Abstract
Brown planthopper (BPH), Nilaparvata lugens (Stål) and white-backed planthopper (WBPH), Sogatella furcifera (Horváth) are the most destructive sucking insect pests of rice in all rice growing parts of the world. For their accurate identification at early stages, we have developed two species-specific markers (SNL4F and SNL4R for BPH; SNF2F and SNF2R for WBPH) based on mitochondrial cytochrome oxidase I (COI) for their easy detection using Polymerase Chain Reaction (PCR). The markers were developed based on nucleotide differences in COI gene and were subjected to various tests based on PCR-based gel images. The designed primers were cross-checked with five other species, which confirmed their specificity. The primers were also found to be efficient in identification of their respective species (BPH and WBPH) in all the individuals sampled from different regions of India. The lowest detection sensitivity of both the primers was up to 1 ng/µl DNA after testing them through a series of varied DNA concentrations. The species-specific primers developed in this study will help in easy and rapid identification of BPH and WBPH in all the stages of their development and in turn facilitate their timely management.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13205-023-03693-x.
Keywords: Brown planthopper, White-backed planthopper, Species-specific primers, Species identification, Polymerase chain reaction, COX I
Introduction
Planthoppers are one of the main insect pests attacking rice crop that incur quality and yield losses in all rice growing regions of the world. Among planthoppers, brown planthopper (BPH), Nilaparvata lugens (Stål), and white-backed planthopper (WBPH), Sogatella furcifera (Horvath) (Hemiptera: Delphacidae) are distinguished as the most damaging ones. They are the sap-sucking insects that cause the yellowing of leaves, reduced tillers/plant height, and unfilled grains that can lead to ‘hopper burn’ under severe attack (Padmavathi et al. 2020). Apart from that, they are the vectors of destructive viral diseases such as, rice grassy stunt, rice ragged stunt in India (Chowdhury et al. 2010). Other planthoppers which are present in rice ecosystem are small brown planthopper, Laodelphax striatellus (Ramya and Meshram 2019), Sogatella vibix, and Sogatella kolophon (Delphacidae; Hemipetra), which are present sporadically in the country (Kumaresan et al. 2016). These species are morphologically more or less similar in general structures and coloration leading to their misidentification. As different planthoppers attack rice at different life stages and plant parts, their correct identification will help in achieving better control. They are identifiable only by following taxonomic keys (Dupo and Barrion 2009). However, these keys are not suitable when insect is at nymphal stage and especially when the differentiation is based on genitalia features; they are also hard to identify by non-professionals. Moreover, small brown planthopper is very much similar to BPH morphologically, and both are known to cause economic damage to rice (Wen et al. 2021). Similarly, BPH and WBPH are hard to distinguish by non-specialists, especially at nymphal stages.
Rapid, timely, and accurate identification of insects is desired but difficult as they are numerous in numbers and diversity. As the world is witnessing rapid climate change events, the natural habitable boundaries of planthoppers are extending up to non-traditional rice growing areas. This expansion is a result of increase in their overwintering ranges from their conventional East and South Asian/Australian countries (Hu et al. 2015). This shift from their natural geographical range will raise concern for their more frequent introductions into new areas of Asia and Pacific demanding stringent biosecurity and phytosanitary practices, which will become plausible only if the insect is correctly identified at earlier stages (Heong and Hardy 2009).
The planthoppers are highly migratory in nature; with the onset of spring season, they migrate from the tropical areas to the temperate/sub-temperate areas every year (Tyagi et al. 2022b; Narayana et al. 2020; Hu et al. 2017; Krishnaiah 2014; Otuka et al. 2008). The movement of BPH is confirmed by its unsuitability to colder regions as well as unavailability of alternate graminaceous hosts to survive (Tyagi et al. 2022a). The management of migratory insects is greatly dependent on their timely forecasting and identification. The existence of macropterous and brachypterous wing morphs of both the insects create even more confusion for their accurate identification, if based on morphology alone (Narayana et al. 2022; Li et al. 2016).
Unlike traditional systematic approaches using morphological characters detection, using molecular marker is efficient as it does not depend on polymorphism, sex, and life stages for the species in question (Asokan et al. 2011). Species-specific markers are easy, accurate, and economic tools of species discrimination and determination of phylogenetic relationship, which produce specific amplicon of the target species, eliminating the need for sequencing (Latip et al. 2010). In previous studies, the identification of planthoppers was mainly focused on BPH (Liu et al. 2018), while only a few included WBPH and Small brown planthopper species (Rahman et al. 2023; Yashiro and Sanada-Morimura 2021; Seo et al. 2017; Wang et al. 2013). However, three-fold testing of robustness of markers for efficiency, specificity, and sensitivity has hardly been achieved in a single study and we have tried to address this limitation.
Considering that, development of suitable species-specific markers will greatly aid in identifying a given species quickly with a high degree of accuracy, especially when there is an overall dearth of taxonomists. The identification of planthoppers was mainly achieved by targeting the varied regions of Mitochondrial cytochrome oxidase gene I (COI) and Internal Transcribed Spacer (ITS) genes, particularly ITS2, in nuclear ribosomal DNA (rDNA) (Yashiro and Sanada-Morimura 2021; Wang et al. 2013). Mitochondrial cytochrome oxidase gene I (COI) has been widely used in the molecular systematics to understand host associated genetic differences, biotypes, cryptic/sister/subspecies due to its robust interspecific differentiation, aiding in species-level detection of insects (Arya et al. 2022; Simon et al. 1994; Jung et al. 2011). Therefore, in the present study, we have developed species-specific markers for BPH and WBPH identification based on COI gene.
Materials and methods
DNA extraction and amplification
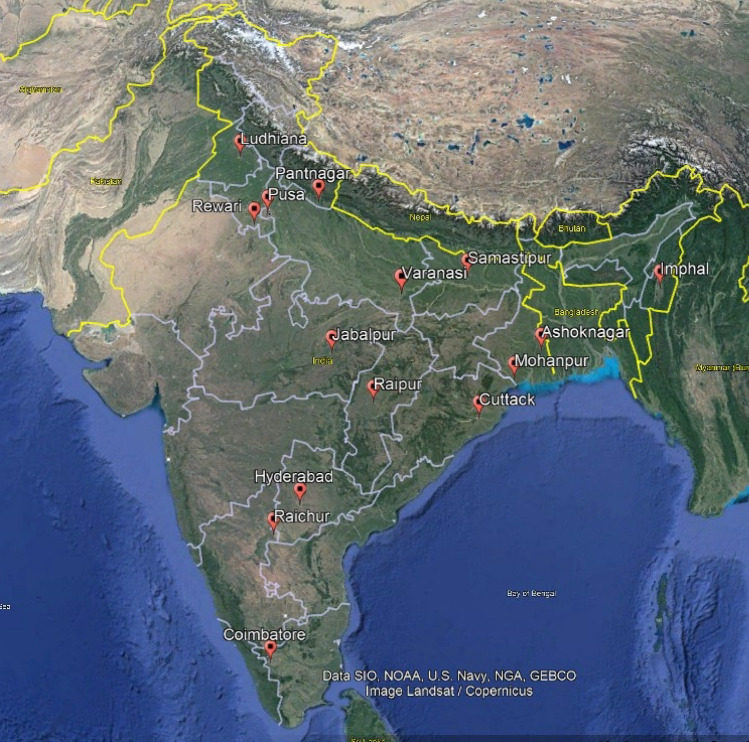
BPH and WBPH were collected from 15 traditional rice growing parts representing the entire country using sweep net in morning hours between 8 and 11 am when the wind speed is slow (Fig. 1, Table 2). The hoppers were preserved in 90% ethanol during transit and were later sorted out under microscope using morphological keys (Dupo and Barrion 2009). The samples were stored at − 20 °C until used for DNA extraction. DNA from individual specimens was extracted using DNAeasy® Blood and Tissue Kit (Qiagen) according to manufacturer’s protocol. Quality of DNA was ensured through Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, USA). The COI-specific amplification of DNA through Polymerase Chain Reaction (PCR) was achieved in Bio-Rad T100™ Thermal cycler. The PCR conditions are as follows: initial denaturation at 94 °C for 5 min; 35 cycles of denaturation at 94 °C for 30 s; annealing at 46 °C for 40 s; elongation at 72 °C for 40 s and a final elongation at 72 °C for 8 min. Total reaction volume was 25 µl, which consisted of 12.5 µl of TaKaRa Emerald Amp® GT master mix, 8.5 µl of nuclease free water and 1 µl each of forward and reverse COI primers with 2 μl of template DNA (50 ng/µl). The universal barcode specific primers (LCO-1490–5′GGT CAA CAA ATC ATA AAG ATA TTG G-3′; HCO-2198–5′-TAA ACT TCA GGG TGA CCA AAA AAT CA-3′) were used for amplification (Folmer et al. 1994).
Fig. 1.
The 15 Indian sites from where the planthopper populations have been collected for the development and validation of species-specific markers for N. lugens and S. furcifera species
Table 2.
Details of N. lugens and S. furcifera samples used in the study
| S. no. | Species | Location | Coordinates | Accession numbers# |
|---|---|---|---|---|
| 1 | N. lugens | Varanasi, Uttar Pradesh | 25.3176 °N, 82.9739 °E | OK076689, OK076690, OK036953 |
| 2 | Ludhiana, Punjab | 30.9010 °N, 75.8573 °E | OK076691, OK076692, OK076693 | |
| 3 | Samastipur, Bihar | 25.8560 °N, 85.7868 °E | OK076694, K076695, OL677401 | |
| 4 | Cuttack, Odisha | 20.4625 °N, 85.8830 °E | MZ813272, MZ823620, MZ828638, | |
| 5 | Raichur, Karnataka | 16.2160 °N, 77.3566 °E | MZ618359, MZ666290, MZ666238 | |
| 6 | Rewari, Haryana | 28.1920 °N, 76.6191 °E | OM268979, OM268980, OM535915 | |
| 7 | Jabalpur, Madhya Pradesh | 23.1815 °N, 79.9864 °E | OM268983, OM268984, OM268985 | |
| 8 | Coimbatore, Tamil Nadu | 11.0168 °N, 76.9558 °E | OL677400, OM535919, OM535920 | |
| 9 | Imphal, Manipur | 24.8170 °N, 93.9368 °E | OM268981, OM268982, OM283785 | |
| 10 | Raipur, Chhattisgarh | 21.2514 °N, 81.6296 °E | OK428850, OM268988, OM268989 | |
| 11 | Mohanpur, West Bengal | 21.8398 °N, 87.4232 °E | OM268986, OM268987, OM535921 | |
| 12 | North 24 Parganas, West Bengal | 22.7100 °N, 88.7109 °E | OM268974, OM268975, OM535912 | |
| 13 | Pantnagar, Uttarakhand | 29.0222 °N, 79.4908 °E | OL677396, OL677397, OL677398 | |
| 14 | Pusa, New Delhi | 28.6377 °N, 77.1571 °E | MW751978, MW751979 | |
| 15 | Hyderabad, Telangana | 17.3850 °N, 78.4867 °E | OM268976, OM268977, OM535913 | |
| 1 | S. furcifera | Varanasi, Uttar Pradesh | 25.3176 °N, 82.9739 °E | – |
| 2 | Coimbatore, Tamil Nadu | 11.0168 °N, 76.9558 °E | MZ666131, OM269032 | |
| 3 | Mohanpur, West Bengal | 21.8398 °N, 87.4232 °E | OM269033 | |
| 4 | Rewari, Haryana | 28.1920 °N, 76.6191 °E | OM269031 | |
| 5 | Jabalpur, Madhya Pradesh | 23.1815 °N, 79.9864 °E | OM269030 | |
| 6 | Hyderabad, Telangana | 17.3850 °N, 78.4867 °E | – | |
| 7 | Bangalore, Karnataka | 12.9716 °N, 77.5946 °E | MH670907* |
#Accession number of planthopper samples used in specificity tests
*COI sequence retrieved from NCBI database to design WBPH-specific marker
The amplified DNA products were purified using QIAquick®(Qiagen) PCR Purification Kit according to manufactures instructions and then sent for sequencing at M/s Eurofins Genomics India Pvt. Ltd. using Sanger’s sequencing method. Species identity was confirmed using BLASTn (http://www.ncbi.nlm.nih.gov) search by comparing with already submitted sequences in NCBI. The sequences were edited and trimmed in BioEdit.7.0 program (Hall 2004) after checking for InDels and stop codons. Corresponding COI sequences of BPH and WBPH were deposited with the National Centre for Biotechnology Information (NCBI) GenBank database and finally accession numbers were obtained (Table 2). To design WBPH specific marker, a previously submitted sequence was also retrieved from NCBI database (accession number: MH670907).
Primers development
The species-specific primers for BPH and WBPH were developed based on the differences in COI sequences of both the species, which were determined using the sequence alignment editor BioEdit.7.0. (Supplementary File). The species-specific primers were designed using Primer3 (Untergasser et al. 2012), which provided possible primer pairs for PCR templates by considering the following criteria: (1) length of primer between 18 and 30 bp; (2) absolute value of Delta G less than 9; (3) 3′ end contains one or more specific bases; (4) absence of hairpin structure; (5) GC% content from 40 to 70%; (6) false priming less than 100%. The species specificity of the designed primers was confirmed using Primer BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/). All the primers were synthesized by M/s Eurofins Genomics India Pvt. Ltd. Bengaluru, India.
Primers selection and sensitivity test
Four sets of forward and reverse primer pairs of BPH and three sets for WBPH were designed based on the variable regions of their aligned sequences (Table 1). The designed primers were validated on identified BPH and WBPH populations. PCR amplification was carried in a 20 µl reaction mixture consisting of 7.8 µl Nuclease free water, 9 µl of TaKaRa Emerald Amp GT master mix, 0.6 µl of each forward and reverse species-specific COI primer with 2 μl of template DNA (50 ng/µl). The conditions for PCR were same as mentioned in “DNA extraction and amplification” except for the annealing temperature, which was confirmed at 64 °C for 40 s. The final annealing temperature was determined by varying annealing temperatures from 45 to 65 °C by evaluating species specificity of different sets of BPH and WBPH primers (Table 1).
Table 1.
Details of species-specific primers developed for N. lugens and S. furcifera
| Species | Primer name | Sequence (5′–3′) | bp | Tm(°C) | GC | Amplicon size (bp) | Specificity at Ta (°C) | |||
|---|---|---|---|---|---|---|---|---|---|---|
| 46 | 50 | 55 | 64 | |||||||
| N. lugens | SNL4F | TTCTGACTTTTACCCCCATCTTTA | 24 | 57.59 | 37.50 | 307 | ✗ | ✗ | ✗ | ✓ |
| SNL4R | CAGCTAGAACAGGAAGGGATAGAA | 24 | 61.01 | 45.83 | ✗ | ✗ | ✗ | ✓ | ||
| SNBPH1F | TATTTGGTCAGGATTTATAGGA | 22 | 52.80 | 31.82 | 430 | ✗ | ✗ | ✗ | ✗ | |
| SNBPH1R | TGAAATAAAATTAATTGCAC | 20 | 45.00 | 20.00 | ✗ | ✗ | ✗ | ✗ | ||
| SNBPH2F | GTAGTATAATTATCCGATCAGA | 22 | 52.80 | 31.82 | 491 | ✗ | ✗ | ✗ | ✗ | |
| SNBPH2R | AAGGAGTAAAATAGCTGTAAT | 21 | 50.06 | 28.57 | ✗ | ✗ | ✗ | ✗ | ||
| SNLF3F | TTCTGACTTTTACCCCCATCTTTA | 24 | 57.59 | 37.50 | 302 | ✗ | ✗ | ✗ | ✗ | |
| SNLR3R | AGAACAGGAAGGGATAGAAGGAGT | 24 | 61.01 | 45.83 | ✗ | ✗ | ✗ | ✗ | ||
| S. furcifera | SNF2F | TCGATCTGAACTAACCCAACCT | 22 | 58.39 | 45.45 | 349 | ✗ | ✗ | ✗ | ✓ |
| SNF2R | GGCAATGTGGAGGGAGAAAAT | 21 | 57.87 | 47.62 | ✗ | ✗ | ✗ | ✓ | ||
| SNWBPH1F | GGTATGATCCGGACTAATTGGT | 22 | 58.39 | 45.45 | 510 | ✗ | ✗ | ✗ | ✗ | |
| SNWBPH1R | ACTGCGGTAATTAAAACTGAT | 21 | 52.01 | 33.33 | ✗ | ✗ | ✗ | ✗ | ||
| SNWBPH2F | AAGAATTTTAATTCGATCTGA | 21 | 48.11 | 23.81 | 401 | ✗ | ✗ | ✗ | ✗ | |
| SNWBPH2R | TTGAAATGAAATTGATAGCTCC | 22 | 52.80 | 31.82 | ✗ | ✗ | ✗ | ✗ | ||
Tm melting temperature, Ta annealing temperature, GC Guanine-Cytosine content
To validate primers specificity, cross-amplification assay was done through PCR for the primer sets developed in the current study on six different species in five replications (Figs. 2, 3). The efficacy of the markers was assessed with BPH and WBPH samples collected from different locations of India (Table 2). Three individuals from each population were assayed for PCR. The sensitivity of markers was evaluated using varying concentrations of template DNA, i.e., 100, 50, 30, 10, 1, and 0.1 ng/µl, through PCR assay, while keeping the concentration of primers similar. The varied concentrations were obtained from serial dilutions of a single female DNA with elution buffer. Each test of efficacy and sensitivity was replicated thrice. The blank consisted of Nuclease free water instead of DNA with the same reaction mixture. COI fragments resulting from species-specific markers for BPH and WBPH were further sequenced and tested through BLASTn.
Fig. 2.
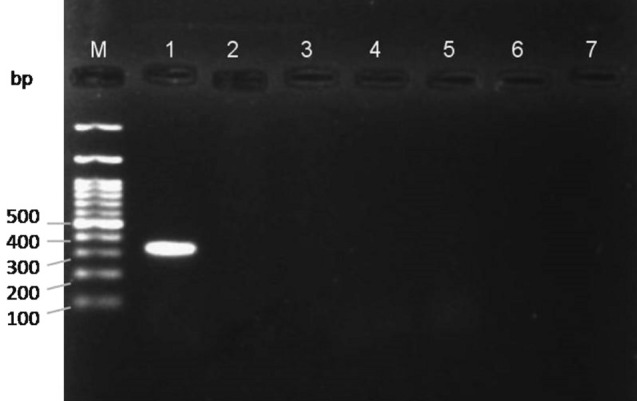
Gel image showing specificity of N. lugens specific primer pair among five other insect species. M—100 bp DNA ladder (BR Biochem); 1: N. lugens; 2: S. furcifera; 3: Aphis gossypii; 4: Callosobruchus maculatus; 5: Vespula vulgaris; 6: Bactrocera correcta; 7: Blank with Nuclease free water
Fig. 3.
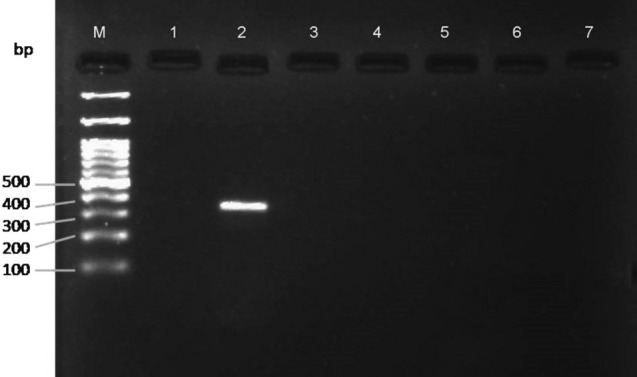
Gel image showing specificity of S. furcifera specific primer pair among five other insect species. M—100 bp DNA ladder (BR Biochem); 1: N. lugens; 2: S. furcifera; 3: Aphis gossypii; 4: Callosobruchus maculatus; 5: Vespula vulgaris; 6: Bactrocera correcta; 7: Blank with Nuclease free water
Results
To make sure that we were using quality Mitochondrial DNA for marker designing, the extracted DNA samples were screened through spectrophotometry (NanoDrop™ 2000/2000c Spectrophotometer), resulting in absorbance values from 1.8 to 2, as well as through PCR amplification, displaying gel bands at 658 bp, indicating that they were of high quality. Using nucleotide polymorphisms in BPH (OM268976) and WBPH (MH670907) sequences with their respective species, which were earlier submitted in NCBI database (Table 2), four and three pairs of primers were developed, respectively, for BPH and WBPH (Supplementary File).
Species-specific markers for BPH and WBPH showed amplicons of sizes 307 and 349 bp, respectively, with uniform bands in all identified and tested individuals from different locations (Fig. 1). From the primers sets developed for each species, only one pair for BPH (SNL4F & SNL4R) and one pair for WBPH (SNF2F & SNF2R) were deemed fit after going through a series of tests in their capability of amplifying specific COI fragment corresponding to their species only. The optimum annealing temperature was confirmed at 64 °C by observing amplification pattern of PCR products in a gradient of annealing temperature from 45 to 65 °C (Table 1).
Markers’ specificity
The specificity of the BPH markers (SNL4F and SNL4R) when tested with six other species resulted in a single clear band for BPH only, rendering blank for all other species using uniplex PCR (Fig. 2). Similarly, there was a single clear band observed for WBPH while using specific marker pair SNF2F and SNF2R (Fig. 3). The amplified bands were in correspondence with their amplicon size, viz, 307 and 349 bp, for BPH and WBPH, respectively. There was absence of any secondary or unwanted band at the standard annealing temperature of 64 °C in all the replications. To confirm that amplified products were from the target COI gene, each product was sequenced in both forward and reverse directions, resulting in the sequences of their respective species only, after checking through NCBI BLAST homology search. The sequences obtained using species-specific markers for both the species showed highest hits for their respective species in Primer-BLAST.
Markers’ efficacy
BPH individuals from 15 locations of India were tested with BPH specific marker to test whether the marker is efficient to identify them. The populations from different locations are described in Table 2 along with their accession numbers. The results clearly depicted a single, uniform band in all the individuals at 307 bp, excluding control (Fig. 4). Likewise, six individuals of WBPH collected from different locations of India revealed a single, clearly uniform band in all the individuals at 349 bp without any non-specific bands (Fig. 5). The results of efficacy were consistent in all the replications for both the species.
Fig. 4.
Validation of N. lugens specific primer pair on the individuals collected from different locations of India. M—100 bp DNA ladder (BR Biochem); 1–15: BPH from different geographical locations corresponding to Table 2; 16: Blank with Nuclease free water
Fig. 5.
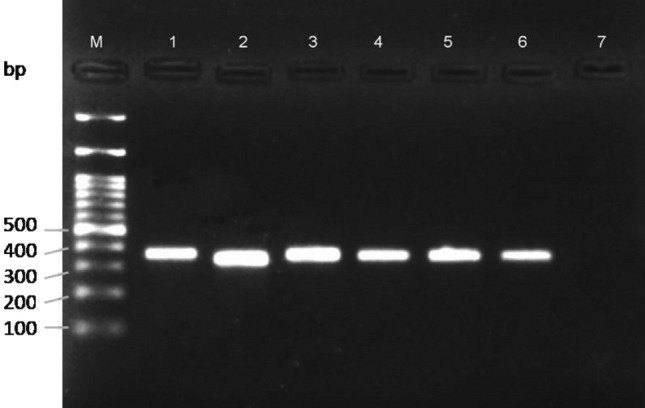
Validation of S. furcifera specific primer pair on the individuals collected from different locations of India. M—100 bp DNA ladder (BR Biochem); 1–6: WBPH from different geographical locations corresponding to Table 2; 7: Blank with Nuclease free water
Markers’ sensitivity
For this purpose, DNA concentration of 200 ng/µl from one female BPH and DNA concentration of 150 ng/µl from another female WBPH were used in making serial dilutions of template DNA (100, 50, 30, 10, 1, and 0.1 ng/µl) by adding Elution buffer (Fig. 6; Fig. 7). The females were differentiated morphologically from males based on the shape of terminal abdominal segments. The PCR assay results with different concentrations of DNA while using same volume of species-specific primers showed a decreasing pattern of banding intensity. The detection was clearly visible up to 10 ng/µl with the presence of strong intensity bands, whereas, a light band at 1 ng/µl can be seen for both BPH and WBPH, targeting required COI fragments at 307 and 309 bp, respectively, in all the replicates. The study suggested that the lower detection limit for both species-specific markers was 1 ng/µl and there was no amplification observed at 0.1 ng/µl.
Fig. 6.

Gel image showing sensitivity of N. lugens specific primer pair on different concentrations of template DNA. M—100 bp DNA ladder (BR Biochem); 1: 100 ng/µl; 2: 50 ng/µl; 3: 30 ng/µl; 4: 10 ng/µl; 5: 1 ng/µl; 6: 0.1 ng/µl; 7: Blank with Nuclease free water
Fig. 7.
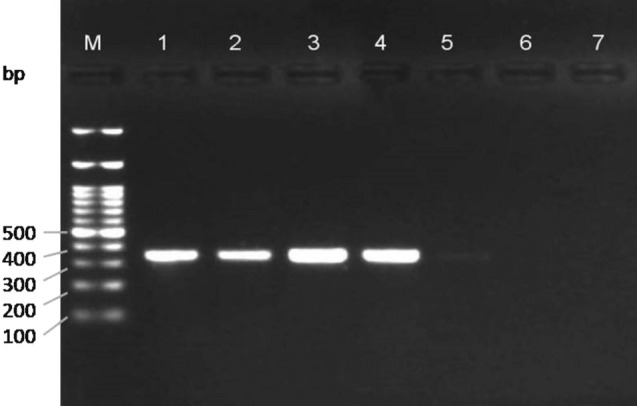
Gel image showing sensitivity of S. furcifera specific primer pair on different concentrations of template DNA. M—100 bp DNA ladder (BR Biochem); 1: 100 ng/µl; 2: 50 ng/µl; 3: 30 ng/µl; 4: 10 ng/µl; 5: 1 ng/µl; 6: 0.1 ng/µl; 7: Blank with Nuclease free water
Discussion
Species-specific markers can make accurate and rapid species identification possible, which is highly useful for non-trained personnel in the field of entomology. They are the future of taxonomy, pest forecasting, and biosecurity (Tsai et al. 2020; Rebijith et al. 2012b; Juric et al. 2015). Keeping these things in mind we have developed species-specific marker pairs each for BPH and WBPH for their easy and precise identification. Species-specific marker dependant detection is based on PCR, which is inexpensive, stable, swift, sensitive, efficient, and is suitable for quarantine facilities (Zhang et al. 2016). The PCR-based taxonomic identification is free from the trouble of sequencing, restriction digestion or slide preparation, which can detect species at any life stage even with parasitized/damaged/decayed specimens (Srinivasa et al. 2019; Wang et al. 2019b) or museum specimens (Townson et al. 1999). The reduced costs of sequencing as well as elimination of DNA purification step are the additional advantages of using species-specific markers.
The morphological ambiguity between BPH, WBPH and other related species have been hampering their management for a long time. Insecticide resistance, host-range or natural enemy interactions vary from species to species, and thus, single tactic, say application of a certain insecticide would not work for all the species causing problem of resistance and resurgence (Rebijith et al. 2012b). The use of species-specific markers is more valued at quarantine stations where early detection is paramount for restricting any chance introduction (Wang et al. 2019a; Shim et al. 2016). Information on DNA barcoding-based species identification through molecular markers is valued for enhancing biosecurity via planning pest control programs for the management of introduced germplasm (Marullo et al. 2020). These markers will aid in decision making by the traditional taxonomists when the morphology-based identification is difficult.
In current study, polymorphism present in COI sequences of BPH and WBPH was utilized in developing species-specific markers. The markers developed in the study were able to distinguish their respective species clearly from others by forming specific COI bands of their own amplicon sizes. The markers showed amplification in all the samples that were collected from different locations of India proving their robustness. Their sensitivity to amplify as low as 1 ng/µl template DNA showed their ability to recognize the species even when only a trace amount of DNA is available.
The COI based barcoding of insects and development of species-specific markers based on it have been achieved successfully in insects like aphids (Rebijith et al. 2012a), fruit flies (Zheng et al. 2019; Jiang et al. 2013), mealy bugs (Wang et al. 2019a), Tribolium spp. (Zhang et al. 2016), Cryptolestes spp. (Varadínová et al. 2015). Very recently, identification of three planthopper species was achieved by developing six species-specific primers based on partial COI sequences, which showed positive results for conventional, multiplex PCR, and loop-mediated isothermal amplification (LAMP) assays (Rahman et al. 2023).
Newer image-based techniques, such as spectrometry and scanning electron microscopy are also gaining momentum in species discrimination; their implementation, though, need much sophisticated system, and as for now they are unavailable for BPH and WBPH (Liu et al. 2018). Other non-COI-based molecular markers such as internal transcribed spacers (ITS) DNA sequence and direct multiplex PCR have shown potential in the identification of planthoppers, Fall Armyworm and other insects (Liu et al. 2018; Tsai et al. 2020). Yashiro and Sanada-Morimura (2021) developed a multiplex PCR assay utilizing markers based on 5.8S-ITS2 rDNA for the identification of S. furcifera, N. lugens, and L. striatellus individuals. Furthermore, multiplex real-time PCR technique employing ITS2 region was used for the detection and quantification of S. furcifera, N. lugens, and L. striatellus in the guts of spiders for determining predatory relationships with planthoppers in rice ecosystems (Wang et al. 2013).
In way forward, it is recommended to test the markers efficacy on the species collected outside from geographical range of India. Use of multiplex PCR in amplifying genes other than COI such as, ITS1, ITS2 and nuclear 12S-16S-18S ribosomal RNA for the development of molecular markers is encouraged. It is also advisable to remove allochthonous micro-organisms attached with the insect body by washing 2–3 times with ethanol as they can obstruct the truthful identification by forming non-specific bands (Kersting et al. 2018).
Conclusions
To conclude, the use of PCR-based species-specific markers for species discrimination and identification is not only easy and quick but it is also irrespective of the life stage, sex, and expertise level or specimen condition. In light of this, we have developed two COI-based marker pairs specific to each BPH (SNL4F and SNL4R) and WBPH (SNF2F and SNF2R) to eliminate the identification error between them and also between their sister/cryptic species such as N. bakeri, N. maeander, N. muiri, S. vibix, and S. kolophon. These markers were specific, efficient, and showed detection sensitivity with as low as 1 ng/µl DNA. This work will be encouraged at quarantine stations where early detection is highly appreciated. Species-specific markers operate as guiding tools for the monitoring programs in order to develop sound management strategies.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
Corresponding author greatly acknowledge the seed grant from Banaras Hindu University under Institute of Eminence scheme (IoE) (Grant No.6031). We are also grateful to Dr. B.K. Sarma (Professor, Dept. of Mycology & Plant Pathology, Institute of Agricultural Sciences, Banaras Hindu University) for allowing us to use his laboratory.
Author contributions
SN and ST conceived the idea. ST, SN and RNS collected the samples from different parts of India. ST, SN and VN performed the experiments and analyses and ST, SN, VN, RNS wrote the manuscript. All authors reviewed and gave suggestions to improve the manuscript.
Funding
This study was funded by seed grant from Banaras Hindu University under Institute of Eminence scheme (IoE) (Grant No. 6031).
Data availability
The data of MtCOI sequences for the accession numbers mentioned in Table 2, which can be retrieved from NCBI database.
Declarations
Conflict of interest
All authors declare no conflict of interest.
Contributor Information
Saniya Tyagi, Email: saniya.tyagi13@bhu.ac.in.
Narayana Srinivasa, Email: srinivasa@bhu.ac.in.
R. N. Singh, Email: singhrnacarology58@gmail.com
N. Vinay, Email: vinaynchethu@gmail.com
References
- Arya V, Narayana S, Tyagi S, Raju SV, Srivastava CP, Sinha T, Divekar P. DNA barcoding of fruit flies associated with cucurbit ecosystem and combination of Cue-Lure and Methyl Eugenol in trap is not effective for mass trapping of responsive fruit flies. Phytoparasitica. 2022;26:1–3. [Google Scholar]
- Asokan R, Rebijith KB, Shakti KS, Sidhu AS, Siddharthan S, Praveen KK, Ellango R, Ramamurthy VV. Molecular identification and phylogeny of Bactrocera species (Diptera: Tephritidae) Fla Entomol. 2011;94:1026–1035. doi: 10.1653/024.094.0441. [DOI] [Google Scholar]
- Chowdhury AK. Present status and research on rice virus diseases in India. J Mycopathological Res. 2010;48(2):187–191. [Google Scholar]
- Dupo ALB, Barrion AT. Planthoppers: new threats to the sustainability of intensive rice production systems in Asia, International Rice Research Institute (IRRI) Manila: Phillipines; 2009. Taxonomy and general biology of delphacid planthoppers in rice agro ecosystems; pp. 3–155. [Google Scholar]
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit 1 from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- Hall T (2004) BioEdit version 7.0.0. Distributed by the author, website: www.mbio.ncsu.edu/BioEdit/bioedit.html
- Heong KL, Hardy B (2009) Planthoppers: New Threats to the Sustainability of Intensive Rice Production Systems in Asia. International Rice Research Institute, Metro Manila, Philippines, ISBN 9712202518
- Hu C, Hou M, Wei G, Shi B, Huang J. Potential overwintering boundary and voltinism changes in the brown planthopper, Nilaparvata lugens, in China in response to global warming. Clim Change. 2015;132:337–352. doi: 10.1007/s10584-015-1427-x. [DOI] [Google Scholar]
- Hu G, Lu MH, Tuan HA, Liu WC, Xie MC, McInerney CE, Zhai BP. Population dynamics of rice planthoppers, Nilaparvata lugens and Sogatella furcifera (Hemiptera, Delphacidae) in Central Vietnam and its effects on their spring migration to China. Bull Entomol Res. 2017;107:369–381. doi: 10.1017/S0007485316001024. [DOI] [PubMed] [Google Scholar]
- Jiang F, Li ZH, Deng YL, Wu JJ, Liu RS, Buahom N. Rapid diagnosis of the economically important fruit fly, Bactrocera correcta (Diptera: Tephritidae) based on a species-specific barcoding cytochrome oxidase I marker. Bull Entomol Res. 2013;103:363–371. doi: 10.1017/S0007485312000806. [DOI] [PubMed] [Google Scholar]
- Jung S, Duwal RK, Lee S. CO-I barcoding of true bugs (Insecta, Heteroptera) Mol Ecol Res. 2011;11:266–270. doi: 10.1111/j.1755-0998.2010.02945.x. [DOI] [PubMed] [Google Scholar]
- Juric I, Salzburger W, Luka H, Balmer O. Molecular markers for Diadegma (Hymenoptera: Ichneumonidae) species distinction and their use to study the effects of companion plants on biocontrol of the diamondback moth. Bio Control. 2015;60:179–187. [Google Scholar]
- Kersting S, Rausch V, Bier FF, von Nickisch-Rosenegk M. A recombinase polymerase amplification assay for the diagnosis of atypical pneumonia. Anal Biochem. 2018;550:54–60. doi: 10.1016/j.ab.2018.04.014. [DOI] [PubMed] [Google Scholar]
- Krishnaiah NV. A global perspective of rice brown planthopper management I-crop-climatic requirement. Intl J Mol Zool. 2014;30:4. [Google Scholar]
- Kumaresan N, Ilango K, Gopinath LR, Bhuvaneswari R, Archaya S. Dynamics of plant hoppers diversity in Kolli Hills, Tamilnadu, India. Int J Fauna Biol Stu. 2016;3:93–97. [Google Scholar]
- Latip SNH, Muhamad R, Manjeri G, Tan SG. Development of microsatellite markers for Helopeltis theivora Waterhouse (Hemiptera: Miridae) Afr J Biotechnol. 2010;9:4478–4481. [Google Scholar]
- Li XY, Chu D, Yin YQ, Zhao XQ, Chen AD, Khay S, Douangboupha B, Kyaw MM, Kongchuensin M, Ngo VV, Nguyen CH. Possible source populations of the white-backed planthopper in the Greater Mekong Sub region revealed by mitochondrial DNA analysis. Sci Rep. 2016;6:1–10. doi: 10.1038/srep39167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Luo J, Liu R, Zhang C, Duan D, Chen H, Bei W, Tang J. Identification of Nilaparvata lugens and Its Two Sibling Species (N. bakeri and N. muiri) by Direct Multiplex PCR. J Econ Entomol. 2018;111:2869–2875. doi: 10.1093/jee/toy232. [DOI] [PubMed] [Google Scholar]
- Marullo R, Mercati F, Vono G. DNA Barcoding: a reliable method for the identification of thrips species (Thysanoptera, Thripidae) collected on sticky traps in onion fields. InSects. 2020;11:489. doi: 10.3390/insects11080489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayana S, Chander S, Twinkle CRK. Genetic homogeneity in brown planthopper, Nilaparvata lugens (Stål) as revealed from mitochondrial cytochrome oxidase I. Curr Sci. 2020;119:1045–1050. doi: 10.18520/cs/v119/i6/1045-1050. [DOI] [Google Scholar]
- Narayana S, Chander S, Doddachowdappa S, Sabtharishi S, Divekar P. Seasonal variation in population and biochemical contents of brown planthopper, Nilaparvata lugens (Stål) J Environ Biol. 2022;4:352–358. [Google Scholar]
- Otuka A, Matsumura M, Watanabe T, Van Dinh T. A migration analysis for rice planthoppers, Sogatella furcifera (Horváth) and Nilaparvata lugens (Stål) (Homoptera: Delphacidae), emigrating from northern Vietnam from April to May. Appl Entomol Zool. 2008;43:527–534. doi: 10.1303/aez.2008.527. [DOI] [Google Scholar]
- Padmavathi G, Lakshmi VJ, Rao LS, Madhav MS. Identification of new quantitative trait loci for resistance to brown planthopper in rice landrace. Sinnasivappu Extended Summaries. 2020;8:143. [Google Scholar]
- Rahman M-M, Nam H, Choi N, Kim J. Development of molecular-based species identification and optimization of reaction conditions for molecular diagnosis of three major Asian planthoppers (Hemiptera: Delphacidae) InSects. 2023;14:124. doi: 10.3390/insects14020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramya N, Meshram NM. New record of small brown planthopper Laodelphax striatellus from Delhi. Indian J Entomol. 2019;81:532–535. doi: 10.5958/0974-8172.2019.00115.9. [DOI] [Google Scholar]
- Rebijith KB, Asokan R, Krishna V, Kumar NK, Ramamurthy VV. Development of species-specific markers and molecular differences in mitochondrial and nuclear DNA sequences of Aphis gossypii and Myzus persicae (Hemiptera: Aphididae) Fla Entomol. 2012;1:674–682. doi: 10.1653/024.095.0318. [DOI] [Google Scholar]
- Rebijith KB, Asokan R, Kumar NK, Srikumar KK, Ramamurthy VV, Bhat PS. DNA barcoding and development of species-specific markers for the identification of tea mosquito bugs (Miridae: Heteroptera) in India. Environ Entomol. 2012;41:1239–1245. doi: 10.1603/EN12096. [DOI] [PubMed] [Google Scholar]
- Seo BY, Park CG, Koh YH, Jung JK, Cho J, Kang C. ITS2 DNA sequence analysis for eight species of delphacid planthoppers and a loop-mediated isothermal amplification method for the brown planthopper-specific detection. Korean J Appl Entomol. 2017;56:377–385. [Google Scholar]
- Shim JK, Khaing TM, Seo HE, Ahn JY, Jung DO, Lee JH, Lee KY. Development of species-specific primers for rapid diagnosis of Tetranychus urticae, T. kanzawai, T. phaselus and T. truncatus (Acari: Tetranychidae) Entomol Res. 2016;46:162–169. doi: 10.1111/1748-5967.12154. [DOI] [Google Scholar]
- Simon C, Frati F, Bechenbach A, Crespi B. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequence and compilation of conserved polymerase chain reaction primers. Ann Entomol Soc Am. 1994;87:651–701. doi: 10.1093/aesa/87.6.651. [DOI] [Google Scholar]
- Srinivasa N, Chander S, Chandel RK, Sagar D. Gonatopus spp. parasitoids on rice plant hoppers. Indian J Entomol. 2019;81:352–354. doi: 10.5958/0974-8172.2019.00051.8. [DOI] [Google Scholar]
- Townson H, Harbach RE, Callan TA. DNA identification of museum specimens of the Anopheles gambiae complex: an evaluation of PCR as a tool for resolving the formal taxonomy of sibling species complexes. Syst Entomol. 1999;24:95–100. doi: 10.1046/j.1365-3113.1999.00084.x. [DOI] [Google Scholar]
- Tsai CL, Chu I, Chou MH, Chareonviriyaphap T, Chiang MY, Lin PA, Lu KH, Yeh WB. Rapid identification of the invasive fall armyworm Spodoptera frugiperda (Lepidoptera, Noctuidae) using species-specific primers in multiplex PCR. Sci Rep. 2020;10:1–8. doi: 10.1038/s41598-020-73786-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi S, Narayana S, Singh RN, Gowda GB. Molecular insights into wing polymorphism and migration patterns of rice planthoppers. In: Chakravarthy AK, editor. Genetic methods and tools for managing crop pests. Singapore: Springer; 2022. pp. 449–453. [Google Scholar]
- Tyagi S, Narayana S, Singh RN, Srivastava CP, Twinkle S, Das SK, Jeer M. Migratory behaviour of brown planthopper, Nilaparvata lugens (Stål) (Hemiptera: Delphacidae), in India as inferred from genetic diversity and reverse trajectory analysis. 3Biotech. 2022;12:266. doi: 10.1007/s13205-022-03337-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. Primer3-new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115–e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadínová Z, Wang YJ, Kučerová Z, Stejskal V, Opit G, Cao Y, Li FJ, Li ZH. COI barcode based species-specific primers for identification of five species of stored-product pests from genus Cryptolestes (Coleoptera: Laemophloeidae) B Entomol Res. 2015;105:202–209. doi: 10.1017/S0007485315000024. [DOI] [PubMed] [Google Scholar]
- Wang GH, Wang XQ, Qiao F, Zhu ZR, Cheng JA. Development and preliminary application of a triplex real-time polymerase chain reaction assay for evaluating predation on three planthoppers in a rice ecosystem. Mol Ecol Resour. 2013;13(5):811–819. doi: 10.1111/1755-0998.12127. [DOI] [PubMed] [Google Scholar]
- Wang YS, Dai TM, Tian H, Wan FH, Zhang GF. Phenacoccus madeirensis green (Hemiptera: Pseudococcidae): new geographic records and rapid identification using a species-specific PCR assay. Crop Prot. 2019;116:68–76. doi: 10.1016/j.cropro.2018.10.003. [DOI] [Google Scholar]
- Wang YS, Hu TI, Wan FH, Zhang GF. Species-specific COI primers for rapid identification of a globally significant invasive pest, the cassava mealybug Phenacoccus manihoti Matile-Ferrero. J Integr Agric. 2019;18:1042–1049. doi: 10.1016/S2095-3119(18)62043-X. [DOI] [Google Scholar]
- Wen S, Xue Y, Du R, Liu C, Wang X, Wang Y, Liu C, Wang S, Wang J, Xia X. Toxicity and sublethal effects of triflumezopyrim on the development and detoxification enzymatic activities in the small brown planthopper (SBPH), Laodelphax striatellus (Fallen) Crop Prot. 2021;150:105813. doi: 10.1016/j.cropro.2021.105813. [DOI] [Google Scholar]
- Yashiro T, Sanada-Morimura SA. Rapid multiplex PCR assay for species identification of Asian Rice Planthoppers (Hemiptera: Delphacidae) and its application to early-Instar Nymphs in paddy fields. PLoS One. 2021;16:e0250471. doi: 10.1371/journal.pone.0250471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Wang YJ, Guo W, Luo D, Wu Y, Kučerová Z, Stejskal V, Opit G, Cao Y, Li FJ, Li ZH. DNA barcoding, species-specific PCR and real-time PCR techniques for the identification of six Tribolium pests of stored products. Sci Rep. 2016;6:1–1. doi: 10.1038/srep28494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Zhang Y, Yang W, Zeng Y, Jiang F, Qin Y, Zhang J, Jiang Z, Hu W, Guo D, Wan J. New species-specific primers for molecular diagnosis of Bactrocera minax and Bactrocera tsuneonis (Diptera: Tephritidae) in China based on DNA barcodes. InSects. 2019;10:447. doi: 10.3390/insects10120447. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data of MtCOI sequences for the accession numbers mentioned in Table 2, which can be retrieved from NCBI database.