased in A-485-treated cells compared to untreated EVTs, while expression of EVT markers (HLA-G, MMP2, integrin subunit alpha 1 (ITGA1) (4)) and EMT-related transcription factors (achaete-scute family bHLH transcription factor 2 (ASCL2) (11, 28), snail family transcriptional repressor 1 (SNAI1) (29)) were reduced (Fig. 1B). In addition, transwell invasion assays showed that the invasive capacity of A-485-treated cells was reduced compared to untreated EVTs, similarly as TSCs (Fig. 1C), which further demonstrates their TSC-like identity.
Fig. 1.
The CREBBP/EP300 inhibitor A-485 impairs EVT differentiation. A-485 (1 µM) was added to the EVT differentiation medium and this condition was compared to untreated TSCs and EVTs. (A) Representative phase-contrast and immunofluorescence images of cells stained for EpCAM (TSC marker, green), HLA-G (EVT marker, magenta) and 4′,6-diamidino-2-phenylindole (DAPI) (nuclei, white). Cells in TSC medium were analyzed on day 3 and cells in EVT medium ± A-485 on day 6 after seeding. (Scale bars: 100 µm.) (B) TSC and EVT/EMT marker mRNA expression as measured by qPCR. Cells in TSC medium were analyzed on day 3 and cells in EVT medium ± A-485 on day 8 after seeding. Bars represent mean log2 fold-change (FC) ± SD, relative to TSC medium (n = 3 independent experiments). (C) Transwell invasion assay. Bars represent mean normalized number of invaded cells ± SD (n = 3 independent experiments). Color legend corresponds to (B and C). *P < 0.05, **P < 0.01, ***P < 0.001.
To examine whether cells treated with A-485 in EVT medium for 7 d retained stem cell characteristics, we passaged untreated and A-485-treated cells into TSC medium without inhibitor for 2 d (SI Appendix, Fig. S3A). A-485-treated cells grew similarly as control TSCs, forming proliferating cell patches, and showed high viability (SI Appendix, Fig. S3 B and C). In contrast, untreated EVT cells did not form proliferating cell patches and viability of these cells was decreased. Subsequently, we assessed whether these cells could still form organoids and differentiate into STBs. For A-485-treated cells, the number of organoids formed as well as size of these organoids were comparable to control organoids (SI Appendix, Fig. S3 B and D). The number and size of organoids formed from untreated EVT cells, however, were decreased compared to control organoids. Judging from morphology and marker expression, the A-485-treated cells in monolayer were still able to undergo STB differentiation, forming multinucleated structures and having a similar marker expression profile as control STBs after incubation in STB medium (SI Appendix, Fig. S3 B and E). In contrast, as expected, the capacity of untreated EVT cells to differentiate into STBs was low, since cell morphology was less STB-like and CGB expression was reduced, whereas these cells still highly expressed the EVT markers HLA-G and MMP2 after STB-inducing conditions. This experiment shows that CREBBP/EP300 inhibition under EVT-inducing conditions rescues the ability of at least a large proportion of these cells to self-renew, give rise to different trophoblast cell types and to form organoids, thus preserving their stem cell characteristics.
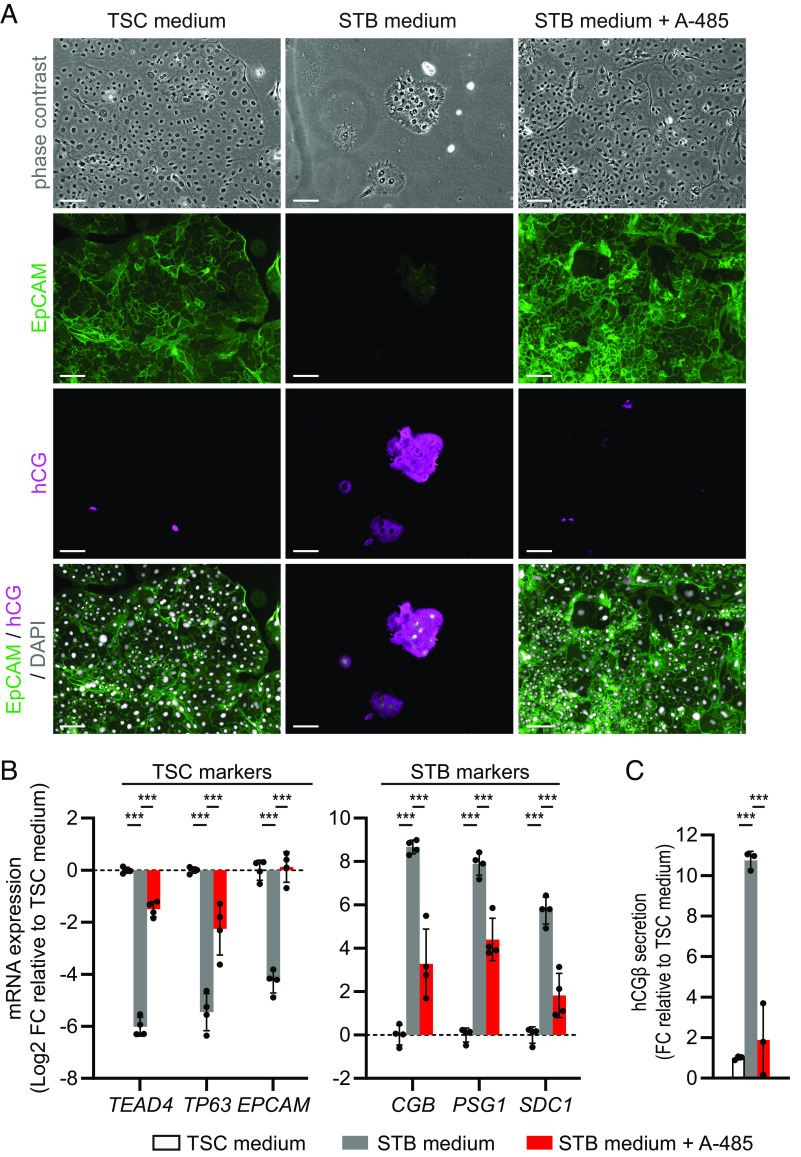
Under STB-inducing conditions, A-485-treated cells also formed proliferating cell patches and resembled TSCs more than STBs (Fig. 2A). Immunostaining showed that the A-485-treated cells were negative for the STB marker hCG and positive for the TSC marker EpCAM. In addition, A-485-treated cells displayed higher mRNA expression of TSC markers (TEAD4, TP63, EPCAM) and lower expression of STB markers (CGB, pregnancy specific beta-1-glycoprotein 1 (PSG1), SDC1) compared to untreated STBs (Fig. 2B). In line with this, secretion of hCGβ into the medium was reduced in A-485-treated cells compared to untreated STBs (Fig. 2C).
Fig. 2.
The CREBBP/EP300 inhibitor A-485 impairs STB differentiation. A-485 (1 µM) was added to STB differentiation medium and this condition was compared to untreated TSCs and STBs. Cells in TSC medium were analyzed on day 3 and cells in STB medium ± A-485 on day 5 after seeding. (A) Representative phase-contrast and immunofluorescence microscopy images of cells stained for EpCAM (TSC marker, green), hCG (STB marker, magenta) and DAPI (nuclei, white). (Scale bars: 100 µm.) (B) qPCR analysis of TSC marker and STB marker mRNA expression. Bars represent mean log2 fold-change (FC) ± SD relative to TSC medium (n = 4 independent experiments). (C) ELISA for hCGβ secretion into the medium within 24 h. TSC medium was collected between day 2 to 3 and STB medium ± A-485 between day 4 to 5. Bars represent mean fold-change (FC) ± SD relative to TSC medium (n = 3 independent experiments). Color legend corresponds to (B and C). ***P < 0.001.
The EP300/CREBBP bromodomain inhibitor CPI-637 (30) showed similar inhibitory effects as A-485 on both EVT and STB differentiation (SI Appendix, Fig. S4). Together, these results show that CREBBP/EP300 inhibition keeps trophoblasts in an undifferentiated state under differentiation-inducing conditions, suggesting that CREBBP/EP300 activity is essential for suppression of the undifferentiated state and thereby induction of both EVT and STB differentiation.
EP300 but not CREBBP Knockdown Inhibits Trophoblast Differentiation.
Since the pharmacological inhibitors target both CREBBP and EP300, we next performed specific siRNA-mediated knockdown to discriminate between the contribution of the individual proteins. TSCs were transfected with EP300 siRNA, CREBBP siRNA, or nontargeting siRNA as a control, and EVT differentiation was induced the following day. Given the relatively short-term effect of transfected siRNA, we harvested the siRNA-transfected cells at day 4 of EVT differentiation, when first signs of mesenchymal morphology typically appear (SI Appendix, Fig. S5A). We selected MMP2 and ITGA1 as relevant EVT markers in this early stage, based on their early onset expression during monolayer EVT differentiation (SI Appendix, Fig. S5B). HLA-G, ASCL2 and SNAI1 were upregulated in a later stage, and TSC markers TEAD4, TP63, EPCAM and ETS proto-oncogene 1, transcription factor (ETS1) all reduced in expression simultaneously.
The EP300 and CREBBP siRNA transfections resulted in a knockdown of approximately 40 and 30%, respectively, on mRNA level (Fig. 3A) and approximately 40% on protein level (SI Appendix, Fig. S6A) compared to the nontargeting siRNA-transfected control. Even though knockdown efficiency was modest, EVT differentiation was blocked in EP300 siRNA-transfected cells, as these cells grew in TSC-like patches under EVT-inducing conditions (Fig. 3B). In line with this, expression of the TSC marker TEAD4 was increased and expression of EVT markers MMP2 and ITGA1 were decreased in EP300 siRNA-transfected cells compared to nontargeting siRNA-transfected cells (Fig. 3C). Interestingly, CREBBP knockdown appeared to have no effect on EVT differentiation, as cells transfected with CREBBP siRNA displayed similar morphology and marker expression as the nontargeting siRNA-transfected controls (Fig. 3 B and C).
Fig. 3.
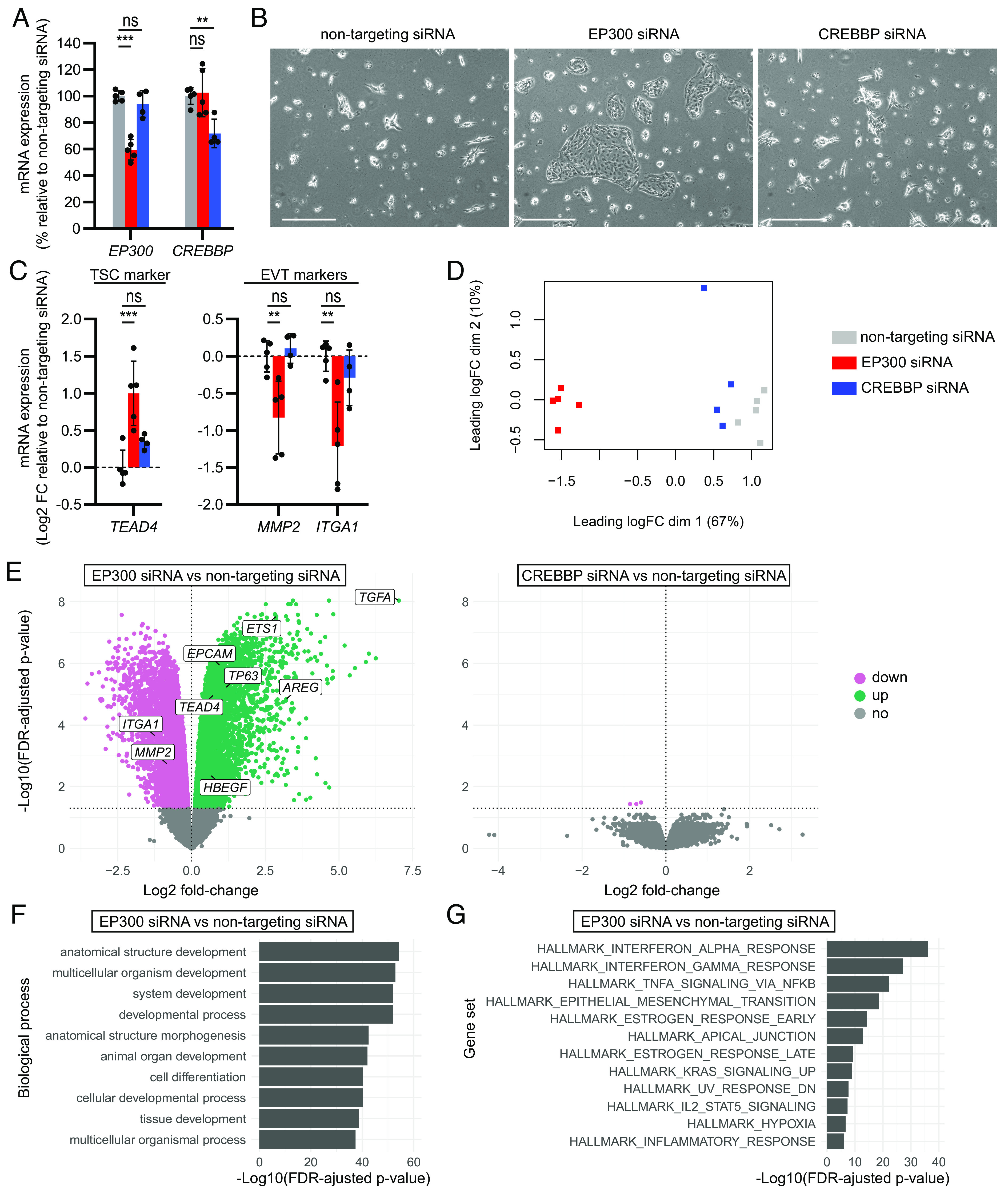
EP300 but not CREBBP knockdown by siRNA inhibits EVT differentiation. TSCs were transfected with EP300 siRNA_1, CREBBP siRNA_1 or nontargeting siRNA (n = 5, n = 4 and n = 5 independent experiments, respectively), and EVT differentiation was initiated on the next day. Cells were harvested for RNA sequencing on post-transfection day 5, EVT differentiation day 4. (A) EP300 and CREBBP mRNA expression as percentage relative to the nontargeting siRNA condition, measured by qPCR. (B) Representative phase-contrast images. (Scale bars: 500 µm.) (C) TSC and EVT marker expression as log2 fold-change (FC) relative to the nontargeting siRNA condition, measured by qPCR. All bars represent mean ± SD. **P < 0.01, ***P < 0.001, ns: not significant. (D) Multidimensional scaling plot showing sample clustering based on batch effect-corrected log2 counts per million. (E) Volcano plots showing differentially up- and downregulated genes (FDR-adjusted P value < 0.05) for EP300 siRNA (Left) and CREBBP siRNA (Right) conditions compared to the nontargeting siRNA control. (F) Top overrepresented biological process GO terms and (G) enriched gene sets within differentially expressed genes (FDR-adjusted P value < 0.05 and absolute log2 fold-change > 0.6) of EP300 siRNA compared to nontargeting siRNA conditions.
We performed bulk RNA sequencing on the siRNA-transfected cells to get more insight into genes and pathways regulated by EP300 and CREBBP during EVT differentiation. Sample clustering by multidimensional scaling (Fig. 3D) and hierarchical clustering of the top 500 most variable genes across samples (SI Appendix, Fig. S6B) revealed two clear clusters: one containing the EP300 siRNA-transfected samples and the other one containing the nontargeting and CREBBP siRNA-transfected samples. For the EP300 siRNA group we found 9,943 differentially expressed genes (DEGs) compared to the nontargeting siRNA-transfected controls (FDR-adjusted P value < 0.05) (Fig. 3E). In line with the TSC-like morphology, upregulated DEGs included the TSC markers TEAD4, TP63, EPCAM and ETS1 and downregulated DEGs included the EVT markers ITGA1 and MMP2 (Fig. 3E and SI Appendix, Fig. S6C). In sharp contrast, transfection with CREBBP siRNA resulted in only 3 DEGs compared to the nontargeting siRNA-transfected controls, including CREBBP itself (Fig. 3E), demonstrating no obvious effects of CREBBP knockdown during EVT differentiation. Gene ontology (GO) analysis of DEGs in the EP300 siRNA group compared to nontargeting siRNA-transfected controls revealed over representation of several biological process GO terms related to development and differentiation (Fig. 3F). Gene set enrichment analysis demonstrated an enrichment for several inflammation-related gene sets and the epithelial–mesenchymal transition gene set (Fig. 3G), probably reflecting the reduced EVT and increased TSC-like signature.
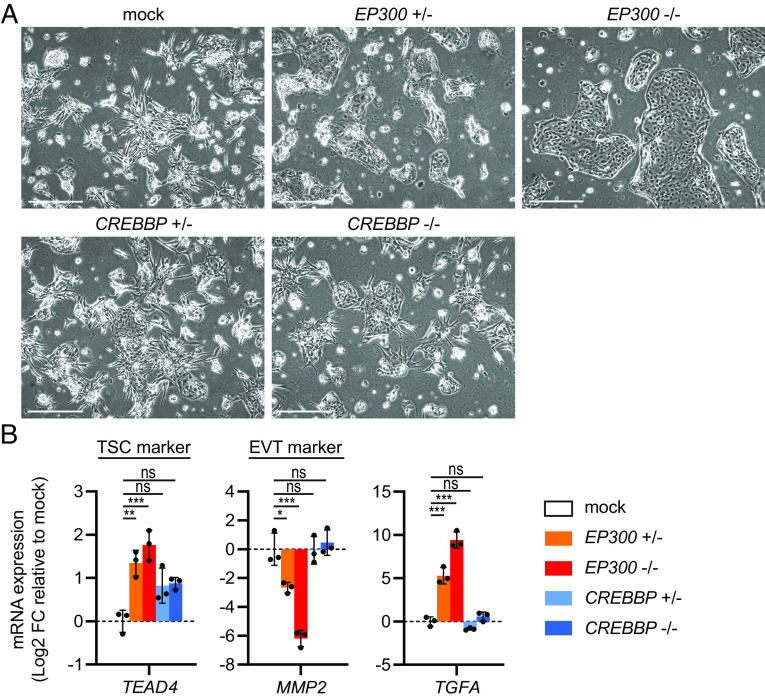
The effects of EP300 and CREBBP siRNA-mediated knockdown on EVT differentiation could be reproduced using a distinct siRNA sequence (SI Appendix, Fig. S7) as well as in the TSC_2 and TSC_3 lines (SI Appendix, Fig. S8). Moreover, we validated these findings using heterozygous and homozygous knockout TSCs for EP300 and CREBBP, in which premature stop codons were introduced by CRISPR/Cas9-mediated mutagenesis (SI Appendix, Table S1 and Fig. S9). As expected, the EP300 heterozygous and homozygous knockout TSCs, but not the CREBBP knockouts, recapitulated the reduced EVT differentiation (Fig. 4 A and B). Together, these findings suggest that EP300 is required for EVT differentiation, whereas CREBBP is not as important in this process.
Fig. 4.
CRISPR/Cas9-mediated knockout of EP300 but not CREBBP inhibits EVT differentiation. Heterozygous and homozygous knockout TSCs for EP300 or CREBBP were induced to undergo EVT differentiation. Cells were harvested for analysis on EVT differentiation day 6. (A) Representative phase-contrast images. (Scale bars: 500 µm.) (B) mRNA expression levels of TSC marker TEAD4, EVT marker MMP2, and TGFA, measured by qPCR. Bars represent mean log2 fold-change (FC) ± SD relative to the mock control (n = 3 independent experiments). *P < 0.05, **P < 0.01, ***P < 0.001, ns: not significant.
Upregulation of Epidermal Growth Factor Receptor (EGFR) Ligands Partially Explains the Inhibitory Effect of EP300 Knockdown on Trophoblast Differentiation.
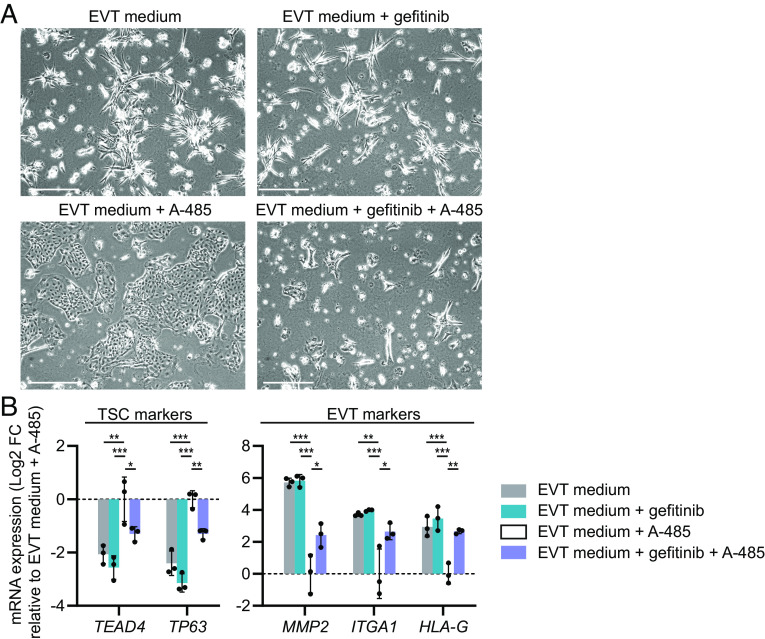
The most outstanding upregulated DEG upon EP300 siRNA-mediated knockdown was transforming growth factor alpha (TGFA, encoding TGF-α) (log2 fold-change = 7.03, FDR-adjusted P value = 9 × 10−9) (Fig. 3E and SI Appendix, Fig. S6D). TGFA was also upregulated in the TSC_2 and TSC_3 lines transfected with EP300 siRNA (SI Appendix, Fig. S8 C and F). In addition, inhibition of EVT differentiation by CRISPR/Cas9-mediated EP300 knockout (Fig. 4B) or pharmacological CREBBP/EP300 inhibition (SI Appendix, Fig. S10) was also accompanied by a pronounced TGFA upregulation, indicating that this was not a cell line- or method-specific effect. TGF-α is related to EGF, both acting as ligands for the EGFR (31). Similarly as TGFA, other ligands for the EGFR, including amphiregulin (AREG) and heparin-binding EGF-like growth factor (HBEGF), were also upregulated upon EP300 siRNA-mediated knockdown (Fig. 3E and SI Appendix, Fig. S6D). EGF is required for proliferation of various epithelial stem cells, including TSCs, and is a component of the TSC culture medium (4). Therefore, upregulation of EGFR ligands may explain why EP300 knockdown under EVT-inducing conditions resulted in a TSC-like cell signature. To test whether EGF or TGF-α could mimic the effect of EP300 knockdown on EVT differentiation, we supplemented EVT medium with EGF or TGF-α in the same concentration as in TSC medium, and analyzed the cells at EVT differentiation day 4. Indeed, addition of EGF or TGF-α also resulted in TSC-like proliferating cell patches in EVT medium, expressing increased levels of TSC markers and reduced levels of early EVT markers compared to the EVT control (Fig. 5 A and B). A similar, yet slightly less pronounced effect of EGF and TGF-α supplementation was seen on STB differentiation (SI Appendix, Fig. S11). The opposite effect was observed in the presence of the EGFR inhibitor gefitinib, as addition of gefitinib prevented, although not completely, the A-485-mediated inhibition of EVT differentiation (Fig. 6 A and B). Together, these results suggest that EP300 suppresses proliferation by downregulating EGFR ligands, and that this mechanism enables differentiation.
Fig. 5.
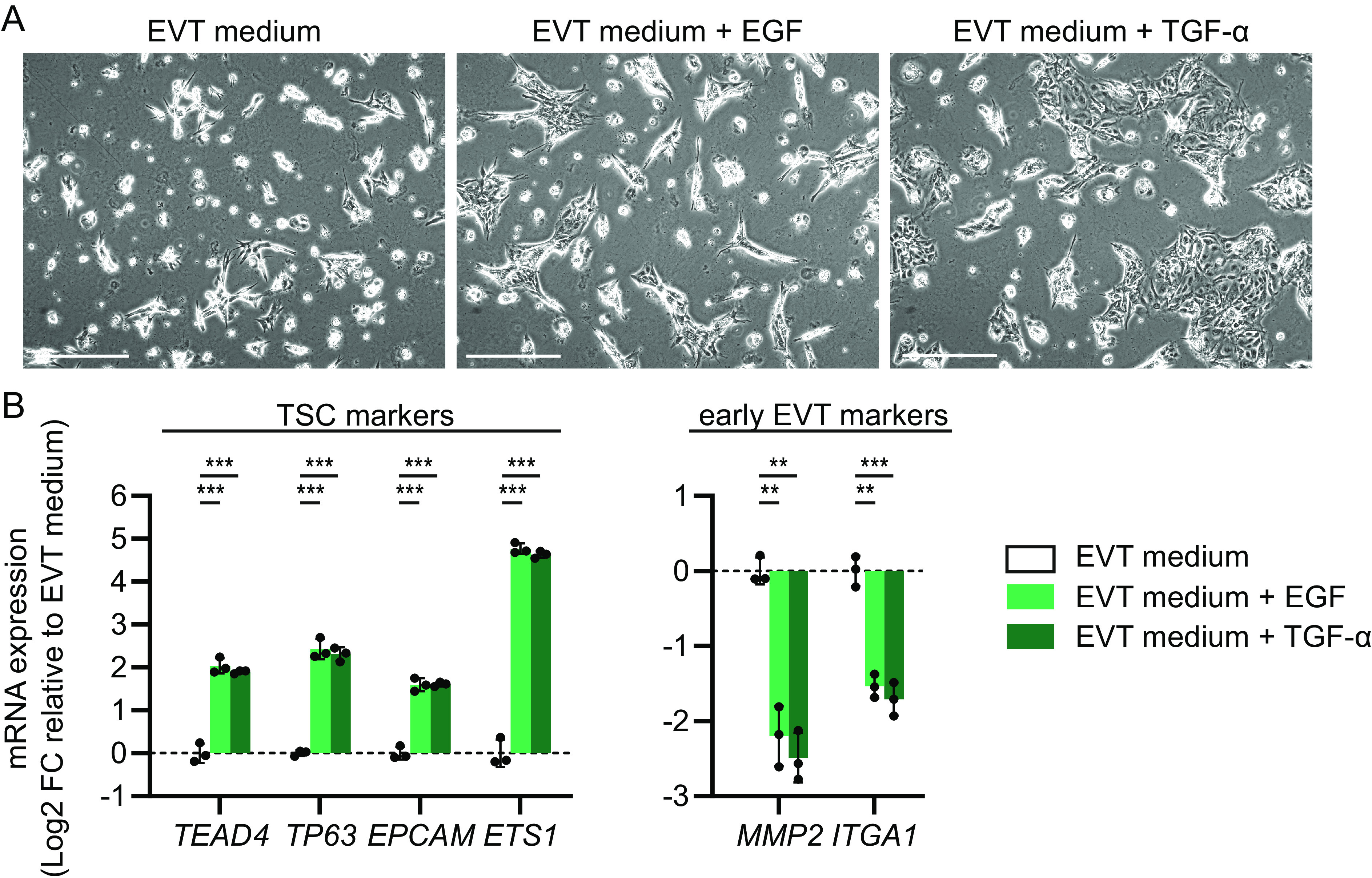
EGF or TGF-α supplementation increases TSC proliferation under EVT-inducing conditions. Human recombinant EGF or TGF-α (50 ng/mL) were added to EVT medium. Cells were analyzed on day 4 of the EVT differentiation protocol. (A) Representative phase-contrast images of untreated cells and cells treated with EGF or TGF-α in EVT medium. (Scale bars: 500 µm.) (B) mRNA expression of TSC and EVT markers, measured by qPCR. Bars represent mean log2 fold-change (FC) ± SD relative to EVT medium control condition (n = 3 independent experiments). **P < 0.01, ***P < 0.001.
Fig. 6.
The EGFR inhibitor gefitinib partially rescues the A-485-mediated inhibition of EVT differentiation. TSCs were pre-treated with gefitinib (2 µM) for 1 day in EVT medium, after which the CREBBP/EP300 inhibitor A-485 (1 µM) was added. Controls were untreated, or treated with only gefitinib or A-485 in EVT medium. Cells were harvested on EVT differentiation day 6. (A) Representative phase-contrast images. (Scale bars: 500 µm.) (B) mRNA expression of TSC and EVT markers, measured by qPCR. Bars represent mean log2 fold-change (FC) ± SD, relative to EVT medium + A-485 (n = 3 independent experiments). *P < 0.05, **P < 0.01, ***P < 0.001.
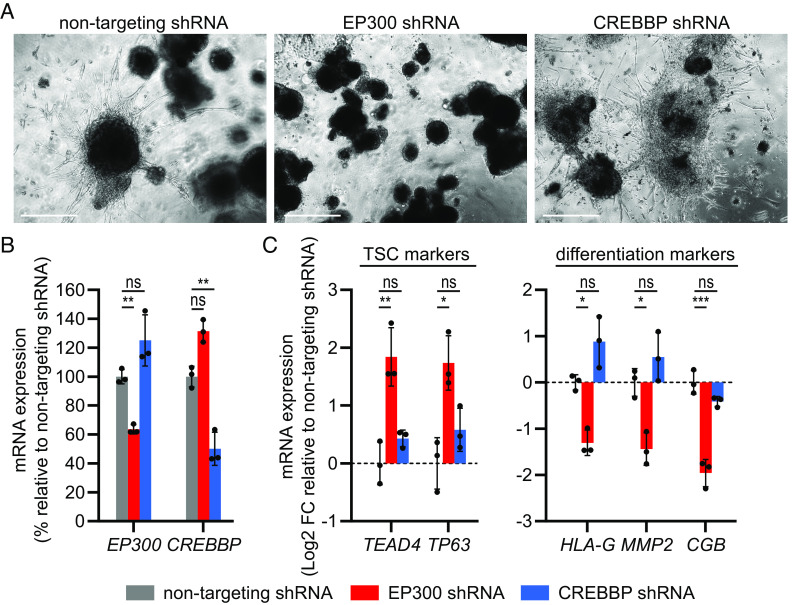
EP300 but not CREBBP Knockdown by shRNA Inhibits Differentiation within Trophoblast Organoids.
Compared to monolayer cell cultures, organoids better recapitulate human physiology and developmental biology as they self-organize into three-dimensional structures that may contain different cell types (32). To investigate the effects of EP300 and CREBBP knockdown in trophoblast organoids, we transduced TSCs with lentivirus carrying EP300 or CREBBP shRNA for stable knockdown, or nontargeting shRNA as control. We generated organoids from these cells and induced EVT differentiation, after which organoids were imaged and harvested for qPCR analysis. CREBBP shRNA-transduced organoids clearly showed the appearance of invasive EVT protrusions, similarly as nontargeting shRNA-transduced organoids, whereas the EP300 shRNA-transduced organoids did not form any clear EVT outgrowth (Fig. 7A). Again, the knockdown efficiency was modest: approximately 40% for EP300 and 50% for CREBBP on mRNA level, compared to the nontargeting shRNA control (Fig. 7B). The mRNA expression levels of TSC markers (TEAD4, TP63) and differentiation markers (EVT markers HLA-G and MMP2, STB marker CGB) were similar in CREBBP shRNA-transduced organoids compared to nontargeting shRNA-transduced organoids (Fig. 7C), indicating normal EVT differentiation. In contrast, EP300 shRNA-transduced organoids showed increased expression of TSC markers and decreased expression of differentiation markers compared to nontargeting shRNA-transduced organoids. These findings are consistent with the effects of siRNA-mediated EP300 and CREBBP knockdown in monolayer cells, and indicate that sufficient EP300 levels are required for trophoblast differentiation, whereas CREBBP seems to be less relevant.
Fig. 7.
EP300 but not CREBBP knockdown by shRNA inhibits differentiation in trophoblast organoids. TSCs transduced with nontargeting, EP300 or CREBBP shRNA were used to generate organoids and induce organoid-EVT differentiation. Organoids were analyzed on EVT differentiation day 12. (A) Representative phase-contrast images. (Scale bars: 500 µm.) (B) EP300 and CREBBP knockdown on mRNA level. Bars represent mean ± SD as percentage relative to the mean of nontargeting shRNA (n = 3 independent experiments). (C) mRNA expression of TSC and differentiation markers. Bars represent mean log2 fold-change (FC) ± SD relative to the mean of nontargeting shRNA (n = 3 independent experiments). Color legend corresponds to (B and C). *P < 0.05, **P < 0.01, ***P < 0.001, ns: not significant.
Discussion
In the present study, we investigated the role of CREBBP and EP300 in human trophoblast differentiation. We showed that treatment with pharmacological CREBBP/EP300 inhibitors blocks the capacity of TSCs to differentiate into functional STBs and EVTs, and results in an expansion of the TEAD4/TP63/EPCAM-positive TSC population under differentiation-inducing conditions. Separate inhibition by RNA interference or CRISPR/Cas9-mediated mutagenesis demonstrated that EP300 but not CREBBP knockdown interferes with differentiation within monolayer trophoblast cells and trophoblast organoids. Transcriptome sequencing revealed the EGFR ligand TGFA as top DEG, being strongly upregulated upon EP300 knockdown. Moreover, supplementing differentiation medium with TGF-α or EGF likewise inhibited STB and EVT differentiation, whereas inhibiting the EGFR had the opposite effect. These findings suggest that EP300 facilitates trophoblast differentiation by at least inhibiting EGFR signaling, which may be a key pathway in normal and pathological human placentation.
Our finding that EP300 but not CREBBP is required for human trophoblast differentiation is consistent with and may explain the increased incidence of complications in pregnancies of Rubinstein–Taybi syndrome-affected fetuses, which is only seen for EP300 mutations and not for CREBBP mutations (24, 25). This suggests that EP300 mutations in the fetus, and thus also in the placenta, which is of fetal origin, result in increased trophoblast proliferation and reduced differentiation into specialized trophoblast cell types. This might hamper normal placenta functioning and spiral artery remodeling, which is clinically manifested as an increased incidence of pre-eclampsia and FGR in these pregnancies.
Using RNA interference, we obtained CREBBP and EP300 levels that approach the levels of Rubinstein–Taybi syndrome patients. These patients carry a heterozygous mutation in CREBBP or EP300, resulting in a reduction of approximately 50% of protein levels (33). In our experiments, around 40% EP300 knockdown on protein level was already sufficient for inhibiting differentiation. Since we found no effect of CREBBP knockdown using two different approaches, siRNA in monolayer cells and shRNA in organoids, we believe that this was not due to inefficient CREBBP knockdown. Moreover, we validated our results using CRISPR/Cas9-mediated knockout TSCs for EP300 or CREBBP. Although CREBBP and EP300 have high sequence homology and many overlapping functions (20–22), they also have some structural and functional differences (19, 34, 35). Our data may also indicate a differential role for CREBBP and EP300 in the context of trophoblasts. However, another explanation could be that EP300 and CREBBP have similar functions, but that EP300 plays a more crucial role in TSCs simply because its expression levels are higher [at least mRNA levels, according to our current RNA sequencing data and data of Okae et al. (4)]. Knocking down CREBBP would then not have any effects, as EP300 levels make up the majority of the combined CREBBP/EP300 levels and are therefore still sufficient. This is supported by the low number of DEGs found in our RNA sequencing data upon CREBBP knockdown.
In our experiments, trophoblast differentiation was strongly affected by targeting the CREBBP/EP300 lysine acetyltransferase domain and bromodomain using the pharmacological inhibitors A-485 and CPI-637, respectively. A-485 directly blocks lysine acetyltransferase activity, whereas CPI-637 interferes with the recognition of acetylated lysine residues (26, 30). Histone acetylation and deacetylation play an important role in the switch between proliferation and differentiation (36) and in EMT (37). In trophoblasts, changes in acetylation status of several histone lysine residues (H2BK5, H3K9, H3K27) are associated with differentiation (38, 39). Weinert et al. (2018) have determined the CREBBP and EP300 acetylome in mouse embryonic fibroblasts, and found that they acetylate all four core histones as well as many nonhistone substrates (40). These substrates include transcriptional effectors in several signaling pathways related to development and differentiation, such as the Wnt/β-catenin, hippo, hedgehog, notch and TGF-β pathway. Although the large number of interactions (as also implicated by the large number of DEGs that we found upon EP300 knockdown) makes it complicated, EP300 might regulate human trophoblast differentiation via acetylation of transcriptional effectors or the histones surrounding genes involved in one or more of these pathways. It would be interesting to further investigate the molecular basis for the role of CREBBP and EP300 in trophoblast differentiation using epigenomic profiling techniques.
We found a prominent upregulation of TGFA upon EP300 knockdown, as well as an upregulation of other EGFR ligands, suggesting that EP300 mediates at least part of its effect on trophoblast differentiation by interfering with the EGFR signaling pathway. EGFR signaling is required for proliferation of various epithelial stem cells, including TSCs (4). Accordingly, addition of TGF-α or EGF under differentiation-inducing conditions resulted in an expansion of TSC-like cells in our experiments, mimicking the effect of EP300 knockdown, whereas blocking the EGFR could partially rescue EVT differentiation. Single-cell RNA sequencing data suggest that EGFR-ligand interactions take place within the human maternal–fetal interface (41). TGFA is predominantly expressed by the decidua, whereas trophoblasts express its receptor EGFR (42), suggesting that the decidua is responsible for providing proliferation signaling cues to trophoblasts in vivo. Upregulation of TGFA in our TSCs therefore seems to be a mechanism specifically induced by EP300 knockdown and suggests that normally EP300 suppresses TGFA expression in trophoblasts. As direct effects of acetyltransferases are generally activating, EP300 might induce the transcription of a negative regulator of the EGFR ligands, thereby indirectly affecting their expression. Since EGFR signaling is implicated in cell cycle progression and DNA synthesis (43, 44), inhibition of EGFR ligands could be a mechanism by which EP300 causes exit from the cell cycle and thereby facilitates differentiation. However, given the large number of potential EP300 targets (40), this may not be the only mechanism by which EP300 regulates differentiation. Overall, EP300 seems to be important for suppression of the stem-cell state, rather than activation of specific differentiation pathways, which may explain why EP300 was important for differentiation into both trophoblast lineages.
In conclusion, we demonstrated the importance of EP300 in human trophoblast differentiation. Our findings point towards a crucial role for EP300 in normal human placentation as well as the pathogenesis of FGR and pre-eclampsia by decreased EP300 levels. This study improves our understanding of early placental development on a molecular level, and offers an explanation for the complications seen in pregnancies of Rubinstein–Taybi syndrome-affected fetuses.
Methods
Isolation, Culture, and Differentiation of TSCs and Trophoblast Organoids.
Human first-trimester placenta collections were approved by the Academic Medical Center Biobank review committee (AMC 2016_246). Placental tissue (6 to 8 wk gestation) was obtained from the Human Immune System Mouse Facility of the Academic Medical Center, Amsterdam and de-identified prior to use in the current study. All material has been collected from donors from whom a written informed consent for the use of the material for research purposes had been obtained by the clinic. TSCs were isolated from placental tissue, cultured and differentiated into STB and EVT lineages as described by Okae et al. (4). Trophoblast organoids were generated from TSCs by seeding them in Matrigel droplets. They were cultured and differentiated into EVT according to protocols described by others (6, 7). For details of experimental procedures, see SI Appendix, Methods and Tables S1–S7.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This work made use of the Dutch national e-infrastructure with the support of the SURF Cooperative using grant no. EINF-2484.
Author contributions
A.J.v.V. and G.B.A. designed research; A.J.v.V., R.K., G.J.M.V., S.A.L.C., D.D., and J.A.V. performed research; A.J.v.V. and R.K. analyzed data; M.v.D., C.R.-S., A.M.M.v.P., and G.B.A. critically evaluated research; and A.J.v.V. and G.B.A. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
Transcriptome sequencing data have been deposited in the NCBI Sequence Read Archive (SRA) under BioProject ID PRJNA863080 (45).
Supporting Information
References
- 1.Turco M. Y., Moffett A., Development of the human placenta. Development 146, dev163428 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Cartwright J. E., Fraser R., Leslie K., Wallace A. E., James J. L., Remodelling at the maternal-fetal interface: Relevance to human pregnancy disorders. Reproduction 140, 803–813 (2010). [DOI] [PubMed] [Google Scholar]
- 3.DaSilva-Arnold S., James J. L., Al-Khan A., Zamudio S., Illsley N. P., Differentiation of first trimester cytotrophoblast to extravillous trophoblast involves an epithelial-mesenchymal transition. Placenta 36, 1412–1418 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Okae H., et al. , Derivation of human trophoblast stem cells. Cell Stem Cell 22, 50–63.e56 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Lee C. Q., et al. , What Is trophoblast? A combination of criteria define human first-trimester trophoblast. Stem Cell Rep. 6, 257–272 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haider S., et al. , Self-renewing trophoblast organoids recapitulate the developmental program of the early human placenta. Stem Cell Rep. 11, 537–551 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turco M. Y., et al. , Trophoblast organoids as a model for maternal-fetal interactions during human placentation. Nature 564, 263–267 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haider S., et al. , Notch1 controls development of the extravillous trophoblast lineage in the human placenta. Proc. Natl. Acad. Sci. U.S.A. 113, E7710–E7719 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meinhardt G., et al. , Pivotal role of the transcriptional co-activator YAP in trophoblast stemness of the developing human placenta. Proc. Natl. Acad. Sci. U.S.A. 117, 13562–13570 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saha B., et al. , TEAD4 ensures postimplantation development by promoting trophoblast self-renewal: An implication in early human pregnancy loss. Proc. Natl. Acad. Sci. U.S.A. 117, 17864–17875 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Varberg K. M., et al. , ASCL2 reciprocally controls key trophoblast lineage decisions during hemochorial placenta development. Proc. Natl. Acad. Sci. U.S.A. 118, e2016517118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hornbachner R., et al. , MSX2 safeguards syncytiotrophoblast fate of human trophoblast stem cells. Proc. Natl. Acad. Sci. U.S.A. 118, e2105130118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang L. J., et al. , Functional antagonism between DeltaNp63alpha and GCM1 regulates human trophoblast stemness and differentiation. Nat. Commun. 13, 1626 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haider S., et al. , Transforming growth factor-beta signaling governs the differentiation program of extravillous trophoblasts in the developing human placenta. Proc. Natl. Acad. Sci. U.S.A. 119, e2120667119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.West R. C., Ezashi T., Schoolcraft W. B., Yuan Y., Beyond fusion: A novel role for ERVW-1 in trophoblast proliferation and type I interferon receptor expression. Placenta 126, 150–159 (2022). [DOI] [PubMed] [Google Scholar]
- 16.Ray S., et al. , Hippo signaling cofactor, WWTR1, at the crossroads of human trophoblast progenitor self-renewal and differentiation. Proc. Natl. Acad. Sci. U.S.A. 119, e2204069119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shilei B., et al. , Downregulation of CDC42 inhibits the proliferation and stemness of human trophoblast stem cell via EZRIN/YAP inactivation. Cell Tissue Res. 389, 573–585 (2022). [DOI] [PubMed] [Google Scholar]
- 18.van Uitert M., et al. , Meta-analysis of placental transcriptome data identifies a novel molecular pathway related to preeclampsia. PLoS One 10, e0132468 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan H. M., La Thangue N. B., p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J. Cell Sci. 114, 2363–2373 (2001). [DOI] [PubMed] [Google Scholar]
- 20.Ogryzko V. V., Schiltz R. L., Russanova V., Howard B. H., Nakatani Y., The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87, 953–959 (1996). [DOI] [PubMed] [Google Scholar]
- 21.Kwok R. P., et al. , Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature 370, 223–226 (1994). [DOI] [PubMed] [Google Scholar]
- 22.Bannister A. J., Kouzarides T., The CBP co-activator is a histone acetyltransferase. Nature 384, 641–643 (1996). [DOI] [PubMed] [Google Scholar]
- 23.Van Gils J., Magdinier F., Fergelot P., Lacombe D., Rubinstein-taybi syndrome: A model of epigenetic disorder. Genes (Basel) 12, 968 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fergelot P., et al. , Phenotype and genotype in 52 patients with Rubinstein-Taybi syndrome caused by EP300 mutations. Am. J. Med. Genet. A 170, 3069–3082 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Cohen J. L., et al. , EP300-related Rubinstein-Taybi syndrome: Highlighted rare phenotypic findings and a genotype-phenotype meta-analysis of 74 patients. Am. J. Med. Genet. A 182, 2926–2938 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Lasko L. M., et al. , Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours. Nature 550, 128–132 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee C. Q. E., et al. , Integrin alpha2 marks a niche of trophoblast progenitor cells in first trimester human placenta. Development 145, dev162305 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zuo Q., et al. , ASCL2 expression contributes to gastric tumor migration and invasion by downregulating miR223 and inducing EMT. Mol. Med. Rep. 18, 3751–3759 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jordan N. V., Johnson G. L., Abell A. N., Tracking the intermediate stages of epithelial-mesenchymal transition in epithelial stem cells and cancer. Cell Cycle 10, 2865–2873 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taylor A. M., et al. , Fragment-based discovery of a selective and cell-active benzodiazepinone CBP/EP300 bromodomain inhibitor (CPI-637). ACS Med. Chem. Lett. 7, 531–536 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harris R. C., Chung E., Coffey R. J., EGF receptor ligands. Exp. Cell Res. 284, 2–13 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Kim J., Koo B. K., Knoblich J. A., Human organoids: Model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 21, 571–584 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dutto I., et al. , Mutations in CREBBP and EP300 genes affect DNA repair of oxidative damage in Rubinstein-Taybi syndrome cells. Carcinogenesis 41, 257–266 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Thomas P. D., Kahn M., Kat3 coactivators in somatic stem cells and cancer stem cells: Biological roles, evolution, and pharmacologic manipulation. Cell Biol. Toxicol. 32, 61–81 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fauquier L., et al. , CBP and P300 regulate distinct gene networks required for human primary myoblast differentiation and muscle integrity. Sci. Rep. 8, 12629 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lehrmann H., Pritchard L. L., Harel-Bellan A., Histone acetyltransferases and deacetylases in the control of cell proliferation and differentiation. Adv. Cancer Res. 86, 41–65 (2002). [DOI] [PubMed] [Google Scholar]
- 37.Skrypek N., Goossens S., De Smedt E., Vandamme N., Berx G., Epithelial-to-mesenchymal transition: Epigenetic reprogramming driving cellular plasticity. Trends Genet. 33, 943–959 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Jaju Bhattad G., et al. , Histone deacetylase 1 and 2 drive differentiation and fusion of progenitor cells in human placental trophoblasts. Cell Death Dis. 11, 311 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kwak Y. T., Muralimanoharan S., Gogate A. A., Mendelson C. R., Human trophoblast differentiation is associated with profound gene regulatory and epigenetic changes. Endocrinology 160, 2189–2203 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weinert B. T., et al. , Time-resolved analysis reveals rapid dynamics and broad scope of the CBP/p300 acetylome. Cell 174, 231–244.e12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arutyunyan A., et al. , Spatial multiomics map of trophoblast development in early pregnancy. Nature 616, 143–151 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun T., et al. , The maternal-fetal interface of successful pregnancies and impact of fetal sex using single cell sequencing. bioRxiv [Preprint] (2019). 10.1101/641118 (Accessed 25 August 2022). [DOI]
- 43.Wang C., et al. , Transforming growth factor alpha (TGFalpha) regulates granulosa cell tumor (GCT) cell proliferation and migration through activation of multiple pathways. PLoS One 7, e48299 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oda K., Matsuoka Y., Funahashi A., Kitano H., A comprehensive pathway map of epidermal growth factor receptor signaling. Mol. Syst. Biol. 1, 2005 0010 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Keijser R., van Voorden A. J., Afink G. B., EP300 facilitates human trophoblast stem cell differentiation. NCBI Sequence Read Archive (SRA). https://www.ncbi.nlm.nih.gov/bioproject/PRJNA863080. Deposited 28 July 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
Transcriptome sequencing data have been deposited in the NCBI Sequence Read Archive (SRA) under BioProject ID PRJNA863080 (45).