In 1985, development of a computer system called “Deep Thought” began at Carnegie Mellon University with the lofty objective of developing an autonomous system capable of outperforming the world’s top chess grandmasters. Later renamed “Deep Blue,” this chess-playing expert system defeated world champion Gary Kasparov in 1997 in a six-game match. However, it was not until 2017 that a deep artificial neural network algorithm known as “AlphaZero” achieved super-human performance in several challenging games, including Chess, Shogi, and Go (1). Such triumphs in computer-based technologies are common today as artificial intelligence (AI) applications, such as ChatGPT and DALL-E, are mimicking human capabilities, even passing medical board examinations (2). The term AI is used to describe the general ability of computers to emulate various characteristics of human intelligence, including pattern recognition, inference, and sequential decision-making, among others. Machine learning (ML) is a subset of AI that can learn the complex interactions or temporal relationships among multivariate risk factors without the need to hand-craft such features via expert knowledge (3). Retrospective studies have demonstrated ML applications are particularly useful for their diagnostic and prognostic capabilities leveraging vast quantity of data available in the ICU (4, 5). Certain ML algorithms have approached human performance at narrow tasks such as predicting resuscitation strategies in sepsis (6), need for mechanical ventilation (7), mortality in critically ill patients (8), and ICU length of stay (9).
Sepsis is an attractive target for ML approaches as it is an inherently complex, common, costly, and deadly condition. Prediction of sepsis is the most common ML application described, although recent advances include approaches to optimize therapeutics and resuscitation strategies (6, 10). Given the potential to improve patient-centered outcomes and excitement about newer analytic approaches, it is no surprise that the number of ML algorithms aimed to improve sepsis care is increasing at a rapid rate. However, errors in sepsis prediction are often highlighted both in anecdotal and health system-wide failures that can be traced to poor implementation approaches, rudimentary ML algorithms, application of algorithms outside their intended use, or without proper maintenance. Noting these criticisms, what can be done at this point to demonstrate value of these predictive models? We believe that a revised focus on data enrichment, proper implementation, and rigorous testing is required to bring the promise of AI to the ICU.
DATA AVAILABILITY AND AUGMENTATION—THINKING OUTSIDE THE EHR
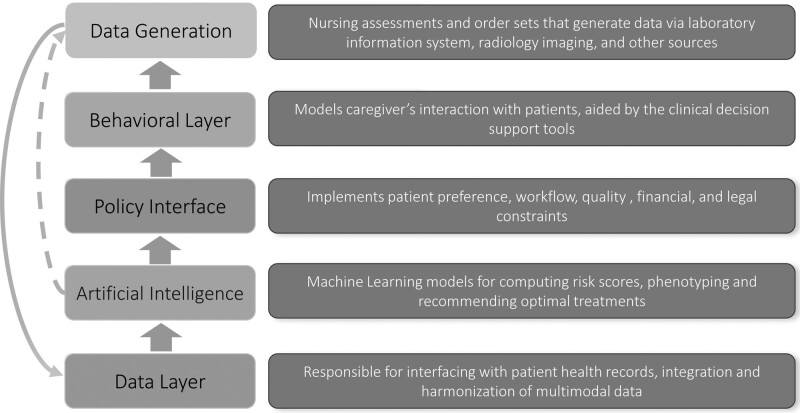
Timely data are needed for any model to improve sepsis care and is the basis of any predictive model as shown in Figure 1. To date, most ML algorithms in clinical use are limited to input features that are limited to data available in the electronic health record (EHR), such as vital signs, demographic data, laboratory results, and occasionally imaging studies. Importantly, the frequency of EHR measurements is commonly a function of level of care, workflow practices, and patients’ severity of illness (11–13). This is particularly important for patients in the hospital wards, where data are sparse and delays in sepsis identification are common (12). In other words, the most data and accurate predictions are occurring where patients are already known to have, or be at risk for, sepsis. Thus, it has been suggested that such systems are essentially looking over clinician’s shoulders. In other words, these models are using clinical behavior (e.g., ordering of a serum lactate level) as the expression of preexisting intuition and suspicion to generate a prediction (14). There are several potential solutions, none of which has been widely studied to date in a prospective setting. First, data enrichment, the process of incorporating updated data elements from sources outside the electronic health record, could be used. For example, multimodal data from bedside monitors, IV pumps, mechanical ventilators, and imaging studies could all be collected. Development of wearable biopatches may allow for incorporation of near instantaneous data. Second, under most existing protocols, AI is not actively involved in data generation. The use of smart laboratories—diagnostic studies suggested or even ordered by an AI system at times of particularly low predictive certainty—and/or additional nursing assessments may help improve accuracy of these algorithms, although the challenge is to “choose wisely” and keeps costs and workflow impediments minimized. Testing of this approach is indicated prior to widespread implementation to ensure that any costs—whether it be direct financial costs, cognitive burden on provider, medicolegal risk, or patient discomfort—is minimized while adding value to care. Although the infrastructure of this approach does require coordination between various key stakeholders, the benefit of a real-time predictive score may outweigh potential costs (15). Indeed, an additional timely laboratory draw has the potential to avoid many downstream costs or could replace commonly used low value strategies (e.g., routine “morning laboratories”). Figure 1 depicts an augmented (via dashed lines) healthcare information generation and processing stack in which the AI systems may initiate data generation to improve predictive accuracy and reduce diagnostic uncertainty and delays.
Figure 1.
Conceptualization of healthcare information generation and processing stack.
BUILDING THE FRAMEWORK: DEVELOPING EFFECTIVE STRATEGIES TO BRING MODELS TO THE BEDSIDE
Even the most promising AI systems in medicine need to be implemented clinically, evaluated with adequate safety nets in place, and iteratively improved over time to be successful. Yet, implementation of these models into clinical practice is too often an afterthought compared with the investment of model development. This “implementation gap” between what has been developed and what is in use continues to expand (16). We propose three strategies for implementation of AI to optimize this “policy layer” as shown in Figure 1:
Real-time case reviews: These reviews are intended to obtain clinical feedback on the performance of AI systems from the perspective of clinical utility (timing and appropriateness of the alerts) and to fine-tune the policy layer of the overall clinical decision support tool. As an example, the policy layer may include suppression of alerts on all patients already promptly recognized as septic and thus receiving early antibiotics. This change also results in a cohort of patients with phenotypically different characteristics (i.e., only unrecognized sepsis) from the retrospective data (all sepsis) for the AI system to manage. The iterative improvements and changes to the predictive model gained by these prospective validation steps are crucially valuable yet overlooked step in the implementation of predictive scores into clinical practice.
Silent trial: An approach in which key stakeholders evaluate an AI model, that is integrated into the electronic health record, on patients in real-time, yet the model is not involved in clinical care or interacting with eventual end-users. The silent trial provides an opportunity to study alert rates, user interface design and usability, and educate the end-users. These silent trials can result in improvement in a model’s predictive ability and also markedly reduce false alarms (17).
A/B testing or rapid-cycle randomized testing (18): A simple controlled experiment in which users are randomly prescribed a control (A) or a variation (B) of a design, where the variation is limited to a single isolated feature. This is common outside of medicine where companies such as Amazon and Microsoft use A/B testing to test two versions of content against each other in an online experimental setting to improve click-through rate (19). A/B testing may enhance user interface for predictive models and improve end-user response, thus increasing effective detection rate (e.g., minimizing “snooze” rates).
We believe all of these strategies should be employed in a learning health system as part of local quality improvement efforts and may not require Institutional Review Board oversight (18, 20).
Finally, testing the influence of the “behavioral layer” (Fig. 1) requires pilot implementation studies in a clinical setting, in which clinical actions (such as the practice of “snoozing” of alerts) and workflow-related factors (e.g., frequency and timing of ordering of laboratories) can impact the performance of the AI system (e.g., false negatives due to lack of timely data availability). As such, even “negative” implementation studies of sepsis predictive scores can uncover unanticipated implementation and process deficiencies that may provide insights into strategies to improve care and future study design.
SHOWING BENEFIT THROUGH APPROPRIATE TESTING—THE NEED FOR PROSPECTIVE STUDIES
To date, there have been only two published randomized clinical investigations evaluating the benefit of a sepsis ML model to improve patient-centered outcomes despite nearly 500 published articles in this area (21, 22). Importantly, of the remaining publications, only a handful are prospective evaluation of predictive algorithms in clinical use, and the vast majority are retrospective in nature (23–25). This situation partially occurs not only due to significant technological and cultural challenges, as well as cost, in real-time implementation of such systems, but also because the publication bar for ML in medicine has been too low. Only recently have some editors developed guidelines to push investigators to submit forward-looking investigations in ML applications that focus on clinical utility (26).
These retrospective studies have flaws that limit generalizability. First, there is significant heterogeneity among established sepsis criteria which ML models are trained and subsequently validated. Recent publications have shown poor overlap between different automated sepsis criteria, and this may significantly impact model usability at hospitals that use a different definition of sepsis (27). In other words, a model trained on administrative diagnoses of sepsis may be poorly received at a hospital where providers rely on the Center of Medicare and Medicaid sepsis definitions. Next, many ML studies in sepsis focus on traditional statistical performance metrics, such as area under the receiver operating characteristic curve (AUCroc) to show benefit. However, clinicians do not measure the success of an algorithm by noting a high AUCroc, but rather how such algorithms improve clinical care and workflow. More realistic and pertinent patient-focused metrics with provider and patient input, such as time to antibiotic administration, decreased hospital length of stay, and mortality benefits are needed for algorithm assessment and comparative analyses in prospective fashion—and for physician and health system buy-in (28, 29). Finally, only since 2019 have mature interoperability standards (such as Fast Healthcare Interoperability Resources R4) and Health Insurance Portability and Accountability Act-compliant cloud computing resources been widely adopted to allow interoperable, secure, scalable, and reliable real-time access to electronic health records and bedside monitoring devices which can facilitate multicenter prospective implementation trials (30, 31). The result has been an exponential growth in retrospective studies and a paucity of high-quality clinical evidence focused on patient-centered outcomes. The Epic Sepsis Score provides a cautionary tale for the approach of implementation of a predictive model. The shortcomings of this model were highlighted by a team at the University of Michigan who reported test characteristics significantly lower than reported by Epic as well as an unacceptably high number of false positives in real-time use (32).
TOWARD CLINICIAN-AI SYMBIOSIS AND CONTINUOUS LEARNING
We recognize healthcare workers are faced with an increasing number of daily tasks to complete during patient care activities. Ineffective or poorly implemented predictive scores result in frustration and mistrust of these models and may cause inadvertent harm through inappropriate or unnecessary antibiotics or fluid administration or exacerbate cognitive overload (13, 33, 34). Widespread efforts are needed to help decrease the number of false alerts while maintaining algorithm integrity and generalizability. Granular nationwide datasets and novel approaches, such as transfer learning and conformal prediction, may afford sepsis predictive models increasing ability to generalize across institutions accurately (35–37). Recently, we demonstrated that a sepsis predictive algorithm can decrease false alarms by detecting unfamiliar patients/situations arising from erroneous data, missingness, distributional shift, and data drifts although such an approach is largely untested in prospective fashion (8). In such scenarios, the AI system refrains from making spurious predictions by saying “I don’t know.” An actionable next step in this situation may be to use smart laboratories or additional nursing assessment to decrease diagnostic uncertainty or trigger the need to update the model (i.e., algorithm change protocol) (38). Although the optimal mode of clinician-AI symbiosis remains a ripe area of research, cultural and regulatory shifts in defining the “reasonable and necessary” causes for deploying diagnostic laboratories and devices by various members of the care team (including bedside nurses, clinicians, or the AI agent) may be required to enhance this partnership (39). Finally, we need to recognize the limit of AI: it may alert us to a patient with sepsis, but—unlike chess—it does not yet know the next move.
The predictive ability of machine learning algorithms is improving rapidly due to larger datasets (40, 41), new approaches to fine-tune algorithms, and continually improved computing power. But, provider and patient input and consideration of human behavior are keys. As an example, an early version of Deep Blue failed to beat Kasparov, and the system had to be trained with data from other grandmasters to prepare for subsequent matches. Although hardware improvements were also needed to achieve super-human performance, the latest iteration of such systems (1) had to embrace more powerful ML algorithms and were programmed to learn continually. We worry that provider mistrust and frustration with such algorithms will only grow and may unfortunately lead to premature dismissal of these tools. Table 1 highlights concerns, responses, and potential solutions to ensure adoption of AI in sepsis and critically ill patients in general. Who is responsible ultimately for implementing these solutions remains an area of debate. Like much in this area, this will require input from multiple stakeholders including patients, healthcare workers, AI developers, and administrators with support from national funding agencies to provide appropriate incentives. Ultimately, we hope that clinicians come to see these algorithms as a trusted partner—like advice from a master clinician—offering an opinion based on all the relevant data and years of past data drawn from deep knowledge of the institution and similar past patients.
TABLE 1.
Potential Reasons Why Artificial Intelligence Has Not Been Embraced by the Critical Care Community
| Concern | Rebuttal | Potential Solution | |
|---|---|---|---|
| Patient factors | |||
| Lack of awareness by patients and families | Newer technology in healthcare without significant lay exposure | Public education and media explanation of AI in healthcare | |
| Reluctance to have AI in care | Noninvasive and meant to augment clinical care, not replace physicians | Explanation of use of algorithm and potential benefits during clinical care | |
| Privacy concerns | Newer systems use HIPAA-compliant cloud computing resources | Emphasis on HIPAA compliance approaches in clinical use | |
| Clinician factors | |||
| Lack of awareness by clinicians | Minimal teaching in this area during medical education | Improved medical education in this area, engagement of clinicians in implementation | |
| Mistrust of AI approaches | Older algorithms lack sophistication, abilities and performance of newer deep learning algorithms | More research and education demonstrating benefit in clinical care. | |
| Concerns about medicolegal aspects using AI | Field is young without clear precedent | U.S. Food and Drug Administration and other regulatory approval; clear “intended use” for AI algorithms | |
| Lack of definitive multicenter randomized trials | Data are evolving; field is young and dynamic | Federal funding agencies should support grant funding on mature algorithms | |
| Technology factors | |||
| Suboptimal predictive abilities of AI algorithms | Powered by big data and multimodal data, these systems are rapidly improving | Newer deep learning algorithms, advances to augment data availability (e.g., biopatches, data by smart laboratories) | |
| Lack of infrastructure for real-time predictive scores | Newer cloud computing and interoperability technologies are lowering the infrastructure barriers | Incorporating cloud computing, healthcare interoperability standards, software engineering, and hospital information technology education into clinical AI curriculum | |
| Systems factors | |||
| Poor implementation approaches | Historically, this has been overlooked in favor of algorithm development | Implementation science and multidisciplinary teams improve use of algorithms | |
| Lack of administrative support to properly implement AI algorithms into clinical use | Although there are upfront costs, potential benefits likely outweigh this | Studies demonstrating improved outcomes and cost-effectiveness; emphasis on interoperability standards | |
| Misalignment of patient care, quality improvement, and financial incentives | Value-based care and the changing landscape of digital health reimbursement | Closer collaboration of AI experts, hospital quality improvement, value-based care, and finance teams | |
AI = artificial intelligence, HIPPA = Health Insurance Portability and Accountability Act.
Footnotes
Dr. Wardi received funding from Northwest Anesthesia and Medicolegal consulting. Drs. Wardi, Malhotra, and Nemati received support for article research from the National Institutes of Health (NIH). Dr. Malhotra’s institution received funding from ResMed; he received funding from the NIH, Livanova, Eli Lilly, Zoll, and Jazz. Drs. Malhotra and Nemati disclosed that they are cofounders and equity shareholders in Healcisio, Inc. Dr. Josef received funding from Healcisio, Inc. Dr. Longhurst disclosed that he is an equity shareholder in Doximity. Dr. Owens has disclosed that he does not have any potential conflicts of interest.
REFERENCES
- 1.Silver D, Hubert T, Schrittwieser J, et al. : A general reinforcement learning algorithm that masters Chess, Shogi, and Go through self-play. Science. 2018; 362:1140–1144 [DOI] [PubMed] [Google Scholar]
- 2.Kung TH, Cheatham M, ChatGPT; et al. : Performance of ChatGPT on USMLE: Potential for AI-assisted medical education using large language models. PLOS Digit Health. 2023; 2:e0000198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.LeCun Y, Bengio Y, Hinton G: Deep learning. Nature. 2015; 521:436–444 [DOI] [PubMed] [Google Scholar]
- 4.Celi LA, Mark RG, Stone DJ, et al. : “Big data” in the intensive care unit. Closing the data loop. Am J Respir Crit Care Med. 2013; 187:1157–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rush B, Stone DJ, Celi LA: From big data to artificial intelligence: Harnessing data routinely collected in the process of care. Crit Care Med. 2018; 46:345–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Komorowski M, Celi LA, Badawi O, et al. : The artificial intelligence clinician learns optimal treatment strategies for sepsis in intensive care. Nat Med. 2018; 24:1716–1720 [DOI] [PubMed] [Google Scholar]
- 7.Shashikumar SP, Wardi G, Paul P, et al. : Development and prospective validation of a deep learning algorithm for predicting need for mechanical ventilation. Chest. 2021; 159:2264–2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Awad A, Bader-El-Den M, McNicholas J, et al. : Early hospital mortality prediction of intensive care unit patients using an ensemble learning approach. Int J Med Inf. 2017; 108:185–195 [DOI] [PubMed] [Google Scholar]
- 9.Houthooft R, Ruyssinck J, van der Herten J, et al. : Predictive modelling of survival and length of stay in critically ill patients using sequential organ failure scores. Artif Intell Med. 2015; 63:191–207 [DOI] [PubMed] [Google Scholar]
- 10.Kanjilal S, Oberst M, Boominathan S, et al. : A decision algorithm to promote outpatient antimicrobial stewardship for uncomplicated urinary tract infection. Sci Transl Med. 2020; 12:eaay5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Agniel D, Kohane IS, Weber GM: Biases in electronic health record data due to processes within the healthcare system: Retrospective observational study. BMJ. 2018; 361:k1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moskowitz A, McSparron J, Stone DJ, et al. : Preparing a new generation of clinicians for the era of big data. Harv Med Stud Rev. 2015; 2:24–27 [PMC free article] [PubMed] [Google Scholar]
- 13.Gal DB, Han B, Longhurst C, et al. : Quantifying electronic health record data: A potential risk for cognitive overload. Hosp Pediatr. 2021; 11:175–178 [DOI] [PubMed] [Google Scholar]
- 14.Beaulieu-Jones BK, Yuan W, Brat GA, et al. : Machine learning for patient risk stratification: Standing on, or looking over, the shoulders of clinicians? NPJ Digit Med. 2021; 4:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rogers P, Boussina AE, Shashikumar SP, et al. : Optimizing the implementation of clinical predictive models to minimize national costs: Sepsis case study. J Med Internet Res. 2023; 25:e43486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seneviratne MG, Shah NH, Chu L: Bridging the implementation gap of machine learning in healthcare. BMJ Innov. 2020; 6:45 [Google Scholar]
- 17.Kwong JCC, Erdman L, Khondker A, et al. : The silent trial - The bridge between bench-to-bedside clinical AI applications. Front Digit Health. 2022; 4:929508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horwitz LI, Kuznetsova M, Jones SA: Creating a learning health system through rapid-cycle, randomized testing. N Engl J Med. 2019; 381:1175–1179 [DOI] [PubMed] [Google Scholar]
- 19.Kohavi R, Longbotham R: Online controlled experiments and A/B testing. In: Encyclopedia of Machine Learning and Data Mining. First Edition. Sammut C, Webb GI. (Eds). New York, NY, Springer, 2017, pp 922–929 [Google Scholar]
- 20.El-Kareh R, Brenner DA, Longhurst CA: Developing a highly reliable learning health system. Learn Health Syst. 2022:e10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shimabukuro DW, Barton CW, Feldman MD, et al. : Effect of a machine learning-based severe sepsis prediction algorithm on patient survival and hospital length of stay: A randomised clinical trial. BMJ Open Respir Res. 2017; 4:e000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tarabichi Y, Cheng A, Bar-Shain D, et al. : Improving timeliness of antibiotic administration using a provider and pharmacist facing sepsis early warning system in the emergency department setting: A randomized controlled quality improvement initiative. Crit Care Med. 2022; 50:418–427 [DOI] [PubMed] [Google Scholar]
- 23.Adams R, Henry KE, Sridharan A, et al. : Prospective, multi-site study of patient outcomes after implementation of the TREWS machine learning-based early warning system for sepsis. Nat Med. 2022; 28:1455–1460 [DOI] [PubMed] [Google Scholar]
- 24.Ginestra JC, Giannini HM, Schweickert WD, et al. : Clinician perception of a machine learning-based early warning system designed to predict severe sepsis and septic shock. Crit Care Med. 2019; 47:1477–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giannini HM, Ginestra JC, Chivers C, et al. : A machine learning algorithm to predict severe sepsis and septic shock: Development, implementation, and impact on clinical practice. Crit Care Med. 2019; 47:1485–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leisman DE, Harhay MO, Lederer DJ, et al. : Development and reporting of prediction models: Guidance for authors from editors of respiratory, sleep, and critical care journals. Crit Care Med. 2020; 48:623–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu SC, Betthauser KD, Gupta A, et al. : Comparison of sepsis definitions as automated criteria. Crit Care Med. 2021; 49:e433–e443 [DOI] [PubMed] [Google Scholar]
- 28.Reyna MA, Josef CS, Jeter R, et al. : Early prediction of sepsis from clinical data: The PhysioNet/computing in cardiology challenge 2019. Crit Care Med. 2020; 48:210–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reyna MA, Nsoesie EO, Clifford GD: Rethinking algorithm performance metrics for artificial intelligence in diagnostic medicine. JAMA. 2022; 328:329–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kasthurirathne SN, Mamlin B, Kumara H, et al. : Enabling better interoperability for HealthCare: Lessons in developing a standards based application programing interface for electronic medical record systems. J Med Syst. 2015; 39:182. [DOI] [PubMed] [Google Scholar]
- 31.Khalilia M, Choi M, Henderson A, et al. : Clinical predictive modeling development and deployment through FHIR web services. AMIA Annu Symp Proc AMIA Symp. 2015; 2015:717–726 [PMC free article] [PubMed] [Google Scholar]
- 32.Wong A, Otles E, Donnelly JP, et al. : External validation of a widely implemented proprietary sepsis prediction model in hospitalized patients. JAMA Intern Med. 2021; 181:1065–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alami H, Lehoux P, Auclair Y, et al. : Artificial intelligence and health technology assessment: Anticipating a new level of complexity. J Med Internet Res. 2020; 22:e17707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gettinger A, Zayas-Cabán T: HITECH to 21st century cures: Clinician burden and evolving health IT policy. J Am Med Inform Assoc JAMIA. 2021; 28:1022–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wardi G, Carlile M, Holder A, et al. : Predicting progression to septic shock in the emergency department using an externally generalizable machine-learning algorithm. Ann Emerg Med. 2021; 77:395–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holder AL, Shashikumar SP, Wardi G, et al. : A locally optimized data-driven tool to predict sepsis-associated vasopressor use in the ICU. Crit Care Med. 2021; 49:e1196–e1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shashikumar SP, Wardi G, Malhotra A, et al. : Artificial intelligence sepsis prediction algorithm learns to say “I don’t know.”. NPJ Digit Med. 2021; 4:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gilbert S, Fenech M, Hirsch M, et al. : Algorithm change protocols in the regulation of adaptive machine learning-based medical devices. J Med Internet Res. 2021; 23:e30545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neumann PJ, Chambers JD: Medicare’s enduring struggle to define “reasonable and necessary” care. N Engl J Med. 2012; 367:1775–1777 [DOI] [PubMed] [Google Scholar]
- 40.Bradwell KR, Wooldridge JT, Amor B, et al. : Harmonizing units and values of quantitative data elements in a very large nationally pooled electronic health record (EHR) dataset. J Am Med Inform Assoc JAMIA. 2022; 29:1172–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Denny JC, Rutter JL, Goldstein DB, et al. ; All of Us Research Program Investigators: The “all of us” research program. N Engl J Med. 2019; 381:668–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Timely data are needed for any model to improve sepsis care and is the basis of any predictive model as shown in Figure 1. To date, most ML algorithms in clinical use are limited to input features that are limited to data available in the electronic health record (EHR), such as vital signs, demographic data, laboratory results, and occasionally imaging studies. Importantly, the frequency of EHR measurements is commonly a function of level of care, workflow practices, and patients’ severity of illness (11–13). This is particularly important for patients in the hospital wards, where data are sparse and delays in sepsis identification are common (12). In other words, the most data and accurate predictions are occurring where patients are already known to have, or be at risk for, sepsis. Thus, it has been suggested that such systems are essentially looking over clinician’s shoulders. In other words, these models are using clinical behavior (e.g., ordering of a serum lactate level) as the expression of preexisting intuition and suspicion to generate a prediction (14). There are several potential solutions, none of which has been widely studied to date in a prospective setting. First, data enrichment, the process of incorporating updated data elements from sources outside the electronic health record, could be used. For example, multimodal data from bedside monitors, IV pumps, mechanical ventilators, and imaging studies could all be collected. Development of wearable biopatches may allow for incorporation of near instantaneous data. Second, under most existing protocols, AI is not actively involved in data generation. The use of smart laboratories—diagnostic studies suggested or even ordered by an AI system at times of particularly low predictive certainty—and/or additional nursing assessments may help improve accuracy of these algorithms, although the challenge is to “choose wisely” and keeps costs and workflow impediments minimized. Testing of this approach is indicated prior to widespread implementation to ensure that any costs—whether it be direct financial costs, cognitive burden on provider, medicolegal risk, or patient discomfort—is minimized while adding value to care. Although the infrastructure of this approach does require coordination between various key stakeholders, the benefit of a real-time predictive score may outweigh potential costs (15). Indeed, an additional timely laboratory draw has the potential to avoid many downstream costs or could replace commonly used low value strategies (e.g., routine “morning laboratories”). Figure 1 depicts an augmented (via dashed lines) healthcare information generation and processing stack in which the AI systems may initiate data generation to improve predictive accuracy and reduce diagnostic uncertainty and delays.
Figure 1.
Conceptualization of healthcare information generation and processing stack.