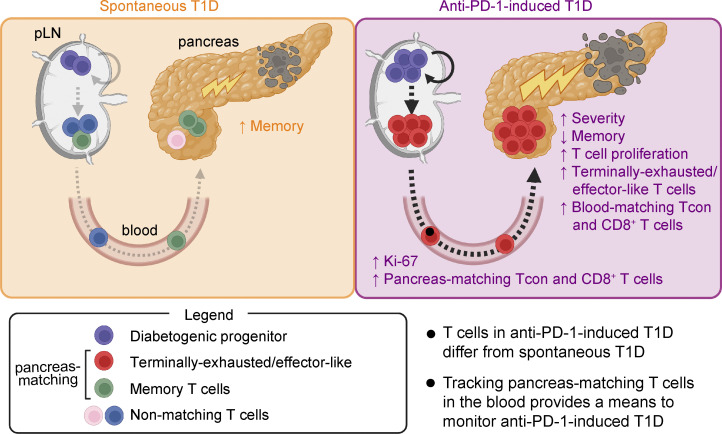
Collier et al. identify unique features of diabetogenic T cells in the pancreas and peripheral tissues between anti-PD-1-induced type 1 diabetes (T1D) and spontaneous T1D in non-obese diabetic mice and provide a rationale for monitoring immune-related adverse events in the blood.
Abstract
Immune-related adverse events (irAEs) are a notable complication of PD-1 cancer immunotherapy. A better understanding of how these iatrogenic diseases compare with naturally arising autoimmune diseases is needed for treatment and monitoring of irAEs. We identified differences in anti-PD-1-induced type 1 diabetes (T1D) and spontaneous T1D in non-obese diabetic (NOD) mice by performing single-cell RNA-seq and TCR-seq on T cells from the pancreas, pancreas-draining lymph node (pLN), and blood of mice with PD-1-induced T1D or spontaneous T1D. In the pancreas, anti-PD-1 resulted in expansion of terminally exhausted/effector-like CD8+ T cells, an increase in T-bethi CD4+FoxP3− T cells, and a decrease in memory CD4+FoxP3− and CD8+ T cells in contrast to spontaneous T1D. Notably, anti-PD-1 caused increased TCR sharing between the pancreas and the periphery. Moreover, T cells in the blood of anti-PD-1-treated mice expressed markers that differed from spontaneous T1D, suggesting that the blood may provide a window to monitor irAEs rather than relying exclusively on the autoimmune target organ.
Graphical Abstract
Introduction
Inhibitors of the programmed death-1 (PD-1) pathway have shown tremendous potential in promoting antitumor immunity in cancer patients and are rapidly becoming a standard of care for many cancer types (Sharpe and Pauken 2018; Ribas and Wolchok 2018). Although PD-1 pathway blockade is generally well tolerated, a subset of cancer patients develops off-tumor inflammatory responses referred to as immune-related adverse events (irAEs). These irAEs can vary widely in tissue impacted, severity, and time of onset. Some irAEs can occur early after treatment initiation, while others occur months to years following treatment (Haanen et al., 2018; Postow et al. 2018). It is generally thought that these irAEs arise due to the physiological role of the PD-1 receptor in regulating peripheral tolerance and/or resolution of inflammation (Sharpe and Pauken 2018), although the precise mechanisms driving these clinical events likely vary widely (Postow et al. 2018; Pauken et al., 2019; Esfahani et al., 2020). Serious irAEs (e.g., myocarditis) can lead to delay or discontinuation of checkpoint therapy, lifelong conditions (e.g., diabetes, adrenal insufficiency), or death. Combination immunotherapy (e.g., anti-PD-1 plus anti-cytotoxic T-lymphocyte associated protein 4 [anti-CTLA-4]) significantly increases the incidence of irAEs and particularly the risk of high-grade irAEs (Wang et al., 2018; Almutairi et al., 2020; Johnson et al., 2016). The diversity in clinical presentation of irAEs suggests distinct underlying mechanisms may drive these responses to checkpoint blockade. A deeper understanding of anti-PD-1 toxicity is essential to advance our understanding of irAEs to develop strategies to prevent or treat irAEs without compromising the efficacy of PD-1 cancer immunotherapy.
The PD-1 coinhibitory receptor is expressed upon T cell activation and negatively regulates T cell receptor (TCR) and CD28 signaling following engagement of its ligands programmed death ligand (PD-L)1 (B7-H1) and PD-L2 (B7-H2). PD-1 or PD-L1 deficiency results in profoundly accelerated autoimmunity (Nishimura et al., 2001; Lucas et al., 2008; Nishimura et al., 1999; Wang et al., 2005), highlighting the critical roles of PD-1 and PD-L1 in the maintenance of tolerance. The non-obese diabetic (NOD) mouse model has been extremely useful for elucidating the roles of the PD-1 pathway in tolerance and autoimmunity, as the NOD mouse develops type 1 diabetes (T1D) by mechanisms that resemble T1D in humans. During spontaneous T1D progression, CD4+ and CD8+ T cells contribute to disease pathogenesis, with insulin-producing β cells in the pancreatic islets being the main autoimmune target. Loss of PD-1 signals (either by genetic ablation of PD-1 or PD-L1, or treatment with anti-PD-1 or anti-PD-L1 antibodies) dramatically accelerates T1D onset in these mice (Guleria et al., 2007; Wang et al., 2005; Ansari et al., 2003; Hu et al., 2020), similar to what is observed in a subset of cancer patients (Akturk et al., 2019).
Elegant studies have used the NOD mouse model to interrogate the importance of the pancreas-draining lymph node (pLN) and the pancreas at different stages of disease. Priming of autoreactive T cells occurs in the pLN around 3 wk of age, and the pLN is essential for T1D progression and anti-PD-1-induced T1D at this age (Guleria et al., 2007; Gagnerault et al., 2002). However, in older NOD mice (e.g., 11 wk), the pLN is dispensable for T1D onset, indicating that the T cells already in the pancreas are sufficient to induce T1D onset following PD-1 blockade (Guleria et al., 2007). Although these studies demonstrate the sufficiency of the cells in the pancreas, they do not necessarily exclude the pLN as a reservoir of potentially pathogenic cells.
Much has been learned about the PD-1 pathway from studies of chronic infection and cancer, where sustained expression of PD-1 and PD-L1 promotes T cell exhaustion—a state of reduced cytokine production, proliferation, and effector function induced by chronic antigen stimulation (Philip and Schietinger 2019; McLane et al. 2019; Collier et al., 2021). Exhausted T cells express the HMG-box transcription factor TOX and multiple co-inhibitory receptors (IRs; e.g., T cell immunoglobulin mucin-3 or TIM-3, lymphocyte-activation protein 3 or LAG-3, and CD160), but are heterogeneous; there are PD-1+ T cell factor 1 (TCF1)+ progenitor-exhausted T cells with stem-like capabilities and PD-1hi terminally exhausted T cells (McLane et al. 2019). PD-1 blockade preferentially induces expansion of the progenitor-exhausted T cell population in the context of chronic infection and cancer (McLane et al. 2019), which has placed considerable focus on this subset. Importantly, there is a differential distribution of these subsets anatomically. PD-1+ TCF1+ progenitor-exhausted T cells tend to be more abundant in secondary lymphoid tissues such as the spleen and tissue-draining LN, while the PD-1hi terminally exhausted T cell subset tends to be more abundant in non-lymphoid tissues and tumors (Dammeijer et al., 2020; Connolly et al., 2021; Schenkel et al., 2021; Li et al., 2022). Recent studies have described an exhausted-like state in autoimmunity (Collier et al., 2021; Grebinoski et al., 2022; Zakharov et al., 2020; Hu et al., 2020) and have shown that diabetogenic CD8+ T cell precursors are present within the pLN, differentiate from stem-like progenitor T cells, and can traffic to the pancreas to mediate disease (Gearty et al., 2022). Insulin-tetramer+ T cells with stem-like capacity have been identified in the peripheral blood of T1D patients (Abdelsamed et al., 2020), and diabetogenic stem-like or progenitor-exhausted T cells have been identified in the pLN and pancreas of NOD mice (Zakharov et al., 2020; Hu et al., 2020; Grebinoski et al., 2022; Gearty et al., 2022). New data showing that PD-1 plays an important role in controlling the generation of stem-like T cells in tumor-draining LNs in cancer models (Dammeijer et al., 2020), together with the preferential effects of PD-1 inhibitors in promoting proliferation of progenitor-like exhausted subsets, raise the question of the role of the pLN in anti-PD-1-induced T1D.
Understanding where PD-1 inhibitors act is important not only for designing therapeutic approaches to mitigate irAEs but also for developing strategies to monitor irAEs. In cancer patients, there is increasing evidence that the peripheral blood, which acts as a conduit of immune cell trafficking between the tumor-draining LN and the tumor, can be an extremely useful site for monitoring the development of protective anti-tumor responses. Indeed, early changes in proliferation (measured by Ki-67) in CD8+ T cell subsets can correlate with outcome following PD-1 blockade in advanced melanoma (Huang et al., 2017; Huang et al., 2019). Determining whether a similar approach can be taken to track irAE development relies on clarifying the role of the peripheral immune system in the development of anti-PD-1-induced pathogenic immune responses.
In this study, we investigated how anti-PD-1-induced T1D compares to spontaneous T1D, and which anatomical locations may be useful for monitoring diabetogenic T cell responses. We used a combination of paired single-cell RNA sequencing (scRNA-seq) and single-cell TCR sequencing (scTCR-seq), flow cytometry, and spatial proteomics to compare peripheral vs. pancreas-residing CD4+ and CD8+ T cells following development of spontaneous T1D vs. anti-PD-1-induced T1D in the NOD mouse model. In the pancreas, diabetogenic CD8+ T cells in PD-1-induced T1D showed a significant increase in proliferation and terminally exhausted/effector-like T cells, and a decrease in memory-like subsets compared with CD8+ T cells in spontaneous T1D. A gene signature derived from terminally exhausted/effector-like CD8+ T cells in the NOD pancreas was enriched in cancer patients who developed anti-PD-1-induced colitis. CD4+ T cells in the pancreas of anti-PD-1-treated mice also showed an increase in proliferation, a gain in a progenitor-like phenotype, and a decrease in memory-like CD4+ T cell subsets. Using the TCR as a molecular barcode (Pauken et al., 2021; Pauken et al., 2022), we identified both CD4+ and CD8+ T cell populations in the peripheral blood and pLN that had clonal relationships with T cells found with the pancreas. T cells in the blood and pLN that had overlapping TCRs with the pancreas were referred to as pancreas-matching (PM) cells, and T cells in the pancreas with TCRs matching to the periphery were called peripheral-matching cells. In anti-PD-1-induced T1D, the percentage of peripheral matching to non-matching CD8+ T cells in the pancreas dramatically increased relative to spontaneous T1D and non-diabetic controls, consistent with an increase in pancreas-specific CD8+ T cells in the pLN and blood by flow cytometry. Importantly, while some transcripts (Gzmk, Cxcr3, Tigit, Itga4, Itgb1, and Klrk1) were shared in matching PM T cells in the blood between spontaneous and anti-PD-1-induced T1D, there were transcripts unique to anti-PD-1-induced T1D (Klrd1, Tox, Cd48, Cx3cr1, and Ifng). Our identification of novel features of PM T cells in the blood indicates the potential of using these circulating T cells for monitoring the development and progression of T1D in patients receiving PD-1 cancer immunotherapy.
Results
Single-cell transcriptional landscape of CD4+ and CD8+ T cells during spontaneous and anti-PD-1-induced T1D in NOD mice
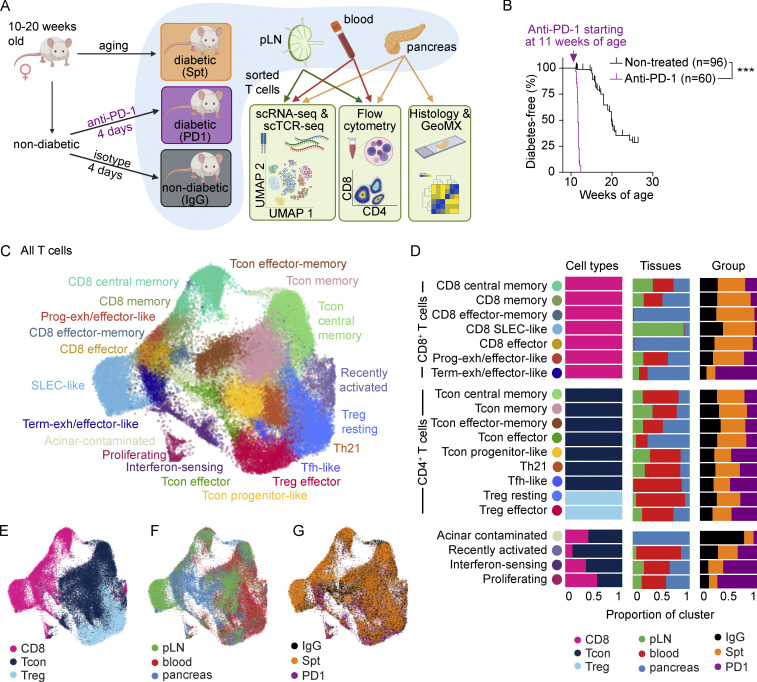
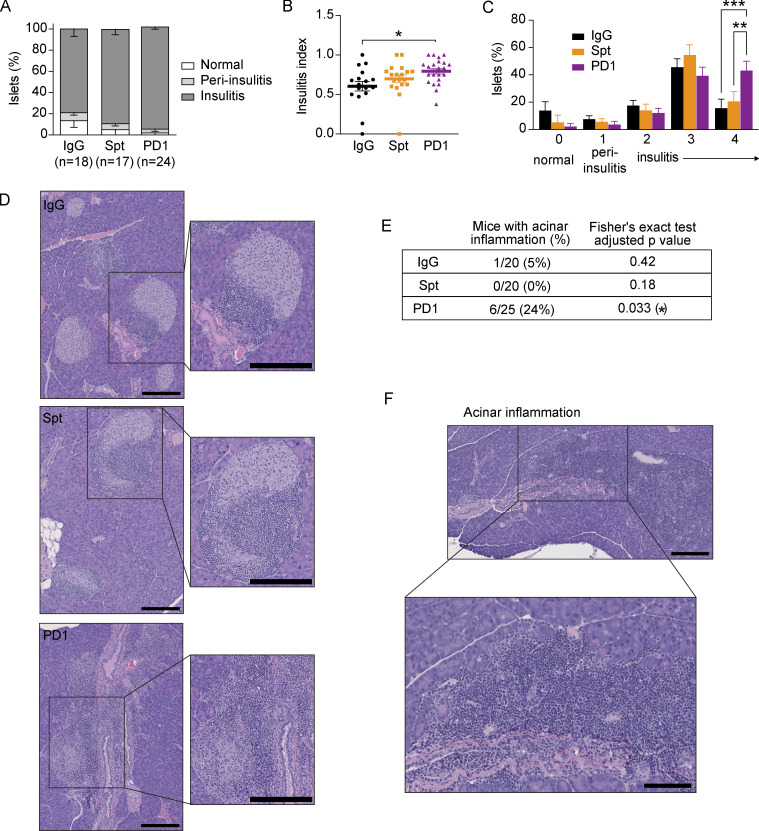
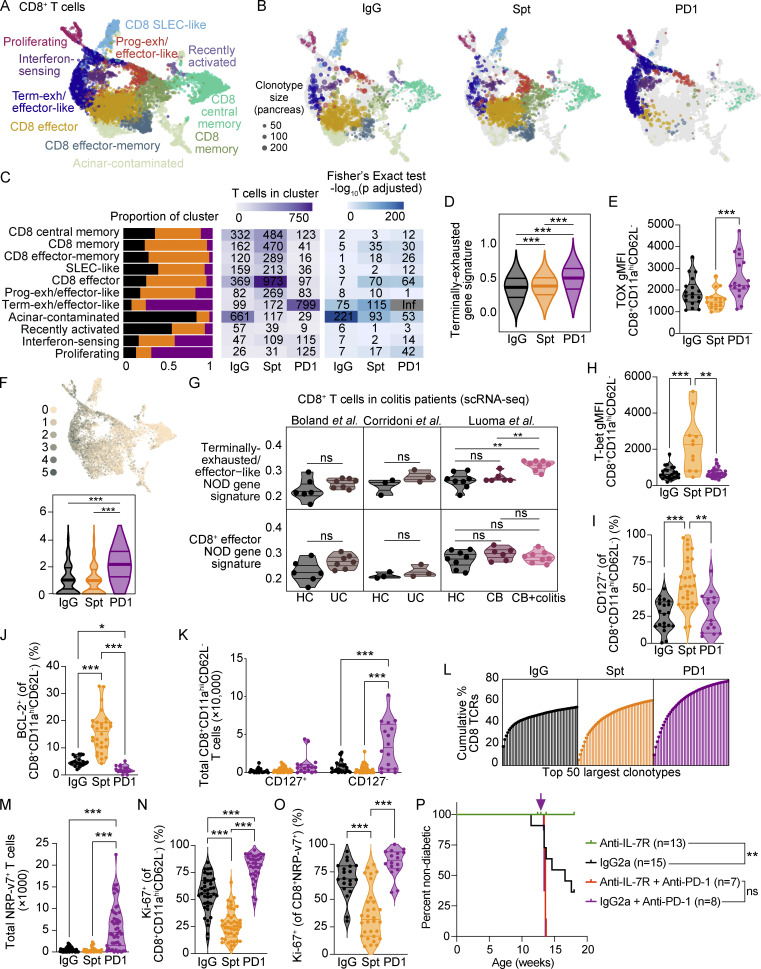
To compare T cells in both spontaneous and anti-PD-1-induced autoimmune diabetes, we examined three groups of female NOD mice between 10 and 20 wk of age: (1) mice developing spontaneous autoimmune diabetes (abbreviated: Spt); (2) non-diabetic mice treated with anti-PD-1 antibody until diabetes onset (abbreviated: PD1); and control mice treated with isotype control antibody (abbreviated: IgG) that did not develop diabetes during the time period of investigation (Fig. 1 A). Consistent with previous reports (Hu et al., 2020; Guleria et al., 2007; Ansari et al., 2003), 11-wk-old NOD mice treated with anti-PD-1 showed a significantly accelerated onset of autoimmune diabetes (Fig. 1 B) and more severe insulitis (Fig. S1, A–D) compared with non-treated NOD mice (Fig. 1 B). Additionally, in the anti-PD-1-treated mice, inflammatory lesions appeared to expand to affect adjacent acinar cells in the pancreas. There was a significant increase in the incidence of inflammation in the exocrine pancreas of mice treated with anti-PD-1 (24%, P = 0.033) compared with non-diabetic (5%) or spontaneously diabetic mice (0%; Fig. S1, E and F). Collectively, these data highlight that pancreatic inflammation is far more extensive in anti-PD-1-induced T1D compared with spontaneous T1D and affects β-islets and exocrine tissue.
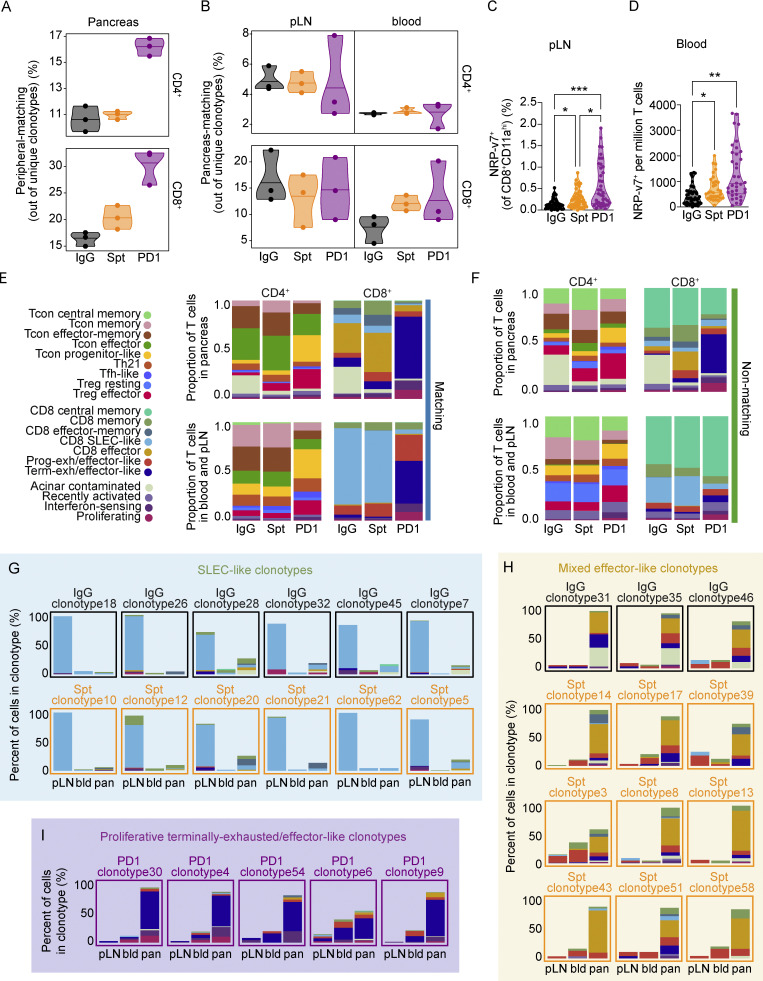
Figure 1.
Comparison of single-cell transcriptional landscape of CD4+ and CD8+ T cells during spontaneous and anti-PD-1-induced T1D in NOD mice. (A) Study design for multisite analysis of IgG, Spt, or PD1 mice. (B) Percent diabetes-free NOD mice following administration of two doses of anti-PD-1 antibody (purple) or isotype control antibody (black) every other day beginning at 11 wk of age. Kaplan–Meier survival curves and results from a Mantel-Cox log-rank test (P < 1 × 10−15) are shown. (C) Clustering and UMAP visualization of integrated CD4+ and CD8+ T cell data from paired blood, pLN, and pancreas from three mice per treatment group (IgG, PD1, and Spt). Colors denote transcriptional clusters, labeled with functional annotations. See Materials and methods for more details on functional annotations, and Table S1 for full list of upregulated genes per cluster. (D) Stacked bar plots showing the proportion of cell types (Tcon, Treg, and CD8+ T cells), tissues, and treatment groups within each cluster. Definitions used to classify cells as CD8+, Tcon, or Treg are indicated in the Materials and methods. The term “Tcon” specifically refers to CD4+ Foxp3− T cells. Most clusters were exclusively one cell type (e.g., CD8+ T cells, Tcons, or Tregs). However, some clusters (e.g., bottom four rows) contained a mix of CD8+ T cells and Tcons. (E–G) UMAP visualization of the distribution of all T cells based on (E) cell type classification, (F) tissues, and (G) treatment groups. Asterisks indicating significance: ***, P < 0.001.
Figure S1.
Histological differences in the pancreas of spontaneous and anti-PD-1-induced T1D in NOD mice. (A) The percentage of β-islets that were classified as normal (score 0) or exhibited either peri-insulitis (score 1) or insulitis (score 2–4). (B) The insulitis index for individual mice calculated according to the equation described in the Materials and methods. Bars show the mean and error bars represent ± SEM. Significance determined using a non-parametric Kruskall–Wallis test with Dunn’s posthoc test for multiple comparisons, P = 0.0103. (C) Percentage of islets within individual mice from A with peri-insulitis or insulitis or that appear normal. Significance determined using a two-way ANOVA with Holm-Sidak’s test for multiple comparisons, Insulitis score 4, IgG-PD1: P = 0.0008; Spt-PD1: 0.0057. Data are shown as means ± SEM. (D) Example H&E images with insets of β-islets with insulitis from the pancreata of IgG, Spt, or PD1 mice. The length of black bars in the lower right of images represents 200 µm. (E) Table showing the number of mice with acinar inflammation out of the total number of samples examined, with percentage in brackets. Statistical significance was determined using a Fisher’s exact test and P values adjusted using the Benjamini and Hochberg method are shown. (F) Example H&E image with inset showing acinar inflammation in pancreas of a mouse with anti-PD-1-induced T1D (PD1). The length of black bars in the lower right of the images represents 100 and 200 µm in the inset. Asterisks indicating significance: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Considering the differences in both the timing of onset and severity of pancreatic inflammation in anti-PD-1 and spontaneous T1D in NOD mice, we hypothesized that the state, functional capacity, and source of the diabetogenic leukocytes that drive pathogenesis in anti-PD-1-treated mice would differ significantly from spontaneous T1D. In this study, we focused our analyses on the T cell compartment (both CD4+ and CD8+ T cells) rather than other immune populations such as B cells since previous work has shown that both CD4+ and CD8+ T cells are necessary for the rapid onset of T1D observed in NOD mice following PD-1 blockade, while antibodies were dispensable (Guleria et al., 2007). To begin to interrogate the drivers of pathogenesis in these two settings, we compared 16-wk-old mice treated with anti-PD-1 or isotype control and mice that spontaneously developed diabetes at 18 wk of age. We performed scRNA-seq and scTCR-seq on total activated T cells (TCR-β+CD11ahi) from key anatomical locations involved in the initiation, progression, and onset of T1D. These sites included (1) the pLN, the site of initial T cell priming and a critical source of pathogenic TCF-1+ stem-like T cells (Gearty et al., 2022; Hu et al., 2020), (2) the peripheral blood, the site of T cell trafficking to the pancreas, and (3) the pancreas, the autoimmune target organ and primary site of autoimmune pathology. We specifically chose to compare spontaneous and anti-PD-1-induced T1D at this 16-wk time point because we sought to identify differences in the mechanisms underlying these two processes at the time of disease onset. While PD-1 inhibitors do precipitate T1D onset in younger NOD mice (Ansari et al., 2003), studies have established that the cellular landscape in the pancreas of older vs. younger NOD mice differs significantly (Zakharov et al., 2020; Anderson and Bluestone 2005). Consequently, to provide the most well-controlled comparison, we utilized age-matched NOD mice in this study that were either prediabetic (isotype control treated), spontaneously diabetic, or diabetic following a short treatment regimen of anti-PD-1 (see Materials and methods).
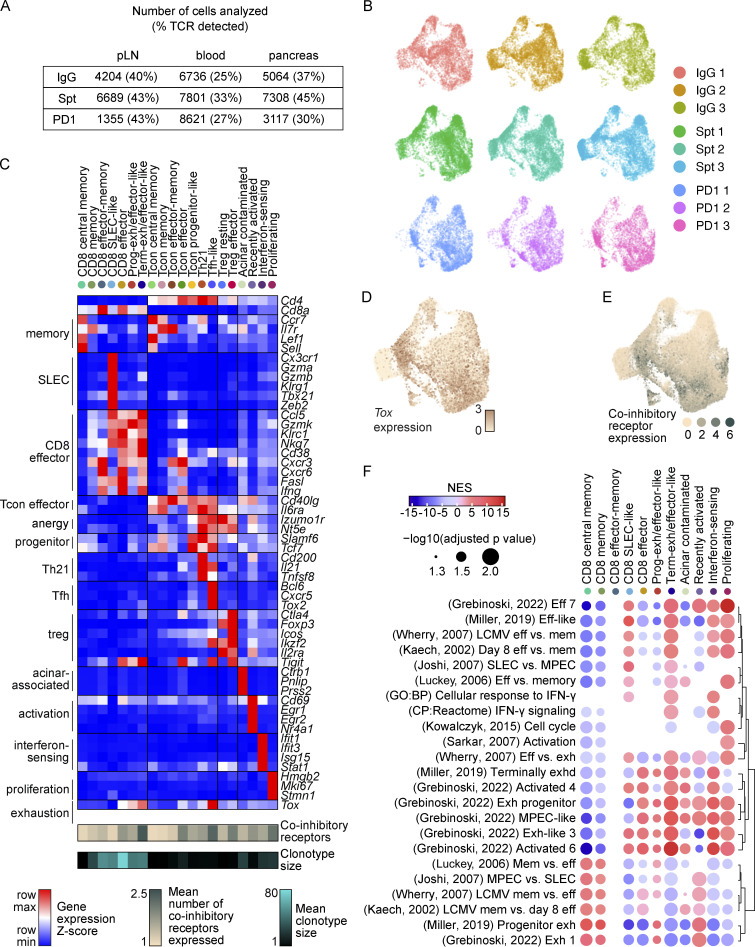
To identify putative diabetogenic T cells in the periphery (e.g., pLN and blood), we used the TCR as a molecular barcode (Pauken et al., 2021) to distinguish clones in the periphery with matching TCR sequences to clones found in the pancreas. A total of 35,335 CD4+ and 15,558 CD8+ T cells were analyzed based on expression of Cd4, Cd8a, and Foxp3, with 25–45% of TCRs being detected in at least 1,300 T cells from each treatment group per tissue (Fig. S2 A), using three mice per treatment group (Fig. S2 B). These cells were distributed across 20 transcriptional clusters that were defined based on upregulated gene expression (Table S1), expression of key markers (Fig. S2, C–E), and enrichment of key signatures (Fig. S2 F) used to define memory, effector, exhausted, progenitor-like, and other functional clusters from the literature (see Materials and methods). Based on these transcriptional clusters, we defined seven CD8+ T cell populations (Cd8ahiCd4lo), seven conventional CD4+ T cell (Tcon) populations (Cd8aloCd4hiFoxp3lo), two populations of regulatory T cells (Treg) (Cd8aloCd4hiFoxp3hi), and four clusters containing both CD4+ and CD8+ T cells (Fig. 1, C–E) across pLN, blood, and pancreas (Fig. 1, D and F). Each tissue showed a distinct enrichment of these different transcriptional clusters, consistent with previous work examining these different anatomical compartments (Gearty et al., 2022; Grebinoski et al., 2022; Zakharov et al., 2020; Hu et al., 2020). For example, for CD8+ T cells, the pancreas showed enrichment in CD8 effector-memory, CD8 effector, and terminally exhausted/effector-like clusters relative to the pLN and blood. Conversely, the blood and pLN showed more of enrichment in CD8 central memory, CD8 memory, CD8 short-lived effector cell (SLEC)-like, and progenitor-exhausted/effector-like compared with the pancreas (Fig. 1 D). When examining the distribution of cells based on treatment group, we observed that terminally exhausted/effector-like T cells, interferon-sensing, and proliferating clusters were expanded to a greater extent in anti-PD-1-induced T1D compared with spontaneous T1D or non-diabetic mice. Central memory and memory-like clusters were reduced in number in anti-PD-1-induced T1D compared with spontaneous T1D (Fig. 1, D and G). Collectively, these data show that anti-PD-1-induced T1D differs from spontaneous T1D by preferential expansion of terminally exhausted/effector-like T cells, interferon-sensing, and proliferating clusters, leading us to further examine these differences in T cell states in spontaneous T1D vs. anti-PD-1-induced T1D.
Figure S2.
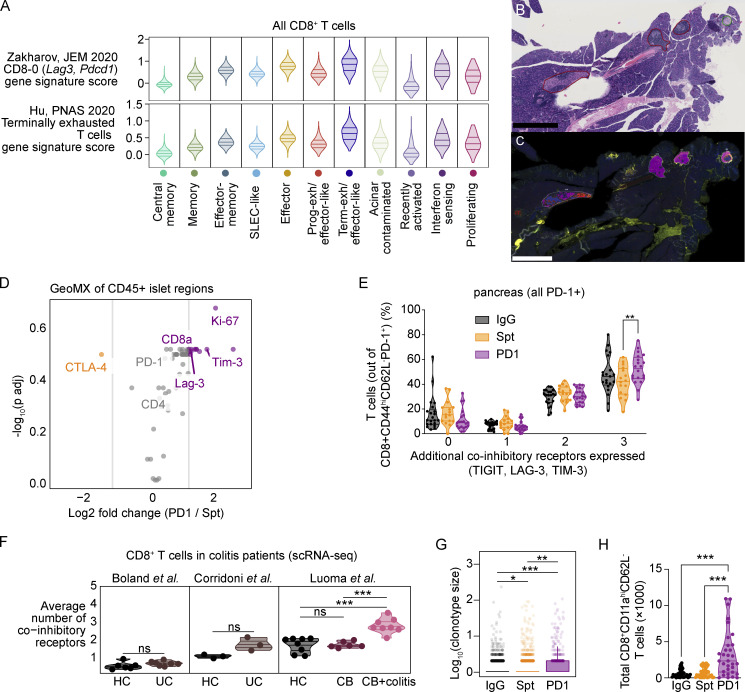
Classification of CD4+ and CD8+ T cells based on transcriptional and TCR features. (A) Table indicating the number of T cells recovered in each tissue and treatment group with percentage of cells in which the TCR was detected in parentheses. (B) UMAP visualization of all T cells from individual mice. (C) Heatmap showing gene expression of selected markers used for cluster annotation. Included are the mean clonotype size for each cluster and the mean number of co-inhibitory receptors co-expressed by cells in each cluster (Tigit, Ctla4, Cd160, Cd244a, Pdcd1, and Havcr2). (D) UMAP visualization of expression of Tox transcript expression across all T cells. (E) UMAP visualization of the distribution of expression of multiple co-inhibitory receptors (Tigit, Ctla4, Cd160, Cd244a, Pdcd1, Havcr2). (F) Dot plot visualization of the Normalized Enrichment Score (NES) calculated by gene set enrichment analysis using pre-ranked genes from CD8+ T cell clusters for selected gene signatures from Joshi et al. (2007); Miller et al. (2019); Grebinoski et al. (2022); Kaech et al. (2002); Luckey et al. (2006); Wherry et al. (2007); Sarkar et al. (2008); Kowalczyk et al. (2015). The size of the dot corresponds to the log10 transformed value of the adjusted P value and the color corresponds to the NES. Dots are only shown for P values <0.05.
Pancreatic CD8+ T cells in PD-1-induced T1D show a shift toward a terminally exhausted gene signature, a decrease in memory subsets, and significantly greater expansion of CD8+CD11ahi T cells compared with spontaneous T1D
CD8+ T cells are essential for anti-PD-1/PD-L1-induced T1D in NOD mice (Guleria et al., 2007; Hu et al., 2020) and are the late-stage effectors of β-islet destruction in spontaneous T1D (Anderson and Bluestone 2005). In view of the importance of CD8+ T cells within the pancreas tissue for mediating immune destruction following PD-1 blockade (Guleria et al., 2007), we first evaluated how the states of diabetogenic CD8+ T cells in the pancreas compared between treatment groups (Fig. 2 A). Effector-like, memory, and effector-memory subsets were present in pancreatic CD8+ T cells from both non-diabetic and spontaneously diabetic NOD mice. However, the transcriptional landscape dramatically shifted in pancreatic CD8+ T cells of anti-PD-1-induced T1D toward the terminally exhausted/effector-like transcriptional cluster (Fig. 2, B and C, Fisher’s exact test: P ≤ 1.0 × 10−324). CD8+ T cells in anti-PD-1-induced T1D showed a clear increase in the expression of a terminally exhausted gene signature derived from CD8+ T cells in chronic lymphocytic choriomeningitis virus (LCMV; Miller et al., 2019; Fig. 2 D). We validated these findings using flow cytometry. One of the key defining features of terminally exhausted T cells is the transcription factor TOX (Khan et al., 2019; Alfei et al., 2019). Tox gene expression was increased in the terminally exhausted/effector-like CD8+ T cell cluster (Fig. S2, C and D) and there was a modest increase in TOX protein expression in pancreatic CD8+ T cells in anti-PD-1-induced T1D compared with spontaneous T1D (Fig. 2 E). Another feature of terminally exhausted T cells is the simultaneous expression of multiple co-inhibitory receptors. Co-expression of Tigit, Ctla4, Cd160, Cd244a, Pdcd1, and Havcr2 transcripts was increased in the CD8+ T cells in the pancreas of anti-PD-1-induced T1D (Fig. 2 F) compared with spontaneous T1D. This subset of terminally exhausted T cells was also enriched for gene signatures of terminally exhausted T cells described in the pancreas of NOD mice by others (Fig. S3 A; Hu et al., 2020; Zakharov et al., 2020). We also performed GeoMX protein analysis of selected proteins (Table S2) on INS−CD45+ and INS+CD45− regions within β-islets of the pancreas (Fig. S3, B and C) to validate the increase in proliferating and exhausted-like CD8+ T cells in T cells infiltrating β-islets in the pancreas. Although these findings did not reach statistical significance, the fold-changes in expression of CD8a and the co-inhibitory receptors TIM-3 and LAG-3 were increased in anti-PD-1-induced T1D compared with spontaneous T1D (Fig. S3 D). By flow cytometry, we observed a significant increase in the frequency of CD8+CD11ahiPD-1+ T cells in the pancreas that expressed three co-inhibitory receptors (in addition to PD-1) in the anti-PD-1-treated mice compared with the spontaneously diabetic mice (Fig. S3 E). These data support a model where anti-PD-1 treatment promotes the acquisition of an exhaustion-like program in CD8+ T cells in the pancreas.
Figure 2.
CD8+ T cells in the pancreas show a gain in proliferation, gain in exhaustion-like features, and decrease in memory-like features in anti-PD-1-induced T1D compared with spontaneous T1D. (A) UMAP visualization of integrated CD8+ T cells detected in the pancreas. Points represent individual cells and colors denote cluster classification as labeled. (B) UMAP visualization of integrated CD8+ T cells detected in the pancreas of each treatment group. Dots represent individual cells with the color representing the cluster classification and size corresponding to the size of the clonotype. Functional annotations for each cluster shared with labels in A. (C) Stacked bar plots denoting the percentage of cells from each treatment group within individual clusters (left panel). Heatmap showing the number of cells detected in each cluster and treatment group (middle panel) and −log10 adjusted P values from a Fisher’s exact test (right panel). (D) Violin plots quantifying the expression of a gene signature score of terminally exhausted T cells (Miller et al., 2019) in all CD8+ T cells in the pancreas. IgG-Spt, P = 1.21 × 10−4; IgG-PD1, P = 4.74 × 10−77; Spt; PD1, P = 1.74 × 10−71. (E) Violin plots showing gMFI of TOX in CD8+CD11ahiCD62L− T cells in the pancreas by flow cytometry analysis. Spt-PD1, P = 0.0008. (F) UMAP visualization of expression of multiple co-inhibitory receptor transcripts (Tigit, Ctla4, Cd160, Cd244a, Pdcd1, and Havcr2; top) and violin plots of the number of co-inhibitory receptors expressed by individual cells in each treatment group (bottom). IgG-PD1, P = 6.46 × 10−158; Spt-PD1, P = 2.12 × 10−203. (G) Violin plots showing the mean gene signature scores from NOD in CD8+ T cells from the colons of HC, UC (Boland et al., 2020; Corridoni et al., 2020), CB, and CB+colitis patients (Luoma et al., 2020). HC-CB+colitis, P = 0.002; CB-CB+colitis, P = 0.002. Gene signatures derived from terminally exhausted/effector-like T cells (top) or effector CD8+ T cells from NOD pancreas (bottom). Each dot shows the average gene signature score of the cells in an individual person. Gene signature from terminally exhausted/effector-like T cells from NOD can be found in Table S3, and effector CD8+ T cells from NOD can be found in Table S4. (H)Violin plots showing gMFI of T-bet in CD8+CD11ahiCD62L− T cells in the pancreas by flow cytometry analysis. IgG-Spt, P = 0.0008; Spt-PD1, P = 0.0020. (I) Violin plots quantifying percent CD127+ staining within CD8+CD11ahiCD62L− T cells in the pancreas measured by flow cytometry. IgG-Spt, P <0.0001; Spt-PD1, P = 0.0012. (J) Violin plots showing percent BCL-2+ in CD8+CD11ahiCD62L− T cells in the pancreas by flow cytometry analysis. IgG-Spt, P < 0.0001; IgG-PD1, P = 0.0298; Spt-PD1, P < 0.0001. (K) Violin plots quantifying the total number of CD127+ and CD127− CD8+CD11ahi CD62L− T cells detected in the pancreas measured by flow cytometry. For CD127− T cells: IgG-PD1 P < 0.001; Spt-PD1 P < 0.0001. (L) Plot of the top 50 most expanded clonotypes, each represented as a vertical line and terminal point, within each treatment group shown as the cumulative total percentage of TCRs detected in CD8+ T cells within that treatment group. (M) Violin plots quantifying the total number of NRP-v7+ T cells detected in the pancreas measured by flow cytometry. IgG-PD1, P < 0.0001; Spt-PD1, P < 0.0001. (N) Violin plot quantifying the percent Ki-67+ staining within CD8+CD11ahiCD62L− T cells in the pancreas measured by flow cytometry. IgG-Spt, P < 0.0001; IgG-PD1, P = 0.0002; Spt-PD1, P < 0.0001. (O) Violin plots quantifying percent Ki-67+ staining within CD8+ NRP-v7+ T cells detected in the pancreas by flow cytometry. IgG-Spt, P = 0.0008; Spt-PD1, P < 0.0001. (P) Percent diabetes-free NOD mice administered isotype control antibody (black) or anti-IL-7Rα antibody (green) twice a week beginning at 10 wk of age. Following 3 wk of treatment with isotype control antibody (purple) or anti-IL-7Rα antibody (red), mice were injected with anti-PD-1 antibody. Kaplan–Meier survival curves are shown. IgG2a vs. anti-IL-7R, P = 0.007. Flow cytometry results are pooled from 3 to 25 independent experiments and significance determined using a non-parametric Kruskall–Wallis test with Dunn’s posthoc test for multiple comparisons. Bars in violin plots represent the first quartile, median, and third quartile. Significant comparisons are indicated with asterisks: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
Figure S3.
Co-inhibitory receptor expression on CD8+ T cells in colitis patients and NOD mice. (A) Violin plots of published gene signature module scores of CD8+ T cells from all tissues by cluster. Top: Zakharov et al., 2020; bottom: Hu et al., 2020. Horizontal lines represent the first quartile, median, and third quartile. (B) Scanned H&E showing representative annotation of pancreatic β-islets scored by a board-certified pathologist (encircled green = peri-insulitis, red = insulitis). The length of black bars in the lower left of the images represents 500 µm. (C) GeoMx DSP scan of mouse pancreas sample labeled with INS (red), CD45 (yellow), Pan-CK (green), and DNA (blue). Segmentation into INS+ (blue) and CD45+ (magenta) areas shown in encircled regions. The length of black bars in the lower left of the images represents 500 µm. (D) GeoMX DSP analysis showing fold-change of normalized protein staining values for indicated markers. Statistical significance was determined using a mixed linear model with Benjamini-Hochberg correction. (E) Violin plots showing the percentage of CD8+CD11ahiCD62L−PD-1+ T cells that express additional co-inhibitory receptors (TIGIT, LAG-3, and/or TIM-3) in the pancreas measured by flow cytometry. Statistical significance determined using a two-way ANOVA with Holm-Sidak’s test for multiple comparisons. For co-expression of three IRs in addition to PD-1, Spt-PD1 P = 0.0046. (F) Violin plots showing the mean expression of multiple co-inhibitory receptor transcripts (Tigit, Ctla4, Cd160, Pdcd1, and Havcr2) expressed by CD8+ T cells from the colons of HC, UC (Boland et al., 2020; Corridoni et al., 2020), CB, and CB+C (Luoma et al., 2020). Each dot shows the average number of co-inhibitory receptors expressed by the cells in an individual patient. HC-CB+colitis, P = 0.0003; CB-CB+colitis, P = 0.000233. (G) Box plot quantifying clonal expansion of CD8+ T cells in the pancreas detected by scTCR-seq. Boxes show the first quartile, median, and third quartile, while the whiskers cover 1.5× the interquartile range. Significance determined using a Wilcoxon rank sum test and P values adjusted using the Benjamini and Hochberg method are shown. IgG-Spt, P = 1.00 × 10−2; IgG-PD1, P = 7.26 × 10−7; Spt-PD1, P = 3.00 × 10−3. (H) Violin plots quantifying the total number of CD8+CD11ahiCD62L− T cells detected in the pancreas measured by flow cytometry. IgG-PD1, P < 0.0001; Spt-PD1, P = 0.0003. Significant comparisons are indicated with asterisks: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
We next determined if similar transcriptional changes could be found in CD8+ T cells from patients with irAEs. The number of published patient datasets for irAEs that have comparable tissue samples from patients with spontaneous autoimmunity is extremely limited, especially for PD-1 monotherapy. We selected three datasets from patients with colitis (Boland et al., 2020; Corridoni et al., 2020; Luoma et al., 2020) since these studies have the best control populations (e.g., healthy controls and non-checkpoint-induced colitis). It should be noted that these datasets contained anti-CTLA-4 monotherapy and anti-PD-1/anti-CTLA-4 combination therapy, which mechanistically may differ from anti-PD-1-induced T1D. Nevertheless, we used these datasets to assess whether shared gene programs could be identified in these datasets as a common mechanism driving irAEs. We analyzed 16,664 CD8+ T cells from a scRNA-seq dataset from the colons of healthy controls (HC) and patients treated with anti-CTLA-4 monotherapy or combination therapy with anti-CTLA-4 and PD-1/PD-L1 that did (CB+colitis) or did not develop colitis (CB; Luoma et al., 2020). To compare with a spontaneous form of colitis, we also analyzed 3,984 CD8+ T cells from two datasets that examined patients with ulcerative colitis (UC) and HC (Boland et al., 2020; Corridoni et al., 2020). For this comparison, we derived a gene signature from the terminally exhausted/effector-like cluster in the pancreas of NOD mice (Table S3), which included genes associated with exhaustion (TOX, HAVCR2, TIGIT, and LAG3) and cytotoxicity (GZMB, GZMK, and NKG7). This terminally exhausted/effector-like gene signature from NOD was enriched in CD8+ T cells in CB+colitis, but not in UC, CB, or HC (Fig. 2 G). We also quantified the simultaneous expression of multiple co-inhibitory receptors (HAVCR2, TIGIT, PDCD1, LAG3, CD160, and CTLA4) by CD8+ T cells from the patients and HC. The CB+colitis patients showed an increase in CD8+ T cells expressing multiple co-inhibitory receptors, consistent with our findings in pancreatic CD8+ T cells of NOD mice with anti-PD-1-induced T1D (Fig. S3 F). To determine whether this finding could be simply attributed to an increase in effector function rather than exhaustion, we derived an additional gene signature from NOD CD8+ effector T cells (Table S4)—prevalent in spontaneous T1D and non-diabetic mice—and found that this signature was not enriched in any of the samples relative to controls (Fig. 2 G). Together, these data illustrate that anti-PD-1-induced T1D is associated with major changes in pancreatic CD8+ T cells, including a shift toward an exhausted/effector-like phenotype that is also observed in patients with checkpoint-induced colitis.
Our scRNA-seq data also showed that compared with spontaneous T1D, the proportion of effector and memory CD8+ T cell clusters were reduced in anti-PD-1-induced T1D. To validate key findings from these scRNA-seq data, we performed flow cytometry to assess protein expression for critical markers associated with these phenotypes. We measured a significant increase in protein expression of T-bet, a marker of effector CD8+ T cells, in pancreatic CD8+ T cells in mice with spontaneous T1D compared with non-diabetic controls and anti-PD-1-treated diabetic mice (Fig. 2 H; Sullivan et al., 2003). In addition, we observed a reduction in the number of CD8+ T cells in the memory (Fisher’s exact test: P = 6.0 × 10−31) and effector-memory clusters (Fisher’s exact test: P = 7.2 × 10−27) in anti-PD-1-induced T1D (Fig. 2 C). We validated the decrease in combined memory and effector-memory CD8+ T cell subsets using flow cytometry to measure protein expression of CD127 (encoded by Il7r) and BCL-2, an anti-apoptotic protein important in memory T cell survival (Grayson et al., 2000), within the CD8+CD11ahiCD62L− population. The percentages of CD127+ (Fig. 2 I) and BCL-2+ CD8+CD11ahiCD62L− T cells (Fig. 2 J) were significantly increased in the pancreas of NOD mice with spontaneous T1D compared with anti-PD-1-induced T1D. There were no differences in the total numbers of CD127+ CD8+CD11ahiCD62L− T cells in the pancreas across the treatment groups, but there was a significant expansion of the CD127− collective population of non-memory effector T cells in anti-PD-1-induced T1D compared with spontaneous T1D and non-diabetic controls (Fig. 2 K). Together, these results indicate that the proportion of memory and effector-memory T cell subsets are reduced in anti-PD-1-induced T1D compared with spontaneous T1D.
The most notable change observed in the CD8+ T cells in anti-PD-1-induced T1D was the significant expansion of CD8+ T cells in the pancreas compared with spontaneous T1D. The number of T cells within the proliferative cluster was dramatically increased in anti-PD-1-induced T1D (Fisher’s exact test: P = 2.4 × 10−42; Fig. 2 C). Using the scTCR-seq data to identify individual clonotypes, we compared clonotype expansion across treatment groups. The top 50 most highly expanded clonotypes cumulatively represented 78% of the TCRs detected in the PD-1 group, but the top 50 clonotypes represented only 54 and 60% of the total TCRs detected in IgG and Spt groups, respectively (Fig. 2 L). Overall expansion of all clonotypes was significantly increased in anti-PD-1-induced T1D compared with non-diabetic or spontaneously diabetic NOD (Fig. S3 G). We validated these findings by flow cytometry: the total numbers of effector CD8+CD11ahiCD62L− T cells (Fig. S3 H) and diabetogenic NRP-v7+ T cells (Fig. 2 M) in the pancreas were significantly increased in anti-PD-1-induced T1D compared with spontaneous T1D and non-diabetic NOD mice. The percentages of Ki-67+ T cells in the effector CD8+CD11ahiCD62L− T cell subset (Fig. 2 N) and NRP-v7+ T cells (Fig. 2 O) were also significantly increased in anti-PD-1-induced T1D. Together, these data demonstrate that there is a significantly greater expansion of non-memory effector CD8+ T cells (CD127− CD8+CD11ahiCD62L−) in the pancreas of anti-PD-1-induced T1D compared with spontaneous T1D.
Our data suggest that distinct populations of diabetogenic CD8+ T cells are driving the onset of clinical disease in spontaneous T1D vs. anti-PD-1-induced T1D. To understand the contribution of different T cell states, we focused on examining the functional significance of enrichment of exhausted/effector like cells in anti-PD-1-induced T1D versus the enrichment of memory T cells in spontaneous T1D. Unlike memory T cells, exhausted T cells do not depend on IL-7 for persistence, as shown by injection of an anti-IL-7Rα blocking antibody in mice with chronic LCMV infection (Shin et al., 2007). We leveraged the differential dependence of memory and exhausted T cells on IL-7 to determine if there is a functional difference in the CD8+ T cell populations identified in our scRNA-seq data in spontaneous T1D and anti-PD-1-induced T1D. We hypothesized if anti-PD-1-induced T1D is dependent on terminally exhausted T cells, unlike spontaneous T1D, then the effects of anti-IL-7Rα would be distinct in anti-PD-1-induced T1D and spontaneous T1D where memory T cells are enriched. Injection of an anti-IL-7Rα antibody (clone A7R34 or 28G9) into NOD mice at 10 wk of age effectively prevents onset of spontaneous T1D (Lee et al., 2012; Penaranda et al., 2012). We injected anti-IL-7Rα or isotype control antibody into NOD mice starting at 10 wk of age and found that anti-IL-7Rα effectively prevents onset of spontaneous T1D, as expected (Fig. 2 P; Lee et al., 2012; Penaranda et al., 2012). In contrast, administration of anti-IL-7Rα to anti-PD-1-treated mice did not alter the timing of onset of T1D compared with anti-PD-1-treated control mice (Fig. 2 P). These findings are consistent with our hypothesis that anti-PD-1-induced T1D is driven by IL-7-independent exhausted T cells and demonstrate that the differences seen in T cell populations in spontaneous T1D and anti-PD-1-induced T1D are functionally significant to diabetes pathogenesis, pointing to distinct mechanisms of diabetogenesis.
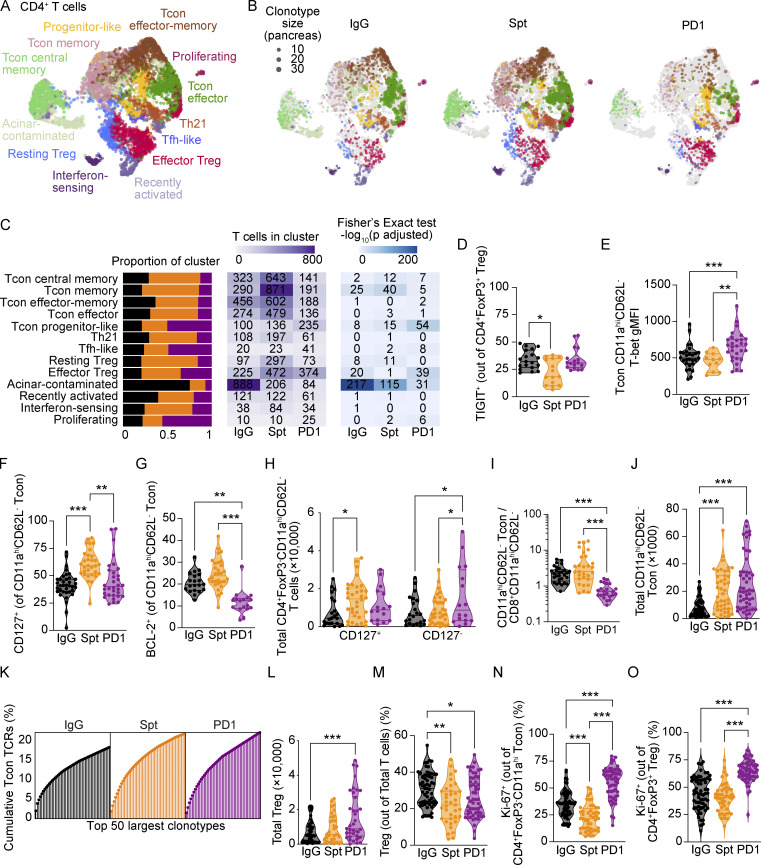
Pancreatic CD4+ T cells show increased proliferation in anti-PD-1-induced T1D compared with spontaneous T1D
CD4+ T cells are essential mediators of both spontaneous (Shizuru et al., 1988) and anti-PD-1-induced T1D (Hu et al., 2020) in NOD mice. CD4+ T cells are posited to have roles in initiation of disease, providing T cell help, and promoting autoantibody production by B cells (Walker and von Herrath 2016). Considering the important role of CD4+ T cells in T1D pathogenesis, we next compared the transcriptional states of CD4+ T cells between spontaneous T1D and anti-PD-1-induced T1D (Fig. 3 A). Similar to CD8+ T cells, CD4+ T cells in the pancreas of anti-PD-1-treated mice differed substantially from CD4+ T cells in spontaneously diabetic mice. In anti-PD-1-treated mice, we observed a shift toward proliferating CD4+ T cells (Fisher’s exact test: P = 9.5 × 10−7), progenitor-like Tcon (Fisher’s exact test: P = 2.7 × 10−54), and TIGIT+ effector Treg (Fisher’s exact test: P = 1.7 × 10−39; Fig. 3, B and C). We then performed flow cytometry to assess protein expression of key markers of these CD4+ T cell populations. There was a modest shift toward TIGIT+ Treg in anti-PD-1-induced T1D compared with spontaneous T1D, but this was not statistically significant by flow cytometry analysis (Fig. 3 D). Protein expression of T-bet in Tcon (CD4+FoxP3−CD11ahiCD62L−) was increased in anti-PD-1-induced T1D, supporting the conclusion that there is a shift toward CD4+ T helper (Th)1 cells in this setting (Fig. 3 E). Analogous to our findings in CD8+ T cells, we observed reduced memory (Fisher’s exact test: P = 2.3 × 10−5) and effector-memory Tcon subsets (Fisher’s exact test: P = 2.3 × 10−5; Fig. 3 C) in anti-PD-1-induced T1D compared to spontaneous T1D. To validate these scRNA-seq findings by flow cytometry, we looked at protein expression of key markers in both memory and effector-memory Tcon subsets: CD127 and BCL-2. We confirmed that the percentages of CD127+ Tcon (Fig. 3 F) and BCL-2+ Tcon (Fig. 3 G) were decreased in the pancreas of NOD mice with anti-PD-1-induced T1D compared with spontaneous T1D. The total number of CD127+ Tcon was reduced in anti-PD-1-induced T1D compared with spontaneous T1D (Fig. 3 H). However, the total number of CD127− Tcon was selectively increased in anti-PD-1-induced T1D (Fig. 3 H), indicating that non-memory effector Tcon are increased in anti-PD-1-induced T1D compared with spontaneous T1D and non-diabetic controls. Together, these results indicate that there is an increase in Th1 and CD127− non-memory effector Tcon in anti-PD-1-induced T1D.
Figure 3.
CD4+ T cells in the pancreas show increased proliferation in anti-PD-1-induced T1D. (A) UMAP visualization of integrated CD4+ T cells detected in the pancreas. Points represent individual cells and colors denote cluster classification as labeled. (B) UMAP visualization of integrated CD4+ T cells detected in the pancreas of each treatment group. Dots represent individual cells with the color representing the cluster classification and size corresponding to the size of the clonotype. Colors denote functional annotations that correspond to the same labels shown in A. (C) Stacked bar plots denoting the percentage of cells from each treatment group within individual clusters (left panel). Heatmap showing the number of cells detected in each cluster and treatment group (middle panel) and −log10 adjusted P values from a Fisher’s exact test (right panel). (D) Violin plots quantifying percent TIGIT+ staining within CD4+FoxP3+ Treg in the pancreas measured by flow cytometry. IgG-Spt, P = 0.0235. (E) Violin plots showing gMFI of T-bet in CD4+FoxP3−CD11ahiCD62L− Tcon in the pancreas by flow cytometry analysis. IgG-PD1, P = 0.0008; Spt-PD1, P = 0.0020. (F) Violin plots quantifying percent CD127+ staining within CD4+FoxP3−CD11ahiCD62L− Tcon in the pancreas measured by flow cytometry. IgG-Spt, P < 0.0001; Spt-PD1, P = 0.0012. (G) Violin plots showing percent BCL-2+ in CD4+FoxP3−CD11ahiCD62L− Tcon in the pancreas by flow cytometry analysis. IgG-PD1, P = 0.0010; Spt-PD1, P < 0.0001. (H) Violin plots quantifying the total number of CD4+FoxP3−CD11ahiCD127+ and CD4+FoxP3−CD11ahiCD127− Tcon detected in the pancreas measured by flow cytometry. For CD127+ Tcon: IgG-Spt P = 0.04. For CD127− Tcon: IgG-PD1 P = 0.04; Spt-PD1, P = 0.04. (I) Ratio of CD4+FoxP3−CD11ahiCD62L− Tcon to CD8+CD11ahiCD62L− T cells in the pancreas measured by flow cytometry. IgG-PD1, P < 0.0001; Spt-PD1, P < 0.0001. (J) Violin plots quantifying the total number of CD4+FoxP3−CD11ahi Tcon detected in the pancreas measured by flow cytometry. IgG-Spt, P = 0.0001; IgG-PD1, P < 0.0001. (K) Plot of the top 50 largest clonotypes, each represented as a line and terminal point, within each treatment group shown as a cumulative total percentage of TCRs detected in Tcon within that treatment group. (L) Violin plots quantifying the total number of CD4+FoxP3+ Treg detected in the pancreas measured by flow cytometry. IgG-PD1, P < 0.0001. (M) Violin plots quantifying the number of CD4+FoxP3+ Treg as a percentage of total TCR-β+ T cells detected in the pancreas. IgG-Spt, P = 0.0012; IgG-PD1, P = 0.0443. (N) Violin plots quantifying percent Ki-67+ staining within CD4+FoxP3−CD11ahiCD62L− Tcon in the pancreas measured by flow cytometry. IgG-Spt, P = 0.0004; IgG-PD1, P < 0.0001; Spt-PD1, P < 0.0001. (O) Violin plots quantifying percent Ki-67+ staining within CD4+FoxP3+ Treg in the pancreas measured by flow cytometry. IgG-PD1, P < 0.0001; Spt-PD1, P < 0.0001. Flow cytometry results are pooled from 3 to 25 independent experiments and significance determined using a non-parametric Kruskall–Wallis test with Dunn’s posthoc test for multiple comparisons. Bars in violin plots represent the first quartile, median, and third quartile. Significant comparisons are indicated with asterisks: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
CD4+ Tcon cells were significantly outnumbered by CD8+ T cells in anti-PD-1-induced T1D, underscoring the marked expansion of CD8+ T cells compared with Tcon in anti-PD-1-induced T1D (Fig. 3 I). By flow cytometry, the total number of Tcon was increased in the pancreas of spontaneous T1D and anti-PD-1-induced T1D compared to non-diabetic NOD mice (Fig. 3 J). The top 50 most expanded Tcon clonotypes made up 22% of the TCRs detected in both spontaneous and anti-PD-1-induced T1D by scTCR-seq, but only 18% in non-diabetic mice, indicating that Tcon exhibit a modest increase in expansion in the pancreas of diabetic mice (Fig. 3 K). The top 50 clonotypes of CD8+ T cells make up between 54 and 78% of the cumulative TCRs detected in each treatment group (Fig. 2 L), suggesting that there is a relatively greater diversity in CD4+ clonotypes compared with CD8+ T cell clonotypes.
The total number of Treg was increased in anti-PD-1-induced T1D (Fig. 3 L); however, the percentage of Treg within the T cell population was reduced in both spontaneous and anti-PD-1-induced T1D (Fig. 3 M) compared with non-diabetic mice. Our scRNA-seq results indicate an increase in the number of proliferating CD4+ T cells in anti-PD-1-induced T1D compared with spontaneous T1D, although it was not clear whether these were Tcon or Treg, or a combination of both (Fig. 3 C). Using flow cytometry, we validated that the percentages of Ki-67+ Tcon (Fig. 3 N) and Treg (Fig. 3 O) were significantly increased in anti-PD-1-induced T1D compared with spontaneous T1D. Overall, these data suggest that the expansion of Tcon and Treg in anti-PD-1-induced T1D is less than CD8+ T cells and the percentage of Treg within the T cell population is reduced in both anti-PD-1-induced T1D and spontaneous T1D relative to non-diabetic mice.
Anti-PD-1 induces increased TCR sharing between the pancreas and periphery, and these shared clones preferentially exhibit a proliferative terminally exhausted phenotype in the pancreas
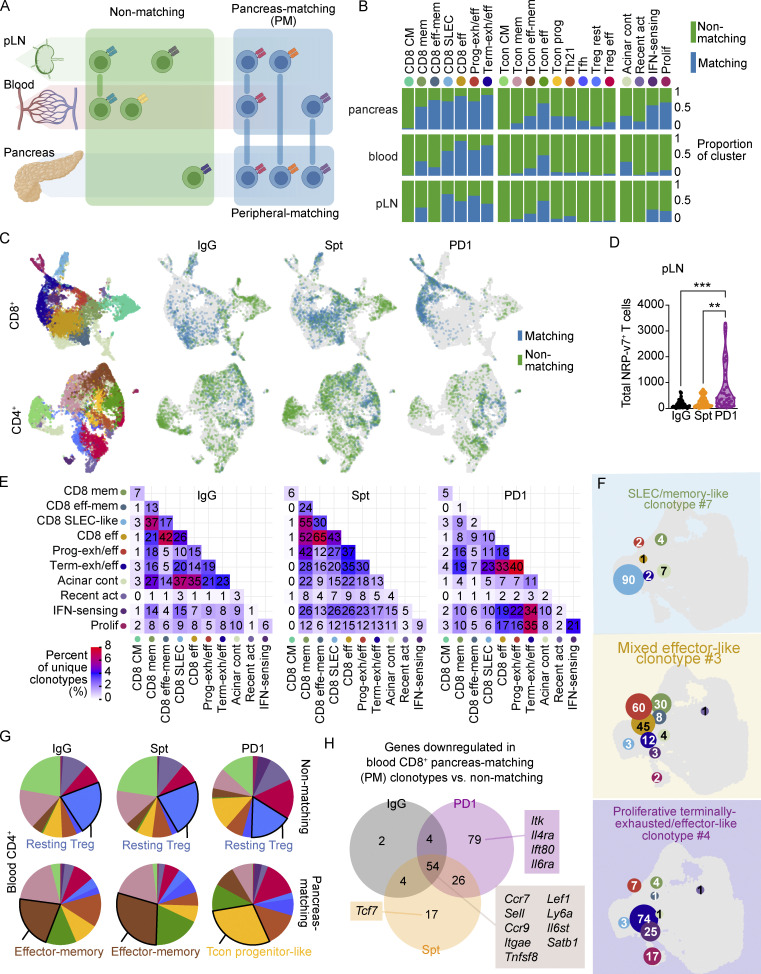
Pathogenic diabetogenic T cells differentiate in the pLN from TCF1hi stem-like progenitor T cells into TCF1lo T cells before trafficking to the pancreas (Gearty et al., 2022), but how this process is affected by PD-1 blockade is not clear. Work in cancer and chronic infection has shown that the stem-like or progenitor population of exhausted CD8+ T cells preferentially proliferates in response to PD-1 blockade compared with the terminally exhausted population. Therefore, we hypothesized that in T1D, treatment with anti-PD-1 would promote a proliferative burst in these stem-like T cells present in the pLN and cause an influx of diabetogenic T cells into the blood as they traffic to the pancreas to induce T1D. To test this hypothesis, we used the TCR as a molecular barcode as an unbiased way to identify any T cell clones in the periphery that have a shared lineage relationship to T cells in the pancreas. This approach allows a broader view of the diabetogenic T cell repertoire than would be possible by using MHC tetramer reagents, which are limited to well-defined antigens. Using the TCR sequence, we distinguished peripheral-matching from non-matching T cells in the pancreas based on whether cells in the clonotype could be identified in at least one peripheral tissue (blood and/or pLN). T cell clones in the periphery (e.g., blood or pLN) that were also identified in the pancreas were classified as PM T cells (Fig. S4 A). In general, CD8+ T cell clusters showed more sharing between the pancreas and periphery than the CD4+ Tcon and Treg clusters (Fig. S4, B and C). In anti-PD-1-induced T1D, the percentage of peripheral-matching to non-matching unique CD4+ and CD8+ clonotypes was increased in the three mice examined compared with spontaneous T1D or non-diabetic mice (Fig. 4 A). In anti-PD-1-induced T1D, there was an increase in the number of unique PM CD4+ and CD8+ T cell clonotypes in the blood, but we did not detect any differences between the treatment groups in the pLN (Fig. 4 B). By flow cytometry, we examined the frequency of NRP-v7+ T cells in both the pLN and blood as a surrogate of PM CD8+ T cells. The total number (Fig. S4 D) and percentage of NRP-v7+ T cells in the pLN were significantly increased in anti-PD-1-induced T1D (Fig. 4 C) compared with spontaneous T1D. The frequency of NRP-v7+ T cells in the blood was also significantly increased in anti-PD-1-induced T1D (Fig. 4 D) compared with spontaneous T1D. The discrepancy in the scRNA-seq and flow cytometry findings may be reflective of sensitivity differences between these two methods. TCR sequence diversity in the blood and pLN is incredibly high relative to the pancreas, so we may not have had sufficient numbers to observe changes in the abundance of the PM population using this method due to the large number of non-matching TCRs in the periphery. When comparing matching vs. non-matching T cells, between tissue compartments and treatment groups, we found that the largest differences in transcriptional states were present in the matching groups (Fig. 4 E), while minimal differences were observed in the non-matching groups (Fig. 4 F). There was a modest increase in the proportion of progenitor-like Tcon in peripheral-matching CD4+ T cells that reflected a comparable increase in the PM T cells measured in anti-PD-1-induced T1D compared with spontaneous T1D. In marked contrast, the distribution of subsets in matching CD8+ T cells was vastly different in anti-PD-1-induced T1D compared with the other treatment groups. Here, the matching CD8+ T cells in both the pancreas and periphery were largely dominated by the terminally exhausted/effector-like cluster, unlike spontaneous T1D and non-diabetic control. Collectively, these data illustrate that anti-PD-1 increases TCR sharing between the periphery and the pancreas, accompanied by an increase in the proportion of terminally exhausted/effector-like T cells within the matching CD8+ T cell population.
Figure S4.
Clonotype matching across tissues and clusters differs in anti-PD-1-induced T1D compared to spontaneous T1D. (A) Diagram depicting the distribution of PM, peripheral-matching, and non-matching across the pLN, blood, and pancreas based on clonotype. (B) Stacked bar plots showing the proportion of matching and non-matching T cells within individual clusters across the pLN, blood, and pancreas. (C) UMAP visualization of CD8+ (top) and CD4+ (bottom) peripheral-matching (blue) and non-matching (green) T cells in the pancreas of each treatment group. Legend for the leftmost UMAP is shared with B. (D) Violin plots quantifying the total number of NRP-v7+ CD8+ T cells in the pLN measured by flow cytometry. IgG-PD1, P < 0.0001; Spt-PD1, P = 0.0071. (E) Heatmaps showing the number of unique clonotypes shared across clusters in the different treatment groups. The color of tiles corresponds to the percentage of unique clonotypes shared across two clusters. (F) UMAP visualization of the number of cells in each cluster within selected individual clonotypes. The color of the circle represents the cluster (legend shared with B), the size of the circle represents the relative number of cells in that cluster, and the number represents the actual number of cells in that cluster. Circles are located at the mean centroid of the UMAP coordinates of the total cells within each cluster. (G) Pie charts displaying the cluster proportions of non-matching (top) and PM (bottom) CD4+ T cells in the blood. The cluster that constitutes the largest proportion is annotated with text. (H) Venn diagram showing the number of down-regulated genes in blood PM vs. non-matching CD8+ T cells between the three treatment groups. Selected genes within each group are listed. Full list can be found in Table S5. Significant comparisons are indicated with asterisks: **, P < 0.01; ***, P < 0.001.
Figure 4.
pLN and blood CD4+ and CD8+ T cells having shared clonality with cells in the pancreas can be identified using the TCR as a barcode. (A) Violin plots showing the percentage of peripheral-matching CD4+ (top) and CD8+ (bottom) clonotypes out of the total unique clonotypes detected in the pancreas. Each dot represents the cells from one individual mouse (n = 3 mice per group). (B) Violin plots showing the percentage of PM CD4+ (top) and CD8+ (bottom) clonotypes out of total unique clonotypes detected in the pLN (left) and blood (right). Each dot represents the cells from one individual mouse (n = 3 mice per group). (C) Violin plots quantifying the percentage of NRP-v7+ CD8+ T cells out of CD8+CD11ahi T cells detected in the pLN measured by flow cytometry. IgG-PD1, P < 0.0001; Spt-PD1, P = 0.0347; IgG-Spt, P = 0.0354. (D) Violin plots quantifying the total number of NRP-v7+ CD8+ T cells per million T cells in the blood measured by flow cytometry. IgG-Spt, P = 0.0287; IgG-PD1, P < 0.0001. (E) Stacked bar plots showing the cluster composition of matching CD4+ and CD8+ T cells across the indicated treatment groups in the pancreas (top) and combined blood and pLN (bottom). (F) Stacked bar plots showing the cluster composition of non-matching CD4+ and CD8+ T cells across the indicated treatment groups in the pancreas (top) and combined blood and pLN (bottom). Legend is shared with E. (G) Stacked bar plots of the percentage of cells detected in clusters within individual SLEC-like clonotypes across pLN, blood (bld), and pancreas (pan). The color of text and border surrounding the stacked bar plot corresponds to the treatment group of the clonotype. (H) Stacked bar plots of the percentage of cells detected in clusters within individual mixed effector-like clonotypes across pLN, blood (bld), and pancreas (pan). The color of the text and border surrounding the stacked bar plot corresponds to the treatment group of the clonotype. (I) Stacked bar plots of the percentage of cells detected in clusters within individual proliferative terminally exhausted/effector-like clonotypes across pLN, blood (bld), and pancreas (pan). (G–I) Legend shared with E. Flow cytometry results are pooled from 3 to 25 independent experiments and significance determined using a non-parametric Kruskall–Wallis test with Dunn’s posthoc test for multiple comparisons. Bars in violin plots represent the first quartile, median, and third quartile. Significant comparisons are indicated with asterisks: *, P < 0.05, **, P < 0.01; ***, P < 0.001.
We next examined the transcriptional profiles of individual clonotypes across the tissues. Within the top 29 expanded clonotypes shared across all three tissues, there were three distinct phenotypes observed: SLEC-like clonotypes, mixed effector-like clonotypes, and proliferative terminally exhausted/effector-like clonotypes. SLEC-like clonotypes (Fig. 4 G) were predominantly in the SLEC-like T cell cluster. The T cells in these clonotypes were not significantly expanded in the pancreas compared with the pLN, suggesting that they may be short-lived. Mixed effector-like clonotypes (Fig. 4 H) were expanded in the pancreas compared to the periphery. Transcriptionally, these T cells were memory or progenitor-exhausted/effector-like in the periphery and memory, effector, and exhausted/effector-like in the pancreas. Both SLEC-like clonotypes and mixed effector-like clonotypes were identified in non-diabetic and spontaneously diabetic mice, but mixed effector-like clonotypes were more numerous in spontaneously diabetic compared with non-diabetic mice. Given that mixed effector-like clonotypes were more highly expanded in the pancreas compared with the periphery and were more numerous in spontaneously diabetic mice, these clonotypes may be associated with diabetogenesis. The proliferative terminally exhausted/effector-like clonotypes (Fig. 4 I) contained significant proportions of interferon-sensing, proliferating, and terminally exhausted/effector-like subsets. Within the top 29 expanded clonotypes shared across all the tissues, the proliferative terminally exhausted/effector-like clonotypes were exclusively identified in anti-PD-1-induced T1D. The numbers of unique clonotypes shared across the interferon-sensing, proliferating, and terminally exhausted/effector-like clusters across all the tissues were higher in anti-PD-1-induced T1D. Clonotypes shared between the effector and memory subsets were higher in spontaneous T1D (Fig. S4 E). Examples of individual clonotypes from each of the three phenotypes illustrate the distinct differences in distribution of CD8+ T cells across different clusters (Fig. S4 F). In summary, these data suggest that diabetogenic CD8+ T cells are significantly increased in the pLN and blood in anti-PD-1-induced T1D, supporting the potential of monitoring the periphery for T cells associated with irAEs.
Circulating blood CD4+ and CD8+ T cells with matching TCRs to cells in the pancreas show unique transcriptional features in anti-PD-1-induced T1D compared with spontaneous T1D
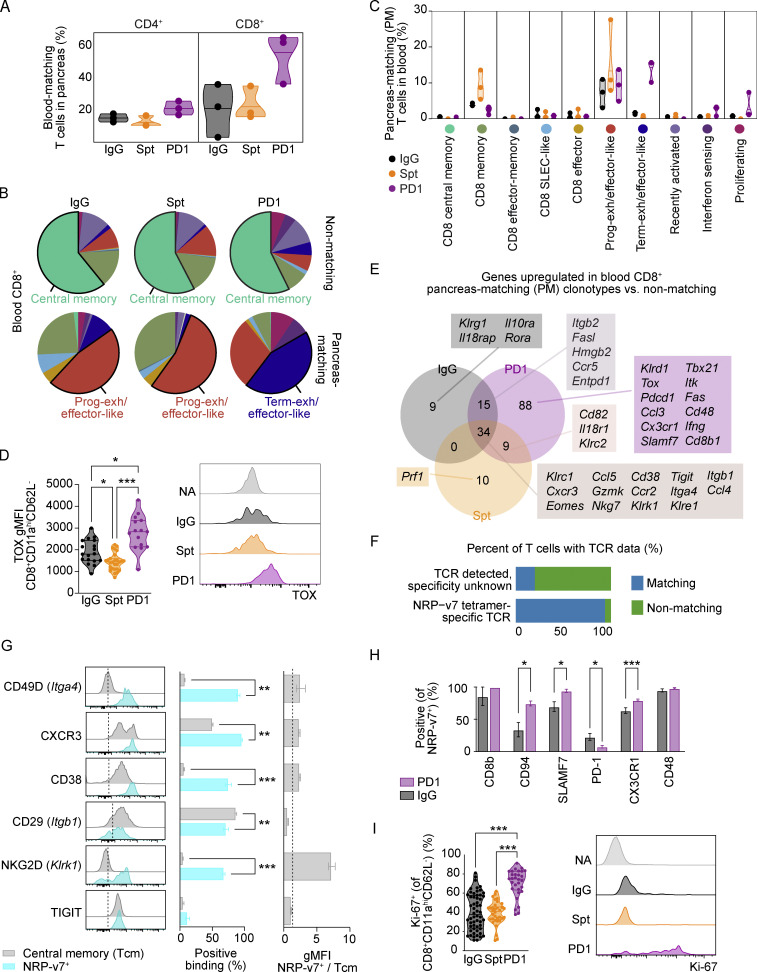
Our findings suggest that mobilization of T cells from the pLN to the pancreas may be crucial for anti-PD-1-induced T1D. Since the blood is a critical conduit of immune cell trafficking between LNs and tissues (Masopust and Schenkel 2013), we speculated that tracking changes in diabetogenic T cell populations in the blood would be a useful metric for tracking irAEs. Indeed, we measured an increase in blood-matching CD8+ T cells in the pancreas of mice with anti-PD-1-induced T1D compared with non-diabetic controls and spontaneously diabetic mice (Fig. 5 A). The transcriptional profiles of non-matching CD4+ T cells in the blood were comparable across the treatment groups, but within the PM T cells in the blood, the proportion of progenitor-like Tcon was increased in anti-PD-1-induced T1D (Fig. S4 G). Similarly, the proportion of terminally exhausted/effector-like CD8+ T cells in the PM T cells was markedly increased in anti-PD-1-induced T1D (Fig. 5 B) across all the mice examined (Fig. 5 C). In contrast, the transcriptional profiles of non-matching CD8+ T cells in the blood were relatively consistent between treatment groups. Flow cytometry analysis of the blood showed a significant increase in TOX expression by CD8+CD11ahiCD62L− effector T cells in anti-PD-1-induced T1D, consistent with an increased prevalence of terminally exhausted/effector-like CD8+ T cells in the blood (Fig. 5 D).
Figure 5.
Circulating blood CD4+ and CD8+ T cells with matching TCRs to pancreatic CD4+ and CD8+ T cells show unique transcriptional features in anti-PD-1-induced T1D compared to spontaneous T1D. (A) Violin plots quantifying the percentage of blood-matching T cells in the pancreas out of all CD4+ (left) or CD8+ (right) T cells. Each dot represents the cells from one individual mouse (n = 3 mice per group). (B) Pie charts displaying the cluster proportions of non-matching (top) and PM (bottom) CD8+ T cells in the blood. The cluster that constitutes the largest proportion is annotated with text. The colors for the pie slices correspond with the clusters identified in Fig. 2 A, and the corresponding names and colors for each cluster are shared with C. (C) Violin plots showing the frequency of PM CD8+ T cells in the blood on a cluster-by-cluster basis, separated by treatment group. The clusters are those that were identified in Fig. 2 A. (D) Violin plots showing gMFI of TOX in CD8+CD11ahiCD62L− T cells in the pancreas by flow cytometry analysis (left). IgG-Spt, P = 0.0393; IgG-PD1, P = 0.0408; Spt-PD1, P < 0.0001. Representative flow cytometry TOX staining on naive (NA; CD8+CD11a−CD62L+) and effector CD8+CD11ahiCD62L− T cells (right) from indicated treatment groups. (E) Venn diagram showing the number of upregulated genes in PM vs. non-PM CD8+ T cells between the three treatment groups. A subset of selected genes within each group is listed. Full list available in Table S5. (F) Bar plot showing the percentage of T cells with known TCR data but with unknown specificity, or TCRs that matched to NRP-v7+ sorted TCR sequences from Gearty et al. (2022). (G) Flow cytometry results of the indicated markers on NRP-v7+ CD8+ T cells and central memory (CD8+CD11ahiCD62L+) T cells in the blood of NOD mice with anti-PD-1-induced T1D. From left to right: Representative flow cytometry histograms; percent positive central memory T cells and NRP-v7+ T cells; and the ratio gMFI of NRP-v7+ over central memory T cells (CD8+CD11ahiCD62L+). Statistical significance determined using a nonparametric Mann–Whitney test with Holm-Sidak’s correction for multiple comparisons. For P values between central memory T cells and NRP-v7+ (middle panel), CD49D P = 0.0023; CXCR3 P = 0.0023; CD38 P < 0.0001; CD29 P = 0.0023, NKG2D P < 0.0001. (H) Bar plots illustrating the positive staining of indicated markers on NRP-v7+ T cells in the blood of anti-PD-1-induced T1D and non-diabetic mice. Significance between PD1 and IgG for the following markers: CD94 P = 0.0277; SLAMF7 P = 0.0202; PD-1 P = 0.0294; CX3CR1 P = 0.0001. (I) Violin plots quantifying percent Ki-67+ staining within CD8+CD11ahiCD62L− T cells in the blood measured by flow cytometry. IgG-PD1, P < 0.0001; Spt-PD1, P < 0.0001. Representative flow cytometry Ki-67 staining on naive (NA; CD8+CD11a−CD62L+) and effector CD8+CD11ahiCD62L− T cells (right). Flow cytometry results are pooled from 3 to 25 independent experiments and significance determined using a non-parametric Kruskall–Wallis test with Dunn’s posthoc test for multiple comparisons. Bars in violin plots represent the first quartile, median, and third quartile. Significant comparisons are indicated with asterisks: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
We next investigated markers that could be used to identify PM T cells in the blood as a potential biomarker of anti-PD-1-induced T1D. We used a Wilcoxon Rank Sum test to identify differentially expressed genes between PM and non-matching CD8+ T cells in the blood and the overlap of these genes (Table S5). There were 34 shared genes upregulated in PM relative to non-matching CD8+ T cells across the treatment groups (Fig. 5 E). These shared genes encoded chemokine receptors (Cxcr3, Ccr2), integrins (Itga4, Itgb1), and effector molecules (Ccl4, Ccl5, Gzmk, Klrk1, Klre1, Nkg7). Downregulated genes shared between the treatment groups included genes associated with central memory T cells: Sell, Ccr7, and Lef1 (Fig. S4 H and Table S5). We examined expression of some of these extracellular markers on blood CD8+ T cells from NOD mice with anti-PD-1-induced T1D by flow cytometry. Since central memory T cells were predominantly non-matching (Fig. 5 B), CD8+CD44hiCD62L+ T cells were used as a non-matching population. NRP-v7+ T cells were used as the PM population, as 93% of the TCRs detected in NRP-v7+ sorted T cells from the literature were identified in our scTCR-seq dataset classified as PM (Fig. 5 F and Table S6). Of the markers examined, CD49D (Itga4), CD38, and NKG2D (Klrk1) were expressed on 60–80% of NRP-v7+ T cells with a large fold-change difference in geometric mean fluorescence intensity (gMFI) that distinguished the NRP-v7+ from central memory T cells (Fig. 5 G). We identified 88 genes specific to PM CD8+ T cells in anti-PD-1-induced T1D (Fig. 5 E), which included genes associated with exhaustion (Pdcd1, Tox) and effector function (Fas, Ifng; Table S5). We assessed a subset of the extracellular markers by flow cytometry (CD8b, CD94, SLAMF7, PD-1, CX3CR1, and CD48). NRP-v7+ cells in anti-PD-1 treated NOD had significantly lower expression of PD-1 and higher expression of CD94, SLAMF7, and CX3CR1 compared with NRP-v7+ cells in control-treated and non-diabetic mice (Fig. 5 H). Lastly, we speculated that proliferation would be useful for tracking PM cells in the blood since our transcriptional data showed that CD8+ T cell proliferation was significantly increased in the anti-PD-1-induced T1D group. Measurement of Ki-67 staining by flow cytometry in blood effector CD8+CD44hiCD62L− T cells revealed a significant increase in the percentage of Ki-67+ T cells within this population in anti-PD-1-induced T1D (70%) compared with non-diabetic (40%, P < 0.0001) and spontaneously diabetic mice (39%, P < 0.0001; Fig. 5 I). Collectively, these data suggest that markers such as CD49D, CD38, and/or NKG2D may distinguish PM CD8+ T cells in the blood from non-matching CD8+ T cells and that these markers together with increased Ki-67 expression may be useful for tracking irAEs following anti-PD-1 treatment.
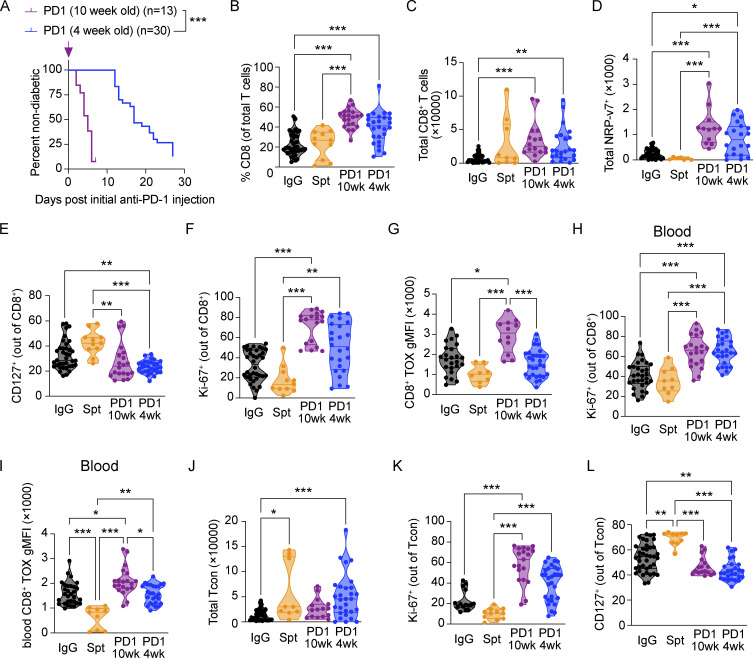
While our data in older NOD mice provide a strong rationale for incorporating analyses of the peripheral immune system to track irAE development, this represents only one late-stage time point. To assess the utility of using this method to detect disease pathogenesis in earlier stage disease, we compared young 4-wk-old NOD mice treated with anti-PD-1 to older NOD mice treated with anti-PD-1 (Fig. S5 A). Previous work has shown that younger mice are sensitive to anti-PD-1-induced acceleration of T1D, but T1D takes longer to develop than in older NOD mice (Ansari et al., 2003). Anti-PD-1-induced T1D in young and old mice exhibited a comparable increase in the percentages (Fig. S5 B) and total numbers (Fig. S5 C) of CD8+ T cells and NRP-v7+ (Fig. S5 D) T cells in the pancreas relative to non-diabetic mice, demonstrating that the increase in pancreatic CD8+ T cells in anti-PD-1-induced T1D in older mice is also observed in younger mice. The percentages of CD127+ (Fig. S5 E) and Ki-67+ (Fig. S5 F) CD8+ T cells in the pancreas of anti-PD-1-induced T1D was not significantly different between young and old mice, suggesting that the proportion of memory T cells and proliferative T cells was comparable between the two conditions. CD8+ T cells in young mice showed reduced expression of TOX in the pancreas relative to anti-PD-1-induced T1D in older mice (Fig. S5 G). In the blood, the percentage of Ki-67+ CD8+ T cells was similarly increased in old and young mice with anti-PD-1-induced T1D relative to non-diabetic mice (Fig. S5 H), indicating that the effects of PD-1 blockade on CD8+ T cell proliferation in the peripheral blood are consistent despite the differences in the kinetics of onset of anti-PD-1-induced T1D. In anti-PD-1-induced T1D, TOX expression in CD8+ T cells in the blood is modestly reduced in young mice compared with old mice, consistent with observations in the pancreas (Fig. S5 I). We next examined CD4+ T cells in the pancreas of young and old mice with anti-PD-1-induced T1D and found no significant difference between the total number of Tcon in the pancreas (Fig. S5 J), the percentage of Ki-67+ Tcon (Fig. S5 K), and the percentage of CD127+ Tcon (Fig. S5 L), suggesting that the numbers and phenotype of Tcon are comparable between the two conditions. Together, these data indicate that the differences in the kinetics of anti-PD-1-induced T1D in young or older treated mice do not affect the number or proportion of T cells in the pancreas at time of diabetes onset and highlight the utility of incorporating analyses of the peripheral immune system in both early stage and late-stage disease.
Figure S5.
Anti-PD-1-induced T1D in young NOD recapitulates features of anti-PD-1-induced T1D in older NOD mice. (A) Percent diabetes-free NOD mice administered anti-PD-1 antibody starting at 4 wk of age (blue) or 10 wk of age (purple). Kaplan–Meier survival curves and results from a Mantel–Cox log-rank test (P < 0.001) are shown. (B) Violin plots quantifying the percentage of CD8+CD11ahiCD62L− T cells in the pancreas out of total T cells measured by flow cytometry. IgG-PD1 (10 wk), P < 0.001; PD1 (10 wk)-Spt, P < 0.001; IgG-PD1 (4 wk), P < 0.001. (C) Violin plots quantifying the total number of CD8+CD11ahiCD62L− T cells in the pancreas measured by flow cytometry. IgG-PD1 (10 wk), P < 0.001; IgG-PD1 (4 wk), P = 0.004. (D) Violin plots quantifying the total number of NRP-v7+ CD8+CD11ahiCD62L− T cells in the pancreas measured by flow cytometry. IgG-PD1 (10 wk), P < 0.001; IgG-PD1 (4 wk), P = 0.01; PD1 (10 wk)-Spt, P < 0.001; PD1 (4 wk)-Spt, P < 0.001. (E) Violin plots quantifying the percentage of CD127+ CD8+CD11ahiCD62L− T cells in the pancreas measured by flow cytometry. IgG-PD1 (4 wk), P = 0.007; PD1 (10 wk)-Spt, P = 0.002; PD1 (4 wk)-Spt, P < 0.001. (F) Violin plots quantifying the percentage of Ki-67+ CD8+CD11ahiCD62L− T cells in the pancreas measured by flow cytometry. IgG-PD1 (10 wk), P < 0.001; PD1 (10 wk)-Spt, P < 0.001; PD1 (4 wk)-Spt, P = 0.001. (G) Violin plots showinggMFI) of TOX in CD8+CD11ahiCD62L− T cells in the pancreas by flow cytometry analysis. IgG-PD1, P = 0.01; PD1 (10 wk)-PD1 (4 wk), P < 0.001; PD1 (10 wk)-Spt, P < 0.001. (H) Violin plots quantifying the percentage of Ki-67+ CD8+CD11ahiCD62L− T cells in the blood measured by flow cytometry. IgG-PD1 (10 wk), P < 0.001; PD1 (10 wk)-Spt, P < 0.001; PD1 (4 wk)-Spt, P = 0.001; IgG-PD1 (4 wk) P < 0.001. (I) Violin plots showing gMFI of TOX in CD8+CD11ahiCD62L− T cells in the blood by flow cytometry analysis. IgG-PD1 (10 wk), P = 0.02; IgG-Spt, P < 0.001; PD1 (10 wk)-PD1 (4 wk), P = 0.02; PD1 (10 wk)-Spt, P < 0.001; PD1 (4 wk)-Spt, P = 0.001. (J) Violin plots quantifying the total number of CD4+CD11ahiCD62L−FoxP3− Tcon in the pancreas measured by flow cytometry. IgG-PD1 (4 wk), P < 0.001; IgG-Spt, P = 0.04. (K) Violin plots quantifying the percentage of Ki-67+ CD4+CD11ahiCD62L−FoxP3− Tcon in the pancreas measured by flow cytometry. IgG-PD1 (10 wk), P < 0.001; PD1 (10 wk)-Spt, P < 0.001; PD1 (4 wk)-Spt, P < 0.001. (L) Violin plots quantifying the percentage of CD127+ CD4+CD11ahiCD62L−FoxP3− Tcon in the pancreas measured by flow cytometry. IgG-PD1 (4 wk), P = 0.003; IgG-Spt, P = 0.008; PD1 (10 wk)-Spt, P < 0.001; PD1 (4 wk)-Spt, P < 0.001. Significant comparisons are indicated with asterisks: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Discussion
A better understanding of how PD-1-induced irAEs compare with naturally arising autoimmune diseases is needed for the treatment and monitoring of these irAEs. Using paired single-cell RNA-seq and TCR-seq of CD4+ and CD8+ T cells isolated from the blood, pLN, and pancreas, we identified similarities and differences between spontaneous T1D and anti-PD-1-induced T1D in the NOD mouse model. We found notable phenotypic and transcriptional differences in both CD4+ and CD8+ T cell compartments between spontaneous T1D and anti-PD-1-induced T1D, including a shift toward a terminally exhausted/effector-like state and reduced memory T cell populations in anti-PD-1-induced T1D. Using the TCR as a molecular barcode to identify diabetogenic T cells, we determined that anti-PD-1 treatment causes a dramatic increase in the frequency of peripheral-matching CD4+ and CD8+ T cells in the pancreas with a concomitant increase in pancreas-specific CD8+ T cells in both the pLN and the peripheral blood by flow cytometry. Moreover, we identified markers on the diabetogenic T cell population in the blood that differed between spontaneous T1D and PD-1-induced T1D, providing proof-of-concept that tracking blood T cells could provide a means to monitor irAEs in patients. Together, these data challenge the existing dogma that the pancreas is the only site that can be used to monitor autoimmune diabetes and provide a rationale for incorporating peripheral tissues such as blood into clinical studies rather than exclusively focusing on the autoimmune target organ.
The dramatic increase in pancreatic CD8+ T cells with TCR sequences matching with those in the periphery in anti-PD-1-induced T1D suggests that anti-PD-1 may affect T cell differentiation and expansion in the pLN and recruitment from the pLN to the pancreas. Our findings support a model whereby PD-1 inhibitors act on diabetogenic CD4+ Tcon and CD8+ T cells in the periphery, inducing a rapid mobilization of these cells to the pancreas even in older NOD mice. Mechanistically, these new recruits to the pancreas as well as the CD4+ and CD8+ T cells that were already in the pancreas prior to PD-1 blockade may functionally contribute to T1D onset. Indeed, work from Guleria and colleagues established that pancreatic T cells in older NOD mice are sufficient to induce T1D following PD-1 blockade (Guleria et al., 2007). However, just because these T cells can induce T1D if properly stimulated does not exclude the possibility that PD-1 blockade can also act on T cells in the periphery, and our data support a model where PD-1 inhibitors act on cells in the periphery as well. We speculate that anti-PD-1 may act, at least in part, by inducing proliferation of the progenitor-like population of T cells in the pLN, causing these cells to differentiate into terminally exhausted and/or effector-like cells and to traffic into the pancreas to mediate T1D. This effect may not be limited to the pLN, as we and other groups have shown that a small population of progenitor-exhausted-like T cells exist in the pancreatic islets (Grebinoski et al., 2022; Zakharov et al., 2020). Our findings are consistent with studies indicating that PD-1 inhibitors preferentially act to induce the proliferation of progenitor-like exhausted cells during chronic infections and cancer (Ciecko et al., 2021; Miller et al., 2019; Im et al., 2016).
Clinically, the blood offers a number of advantages for immune monitoring, including that it can be sampled repeatedly and in a far less invasive manner than sampling the autoimmune target organ. However, reliable blood biomarkers for monitoring patients on checkpoint blockade therapy for irAEs are lacking. Our TCR-seq data showed that the increase in terminally exhausted/effector-like T cells in the pancreas was reflected in the blood in mice with anti-PD-1-induced T1D, highlighting this population as a potential biomarker for irAEs. We identified 34 differentially expressed genes for surface markers associated with matching CD8+ T cells in the blood across the treatment groups including Itga4 (CD49D), Cxcr3, Cd38, Itgb1 (CD29), Klrk1 (NKG2D), and Tigit. To validate these markers by flow cytometry, we analyzed blood NRP-v7+ T cells, which should match T cell clones in the pancreas, and CD8+CD11ahiCD62L+ central memory T cells as a non-matching control, as this cluster was highly represented in non-matching T cells. CD49D, CD38, and NKG2D were more highly expressed by NRP-v7+ T cells compared with central memory T cells, suggesting that they may be suitable markers for this terminally exhausted/effector-like population. These studies provide proof-of-concept for use of markers on T cells in the blood as a biomarker of PM T cells that mediate irAEs in cancer patients. Further studies are required to assess the utility of CD49D, CD38, and NKG2D expression on activated CD8+ T cells in the blood as potential biomarkers of PM T cells that mediate irAEs. Additional studies comparing blood and organ-matching cells in other irAEs may reveal disease-specific biomarkers. The percentage of activated CD38+ T cells in the peripheral blood is increased in cancer patients treated with anti-PD-1/PD-L1 (Kim et al., 2020; Huang et al., 2017), but whether this correlates with the incidence of irAEs is not clear. The significant increase in the percentage of Ki-67+ T cells within effector Tcon, Treg, and CD8+ T cells in the blood also may serve as a biomarker of anti-PD-1-induced T1D. Given that checkpoint blockade–induced colitis in patients is associated with an increase in Ki-67+ Tcon, Treg, and CD8+ T cells in the colon (Luoma et al., 2020), we speculate that the observed increase in proliferation in tissues affected by irAEs may be extended to a systemic response that can be monitored in the peripheral blood. It should be noted that our study specifically examined changes in the blood at the onset of T1D. Therefore, additional or alternative markers may be involved in tracking the early development of irAEs, and future work identifying these markers could be extremely useful for early detection. For such studies, obtaining a blood sample for analysis prior to immunotherapy will be important for establishing a baseline as a comparison point. When to sample for early detection of irAEs is less clear. In cancer, the major proliferative burst in CD8+ T cells in the blood occurs early following the first treatment with anti-PD-1 (Huang et al., 2017; Huang et al., 2019). However, whether all of those T cells were tumor-specific or if some of those T cells could have contributed to irAEs is unclear. Considering the high level of variability in the timing of onset of irAEs following checkpoint blockade (Pauken et al., 2019; Brahmer et al., 2018), identifying the precise time window in which blood can predict these events would be tremendously useful.
A major outstanding question is how to reliably distinguish pathogenic T cells that are responsible for irAEs from protective T cells that are driving the productive anti-tumor response. In the present study, we utilized the TCR as a molecular barcode to identify putative diabetogenic T cells in the periphery based on shared TCR sequences with T cells found in the pancreas tissue. However, many of the markers that we identified here have also been observed by us (Pauken et al., 2021) and others (Huang et al. 2017; Lucca et al., 2021; Huang et al. 2019) to be associated with anti-tumor T cell responses. Consequently, without having the TCR sequences to classify cells as tumor-matching or PM, it is difficult to evaluate whether a cell is protective or pathogenic simply based on the surface markers expressed. One of the limitations of using the TCR as a barcode is that a reference list of TCRs is needed. For example, in this study, we used the list of TCR sequences recovered from the pancreas as a starting point to identify cells of interest in the periphery. Similarly, in tumor studies, the list of TCR sequences recovered from the tumor was used as a starting point to identify cells of interest in the blood (Pauken et al., 2021; Lucca et al., 2021; Gueguen et al., 2021). In patients, getting paired blood and tumor tissue is often challenging, and extending this to adverse events—where tissue from the organ impacted would be needed—would be technically difficult. A public database of TCR sequences known to be associated with autoimmunity vs. anti-tumor immunity could have clinical utility. However, many TCR sequences are private to an individual, meaning they are only found in that person (Striebich et al., 1998; Klarenbeek et al., 2012; Chang et al., 2021). This would restrict the usefulness of TCR databases to those TCR sequences that are public or shared across individuals (Munson et al., 2016; Beausang et al., 2017; Wang et al., 2017; Levy et al., 2016), and limit the proportion of the TCR repertoire that could be assessed in a single individual. Alternatively, consensus-based approaches for identifying TCRs based on the antigen they are likely to bind could increase the utility of these databases since they would rely on identification of shorter consensus motifs rather than the entire TCR sequence (Glanville et al., 2017; Sidhom et al., 2021; Dash et al., 2017). Ultimately, developing methods to reliably distinguish pathogenic from protective TCR sequences, particularly in the blood, would advance monitoring of irAEs in patients.
In summary, we have identified differences in spontaneous and anti-PD-1-induced T1D. Anti-PD-1-induced T1D is associated with an increase in terminally exhausted/effector-like T cells that match the blood, while spontaneous T1D is largely dominated by effector and memory T cell subsets. Moreover, our results suggest that the pancreas is not the only anatomical site that is important in anti-PD-1-induced T1D. Rather, anti-PD-1 treatment can act on diabetogenic T cells in the pLN and blood, and the blood provides a more clinically tractable site for monitoring irAE development than sampling the pancreas directly. IrAEs in cancer patients have been tremendously diverse in terms of tissue impacted, severity of symptoms, time of onset, and pre-existing condition, and there are likely many different mechanisms that contribute to irAE development and symptom onset (Postow et al. 2018). In our study, we focused on the T cell compartment since both CD4+ and CD8+ T cells have been shown to be critical for PD-1-induced T1D in NOD mice (whereas antibodies were dispensable; Guleria et al., 2007). Similarly, a number of checkpoint-induced irAEs are associated with T cell infiltration of the affected tissue, including myocarditis (Johnson et al., 2016; Zhu et al., 2022) and colitis (Luoma et al., 2020), while autoantibodies have been less useful at least in preliminary studies of T1D (Tsang et al., 2019; Johansen et al., 2019). However, considering the staggering diversity of irAEs, expanding the database of irAE samples from patients will be essential for clarifying the underlying etiology of these diseases, and this mechanistic understanding will inform monitoring and treatment strategies. Additionally, larger-scale studies incorporating multiple cell types and/or effector molecules will be useful for precisely identifying the roles of different leukocyte populations in checkpoint-induced irAE patients. For example, B cells and autoantibodies play a critical role in several naturally occurring autoimmune diseases (e.g., rheumatoid arthritis, Grave’s disease, lupus, etc.). Additionally, increasing evidence suggests a role for myeloid populations in autoimmune pathology. Of note, the composition of the myeloid compartment is significantly different in older vs. younger NOD, including the proportion of Cxcl6hi macrophages that express high levels of IFNγ-inducible genes being higher in the pancreas of older NOD mice (Zakharov et al., 2020). Future studies investigating how these different myeloid populations contribute to checkpoint-induced pathologies are of interest. Larger-scale clinical studies examining diverse leukocyte populations in patients with irAEs will be extremely useful for clarifying the underlying etiology behind these diseases and will be necessary to refine immunotherapy approaches to mitigate these events.
Materials and methods
Experimental model details
Mice and antibody treatment
WT female NOD/ShiLtJ mice were purchased from The Jackson Laboratory (stock number 001976) at 8 wk of age and sacrificed between 10 and 20 wk of age. The average age of mice across all experiments was 16.2 ± 2.1 wk of age. For scRNA-seq analysis, 16-wk-old NOD mice were given two intraperitoneal (i.p.) doses of 200 μg of anti-PD-1 antibody (clone: 29F.1A12, from BioXCell) or isotype control antibody (clone: 2A3, from BioXCell) in 200 μl per dose on days −4 and −2. For time-course experiments, mice were injected with anti-PD-1 three times a week starting at 10–11 wk of age, or 4 wk of age for the young cohort of mice (Fig. S5). For anti-IL-7R experiments, 200 μg of anti-IL-7Rα antibody (clone A7R34) or Rat IgG2a isotype control antibody (clone 2A3) were injected i.p. twice per week into NOD mice starting at 10 wk of age. At 13 wk of age, a subset of these mice was also injected with 100 µg anti-PD-1 antibody (clone 29F.1A12) i.p. three times a week for five additional weeks or until diabetes onset. For the time-course, histological, and flow cytometry analyses, antibody-treated mice were within 3 wk of age from spontaneously diabetic mice analyzed within an experiment. Age-matched mice were chosen for these experiments since previous work has shown that the cellular landscape of leukocytes in the pancreas changes dramatically with age (Zakharov et al., 2020), including both the activation state of the T cell populations and the composition of the myeloid compartment. Consequently, age-matched mice were used to limit the variables between experimental groups. Mice were analyzed when they became diabetic. Mice were considered diabetic upon detection of two positive (≥250 mg/dl) reagent strips for glucose urinalysis (VWR) measured on two consecutive days. Mice were maintained at Harvard Medical School in specific pathogen-free facilities under housing, husbandry, and diet conditions in accordance with Institutional Animal Care and Use Committee and National Institutes of Health guidelines. All experimental procedures performed were approved by the Institutional Animal Care and Use Committee at Harvard Medical School.
Method details
Lymphocyte isolation from tissues
Peripheral blood from the retro-orbital sinus was collected into 4% sodium citrate (Sigma-Aldrich) to prevent clotting. Roswell Park Memorial Institute (RPMI) + 10% fetal bovine serum (FBS) were added and the mixture was centrifuged at 400 × g through histopaque-1083 to isolate white blood cells (Sigma-Aldrich). The white blood cell layer at the interface between the histopaque and remaining medium was collected and washed with RPMI + 10% FBS before staining for flow cytometry analysis or sorting for scRNA-seq. Single-cell suspensions from the pLN were generated by smashing the tissue on a 70-µm filter using the plunger of a 3-cc syringe.
The pancreas was dissected and finely minced using scissors. The dissociated tissue was digested with collagenase type I (400 U/ml; Worthington Biochemical) and DNAse I (10 μg/ml; Sigma-Aldrich) for 15–20 min at 37°C with gentle shaking. Samples were further dissociated using a GentleMACS (Miltenyi) for flow cytometry or by physical pressing with a syringe plunger through a 70-µm filter for scRNA-seq, and lymphocytes were enriched using centrifugation through a Percoll gradient (30 and 70%) at 400 × g. The enriched lymphocyte layer at the 30/70% interface was collected and washed with RPMI + 10% FBS before staining for flow cytometry or sorting for scRNA-seq.
Flow cytometry and cell sorting
Single-cell suspensions of the pLN, blood, and pancreas were labeled with LIVE/DEAD Fixable Near-IR Cell Stain in phosphate-buffered saline (PBS; Thermo Fisher Scientific) to exclude dead cells from downstream analyses. Cells were preincubated with TruStain Fc Receptor Block (anti-mouse CD16/CD32, clone 93; BioLegend) and then labeled with extracellular antibodies from BioLegend: CD8α (53-6.7), PD-1 (RMP1-30), CD29 (HMβ 1-1; BioLegend), CD48 (HM48-1), CD94 (18d3), NKG2D (CX5), CD39 (Duha59), NKG2A (16A11), TIM-3 (RMT3-23), SLAMF7 (4G2), TIGIT (IG9), CX3CR1 (SA011F11), CD62L (MEL-14), CD44 (IM7), CD127 (A7R34), CD49D (R1-2), CD38 (90), CXCR3 (CXCR3-173); BD Biosciences: TCR-β (H57-597), CD4 (GK1.5), CD11a (M17/4), CD8b (H35-17.2); and Bio-Rad: LAG-3 (C9B7W). Notably, detection of PD-1 using the RMP1-30 clone is not blocked by the anti-PD-1 antibody (clone 29F.1A12) used in vivo. For intracellular analysis, cells were fixed for 1 h at 4°C with Foxp3/Transcription Factor Fixation/Permeabilization buffer kit (Thermo Fisher Scientific) and incubated in permeabilization buffer containing antibodies to intracellular proteins overnight. Intracellular antibodies were from BioLegend: BCL-2 (100), FoxP3 (FJK-16s), TOX (TXRX10), T-bet (4B10); and BD Biosciences: Ki-67 (clone B56). Biotinylated NRP-v7 tetramer (National Institutes of Health Tetramer Core Facility) was conjugated with PE-streptavidin by addition of 10 aliquots of 31.8 μl of PE-streptavidin (Thermo Fisher Scientific) added every 10 min at 4°C. Flow cytometry labeling (without inclusion of Feature Barcoding antibodies from BioLegend) was performed in PBS supplemented with 2% FBS. For Cellular Indexing of Transcriptomes and Epitopes by Sequencing (CITE-seq), cells from individual mice were labeled with hashtag TotalSeq-C antibodies 1, 2, 3, or 4 (TotalSeq C0301-C0304; from BioLegend). Labeling with Feature Barcoding and hashtag antibodies was performed in cell-staining buffer (BioLegend). PE-labeled NRP-v7 tetramer was detected using anti-PE (TotalSeq C0911; from BioLegend). Samples were acquired on a FACSymphony (BD Biosciences) and analyzed with FlowJo 10.8.1 (Treestar). Flow cytometry-based sorting for scRNA-seq was performed using a FACSAria (BD Biosciences). For scRNA-seq, live TCR-β+CD11ahigh lymphocytes were sorted.
scRNA-seq
Gene expression and TCR libraries were generated using the Chromium Single-cell 5′ Library, V(D)J Reagent Kit, and 5′ Feature Barcode Library Kit (10X Genomics) according to the manufacturer’s recommendations. Following sorting as described above, ∼10,000 cells per sample were combined and 35,000 cells were loaded into each channel of the Chromium Chip. Feature Barcoding, TCR, and gene expression libraries were pooled and sequenced on a NovaSeq 3000 sequencer (Illumina) by the Biopolymers Facility at Harvard Medical School. The pooled libraries were sequenced using the 26 × 8 × 91-bp parameters recommended by 10X Genomics. Based on approximate cell numbers expected, we sequenced a minimum of 20,000 reads per cell for gene expression libraries and 5,000 reads per cell for TCR and Feature Barcoding libraries. Raw reads were processed using Cell Ranger Multi version 7.0 to generate raw counts matrices of gene expression and CSV files corresponding to TCR clonality. The mm10 mouse reference genome was used for alignment.
Processing of gene expression and CITE-seq data
All analyses were performed in R version 4.0.5 and Seurat version 4.0.2 with additional utilization of the packages dplyr, data.table, ggplot2, viridis, gridExtra, presto, ggtree, readxl, fgsea, msigdbr, rstatix, reshape, RColorBrewer, ggpubr, ggrepel, ggprism, and tibble. Seurat objects were created with the min.cells parameter set to 20 and minimum features set to 200. Demultiplexing was done using MULTIseqDemux with automatic thresholding. Cells were filtered if they were classified as doublets or negative for hashtag antibody based on the demultiplexing results, or if they had fewer than 200 features or >10% mitochondrial RNA was detected. The data for the different tissues run across different sequencing lanes were integrated for clustering and uniform manifold approximation and projection (UMAP) visualization purposes using the FindIntegrationAnchors and IntegrateData using 20 dimensions. Data were normalized using the default Seurat function (generating log-transformed transcripts-per-10K read measurements) followed by scaling, and FindVariableGenes was run using the default “vst” method.
For the analysis of murine data, the FindClusters function was run at high resolution to generate clusters containing 50 or more cells. Clusters were removed if >5% of cells expressed genes associated with myeloid or natural killer cell lineages: Lyz2, H2-Aa, Xcl1, and/or Cd74. Clusters were classified as CD4+ and CD8+ T cells based on the average ratio of expression of Cd4 and Cd8a and subsequently reclassified as necessary based on the classification of the majority of cells in clonotypes >3 cells. CD8+ T cells were reclassified as CD4+ T cells based on Foxp3 expression >0.5. CD8+ and CD4+ T cells were reclustered separately at a resolution of 0.8–1.1 to obtain 11 and 13 biologically meaningful clusters each, respectively, and subsequently merged for visualization. Clusters with more than 20 cells were retained.
For human data analyses, paired scRNA-seq and scTCR-seq data were downloaded from GEO: GSE125527 (Boland et al., 2020), GSE148837 (Corridoni et al., 2020), and GSE144469 (Luoma et al., 2020). The FindClusters function was run at a resolution of 0.3–0.8 and the ratio of the mean CD4/CD8A expression in each cluster was used to distinguish CD8+ and CD4+ T cells. Mean expression of FOXP3 >0.5 in a cluster was used to distinguish Treg from Tcon. Clusters that expressed >20% CD44 were retained. Individual clusters were not annotated for subsequent analyses. Mean expression of gene signatures for each individual patient for CD8 and Tcon was used for visualization.
A Wilcox rank-sum test implemented in the FindAllMarkers function in Seurat was used to identify upregulated genes associated with each individual cluster and used for cluster annotation. To calculate gene signature scores, gene signatures were taken from the literature or from differentially expressed genes between clusters using the FindMarkers function, and scores were calculated for each cluster using the AddModuleScore function in Seurat as described in Tirosh et al. (2016). For gene set enrichment analyses, a rapid Wilcox rank-sum test and area under the curve statistic were computed using the “presto” package in R to rank genes. All gene signatures within the C2, C5, C7, and Hallmark categories from the Molecular Signatures Database were analyzed (Subramanian et al., 2005) in addition to gene signatures derived from the literature as cited in the text. Hierarchical clustering was performed on gene signatures using the “ggtree” package.
scTCR analysis
A single α and β chain was selected for each clonotype by selecting a single productive chain for each according to the sequence with the highest unique molecular identifiers, or highest reads (in case of a tie). Clonotypes were defined for CD4+ and CD8+ T cells separately for each mouse or person based on matching VDJ genes and TCR cdr3 nucleotide sequences for both α and β chains. Clonotype expansion was measured by counting the number of cells in the clonotype. A clonotype present in the periphery was considered PM if at least one cell of the clonotype was identified in the pancreas, or blood-matching if at least one cell of the clonotype was identified in the blood and pancreas. Cells were classified as NRP-v7+ tetramer-specific if the CDR3 amino acid sequences for the α and β chains were an exact match to any of the NRP-v7 tetramer-sorted T cells from Gearty et al. (2022). Cells were excluded from TCR analyses if they did not have both an α and a β chain detected.
Functional annotations of Seurat clusters
Seurat clusters were manually annotated using a combination of upregulated genes for each cluster (Table S1), visual inspection of key markers using UMAP visualization, expression of multiple coinhibitory receptors, and gene signature analysis. Key markers used for aiding in annotation included Foxp3, Sell, Tcf7, Lef1, Ccr7, Il7r, Klrg1, Cx3cr1, S1pr5, Il21, Gzmk, Prf1, Mki67, Slamf6, Pdcd1, Tigit, Ctla4, Cxcr6, Cd69, Bcl6, Cxcr5, Cxcr3, Prss2, Ifit1, Ift3, Stmn1, and Tox. Expression of genes associated with memory distinguished subsets of central memory (Sell, Ccr7, Lef1, and S1pr1) and memory (Il7r) subsets of CD8+ T cells and Tcon. Treg clusters expressed Foxp3 and Ikzf2, and expression of Tigit and Ctla4 distinguished effector Treg (Tigithi) from resting Treg cells. Effector T cells were identified based on expression of effector molecules (Gzmk, Nkg7, and Fasl), cytokines (Ccl5, Ccl4, and Ifng), and chemokine receptors (Cxcr3 and Cxcr6). A subset of SLEC-like CD8+ T cells, previously described in acute infection, were defined based on high expression of Cx3cr1, Zeb2, Klrg1, Gzma, and Tbx21 and low expression of Il7r. KLRG1hiIL-7rloT-bethi SLEC in acute infection have low memory cell potential, are non-self-renewing, and decline in number as the infection progresses (Joshi et al., 2007; Omilusik et al., 2015). T follicular helper (Tfh)-like cells, a subset of Tcon that are important in germinal center formation and provide B cell help for antibody production, were identified by high expression of Cxcr5, Tox2, and Bcl6 (Xu et al., 2019; Crotty 2011). We also annotated a cluster of Th21 cells, an additional subset of Tcon characterized by expression of transcript for the cytokine Il21, which has been shown to be critical for NOD diabetes (Spolski et al., 2008). The Th21 and Tfh-like clusters also expressed high levels of genes associated with anergy (Nt52, Izumo1r). Clusters of progenitor-like T cells were distinguished by high expression of Slamf6 and Tcf7, genes associated with self-renewal capacity in exhausted CD8+ T cells and persistent Tcon that mediate autoimmunity (Collier et al., 2021; Xia et al., 2022; Im et al., 2016). Four clusters contained a mixture of both CD8+ and CD4+ T cells with a common transcriptional profile: acinar-contaminated cells that expressed transcripts encoding pancreatic enzymes (Ctrb1, Pnlip, and Prss2); recently activated T cells (Cd69, Egr1, and Egr2); interferon-sensing T cells (Ifit1, Ifit3, and Isg15); and proliferating cells (Mki67 and Stmn1). Effector CD8+ T cell populations with exhaustion-like transcriptional features were defined based on expression of the transcription factor Tox (Fig. S2, C and D) and simultaneous expression of multiple co-inhibitory receptors (Tigit, Ctla4, Cd160, Cd244a, Pdcd1, and Havcr2; Fig. S2, C and E). A gene signature derived from terminally exhausted T cells in chronic viral infection (Miller et al., 2019) was increased in these clusters (Fig. S2 F). A progenitor-exhausted-like T cell cluster was distinguished from terminally exhausted T cells based on increased expression of Slamf6 and Tcf7 (Fig. S2 C). We defined these clusters as progenitor-exhausted/effector-like and terminally exhausted/effector-like, as these cells exhibited an effector-like transcriptional profile in addition to features of exhaustion. These clusters highly expressed a significant number of genes associated with effector function (e.g., Ccl5, Gzmk, Ccl4, Ifng, Fasl, and Klrc1), were clonally expanded (Fig. S2 C), and enriched in published gene signatures associated with CD8+ effector T cells (Fig. S2 F). Changes in the number of cells within clusters between treatment groups were calculated using Fisher’s exact test, and resultant P values were adjusted using the Benjamini and Hochberg method. Fisher’s exact test examines the difference in the proportion of each cluster within each treatment group against all the other clusters in the other treatment groups combined using a 2 × 2 contingency table.
Immunohistochemistry
Mouse pancreata fixed in 10% formalin and stored in 70% ethanol were paraffin-embedded, sectioned, and stained with H&E at the Brigham Pathology Core or the Dana-Farber/Harvard Cancer Center Rodent Histopathology Core. Semiquantitative scoring of tissue interactions was performed by a blinded board-certified pathologist using conventional microscopy on H&E-stained slides. One section of pancreas was scored for insulitis or acinar inflammation for each mouse. Using the NOD Insulitis Scoring Rubric, pancreatic islets were assigned a score of 0 if no infiltration of leukocytes was detected, 1 if leukocytes touched the perimeter of an islet without penetration, 2 for leukocyte penetration up to 25% of islet mass, 3 for leukocyte penetration up to 75% of islet mass, and 4 for end-stage insulitis where <25% of islet mass remained. From this, the percentages of normal, peri-insulitis, and insulitis islets were calculated. The insulitis index was calculated for each individual mouse according to the formula:
where n0 is the number of score 0 islets, n1 is the number of score 1 islets, n2 is the number of score 2 islets, n3 is the number of score 3 islets, and n4 is the number of score 4 islets.
For anti-insulin immunohistochemical staining, 5 µm formalin-fixed paraffin embedded sections were baked, deparaffinized with xylene, and rehydrated by passing slides through decreasing concentrations of ethanol. Two washes of 100% ethanol, two washes of 95% ethanol, and two washes in distilled water for 10 min per wash. Heat-induced antigen retrieval was completed using Target Antigen Retrieval Solution (Dako) in a pressure cooker for 45 min. After cooling to room temperature, the sections were washed with PBS and blocked with 10% goat serum for 1 h. Sections were incubated at 4°C overnight at 1:30,000 with primary rabbit anti-insulin antibody (ab181547; Abcam) and at room temperature for 1 h at 1:200 with the secondary antibody goat anti-rabbit IgG HRP (PI-1000; Vector Laboratories) before application of 3,3'-diaminobenzidine (3468; Dako), counterstaining with Harris hematoxylin (245-697; Thermo Fisher Scientific), treatment with defining solution (310-350; Thermo Fisher Scientific), bluing (HXB00242E; Statlab), and cover-slipping. Slides were scanned using a NanoZoomer S60 Digital slide scanner (Hamamatsu, Japan) and images captured using the NDP View 2 software package (Hamamatsu). Histology images were edited with ImageJ (version 1.53 s).
GeoMx digital spatial profiling (DSP)
DSP was performed using the GeoMx platform (NanoString; Merritt et al., 2020). To assist in identifying islets during region of interest (ROI) selection, H&E-stained slides were scanned using a NanoZoomer S60 Digital slide scanner (Hamamatsu). The NDP View 2 software package version 2.9.29 (Hamamatsu) was used to annotate previously scored islets on the scanned H&E images. For DSP, the islets were binned into three categories: normal (score 0), peri-insulitis (score 1–2), and insulitis (score 3–4).
An anti-INS-AF647 antibody (NBP2-34738AF647; Novus Biologics) was optimized for use as a morphology marker and used for GeoMx at a final concentration of 1:25,000. Immunofluorescence for INS (pancreatic β-cells), pan-Cytokeratin (epithelial cells), CD45 (leukocytes), and SYTO 13 (DNA stain) guided selection of ROIs. Samples were incubated with 63 oligonucleotide-conjugated and photocleavable (Abcam) antibodies (Table S2) plus negative and positive controls at 4°C in a humidity chamber overnight. After incubation and imaging, ROIs were segmented into INS+ and CD45+ areas. Oligo tags were released from the areas of illumination via targeted exposure to ultraviolet radiation, followed by hybridization and counting using the NanoString GeoMx nCounter system (Merritt et al., 2020).
DSP data analysis
The GeoMx DSP Data Analysis Suite (v2.4.0.421) was used to evaluate raw count data output from the nCounter platform. Initial quality control (QC) was performed using default parameters. 24 segments flagged with warnings other than High Positive Control Normalization were removed from further analysis. One segment that failed only QC parameter High Positive Control Normalization but had a Positive Norm Factor of 23.75 was removed from further analysis. An additional segment that did not fail any QC parameter but clustered separately from all other samples and accounted for 14.5% of the variance along PC2 by principal component analysis was also removed from further analysis. As the counts for the three housekeeping control antibodies (Histone H3, GAPDH, and S6) were sufficiently high and concordant, we used the geometric mean of all three for normalization. For statistical testing, the negative controls (Ms IgG1, Ms IgG2a, and Rb IgG1), the housekeeping genes (Histone H3, GAPDH, and S6), and the markers used to stain the slides during ROI selection (CD45, Pan-CK) were removed, leaving 62 protein targets.
Quantification and statistical analysis
Statistical analyses for flow cytometry data were performed with Prism software (GraphPad). Outliers for all flow cytometry data were identified using the Robust regression and Outlier removal method at a Q value of 1% and only cleaned values were retained for analysis. For three-way comparisons in flow cytometry data, significance was determined using a non-parametric Kruskall–Wallis test with Dunn’s posthoc test for multiple comparisons. For statistical significance across multiple levels, a two-way ANOVA with Holm–Sidak test for multiple comparisons was used. For two-way comparisons, statistical significance was determined using a nonparametric Mann–Whitney test with Holm-Sidak’s correction for multiple comparisons. For percent diabetes-free curves, a Kaplan–Meier curve analysis was performed and results from the log-rank (Mantel–Cox) test are shown. A P value <0.05 was considered statistically significant. For insulitis index, significance was determined using non-parametric Kruskall–Wallis test with Dunn’s posthoc test for multiple comparisons. Statistical significance of acinar inflammation was determined using a Fisher’s exact test and P values were adjusted using the Benjamini and Hochberg method. Asterisks indicating significance in the figures correspond to *P < 0.05, **P < 0.01, and ***P < 0.001. Statistical tests used for computational analyses are indicated in the corresponding figure legends and Materials and methods sections. Exact P values are listed in the figure legends.
Online supplemental material
Fig. S1 describes the histological differences in the pancreas of spontaneous and anti-PD-1-induced T1D in NOD mice. Fig. S2 describes the classification of CD4+ and CD8+ T cells based on transcriptional and TCR features. Fig. S3 illustrates co-inhibitory receptor expression on CD8+ T cells in colitis patients and NOD mice. Fig. S4 demonstrates the differences in clonotype matching across tissues and clusters in anti-PD-1-induced T1D compared to spontaneous T1D. Fig. S5 demonstrates that anti-PD-1-induced T1D in young NOD recapitulates features of anti-PD-1-induced T1D in older NOD mice. Table S1 describes upregulated genes per transcriptional cluster identified in T cells isolated from the pLN, blood, and pancreas of IgG, Spt, and PD1 NOD mice. Table S2 lists the selected protein markers used in the GeoMX analysis. Table S3 describes the genes comprising the signature derived from the NOD terminally exhausted/effector-like cluster. Table S4 describes the genes comprising the gene signature derived from the NOD effector cluster. Table S5 describes the genes associated with PM T cells in the blood that are shared or unique between treatment groups. Table S6 describes the TCR data detected for all T cells.
Supplementary Material
describes upregulated genes per transcriptional cluster identified in T cells isolated from the pLN, blood, and pancreas of IgG, Spt, and PD1 NOD mice.
lists the selected protein markers used in the GeoMX analysis.
describes the genes comprising the signature derived from the NOD terminally exhausted/effector-like cluster.
describes the genes comprising the gene signature derived from the NOD effector cluster.
describes the genes associated with PM T cells in the blood that are shared or unique between treatment groups.
describes the TCR data detected for all T cells.
Acknowledgments
We would like to thank S. Kissler for critical reading of the manuscript. We would like to thank the Biopolymers Facility at Harvard Medical School for sequencing of single-cell libraries, and the Harvard Medical School Immunology Flow Cytometry Core Facility for cell sorting and flow cytometry support.
This work was supported by funding from the National Institutes of Health, grants P01 AI56299, P01AI039671, P01 AI108545, and Vertex.
Author contributions: J.L. Collier, K.E. Pauken, and A.H. Sharpe conceived the study. J.L. Collier, D.G. Patterson, K.E. Pauken, and T.S. Conway performed mouse experiments. J.L. Collier performed computational and statistical analyses. S.C. Markson provided input on the computational and statistical analysis. C.A.A. Lee, K.N. Mucciarone, C.G. Lian, and G.T. Murphy performed histologic analyses. M.E. Fung performed library generation for sequencing. J.A. France monitored mice for diabetes development during experiments. J.L. Collier, K.E. Pauken, and A.H. Sharpe contributed to data interpretation. J.L. Collier, K.E. Pauken, and A.H. Sharpe contributed to writing the manuscript. All others reviewed the manuscript. A.H. Sharpe supervised the study.
Data availability
Requests for resources, reagents, and further information regarding this study should be directed to and will be fulfilled by the corresponding author. This study did not generate new unique reagents. The scRNA-seq and scTCR-seq data generated during this study are available at GEO under the following accession number: GSE228233. Code generated for this study is available at: https://github.com/JennaCollier/NOD_spt_pd1.
References
- Abdelsamed, H.A., Zebley C.C., Nguyen H., Rutishauser R.L., Fan Y., Ghoneim H.E., Crawford J.C., Alfei F., Alli S., Ribeiro S.P., et al. 2020. Beta cell-specific CD8+ T cells maintain stem cell memory-associated epigenetic programs during type 1 diabetes. Nat. Immunol. 21:578–587. 10.1038/s41590-020-0633-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akturk, H.K., Kahramangil D., Sarwal A., Hoffecker L., Murad M.H., and Michels A.W.. 2019. Immune checkpoint inhibitor-induced Type 1 diabetes: A systematic review and meta-analysis. Diabet. Med. 36:1075–1081. 10.1111/dme.14050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfei, F., Kanev K., Hofmann M., Wu M., Ghoneim H.E., Roelli P., Utzschneider D.T., von Hoesslin M., Cullen J.G., Fan Y., et al. 2019. TOX reinforces the phenotype and longevity of exhausted T cells in chronic viral infection. Nature. 571:265–269. 10.1038/s41586-019-1326-9 [DOI] [PubMed] [Google Scholar]
- Almutairi, A.R., McBride A., Slack M., Erstad B.L., and Abraham I.. 2020. Potential immune-related adverse events associated with monotherapy and combination therapy of Ipilimumab, Nivolumab, and Pembrolizumab for advanced melanoma: A systematic review and meta-analysis. Front. Oncol. 10:91. 10.3389/fonc.2020.00091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, M.S., and Bluestone J.A.. 2005. The NOD mouse: A model of immune dysregulation. Annu. Rev. Immunol. 23:447–485. 10.1146/annurev.immunol.23.021704.115643 [DOI] [PubMed] [Google Scholar]
- Ansari, M.J., Salama A.D., Chitnis T., Smith R.N., Yagita H., Akiba H., Yamazaki T., Azuma M., Iwai H., Khoury S.J., et al. 2003. The programmed death-1 (PD-1) pathway regulates autoimmune diabetes in nonobese diabetic (NOD) mice. J. Exp. Med. 198:63–69. 10.1084/jem.20022125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beausang, J.F., Wheeler A.J., Chan N.H., Hanft V.R., Dirbas F.M., Jeffrey S.S., and Quake S.R.. 2017. T cell receptor sequencing of early-stage breast cancer tumors identifies altered clonal structure of the T cell repertoire. Proc. Natl. Acad. Sci. USA. 114:E10409–E10417. 10.1073/pnas.1713863114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland, B.S., He Z., Tsai M.S., Olvera J.G., Omilusik K.D., Duong H.G., Kim E.S., Limary A.E., Jin W., Milner J.J., et al. 2020. Heterogeneity and clonal relationships of adaptive immune cells in ulcerative colitis revealed by single-cell analyses. Sci. Immunol. 5:eabb4432. 10.1126/sciimmunol.abb4432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahmer, J.R., Lacchetti C., Schneider B.J., Atkins M.B., Brassil K.J., Caterino J.M., Chau I., Ernstoff M.S., Gardner J.M., Ginex P., et al. 2018. Management of immune-related adverse events in patients treated with immune checkpoint inhibitor therapy: American society of clinical oncology clinical practice guideline. J. Clin. Oncol. 36:1714–1768. 10.1200/JCO.2017.77.6385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, C.-M., Feng P.-H., Wu T.-H., Alachkar H., Lee K.-Y., and Chang W.-C.. 2021. Profiling of T Cell repertoire in SARS-CoV-2-infected COVID-19 patients between mild disease and pneumonia. J. Clin. Immunol. 41:1131–1145. 10.1007/s10875-021-01045-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciecko, A.E., Schauder D.M., Foda B., Petrova G., Kasmani M.Y., Burns R., Lin C.-W., Drobyski W.R., Cui W., and Chen Y.-G.. 2021. Self-renewing islet TCF1+ CD8 T cells undergo IL-27-controlled differentiation to become TCF1− terminal effectors during the progression of Type 1 diabetes. J. Immunol. 207:1990–2004. 10.4049/jimmunol.2100362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier, J.L., Weiss S.A., Pauken K.E., Sen D.R., and Sharpe A.H.. 2021. Not-so-opposite ends of the spectrum: CD8+ T cell dysfunction across chronic infection, cancer and autoimmunity. Nat. Immunol. 22:809–819. 10.1038/s41590-021-00949-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly, K.A., Kuchroo M., Venkat A., Khatun A., Wang J., William I., Hornick N.I., Fitzgerald B.L., Damo M., Kasmani M.Y., et al. 2021. A reservoir of stem-like CD8+ T cells in the tumor-draining lymph node preserves the ongoing antitumor immune response. Sci. Immunol. 6:eabg7836. 10.1126/sciimmunol.abg7836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corridoni, D., Antanaviciute A., Gupta T., Fawkner-Corbett D., Aulicino A., Jagielowicz M., Parikh K., Repapi E., Taylor S., Ishikawa D., et al. 2020. Single-cell atlas of colonic CD8+ T cells in ulcerative colitis. Nat. Med. 26:1480–1490. 10.1038/s41591-020-1003-4 [DOI] [PubMed] [Google Scholar]
- Crotty, S. 2011. Follicular helper CD4 T cells (TFH). Annu. Rev. Immunol. 29:621–663. 10.1146/annurev-immunol-031210-101400 [DOI] [PubMed] [Google Scholar]
- Dammeijer, F., van Gulijk M., Mulder E.E., Lukkes M., Klaase L., van den Bosch T., van Nimwegen M., Lau S.P., Latupeirissa K., Schetters S., et al. 2020. The PD-1/PD-L1-checkpoint restrains T cell immunity in tumor-draining lymph nodes. Cancer Cell. 38:685–700.e8. 10.1016/j.ccell.2020.09.001 [DOI] [PubMed] [Google Scholar]
- Dash, P., Fiore-Gartland A.J., Hertz T., Wang G.C., Sharma S., Souquette A., Crawford J.C., Clemens E.B., Nguyen T.H.O., Kedzierska K., et al. 2017. Quantifiable predictive features define epitope-specific T cell receptor repertoires. Nature. 547:89–93. 10.1038/nature22383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esfahani, K., Elkrief A., Calabrese C., Lapointe R., Hudson M., Routy B., Miller W.H. Jr, and Calabrese L.. 2020. Moving towards personalized treatments of immune-related adverse events. Nat. Rev. Clin. Oncol. 17:504–515. 10.1038/s41571-020-0352-8 [DOI] [PubMed] [Google Scholar]
- Gagnerault, M.-C., Luan J.J., Lotton C., and Lepault F.. 2002. Pancreatic lymph nodes are required for priming of beta cell reactive T cells in NOD mice. J. Exp. Med. 196:369–377. 10.1084/jem.20011353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gearty, S.V., Dündar F., Zumbo P., Espinosa-Carrasco G., Shakiba M., Sanchez-Rivera F.J., Socci N.D., Trivedi P., Lowe S.W., Lauer P., et al. 2022. An autoimmune stem-like CD8 T cell population drives type 1 diabetes. Nature. 602:156–161. 10.1038/s41586-021-04248-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glanville, J., Huang H., Nau A., Hatton O., Wagar L.E., Rubelt F., Ji X., Han A., Krams S.M., Pettus C., et al. 2017. Identifying specificity groups in the T cell receptor repertoire. Nature. 547:94–98. 10.1038/nature22976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson, J.M., Zajac A.J., Altman J.D., and Ahmed R.. 2000. Cutting edge: Increased expression of bcl-2 in antigen-specific memory CD8+ T cells. J. Immunol. 164:3950–3954. 10.4049/jimmunol.164.8.3950 [DOI] [PubMed] [Google Scholar]
- Grebinoski, S., Zhang Q., Cillo A.R., Manne S., Xiao H., Brunazzi E.A., Tabib T., Cardello C., Lian C.G., Murphy G.F., et al. 2022. Autoreactive CD8+ T cells are restrained by an exhaustion-like program that is maintained by LAG3. Nat. Immunol. 23:868–877. 10.1038/s41590-022-01210-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueguen, P., Metoikidou C., Dupic T., Lawand M., Goudot C., Baulande S., Lameiras S., Lantz O., Girard N., Seguin-Givelet A., et al. 2021. Contribution of resident and circulating precursors to tumor-infiltrating CD8+ T cell populations in lung cancer. Sci. Immunol. 6:eabd5778. 10.1126/sciimmunol.abd5778 [DOI] [PubMed] [Google Scholar]
- Guleria, I., Gubbels Bupp M., Dada S., Fife B., Tang Q., Ansari M.J., Trikudanathan S., Vadivel N., Fiorina P., Yagita H., et al. 2007. Mechanisms of PDL1-mediated regulation of autoimmune diabetes. Clin. Immunol. 125:16–25. 10.1016/j.clim.2007.05.013 [DOI] [PubMed] [Google Scholar]
- Haanen, J.B.A.G., Carbonnel F., Robert C., Kerr K.M., Peters S., Larkin J., Jordan K., and ESMO Guidelines Committee . 2018. Management of toxicities from immunotherapy: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 29:iv264–iv266. 10.1093/annonc/mdy162 [DOI] [PubMed] [Google Scholar]
- Hu, H., Zakharov P.N., Peterson O.J., and Unanue E.R.. 2020. Cytocidal macrophages in symbiosis with CD4 and CD8 T cells cause acute diabetes following checkpoint blockade of PD-1 in NOD mice. Proc. Natl. Acad. Sci. USA. 117:31319–31330. 10.1073/pnas.2019743117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, A.C., Orlowski R.J., Xu X., Mick R., George S.M., Yan P.K., Manne S., Kraya A.A., Wubbenhorst B., Dorfman L., et al. 2019. A single dose of neoadjuvant PD-1 blockade predicts clinical outcomes in resectable melanoma. Nat. Med. 25:454–461. 10.1038/s41591-019-0357-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, A.C., Postow M.A., Orlowski R.J., Mick R., Bengsch B., Manne S., Xu W., Harmon S., Giles J.R., Wenz B., et al. 2017. T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature. 545:60–65. 10.1038/nature22079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im, S.J., Hashimoto M., Gerner M.Y., Lee J., Kissick H.T., Burger M.C., Shan Q., Hale J.S., Lee J., Nasti T.H., et al. 2016. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature. 537:417–421. 10.1038/nature19330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen, A., Christensen S.J., Scheie D., Højgaard J.L.S., and Kondziella D.. 2019. Neuromuscular adverse events associated with anti-PD-1 monoclonal antibodies: Systematic review. Neurology. 92:663–674. 10.1212/WNL.0000000000007235 [DOI] [PubMed] [Google Scholar]
- Johnson, D.B., Balko J.M., Compton M.L., Chalkias S., Gorham J., Xu Y., Hicks M., Puzanov I., Alexander M.R., Bloomer T.L., et al. 2016. Fulminant myocarditis with combination immune checkpoint blockade. N. Engl. J. Med. 375:1749–1755. 10.1056/NEJMoa1609214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi, N.S., Cui W., Chandele A., Lee H.K., Urso D.R., Hagman J., Gapin L., and Kaech S.M.. 2007. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity. 27:281–295. 10.1016/j.immuni.2007.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech, S.M., Hemby S., Kersh E., and Ahmed R.. 2002. Molecular and functional profiling of memory CD8 T cell differentiation. Cell. 111:837–851. 10.1016/s0092-8674(02)01139-x [DOI] [PubMed] [Google Scholar]
- Khan, O., Giles J.R., McDonald S., Manne S., Ngiow S.F., Patel K.P., Werner M.T., Huang A.C., Alexander K.A., Wu J.E., et al. 2019. TOX transcriptionally and epigenetically programs CD8+ T cell exhaustion. Nature. 571:211–218. 10.1038/s41586-019-1325-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, K.H., Hur J.Y., Cho J., Ku B.M., Koh J., Koh J.Y., Sun J.-M., Lee S.H., Ahn J.S., Park K., et al. 2020. Immune-related adverse events are clustered into distinct subtypes by T-cell profiling before and early after anti-PD-1 treatment. OncoImmunology. 9:1722023. 10.1080/2162402X.2020.1722023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klarenbeek, P.L., de Hair M.J.H., Doorenspleet M.E., van Schaik B.D.C., Esveldt R.E.E., van de Sande M.G.H., Cantaert T., Gerlag D.M., Baeten D., van Kampen A.H., et al. 2012. Inflamed target tissue provides a specific niche for highly expanded T-cell clones in early human autoimmune disease. Ann. Rheum. Dis. 71:1088–1093. 10.1136/annrheumdis-2011-200612 [DOI] [PubMed] [Google Scholar]
- Kowalczyk, M.S., Tirosh I., Heckl D., Rao T.N., Dixit A., Haas B.J., Schneider R.K., Wagers A.J., Ebert B.L., and Regev A.. 2015. Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res. 25:1860–1872. 10.1101/gr.192237.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, L.-F., Logronio K., Tu G.H., Zhai W., Ni I., Mei L., Dilley J., Yu J., Rajpal A., Brown C., et al. 2012. Anti-IL-7 receptor-α reverses established type 1 diabetes in nonobese diabetic mice by modulating effector T-cell function. Proc. Natl. Acad. Sci. USA. 109:12674–12679. 10.1073/pnas.1203795109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy, E., Marty R., Gárate Calderón V., Woo B., Dow M., Armisen R., Carter H., and Harismendy O.. 2016. Immune DNA signature of T-cell infiltration in breast tumor exomes. Sci. Rep. 6:30064. 10.1038/srep30064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z., Tuong Z.K., Dean I., Willis C., Gaspal F., Fiancette R., Idris S., Kennedy B., Ferdinand J.R., Peñalver A., et al. 2022. In vivo labeling reveals continuous trafficking of TCF-1+ T cells between tumor and lymphoid tissue. J. Exp. Med. 219:e20210749. 10.1084/jem.20210749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas, J.A., Menke J., Rabacal W.A., Schoen F.J., Sharpe A.H., and Kelley V.R.. 2008. Programmed death ligand 1 regulates a critical checkpoint for autoimmune myocarditis and pneumonitis in MRL mice. J. Immunol. 181:2513–2521. 10.4049/jimmunol.181.4.2513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucca, L.E., Axisa P.-P., Lu B., Harnett B., Jessel S., Zhang L., Raddassi K., Zhang L., Olino K., Clune J., et al. 2021. Circulating clonally expanded T cells reflect functions of tumor-infiltrating T cells. J. Exp. Med. 218:e20200921. 10.1084/jem.20200921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luckey, C.J., Bhattacharya D., Goldrath A.W., Weissman I.L., Benoist C., and Mathis D.. 2006. Memory T and memory B cells share a transcriptional program of self-renewal with long-term hematopoietic stem cells. Proc. Natl. Acad. Sci. USA. 103:3304–3309. 10.1073/pnas.0511137103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luoma, A.M., Suo S., Williams H.L., Sharova T., Sullivan K., Manos M., Bowling P., Hodi F.S., Rahma O., Sullivan R.J., et al. 2020. Molecular pathways of colon inflammation induced by cancer immunotherapy. Cell. 182:655–671.e22. 10.1016/j.cell.2020.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masopust, D., and Schenkel J.M.. 2013. The integration of T cell migration, differentiation and function. Nat. Rev. Immunol. 13:309–320. 10.1038/nri3442 [DOI] [PubMed] [Google Scholar]
- McLane, L.M., Abdel-Hakeem M.S., and Wherry E.J.. 2019. CD8 T cell exhaustion during chronic viral infection and cancer. Annu. Rev. Immunol. 37:457–495. 10.1146/annurev-immunol-041015-055318 [DOI] [PubMed] [Google Scholar]
- Merritt, C.R., Ong G.T., Church S.E., Barker K., Danaher P., Geiss G., Hoang M., Jung J., Liang Y., McKay-Fleisch J., et al. 2020. Multiplex digital spatial profiling of proteins and RNA in fixed tissue. Nat. Biotechnol. 38:586–599. 10.1038/s41587-020-0472-9 [DOI] [PubMed] [Google Scholar]
- Miller, B.C., Sen D.R., Al Abosy R., Bi K., Virkud Y.V., LaFleur M.W., Yates K.B., Lako A., Felt K., Naik G.S., et al. 2019. Subsets of exhausted CD8+ T cells differentially mediate tumor control and respond to checkpoint blockade. Nat. Immunol. 20:326–336. 10.1038/s41590-019-0312-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munson, D.J., Egelston C.A., Chiotti K.E., Parra Z.E., Bruno T.C., Moore B.L., Nakano T.A., Simons D.L., Jimenez G., Yim J.H., et al. 2016. Identification of shared TCR sequences from T cells in human breast cancer using emulsion RT-PCR. Proc. Natl. Acad. Sci. USA. 113:8272–8277. 10.1073/pnas.1606994113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, H., Nose M., Hiai H., Minato N., and Honjo T.. 1999. Development of lupus-like autoimmune diseases by disruption of the PD-1 gene encoding an ITIM motif-carrying immunoreceptor. Immunity. 11:141–151. 10.1016/S1074-7613(00)80089-8 [DOI] [PubMed] [Google Scholar]
- Nishimura, H., Okazaki T., Tanaka Y., Nakatani K., Hara M., Matsumori A., Sasayama S., Mizoguchi A., Hiai H., Minato N., and Honjo T.. 2001. Autoimmune dilated cardiomyopathy in PD-1 receptor-deficient mice. Science. 291:319–322. 10.1126/science.291.5502.319 [DOI] [PubMed] [Google Scholar]
- Omilusik, K.D., Best J.A., Yu B., Goossens S., Weidemann A., Nguyen J.V., Seuntjens E., Stryjewska A., Zweier C., Roychoudhuri R., et al. 2015. Transcriptional repressor ZEB2 promotes terminal differentiation of CD8+ effector and memory T cell populations during infection. J. Exp. Med. 212:2027–2039. 10.1084/jem.20150194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauken, K.E., Dougan M., Rose N.R., Lichtman A.H., and Sharpe A.H.. 2019. Adverse events following cancer immunotherapy: Obstacles and opportunities. Trends Immunol. 40:511–523. 10.1016/j.it.2019.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauken, K.E., Lagattuta K.A., Lu B.Y., Lucca L.E., Daud A.I., Hafler D.A., Kluger H.M., Raychaudhuri S., and Sharpe A.H.. 2022. TCR-Sequencing in cancer and autoimmunity: Barcodes and beyond. Trends Immunol. 43:180–194. 10.1016/j.it.2022.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauken, K.E., Shahid O., Lagattuta K.A., Mahuron K.M., Luber J.M., Lowe M.M., Huang L., Delaney C., Long J.M., Fung M.E., et al. 2021. Single-cell analyses identify circulating anti-tumor CD8 T cells and markers for their enrichment. J. Exp. Med. 218:e20200920. 10.1084/jem.20200920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penaranda, C., Kuswanto W., Hofmann J., Kenefeck R., Narendran P., Walker L.S.K., Bluestone J.A., Abbas A.K., and Dooms H.. 2012. IL-7 receptor blockade reverses autoimmune diabetes by promoting inhibition of effector/memory T cells. Proc. Natl. Acad. Sci. USA. 109:12668–12673. 10.1073/pnas.1203692109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip, M., and Schietinger A.. 2019. Heterogeneity and fate choice: T cell exhaustion in cancer and chronic infections. Curr. Opin. Immunol. 58:98–103. 10.1016/j.coi.2019.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postow, M.A., Sidlow R., and Hellmann M.D.. 2018. Immune-related adverse events associated with immune checkpoint blockade. N. Engl. J. Med. 378:158–168. 10.1056/NEJMra1703481 [DOI] [PubMed] [Google Scholar]
- Ribas, A., and Wolchok J.D.. 2018. Cancer immunotherapy using checkpoint blockade. Science. 359:1350–1355. 10.1126/science.aar4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar, S., Kalia V., Haining W.N., Konieczny B.T., Subramaniam S., and Ahmed R.. 2008. Functional and genomic profiling of effector CD8 T cell subsets with distinct memory fates. J. Exp. Med. 205:625–640. 10.1084/jem.20071641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenkel, J.M., Herbst R.H., Canner D., Li A., Hillman M., Shanahan S.-L., Gibbons G., Smith O.C., Kim J.Y., Westcott P., et al. 2021. Conventional type I dendritic cells maintain a reservoir of proliferative tumor-antigen specific TCF-1+ CD8+ T cells in tumor-draining lymph nodes. Immunity. 54:2338–2353.e6. 10.1016/j.immuni.2021.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe, A.H., and Pauken K.E.. 2018. The diverse functions of the PD1 inhibitory pathway. Nat. Rev. Immunol. 18:153–167. 10.1038/nri.2017.108 [DOI] [PubMed] [Google Scholar]
- Shin, H., Blackburn S.D., Blattman J.N., and Wherry E.J.. 2007. Viral antigen and extensive division maintain virus-specific CD8 T cells during chronic infection. J. Exp. Med. 204:941–949. 10.1084/jem.20061937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shizuru, J.A., Taylor-Edwards C., Banks B.A., Gregory A.K., and Fathman C.G.. 1988. Immunotherapy of the nonobese diabetic mouse: Treatment with an antibody to T-helper lymphocytes. Science. 240:659–662. 10.1126/science.2966437 [DOI] [PubMed] [Google Scholar]
- Sidhom, J.-W., Larman H.B., Pardoll D.M., and Baras A.S.. 2021. Author correction: DeepTCR is a deep learning framework for revealing sequence concepts within T-cell repertoires. Nat. Commun. 12:2309. 10.1038/s41467-021-22667-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spolski, R., Kashyap M., Robinson C., Yu Z., and Leonard W.J.. 2008. IL-21 signaling is critical for the development of type I diabetes in the NOD mouse. Proc. Natl. Acad. Sci. USA. 105:14028–14033. 10.1073/pnas.0804358105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Striebich, C.C., Falta M.T., Wang Y., Bill J., and Kotzin B.L.. 1998. Selective accumulation of related CD4+ T cell clones in the synovial fluid of patients with rheumatoid arthritis. J. Immunol. 161:4428–4436. 10.4049/jimmunol.161.8.4428 [DOI] [PubMed] [Google Scholar]
- Subramanian, A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P.. 2005. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, B.M., Juedes A., Szabo S.J., von Herrath M., and Glimcher L.H.. 2003. Antigen-driven effector CD8 T cell function regulated by T-bet. Proc. Natl. Acad. Sci. USA. 100:15818–15823. 10.1073/pnas.2636938100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh, I., Izar B., Prakadan S.M., Wadsworth M.H. II, Treacy D., Trombetta J.J., Rotem A., Rodman C., Lian C., Murphy G., et al. 2016. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 352:189–196. 10.1126/science.aad0501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang, V.H.M., McGrath R.T., Clifton-Bligh R.J., Scolyer R.A., Jakrot V., Guminski A.D., Long G.V., and Menzies A.M.. 2019. Checkpoint inhibitor-associated autoimmune diabetes is distinct from Type 1 diabetes. J. Clin. Endocrinol. Metab. 104:5499–5506. 10.1210/jc.2019-00423 [DOI] [PubMed] [Google Scholar]
- Walker, L.S.K., and von Herrath M.. 2016. CD4 T cell differentiation in type 1 diabetes. Clin. Exp. Immunol. 183:16–29. 10.1111/cei.12672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D.Y., Salem J.-E., Cohen J.V., Chandra S., Menzer C., Ye F., Zhao S., Das S., Beckermann K.E., Ha L., et al. 2018. Fatal toxic effects associated with immune checkpoint inhibitors: A systematic review and meta-analysis. JAMA Oncol. 4:1721–1728. 10.1001/jamaoncol.2018.3923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Yoshida T., Nakaki F., Hiai H., Okazaki T., and Honjo T.. 2005. Establishment of NOD-Pdcd1−/− mice as an efficient animal model of type I diabetes. Proc. Natl. Acad. Sci. USA. 102:11823–11828. 10.1073/pnas.0505497102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, T., Wang C., Wu J., He C., Zhang W., Liu J., Zhang R., Lv Y., Li Y., Zeng X., et al. 2017. The different T-cell receptor repertoires in breast cancer tumors, draining lymph nodes, and adjacent tissues. Cancer Immunol. Res. 5:148–156. 10.1158/2326-6066.CIR-16-0107 [DOI] [PubMed] [Google Scholar]
- Wherry, E.J., Ha S.-J., Kaech S.M., Haining W.N., Sarkar S., Kalia V., Subramaniam S., Blattman Joseph N, Barber D.L., and Ahmed R.. 2007. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 27:670–684. 10.1016/j.immuni.2007.09.006 [DOI] [PubMed] [Google Scholar]
- Xia, Y., Sandor K., Pai J.A., Daniel B., Raju S., Wu R., Hsiung S., Qi Y., Yangdon T., Okamoto M., et al. 2022. BCL6-dependent TCF-1+ progenitor cells maintain effector and helper CD4+ T cell responses to persistent antigen. Immunity. 55:1200–1215.e6. 10.1016/j.immuni.2022.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, W., Zhao X., Wang X., Feng H., Gou M., Jin W., Wang X., Liu X., and Dong C.. 2019. The transcription factor Tox2 drives T follicular helper cell development via regulating chromatin accessibility. Immunity. 51:826–839.e5. 10.1016/j.immuni.2019.10.006 [DOI] [PubMed] [Google Scholar]
- Zakharov, P.N., Hu H., Wan X., and Unanue E.R.. 2020. Single-cell RNA sequencing of murine islets shows high cellular complexity at all stages of autoimmune diabetes. J. Exp. Med. 217:e20192362. 10.1084/jem.20192362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, H., Galdos F.X., Lee D., Waliany S., Huang Y.V., Ryan J., Dang K., Neal J.W., Wakelee H.A., Reddy S.A., et al. 2022. Identification of pathogenic immune cell subsets associated with checkpoint inhibitor-induced myocarditis. Circulation. 146:316–335. 10.1161/CIRCULATIONAHA.121.056730 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
describes upregulated genes per transcriptional cluster identified in T cells isolated from the pLN, blood, and pancreas of IgG, Spt, and PD1 NOD mice.
lists the selected protein markers used in the GeoMX analysis.
describes the genes comprising the signature derived from the NOD terminally exhausted/effector-like cluster.
describes the genes comprising the gene signature derived from the NOD effector cluster.
describes the genes associated with PM T cells in the blood that are shared or unique between treatment groups.
describes the TCR data detected for all T cells.
Data Availability Statement
Requests for resources, reagents, and further information regarding this study should be directed to and will be fulfilled by the corresponding author. This study did not generate new unique reagents. The scRNA-seq and scTCR-seq data generated during this study are available at GEO under the following accession number: GSE228233. Code generated for this study is available at: https://github.com/JennaCollier/NOD_spt_pd1.