Abstract
Background
Neurodevelopmental disorders (NDDs) are associated with altered development of the brain especially in childhood. Copy number variants (CNVs) play a crucial role in the genetic aetiology of NDDs by disturbing gene expression directly at linear sequence or remotely at three-dimensional genome level in a tissue-specific manner. Despite the substantial increase in NDD studies employing whole-genome sequencing, there is no specific tool for prioritising the pathogenicity of CNVs in the context of NDDs.
Methods
Using an XGBoost classifier, we integrated 189 features that represent genomic sequences, gene information and functional/genomic segments for evaluating genome-wide CNVs in a neuro/brain-specific manner, to develop a new tool, neuroCNVscore. We used Human Phenotype Ontology to construct an independent NDD-related set.
Results
Our neuroCNVscore framework (https://github.com/lxsbch/neuroCNVscore) achieved high predictive performance (precision recall=0.82; area under curve=0.85) and outperformed an existing reference method SVScore. Notably, the predicted pathogenic CNVs showed enrichment in known genes associated with autism.
Conclusions
NeuroCNVscore prioritises functional, deleterious and pathogenic CNVs in NDDs at whole genome-wide level, which is important for genetic studies and clinical genomic screening of NDDs as well as for providing novel biological insights into NDDs.
Keywords: Neurodevelopmental disorder, Copy number variant, Pathogenicity, Tissue specificity, Gene expression
WHAT IS ALREADY KNOWN ON THIS TOPIC
Copy number variants (CNVs) are important in the genetic aetiology of neurodevelopmental disorders (NDDs). Systematic identification of CNV pathogenicity by virtue of their size, number and impact on genome is challenge. Several tools are available to evaluate CNVs or structural variants, but none on CNVs specific for NDDs.
WHAT THIS STUDY ADDS
NeuroCNVscore is a useful tool in prioritising functional and/or pathogenic CNVs in NDDs at whole genome-wide level in a neuro/brain-specific manner.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
Given the expanding studies on NDDs and the usage of sequencing in clinical practice, our neuroCNVscore speeds up the screening on pathogenic CNVs, which facilitates the clinical diagnoses of CNVs with unknown significant, and thus may provide novel biological insights into NDDs.
Introduction
Neurodevelopmental disorders (NDDs) are characterised by the inability to achieve cognitive, emotional and motor developmental milestones including autism spectrum disorder (ASD), attention deficit hyperactivity disorder (ADHD) and schizophrenia. It is estimated to affect over 11.3%, and 15% of the population in low-income and middle-income countries1 and USA,2 respectively. NDD’s heritability is high that has been estimated from twin and family studies as 50%–90% in ASD,3 88% in ADHD4 and 85% in schizophrenia.5 Genomic alterations are commonly found in children with NDDs. However, the explained genetic aetiology of NDDs accounts for only a small proportion.
Copy number variants (CNVs) are structural variants (SVs) in the genome that involve the gain or loss of large segments of DNA, which have been implicated in NDDs.6 7 Systematic identification of CNV pathogenicity by virtue of their number, size and impact on the genome is still a challenge. It is approximately 1000 CNVs per genome ranging in size from 50 base pairs (bp) to several mega bases (Mb). CNVs make effects by altering the dosage of gene regions8 as well as by perturbing non-coding areas.7 9 Growing number of studies by whole genome sequencing (WGS) and the complexity of identifying pathogenic CNVs call for computational prediction tools.
Many assessing tools have been developed to evaluate the pathogenicity of single nucleotide variants,10 11 but fewer studies have systematically focused on assessing the pathogenic CNVs, especially none in NDD-related CNVs. Recently, SVScore,12 SVFX,13 SVPath14 and AnnotSV15 have been developed to interpret the SVs by integrating results from prediction matrices of SNPs, using cancer-related SVs as inputs, counting SVs with overlapped exons, or integrating multiple sources to annotate SVs. However, the aggregated effects on SNPs, somatic impacts of SVs or only overlapping exons without tissue-specific information may bias the effects of CNVs. As germline variations are the major focus in NDDs, a specific tool is needed for assessing the effects of CNVs on NDDs.
We here present a novel supervised machine learning framework, named as neuroCNVScore (https://github.com/lxsbch/neuroCNVscore), to score the pathogenicity of CNVs related to NDDs. We hypothesise that the computational prediction on pathogenic CNVs would benefit from a set of comprehensive tissue-specific features covering the whole genomic regions. Hence, we employed germline CNVs obtained from published NDD studies,16–19 and curated gene lists together with a comprehensive set of neuro/brain-specific data on non-coding regions from ENCODE,20 Roadmap,21 EpiMap22 and PsychENCODE23 to train our models. Moreover, we constructed an independent dataset associated with NDDs by filtering the phenotypes from Human Phenotype Ontology (HPO, https://hpo.jax.org/) to evaluate the performance of our trained models. The performance of neuroCNVScore was compared with a reference method SVScore.12 This neuroCNVScore is designed for assessing the pathogenicity of CNVs in NDDs generated from association studies or genetic tests.
Methods
Data collection and preprocessing/harmonisation
We developed neuroCNVscore, which used XGBoost and comprehensive genome-wide features to evaluate the likelihood that a given CNV contributes to the development or manifestation of NDDs. To assess the pathogenicity associated with CNV in NDDs, we gathered training set (identified by genomic coordinates) from several case–control NDD studies. We assigned CNVs from cases as likely pathogenic (LP). In contrast, the CNVs from unaffected individuals and parents served as the control. Together, we collected 86 694 CNVs in the LP set and 786 058 in the control set from four data sources, respectively (figure 1).
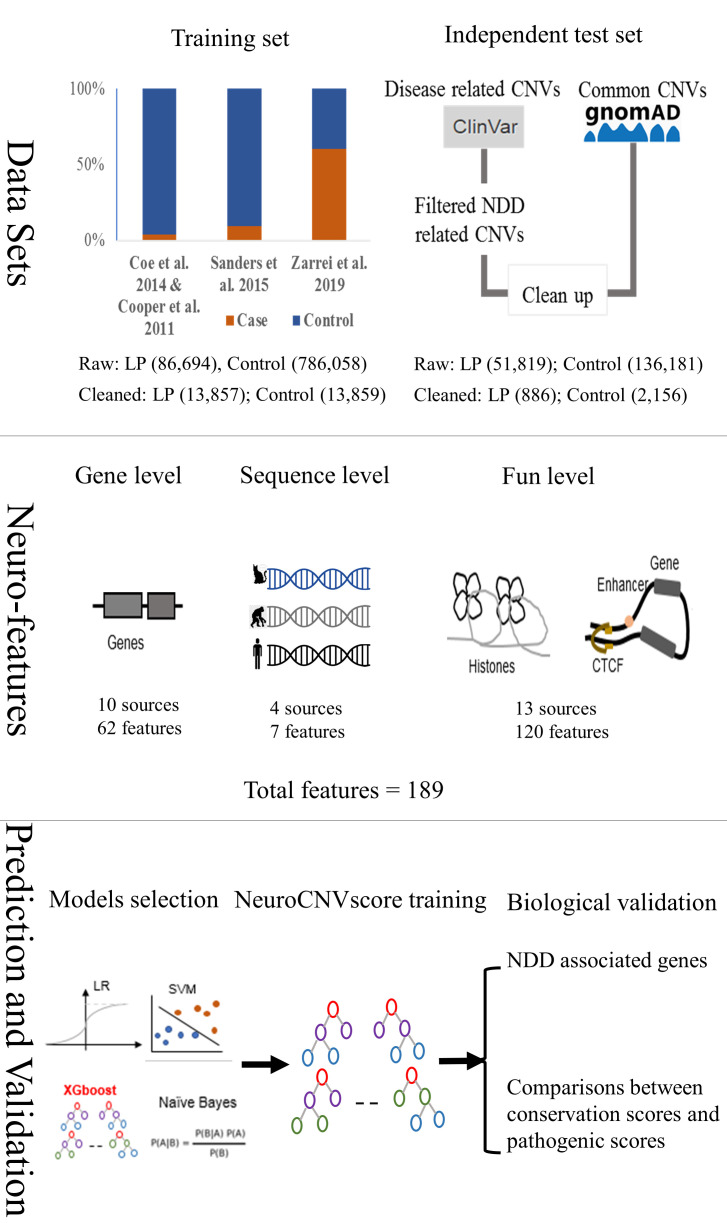
Figure 1.
The flow chart of neuroCNVscore development and evaluation in this study. In data sets, the sources of training set and test set are listed. The training set was derived from four neurodevelopmental disorders (NDDs) studies under the case–control design, while the validation set was from ClinVar and GnomAD. The numbers of raw and cleaned CNVs in the brackets are indicated. In neurofeatures, comprehensive neuro/brain-related features were gathered at gene, sequence and functional/genomic segments levels. In prediction and validation, biological validations were performed in two ways: (1) correlation analyses between phyloP46way and the pathogenic scores generated by the new model where phyloP46way was excluded from the feature matrix; (2) utilisation of an independent set of NDD-related gene lists including PSD genes to cognition, CHD8 targets and ASD risk genes. CNV, copy number variantl LP, likely pathogenic; PSD, postsynaptic density.
Initial data filtering and harmonisation were performed on all autosomal chromosome CNVs in three major steps. First, we excluded CNVs with a size smaller than 50 bp, and the remaining CNVs were categorised into two groups based on their impact on the genome: copy number loss and copy number gain. Next, we deleted CNVs which had 90% reciprocal overlap between LP and control. Finally, we applied an empirical cumulative distribution function with bin size of 60 to generate size matched LP and control to overcome the amount of disparity between groups. For each CNV type, we sampled an equal number of LP CNVs ensuring the matching of control CNVs in each bin. For the training process, we retained 13 857 cleaned LP CNVs and 13 859 cleaned control CNVs.
Next, we constructed an independent test set by assembling 51 819 disease associated variations from ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/) and 136 181 common CNVs from GnomAD 2.1 (http://www.gnomad-sg.org/). For the NDD-related set, we retained CNVs with length >50 bp, germline, pathogenic and the term of HPO: 0012759 (neurodevelopmental abnormality associated genes). For common CNVs, we kept CNVs with quality record PASS, and allele frequency >0.1. To avoid overestimation, we removed those CNVs with 90% reciprocal overlap within the training dataset under the same variant type.
Finally, we collected several NDD-related gene lists to evaluate the biological validity and robustness of neuroCNVscore including CHD8 target genes,24 human postsynaptic density proteins25 and ASD risk genes (FDR (false discovery rate)<0.3).18 The overall workflow is outlined in figure 1.
A comprehensive tissue-specific feature collection and feature matrix construction
For each CNV, a broad range of features was compiled into a feature matrix. We leveraged 189 features in total from three different levels: (1) gene level (Gen), (2) functional/genomic segment level (Fun) and (3) sequence level (Seq). The description of features is shown in online supplemental table S1.
bmjpo-2023-001966supp001.pdf (165.1KB, pdf)
In brief, a set of gene level features (N=62) that contain gene entity, dosage sensitivity and neurodevelopmental phenotype were collected. Since non-coding CNVs may disrupt regulatory regions to compromise gene expression and translation in a linear or three-dimensional (3D) manner, we obtained a regulatory cascade catalogue (N=120 at functional/genomic segment level). This catalogue integrated multiomics data encompassing experimentally identified or computational predicted regulatory regions with a focus on tissue-specific annotation. Finally, the sequence level features (N=7) composed of information of GC content, cross-species conservation score (phylop46way and phastcon46way which are derived from phyloP or Hidden Markov Model via multiple alignment of 45 vertebrate genomes to the human genome), heterochromatin positions, collapsed repeat regions (DacMapExclude, DukeMapExclude are genomic regions calculated by different algorithms) retrieved from the UCSC genome browser (http://genome.ucsc.edu/), and human accelerated regions accessed by Doan et al.26 These features were instrumental in identifying functional genomic regions and/or filtering out the genomic regions which may cause artefacts from downstream segments.
Based on a variety of features, annotations were performed in three distinct ways: (1) counting the number of overlapped features with a given CNV, (2) assessing a discrete value that denotes the number of the features which has >50% reciprocal overlapped regions with a given CNV and (3) calculating the average value of overlapped regions between the feature and a given CNV. After initial annotation, we divided the entire feature matrix based on the length of each CNV and then applied min-max scaling. Considering the differences in features, for example, triplosensitivity is a measurement only for the copy number gain, we kept 172 features out of 189 for the copy number loss model and 172 features out of 189 in the copy number gain model, respectively.
Design of XGBoost model and the training strategy
To choose an appropriate model, we compared the performances among different algorithms (Naïve Bayes, logistic regression, support vector machine (SVM) and XGBoost), and we found that XGBoost had the best performance in the python framework from Scikit 0.22.1 with the binary logistic objective function. A total of 80%/20% of the variant sets were used as training/test sets, respectively. Next, we trained the XGBoost model with optimised parameters by using grid search and evaluated our models through an independent test set. Additionally, we assessed the performance by comparing our model with SVScore, which can evaluate various types of SV including CNV.
Statistics
Statistical analyses were performed using Python (V.2.7). The performance was measured by precision recall (PR) and receiver operating characteristic (ROC) curves. For individual feature comparison, we applied two-tailed Wilcoxon rank-sum tests. All genomic data is in GRCh37 genome build. Figures were generated by the ggplot package in R (V.3.6.1) or matplotlib in Python.
Patient and public involvement
Patients or the public were not involved in the design, or conduct, or reporting, or dissemination plans of our research.
Results
Feature analyses pinpoint comprehensive feature sets
To understand the characteristics of CNVs in NDDs, we investigated the distribution of features between LP and control sets. In total, we observed 121 and 106 significant features at the threshold of p=0.05 in copy number loss and copy number gain models, respectively (online supplemental table S2). These findings demonstrated that a large spectrum of features has significant differences between sets.
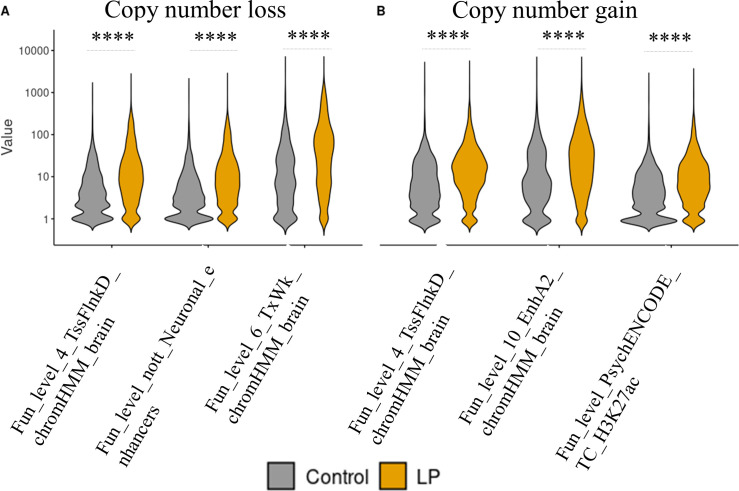
Among these significant features, functional/genomic segment features ranked higher than the others. Most of the highly ranked features were related to histone modification markers (eg, H3K27me3, H3K27ac) and 3D chromatin-related features (eg, enhancers) (figure 2). This is as expected since non-coding regions account for 98% of the human genome and CNVs can affect the gene function by interrupting the regulatory regions.
Figure 2.
Comparisons of top three features between control and LP (likely pathogenic) sets. The top three significant features between control and LP sets in copy number loss (A) and copy number gain (B). The x-axis shows the types of significant features. Fun_level, function/genomic segment level. The y-axis displays the values of log- transformed feature matrices. Unpaired t-tests were applied and significant levels were. ****p<0.0001.
Comparisons among four algorithms reveal the superior performance of XGBoost
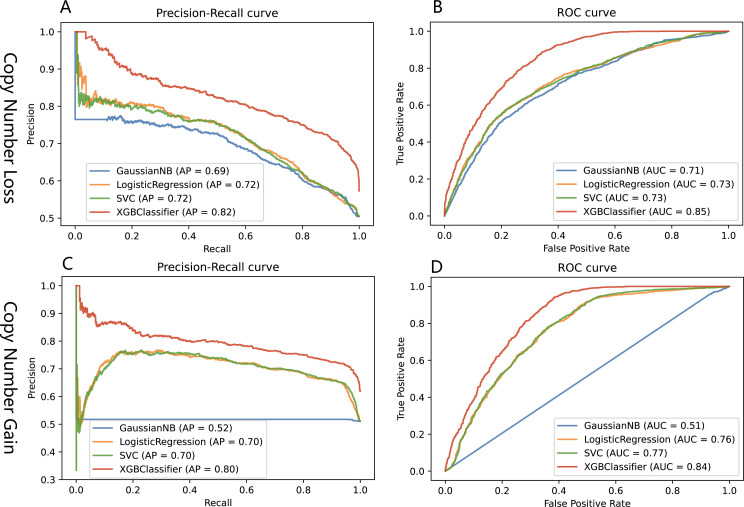
To find an optimal model for identifying pathogenic CNVs, we evaluated the predictive performance of Naïve Bayes, logistic regression, SVM and XGBoost on the test sets (figure 3). The XGBoost model showed the highest performance (average precision (AP) and area under curve (AUC) were 0.82, 0.85 for copy number loss; AP and AUC were 0.80, 0.84 for copy number gain). Therefore, we applied the XGBoost model to construct our neuroScoreCNV framework.
Figure 3.
Performances of Naïve Bayes, logistic regression, support vector machine (SVM) and XGBoost algorithms in evaluating CNVs. XGBoost showed superior performance demonstrated by precision-recall curves and receiver operating characteristic (ROC) curves for both copy number loss (A, B) and copy number gain (C, D). AP, average precision; AUC, area under curve; CNVs, copy number variants.
Accuracy assessments reveal better performance of neuroScoreCNV than SVScore
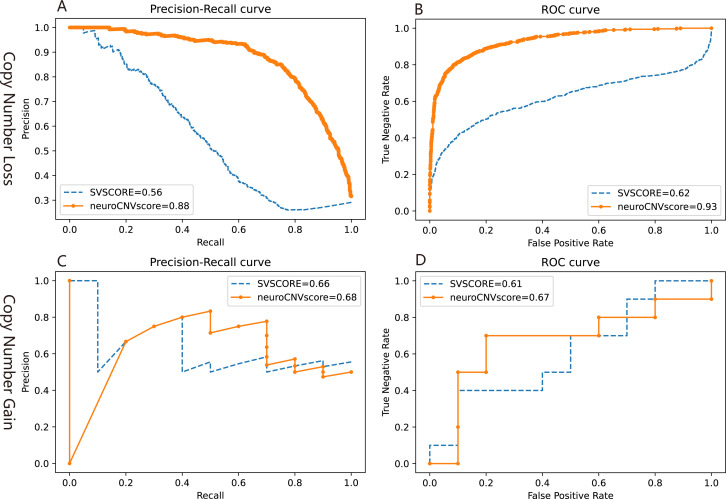
We evaluated the performance of neuroScoreCNV and SVScore by an independent set as described in the flow chart (figure 1). NeuroScoreCNV achieved relatively better performance evaluated by both AP and AUC values compared with SVScore (figure 4). The different performances between models are in agreement with a previous study.13
Figure 4.
Performances of neuroCNVscore and SVScore in an independent set as described in the flow chart of figure 1. Precision-recall (A) and ROC (B) curves were calculated with copy number loss from the independent dataset; precision-recall (C) and ROC (D) curves were calculated with copy number gain from the independent dataset. CNV, copy number variants; ROC, receiver operating characteristic; SVs, structural variants.
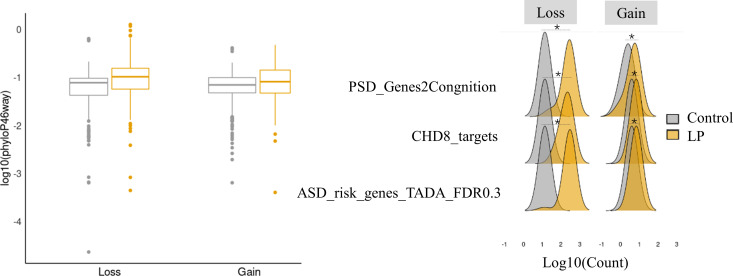
Moreover, we investigated the biological validity and robustness from two aspects. It was shown that interruptions at conserved regions could cause diseases since these regions are normally functional.27 Therefore, we first computed the CNV pathogenic scores generated with the new feature matrices in which a conservation score (ie, PhyloP46way, one of the commonly used conservation score that considering individual base conservation) was excluded. We observed that higher CNV pathogenic scores (≥0.7) tended to have higher conservation scores, as indicated by the correlation between log10(PhyloP46way) and the new pathogenic scores (figure 5A, B). Then, we checked if our predicted scores were capable of prioritising CNVs with known NDD-associated genes. LP CNVs covered significantly (p<0.05) more NDD-related genes than the control group (figure 5B). Overall, our approach achieved higher performance in discriminating LP CNVs from control or benign CNVs.
Figure 5.
Biological validation of neuroCNVscore. The plot (A) shows the comparisons between PhyloP scores (log10(PhyloP46way)) and pathogenic scores generated by excluding PhyloP46way from the original neuroCNVscore model, regions with higher pathogenic scores tend to have higher PhyloP scores. The number of NDD-related genes (B) between the predicted LP and control groups in both copy number loss and copy number gain models shows that more NDD-related genes are found in LP groups. For better presentation, log transformations were applied to PhyloP46way scores and the gene counts. *p<0.05. CNV, copy number variant; LP, likely pathogenic; NDD, neurodevelopmental disorder.
Feature importancy highlights the important role of regulatory regions in NDDs
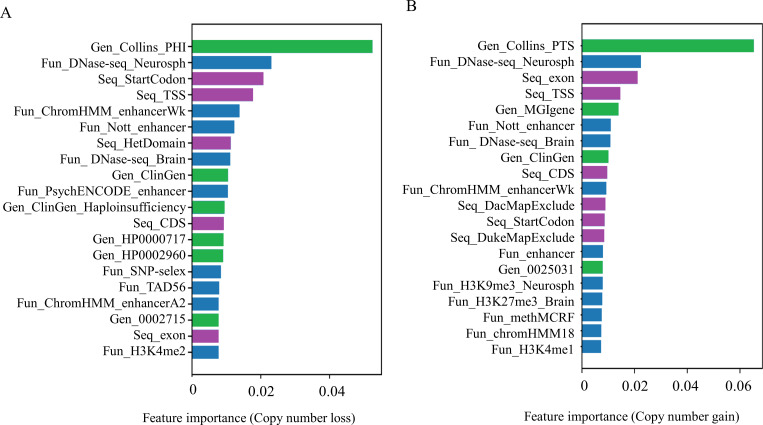
We categorised model features into three groups: functional/genomic level (Fun), gene level (Gen) and sequence level (Seq) and computed the feature importancy by permutation (figure 6, online supplemental table S3). The most important features were genes with haploinsufficiency scores (PHI) and triplosensitivity scores (PTS). PHI reflects the probability of one single functional copy to be sufficient to maintain function, whereas PTS suggests the probability of an additional copy of a gene for generating phenotypes. PHI and PTS are important parameters for evaluating the pathogenicity in clinical diagnoses based on the ACMG guidelines.28 This is also true in neuroCNVScore. In NDDs, several studies found pathogenic CNVs were sensitive to dosage.29
Figure 6.
Top 20 features obtained from feature importance analyses. Highly important features of copy number loss model (A) and copy number gain model (B) are listed. All the feature names were colour-coded and formatted as following: feature type (Fun_/Gen_/Seq_feature names (original sources)_tissue type (if applicable). Fun: Function, in blue; Gen: Gene, in green; Seq: Sequence, in purple.
Additionally, we noticed several prominent phenotypes such as HPO: 000717 (autism associated genes), HPO: 0002960 (autoimmunity associated genes) and HPO: 0025031 (abnormality of the digestive system associated genes). It is known that immune system abnormalities and/or gastrointestinal symptoms can co-occur with ASD30 and schizophrenia.31 Compelling evidence has demonstrated the importance of autoimmune response in ASD.32 Purified IgG containing antibodies from the mothers of children with ASD can cause abnormal behaviours in animal models.33 34
Among the important features at the functional/genomic segment level, we observed several key players in 3D chromatin conformation including enhancers and topologically associated domains. Meanwhile, DNase-Seq which suggests active regulatory elements at open chromatin was also an important feature. The emerging evidence has highlighted the role of 3D chromatin conformation in relation to NDDs.23 35 Collectively, studying the interaction between CNVs and the higher order of chromatin conformation could provide novel insights into the aetiology of NDDs and explain the missing heredity of NDDs.
Discussion
In this study, we have introduced a novel framework, neuroCNVscore, to evaluate the pathogenicity of CNVs in NDDs. NeuroCNVscore outperformed a commonly used tool SVScore on independent datasets from ClinVar and gnomAD. Importantly, neuroCNVscore has the unique ability to prioritise the functional, deleterious and pathogenic CNVs derived from either NDD’s association studies or clinical diagnoses, which may provide biological insights into NDDs, especially at the three-dimensional genome level.
There are several factors contribute to the accuracy and robustness of neuroCNVscore. First, we used a high-quality set of germline CNVs from published NDD studies as the training set, ensuring the high reliability of this model. Second, we validated our models by using an independent dataset associated with NDD, which outperformed a published tool, SVScore. Furthermore, we curated a comprehensive feature collection (N=189) at gene, functional genomic and sequence levels. Specifically, we incorporated a significant amount of tissue-specific functional genomic data, enabling the identification of disrupted genes and regulatory elements that act in a tissue-specific manner during development. This is especially important for the studies in NDD since brain tissue is normally hard to access.
While the neuroCNVscore performed well, it may be improved by incorporating expert-curated CNVs from WGS studies in NDDs and healthy controls. Along with the increased knowledge and functional genomics data on non-coding regions, additional informative features can be integrated into the model to better address the underlying mechanisms. Moreover, we developed neuroCNVscore based on XGBoost, but it is worth exploring deep learning algorithms in future investigation.
In summary, our neuroCNVscore is a useful tool for generating hypotheses in genome-wide association studies in NDDs and could facilitate the understanding of genetic aetiology of NDDs.
Supplementary Material
Acknowledgments
We thank MacArthur's Lab for sharing the comprehensive collections of gene lists. We thank Dr. Sree Rohit Raj Kolora for reviewing, revising the manuscript and useful discussion.
Footnotes
Contributors: XL designed the study, performed the analysis and drafted the manuscript. WX and FL participated in the design and interpretation of the data and revised the manuscript. PZ, RG and YZ participated in the interpretation of data. CH coordinated the project and supervised the study. XN coordinated the project and acquisition the funding. WL coordinated the project, supervised the study, critically reviewed and revised the manuscript. All authors read and approved the final manuscript. WL is the guarantor of this manuscript.
Funding: This work was partially supported by the Ministry of Science and Technology of China (2019YFA0802104; 2016YFC1000306); the National Natural Science Foundation of China (31830054); the Beijing Natural Science Foundation (5222007) and the Beijing Municipal Health Commission (JingYiYan 2018-5).
Competing interests: None declared.
Patient and public involvement: Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data sharing not applicable as no datasets generated and/or analysed for this study. All features analysed during this study are collected from public datasets. Sources can be found from https://github.com/macarthur-lab/gene_lists. All CNV training data are included in these publications 16–19 and testing data are from the ClinVar database. The source code is available at https://github.com/lxsbch/neuroCNVscore.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study has been approved by the Ethics Committee of Beijing Children’s Hospital, Capital Medical University (2018-k-62). No ethical issues are involved in this study as this paper only used the data deposited in the public accessible databases.
References
- 1. Bitta M, Kariuki SM, Abubakar A, et al. Burden of neurodevelopmental disorders in low and middle-income countries: a systematic review and meta-analysis. Wellcome Open Res 2017;2:121. 10.12688/wellcomeopenres.13540.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. America’s Children and the Environment . Health: neurodevelopmental disorders – report contents; 2019.
- 3. Gaugler T, Klei L, Sanders SJ, et al. Most genetic risk for autism resides with common variation. Nat Genet 2014;46:881–5. 10.1038/ng.3039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Larsson H, Chang Z, D’Onofrio BM, et al. The heritability of clinically diagnosed attention deficit hyperactivity disorder across the lifespan. Psychol Med 2014;44:2223–9. 10.1017/S0033291713002493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cardno AG, Marshall EJ, Coid B, et al. Heritability estimates for psychotic disorders: the maudsley twin psychosis series. Arch Gen Psychiatry 1999;56:162–8. 10.1001/archpsyc.56.2.162 [DOI] [PubMed] [Google Scholar]
- 6. Marshall CR, Howrigan DP, Merico D, et al. Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat Genet 2017;49:27–35. 10.1038/ng.3725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brandler WM, Antaki D, Gujral M, et al. Paternally inherited cis-regulatory structural variants are associated with autism. Science 2018;360:327–31. 10.1126/science.aan2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Coe BP, Stessman HAF, Sulovari A, et al. Neurodevelopmental disease genes implicated by de novo mutation and copy number variation morbidity. Nat Genet 2019;51:106–16. 10.1038/s41588-018-0288-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Devanna P, Chen XS, Ho J, et al. Next-gen sequencing identifies non-coding variation disrupting miRNA-binding sites in neurological disorders. Mol Psychiatry 2018;23:1375–84. 10.1038/mp.2017.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods 2010;7:248–9. 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jaganathan K, Kyriazopoulou Panagiotopoulou S, McRae JF, et al. Predicting splicing from primary sequence with deep learning. Cell 2019;176:535–48. 10.1016/j.cell.2018.12.015 [DOI] [PubMed] [Google Scholar]
- 12. Ganel L, Abel HJ, et al. , FinMetSeq Consortium . Svscore: an impact prediction tool for structural variation. Bioinformatics 2017;33:1083–5. 10.1093/bioinformatics/btw789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kumar S, Harmanci A, Vytheeswaran J, et al. SVFX: a machine learning framework to quantify the pathogenicity of structural variants. Genome Biol 2020;21:274. 10.1186/s13059-020-02178-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang Y, Wang X, Zhou D, et al. Svpath: an accurate pipeline for predicting the pathogenicity of human exon structural variants. Brief Bioinform 2022;23:bbac014. 10.1093/bib/bbac014 [DOI] [PubMed] [Google Scholar]
- 15. Geoffroy V, Guignard T, Kress A, et al. AnnotSV and knotAnnotSV: a web server for human structural variations annotations, ranking and analysis. Nucleic Acids Res 2021;49:W21–8. 10.1093/nar/gkab402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Coe BP, Witherspoon K, Rosenfeld JA, et al. Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nat Genet 2014;46:1063–71. 10.1038/ng.3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cooper GM, Coe BP, Girirajan S, et al. A copy number variation morbidity map of developmental delay. Nat Genet 2011;43:838–46. 10.1038/ng.909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sanders SJ, He X, Willsey AJ, et al. Insights into autism spectrum disorder genomic architecture and biology from 71 risk Loci. Neuron 2015;87:1215–33. 10.1016/j.neuron.2015.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zarrei M, Burton CL, Engchuan W, et al. A large data resource of genomic copy number variation across neurodevelopmental disorders. NPJ Genom Med 2019;4:26. 10.1038/s41525-019-0098-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Davis CA, Hitz BC, Sloan CA, et al. The encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res 2018;46:D794–801. 10.1093/nar/gkx1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kundaje A, Meuleman W, Roadmap Epigenomics Consortium . Integrative analysis of 111 reference human epigenomes. Nature 2015;518:317–30. 10.1038/nature14248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Boix CA, James BT, Park YP, et al. Regulatory genomic circuitry of human disease loci by integrative epigenomics. Nature 2021;590:300–7. 10.1038/s41586-020-03145-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang D, Liu S, Warrell J, et al. Comprehensive functional genomic resource and integrative model for the human brain. Science 2018;362:eaat8464. 10.1126/science.aat8464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sugathan A, Biagioli M, Golzio C, et al. CHD8 regulates neurodevelopmental pathways associated with autism spectrum disorder in neural progenitors. Proc Natl Acad Sci U S A 2014;111:E4468–77. 10.1073/pnas.1405266111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bayés A, van de Lagemaat LN, Collins MO, et al. Characterization of the proteome, diseases and evolution of the human postsynaptic density. Nat Neurosci 2011;14:19–21. 10.1038/nn.2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Doan RN, Bae B-I, Cubelos B, et al. Mutations in human accelerated regions disrupt cognition and social behavior. Cell 2016;167:341–54. 10.1016/j.cell.2016.08.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kellis M, Wold B, Snyder MP, et al. Defining functional DNA elements in the human genome. Proc Natl Acad Sci U S A 2014;111:6131–8. 10.1073/pnas.1318948111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American college of medical genetics and genomics and the association for molecular pathology. Genet Med 2015;17:405–24. 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Han X, Chen S, Flynn E, et al. Distinct epigenomic patterns are associated with haploinsufficiency and predict risk genes of developmental disorders. Nat Commun 2018;9:2138. 10.1038/s41467-018-04552-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hughes HK, Mills Ko E, Rose D, et al. Immune dysfunction and autoimmunity as pathological mechanisms in autism spectrum disorders. Front Cell Neurosci 2018;12:405. 10.3389/fncel.2018.00405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Severance EG, Prandovszky E, Castiglione J, et al. Gastroenterology issues in schizophrenia: why the gut matters. Curr Psychiatry Rep 2015;17:27. 10.1007/s11920-015-0574-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wu S, Ding Y, Wu F, et al. Family history of autoimmune diseases is associated with an increased risk of autism in children: a systematic review and meta-analysis. Neurosci Biobehav Rev 2015;55:322–32. 10.1016/j.neubiorev.2015.05.004 [DOI] [PubMed] [Google Scholar]
- 33. Bauman MD, Iosif A-M, Ashwood P, et al. Maternal antibodies from mothers of children with autism alter brain growth and social behavior development in the rhesus monkey. Transl Psychiatry 2013;3:e278. 10.1038/tp.2013.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hertz-Picciotto I, Croen LA, Hansen R, et al. The CHARGE study: an epidemiologic investigation of genetic and environmental factors contributing to autism. Environ Health Perspect 2006;114:1119–25. 10.1289/ehp.8483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Won H, de la Torre-Ubieta L, Stein JL, et al. Chromosome conformation elucidates regulatory relationships in developing human brain. Nature 2016;538:523–7. 10.1038/nature19847 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
bmjpo-2023-001966supp001.pdf (165.1KB, pdf)
Data Availability Statement
Data sharing not applicable as no datasets generated and/or analysed for this study. All features analysed during this study are collected from public datasets. Sources can be found from https://github.com/macarthur-lab/gene_lists. All CNV training data are included in these publications 16–19 and testing data are from the ClinVar database. The source code is available at https://github.com/lxsbch/neuroCNVscore.