Transfer RNA (tRNA), well-studied as the key adapter molecule in protein translation, is now known to be processed into a large collection of smaller stable RNAs. High-throughput small-RNA sequencing methods, including those specialized for tRNAs such as ARM-Seq1, Pandora-seq2, and others, have led to the identification of these tRNA-derived RNAs (tDRs) across all domains of life3,4. Functional studies have revealed a plethora of regulatory roles associated with these small noncoding RNAs, including modulation of ribosome biogenesis, protein synthesis, gene silencing, stress response and plant nodulation5. Moreover, differential abundance of tDRs observed across tissues and cellular conditions suggests the possibility of leveraging tDRs as biomarkers for early diagnosis of cancer, heart disease, neurological disorders and other diseases6. Historically, they have been rudimentarily categorized by their positions in source tRNAs: namely, 5′ or 3′ fragments, 5′ or 3′ halves, internal fragments, 5′ leaders or 3′ trailers7. A constellation of diverse yet often overlapping names and acronyms has emerged, including “tRFs”, “tsRNAs”, “tiRNAs”, “SHOT-RNAs” and “tRNA halves”, among others. Several databases3,8 have created their own independent serial or hexadecimal numbering systems, but, for various reasons, these names have not been widely adopted. Here we propose a consistent, uniform naming system (Fig. 1a) that is biologically informative and builds upon an existing, widely accepted naming scheme for full-length tRNAs9. To make the naming system readily accessible, we have also developed tDRnamer (Supplementary Fig. 1 and Supplementary Table 1), a resource providing a user-friendly, web-based interface, as well as a downloadable software version for efficient bulk processing. Given one or more sequences, the tool deterministically generates standardized tDR names within an information-rich display; alternatively, if given tDRnamer-generated names, it can produce the exact corresponding sequences. Additional graphical visualizations (Fig. 1c,d and Supplementary Figs. 2 and 3) are provided to further highlight the relatedness of similar tDRs and their positions within source tRNA(s).
Fig. 1. Standardized naming for tRNA-derived RNAs (tDRs) illustrated by seven tDRs potentially derived from up to eight human tRNAPro transcripts.
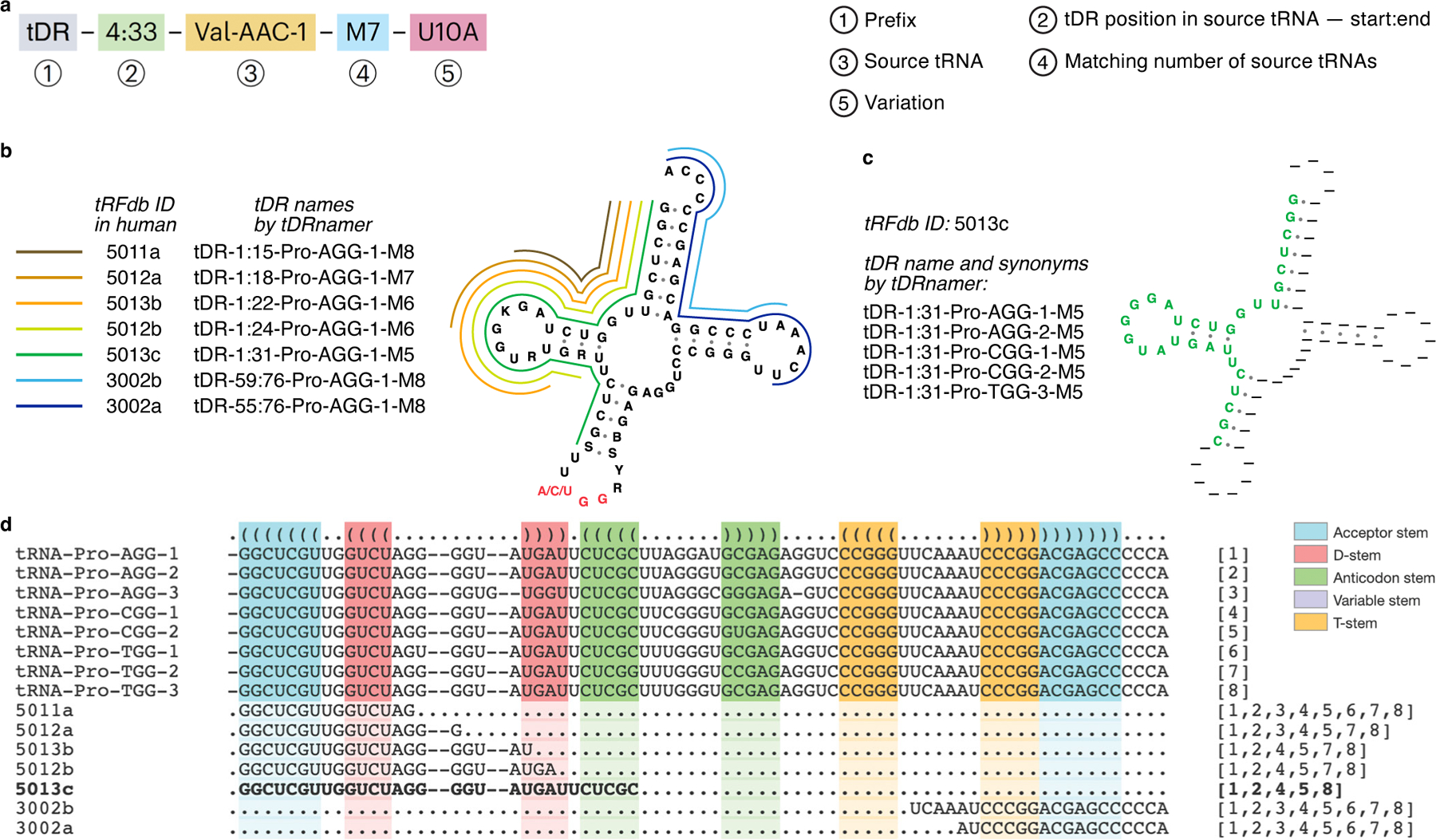
a. The standardized tDR name contains three required components, plus up to two optional notations: (1) the “tDR” prefix; (2) start and end positions of the tDR relative to its source tRNA in Sprinzl numbering10 (if present, 5′ leader and 3′ trailer positions are preceded with “L” or “T” respectively); (3) the source tRNA name from GtRNAdb9; (4) “M” with the total number of matching source tRNA transcripts, when more than one exists; and (5) nucleotide variation annotation, if the tDR does not precisely match a reference source tRNA – it consists of the reference source tRNA nucleotide, followed by the relative position in tDR, and finally the substituted nucleotide found in the tDR. b. The tDR names for seven tRFdb entries with tRNAPro(AGG) as the source tRNA are listed. Colored lines around the tRNAPro secondary structure represent the positions of the derived tDRs. c. An example tDR (tRFdb ID 5013c) is located at the 5′ end of tRNAPro from position 1 to 31, as illustrated in green within the full tRNA secondary structure, generated by tDRnamer. The tDR name and synonyms show that its source tRNAs may include two different transcripts of tRNAPro(AGG), two different transcripts of tRNAPro(CGG) and/or one tRNAPro(UGG). d. tDRs are grouped and aligned with all potential source tRNAPro by tDRnamer. Color blocks represent stems in tRNA secondary structure. Bracketed numbers at the end of each tDR (for example, “[1,2,4,5,8]” for 5013c) refer to the five possible source tRNAs shown at the top of the alignment.
The tDR naming system (Fig. 1a) consists of three required components, plus up to two supplemental notations in special cases. First, each name starts with the general prefix “tDR”, which is neutral to any functional role. If the tDR is derived from mitochondrial or plastid tRNAs, “mtDR” and “ptDR” is used. Second, the tDR’s precise start and end positions, relative to its source tRNA, are specified using the standardized Sprinzl tRNA position numbering10 (for example, “tDR-4:33”). This gives stems and loops consistent base numbers across all tRNAs (for example, the anticodon in every tRNA is at position 34–36). Third, the identifier of the most similar source tRNA found in the Genomic tRNA Database (GtRNAdb)9 is appended to the tDR name (for example, “Val-AAC-1”). Because some portions of different tRNAs may be identical, tDRs can often be derived from multiple indistinguishable parent tRNAs. In this case, a fourth component is added to the tDR name to indicate the number of potential source tRNAs (for example, “M7”). Finally, nucleotide differences may exist between tDRs and the reference tRNA set due to natural genetic variation, or may also be caused by misincorporation by reverse transcriptase at certain RNA modifications. To describe these tDRs succinctly, a name component can be added consisting of the original and substituted bases with their relative positions (for example, “U10A”).
To demonstrate the application of this new naming system with tDRnamer, we created systematic names for existing entries in tRFdb3 (Supplementary Data 1), immediately yielding useful observations. One notable case involves a group of seven tDRs potentially derived from multiple unique transcripts of tRNAPro (Fig. 1b). For tDR-1:31-Pro-AGG-1-M5 (tRFdb ID: 5013c), one can see five possible synonyms corresponding to five matching source tRNAs (Fig. 1c–d and Supplementary Table 2). In comparison to prior nomenclatures3,8, this information is necessarily absent. The graphic (Fig.1b) and alignment (Fig. 1d) show most of these tDRs are anchored to the 5′ end of source tRNAs (for example, “1:15”, “1:18”) while the remaining two align to the 3′ end (“59:76”, “55:76”). The alignment further shows that the longest tDR (5013c) exactly matches five potential source tRNAs, precisely enumerated by numbers in brackets following tDR sequences. Additional examples of complex tDR relationships showcase the unique functionality of the tDRnamer web analysis interface (Supplementary Figs. 2 and 3). For instance, three different regions of human tRNA-Gly-GCC-1 are processed into distinct tDRs of multiple lengths (Supplementary Fig. 2), whereas a tDR derived from 3′ trailer sequence of tRNA-Arg-ACG-1-3 includes multiple single-nucleotide variants (Supplementary Fig. 3). These examples demonstrate the breadth and descriptive capabilities of the proposed tDR naming system. As such, tDRnamer should become a valuable analytic tool for recognizing the biological relatedness among tDRs, enabling more focused study of their biogenesis from source tRNAs. The new system will also provide a ready means for detailed comparisons of tDR sequencing results between studies, both retrospectively and prospectively, facilitating advances in this blossoming field of noncoding RNA research.
Supplementary Material
Acknowledgements
This work was supported by the National Human Genome Research Institute, National Institutes of Health (R01HG006753 to T.L.).
Footnotes
Code availability
The source code of tDRnamer and its web server is available at https://github.com/UCSC-LoweLab/tDRnamer and https://github.com/UCSC-LoweLab/tDRnamer-web respectively, both under the GNU General Public License v3.0.
Competing interests
The authors declare no competing interests.
Data availability
The tDRnamer web server is available at http://trna.ucsc.edu/tDRnamer/. Standalone software can be obtained from GitHub at https://github.com/UCSC-LoweLab/tDRnamer and Docker image at https://hub.docker.com/r/ucsclowelab/tdrnamer. Example data can be downloaded from http://trna.ucsc.edu/tDRnamer/data/examples/. Pre-built reference databases for model organisms are available at http://trna.ucsc.edu/tDRnamer/docs/refdb/. The complete set of tRFdb3 data re-annotations by tDRnamer is available at http://trna.ucsc.edu/tDRnamer/data/tRFdb_reannotations/.
References
- 1.Cozen AE et al. Nat Methods 12, 879–84 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shi J et al. Nat. Cell Biol 23, 424–436 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kumar P, Mudunuri SB, Anaya J & Dutta A Nucleic Acids Res 43, D141–D145 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gebetsberger J & Polacek N RNA Biol 10, 1798–1806 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Polacek N & Ivanov P RNA Biol 17, 1057–1059 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim HK, Yeom J & Kay MA Mol. Ther 28, 2340–2357 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Magee R & Rigoutsos I Nucleic Acids Res 48, 9433–9448 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pliatsika V et al. Nucleic Acids Res 46, D152–D159 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan PP & Lowe TM Nucleic Acids Res 44, D184–9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steinberg S, Misch A & Sprinzl M Nucleic Acids Res 21, 3011–5 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The tDRnamer web server is available at http://trna.ucsc.edu/tDRnamer/. Standalone software can be obtained from GitHub at https://github.com/UCSC-LoweLab/tDRnamer and Docker image at https://hub.docker.com/r/ucsclowelab/tdrnamer. Example data can be downloaded from http://trna.ucsc.edu/tDRnamer/data/examples/. Pre-built reference databases for model organisms are available at http://trna.ucsc.edu/tDRnamer/docs/refdb/. The complete set of tRFdb3 data re-annotations by tDRnamer is available at http://trna.ucsc.edu/tDRnamer/data/tRFdb_reannotations/.


