We show that tumor-derived IL-6 blocks the development of type 1 classical dendritic cells through the induction of C/EBPβ in the common dendritic cell progenitor, which supports Zeb2 expression in the −165 kb enhancer, thereby preventing pre-cDC1 specification.
Abstract
Cytokines produced in association with tumors can impair antitumor immune responses by reducing the abundance of type 1 conventional dendritic cells (cDC1), but the mechanism remains unclear. Here, we show that tumor-derived IL-6 generally reduces cDC development but selectively impairs cDC1 development in both murine and human systems through the induction of C/EBPβ in the common dendritic cell progenitor (CDP). C/EBPβ and NFIL3 compete for binding to sites in the Zeb2 −165 kb enhancer and support or repress Zeb2 expression, respectively. At homeostasis, pre-cDC1 specification occurs upon Nfil3 induction and consequent Zeb2 suppression. However, IL-6 strongly induces C/EBPβ expression in CDPs. Importantly, the ability of IL-6 to impair cDC development is dependent on the presence of C/EBPβ binding sites in the Zeb2 −165 kb enhancer, as this effect is lost in Δ1+2+3 mutant mice in which these binding sites are mutated. These results explain how tumor-associated IL-6 suppresses cDC1 development and suggest therapeutic approaches preventing abnormal C/EBPβ induction in CDPs may help reestablish cDC1 development to enhance antitumor immunity.
Graphical Abstract
Introduction
Type 1 conventional dendritic cells (cDC1s) are important for priming CD8+ T cells against many viruses and tumors due to their highly efficient cross-presentation of cell-associated antigens (Böttcher and Reis e Sousa, 2018; Anderson et al., 2018). The specialized ability of this subset for cross-presentation was first recognized by in vitro induction of T cell proliferation of DCs loaded with antigens in vivo (den Haan et al., 2000). The availability of mouse models in which cDC1s fail to develop confirmed their critical role in both antiviral and antitumor immunity (Hildner et al., 2008; Durai et al., 2019). Development of cDC1 relies on a transcriptional cascade involving several transcription factors, including Nfil3, Zeb2, Id2, Batf3, and Irf8 (Grajales-Reyes et al., 2015; Bagadia et al., 2019; Liu et al., 2022). cDC1 development is initiated within the common DC progenitor (CDP; Naik et al., 2007; Onai et al., 2007) and proceeds through stages of specification and commitment for the cDC1 and cDC2 lineages in both mouse and human (Grajales-Reyes et al., 2015; Schlitzer et al., 2015; Breton et al., 2015). In the mouse, committed pre-cDC1 progenitors were identified at homeostasis by two studies that described different approaches for their isolation (Grajales-Reyes et al., 2015; Schlitzer et al., 2015). One study emphasized the importance of the intermediate level of cKit expression for the pre-cDC1 progenitor (Grajales-Reyes et al., 2015), while the other was agnostic to cKit, but emphasized the lack of Ly6C and SiglecH (Schlitzer et al., 2015). The relevance of these differences for identifying cDC1 progenitors in various settings beyond homeostasis has not been examined.
Recent studies have found that the cDC1 subset can be reduced or depleted in the setting of certain tumors (Meyer et al., 2018; Hegde et al., 2020; Lin et al., 2020). In one study, the reduction in cDC1 was caused by loss of the specified pre-cDC1 progenitor using only one of the definitions of the pre-cDC1 progenitor (Grajales-Reyes et al., 2015). Reduced cDC1 development in this study was associated with impaired antitumor immune responses and was present in both the PyMT-B6 mammary tumor model and the KPC (p48-Cre; LSL-KrasG12D Trp53flox/+) model of pancreatic cancer (Meyer et al., 2018). Tumor cell production of granulocyte-colony stimulating factor (G-CSF) was reported as responsible for inhibition of cDC1 development in the PyMT-B6 tumor model, which correlated with reduced interferon regulatory factor 8 (IRF8) expression in the bone marrow (BM) progenitors. Antitumor responses could be restored by increasing cDC1 production through the administration of FMS-like tyrosine kinase 3 ligand (Flt3L; Hegde et al., 2020). However, this study did not identify the precise mechanism by which the pre-cDC1 was eliminated.
Another study also observed significantly reduced cDC1 numbers occurring in the KPC pancreatic cancer model (Pdx1-Cre; KrasLSL-G12D/+ Trp53LSL-R172H/+) that resulted from elevated serum IL-6 (Lin et al., 2020). This study attributed the loss of cDC1 to increased cell death of mature cDC1 in peripheral tissues and not to a defect in cDC1 development. However, this study identified pre-cDC1s as defined by negative Ly6C and SiglecH expression (Schlitzer et al., 2015) and not on the basis of cKit levels (Grajales-Reyes et al., 2015). Thus, while these studies agree that cDC1s are reduced in the KPC pancreatic cancer model, they disagree on the mechanism. However, this disagreement relies on distinct gating schemes to identify pre-cDC1, which were defined only under homeostatic conditions.
We recently identified novel roles for CCAAT-enhancer-binding protein (C/EBP) transcription factors in the divergence of the CDP into pre-cDC1 and pre-cDC2 progenitors (Liu et al., 2022). We showed that C/EBPα and C/EBPβ bind to −165 kb enhancer of Zeb2 (Zinc-finger E-box binding homeobox 2; Huang et al., 2021) and act to support cDC2 development and inhibit pre-cDC1 development by blocking binding of the repressor nuclear factor, IL-3 regulated (NFIL3; Liu et al., 2022). C/EBPβ was identified as both being induced by IL-6 as well as inducing Il6 transcription (Akira et al., 1990). However, the ability of IL-6 to alter C/EBPβ expression in DC progenitors has not been studied and its impact on cDC1 development is unclear. Therefore, we wondered whether the systemic IL-6 observed in certain murine cancer models (Meyer et al., 2018; Lin et al., 2020) could act to induce C/EBPβ in BM progenitors and thereby interfere with NFIL3-dependent cDC1 specification.
Results
Tumor-derived IL-6 inhibits cDC1 development
We first tested whether IL-6 impacted in vitro cDC1 development from sort-purified BM progenitors (Fig. 1, A–D). In mouse CDPs, IL-6 caused a dose-dependent reduction in cDC1 development with complete abrogation at 5 ng/ml (Fig. 1, A and B). IL-6 caused a slight reduction in total cDCs but selectively abrogated cDC1 while sparing cDC2 development (Fig. 1 B). In mouse monocyte-dendritic cell progenitors (MDPs), IL-6 similarly abrogated cDC1 development but also induced Ly6C+ M-CSFR+ MerTK− monocyte development at the expense of total cDCs (Fig. S1 A). We next asked if IL-6 affected human cDC1 development. cDC1 can be generated from CD34+ human cord blood cells cultured with stem cell factor (SCF), Flt3L, granulocyte-macrophage colony-stimulating factor (GM-CSF), and IL-4 (Fig. 1 C). We found that addition of IL-6 caused a dose-dependent inhibition of cDC1 in these conditions (Fig. 1, C and D).
Figure 1.
IL-6 suppresses cDC1 development from murine and human progenitors. (A) Sort-purified CDPs cultured with Flt3L and IL-6 (0, 1, 5, and 25 ng/ml) for 4 d and analyzed. Shown is FACS analysis of MerTK− B220− SiglecH− cells. Data are representative of four independent experiments. (B) The bar-scatter graphs show average cDC frequencies in the single cell gate (left, gray) and cDC1 (right, red) or cDC2 (right, blue) in total cDCs of the indicated conditions (average % ± SD, n = 4). (C) Analysis of human cDC1 differentiated from umbilical cord blood–derived CD34+ progenitors cultured with SCF (20 ng/ml), GM-CSF (20 ng/ml), IL-4 (20 ng/ml), Flt3L (100 ng/ml), and various concentrations of IL-6 (0, 1, and 5 ng/ml) for 14 d. (D) The bar-scatter graphs show average cDC1 frequencies in the single cell gate of the indicated conditions (average % ± SD, n = 3). (E and F) Analysis of cDCs in the (E) spleen and (F) MLNs of Zbtb46egfp/+ mice injected i.p. with PBS, EL4-empty, or EL4-IL-6 tumor (106 cells) on day 14 after injection. The histogram for the splenic cDCs is representative of two independent experiments. (G and H) Sort-purified OT-I and OT-II (2.5 × 105 cells of each) were transferred i.v. into WT or Irf8 +32−/− mice inoculated 14 d earlier with PBS, EL4-empty, or EL4-IL-6 tumors. OVA-loaded splenocytes (5 × 105 cells/mouse) were transferred i.v. after 3 h. (G) In vivo proliferation of OT-I (upper panel) and OT-II (lower panel) on day 3 after transfer. (H) Bar-scatter graphs from G for OT-I (upper) and OT-II (lower) of the indicated conditions (average % ± SD). Individual mice are shown as dots. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (Student’s t test).
Figure S1.
Generation and validation of IL-6 expressing EL4 tumor in vitro and in vivo. (A) Sort-purified MDPs were cultured with Flt3L with or without 25 ng/ml IL-6 and analyzed by FACS after 7 d. Shown is FACS analysis of MerTK− B220− SiglecH− cells. Data is representative of four independent experiments. (B) EL4-empty and EL4-IL-6 cell lines were treated with brefeldin A (Sigma-Aldrich) for 3 h and analyzed by FACS for intracellular staining of IL-6 and EGFP. Rat IgG1 antibody was used for isotype control (ctl) staining. (C) Sort-purified cKithi BM progenitors were cultured for 9 d with Flt3L and the indicated concentration of conditioned media (CM) from EL4-empty or EL4-IL-6 cells or recombinant murine IL-6. (D and F) Related to Fig. 1 E, the bar-scatter graphs show the average (D) frequencies (%) and (F) absolute number of cDC in the spleen ± SD. (E and G) Related to Fig. 1 F, the bar-scatter graphs show the average (E) frequencies (%) and (G) absolute number of cDC in the MLNs ± SD. cDC total (gray and/or white), cDC1 (red), and cDC2 (blue). The numbers of mice for each experimental group were indicated as dots in the bar graphs, PBS (n = 7), EL4-empty (n = 9), EL4-IL-6 (n = 11). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (Student’s t test).
To test whether IL-6 influences cDC1 development in vivo, we generated an EL4 lymphoblast cell line that expresses IL-6 by stable retroviral transduction (EL4-IL-6) and a control EL4 cell line (EL4-empty). IL-6 production was validated by intracellular staining (Fig. S1 B). Conditioned media (CM) from EL4-IL-6, but not EL4-empty, suppressed in vitro cDC1 development as above (Fig. S1 C). We inoculated EL4-IL-6 and EL4-empty into Zbtb46egfp/+ mice, in which EGFP is expressed in cDC1 progenitors and cDC1s, and tested for loss of cDC1 (Fig. 1, E and F; and Fig. S1, D–G). Notably, we found a severe reduction in splenic cDC1 specific to EL4-IL-6–inoculated mice with an overall reduction in total cDC numbers including a partial reduction of cDC2 (Fig. 1 E; and Fig. S1, D and F). A similar and severe reduction in resident cDC1 numbers was also observed in mesenteric lymph nodes (MLNs; Fig. 1 F; and Fig. S1, E and G).
To test the biological relevance of reduced cDC1 numbers caused by EL4-IL-6, we measured in vivo priming of OT-I and OT-II cells in response to immunization with cell-associated antigens (Fig. 1, G and H). We reported recently that cDC1 is required for both CD8 and CD4 T cell priming in response to cell-associated antigens (Ferris et al., 2020). Mice harboring EL4-IL-6 showed a nearly complete loss of in vivo OT-I and OT-II proliferation, in contrast to the robust OT-I and OT-II proliferation in mice bearing EL4-empty tumors or PBS-treated control mice (Fig. 1, G and H). These results indicate that the loss of cDC1 caused by IL-6 causes a biologically relevant defect in T cell priming in response to cell-associated antigens.
IL-6 blocks CDP specification toward pre-cDC1
Two studies identified pre-cDC1 progenitors in BM by FACS using different approaches (Grajales-Reyes et al., 2015; Schlitzer et al., 2015). One study identified the pre-cDC1 as either lin− SiglecH− Flt3+ CD11c+ cKitint MHCIIint CD24+ BM cells, or (using the Zbtb46egfp reporter mouse) as lin− SiglecH− Flt3+ CD11c+ cKitint MHCIIint Zbtb46-EGFP+ BM cells (Grajales-Reyes et al., 2015). By contrast, a second study defined the pre-cDC1 as lin− CD11c+ MHCII− Flt3+ Sirpα− SiglecH− Ly6C− BM cells (Schlitzer et al., 2015). These alternative definitions differ in their use of cKit expression, the level of MHCII expression, and by reliance on either CD24 or Zbtb46-EGFP in place of Sirpα and Ly6C.
Previously we reported that the pre-cDC1 is an MHCIIint BM population (Grajales-Reyes et al., 2015). In agreement, we now find that the Lin− cKitint Zbtb46-EGFP+ BM population, which contains CDPs committed to the cDC1 lineage (Bagadia et al., 2019) as well as the previously defined pre-cDC1 (Grajales-Reyes et al., 2015), is composed of both MHCII+ and MHCII− cells (Fig. S2 A). Thus, defining pre-cDC1 as MHCII− BM cells (Schlitzer et al., 2015) may reduce the yield of pre-cDC1, depending on the stringency for excluding MHCIIint progenitors.
Figure S2.
Reduction of DC and monocyte progenitors in BM by EL4-IL-6. (A) Lin− BM from Zbtb46egfp/+ mice was analyzed by FACS. cKitint Zbtb46-EGFP+ cells were examined for MHCII expression. Numbers represent the percentage of cells in the indicated gate. (B) Zbtb46egfp/+ mice were inoculated i.p. with PBS, EL4-empty, or EL4-IL-6 (106 cells) as described in Fig. 2. BM cells were analyzed by FACS on day 14 after inoculation. Shown are pre-cDC1 progenitors defined as gate R1 in Fig. 2 A (Grajales-Reyes et al., 2015). (C) Bar-scatter graphs for absolute number of pre-cDC1 from Fig. 2, F and G (average # ± SD). Cells were enumerated from total BM cells collected from two femurs and tibias. Numbers of mice for each experimental group are indicated as dots. (D) Zbtb46egfp/+ mice injected i.p. with PBS, EL4-empty, or EL4-IL-6 (106 cells), and the BM progenitors were analyzed on day 14 after inoculation. Shown are analyses for MDP (cKithi Flt3+ M-CSFR+; red gate), CDP (cKitint Flt3+ M-CSFR+ MHCII− CD11c−; blue gate) and monocyte progenitors (cKit+ Flt3− M-CSFR+), mostly cMoPs (black gate). Data shown are representative of two independent experiments. (E) Bar-scatter graphs show the frequency of MDP (red), CDP (blue), and monocyte progenitors (cKithi, gray bars; ckitint, white bars) as a fraction of Lin− BM cells for the mice in D (average % ± SD). Each dot represents an individual mouse for PBS (n = 7), EL4-empty (n = 9), EL4-IL-6 (n = 11). *P < 0.05, **P < 0.01, ****P < 0.0001 (Student’s t test).
We also asked whether cKit expression was an important factor in defining the pre-cDC1 (Fig. 2, A–C). The pre-cDC1 (R1) defined by cKitint expression (Grajales-Reyes et al., 2015) generated 95% cDC1, 5% cDC2, and 0% plasmacytoid DCs (pDCs) when cultured in Flt3L (Fig. 2, A and C). By contrast, the pre-cDC1 (R2), defined as SiglecH− Ly6C− pre-cDCs (Schlitzer et al., 2015), was heterogeneous for cKit expression, containing equal components of cKit− and cKitint populations (Fig. 2 B). We examined the lineage potential of these populations in the R2 gate using cell sorting and Flt3L cultures (Fig. 2 C). The cKitint fraction of this pre-cDC1 (R3) was uniformly positive for Zbtb46-EGFP expression and generated 96% cDC1 in Flt3L cultures, whereas the cKit− fraction (R4) generated pDCs, cDC2, and MerTK+ cells, but no cDC1 (Fig. 2, B and C). Thus, the exclusion of cKit− BM cells and the positive expression of Zbtb46 might be important in defining the pre-cDC1 progenitor, as reported (Grajales-Reyes et al., 2015).
Figure 2.
IL-6 suppresses development of pre-cDC1. (A) pre-cDC1 (R1) in Zbtb46egfp/+ mice defined by Grajales-Reyes et al. (2015). (B) Upper panel: Pre-cDC1 (R2) in Zbtb46egfp/+ mice defined by Schlitzer et al. (2015). Lower panel: Cells in R2 were analyzed for cKit and Zbtb46-EGFP expression (R3 and R4). A bar-scatter graph shows the percentage (average % of R2 ± SD) of cKitint R2 cells (R3) and cKitlo R2 cells (R4). (C) Sort-purified cells from gate R1 in A and R3 and R4 in B were cultured in Flt3L for 3 d and analyzed by FACS. Arrows indicate the order of gating for successive histograms. Results are representative of three independent experiments. (D) Zbtb46egfp/+ mice were inoculated i.p. with PBS, EL4-empty, or EL4-IL-6 tumor cells (106 cells). BM cells were analyzed by FACS after 7, 10, 12, and 14 d as indicated. Numbers represent the percentage of Lin− BM cells in the indicated gate (cKitint Zbtb46-EGFP+). (E) Shown are bar-scatter graphs for mice from D for the frequency of pre-cDC1 in Lin− BM (average % ± SD). (F) Pre-cDC1 progenitor as defined in Grajales-Reyes et al. (2015) (R1 in A) from Zbtb46egfp/+ mice at 14 d after inoculation. (G) Upper panel: Pre-cDC1 progenitor defined in Schlitzer et al. (2015) (R2 in B) from Zbtb46egfp/+ mice at 14 d after inoculation. Lower panel: Analysis of R2 in upper panel for cKit and Zbtb46-EGFP+. (H) Bar-scatter graphs for frequency of pre-cDC1 as defined in F and G (average % of Lin− BM ± SD). Individual mice are indicated as dots. *P < 0.05, **P < 0.01, ****P < 0.0001 (Student’s t test).
We next used Zbtb46egfp reporter mice to test if tumor-derived IL-6 also blocks pre-cDC1 specification in vivo (Fig. 2, D–H). Lin− Kitint Zbtb46-EGFP+ BM cells, containing pre-cDC1, were selectively reduced by 12 d after tumor inoculation with EL4-IL-6, but not EL4-empty cells (Fig. 2, D and E). Importantly, applying the different gating schemes to define the pre-cDC1 led to a discrepancy regarding whether IL-6 blocks pre-cDC1 specification. Using the pre-cDC1 gate defined as Kitint (R1; Grajales-Reyes et al., 2015), we found that EL4-IL-6 induced a 10-fold reduction compared with EL4-empty tumor cells (Fig. 2, F and H; and Fig. S2, B and C). By contrast, using the pre-cDC1 defined as SiglecH− Ly6C− pre-cDCs (R2; Schlitzer et al., 2015), we found that EL4-IL-6 caused no reduction in cell numbers compared with EL4-empty tumor cells (Lin et al., 2020; Fig. 2, G and H; and Fig. S2 C). Notably, the Kitint component of this pre-cDC1 gate (R2) showed a substantial reduction in Zbtb46-EGFP expression in EL4-IL-6 bearing mice (Fig. 2 G). Thus, we confirm that tumor-associated IL-6 reduces cDC1 abundance as reported (Lin et al., 2020), but we find a blockade in pre-cDC1 specification that was not observed previously. We show that the gating system (Schlitzer et al., 2015) previously used to identify pre-cDC1s (Lin et al., 2020) harbored cKit− BM cells with cDC2 and pDC potential that persist in the presence of IL-6. Tumor-derived IL-6 also altered the MDP (Auffray et al., 2009), CDP, and committed monocyte progenitor (cMoP; Hettinger et al., 2013; Kawamura et al., 2017), which were substantially reduced specifically in EL4-IL-6 bearing mice (Fig. S2, D and E).
IL-6 induces C/EBPβ in CDPs in vitro and in vivo
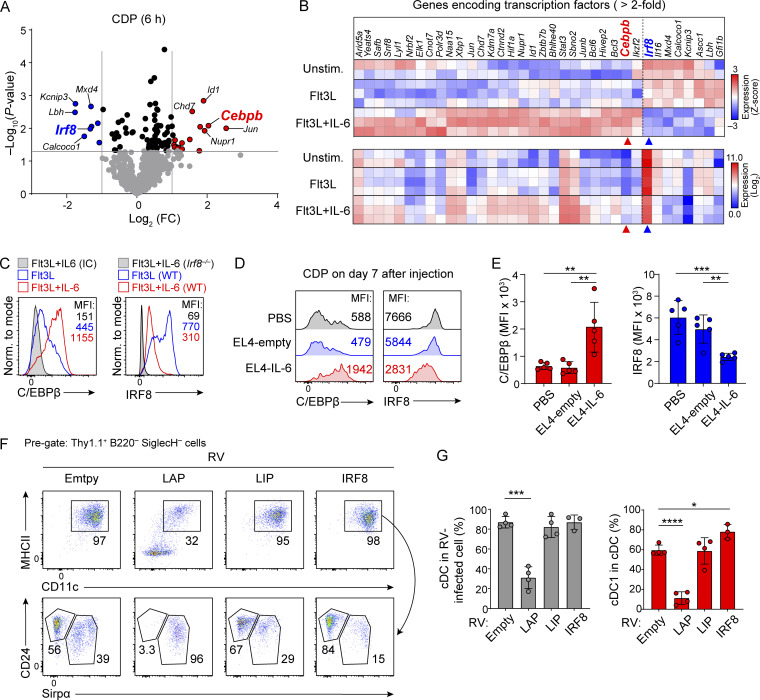
Since pre-cDC1s develop from CDPs (Bagadia et al., 2019), we asked how IL-6 impacts CDP gene expression (Fig. 3 and Fig. S3, A–C). We performed RNA sequencing (RNA-seq) in sort-purified CDPs cultured in vitro in Flt3L with or without IL-6 (Fig. 3 and Fig. S3, A–C). IL-6 increased expression of many genes normally expressed by macrophages (e.g., Socs3), monocytes (e.g., Ms4a6d, Ms4a4, and Fcgr1), and granulocytes (e.g., Mmp19; Gautier et al., 2012; Fig. S3, A and B). In contrast, IL-6 reduced expression of genes normally expressed by cDC1 (e.g., Rnf144b, Clec9a, and Irf8; Miller et al., 2012; Fig. 3 A and Fig. S3, A and B). Notably, Cebpb was among the most highly induced transcription factors with IL-6 treatment (Fig. 3, A and B). Further, C/EBPβ protein expression was also highly induced by IL-6 in CDPs both in vitro (Fig. 3 C) and in vivo in mice harboring EL4-IL-6 tumors, but not EL4-empty tumors (Fig. 3, D and E). By contrast, IL-6 reduced IRF8 expression in CDPs in vitro and in vivo (Fig. 3, A–E). In CDPs cultured in Flt3L alone, IRF8 expression is bimodal, having clear high and low populations. This represents development of both cDC1 and cDC2 cells that express high or low levels of IRF8, respectively. In contrast, CDPs cultured in Flt3L and IL-6 show a uniformly low level of IRF8 expression, consistent with the loss of cDC1 development. Together, these results suggest that IL-6 might block cDC1 development by inducing C/EBPβ.
Figure 3.
IL-6 increases Cebpb expression in CDPs. (A) RNA-seq was performed on CDPs cultured for 6 h with Flt3L alone or with added IL-6. Shown is a volcano plot of differentially expressed transcription factors as the fold change (FC) induced by IL-6. Selected genes with greater than twofold change are highlighted in red (increased) or blue (decreased). (B) Heatmaps of the top 36 differentially expressed transcription factors from A clustered on (upper panel) Z-score or (lower panel) log2 expression value. (C) Intracellular expression of C/EBPβ and IRF8 in sort-purified CDPs treated in vitro with Flt3L (blue) or Flt3L with 25 ng/ml IL-6 (red) for 20 h. IC indicates staining using isotype control antibody. (D) Intracellular expression of C/EBPβ and IRF8 in CDPs from BM of Zbtb46egfp/+ mice analyzed 7 d after tumor inoculation i.p. as indicated. (E) Bar-scatter graphs show the geometric mean fluorescence intensity (MFI) ± SD for experiments shown in E for C/EBPβ (red) and IRF8 (blue). (F) Sort-purified CDPs were transduced in vitro with retroviral vectors (RV) encoding LAP, LIP, or IRF8, cultured in Flt3L for 4 d, and analyzed for FACS. Shown are Thy1.1+ B220− SiglecH− cells for expression of MHCII, CD11c, CD24, and Sirpα. Results are representative of four independent experiments. (G) Scatter plots with error bars for cells described in F for total MHCII+ CD11c+ cells (left panel) or for MHCII+ CD11c+CD24+ Sirpα− cells (right panel; average % ± SD). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (Student’s t test).
Figure S3.
Increases of C/EBPβ expression and bindings at Zeb2 −165 kb in CDP by IL-6. (A) RNA-seq was performed on CDPs cultured for 6 h with Flt3L alone or with added IL-6 (25 ng/ml) as described in Fig. 3. Shown is a volcano plot of differentially expressed genes as fold change (FC) induced by IL-6. Selected genes with greater than threefold changes are highlighted in red (increased) or blue (decreased). (B) Shown are genes from A with greater than threefold decrease (blue) or greater than threefold increase in expression (red) in response to IL-6 in CDPs after 6 h. (C) Heatmap of top 101 differentially expressed genes from A clustered on log2 expression value. (D and E) Characterization of Hoxb8-transduced BM progenitor cell line. (D) FACS analysis of Hoxb8 cells and BM progenitors. (E) Intracellular expression of C/EBPβ in Hoxb8 cells cultured with or without IL-6 (25 ng/ml) for 22 h. Isotype control (IC) is a mouse IgG2a. (F) cDC differentiation from Hoxb8 cell line cultured with or without IL-6 (25 ng/ml) for 7 d. (G and H) CUT&RUN analysis for C/EBPβ bindings to the Zeb2 locus in M-CSFR+ Hoxb8 cells cultured in Flt3L with or without IL-6 (25 ng/ml). (G) A snapshot of UCSC genome browser (chr2:44,912,814-45,315,672). Zeb2 −165 kb enhancer is highlighted in yellow. (H) A scatter plot shows total tag counts normalized to IC experimental group in the Zeb2 −165 kb enhancer. Peak calling was performed with HOMER command findPeaks.
We recently reported that C/EBPβ opposed cDC1 specification by competing with NFIL3 for binding to sites in the Zeb2 −165 kb enhancer (Liu et al., 2022). Cebpb encodes both activating (liver-enriched activator proteins, LAP* and LAP) and inhibitory (liver-enriched inhibitory protein, LIP) isoforms (Descombes and Schibler, 1991), which are produced by alternative translation of the same mRNA (Calkhoven et al., 2000). To identify which isoform inhibits cDC1 development, we compared Cebpb cDNAs that encode either LAP or LIP separately for their impact on cDC1 development (Fig. 3, F and G). We found that expression of LAP, but not LIP, specifically inhibited cDC1 development, suggesting that IL-6–induced C/EBPβ acts to block cDC1 development.
IL-6–induced Cebpb blocks cDC1 development by activating the Zeb2 −165 kb enhancer
To test whether the IL-6 inhibition of cDC1 development operates through the support of Zeb2 expression by C/EBPβ, we examined the requirements for C/EBPβ binding sites in mediating the effects of IL-6. For this analysis, we used recently reported Δ1+2+3 mice, which have simultaneous mutations of the three C/EBP/NFIL3 binding sites in the Zeb2 −165 kb enhancer (Liu et al., 2022). First, we used reporter constructs to examine the effects of IL-6 on the activity of the Zeb2 −165 kb enhancer in CDPs in vitro (Fig. 4, A and B), as previously described (Durai et al., 2019; Liu et al., 2022). Briefly, we used an integrating retroviral reporter in which the EGFP reporter cassette is encoded in the bottom retroviral strand in opposite orientation to the retroviral long terminal repeat. The EGFP reporter is driven by a minimal CMV promoter (CMVmin) placed adjacent to various enhancer elements (Fig. 4 A). Sort-purified CDPs were infected with retrovirus and cultured in vitro with Flt3L with or without the addition of IL-6, and cDC development and EGFP reporter activity are assessed after 2 d. We find that IL-6 strongly increases the EGFP reporter activity of the native −165 kb enhancer (WT) in all populations. By contrast, mutation of the three C/EBP binding sites in the −165 kb enhancer (Δ1+2+3; Liu et al., 2022) reduced overall activity and prevented the increase in activity induced by IL-6 (Fig. 4, A and B).
Figure 4.
IL-6 blocks cDC1 development by activating the Zeb2 −165 kb enhancer. (A) Upper panel: Retroviral reporter construct: Zeb2 −165 kb enhancer (yellow), CMV minimal promoter (red arrow), and EGFP (green). Lower panel: Sort-purified CDPs were transduced with reporter constructs harboring WT or Δ1+2+3 enhancers and cultured for 2 d with 5% Flt3L with or without 25 ng/ml IL-6, and EGFP expression assessed in Thy1.1+ cells. Numbers represent the MFI of cells. LTR, long terminal repeat. (B) A bar-scatter graph of three independent experiments from A for EGFP expression (average MFI ± SD). ***P < 0.001 (Student’s t test). (C) Sort-purified CDPs from WT or Δ1+2+3 mice were cultured in vitro with Flt3L and the indicated concentrations of IL-6 for 4 d. Shown is FACS analysis for MerTK− B220− SiglecH− cells. Numbers indicate the percentage of cells in the indicated gates. Data shown are representative of two independent similar experiments. (D) Sort-purified CDPs from WT or Δ1+2+3 mice were cultured in vitro with Flt3L alone (blue) or with 25 ng/ml IL-6 (red) for 20 h and analyzed for intracellular C/EBPβ expression. IC indicates staining with isotype control antibody. Number indicates MFI. (E and F) WT and Δ1+2+3 mice were inoculated with EL4-empty or EL4-IL-6 tumors and analyzed after 14 d. Shown is FACS analysis of (E) splenocytes pre-gated as MerTK− B220− SiglecH− MHCII+ CD11c+ cells and (F) BM DC progenitors pre-gated as Lin− SiglecH− Flt3+ CD11c+ MHCIIlo-int BM cells for cKit and CD24 expression. (G and H) Sort-purified OT-I and OT-II (2.5 × 105 cells, each) were transferred i.v. into WT or Δ1+2+3 mice inoculated 14 d earlier with EL4 tumors. OVA-loaded splenocytes (5 × 105 cells/mouse, i.v.) were transferred i.v. after 3 h. (G) In vivo proliferation of OT-I (upper panel) and OT-II (lower panel) on day 3 after transfer. (H) Bar-scatter graphs from G for OT-I (upper) and OT-II (lower) of the indicated conditions (average % ± SD). Individual mice are shown as dots. *P < 0.05, **P < 0.01, ****P < 0.0001 (Student’s t test).
Thus far, we found that IL-6 induces expression of C/EBPβ mRNA and protein and increases activity of the Zeb2 −165 kb enhancer. To test if IL-6 also increases C/EBPβ binding to the Zeb2 −165 kb enhancer, we considered using CUT&RUN for C/EBPβ (Skene and Henikoff, 2017). However, application of CUT&RUN to CDPs purified directly from BM was infeasible due to the insufficient numbers of cells that could be isolated (Liu et al., 2022). As an alternative approach, we generated CDP-like cell lines by culturing Hoxb8-transformed BM cells (Wang et al., 2006) in SCF and Flt3L (Fig. S3, D–F). First, such Hoxb8-transformed cells express macrophage colony-stimulating factor receptor (M-CSFR), similar to the CDP (Fig. S3 D). Second, they induce C/EBPβ in response to IL-6 treatment (Fig. S3 E), similar to the CDP (Fig. 3 C). Third, they are able to differentiate into pDCs, cDC1s, and cDC2s (Fig. S3 F), also similar to CDPs. And finally, IL-6 completely abrogates cDC1 development from these Hoxb8 cells (Fig. S3 F), similar to the effect of IL-6 on CDPs (Fig. 1, A and B).
For the above reasons, we used CUT&RUN to test the impact of IL-6 on C/EBPβ binding to the Zeb2 −165 kb enhancer in these Hoxb8-transformed CDP-like cells (Fig. S3, G and H). We first confirmed our previous result that C/EBPβ binds the Zeb2 −165 kb enhancer (Fig. S3 G), as we recently reported (Liu et al., 2022). Next, we carried out a quantitative assessment of the tag-counts for C/EBPβ binding to the −165 kb region using the HOMER command findPeaks (Heinz et al., 2010). We find that IL-6 treatment increased the normalized tag-counts for C/EBPβ binding to this region approximately twofold (Fig. S3 H). While these results may not mimic the exact setting occurring in CDPs in vivo, they suggest that increased binding of C/EBPβ to the Zeb2 −165 kb enhancer could act to prevent efficient NFIL3-driven cDC1 specification (Liu et al., 2022).
We next evaluated the direct impact of IL-6 on ZEB2 protein expression. For this, we used the ZEB2-EGFP fusion protein reporter mouse (Zeb2ZEB2-EGFP/+; Nishizaki et al., 2014) that conveys actual levels of ZEB2 protein (Fig. S4 A). We find that ZEB2-EGFP was gradually lost as CDPs were cultured with Flt3L in vitro over time. By contrast, the addition of IL-6 caused ZEB2-EGFP to be maintained over time (Fig. S4 A). Furthermore, IL-6 could completely inhibit in vitro cDC1 development of CDPs from WT mice. However, in Δ1+2+3 mice, IL-6 reduced cDC1 development only partially but allowed between 50 and 70% of cDC1 to develop (Fig. 4 C). Importantly, we show that IL-6 induced C/EBPβ equally in WT and Δ1+2+3 mice (Fig. 4 D). These results indicate that IL-6 represses cDC1 development by inducing C/EBPβ, which acts by binding the Zeb2 −165 kb enhancer and maintaining ZEB2 expression.
Figure S4.
IL-6 blocks cDC1 development by acting the Zeb2 −165 kb Δ1+2+3 enhancer. (A) CDPs from WT mice or Zeb2ZEB2-EGFP/+ reporter mice were sort-purified and cultured with Flt3L with (red) or without (blue) IL-6 (25 ng/ml) for the indicated time, and analyzed by FACS for EGFP expression. Numbers indicate the MFI for EGFP of cells. The line graph (lower left) shows the ratio of the ZEB2-EGFP MFI and WT MFI at each time point. (B) WT and Δ1+2+3 mice were inoculated i.p. with EL4-empty or EL4-IL-6 (106 cells) and analyzed after 14 d. Shown is FACS analysis of splenocytes gated as MerTK− B220− SiglecH− cells. (C and D) Bar-scatter graphs for (C) the splenic MerTK− B220− SiglecH− cells described in B and (D) the splenic MerTK− B220− SiglecH− MHCII+ CD11c+ cells described in the main Fig. 4 F. (E) WT and Δ1+2+3 mice inoculated in B were analyzed for DC progenitors in the BM. Shown is FACS analysis of Lin− BM cells. Numbers indicate the percentage of cells in the indicated gates. (F) Bar-scatter graphs for cells described in E for WT (red) and Δ1+2+3 mice (gray; average % ± SD). *P < 0.05, **P < 0.01, ***P < 0.001 (Student’s t test).
Finally, we validated this mechanism in vivo in the setting of tumor-derived IL-6. We inoculated WT or Δ1+2+3 mice with EL4-IL-6 or EL4-empty tumors and evaluated peripheral cDC1 development (Fig. 4 E and Fig. S4, B–D). EL4-IL-6 tumors reduced peripheral cDCs and cDC1 in WT mice, but not in Δ1+2+3 mice (Fig. S4, B and C). For this analysis, we also crossed Δ1+2+3 mice onto the Zbtb46egfp background (Satpathy et al., 2012) to enhance analysis of BM pre-cDC1 progenitors. Here, EL4-IL-6 tumors caused a severe reduction in Lin− Kitint Zbtb46-GFP+ BM cells in WT mice, but not in Δ1+2+3 mice (Fig. S4 E). In agreement, pre-cDC1 defined Lin− SiglecH− Flt3+ CD11c+ cKitint MHCIIint CD24+ BM cells (Grajales-Reyes et al., 2015) were also reduced in WT mice, but not in Δ1+2+3 mice (Fig. 4 F and Fig. S4 F). In summary, our results show that tumor-derived IL-6 can systemically alter cDC development and strongly abrogate cDC1 specification by elevation of C/EBPβ expression in BM progenitors.
Finally, we tested whether the repression of T cell priming caused by IL-6 (Fig. 1, G and H) was restricted to its blockade of cDC1 development (Fig. 4, G and H). Again, WT mice harboring EL4-IL-6 tumors showed nearly complete loss of OT-I and OT-II proliferation in response to cell-associated antigen. By contrast, Δ1+2+3 mice harboring EL4-IL-6 tumors, which retain normal cDC1 development (Fig. 4 E), also showed normal priming of both OT-I and OT-II in response to cell-associated antigen (Fig. 4, G and H). Thus, the repression of T cell priming caused by IL-6 in WT mice appears to result from the loss of cDC1 rather than a pleiotropic action of IL-6 on other targets.
Discussion
The purpose of this study is to provide the mechanism for a previously reported phenomenon, specifically the loss of cDC1 associated with certain tumor models that result in systemic IL-6 (Lin et al., 2020; Meyer et al., 2018). Understanding this mechanism may be important because other recent studies have suggested that neutralizing IL-6 may improve the effectiveness of checkpoint blockade immunotherapy (Hailemichael et al., 2022; Li et al., 2022). Since checkpoint blockade reportedly relies on the presence of cDC1 (Gubin et al., 2014), understanding the mechanism by which IL-6 leads to reduced cDC1 may be relevant to various settings of tumor immunotherapy. Further, cDC1s are also important in various responses to intracellular pathogens including many viruses, broadening the potential impact of conditions where IL-6 becomes systemic and alters cDC1 development. Thus, our findings reveal the mechanism of how systemic IL-6 impacts cDC1 development, rather than addressing its impact on spontaneous tumorigenesis.
Here, we show that systemic IL-6 induces C/EBPβ expression in CDPs, causing a blockade of normal NFIL3-dependent specification of the pre-cDC1 progenitor. We recently reported that cDC1 specification requires the suppression of Zeb2 in CDPs (Bagadia et al., 2019) and that the suppression of Zeb2 is mediated by transient NFIL3 expression in a fraction of CDPs (Liu et al., 2022). This suppression of Zeb2 by NFIL3 is mediated by binding to three sites in the Zeb2 −165 kb enhancer that is also bound by C/EBPα and C/EBPβ (Liu et al., 2022). Here, we show that the loss of cDC1 development caused by IL-6 does not occur in Δ1+2+3 mice, in which the three C/EBP binding sites in the Zeb2 −165 kb enhancer have been mutated (Liu et al., 2022). These results demonstrate that IL-6 induction of C/EBPβ in CDPs prevents cDC1 development by supporting Zeb2 enhancer beyond the capacity for repression by normal NFIL3 expression.
Further, our results resolve an apparent discrepancy between previous studies that examined cytokine-induced loss of cDC1 in tumor-bearing mice (Meyer et al., 2018; Lin et al., 2020). One study concluded that the defect in development occurred at the CDP stage in BM, with a loss of pre-cDC1 (Meyer et al., 2018). In contrast, a second study concluded that cDC1s were lost by apoptosis in peripheral tissues and observed no reduction pre-cDC1 in BM (Lin et al., 2020). We noticed that these two studies used different methods for identifying the pre-cDC1, one which included consideration of the intermediate level of cKit expression of the pre-cDC1 (Grajales-Reyes et al., 2015) and one which was indifferent to cKit expression (Schlitzer et al., 2015). By directly comparing these distinct definitions for the pre-cDC1, we are able to resolve this apparent discrepancy. The loss of pre-cDC1 in our study was clearly evident using the former method that identified the pre-cDC1 as a cKitint Zbtb46-EGFP+ BM population (Meyer et al., 2018). In contrast, the loss of pre-cDC1 was obscured in our study when using the latter method that identified the pre-cDC1 as a SiglecH− Ly6C− BM pre-cDC indifferent to cKit expression (Lin et al., 2020). We show that the inclusion of cKit− SiglecH− Ly6C− cells gives an inaccurate indication of pre-cDC1 abundance since these cells persist in the setting of tumor-derived systemic IL-6. These of cKit− SiglecH− Ly6C− cells are not cDC1 progenitors, but instead represent pDC and cDC2 progenitors (Fig. 2 C).
Immune checkpoint blockade has brought major therapeutic benefits to the treatment of cancer (Sharma and Allison, 2015). An important component of effective checkpoint blockade is the abundance of the cDC1 subset (Gubin et al., 2014). The contribution of cDC1 to effective checkpoint blockade therapies is likely due to its capacity for cross-presentation of tumor-derived antigens to CD8 T cells (den Haan et al., 2000; Hildner et al., 2008). Recently, the cDC1 was also shown to act in maintaining the population of tumor-specific CD8 T cells capable of undergoing proliferation (Schenkel et al., 2021), a finding that also extends to the situation of chronic viral infection (Dahling et al., 2022). While cDC1s clearly function in priming CD8 T cell responses in secondary lymphoid tissues, they may also act in the tumor microenvironment to further enhance antitumor CD8 T cells (Spranger et al., 2017). In agreement with recent suggestions (Hailemichael et al., 2022; Li et al., 2022), our results imply that neutralization of IL-6 may help prevent the loss of cDC1 caused by conditions such as systemic IL-6 that act to impair the specification of the pre-cDC1 in the BM.
Materials and methods
Mice
WT C57BL6/J (JAX:000664) mice were from The Jackson Laboratory. Zeb2ZEB2-EGFP fusion protein reporter mice (Nishizaki et al., 2014) were provided by RIKEN BioResource Center through the National BioResource Project of the Ministry of Education, Culture, Sports, Science and Technology, Japan (STOCK Zfhxlbtm2.1Yhi). Zbtb46egfp reporter mice (Satpathy et al., 2012) and Δ1+2+3 mice (Liu et al., 2022) were previously reported and kept in-house. MHCI triple KO mice (Kb−/−Db−/−β2m−/−) were originally provided by T. Hansen (Washington University School of Medicine, St. Louis, MO, USA; Lybarger et al., 2003). C57BL/6-Tg(TcraTcrb)1100Mb/J (OT-I, JAX:003831) and B6.Cg-Tg(TcraTcrb)425Cbn/J (OT-II, JAX:004194) mice were obtained from The Jackson Laboratory. Irf8 +32−/− mice were previously described (Durai et al., 2019) and kept in-house. All mice were maintained in a specific pathogen–free animal facility following institutional guidelines with protocols approved by Animal Studies Committee at Washington University in St. Louis. Experiments were performed with mice between 6 and 12 wk of age.
Flow cytometry and antibodies
BM cells and splenocytes were isolated as previously reported (Kim et al., 2020; Liu et al., 2022). Lineage+ cells were depleted using biotinylated anti-mouse CD3Ɛ, anti-mouse CD19, anti-mouse CD105, anti-mouse Ly6G, anti-mouse TER119, and anti-mouse NK1.1 antibodies, and MagniSort streptavidin negative selection bead (Invitrogen) or Mojosort streptavidin nanobeads (BioLegend) as needed. Progenitors were defined as follows: MDP (Lin− cKithi Flt3+ M-CSFR+), CDP (Lin− cKitint Flt3+ M-CSFR+ MHCII−, CD11c−) and cMoP (Lin− cKit+ Flt3− M-CSFR+). Cells were stained as previously described (Kim et al., 2020; Liu et al., 2022). Biotinylated antibodies used for lineage depletion for BM progenitor analysis and sort-purification of OT-I and OT-II T cells are as follows: biotinylated anti-mouse CD3Ɛ (145-2C11), anti-mouse CD19 (6D5), anti-mouse CD45R/B220 ( RA3-6B2), anti-mouse Ly6G (1A8), anti-mouse TER119 (TER-119), anti-mouse NK1.1 (PK136), anti-mouse CD8b (YTS156.7.7), and anti-mouse CD4 (GK1.5) antibodies were purchased from BioLegend. Biotinylated anti-mouse CD105 antibody (MJ7/18) was obtained from Invitrogen. For fluorochrome-conjugated antibodies, PE/Dazzle594-conjugated anti-mouse CD45R/B220 (RA3-6B2), PE-conjugated anti-mouse SiglecH (551), Brilliant Violet (BV) 510-conjugated anti-mouse MHCII (M5/114.15.2), Alexa Fluor (AF) 647- or PE-Cy7-conjugated anti-mouse CD24 (M1/69), APC-conjugated anti-mouse Sirpα (P84), BV421-conjugated anti-mouse XCR1 (ZET), AF647-conjugated anti-mouse CD11b (M1/70), PE-Cy7- or BV711-conjugated anti-mouse M-CSFR (AFS98), BV421-conjugated anti-mouse Ly6C (HK1.4), PE-conjugated anti-mouse Vα2 (B20.1), APC-Cy7-conjugated anti-mouse CD45.1 (A20), PE-Cy7-conjugated anti-mouse CD45.2 (104), APC- or AF700-conjugated anti-mouse/human CD44 (IM7), AF488-conjugated anti-mouse CD45R/B220 (RA3-6B2), BV605-conjugated anti-mouse CD8a (53-6.7), BV711-conjugated anti-mouse CD4 (RM4-5), and PerCP-Cy5.5-conjugated anti-mouse CD62L (MEL-14) antibodies, and BV605-conjugated streptavidin were obtained from BioLegend. Brilliant Ultra Violet (BUV) 395-conjugated anti-mouse CD45R/B220 (RA3-6B2), BUV395- or PE-conjugated anti-mouse CD90.1/Thy1.1 (OX-7), BUV395-conjugated anti-mouse cKit (2B8), PE-CF594-conjugated anti-mouse Flt3 (A2F10.1), and AF700-conjugated anti-mouse CD24 antibodies were purchased from BD Biosciences. PE-Cy7-conjugated anti-mouse MerTK (DS5MMER), APC-ef780-conjugated anti-mouse CD11c (N418), PerCP-ef710-conjugated anti-mouse Sirpα (P84), and Super Bright 645-conjugated anti-mouse MHCII (M5/114.15.2) antibodies were purchased from Invitrogen. BV605-conjugated streptavidin was obtained from BioLegend. For human DC analysis, PE-Cy7-conjugated anti-human CD141 (M80) and PE-conjugated anti-human CLEC9A (8F9) antibodies were obtained from BioLegend. Antibodies used for intracellular staining of IRF8 and C/EBPβ are as follows: PerCP ef710-conjugated anti-human/mouse IRF8 (V3GYWCH) antibody was purchased from Invitrogen. Anti-mouse C/EBPβ (H-7) antibody was from Santa Cruz. Mouse IgG2a (HOPC-1) antibody was from SouthernBiotech. R-PE-conjugated goat anti-mouse IgG2a was from Jackson ImmunoResearch. Antibodies for intracellular staining of IL-6: PE-conjugated anti-mouse IL-6 (MP5-20F3) and PE-conjugated rat IgG1a isotype control (R3-34) antibodies were obtained from BD Biosciences. AF488-conjugated anti-GFP (FM264G) antibody was from BioLegend. Cells were analyzed on a FACSAria Fusion flow cytometer (BD Biosciences), and data were analyzed with FlowJo version 10 software (TreeStar).
Culture of human cord blood–derived cDCs
All experiments using human cord blood were performed under the approval of the institutional review board of Washington University in Saint Louis (#20110348). Human cord blood cells were cultured using a two-step protocol as previously described (Poulin et al., 2010). Human CD34+ cord blood cells (70008.1; STEMCELL Technologies) were cultured at 5 × 104 cells/ml in StemSpan serum-free medium with penicillin/streptomycin, 100 ng/ml human Flt3L and 20 ng/ml SCF, 20 ng/ml IL-3, and 20 ng/ml IL-6 for 7 d and frozen. For use, cells were thawed and cultured at 105 cells/ml in complete I10F (Iscove’s Modified Dulbecco’s Medium supplemented with 10% heat-inactivated FCS, L-glutamine, sodium pyruvate, MEM non-essential amino acid, penicillin/streptomycin, and 55 µM β-mercaptoethanol) together with 100 ng/ml human Flt3L, 20 ng/ml human SCF, 20 ng/ml human GM-CSF, 20 ng/ml human IL-4, with or without human IL-6 for 14 d. Culture media with cytokines were replaced on day 6. All human cytokines were from STEMCELL Technologies or PeproTech.
Intracellular staining
C/EBPβ and IRF8 in BM cells were analyzed as described (Kim et al., 2020). Briefly, sort-purified CDP was cultured with 5% Flt3L conditioned media with or without addition of 25 ng/ml murine recombinant IL-6 (PeproTech) for 20 h. Cells were suspended in 100 μl 1× Fixation/Permeabilization buffer (005123-43 and 00-5223-56; Invitrogen), incubated for 30 min on ice, and then washed with 200 μl 1× Permeabilization buffer (00-8333-56; Invitrogen) twice. The cells were resuspended in 25 μl 1× Permeabilization buffer containing 2% rat serum (Sigma-Aldrich), incubated on ice, and then stained with anti-mouse C/EBPβ antibody (200 µg/ml, 1:200), mouse IgG2a isotype antibody (100 µg/ml, 1:100), or PerCP-ef710 anti-mouse/human IRF8 antibody (1:200) for 45–60 min. After washing with 200 μl 1× Permeabilization buffer, the cells were further stained with R-PE-conjugated goat anti-mouse IgG2a (1:200 in 25 μl), for the secondary antibody of C/EBPβ staining, for 30 min on ice and then washed with 200 μl 1× Permeabilization buffer.
In vivo T cell–priming assay with cell-associated antigen
WT or Zeb2 Δ1+2+3 mice were i.p. inoculated with PBS, EL4-empty, or EL4-IL-6 (106 cells/100 μl/mouse) and kept for 14 d. OT-I and OT-II T cells were prepared as following procedures: the lymph nodes and spleens were isolated from CD45.1 OT-I TCR or CD45.1 OT-II TCR transgenic mice and dispersed into single-cell suspensions by mechanical separation with slide glasses and 70-μm strainers. RBC was lysed with ACK lysis buffer for 2 min. For lineage depletion, the cells were stained with biotinylated B220, I-A/I-E, Ly6G, CD8b (only for OT-II sorting), and CD4 (only for OT-I sorting) antibodies for 20 min at 4°C, and then the lineage+ cells were depleted by magnetic separation using MagniSort streptavidin negative selection bead according to manufacturer’s instructions. CD45.1+ B220− CD8α+ CD4− Vα2+ CD44− CD62L+ OT-I and CD45.1+ B220− CD8α− CD4+ Vα2+ CD44− CD62L+ OT-II cells were sort-purified and then labeled with 1 μM Cell Trace Violet proliferation dye (Thermo Fisher Scientific). The Cell Trace Violet–labeled OT-I cells (2.5 × 105) and OT-II cells (2.5 × 105) were mixed as 1:1 ratio, suspended in 150 μl PBS, and transferred i.v. into EL4 tumor-bearing mice or PBS-inoculated control mice. Cell-associated OVA antigens were prepared as follows: 2.5 × 107 splenocytes isolated from MHCI triple KO mice were osmotically loaded with either 2.5 mg soluble ovalbumin (Worthington Biochemical Corporation) or PBS, then irradiated at 1,350 rad (13.5 Gy). The PBS- or OVA-loaded splenocytes were i.v. injected (5 × 105 cells/150 μl/mouse) into the mice 2–3 h later after OT-I and OT-II transfer. After 3 d, proliferation and activation of OT-I and OT-II T cells in the spleen were analyzed by Aurora flow cytometry (Cytek).
Plasmids, retroviral packaging, and overexpression
LAP and LIP were amplified from cDNA of Cebpb (pcDNA 3.1(−) mouse C/EBP beta (LAP) was a gift from Peter Johnson (Addgene plasmid #12557; http://n2t.net/addgene:12557; RRID:Addgene_12557; Lee et al., 2010) using forward LAP primer (5′-GTCAGATCTAGACCCATGGAAGTGGCCAAC-3′), forward LIP primer (5′-GCTAGATCTGACGCGCCCGCCATGGCGGCC-3′), and common reverse primer (5′-AATACTCGAGGCAGTGGCCCGCCG-3′), and cloned as a BglII/XhoI fragment into MSCV-based retroviral vector (T2a-Thy1.1 RV; Liu et al., 2022) to generated MSCV-LAP-T2a-Thy1.1 and MSCV-LIP-T2a-Thy1.1. MSCV-Irf8-T2a-Thy1.1 has been described (Kim et al., 2020). IL-6 cDNA was amplified from a cDNA library made from CD11c+ splenocytes and cloned into IRES-EGFP RV (Ranganath et al., 1998) using forward and reverse primers (5′-CATGGATCCGCCACCATGAAGTTCCTCTCTGCAAGA-3′; 5′-GATCCTCGAGCTAGGTTTGCCGAGTAGATCT-3′) to generate MSCV-Il6-IRES-EGFP. MSCV-Neo- ER-Hoxb8 construct was used as previously described (Liu et al., 2022). Relevant restriction enzyme sites in these primers are underlined. Constructs were packaged using Platinum-E cells transfected with TransIT (Mirus bio) as described (Kim et al., 2020).
Cell lines
Platinum-E retroviral packaging cell line (Morita et al., 2000) and EL4 lymphoblast cell line (ATCC, TIB-39) were cultured in complete I10F. EL4-empty and EL4-IL-6 were generated by retroviral transductions with MSCV-IRES-EGFP or MSCV-Il6-IRES-EGFP, respectively, and sort-purified as EGFP+ cells twice.
Generation and characterization of Hoxb8-transformed BM progenitors
To generate an immortalized BM progenitor cell line for CUT&RUN, sort-purified Lin− cKithi Flt3+ M-CSFR− BM progenitors from Zbtb46egfp/+ mice were transduced with MSCV-Neo-ER-Hoxb8-RV and then cultured in complete I10F media supplemented with 1 μM estradiol (Sigma-Aldrich), 5% Flt3L conditioned media, and SCF conditioned media. Hoxb8-transformed cells were enriched by culturing the cells for more than 3 wk. The cells were characterized based on (i) surface markers for BM progenitors, (ii) C/EBPβ intracellular expression, and (iii) differentiation potential into DCs in response to IL-6 treatment, in comparison with each character of CDP. Details of experimental procedures for characterizing Hoxb8-cell lines were stated in the legend for Fig. S3, D–F.
Retroviral reporter analysis
We have previously reported the retroviral reporter vector, Thy1.1 pA EGFP CMVp_min PmeI MCS (multiple cloning sites) RV (Durai et al., 2019), and reporter constructs for the WT Zeb2 −165 kb enhancer (Huang et al., 2021) and the Δ1+2+3 mutations in the Zeb2 −165 kb enhancer (Liu et al., 2022). Reporter plasmids were packaged, and sort-purified BM progenitors were transduced, cultured in Flt3L alone or with 25 ng/ml IL-6 for 2 d, and Thy1.1+ cells were analyzed for EGFP expression.
Tumor inoculation
Mice were inoculated with the indicated EL-4 tumor lines through i.p. injections of 106 cells in 100 μl PBS. Control injections were 100 μl PBS. EL-4 tumor lines were validated for uniform EGFP expression on the day of inoculation by FACS.
RNA-seq
CDPs were sort purified after lineage depletion (CD105, Ter119, Ly6G, NK1.1, CD3Ɛ, CD19, and B220) and cultured with Flt3L with or without IL-6 (25 ng/ml) addition for 6 h. RNA-seq libraries were prepared, sequenced, aligned, and analyzed as previously described (Liu et al., 2022). Genes expressed below 20 counts-per-million were filtered out from analysis.
CUT&RUN
C/EBPβ CUT&RUN was performed in Hoxb8-transformed BM progenitor cell line with a CUTANA ChIC/CUT&RUN Kit (EpiCypher) according to the manufacturer’s instructions, with modifications as previously described (Liu et al., 2022). M-CSFR+ cells in the Hoxb8 cell lines (5 × 106 cells/sample) were sort-purified and stimulated with 5% Flt3L with or without IL-6 (25 ng/ml) addition for 24 h. For C/EBPβ and isotype control CUT&RUN, 0.5 μg of anti-C/EBPβ antibody (sc-7962X; Santa Cruz Biotechnology) and 0.5 μg of mouse IgG2a antibody (0103-01; SouthernBiotech) were used, respectively. Libraries for CUT&RUN were prepared with a NEBNext Ultra II DNA Library Prep Kit for Illumina (New England Biolabs), and DNA fragments with sizes ranging from 150 to 300 bp were obtained using AMPure XP beads (Beckman Coulter) as previously described (Liu et al., 2022). The libraries, each having distinct indexes, were combined and subjected to paired-end sequencing using an Illumina NovaSeq 6000. For data analysis, the following programs and commands were used. The adaptor sequences were removed before alignment to the mouse reference genome using Trimmomatic53 with the arguments as followings: java -jar trimmomatic-0.39.jar PE -threads 30 -phred33 sample1_R1.fastq.gz sample1_R2.fastq.gz -baseout sample1.fastq.gz ILLUMINACLIP:Truseq3.PE.fa:2:15:4:4:true LEADING:20 TRAILING:20 SLIDINGWINDOW:4:15 MINLEN:25. The trimmed reads were aligned and mapped to the mouse reference genome (GRCm38/mm10) using Bowtie2 software version 2.2.5 with the arguments as follows: bowtie2 -p 30 --dovetail --phred33 -x mm10 -1 sample1_1P.fastq.gz -2 sample1_2P.fastq.gz > sample1.sam. Duplicated reads are discarded using “makeTagDirectory” of Homer software package (version 4.9) with the parameter -tbp 1: makeTagDirectory sample1.tags sample1.sam -fragLength given -tbp 1. The datasets were visualized with UCSC genome browser in a bedgraph format using “makeUCSCfile” of Homer: makeUCSCfile sample1.tags -o auto -fragLength given -tbp 1 -fsize 5e7. Peak calling was performed with “findPeaks” of Homer: findPeaks sample1.tag -style factor -size 400 -o sample1.peak -i isotypectl_for_sample1.tag -poisson 1e−10.
Statistical analysis
All statistical analyses, scatter plots with error bars, and volcano plots (Cui and Churchill, 2003) were performed with Prism version 9 (GraphPad Software). Heatmaps were generated using Morpheus, https://software.broadinstitute.org/morpheus.
Online supplemental material
Fig. S1 shows the generation and validation of IL-6–expressing EL4 tumor cells in vitro and in vivo. Fig. S2 shows the reduction of various BM progenitors for DCs and monocytes upon inoculation of IL-6–producing EL4 tumor cells. Fig. S3 is RNA-seq showing changes in gene expression by IL-6 treatment in CDP. It shows that IL-6 increases C/EBPβ expression and bindings at Zeb2 −165 kb in CDP. Fig. S4 shows a mechanism by which IL-6 blocks cDC1 development by acting at the Zeb2 −165 kb Δ1+2+3 enhancer.
Acknowledgments
We thank the Genome Technology Access Center at the McDonnell Genome Institute at Washington University School of Medicine for help with the genomic analysis of RNA-seq.
This work was supported by grants from the National Institutes of Health (R01AI150297, R01CA248919, R21AI164142, R01AI162643, and R21AI163421 to K.M. Murphy, and T32CA009621 to M.Y. Chen), and a gift from the 1440 Foundation to W.E. Gillanders and K.M. Murphy. S.T. Ferris is a Cancer Research Institute Irvington Fellow supported by the Cancer Research Institute.
Author contributions: S. Kim, T.L. Murphy, and K.M. Murphy designed the study. S. Kim, J. Chen, S. Jo, F. Ou, S.T. Ferris, T. Liu, and W.E. Gillanders (of the Dept. of Pathology and Immunology, Washington University) conducted experiments related to FACS analysis of splenocytes and BM progenitors, cell sorting, culture, RNA-seq, and CUT&RUN with advice from R.A. Ohara, D.A. Anderson III, and R. Wu. F. Ou performed culture and analysis of human cord blood–derived DCs with advice from R.A. Ohara, M.Y. Chen, and W.E. Gillanders (of the Dept. of Surgery, Washington University, and The Alvin J. Siteman Cancer Center). S. Jo performed in vivo T cell–priming assay with cell-associated antigen. S. Kim and J. Chen performed computational analysis of RNA-seq and CUT&RUN with advice from T. Liu and D.A. Anderson III. S. Kim, F. Ou, and T.L. Murphy performed all retroviral assays. S. Kim, T.L. Murphy, and K.M. Murphy wrote the manuscript with advice from all authors.
Data availability
RNA-seq data from Figs. 3 and S3 and CUT&RUN data from Fig. S3 are available on the National Center for Biotechnology Information Gene Expression Omnibus database with the accession numbers GSE215751 and GSE229271, respectively. All data in this study are available in the main figures, text, and supplementary materials.
References
- Akira, S., Isshiki H., Sugita T., Tanabe O., Kinoshita S., Nishio Y., Nakajima T., Hirano T., and Kishimoto T.. 1990. A nuclear factor for IL-6 expression (NF-IL6) is a member of a C/EBP family. EMBO J. 9:1897–1906. 10.1002/j.1460-2075.1990.tb08316.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, D.A., III, Murphy K.M., and Briseño C.G.. 2018. Development, diversity, and function of dendritic cells in mouse and human. Cold Spring Harb. Perspect. Biol. 10:a028613. 10.1101/cshperspect.a028613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auffray, C., Fogg D.K., Narni-Mancinelli E., Senechal B., Trouillet C., Saederup N., Leemput J., Bigot K., Campisi L., Abitbol M., et al. 2009. CX3CR1+ CD115+ CD135+ common macrophage/DC precursors and the role of CX3CR1 in their response to inflammation. J. Exp. Med. 206:595–606. 10.1084/jem.20081385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagadia, P., Huang X., Liu T.T., Durai V., Grajales-Reyes G.E., Nitschké M., Modrusan Z., Granja J.M., Satpathy A.T., Briseño C.G., et al. 2019. An Nfil3-Zeb2-Id2 pathway imposes Irf8 enhancer switching during cDC1 development. Nat. Immunol. 20:1174–1185. 10.1038/s41590-019-0449-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher, J.P., and Reis e Sousa C.. 2018. The role of type 1 conventional dendritic cells in cancer immunity. Trends Cancer. 4:784–792. 10.1016/j.trecan.2018.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton, G., Lee J., Liu K., and Nussenzweig M.C.. 2015. Defining human dendritic cell progenitors by multiparametric flow cytometry. Nat. Protoc. 10:1407–1422. 10.1038/nprot.2015.092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calkhoven, C.F., Müller C., and Leutz A.. 2000. Translational control of C/EBPalpha and C/EBPbeta isoform expression. Genes Dev. 14:1920–1932. 10.1101/gad.14.15.1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, X., and Churchill G.A.. 2003. Statistical tests for differential expression in cDNA microarray experiments. Genome Biol. 4:210. 10.1186/gb-2003-4-4-210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahling, S., Mansilla A.M., Knopper K., Grafen A., Utzschneider D.T., Ugur M., Whitney P.G., Bachem A., Arampatzi P., Imdahl F., et al. 2022. Type 1 conventional dendritic cells maintain and guide the differentiation of precursors of exhausted T cells in distinct cellular niches. Immunity. 55:656–670. 10.1016/j.immuni.2022.03.006 [DOI] [PubMed] [Google Scholar]
- den Haan, J.M., Lehar S.M., and Bevan M.J.. 2000. CD8(+) but not CD8(−) dendritic cells cross-prime cytotoxic T cells in vivo. J. Exp. Med. 192:1685–1696. 10.1084/jem.192.12.1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Descombes, P., and Schibler U.. 1991. A liver-enriched transcriptional activator protein, LAP, and a transcriptional inhibitory protein, LIP, are translated from the same mRNA. Cell. 67:569–579. 10.1016/0092-8674(91)90531-3 [DOI] [PubMed] [Google Scholar]
- Durai, V., Bagadia P., Granja J.M., Satpathy A.T., Kulkarni D.H., Davidson J.T. IV, Wu R., Patel S.J., Iwata A., Liu T.T., et al. 2019. Cryptic activation of an Irf8 enhancer governs cDC1 fate specification. Nat. Immunol. 20:1161–1173. 10.1038/s41590-019-0450-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris, S.T., Durai V., Wu R., Theisen D.J., Ward J.P., Bern M.D., Davidson J.T. IV, Bagadia P., Liu T., Briseño C.G., et al. 2020. cDC1 prime and are licensed by CD4+ T cells to induce anti-tumour immunity. Nature. 584:624–629. 10.1038/s41586-020-2611-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier, E.L., Shay T., Miller J., Greter M., Jakubzick C., Ivanov S., Helft J., Chow A., Elpek K.G., Gordonov S., et al. 2012. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol. 13:1118–1128. 10.1038/ni.2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grajales-Reyes, G.E., Iwata A., Albring J., Wu X., Tussiwand R., Kc W., Kretzer N.M., Briseño C.G., Durai V., Bagadia P., et al. 2015. Batf3 maintains autoactivation of Irf8 for commitment of a CD8α(+) conventional DC clonogenic progenitor. Nat. Immunol. 16:708–717. 10.1038/ni.3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubin, M.M., Zhang X., Schuster H., Caron E., Ward J.P., Noguchi T., Ivanova Y., Hundal J., Arthur C.D., Krebber W.J., et al. 2014. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature. 515:577–581. 10.1038/nature13988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailemichael, Y., Johnson D.H., Abdel-Wahab N., Foo W.C., Bentebibel S.E., Daher M., Haymaker C., Wani K., Saberian C., Ogata D., et al. 2022. Interleukin-6 blockade abrogates immunotherapy toxicity and promotes tumor immunity. Cancer Cell. 40:509–523.e6. 10.1016/j.ccell.2022.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde, S., Krisnawan V.E., Herzog B.H., Zuo C., Breden M.A., Knolhoff B.L., Hogg G.D., Tang J.P., Baer J.M., Mpoy C., et al. 2020. Dendritic cell paucity leads to dysfunctional immune surveillance in pancreatic cancer. Cancer Cell. 37:289–307.e9. 10.1016/j.ccell.2020.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz, S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., and Glass C.K.. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 38:576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettinger, J., Richards D.M., Hansson J., Barra M.M., Joschko A.C., Krijgsveld J., and Feuerer M.. 2013. Origin of monocytes and macrophages in a committed progenitor. Nat. Immunol. 14:821–830. 10.1038/ni.2638 [DOI] [PubMed] [Google Scholar]
- Hildner, K., Edelson B.T., Purtha W.E., Diamond M., Matsushita H., Kohyama M., Calderon B., Schraml B.U., Unanue E.R., Diamond M.S., et al. 2008. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. 322:1097–1100. 10.1126/science.1164206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, X., Ferris S.T., Kim S., Choudhary M.N.K., Belk J.A., Fan C., Qi Y., Sudan R., Xia Y., Desai P., et al. 2021. Differential usage of transcriptional repressor Zeb2 enhancers distinguishes adult and embryonic hematopoiesis. Immunity. 54:1417–1432.e7. 10.1016/j.immuni.2021.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura, S., Onai N., Miya F., Sato T., Tsunoda T., Kurabayashi K., Yotsumoto S., Kuroda S., Takenaka K., Akashi K., and Ohteki T.. 2017. Identification of a human clonogenic progenitor with strict monocyte differentiation potential: A counterpart of mouse cMoPs. Immunity. 46:835–848.e4. 10.1016/j.immuni.2017.04.019 [DOI] [PubMed] [Google Scholar]
- Kim, S., Bagadia P., Anderson D.A. III, Liu T.T., Huang X., Theisen D.J., O’Connor K.W., Ohara R.A., Iwata A., Murphy T.L., and Murphy K.M.. 2020. High amount of transcription factor IRF8 engages AP1-IRF composite elements in enhancers to direct type 1 conventional dendritic cell identity. Immunity. 53:759–774.e9. 10.1016/j.immuni.2020.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S., Miller M., Shuman J.D., and Johnson P.F.. 2010. CCAAT/Enhancer-binding protein beta DNA binding is auto-inhibited by multiple elements that also mediate association with p300/CREB-binding protein (CBP). J. Biol. Chem. 285:21399–21410. 10.1074/jbc.M110.128413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W., Wu Z., Meng W., Zhang C., Cheng M., Chen Y., Zou Y., Li K., Lin S., Xiong W., et al. 2022. Blockade of IL-6 inhibits tumor immune evasion and improves anti-PD-1 immunotherapy. Cytokine. 158:155976. 10.1016/j.cyto.2022.155976 [DOI] [PubMed] [Google Scholar]
- Lin, J.H., Huffman A.P., Wattenberg M.M., Walter D.M., Carpenter E.L., Feldser D.M., Beatty G.L., Furth E.E., and Vonderheide R.H.. 2020. Type 1 conventional dendritic cells are systemically dysregulated early in pancreatic carcinogenesis. J. Exp. Med. 217:217. 10.1084/jem.20190673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, T.T., Kim S., Desai P., Kim D.H., Huang X., Ferris S.T., Wu R., Ou F., Egawa T., Van Dyken S.J., et al. 2022. Ablation of cDC2 development by triple mutations within the Zeb2 enhancer. Nature. 607:142–148. 10.1038/s41586-022-04866-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lybarger, L., Yu Y.Y., Miley M.J., Fremont D.H., Myers N., Primeau T., Truscott S.M., Connolly J.M., and Hansen T.H.. 2003. Enhanced immune presentation of a single-chain major histocompatibility complex class I molecule engineered to optimize linkage of a C-terminally extended peptide. J. Biol. Chem. 278:27105–27111. 10.1074/jbc.M303716200 [DOI] [PubMed] [Google Scholar]
- Meyer, M.A., Baer J.M., Knolhoff B.L., Nywening T.M., Panni R.Z., Su X., Weilbaecher K.N., Hawkins W.G., Ma C., Fields R.C., et al. 2018. Breast and pancreatic cancer interrupt IRF8-dependent dendritic cell development to overcome immune surveillance. Nat. Commun. 9:1250. 10.1038/s41467-018-03600-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, J.C., Brown B.D., Shay T., Gautier E.L., Jojic V., Cohain A., Pandey G., Leboeuf M., Elpek K.G., Helft J., et al. 2012. Deciphering the transcriptional network of the dendritic cell lineage. Nat. Immunol. 13:888–899. 10.1038/ni.2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita, S., Kojima T., and Kitamura T.. 2000. Plat-E: An efficient and stable system for transient packaging of retroviruses. Gene Ther. 7:1063–1066. 10.1038/sj.gt.3301206 [DOI] [PubMed] [Google Scholar]
- Naik, S.H., Sathe P., Park H.Y., Metcalf D., Proietto A.I., Dakic A., Carotta S., O’Keeffe M., Bahlo M., Papenfuss A., et al. 2007. Development of plasmacytoid and conventional dendritic cell subtypes from single precursor cells derived in vitro and in vivo. Nat. Immunol. 8:1217–1226. 10.1038/ni1522 [DOI] [PubMed] [Google Scholar]
- Nishizaki, Y., Takagi T., Matsui F., and Higashi Y.. 2014. SIP1 expression patterns in brain investigated by generating a SIP1-EGFP reporter knock-in mouse. Genesis. 52:56–67. 10.1002/dvg.22726 [DOI] [PubMed] [Google Scholar]
- Onai, N., Obata-Onai A., Schmid M.A., Ohteki T., Jarrossay D., and Manz M.G.. 2007. Identification of clonogenic common Flt3+M-CSFR+ plasmacytoid and conventional dendritic cell progenitors in mouse bone marrow. Nat. Immunol. 8:1207–1216. 10.1038/ni1518 [DOI] [PubMed] [Google Scholar]
- Poulin, L.F., Salio M., Griessinger E., Anjos-Afonso F., Craciun L., Chen J.L., Keller A.M., Joffre O., Zelenay S., Nye E., et al. 2010. Characterization of human DNGR-1+ BDCA3+ leukocytes as putative equivalents of mouse CD8alpha+ dendritic cells. J. Exp. Med. 207:1261–1271. 10.1084/jem.20092618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganath, S., Ouyang W., Bhattarcharya D., Sha W.C., Grupe A., Peltz G., and Murphy K.M.. 1998. GATA-3-dependent enhancer activity in IL-4 gene regulation. J. Immunol. 161:3822–3826. 10.4049/jimmunol.161.8.3822 [DOI] [PubMed] [Google Scholar]
- Satpathy, A.T., Kc W., Albring J.C., Edelson B.T., Kretzer N.M., Bhattacharya D., Murphy T.L., and Murphy K.M.. 2012. Zbtb46 expression distinguishes classical dendritic cells and their committed progenitors from other immune lineages. J. Exp. Med. 209:1135–1152. 10.1084/jem.20120030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenkel, J.M., Herbst R.H., Canner D., Li A., Hillman M., Shanahan S.L., Gibbons G., Smith O.C., Kim J.Y., Westcott P., et al. 2021. Conventional type I dendritic cells maintain a reservoir of proliferative tumor-antigen specific TCF-1+ CD8+ T cells in tumor-draining lymph nodes. Immunity. 54:2338–2353.e6. 10.1016/j.immuni.2021.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlitzer, A., Sivakamasundari V., Chen J., Sumatoh H.R., Schreuder J., Lum J., Malleret B., Zhang S., Larbi A., Zolezzi F., et al. 2015. Identification of cDC1- and cDC2-committed DC progenitors reveals early lineage priming at the common DC progenitor stage in the bone marrow. Nat. Immunol. 16:718–728. 10.1038/ni.3200 [DOI] [PubMed] [Google Scholar]
- Sharma, P., and Allison J.P.. 2015. The future of immune checkpoint therapy. Science. 348:56–61. 10.1126/science.aaa8172 [DOI] [PubMed] [Google Scholar]
- Skene, P.J., and Henikoff S.. 2017. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. Elife. 6:e21856. 10.7554/eLife.21856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spranger, S., Dai D., Horton B., and Gajewski T.F.. 2017. Tumor-residing Batf3 dendritic cells are required for effector T cell trafficking and adoptive T cell therapy. Cancer Cell. 31:711–723.e4. 10.1016/j.ccell.2017.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G.G., Calvo K.R., Pasillas M.P., Sykes D.B., Häcker H., and Kamps M.P.. 2006. Quantitative production of macrophages or neutrophils ex vivo using conditional Hoxb8. Nat. Methods. 3:287–293. 10.1038/nmeth865 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
RNA-seq data from Figs. 3 and S3 and CUT&RUN data from Fig. S3 are available on the National Center for Biotechnology Information Gene Expression Omnibus database with the accession numbers GSE215751 and GSE229271, respectively. All data in this study are available in the main figures, text, and supplementary materials.