Recently, IBM announced a new quantum computer with a 400-plus qubit processor, which helps the realm of quantum computing to attain the next step. The newly developed IBM computer with a 433 qubit processor is called “Osprey.” It is one of the breakthroughs in qubit technology, with 400-plus qubits (Figure 1A). This quantum computer will help in the advancements in quantum computation.1 The example pointed out the developmental effort of quantum computers by the industry. The industry, academia, and scientific communities are putting tremendous efforts into developing next-generation computing systems, quantum computers, to resolve the world’s most challenging problems.
Figure 1.
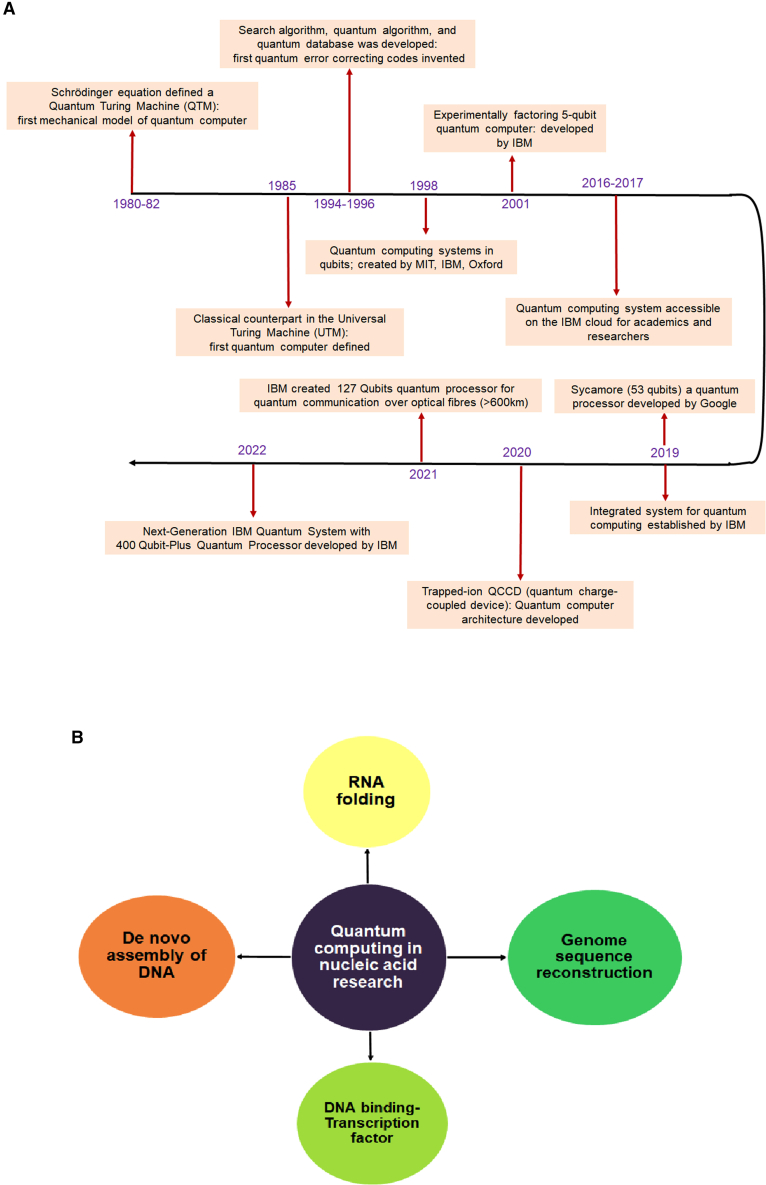
A timeline with milestone achievements of quantum computing
Some problem-solving approaches of quantum computing in nucleic acid research (A). Timeline depicts the milestone achievements of quantum computer. The latest milestone achievement was reported as the IBM computer with the 400-plus Qubits processor. (B) Different emerging and fascinating problem-solving approaches of quantum commuting in nucleic acid research included de novo assembly of DNA, DNA-binding transcription factors, genome sequence reconstruction, RNA folding, etc.
Several advantages
Quantum computers can perform any specific computation much faster than classical computers (ordinary computers). They promise vast information storage and faster processing capacity than classical computers. Using a photonic processor, Madsen et al. carried out Gaussian boson sampling (GBS), which requires only 36μs (Table 1). The researchers also stated that if the work would be conducted with present-day supercomputers and the current best-existing algorithms, it might require more than 9,000 years.2 Another GBS experiment was performed by Bulmer et al. They completed the exponent in minutes. The equal task might need 600 million years for a state-of-the-art classical supercomputer to simulate using the best current classical algorithms.3 Using quantum computers, Arute et al. carried out another experiment that took about 200s to compute. The equivalent problem might require approximately 10,000 years to be completed by a modern classical supercomputer.4 All the above research informed the vivid increase in the speed of quantum computers. Other than the speed, these computers have several other properties. It can compute through the quantum computer’s properties like interference, entanglement, and superposition and finally use a vast computational space.5 In the era of quantum computer revolution, researchers are trying to develop new hardware to improve quantum computing. At the same time, new software are required which is compatible with the hardware for the quantum computers. New algorithms, such as variational quantum eigen solver, quantum phase estimation, quantum Fourier transform, quantum Monte Carlo, quantum walks, quantum approximate optimization algorithm, etc., have been developed to solve next-generation problems.6,7,8 These algorithms help quantum computing to solve problems in different new applications such as chemical product design, biochemical systems, energy applications, metallurgy, etc.9,10,11,12
Table 1.
Some terminologies related to quantum computing on nucleic acid research and their illustration
| Sl no. | Some terminologies related to quantum computing on nucleic acid research | Short description |
|---|---|---|
| 1 | Gaussian boson sampling (GBS) | a model for photonic quantum computation; using GBS, photons are measured from a highly entangled Gaussian state |
| 2 | Grover’s search algorithm | a quantum search-related algorithm; it can speed up an unstructured search problem; it can provide a platform for searching an unsorted database |
| 3 | Replica-exchange Monte Carlo (REMC) | the algorithm helps to run Monte Carlo for a certain number of time steps; it maintains χ-independent replicas of a potential solution. |
| 4 | Qubit | also known as a quantum bit; a qubit is a basic unit of quantum information; it is a two-level (or two-state) quantum-mechanical system; the vectors of the qubit are denoted as ├ ǀ0⟩ and ├ ǀ1⟩ |
Towards practical applications in biological science
Recently, scientists have been trying to solve biological science’s emerging and challenging problems with different next-generation computational techniques, and quantum computing is one of them. These critical problems in the different fields of biological science have been studied from time to time using quantum computing. Chemistry is currently described as one of the potential and promising science subjects with several applications near-term to quantum computers.. At the same time, quantum chemistry is trying to solve the different problems in the area of biochemistry. Quantum chemistry is solving complicated problems of chemistry, especially biochemistry, such as molecular recognition, protein folding, structural biology, advanced drug discovery, etc.9,13,14,15 One of the most promising fields is quantum computational chemistry, a bright field for solving biochemical problems. It uses the knowledge of both computational chemistry and quantum computing.16
Quantum computing in nucleic acid research
Presently, quantum commuting is trying to solve the emerging and fascinating problems of nucleic acid research, such as de novo assembly of DNA, DNA-binding transcription factors, genome sequence reconstruction, RNA folding, and many more (Figure 1B).
De novo assembly of DNA
In the de novo assembly of DNA, its genome sequence is reconstructed where no reference genome is available. De novo assembly is currently used in cancer and transcriptome studies and has promising applications in personalized medicine. Recently, researchers have attempted to address the de novo assembly of DNA using quantum computing. Boev et al. have performed genome assembly using a quantum and quantum-inspired annealing platform. Genome assembly of the φX174 bacteriophage was performed using this method. The work has also shown the effectiveness of the problem-solving capacity of bioinformatics using quantum computing.17
Similarly, Sarkar et al. reconstructed a reference-free DNA sequence through the quantum annealing technique. The researchers have entailed the method of de novo assembly as QuASeR. They have used the reconstructed sequence using a D-Wave quantum annealing simulator, a gate-based quantum simulator, and quantum hardware systems.18 However, there are several limitations of existing quantum hardware. Recent progress of quantum computers may solve and upgrade the quantum hardware.
DNA-binding transcription factors
Another significant problem of biology is determining the DNA transcription factor’s binding sites. Scientists are trying to use quantum computation to study the DNA-binding transcription factor. Li et al. used quantum computation to predict the binding site specificity when they used machine learning models. Using the quantum machine learning approach, researchers used a small number of essential datasets of DNA sequences and trained with the (commercially) available quantum annealer. It helps the transcription factor binding to classify and rank. The results were compared with state-of-the-art standard approaches such as simulated quantum annealing, simulated annealing, etc.19
Genome sequence reconstruction
Genome sequence reconstruction is another exciting area today. The problem has been tried to address using quantum computers. Sarkar et al. have developed QiBAM, a new quantum algorithm for genome sequence reconstruction in quantum computation area. Based on Hamming distances, the algorithm uses approximate pattern matching. It might be the extension of Grover’s search algorithm (Table 1).20
RNA folding
Determination of the secondary structure of RNA is a significant problem in computational biology. Scientists are trying to develop the RNA folding or secondary structure of RNA using the quantum computer. It is an nondeterministic polynomial (NP)-complete computational problem. Fox et al. attempted to develop the secondary structure of RNA using the quantum computer. Here, the researchers developed the binary quadratic model (BQM) to determine the secondary structure of RNA on a D-wave quantum annealer. In this study, the replica-exchange Monte Carlo (REMC) search algorithm was applied to investigate the RNA folding landscape (Table 1).21
Conclusion
Along with the massive progress in bioinformatics and computational biology in the last three decades, several challenges remain unsolved in nucleic acid research. Researchers have shown interest in solving the problem of nucleic acid research using quantum computation. Although quantum technology is in the very early stage of development, the area looks very promising.
With the participation of scientific communities, industry, and academia, the remarkable progress of quantum computing has been noted in the last few years. Simultaneously, huge investments are observed from governments from different countries and the companies such as IBM, Intel, Google, and Microsoft. In the same direction, several start-up companies have been formed along with venture-capital firms.22 Immediately, we need more quantum computing algorithms to unfold nucleic acid research problems. Finally, we urge the scientists from the quantum-computing community to utilize classical technologies and programs such as simulation, artificial intelligence (AI), machine learning (ML), and deep learning (DL) to solve critical problems in the field of nucleic acid research.
Data availability
The authors confirm that the data supporting the findings of this study are available within the article.
References
- 1.Newsroom I.B.M. 2022. IBM Unveils 400 Qubit-Plus Quantum Processor and Next-Generation IBM Quantum System Two.https://newsroom.ibm.com/2022-11-09-IBM-Unveils-400-Qubit-Plus-Quantum-Processor-and-Next-Generation-IBM-Quantum-System-Two [Google Scholar]
- 2.Madsen L.S., Laudenbach F., Askarani M.F., Rortais F., Vincent T., Bulmer J.F.F., Miatto F.M., Neuhaus L., Helt L.G., Collins M.J., et al. Quantum computational advantage with a programmable photonic processor. Nature. 2022;606:75–81. doi: 10.1038/s41586-022-04725-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bulmer J.F.F., Bell B.A., Chadwick R.S., Jones A.E., Moise D., Rigazzi A., Thorbecke J., Haus U.U., Van Vaerenbergh T., Patel R.B., et al. The boundary for quantum advantage in Gaussian boson sampling. Sci. Adv. 2022;8 doi: 10.1126/sciadv.abl9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arute F., Arya K., Babbush R., Bacon D., Bardin J.C., Barends R., Biswas R., Boixo S., Brandao F.G.S.L., Buell D.A., et al. Quantum supremacy using a programmable superconducting processor. Nature. 2019;574:505–510. doi: 10.1038/s41586-019-1666-5. [DOI] [PubMed] [Google Scholar]
- 5.Oliver W.D. Quantum computing takes flight. Nature. 2019;574:487–488. doi: 10.1038/d41586-019-03173-4. [DOI] [PubMed] [Google Scholar]
- 6.Bauer B., Bravyi S., Motta M., Kin-Lic Chan G. Quantum algorithms for quantum chemistry and quantum materials science. Chem. Rev. 2020;120:12685–12717. doi: 10.1021/acs.chemrev.9b00829. [DOI] [PubMed] [Google Scholar]
- 7.Dong Y., Lin L., Tong Y. Ground-state preparation and energy estimation on early fault-tolerant quantum computers via quantum eigenvalue transformation of unitary matrices. PRX Quantum. 2022;3 [Google Scholar]
- 8.Kendon V., Tregenna B. Decoherence can be useful in quantum walks. Phys. Rev. 2003;67 [Google Scholar]
- 9.Cheng H.P., Deumens E., Freericks J.K., Li C., Sanders B.A. Application of quantum computing to biochemical systems: a look to the future. Front. Chem. 2020;8 doi: 10.3389/fchem.2020.587143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spurgeon S.R., Ophus C., Jones L., Petford-Long A., Kalinin S.V., Olszta M.J., Dunin-Borkowski R.E., Salmon N., Hattar K., Yang W.C.D., et al. Towards data-driven next-generation transmission electron microscopy. Nat. Mater. 2021;20:274–279. doi: 10.1038/s41563-020-00833-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andersson M.P., Jones M.N., Mikkelsen K.V., You F., Mansouri S.S. Quantum computing for chemical and biomolecular product design. Current Opinion in Chemical Engineering. 2022;36 [Google Scholar]
- 12.Paudel H.P., Syamlal M., Crawford S.E., Lee Y.-L., Shugayev R.A., Lu P., Ohodnicki P.R., Mollot D., Duan Y. Quantum computing and simulations for energy applications: review and perspective. ACS Eng. Au. 2022;2:151–196. [Google Scholar]
- 13.Baiardi A., Christandl M., Reiher M. Quantum computing for molecular biology. Chembiochem : a European journal of chemical biology. 2023:e202300120. doi: 10.1002/cbic.202300120. [DOI] [PubMed] [Google Scholar]
- 14.Montenegro-Pohlhammer N., Kuppusamy S.K., Cárdenas-Jirón G., Calzado C.J., Ruben M. Computational demonstration of isomer- and spin-state-dependent charge transport in molecular junctions composed of charge-neutral iron(II) spin-crossover complexes. Dalton Trans. 2023;52:1229–1240. doi: 10.1039/d2dt02598a. [DOI] [PubMed] [Google Scholar]
- 15.Jinich A., Sanchez-Lengeling B., Ren H., Harman R., Aspuru-Guzik A. A mixed quantum chemistry/machine learning approach for the fast and accurate prediction of biochemical redox potentials and its large-scale application to 315 000 redox reactions. ACS Cent. Sci. 2019;5:1199–1210. doi: 10.1021/acscentsci.9b00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McArdle S., Endo S., Aspuru-Guzik A., Benjamin S.C., Yuan X. Quantum computational chemistry. Rev. Mod. Phys. 2020;92 [Google Scholar]
- 17.Boev A.S., Rakitko A.S., Usmanov S.R., Kobzeva A.N., Popov I.V., Ilinsky V.V., Kiktenko E.O., Fedorov A.K. Genome assembly using quantum and quantum-inspired annealing. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-88321-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sarkar A., Al-Ars Z., Bertels K. QuASeR: quantum Accelerated de novo DNA sequence reconstruction. PLoS One. 2021;16 doi: 10.1371/journal.pone.0249850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li R.Y., Di Felice R., Rohs R., Lidar D.A. Quantum annealing versus classical machine learning applied to a simplified computational biology problem. npj Quantum Inf. 2018;4 doi: 10.1038/s41534-018-0060-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sarkar A., Al-Ars Z., Almudever C.G., Bertels K.L.M. QiBAM: approximate sub-string index search on quantum accelerators applied to DNA read alignment. Electronics. 2021;10:2433. [Google Scholar]
- 21.Fox D.M., MacDermaid C.M., Schreij A.M.A., Zwierzyna M., Walker R.C. RNA folding using quantum computers. PLoS Comput. Biol. 2022;18 doi: 10.1371/journal.pcbi.1010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeng W., Johnson B., Smith R., Rubin N., Reagor M., Ryan C., Rigetti C. First quantum computers need smart software. Nature. 2017;549:149–151. doi: 10.1038/549149a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article.