Keywords: anophelines, diverged evolution, genome, immune response, microbiota
Abstract
Anophelines are vectors of malaria, the deadliest disease worldwide transmitted by mosquitoes. The availability of genomic data from various Anopheles species allowed evolutionary comparisons of the immune response genes in search of alternative vector control of the malarial parasites. Now, with the Anopheles aquasalis genome, it was possible to obtain more information about the evolution of the immune response genes. Anopheles aquasalis has 278 immune genes in 24 families or groups. Comparatively, the American anophelines possess fewer genes than Anopheles gambiae s. s., the most dangerous African vector. The most remarkable differences were found in the pathogen recognition and modulation families like FREPs, CLIP and C-type lectins. Even so, genes related to the modulation of the expression of effectors in response to pathogens and gene families that control the production of reactive oxygen species were more conserved. Overall, the results show a variable pattern of evolution in the immune response genes in the anopheline species. Environmental factors, such as exposure to different pathogens and differences in the microbiota composition, could shape the expression of this group of genes. The results presented here will contribute to a better knowledge of the Neotropical vector and open opportunities for malaria control in the endemic-affected areas of the New World.
1. Introduction
Malaria is a severe public health problem in several tropical and subtropical areas. It is caused by parasites of the genus Plasmodium, and is of most concern in the African, Asian and American continents. Annually there are 229 million cases that result in nearly half a million deaths worldwide, affecting mainly pregnant women and children [1]. The disease is transmitted to humans by the bite of female Anopheles sp. Among more than 400 known species of Anopheles, 41 are recognized vectors worldwide, 9 of which are found in the Americas, including Anopheles aquasalis, the primary malaria vector of coastal Central and South Americas and the Caribbean Islands [2].
The biological characteristics influenced by variations in the ability of Anopheles vectors to transmit Plasmodium (e.g. molecular components of the immune response, microbiota and intestinal physiology) are well studied and have been characterized in established African and Asian models such as An. gambiae s. s. and An. stephensi [3–5]. The defence mechanisms are critical for establishing the Plasmodium in the vectors, and three-quarters of the total ingested parasites die inside the midgut at the start of their life cycle [6]. Changes in the expression of immunity genes occur and vary according to the invaded organ. Also, in this phenomenon the participation of the immune components changes depending on the anopheline species and the malaria parasite [5,7,8].
Several families of innate immune genes control the mosquito response to pathogens. They can influence vector competence, allowing or preventing the development of Plasmodium's life cycle in the mosquito [9–11]. Also, other components of the humoral immune response in mosquito vectors are effector molecules, recognition molecules of microorganisms, signalling pathways and protease cascades. The hemocytes present in the hemolymph are the primary cellular immune response. They participate in phagocytosis, encapsulation, melanization and production of antagonistic molecules to the pathogens [12,13].
It is assumed that the vector's immune response is predominant in controlling Plasmodium infection and transmission through the synthesis of anti-pathogen molecules, including those expressed after activation of immune signalling pathways. Several components of the IMD (immune deficiency) and TOLL signalling pathways influence the development of murine and human Plasmodium in different anopheline species such as An. gambiae s. s., An. stephensi and An. albimanus [14,15]. For example, the silencing of Caspar, the negative regulator of the REL2 in the IMD pathway, had a marked effect leading to increased production of antispasmodic molecules, such as thioester-containing protein-1 (TEP1), leucine-rich repeat protein-7 (LRR7), Anopheles Plasmodium-responsive Leucine-Rich Repeat-2 (APL2), and fibrillin-9 (FBN9) [14,15]. Consequently, the suppression of the action of Caspar leads to the continuous activation of the REL2 transcription factor and, therefore, to the increase in the expression of effectors and other molecules regulated by the IMD pathway [14,15]. Based on the findings made in the IMD pathway, the genetic manipulation of An. stephensi for the continuous expression of REL2 was carried out, allowing the generation of mosquitoes highly resistant to P. falciparum infection [16].
Likewise, immune mechanisms responsible for microbial homeostasis in the mosquito and molecules related to the structural maintenance of the peritrophic matrix have been described as limiting factors for developing plasmodia [17,18]. In An. gambiae s. s. and An. stephensi mosquitoes, proteins dual oxidase:peroxidase and NADPH-oxidase domains (DUOX) and heme peroxidase 15 (HPX15) participate in catalysing the cross-linking of the mucin layer that delimits the food bolus avoiding the direct contact of the intestinal epithelium with the gut microbiota. The silencing of HPX15 increased the expression of the nitric oxide synthase (NOS) protein, the main effector regulated by the JAK/STAT pathway, eliminating both P. berghei and P. falciparum through the TEP1 protein [19–21]. On the other hand, the silencing of DUOX protein, a substrate for HPX15 protein, increases the expression of NOS, decreasing the number of P. berghei in A. gambiae s. s. [21]. However, the silencing of this protein does not affect the parasite development in An. stephensi since it does not affect the NOS expression [22].
In this sense, pathogen recognition components are so important in the sporogony cycle of the malaria parasite within the mosquito that they have become candidates for the production of transmission-blocking vaccines (TBVs) or genetic modification in the anophelines [23,24]. As has been shown, for example, for the fibrinogen-1 related protein (FREP1), its presence is necessary for the ookinetes of murine and human plasmodia. When the parasite interacts with this protein, it manages to overcome the peritrophic matrix and continues its movement toward the epithelium of the intestine medium [24,25]. The interaction between FREP1 and ookinetes has been demonstrated in African and Asian anopheline species [24,25]. In addition, according to bioinformatic analysis, it is a highly conserved member of the anopheles genus. Therefore it is speculated to have the same relevance in several malaria vectors, making it a good candidate for TBV [25].
Studies about the interaction between mosquito vectors and Plasmodium highlight the importance of understanding the functioning and participation of distinct genes involved in the immune response process. In the future, this knowledge may help raise mechanisms for malaria control. The exciting issue about these data on genetic modification targets or candidates for transmission-blocking vaccines is their conservation among anophelines [19,22,25]. With the annotation of 16 anopheline genomes, it was possible to verify that roughly 60% of orthologous groups are shared within the genus Anopheles. Although, in this genomic comparison, intrinsic evolutionary characteristics of the groups or families of genes related to the immune response in anophelines have been recognized. Phylogenetic comparisons could reveal the existence of agonist or antagonist genes against plasmodia in American anophelines, which were detected in African or Asian anophelines in functional experiments [26].
Nevertheless, most of these results were raised from restricted groups of Old World anophelines, which limits what is known about these genes in the neotropical malaria vectors. In the present study, we used the recently annotated genome of the neotropical mosquito An. aquasalis to study the composition of the immune response gene families and to make phylogenetic comparisons among four anopheline species of mosquitoes whose genomic data is available. This work is based on homology comparisons with the genes of the immune response of An. gambiae s. s., An. albimanus and An. darlingi, in addition to using the neighbour-joining (NJ) methodology to make evolutionary inferences with the immune response components identified in An. aquasalis.
2. Material and methods
2.1. Identification and structural characterization of immune response genes in Anopheles aquasalis
The Anopheles aquasalis mosquito comes from the colony established in the laboratory of Medical Entomology at FIOCRUZ-MG. An. aquasalis protein sequences were obtained from genome annotation (GCA_002846955.1) [27]. Putative An. aquasalis immune response proteins were identified in the genome and annotated proteins by tBLASTn and BLASTp (amino acid identity greater than 40% and e-value < 0.0001) searches against Anopheles gambiae s. s. immune response proteins available at the ImmunoDB database (https://www.ezlab.org/#newick-utils, 6 October 2020) [28].
We used amino acid sequences described by Cao et al. [29] as models for blast searches for serine-proteases with CLIP domains. Orthology was initially confirmed by the reciprocal best match method. Genes identified in the genome but not in the annotated proteins had their code regions predicted by GeneWise, using the splice site modelled option [30]. The protein domains of An. aquasalis were characterized with the Conserved Domains Database (CDD) database or Interproscan online tool [31,32]. Motifs within some relevant protein domains were identified and visualized using MEME, with the discriminate mode option using proteins of the An. gambiae s. s. as a control [33]. Other protein characteristics, such as a signal peptide, were searched for with the SignalP and Macoil programs [34,35].
2.2. Evolutionary analysis of An. aquasalis immune response genes
An. gambiae s. s. immune response proteins were retrieved from the ImmunoDB. Protein sequences from the new world mosquitoes An. darlingi were also downloaded (Coari AdarC3.8), An. albimanus (STECLA AalbS2.6) and from An. gambiae s. s. (PEST PEPTIDES AgamP4.12) from the VectorBase database website (https://vectorbase.org/) [36]. To carry out the phylogenetic analysis of the stat1 and stat2 genes, the reference sequences deposited in the Vectorbase database of 14 anopheline species were used (101). Multiple alignments of the sequences between the proteins of the four mosquito species were performed with the Muscle tool with the UPGMA clustering method, with gap opening and gap extension penalties of −2.9 and 0, respectively [37]. The phylogenetic trees were constructed using the NJ method, in which the distance was estimated by the p-distance method implemented in the MEGA-X program. The pairwise deletion option was applied in the NJ tree construction, and the tree topology's accuracy was evaluated using 1000 bootstrap replicates of the sequence alignment [38]. Finally, gene expansion was visualized through a heatmap using the online tool Heatmapper. Six species of mosquitoes were used, including Culex quinquefasciatus as a species within the Culicidae family, little related to the Anopheles genus, and the An. stephensi mosquito, a species of the Cellia subgenus. Thus, the mean of each family of immune response genes was calculated from the information of the six mosquitoes and the loss or gain of genes was expressed with the standard deviation for each species [39].
3. Results
The data used in this study come from the annotation of the An. aquasalis genome [27]. The assembled An. aquasalis genome (BioProject PRJNA389759) has 162 944 Mb, distributed in 16 504 scaffolds (N50 14 431) and according to BUSCO analysis, has a completeness of 96.2% of complete single-copy genes, 0.04% complete and duplicated, 2.3% fragmented and 1.5% missing genes. A total of 12 446 protein-coding genes were predicted in the genome and used for the characterization of the components of the immune response.
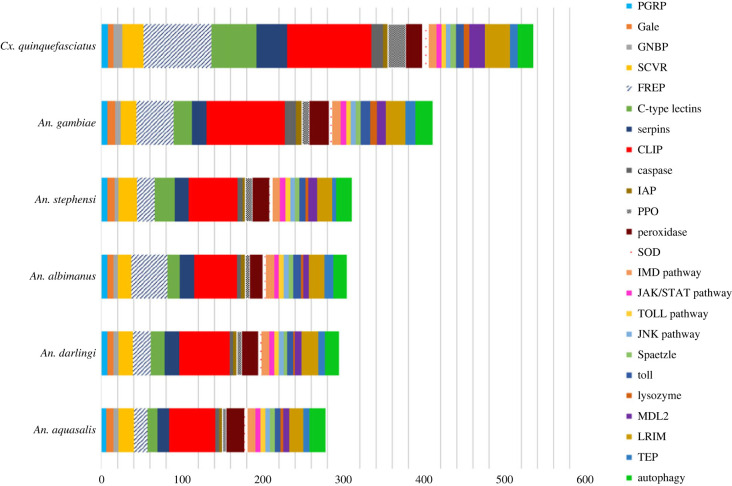
Two hundred and seventy-eight proteins were identified as of putative immune response function in the An. aquasalis genome (figure 1; electronic supplementary material, excel table). These proteins were categorized into 24 families or signalling pathways. Among the sequences, nine genes showed fragmented structures and were manually annotated with the GeneWise program (electronic supplementary material, table S1). Our analysis suggests that An. aquasalis maintained a similar quantity of immune genes compared with the other New World mosquitoes (figure 1; electronic supplementary material, table S1) and to An. stephensi (310 genes), unlike An.gambiae s. s., which had approximately 130 more immune response genes. We observed that such differences compared to the An. gambiae s. s. is concentrated in two gene families, fibrinogen-related protein (FREP) and CLIP-domain serine protease (CLIP). While An. gambiae s. s. has 46 FREP proteins, we found only 17 in An. aquasalis. For CLIP proteins, An. gambiae s. s. has 97 annotated proteins, while An. aquasalis has 57 such genes.
Figure 1.
The number of immune response genes in Anopheles aquasalis and other anopheline mosquitoes (Culicidae: subfamily Anophelinae). Culex quinquefasciatus (Culicidae: subfamily Culicinae) was used as an external group. Each colour in the graph represents an immunity-related gene family.
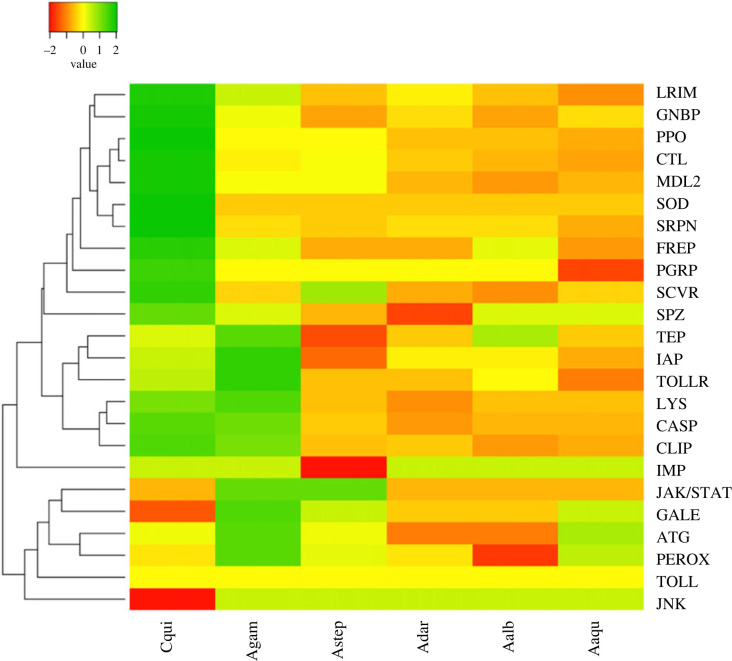
To facilitate the visualization of gene number variation in each immune-related gene family, we performed a heatmap analysis (figure 2). The heatmap shows the expansion and contraction of gene families in the standard deviation fold. The standard deviation represented the specific variation of each family of the immune response by species. Many gene families present a significant variation in gene copy number in anophelines, remarkably FREP (22.0 copies ± 13.9), CLIP (61.0 ± 17.6), and C-type lectins (17.0 ± 5.5). On the other hand, we observed slight variation in peptidoglycan recognition proteins (PGRPs) (7.0 ± 0.5), Serpins (18.0 ± 1.3) and the IMD pathway (10.0 ± 0.5), and no variation in superoxide dismutase (4 in all anophelines), Toll pathway and JNK pathway genes (both with six genes each). Using Culex quinquefasciatus as an external group, we can observe groups of expansions in An. gambiae s. s. (first: TEP, apoptosis inhibitors, Toll, lysozyme, caspase and CLIP; second: JAK/STAT pathway, galectins, autophagy and peroxidases). In New-World anophelines, we observe contractions in leucine-rich immune protein (LRIM), gram-negative binding proteins, prophenoloxidases, C-type lectins and Niemann-Pick C2 proteins (MLD2) compared to An. gambiae s. s. and Cx. quinquefasciatus. Regarding specifically An. aquasalis, our attention is drawn to the loss of peptidoglycan recognition protein, serpins and LRIM proteins, as much as the gain of peroxidase and autophagy-related proteins. In the following sections, we will focus on the results of the classic and conserved pathways and those gene families that showed remarkable gains or losses in An. aquasalis.
Figure 2.
Heatmap of gene copy number variation of immunity-related gene families in Anopheles aquasalis and other anopheline mosquitoes (Culicidae: subfamily Anophelinae). Culex quinquefasciatus (Culicidae: subfamily Culicinae) was used as an external group. Twenty-four gene families were shown. A phylogenetic tree was generated to represent the gene families and the mosquitoes. The standard deviation represented the specific variation of each family of the immune response by species. Thus, values close to two represent copy gain (Green), and values relative to −2, on the contrary, indicate copy loss (brown) and no variation in those values close to zero (yellow). Culex quinquefasciatus (Cqui), An. gambiae s. s. (Agam), An. aquasalis (Aaqu), An. darlingi (Adar) and An. albimanus (Aalb).
3.1. Intracellular signal pathways (IMD, Toll, JAK/STAT and JNK)
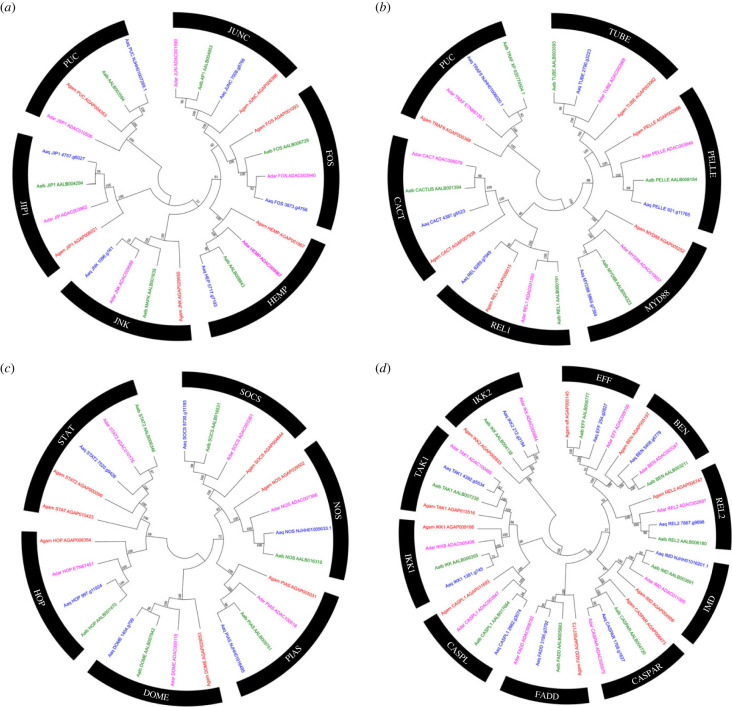
After identifying microorganisms and other foreign organisms, the signalling pathways lead to the transcriptional activation of antagonist molecules. The most studied signalling transduction pathways in anophelines are Toll, IMD, and JAK/STAT, but some works also implicate the JNK pathway in control against some species of Plasmodium [40–42]. Most genes are part of the signalling pathways identified in An. gambiae s. s. are present in An. aquasalis, An. darlingi and An. albimanus, here an orthologous relationship between each individual member of the family was checked (figure 3).
Figure 3.
Evolutionary relationships of classical gene families of the IMD, Toll, JNK, and JAK/STAT immune pathways in Anopheles aquasalis. (a). JNK pathway tree, (b). Toll pathway tree, (c). JAK/STAT pathway tree, (d). IMD pathway tree. Neighbour-joining tree of classical immune systems pathways from the amino acid sequences of the mosquitoes An. aquasalis (blue), An. darlingi (pink), An. albimanus (green) and An. gambiae s. s. (red). Bootstrap values were calculated with 1000 replicates, and their values are presented on each tree branch.
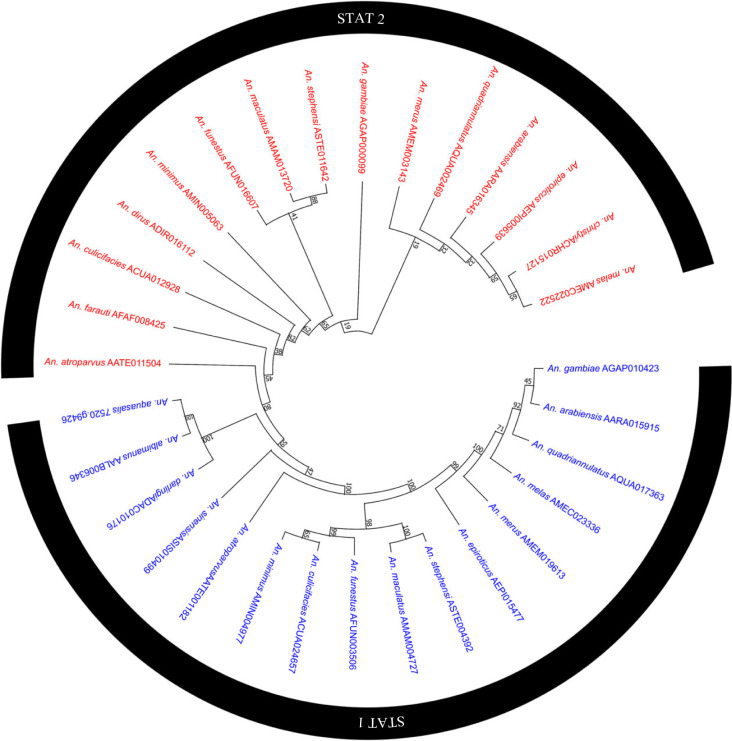
Among the exciting events to highlight was the duplication of the STAT gene (JAK/STAT pathway signalling transcription factor) in An. gambiae s. s. Apparently, after the divergence between the members of the subgenus Nyssorhynchus (American anophelines) and the subgenus Cellia (An. gambiae s. s.), a new copy emerged for the African mosquito. A result that was also observed when carrying out the phylogenetic tree with the sequences of 18, STAT2 arises in several of the species of the Cellia subgenus and even in representatives of the Anopheles subgenus. In addition, the absence of STAT2 was not exclusive to American anophelines; the Asian species An. sinensis, does not have a copy of this gene (figure 4). In An. sinensis, this seems to be the product of a loss, since its congeners, An. stephensi and An. atroparvus, possess a copy of STAT2. On the other hand, in the STAT2 clade, the orthologous relationship was not very clear for An. minimus, An. dirus, An. culicifacies, An. farauti and An. atroparvus, even though their sequences had identity values higher than 78% (electronic supplementary material, table S6).
Figure 4.
Phylogenetic analysis of the STAT1 and STAT2 genes, carried out with the sequences of 18 anophelines. Bootstrap values were calculated with 1000 replicates, and their values are presented on each tree branch.
In the JNK signalling pathway (figure 3c), An. aquasalis and An. albimanus had orthologues of the well-known JNK cascade proteins shared with An. gambiae s. s., but An. darlingi seems to have lost the PUCKERED gene, a negative regulator of this pathway. It is possible that, in An. darlingi, the PUCKERED gene has its role performed by the dual phosphatase gene since both genes have the same regulatory role in insects such as the fruit fly D. melanogaster [43]. Furthermore, the genes of all the An. aquasalis signaling pathways share identity values with An. gambiae s. s. above 41%. (electronic supplementary material, table S2). Within the An. aquasalis annotation, the predictions of the IMD genes, the transcription factor regulator PIAS, TRAF6 (Toll pathway signalling TNF Receptor-Associated Factor6), and the NOS genes were fragmented over several scaffolds or missing. Therefore, their predictions were made manually.
3.2. Serine protease inhibitors
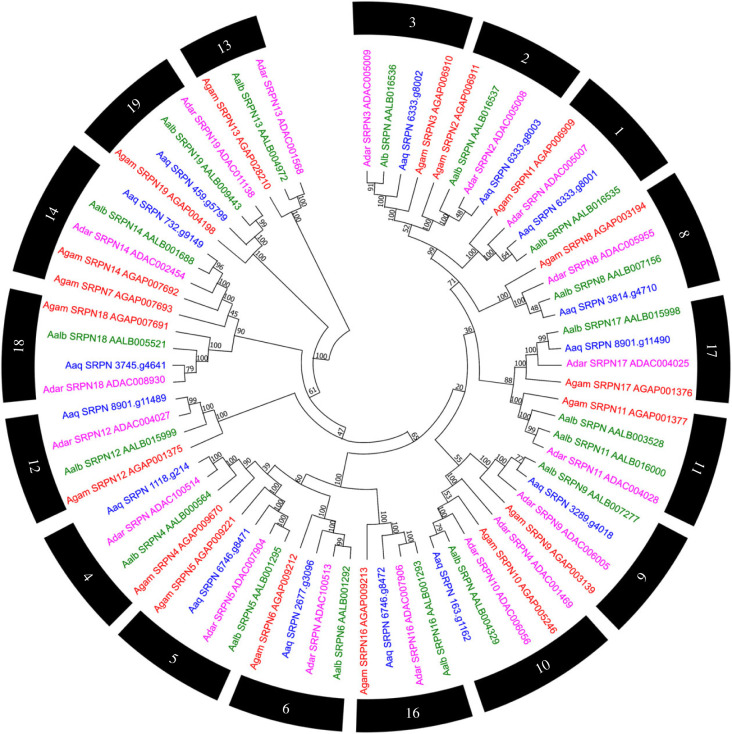
A total of 15 genes of the serine protease inhibitor family (serpins – SRPN) were found within the genome of An. aquasalis, different from the 18 genes identified in An. gambiae s. s. The loss of genes was manually reviewed. The SRPN7 and SRPN13 genes from An. gambiae s. s. had a low identity value (26% and 48%, respectively) in the BLAST alignment against the An. aquasalis genome. Furthermore, this comparison revealed a low score for SRPN7 (102) and low coverage for SRPN13 (12%), making it impossible to annotate these genes manually.
In general, a majority of the serpin genes have single-copy orthologues among the anophelines compared. In clade 11, the expansion of An. albimanus serpins was identified. Most An. aquasalis sequences have values above 52% of identity with An. gambiae s. s. (electronic supplementary material, table S2) (figure 5).
Figure 5.
Evolutionary relationship of SERPINs of Anopheles aquasalis. Neighbour-joining tree of the family of serine protease inhibitors from the amino acid sequences of the mosquitoes An. aquasalis (blue), An. darlingi (pink), An. albimanus (green) and An. gambiae s. s. (red). Bootstrap values were calculated with 1000 replicates, and their values are presented in each tree branch.
3.3. Superoxide dismutase proteins
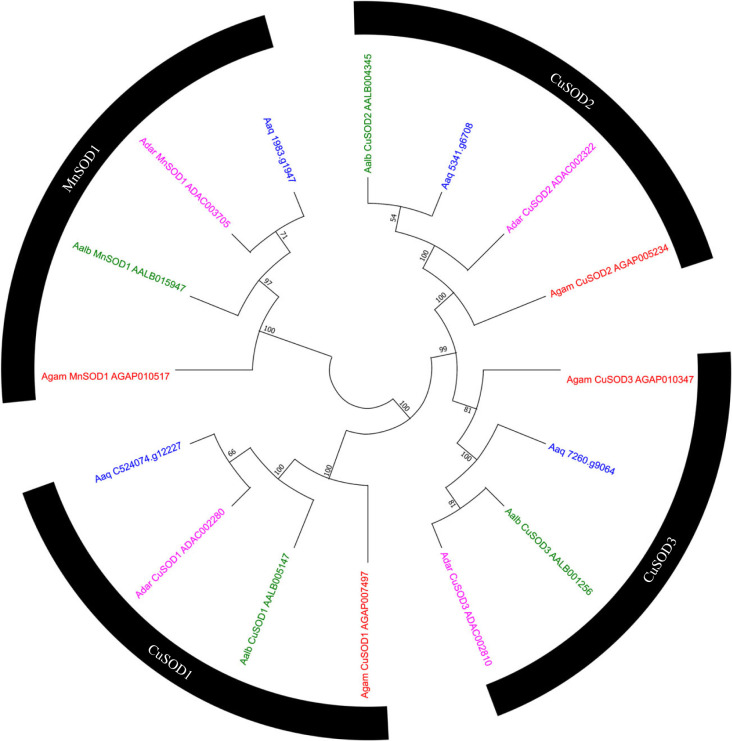
Besides the proteins from the signalling pathways, superoxide dismutase (SOD) was another group of highly conserved genes in the anophelines. Four genes of this family were identified in all mosquitoes with one-to-one orthologues (figure 6), with highly conserved amino acid identity sequence (greater than 90%, Electronic supplementary material, table S2).
Figure 6.
Evolutionary relationship of SODs of Anopheles aquasalis. Neighbour-joining tree of the family of superoxide dismutase family from the amino acid sequences of the mosquitoes An. aquasalis (blue), An. darlingi (pink), An. albimanus (green) and An. gambiae s. s. (red). Bootstrap values were calculated with 1000 replicates, and their values are presented in each tree branch.
3.4. Peptidoglycan recognition proteins
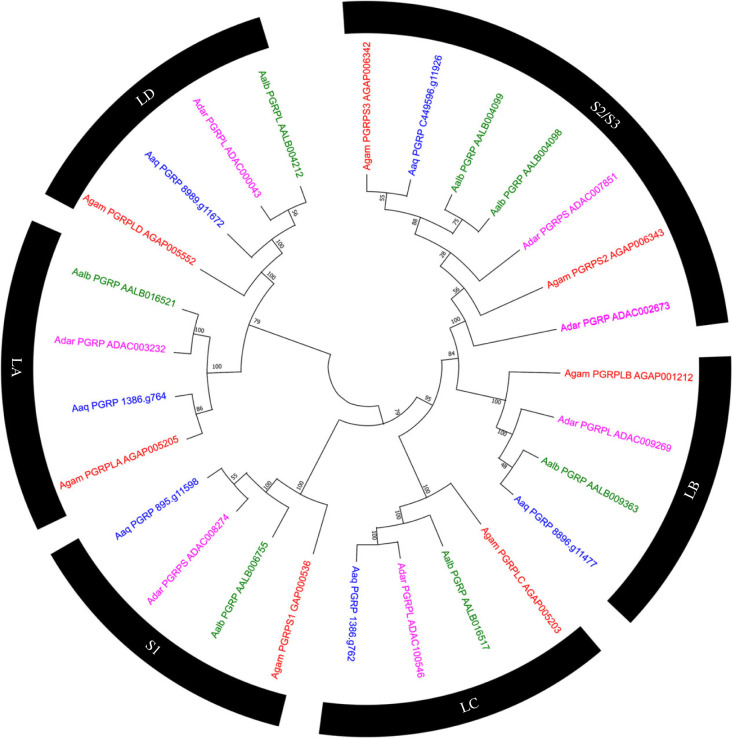
In An. aquasalis six PGRP were identified, a result similar to the finding in other anophelines, where the composition of this family is from five to seven genes [26,44]. Three sequences of An. aquasalis had identity values above 75% compared to their orthologues in the mosquito An. gambiae s. s. and at least the identity values are above 59.8%; in this group, the orthologue of AgamPGRPS2 in An. aquasalis was fragmented across several scaffolds and was manually annotated. Despite having many single-copy orthologous groups among the four species (LD/LA/S1/LB Groups), in the S2/S3 group, it was impossible to identify orthologues with the AgamPGRPS2 and AgamPGRPS3 proteins for American mosquitoes (figure 7).
Figure 7.
Evolutionary relationship of peptidoglycan recognition proteins of Anopheles aquasalis. Neighbour-joining tree of PGRP receptors made with mosquito amino acid sequences. An aquasalis (Blue), An. darlingi (Pink), An. albimanus (Green) and An. gambiae s. s. (Red). Bootstrap values were calculated with 1000 replicates, and their values are presented in each tree branch.
3.5. Proteins with leucine-rich repeats of the immune response
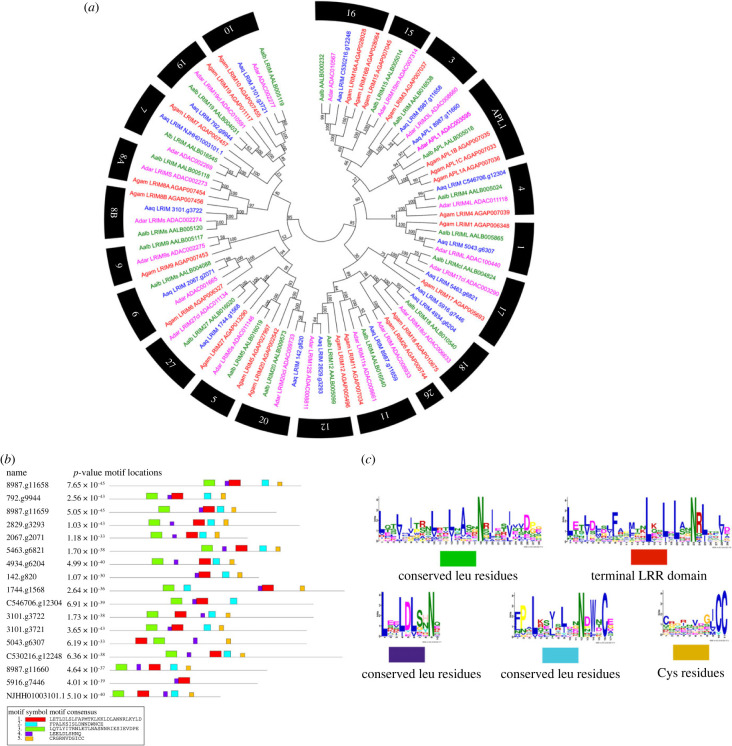
The proteins with leucine-rich repeats of the immune response (LRIM) are abundant in anophelines; in An. aquasalis 17 LRIM genes were described (figure 8a). In general, there were orthologues one by one in several of the groups identified. However, evolutionary analysis suggests the loss of LRIM-5, 8A, 9, 15 and 26 in An. aquasalis. An. gambiae s. s. was the species that had the most gene expansion in this family, mainly in the subgroup of long proteins (figure 8a).
Figure 8.
Analysis of leucine-rich repeats of immune response (LRIM) of Anopheles aquasalis. (a) Evolutionary relationships of anopheline LRIMs. Neighbour-joining tree of LRIM proteins made with An. aquasalis amino acid sequences (Blue), An. darlingi (Pink), An. albimanus (Green) and An. gambiae s. s. (Red). Bootstrap values were calculated with 1000 replicates, and their values were presented in each tree branch. (b) Distribution of motifs in the LRIM proteins of the An. aquasalis mosquito. (c) Composition of the motifs of the LRIM proteins in the An. aquasalis mosquito.
The LRIM family has been subdivided into four subgroups according to their structural characteristics and previous evolutionary analyzes. They are recognized as lengthy, short, without coiled-coil and transmembrane [45,46]. The phylogenetic analysis of the present work indicates the exact origin of the short LRIM and those without coiled-coil, despite the structural differences between the two subfamilies (figure 8b,c; electronic supplementary material, table S1). It was common to find proteins from the two subfamilies between the tree clades without differentiating between the two groups.
Also, the proteins with transmembrane regions (with coiled-coil domains) were represented by groups 15 and 16 in anophelines. In An. aquasalis, only one member of these proteins with transmembrane parts was identified (group 16). The other anophelines had a more significant number of copies of them, as evidenced in An. gambiae s. s.. These proteins are closely related to the subfamily of long LRIM, with both having a common origin. The long proteins were represented by three groups, composed chiefly of single-copy orthologues for the four anopheline species for two of their genes and the expansion of the APL1 group in An. gambiae s. s. and one copy too much in An. aquasalis was evident (figure 8a).
As pointed out previously, in the LRIM proteins, the leucine-rich repeats (LRR) domain has a particular distribution pattern of residues essential for identifying some proteins of this group with poor identity values [46,47]. In this work, the different motifs with the leucine repeats were determined as described in green, blue, and purple colours; the motif with reproductions of Leu, Trp, and a well-preserved Cys at the end of the LRR domain is represented in red; and the motif with cysteines between the LRR and the coiled coils are represented in yellow (figure 8b). Likewise, the distribution of each motif can be observed in figure 7c, highlighting that most of the Leu repeat motifs were before the red and yellow motifs, with the conserved residues of Trp and Cys delimiting the LRR domain.
3.6. Proteins with fibrinogen domains (FREP)
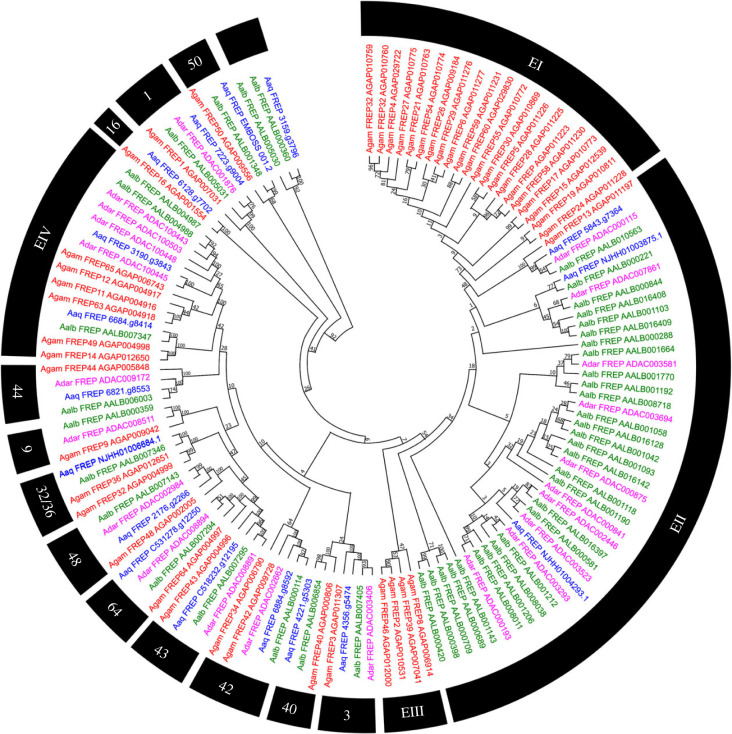
The fibrinogen domain proteins (FREP) genes have heterogeneous abundance in anophelines. In An. gambiae s. s., up to 46 genes have been identified, and the mosquito An. aquasalis has 17 FREP proteins. Our evolutionary analysis showed four significant expansions in the FREP family in anophelines. The expansions EI and EIII are exclusive of An. gambiae s. s. (figure 9). Conversely, the expansion EII excludes An. albimanus, with no apparent orthologues, shared with An. gambiae s. s. Concerning An. aquasalis, there are only 2 FREP genes in EII, suggesting the loss of some genes and evidencing that many independent expansions occurred in An. albimanus. We also observed a fourth event of other additions, EIV, which was more prominent in An. gambiae s. s. and An. darlingi compared to the other anophelines.
Figure 9.
Neighbour-joining tree of proteins with fibrinogen domains (FREP) from the amino acid sequences of Anopheles aquasalis. An. aquasalis (blue), An. darlingi (pink), An. albimanus (green) and An. gambiae s. s. (red) mosquitoes. Bootstrap values were calculated with 1000 replicates, and their values were presented in each tree branch.
4. Discussion
Conservation of proteins at the amino acid and copy number levels is frequently associated with essential genes for the organism's survival and fitness, considering the classical pathways. At first glance, it is possible to observe that the most conserved gene families (concerning copy numbers) were the immune pathways (IMD, JAK/STAT, TOLL and JNK), with 1 : 1 orthologues in almost all anophelines. Proteins of the IMD, JAK/STAT, TOLL and JNK pathways are among the most studied proteins in anophelines. Their importance in the process of vector/parasite interaction has been demonstrated in many insect vectors. The disruption of any of these main pathways by gene knockout or silencing leads to an inability of the vector to defend against pathogens. Interestingly, in anopheline species, duplication in the STAT transcription factor of the JAK/STAT signalling pathway is recognized for some of the members of the Cellia and Anopheles subgenera [48,49]. Our data analysis suggests that the additional STAT copy emerged after the New and Old World Anopheles divergence.
Another highly conserved family of proteins identified here relates to the metabolism of reactive oxygen species (ROS) products, as in the case of superoxide dismutase proteins (SOD). This group is universally present and is believed to have arisen long ago in prokaryotes from increased oxygen production in the atmosphere [50,51]. In An. aquasalis and An. gambiae s. s., SOD3 isoforms have emerged to regulate ROS at specific times and in conditions to combat pathogens in the midgut during blood digestion [52,53]. The emergence of SOD3 isoforms in anophelines became a rapid evolutionary strategy to respond to the increase in free radicals at different times or to act in specific tissues during blood feeding of the female, where under certain circumstances it is exposed to pathogens [52,54].
By way of the conserved orthologues between An. aquasalis and An. gambiae s. s., we observed that some gene families (Autophagy, Galectins and Peroxidases) have similar numbers of genes (in a 1 : 1 orthologous relationships). Among the American anophelines, the only species with the same number of autophagy proteins of An. gambiae s. s. was An. aquasalis. The only exception was the ATG12 gene, which was found in two copies in the An. gambiae s. s. genome [55], while the three Neotropical mosquitoes (An. aquasalis, An. darlingi and An. albimanus) had only one copy of these genes.
Several components responsible for regulating the catabolic process of autophagy have been conserved in several phyla of eukaryotes such as plants, worms, arthropods and mammals [56–58]. Even so, in these organisms, differences in the number of copies have been found in regulating autophagy's induction and nucleation. In addition, the presence of isoforms has been recorded in the autophagy-related proteins (ATG) [56]. In insects, for example, one of the ATG1 isoforms participates in the activation of autophagy under conditions of starvation or excess food, as well as during metamorphosis, by being phosphorylated in the presence of juvenile growth hormone 20E [59]. Even so, the presence of isoforms does not always lead to phenotypic effects, as was found in Arabidopsis thaliana for the ATG12 isoforms and represents a clear example of sub-functionalization. As demonstrated in A. thaliana, where in long periods of starvation and darkness, the silencing of both ATG12B and ATG12A genes resulted in increased mortality. According to the results, any of the two ATG12 is sufficient for the association of ATG8 with autophagic bodies. In addition, its expression changes during the life cycle of the plant, with ATG12A frequently expressed in old plants and ATG12B in young A. thaliana tissues [60].
ATG8 was one of the proteins conserved in all anophelines except for An. darlingi. This protein participates in the final phase of autophagosome formation. In its absence, there is no direction for constructing the pre-autophagosome structure [57,58]. Therefore, ATG8 is ubiquitous in eukaryotes, and its lack in An. darlingi may be due to the existence of another protein that can activate autophagosome formation, or it is necessary to annotate ATG8 in this mosquito [56].
Interestingly, other proteins, such as caspases and apoptosis inhibitors related to apoptosis processes, have expanded copy numbers in An. gambiae s. s.. The emergence of extra copies in An. gambiae s. s. may be related to a greater capacity for regulating the apoptosis process [61–63]. Regarding the galectins, these proteins had more than half of the groups with single-copy orthologues. There was a loss of orthologues in American mosquitoes in two groups and a duplication in An. gambiae s. s.. Previously it was shown that galectins underwent an expansion process almost exclusive to mosquitoes, suggesting an influence of hematophagy and exposure to different pathogens for the diversification of this type of recognition receptors in culicids [64,65].
In An. gambiae s. s. galectins, there are data on their participation in infections made with Gram-positive bacteria and with P. berghei, GALE6–8 also increased its expression during exposure to the alphavirus ONNV, GALE8 being an important antagonist of the virus, here these galectins are well related in a clade in which An. gambiae s. s. exhibited a duplication precisely with GALE8/9 [3,66]. Finally, concerning the peroxidases, it is known that they are related to population control of the gut microbiota, avoiding direct contact with effectors of the immune response or increasing the expression of those with bactericidal properties like the DUOX and HPX2 proteins [18,19,22,67]. Relevant heme-peroxidases [19,20], such as HPX15, were found in all anophelines but, again, An. aquasalis presented a higher number of 1 : 1 orthologues to An. gambiae s. s. than other American anophelines. In summary, by simple parsimony, it is possible to conjecture that An. aquasalis and An. gambiae s. s. present a conserved ancestral catalogue of genes, with unexpected losses in other American anophelines.
Pattern recognition proteins (PRPs) underwent gene gain and loss dynamics ranging from conserved copy number clusters to extensive species-specific diversifications. For example, in the Gram-negative binding protein (GNBP) and PGRP families, conservation was observed for a few orthologues with possible losses in American mosquitoes. Genes identified as PGRPS2 and GNBP2/3 could have implications in limiting the growth of populations of exogenous bacteria functionally recognized in An. gambiae s. s. and Ae. aegytpi [68,69]. The absence of the PGRPS2 protein could suggest differences in Plasmodium control since in An. gambiae s. s. PGRPS2 participates in the reduction of P. falciparum and P. berghei [70]. Some PRP families present removal only for New World anophelines, such as galectins (except for An. aquasalis), C-type lectins, MLD2 and scavenger receptors. Interestingly, for the last two, their functions are not limited exclusively to the immune response, and their diversification could be due to each organism's metabolic or sensory needs [71,72]. Also, it has been demonstrated that An. gambiae s. s. and An. stephensi MLD silencing increases the oocyst number of P. falciparum and P. berghei in the midgut, reinforcing the hypothesis previously raised that MLD possibly prevents sterol uptake by parasites [73,74].
As in other insects, CTL proteins (C-type lectins with a single CRD domain) have the most significant diversification in anophelines. In other mosquitoes, it is known that these proteins participate in the homeostatic balance of the microbiota, preventing the activation of antimicrobial peptides (AMPs) [75]. The diversification of these proteins may be in response to each mosquito Field species' bacterial community [75]. Several CTLs are necessary factors that allow the entry of viruses such as Japanese encephalitis virus (JEV), Dengue virus (DENV) and West Nile virus (WNV) into the host cells of Aedes and Culex mosquitoes, and they are specific for each virus or even for serotypes [76–78]. The CTL evolution could have occurred to protect these insects’ commensal bacteria from the immune and digestion mechanisms activated during the blood meal in the midgut, where viruses and other pathogens take advantage of the production of CTLDs to evade the immune response [79]. This agonist capacity of CTLDs in anophelines was initially recognized in An. gambiae s. s. with P. berghei in the proteins CTL4 and CTLMA2, these two lectins type C protect the malarial ookinetes from melanization [80,81]. However, in the An. albimanus, the protective action of these proteins against plasmodia does not exist, and, according to our analysis based on the phylogenetic data, it is suggested that this functional difference is related to the lack of a true orthologue of CTL4 by the American species [82].
The species-specific diversifications were much more abrupt in the PRP of the FREP, TEP, TOLL and LRIM families in the four studied anopheline species, especially in An. gambiae s. s.. This phenomenon was highly accentuated in the FREP family, a previously recognized group with rapid divergence and relaxed evolutionary constraints in insect lineages such as Diptera [83]. FREPs are essential in maintaining immune homeostasis and bacteria elimination. They also work synergistically against murine and human Plasmodium [23]. TEP is necessary for the pathogen response in mosquitoes and Drosophila, serving as opsonization molecules to control microbes and pathogens, triggering lysis, phagocytosis or melanization [84,85]. There were expansions of TEP in An. gambiae s. s. concerning proteins related to TEP1, which is associated with controlling the microbiota and the fight against pathogens derived from blood meal [64,86]. Interestingly, the only group related to TEP1 conserved among the anophelines of the New World was TEP4, a protein with antibacterial properties [47]. LRIMs are described as effectors against pathogens in An. gambiae s. s., but not in An. albimanus [46,47,87]. In An. aquasalis, only one LRR was recognized under the same conditions. However, most of the An. gambiae s. s. LRIMs were present in the New World mosquitoes being 17 of 27 clades composed of single-copy orthologues. Characteristically, Toll receptors showed the same division of three subgroups identified in the insect genomic comparison [88]. The diversification of TOLL receptors, mainly in clades 1, 5 and 9, is attractive since proteins in these groups are commonly related to immune response and embryonic development. The most conserved TLRs among anophelines are in clades 6, 7, 10 and 11. TOLL11 has been described as an antagonist of P. falciparum in An. gambiae s. s. and has orthologues in the other anopheline species [88–90]. In An. aquasalis and An. albimanus, transcriptomic studies indicate little activity of TOLL receptors against Plasmodium after ingesting a blood meal, even if the TOLL signalling pathway is activated [8,14,91]. Thus, in New World mosquitoes, TLRs are more related to other physiological activities or embryonic development than the immune response. More detailed data are needed for this group to better understand their role in American anophelines.
A few authors have asserted that expansions in the families related to the modulation process of the immune response are widespread [29,64,92]. Its modulatory capacity is due to the particular use of combinations of serine proteases and their inhibitors to activate immune response mechanisms and other physiological processes in which it participates [92]. It is speculated how exposure to specific pathogens could influence the organization and diversification of these protein families. Serine proteases with clip domains suffered more significant species-specific expansions/contractions, especially in An. gambiae s. s. in subfamilies B and E [29], also observed in An. darlingi and An. albimanus. In mosquitoes such as Ae. aegypti and An. gambiae s. s., it is widely studied how serine protease with CLIP domains actively regulates melanization as a defence mechanism against bacteria, fungi and parasites [81,93–96]. In An. gambiae s. s., non-catalytic CLIPs are recruited with the TEP1/LRIM1/APL1C complex. This process mediates the accumulation of the complement system on the surface of microorganisms [95,96]. Also, it helps to regulate and activate other serine proteases in cascades where a substantial amount of these proteins with redundant functions influences the intensity of the response by acting in a synergy [97,98]. Some are products of duplications, where sub-functionalization or loss of function possibly occurred, contributing to the regulation of cascades [29]. Species-specific expansions could be related to regulating effectors in response to specific pathogens. In Ae. aegypti, for example, CLIP29 and CLIP30/31 paralogues activate the TOLL signalling pathway along with the SERPIN2 protein acting against the fungus Beauveria bassiana [93]. The differences observed in protein composition that participate in the melanization process result from the need to regulate the proteolytic cascades to react quickly to pathogen infections or increase the microbiota.
The prophenoloxidase (PPO) system is a hemolymph-based complex of enzymes that, when activated, generate peptides and adhesive proteins, mediating many of the defence functions in arthropods. The expansion of PPO in mosquitoes is well-known. There is a discussion that a series of duplication events have led to the current number of proteins in the culicids [64,99,100]. Even so, losses were identified in the New World's anophelines, especially in An. aquasalis. The PPO-6 clade is associated with the melanization of plasmodia in refractory strains of An. gambiae s. s. and An. stephensi [98,99]. However, PPO-3, derived from PPO-6, is known to cause the melanization of P. berghei in An. gambiae s. s. by the activation of the PGE2 protein. Moreover, in An. dirus, PPO4 has an antispasmodic effect independent of the melanization [101,102].
Overall, the results presented in this study indicate a variable pattern of evolution in the immune response genes of the studied anopheline mosquitoes. Environmental factors, such as exposure to different pathogens and differences in the composition of the microbiota, could shape the expression of this group of genes. This study formulated a catalog of the evolution of immune response genes in An. aquasalis, the primary malaria vector of coastal Central and South America and the Caribbean Islands. The An. aquasalis mosquitoes used in this study are from a well-established colony that has also analysed the interaction process with murine and human malarial Plasmodium strains. The results raised here will contribute knowledge of the study of a Neotropical vector, which may open opportunities for future malaria control in the malaria-endemic areas of the New World.
Acknowledgements
This manuscript is a part of the PhD theses developed by C.C.P.S. and R.M.A., supervised by P.F.P.P. and L.B.K. M.V.G.L, W.M.M., G.C.M., V.S.S., P.F.P.P. and N.F.C.S. are productivity fellows of the CNPq.
Contributor Information
Leonardo Barbosa Koerich, Email: lbkoerich@ufmg.br.
Paulo Filemon Paolucci Pimenta, Email: pfppimenta@gmail.com.
Data accessibility
Data are available upon request from the corresponding author or related data bank cited in the material and methods.
Additional information is provided in electronic supplementary material [103].
Authors' contributions
C.C.P.S.: conceptualization, data curation, formal analysis, validation, writing—original draft; R.M.A.: conceptualization, data curation, formal analysis, investigation, methodology, validation, writing—original draft; R.A.S.: data curation, formal analysis, methodology; I.B.S.: data curation, formal analysis; G.M.A.D.: data curation, formal analysis, methodology; R.S.M.G.: conceptualization, formal analysis, methodology; A.P.D.: data curation, formal analysis, methodology, project administration; S.C.P.L.: conceptualization, formal analysis, investigation, methodology, resources, writing—original draft; M.V.G.L.: conceptualization, investigation, resources, writing—original draft; W.M.M: investigation, methodology, resources, supervision, validation; R.N.-P: conceptualization, data curation, formal analysis, supervision, validation; N.F.C.S.: data curation, investigation, methodology, project administration, supervision, writing—original draft; L.B.K.: conceptualization, data curation, formal analysis, investigation, methodology, project administration, software, validation, writing—review and editing; P.F.P.P.: conceptualization, methodology, project administration, resources, supervision, visualization, writing—review and editing.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Conflict of interest declaration
We declare we have no competing interests.
Funding
The following Brazilian agencies partially supported fellowships for the students and researchers of this study: Foundation of the Institute Oswaldo Cruz (FIOCRUZ), Brazilian Council for Scientific and Technological Development (CNPq), Coordination for Improvement of Higher Education Personnel (CAPES), Minas Gerais and Amazonas State Research Support Foundations (FAPEMIG and FAPEAM - PROESTADO).
References
- 1.World Health Organization. 2022. World malaria report 2022. Geneva, Switzerland: World Health Organization. [Google Scholar]
- 2.Sinka ME, et al. 2012. A global map of dominant malaria vectors. Parasites Vectors 5, 69. ( 10.1186/1756-3305-5-69) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dimopoulos G, Richman A, Müller HM, Kafatos FC. 1997. Molecular immune responses of the mosquito Anopheles gambiae to bacteria and malaria parasites. Proc. Natl Acad. Sci. USA 94, 11 508-11 513. ( 10.1073/pnas.94.21.11508) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levashina EA. 2004. Immune responses in Anopheles gambiae s. s. Insect Biochem. Mol. Biol. 34, 673-678. ( 10.1016/j.ibmb.2004.03.020) [DOI] [PubMed] [Google Scholar]
- 5.Xu X, et al. 2005. Transcriptome analysis of Anopheles stephensi-Plasmodium berghei interactions. Mol. Biochem. Parasitol. 142, 76-87. ( 10.1016/j.molbiopara.2005.02.013) [DOI] [PubMed] [Google Scholar]
- 6.Gouagna LC, Mulder B, Noubissi E, Tchuinkam T, Verhave JP, Boudin C. 1998. The early sporogonic cycle of Plasmodium falciparum in laboratory-infected Anopheles gambiae: an estimation of parasite efficacy. Trop. Med. Int. Heal. 3, 21-28. ( 10.1046/j.1365-3156.1998.00156.x) [DOI] [PubMed] [Google Scholar]
- 7.Dong Y, Aguilar R, Xi Z, Warr E, Mongin E, Dimopoulos G. 2006. Anopheles gambiae immune responses to human and rodent Plasmodium parasite species. PLoS Pathog. 2, 0513-0525. ( 10.1371/journal.ppat.0020052) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martínez-Barnetche J, et al. 2012. Transcriptome of the adult female malaria mosquito vector Anopheles albimanus. BMC Genom. 13, 207. ( 10.1186/1471-2164-13-207) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Habtewold T, Groom Z, Christophides GK. 2017. Immune resistance and tolerance strategies in malaria vector and non-vector mosquitoes. Parasites Vectors 10, 1-12. ( 10.1186/s13071-017-2109-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar A, Srivastava P, Sirisena PDNN, Dubey SK, Kumar R, Shrinet J, Sunil S. 2018. Mosquito innate immunity. Insects 9, 95. ( 10.3390/insects9030095) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sloan MA, Ligoxygakis P. 2017. Immunology of insect vectors: midgut interactions of sandflies and tsetse with kinetoplastid parasites as a paradigm for establishing infection. In Advances in insect physiology, vol. 52, pp. 231-248, 1st edn. New York, NY: Elsevier. ( 10.1016/bs.aiip.2017.04.003. [DOI] [Google Scholar]
- 12.Christensen BM, Forton KF. 1986. Hemocyte-mediated melanization of microfilariae in Aedes aegypti. J Parasitol. 72, 220-225. ( 10.2307/3281595) [DOI] [PubMed] [Google Scholar]
- 13.Hernández-Martínez S, Lanz H, Rodríguez MH, González-Ceron L, Tsutsumi V. 2002. Cellular-mediated reactions to foreign organisms inoculated into the hemocoel of Anopheles albimanus (Diptera: Culicidae). J. Med. Entomol. 39, 61-69. ( 10.1603/0022-2585-39.1.61) [DOI] [PubMed] [Google Scholar]
- 14.Garver LS, Dong Y, Dimopoulos G. 2009. Caspar controls resistance to Plasmodium falciparum in diverse anopheline species. PLoS Pathog. 5, e1000335. ( 10.1371/journal.ppat.1000335) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garver LS, Bahia AC, Das S, Souza-Neto JA, Shiao J, Dong Y, Dimopoulos G. 2012. Anopheles Imd pathway factors and effectors in infection intensity-dependent anti-Plasmodium action. PLoS Pathog. 8, 7-9. ( 10.3390/pathogens8010007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong Y, Das S, Cirimotich C, Souza-Neto JA, McLean KJ, Dimopoulos G. 2011. Engineered Anopheles immunity to Plasmodium infection. PLoS Pathog. 7, e1002458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar S, Molina-Cruz A, Gupta L, Rodrigues J, Barillas-Mury C. 2010. A peroxidase/dual oxidase system modulates midgut epithelial immunity in Anopheles gambiae. Science 327, 1644-1648. ( 10.1126/science.1184008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kajla M, Kakani P, Choudhury TP, Kumar V, Gupta K, Dhawan R, Gupta L, Kumar S. et al. 2017. Anopheles stephensi heme peroxidase HPX15 suppresses midgut immunity to support Plasmodium development. Front Immunol. 8, 240. ( 10.3389/fimmu.2017.00249) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kajla M, Kakani P, Choudhury TP, Gupta K, Gupta L, Kumar S. 2016. Characterization and expression analysis of gene encoding heme peroxidase HPX15 in major Indian malaria vector Anopheles stephensi (Diptera: Culicidae). Acta Trop 158, 107-116. ( 10.1016/j.actatropica.2016.02.028) [DOI] [PubMed] [Google Scholar]
- 20.Kajla M, Choudhury TP, Kakani P, Gupta K, Dhawan R, Gupta L, Kumar S. 2016. Silencing of Anopheles stephensi heme peroxidase HPX15 activates diverse immune pathways to regulate the growth of midgut bacteria. Front. Microbiol. 7, 1-13. ( 10.3389/fmicb.2016.01351) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar S, Gupta L, Yeon SH, Barillas-Mury C. 2004. Inducible peroxidases mediate nitration of Anopheles midgut cells undergoing apoptosis in response to Plasmodium invasion. J. Biol. Chem. 279, 53 475-53 482. ( 10.1074/jbc.M409905200) [DOI] [PubMed] [Google Scholar]
- 22.Kakani P, Kajla M, Choudhury TP, Gupta L, Kumar S. 2019. Anopheles stephensi Dual Oxidase Silencing Activates the Thioester-Containing Protein 1 Pathway to Suppress Plasmodium Development. J. Innate Immun. 127021, 496-505. ( 10.1159/000497417) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong Y, Dimopoulos G. 2009. Anopheles fibrinogen-related proteins provide expanded pattern recognition capacity against bacteria and malaria parasites. J. Biol. Chem. 284, 9835-9844. ( 10.1074/jbc.M807084200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang G, Niu G, Franca CM, Dong Y, Wang X, Butler NS, Dimopoulos G, Li J. 2015. Anopheles midgut FREP1 mediates Plasmodium Invasion. J. Biol. Chem. 290, 16 490-16 501. ( 10.1074/jbc.M114.623165) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niu G, França C, Zhang G, Roobsoong W, Nguitragool W, Wang X, Prachumsri J, Butler NS, Li J. 2017. The fibrinogen-like domain of FREP1 protein is a broad-spectrum malaria transmission-blocking vaccine antigen. J. Biol. Chem. 292, 11 960-11 969. ( 10.1074/jbc.M116.773564) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neafsey DE, et al. 2015. Highly evolvable malaria vectors: the genomes of 16 Anopheles mosquitoes. Science 347, 1-20. ( 10.1126/science.1258522) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alencar RM, et al. In press. Genome analysis of the main malaria vector mosquito of coastal Central and South America and the Caribbean Islands: the brackish-water Anopheles aquasalis. Scientific Reports. [Google Scholar]
- 28.Kriventseva EV, Kuznetsov D, Tegenfeldt F, Manni M, Dias R, Simão FA, Zdobnov EM. 2019. OrthoDB v10: Sampling the diversity of animal, plant, fungal, protist, bacterial and viral genomes for evolutionary and functional annotations of orthologs. Nucleic Acids Res. 47(D1), D807-D811. ( 10.1093/nar/gky1053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao X, Gulati M, Jiang H. 2017. Serine protease-related proteins in the malaria mosquito, Anopheles gambiae Insect Biochem. Mol. Biol. 176, 139-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Birney E, Clamp M, Durbin R. 2004. GeneWise and Genomewise. Genome Res. 14, 988-995. ( 10.1101/gr.1865504) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marchler-Bauer A, et al. 2015. CDD: NCBI's conserved domain database. Nucleic Acids Res. 43(D1), D222-D226. ( 10.1093/nar/gku1221) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blum M, et al. 2021. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 49(D1), D344-D354. ( 10.1093/nar/gkaa977) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME Suite: Tools for motif discovery and searching. Nucleic Acids Res. 37(Suppl. 2), 202-208. ( 10.1093/nar/gkp335) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Almagro Armenteros JJ, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, Brunak S, Von Heijne G, Nielsen H. 2019. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 37, 420-423. ( 10.1038/s41587-019-0036-z) [DOI] [PubMed] [Google Scholar]
- 35.Delorenzi M, Speed T. 2002. An HMM model for coiled-coil domains and a comparison with PSSM-based predictions. Bioinformatics 18, 617-625. ( 10.1093/bioinformatics/18.4.617) [DOI] [PubMed] [Google Scholar]
- 36.Giraldo-Calderón GI, et al. 2015. VectorBase: an updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res. 43(D1), D707-D713. ( 10.1093/nar/gku1117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113. ( 10.1186/1471-2105-5-113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547-1549. ( 10.1093/molbev/msy096) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Babicki S, Arndt D, Marcu A, Liang Y, Grant JR, Maciejewski A, Wishart DS. 2016. Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res. 44, W147-W153. ( 10.1093/nar/gkw419) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bahia AC, et al. 2011. The JAK-STAT pathway controls Plasmodium vivax load in early stages of Anopheles aquasalis infection. PLoS Negl. Trop. Dis. 5, e1317. ( 10.1371/journal.pntd.0001317) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garver LS, de Almeida Oliveira G, Barillas-Mury C. 2013. The JNK pathway is a key mediator of Anopheles gambiae antiplasmodial immunity. PLoS Pathog. 9, e1003622. ( 10.1371/journal.ppat.1003622) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramphul UN, Garver LS, Molina-Cruz A, Canepa GE, Barillas-Mury C. 2015. Plasmodium falciparum evades mosquito immunity by disrupting JNK-mediated apoptosis of invaded midgut cells. Proc. Natl Acad. Sci. USA 112, 1273-1280. ( 10.1073/pnas.1423586112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martín-Blanco E, Gampel A, Ring J, Virdee K, Kirov N, Tolkovsky AM, Martinez-Arias A. 1998. puckered encodes a phosphatase that mediates a feedback loop regulating JNK activity during dorsal closure in Drosophila. Genes Dev. 12, 557-670. ( 10.1101/gad.12.4.557) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mendes C, Felix R, Sousa AM, Lamego J, Charlwood D, Do Rosrio VE, Pinto J, Silveira H. et al. 2010. Molecular evolution of the three short PGRPs of the malaria vectors Anopheles gambiae and Anopheles arabiensis in East Africa. BMC Evol. Biol. 10, 1-11. ( 10.1186/1471-2148-10-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Povelones M, Waterhouse RM, Kafatos FC, Christophides GK. 2009. Leucine-rich repeat protein complex activates mosquito complement in defense against Plasmodium parasites. Science 324, 258-261. ( 10.1126/science.1171400) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Waterhouse RM, Povelones M, Christophides GK. 2010. Sequence-structure-function relations of the mosquito leucine-rich repeat immune proteins. BMC Genom. 11, 1-10. ( 10.1186/1471-2164-11-531) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Povelones M, Upton LM, Sala KA, Christophides GK. 2011. Structure-function analysis of the Anopheles gambiae LRIM1/APL1C complex and its interaction with complement C3-like protein TEP1. PLoS Pathog. 7, e1002023. ( 10.1371/journal.ppat.1002023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gupta L, Molina-Cruz A, Kumar S, Rodrigues J, Dixit R, Zamora RE, Barillas-Mury C. 2009. The STAT pathway mediates late-phase immunity against Plasmodium in the mosquito Anopheles gambiae. Cell Host Microbe. 5, 498-507. ( 10.1016/j.chom.2009.04.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gupta K, Dhawan R, Kajla M, Misra T, Kumar S, Gupta L. 2017. The evolutionary divergence of STAT transcription factor in different Anopheles species. Gene 596, 89-97. ( 10.1016/j.gene.2016.09.022) [DOI] [PubMed] [Google Scholar]
- 50.Zelko IN, Mariani TJ, Folz RJ. 2002. Superoxide dismutase multigene family: A comparison of the CuZn-SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) gene structures, evolution, and expression. Free Radic Biol Med. 33, 337-349. ( 10.1016/S0891-5849(02)00905-X) [DOI] [PubMed] [Google Scholar]
- 51.Shi G, Shen J, Ren F, Yang W. 2021. Molecular cloning, expression, and characterization of BmSOD3 in silkworm (Bombyx mori). Arch Insect Biochem. Physiol. 106, 1-14. ( 10.1002/arch.21777) [DOI] [PubMed] [Google Scholar]
- 52.Bahia AC, et al. 2013. The role of reactive oxygen species in Anopheles aquasalis response to Plasmodium vivax infection. PLoS ONE 8, 1-10. ( 10.1371/journal.pone.0057014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kumar S, Christophides GK, Cantera R, Charles B, Han YS, Meister S, Dimopoulos G, Kafatos FC, Barillas-Mury C. 2003. The role of reactive oxygen species on Plasmodium melanotic encapsulation in Anopheles gambiae. Proc. Natl Acad. Sci. USA 100(Suppl. 2), 14 139-14 144. ( 10.1073/pnas.2036262100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Molina-Cruz A, DeJong RJ, Charles B, Gupta L, Kumar S, Jaramillo-Gutierrez G, Barillas-Mury C. 2008. Reactive oxygen species modulate Anopheles gambiae immunity against bacteria and Plasmodium. J. Biol. Chem. 283, 3217-3223. ( 10.1074/jbc.M705873200) [DOI] [PubMed] [Google Scholar]
- 55.Holt RA, et al. 2002. The genome sequence of the malaria mosquito Anopheles gambiae. Science 298, 129-149. ( 10.1126/science.1076181) [DOI] [PubMed] [Google Scholar]
- 56.Kourtis N, Tavernarakis N. 2009. Autophagy and cell death in model organisms. Cell Death Differ. 16, 21-30. ( 10.1038/cdd.2008.120) [DOI] [PubMed] [Google Scholar]
- 57.Mizushima N, Yoshimori T, Ohsumi Y. 2011. The role of atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27, 107-132. ( 10.1146/annurev-cellbio-092910-154005) [DOI] [PubMed] [Google Scholar]
- 58.Suzuki K, Kubota Y, Sekito T, Ohsumi Y. 2007. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells 12, 209-218. ( 10.1111/j.1365-2443.2007.01050.x) [DOI] [PubMed] [Google Scholar]
- 59.Zhao H, et al. 2023. Atg1 phosphorylation is activated by AMPK and indispensable for autophagy induction in insects. Insect Biochem. Mol. Biol. 152, 103888. ( 10.1016/j.ibmb.2022.103888) [DOI] [PubMed] [Google Scholar]
- 60.Chung T, Phillips AR, Vierstra RD. 2010. ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A and ATG12B loci. Plant J. 62, 483-493. ( 10.1111/j.1365-313X.2010.04166.x) [DOI] [PubMed] [Google Scholar]
- 61.Puglise JM, Estep AS, Becnel JJ. 2016. Expression profiles and RNAi silencing of inhibitor of apoptosis transcripts in Aedes, Anopheles and Culex mosquitoes (Diptera: Culicidae). J. Med. Entomol. 53, 304-314. ( 10.1093/jme/tjv191) [DOI] [PubMed] [Google Scholar]
- 62.Vlachou D, Schlegelmilch T, Christophides GK, Kafatos FC. 2005. Functional genomic analysis of midgut Epithelial responses in Anopheles during Plasmodium invasion. Curr. Biol. 15, 1185-1195. ( 10.1016/j.cub.2005.06.044) [DOI] [PubMed] [Google Scholar]
- 63.Bryant B, Blair CD, Olson KE, Clem RJ. 2008. Annotation and expression profiling of apoptosis-related genes in the yellow fever mosquito, Aedes aegypti. Insect Biochem. Mol. Biol. 38, 331-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Waterhouse RM, et al. 2007. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science 316, 1738-1743. ( 10.1126/science.1139862) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lin Z, Wang JL, Cheng Y, Wang JX, Zou Z. 2020. Pattern recognition receptors from lepidopteran insects and their biological functions. Dev. Comp. Immunol. 108, 103688. ( 10.1016/j.dci.2020.103688) [DOI] [PubMed] [Google Scholar]
- 66.Waldock J, Olson KE, Christophides GK. 2012. Anopheles gambiae antiviral immune response to systemic O'nyong-nyong infection. PLoS Negl. Trop. Dis. 6, e1565. ( 10.1371/journal.pntd.0001565) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Oliveira GDA, Lieberman J, Barillas-Mury C. 2012. Epithelial nitration by a peroxidase/NOX5 system mediates mosquito antiplasmodial immunity. Science 335, 856-859. ( 10.1126/science.1209678) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Warr E, Das S, Dong Y, Dimopoulos G. 2008. The Gram-negative bacteria-binding protein gene family: its role in the innate immune system of Anopheles gambiae and in anti-Plasmodium defence. Insect Mol. Biol. 17, 39-51. ( 10.1111/j.1365-2583.2008.00778.x) [DOI] [PubMed] [Google Scholar]
- 69.Wang S, Beerntsen BT. 2015. Functional implications of the peptidoglycan recognition proteins in the immunity of the yellow fever mosquito, Aedes aegypti. Insect Mol. Biol. 24, 293-310. ( 10.1111/imb.12159) [DOI] [PubMed] [Google Scholar]
- 70.Gendrin M, Turlure F, Rodgers FH, Cohuet A, Morlais I, Christophides GK. 2017. The peptidoglycan recognition proteins PGRPLA and PGRPLB regulate Anopheles immunity to bacteria and affect infection by Plasmodium. J. Innate Immun. 9, 333-342. ( 10.1159/000452797) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nichols Z, Vogt RG. 2008. The SNMP/CD36 gene family in Diptera, Hymenoptera and Coleoptera: Drosophila melanogaster, D. pseudoobscura, Anopheles gambiae, Aedes aegypti, Apis mellifera, and Tribolium castaneum. Insect Biochem. Mol. Biol. 38, 398-415. ( 10.1016/j.ibmb.2007.11.003) [DOI] [PubMed] [Google Scholar]
- 72.Anari M, Stroehlein AJ, Hall RS, Chang BCH, Gasser RB, Young ND. 2020. Expanded complement of Niemann-Pick type C2-like protein genes in Clonorchis sinensis suggests functions beyond sterol binding and transport. Parasites Vectors 13, 1-10. ( 10.1186/s13071-020-3910-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pike A, Vadlamani A, Sandiford SL, Gacita A, Dimopoulos G. 2014. Characterization of the Rel2-regulated transcriptome and proteome of Anopheles stephensi identifies new anti-Plasmodium factors. Insect Biochem. Mol. Biol. 52, 82-93. ( 10.1016/j.ibmb.2014.06.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cui Y, Niu G, Li VL, Wang X, Li J. 2020. Analysis of blood-induced Anopheles gambiae midgut proteins and sexual stage Plasmodium falciparum interaction reveals mosquito genes important for malaria transmission. Sci. Rep. 10, 1-12. ( 10.1038/s41598-020-71186-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pang X, Xiao X, Liu Y, Zhang R, Liu J, Liu Q, Wang P, Cheng G. 2016. Mosquito C-type lectins maintain gut microbiome homeostasis. Nat. Microbiol. 1, 1-11. ( 10.1038/nmicrobiol.2016.23) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu K, et al. 2017. mosGCTL-7, a C-type lectin protein, mediates Japanese encephalitis virus infection in mosquitoes. J. Virol. 91, 1-18. ( 10.1186/s12985-016-0669-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu Y, Zhang F, Liu J, Xiao X, Zhang S, Qin C, Xiang Y, Wang P, Cheng G. 2014. Transmission-blocking antibodies against mosquito C-type lectins for Dengue prevention. PLoS Pathog. 10, e1003931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cheng G, Cox J, Wang P, Krishnan MN, Dai J, Qian F, Anderson JF, Fikrig E. 2010. A C-type lectin collaborates with a CD45 phosphatase homologue to facilitate West Nile virus infection of mosquitoes. Cell 142, 714-725. ( 10.1016/j.cell.2010.07.038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Simões ML, Dong Y, Mlambo G, Dimopoulos G. 2022. C-type lectin 4 regulates broad-spectrum melanization-based refractoriness to malaria parasites. PLOS Biol. 20, e3001515. ( 10.1371/journal.pbio.3001515) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Osta MA, Christophides GK, Kafatos FC. 2004. Effects of Mosquito Genes on Plasmodium Development. Science 303, 2030-2032. ( 10.1126/science.1091789) [DOI] [PubMed] [Google Scholar]
- 81.Schnitger AKD, Yassine H, Kafatos FC, Osta MA. 2009. Two C-type lectins cooperate to defend Anopheles gambiae against Gram-negative bacteria. J. Biol. Chem. 284, 17 616-17 624. ( 10.1074/jbc.M808298200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Simões ML, Mlambo G, Tripathi A, Dong Y, Dimopoulos G. 2017. Immune regulation of plasmodium is Anopheles species specific and infection intensity dependent. MBio 8, 1-13. ( 10.1128/mBio.01631-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang X, Zhao Q, Christensen BM. 2005. Identification and characterization of the fibronogen-like domain of fibronogen-related proteins in the mosquito, Anopheles gambiae, and the fruitfly, Drosophila melanogaster, genomes. BMC Genom. 6, 1-15. ( 10.1186/1471-2164-6-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Blandin S, Shiao SH, Moita LF, Janse CJ, Waters AP, Kafatos FC, Levashina EA. 2004. Complement-like protein TEP1 is a determinant of vectorial capacity in the malaria vector Anopheles gambiae. Cell 116, 661-670. ( 10.1016/S0092-8674(04)00173-4) [DOI] [PubMed] [Google Scholar]
- 85.Blandin SA, Wang-Sattler R, Lamacchia M, Gagneur J, Lycett G, Ning Y, Levashina EA, Steinmetz LM. 2009. Dissecting the genetic basis of resistance to malaria parasites in Anopheles gambiae. Science 326, 147-150. ( 10.1126/science.1175241) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zou Z, Evans JD, Lu Z, Zhao P, Williams M, Sumathipala N, Hetru C, Hultmark D, Jiang H. 2007. Comparative genomic analysis of the Tribolium immune system. Genome Biol. 8, 1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xu X, Dong Y, Abraham EG, Kocan A, Srinivasan P, Ghosh AKet al. . 2005. Anopheles gambiae immune responses to human and rodent Plasmodium parasite species. Mol. Biochem. Parasitol. 142, e52. [Google Scholar]
- 88.Lima LF, Torres AQ, Jardim R, Mesquita RD, Schama R. 2021. Evolution of Toll, Spatzle and MyD88 in insects: the problem of the Diptera bias. BMC Genom. 22, 1-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Redmond SN, et al. 2015. Association mapping by pooled sequencing identifies TOLL 11 as a protective factor against Plasmodium falciparum in Anopheles gambiae. BMC Genom. 16, 1-13. ( 10.1186/s12864-015-2009-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Levin TC, Malik HS. 2017. Rapidly evolving Toll-3/4 genes encode male-specific Toll-like receptors in Drosophila. Mol. Biol. Evol. 34, 2307-2323. ( 10.1093/molbev/msx168) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Santana RAG, et al. 2019. Anopheles aquasalis transcriptome reveals autophagic responses to Plasmodium vivax midgut invasion. Parasites Vectors 12, 1-14. ( 10.1186/s13071-019-3506-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gulley MM, Zhang X, Michel K. 2013. The roles of serpins in mosquito immunology and physiology. J. Insect Physiol. 59, 138-147. ( 10.1016/j.jinsphys.2012.08.015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zou Z, Shin SW, Alvarez KS, Kokoza V, Raikhel AS. 2010. Distinct melanization pathways in the mosquito Aedes aegypti. Immunity 32, 41-53. ( 10.1016/j.immuni.2009.11.011) [DOI] [PubMed] [Google Scholar]
- 94.Nakhleh J, El Moussawi L, Osta MA. 2017. The melanization response in insect immunity. Adv. Insect Physiol. 52, 83-109. ( 10.1016/bs.aiip.2016.11.002) [DOI] [Google Scholar]
- 95.Povelones M, Bhagavatula L, Yassine H, Tan LA, Upton LM, Osta MA, Christophides GK. 2013. The CLIP-domain serine protease homolog SPCLIP1 regulates complement recruitment to microbial surfaces in the malaria mosquito Anopheles gambiae. PLoS Pathog. 9, e1003623. ( 10.1371/journal.ppat.1003623) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yassine H, Kamareddine L, Chamat S, Christophides GK, Osta MA. 2014. A serine protease homolog negatively regulates TEP1 consumption in systemic infections of the malaria vector Anopheles gambiae J Innate Immun. 6, 806-818. ( 10.1159/000363296) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Volz J, Osta MA, Kafatos FC, Müller HM. 2005. The roles of two clip domain serine protease in innate immune responses of the malaria vector Anopheles gambiae J. Biol. Chem. 280, 40 161-40 168. ( 10.1074/jbc.M506191200) [DOI] [PubMed] [Google Scholar]
- 98.Volz J, Müller HM, Zdanowicz A, Kafatos FC, Osta MA. 2006. A genetic module regulates the melanization response of Anopheles to Plasmodium. Cell Microbiol. 8, 1392-1405. ( 10.1111/j.1462-5822.2006.00718.x) [DOI] [PubMed] [Google Scholar]
- 99.Cui L, Luckhart S, Rosenberg R. 2000. Molecular characterization of a prophenoloxidase cDNA from the malaria mosquito Anopheles stephensi. Insect Mol Biol. 9, 127-137. ( 10.1046/j.1365-2583.2000.00169.x) [DOI] [PubMed] [Google Scholar]
- 100.Müller HM, Dimopoulos G, Blass C, Kafatos FC. 1999. A hemocyte-like cell line established from the malaria vector Anopheles gambiae expresses six prophenoloxidase genes. J. Biol. Chem. 274, 11 727-11 735. ( 10.1074/jbc.274.17.11727) [DOI] [PubMed] [Google Scholar]
- 101.Kwon H, Hall DR, Smith RC. 2021. Prostaglandin E2 signaling mediates oenocytoid immune cell function and lysis, limiting bacteria and Plasmodium oocyst survival in Anopheles gambiae s. s. Front. Immunol. 12, 1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang Y, Hao H, Qiu Zw, Xu Wy, Zhang J, Zhou Tl, Zhang X-, Huang F-S. et al. 2009. Involvement of prophenoloxidases in the suppression of Plasmodium yoelii development by Anopheles dirus. Exp. Parasitol. 123, 6-10. ( 10.1016/j.exppara.2009.05.017) [DOI] [PubMed] [Google Scholar]
- 103.Prado Sepulved CC, et al. 2023. Evolution and assembly of Anopheles aquasalis’s immune genes: primary malaria vector of coastal central and South America and the Caribbean islands. Figshare. ( 10.6084/m9.figshare.c.6728300) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Prado Sepulved CC, et al. 2023. Evolution and assembly of Anopheles aquasalis’s immune genes: primary malaria vector of coastal central and South America and the Caribbean islands. Figshare. ( 10.6084/m9.figshare.c.6728300) [DOI] [PMC free article] [PubMed]
Data Availability Statement
Data are available upon request from the corresponding author or related data bank cited in the material and methods.
Additional information is provided in electronic supplementary material [103].