Summary
Pancreatic tissue slices allow functional investigations under close physiological conditions in situ. This approach is particularly advantageous for studying infiltrated and structurally damaged islets as found in T1D. More importantly, slices allow studying the interplay between endocrine and exocrine compartments. We here describe how to perform agarose injections, tissue preparation, and slice procedure for mouse and human tissue. We then describe in detail how to use the slices to perform functional studies using hormone secretion and calcium imaging as readouts.
For complete details on the use and execution of this protocol, please refer to Panzer et al. (2022).1
Subject areas: Cell Biology, Tissue Engineering, Biotechnology and Bioengineering
Graphical abstract

Highlights
-
•
Generation of pancreatic tissue slices for mouse and human tissue
-
•
Measuring endocrine and exocrine hormone release from dynamic slice perifusion
-
•
Quantification of dynamic changes in intracellular calcium levels at single-cell level
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Pancreatic tissue slices allow functional investigations under close physiological conditions in situ. This approach is particularly advantageous for studying infiltrated and structurally damaged islets as found in T1D. More importantly, slices allow studying the interplay between endocrine and exocrine compartments. We here describe how to perform agarose injections, tissue preparation, and slice procedure for mouse and human tissue. We then describe in detail how to use the slices to perform functional studies using hormone secretion and calcium imaging as readouts.
Before you begin
The protocol below describes the specific steps for generating mouse pancreatic tissue slices. The same procedure can be applied to prepare human pancreas tissue slices, with slight modifications in agarose concentration. Step 1 (Agarose injection) does not apply for human tissue. Differences for generating human tissue slices will always be marked for the individual steps. The handling of the slices for imaging and hormone measurements are identical for mouse and human tissue slices.
Before beginning, two essential elements need to be prepared.
-
1.
Preparation of animals: Slices can be generated from any mouse strain, including transgenic and wild-type mice.
Note: For imaging we recommend using transgenic mice with a fluorescent Calcium indicator like GCamP3/6 crossed with a cell specific promotor of interest.
Note: We used female Gcg/tm 1.1(icre)Gkg/J mice (JAX stock #030663), crossed with male mice that express GCaMP3 downstream of a loxP-flanked STOP cassette (JAX stock #029043).
Note: To model beta cell loss, we recommend a single dose STZ injection protocol. Mice were in injected with a single dose of streptozotocin (200 mg/Kg i.v.) or saline at 10 weeks of age and euthanized for slice preparation 2 weeks after treatment.
-
2.
Preparation of buffers and agarose: Buffers can be prepared a day prior to the experiments. The necessary volume needs to be calculated depending on the number of samples, stimuli and protocol length.
Institutional permissions
Mice used in this protocol were housed and maintained at the University of Miami Core facility. Experimental protocols were approved by the University of Miami Institutional Animal Care and Use Committee IACUC) and complied with all ethical regulations. Please note that any experiments on animals must be performed in accordance with institutional and national guidelines and need ethical approval of animal use prior to starting this protocol.
Buffer preparation
Timing: 10–30 min
-
3.
Prepare 1 L of each stock HEPES buffer, with and without BSA, (125 mM NaCl, 5.9 mM KCl, 2.56 mM CaCl2, 1 mM MgCl2, 25 mM HEPES, 0.1% BSA) and adjust pH to 7.4 and sterile filter.
Note: HEPES buffer without BSA can be stored for a month at 4°C while HEPES buffer with BSA should not be stored for longer than 1 week at 4°C. It is recommended to prepare buffers fresh before every experiment.
Note: HEPES buffer without BSA is only used to prepare the low-melting agarose as the BSA would denature during heating. It can be stored for up to one month at 4°C.
-
4.
Prepare your baseline buffer, which is HEPES buffer with BSA and resting glucose concentration (5.5 mM for human slices and 7 mM for mouse slices).
-
5.
Take 50 mL of your baseline buffer and add aprotinin (final concentration 10 μg/mL).
CRITICAL: Baseline buffer with inhibitor (aprotinin) is always used when slices are not perfused (collect during slicing, trimming process, labeling time, etc.). The inhibitor is essential for slice viability. During imaging or hormone secretion buffers without inhibitors are used.
-
6.
Prepare all other solutions needed for experiments by adding glucose and/or stimuli to the HEPES buffer with BSA (e.g., KCl, adrenaline, etc.).
Note: For hormone secretion buffers are supplemented with amino acids (final concentration of 2 mM L-alanine, L-arginine and L-glutamine).
CRITICAL: Always prepare more baseline buffer than you need to avoid slices accidentally running dry and account for priming of tubing’s.
-
7.
Once all buffers are prepared, start priming the perifusion system (step 22–25) for optimal time management.
Prepare low melting point agarose
Timing: 10 min
-
8.Prepare low-melting-point agarose in HEPES buffer without BSA using a microwave (1.2% for mouse and 3.8% for human tissue).
-
a.Weigh 0.6 g of low-melting point agarose in a 50-mL tube.
-
b.Add about 30 mL of HEPES buffer.
-
c.Heat carefully using a microwave with 600–800 W.Note: Only heat for a few seconds at a time to avoid spilling.
-
d.Once powder is dissolved completely, add buffer to 50 mL.
 CRITICAL: Keep agarose solution at 37°C for injection. Do not inject boiling agarose as it will damage the tissue.
CRITICAL: Keep agarose solution at 37°C for injection. Do not inject boiling agarose as it will damage the tissue.
-
a.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Insulin (1:500) | Dako | Cat# A-0546 |
| Glucagon (1:2000) | Sigma | Cat# G2654 |
| Somatostatin (1:200) | Millipore | Cat# MAB354 |
| AlexaFluor® 488 goat anti-guineapig (1:500) | Invitrogen | Cat# A11073 |
| AlexaFluor® 546 goat anti-rat (1:500) | Thermo Fisher | Cat# A11077 |
| AlexaFluor® 647 goat anti-mouse (1:500) | Thermo Fisher | Cat# A21235 |
| Biological samples | ||
| Human pancreas slices | nPOD | www.jdrfnpod.org |
| Human isolated islets | Prodo Laboratories | |
| Chemicals, peptides, and recombinant proteins | ||
| Alanine | Sigma | Cat# A7627 |
| Aprotinin | Sigma | Cat# A1153 |
| Arginine | Sigma | Cat# A5006 |
| Bovine berum albumin | Sigma | Cat# 9048-46-8 |
| Calbryte 520 AM | AAT Bioquest | Cat# 20651 |
| Calcium chloride | Sigma | Cat# C4901 |
| Epinephrine | Sigma | Cat# E4375 |
| Fluo4-AM | Invitrogen | Cat# F14201 |
| Glutamic acid | Tocris | Cat# 0218 |
| Glutamine | Sigma | Cat# G3126 |
| Hepes | Sigma | Cat# 7365-45-9 |
| Kainic acid | Tocris | Cat# 0222 |
| Low melting point agarose | Sigma | Cat# A9414 |
| Magnesium chloride | Millipore | Cat# M8266 |
| Potassium chloride | Sigma | Cat# P9541 |
| Sodium chloride | Millipore | Cat# 567440 |
| Critical commercial assays | ||
| Insulin ELISA (human) | Mercodia | Cat# 10-1113-01 |
| Glucagon ELISA (human kit) | Mercodia | Cat# 10-1271-01 |
| Glucagon ELISA 10 μL (for animal samples) | Mercodia | Cat# 10-1281-01 |
| Experimental models: Organisms/strains | ||
| C57Bl6J (mouse, male, 8–16 weeks) | The Jackson Laboratory | Stock #000664 |
| Gcg/tm 1.1(icre)Gkg/J (mouse, female, 8–16 weeks) | The Jackson Laboratory | Stock #030663 |
| B6.Cg-Gt(ROSA)26Sor(tm38(CAG-GCaMP3)HZE) (mouse, male, 8–16 weeks) | The Jackson Laboratory | Stock #029043 |
| Software and algorithms | ||
| Prism 6 | GraphPad | Prism 9 |
| Fiji | Schindelin et al.2 | https://fiji.sc/ |
| Matlab | MathWorks | R2019a |
| Other | ||
| Semi-automatic vibratome VT1200S, | Leica | Cat#14048142066 |
| SP5 upright laser-scanning confocal microscope | Leica | |
| Perifusion system with automated tray handling | Biorep Technologies | PERI4-02-230-FA |
| Pancreas slice chamber | Biorep Technologies | PERI-PSC-001 |
| Pancreas slice chamber extender kit | Biorep Technologies | PERI-PSC-EXT |
| Pancreas slice chamber perforated plate | Biorep Technologies | PERI-PSC-PP |
Materials and equipment
HEPES buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| NaCl (5 M) | 125 mM | 25 mL |
| KCl (1 M) | 5.9 mM | 5.9 mL |
| CaCl2 (1 M) | 2.56 mM | 2.56 mL |
| MgCl2 (1 M) | 1.2 mM | 1.2 mL |
| HEPES (1 M) | 25 mM | 25 mL |
| BSA | 1 mg/mL | 1 g |
| ddH2O | N/A | 940.34 mL |
| Total | N/A | 1000 mL |
Buffer containing BSA can be stored for up to 1 week at 4°C. All solutions used for experiments described in this protocol are made from this stock buffer.
CRITICAL: Adjust pH to 7.4 using NaOH and sterile filter buffer.
CRITICAL: Pure NaOH solution is corrosive to skin and eyes.
Calculation table for hormone secretion buffer preparation
| Buffer | Minutes applied | # Of chambers | Flow rate | Calculation | Amount needed | Amount to prepare |
|---|---|---|---|---|---|---|
| Baseline (5.5/7 mM) | 120 min | 2 | 100 μL/min | 120 × 2 × 0.1 | 24 mL + 50 mL | 100 mL |
| High glucose (16.7 mM) | 20 min | 2 | 100 μL/min | 20 × 2 × 0.1 | 4 mL | 10 mL |
| Low glucose (1 mM) | 30 min | 2 | 100 μL/min | 30 × 2 × 0.1 | 6 mL | 10 mL |
| Membrane depolarization (30 mM KCl) | 5 min | 2 | 100 μL/min | 5 × 2 × 0.1 | 1 mL | 5 mL |
| Total | 175 min | min × chambers × flow | 125 mL |
This table is based on the perifusion protocol described below (Step 22). Numbers need to be adjusted depending on specific protocol applied.
Note: ‘Amount to prepare’ includes additional buffer volume for priming of the system and a little extra as safety volume (evaporation, spilling, reach of tubings, etc). Volume is increased to an amount for simple preparation and calculation. For baseline buffer 50 mL are added to prepare islet chambers prior to experiment.
CRITICAL: For hormone secretion experiments add amino acids to each buffer (2 mM final concentration of L-alanine, L-arginine and L-glutamine). This is essential for physiological conditions and to elevate baseline secretion levels in order to detect hormones other than insulin. It might be easier to add amino acids to the stock HEPES buffer (in this case 125 mL) before separating into the individual buffers listed above.
Note: We suggest preparing these buffers fresh before every experiment.
Calculation table for calcium imaging buffer preparation
| Buffer | Minutes applied | Flow rate | Calculation | Amount needed | Amount to prepare |
|---|---|---|---|---|---|
| Baseline (5.5/7 mM) | 30 min | 500 μL/min | 30 × 0.5 | 15 mL + 50 mL | 100 mL |
| High glucose (16.7 mM) | 10 min | 500 μL/min | 10 × 0.5 | 5 mL | 10 mL |
| Low glucose (1 mM) | 10 min | 500 μL/min | 10 × 0.5 | 5 mL | 10 mL |
| Glutamate | 3 min | 500 μL/min | 3 × 0.5 | 1.5 mL | 5 mL |
| Kainate | 3 min | 500 μL/min | 3 × 0.5 | 1.5 mL | 5 mL |
| Epinephrine | 3 min | 500 μL/min | 3 × 0.5 | 1.5 mL | 5 mL |
| Membrane depolarization (30 mM KCl) | 3 min | 500 μL/min | 3 × 0.5 | 1.5 mL | 5 mL |
| Total | 62 min | min × flow | 140 mL |
This table is based on the imaging protocol described below (Step 61). Numbers need to be adjusted depending on specific protocol applied.
Note: ‘Amount to prepare’ includes additional buffer volume for priming of the system and a little extra as safety volume (evaporation, etc). Volume is increased to an amount for simple preparation and calculation. For baseline buffer 50 mL are added for labeling slices and flushing the system in between slices imaged.
Step-by-step method details
It is essential to perform steps 1–3 (Agarose injection, Tissue preparation and Slice procedure) in a timely manner to ensure tissue viability. Slices maintain viability much better compared to tissue pieces due to better oxygenation.
Agarose injection
Timing: 10 min
This section describes how to inflate the pancreas with low-melting point agarose to generate slices for following functional experiments. This step does not apply for human tissue.
-
1.
Euthanize the mouse by cervical dislocation.
-
2.
Place the mouse under a stereomicroscope and wet the abdominal fur with 70% ethanol (Figure 1A).
-
3.
Expose the organs by opening the abdomen using surgical scissors and flip over liver for easier access and visibility (Figures 1B and 1C).
-
4.
Clamp off the common bile duct at the ampulla of Vater to avoid drainage into the duodenum (Figure 1D).
-
5.
Clean the duct from fat or connective tissue for easier access (Figure 1E).
-
6.
Cannulate the common bile duct using a 30-gauge needle and 5 mL syringe with Luer-Lok-Tip (Figure 1F).
Note: Needle scan be bend between 30–90 degrees angle for easier injection.
-
7.
Steadily inject 2–3 mL of low-melting agarose until pancreas is fully inflated.
-
8.
Remove the hardened pancreas and place in a dish filled with HEPES buffer (containing baseline glucose concentration; we use 7 mM for mice and 5.5 mM for human tissue slices).
CRITICAL: Full inflation of the pancreas is essential for successful slicing procedure and maximal outcome. This step requires fast handling as agarose will solidify quickly and thus block the syringe. Note that the injection procedure is performed the same way as for islet isolation.
Figure 1.
Pancreas injection procedure of low melting point agarose through the common bile duct related to step 1
(A) Preparation of euthanized mouse for procedure by opening of the abdomen.
(B) Exposure of common bile duct.
(C) Clamping of intestine.
(D) Injection with support of tweezers from below. Pancreas (P) indicated by dashed line.
(E) Partially inflated pancreas.
(F) Fully injected pancreas.
Tissue preparation
Timing: 5 min
This step describes the preparation of the tissue prior to slicing.
Note: For human samples, tissue is emerged with 3.8% low-melting agarose (Figure 3).
-
9.
Gently remove connective, fibrotic, adipose tissue and sections that are not inflated with agarose (Figures 3A and 3B).
-
10.
Cut tissue into small cubes of about 0.5 cm3 size (Figure 3B).
-
11.
Inspect pieces under the stereomicroscope. If necessary, remove thick ducts and fibrotic, adipose tissue.
-
12.
Blot dry tissue pieces shortly before embedding in agarose.
-
13.
Transfer 4–5 tissue blocks into a 35-mm petri dish and fill up with low-melting agarose until tissue is fully submerged (Figures 2A and 3C).
-
14.
Allow agarose to harden before cutting small cubes using a scalpel.
CRITICAL: Proper tissue cleaning and drying is essential to avoid tissue loss during slicing.
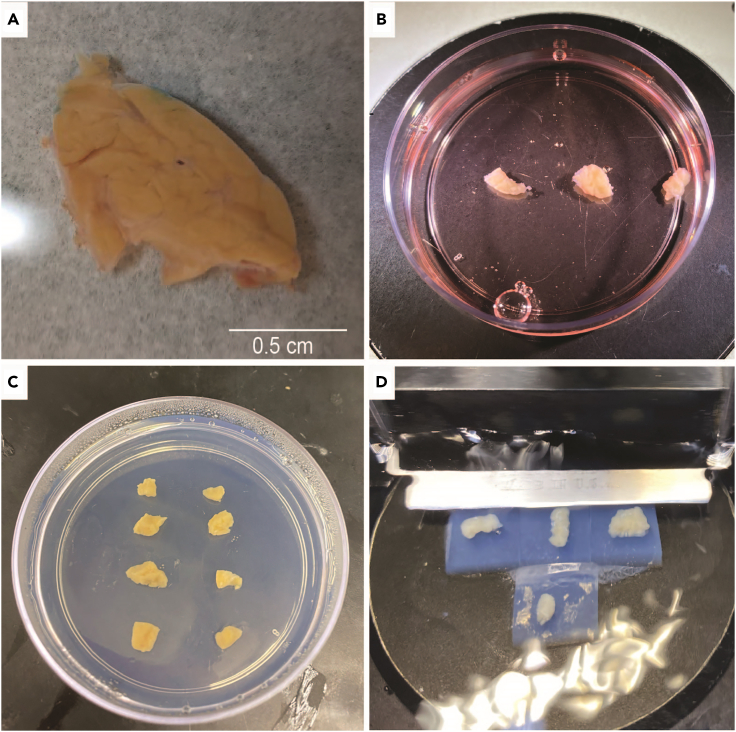
Figure 3.
Human pancreas tissue slice preparation related to step 2 and 3
(A) Pancreas tail piece of roughly 1 cm/ 1–2 g size. Scale bar, 0.5 cm.
(B) Cleaned pancreas pieces.
(C) Pieces embedded in low melting point agarose.
(D) Slicing procedure.
Figure 2.
Mouse pancreas tissue preparation and slicing procedure related to step 2 and 3
(A) Cleaned pancreas pieces.
(B) Agarose embedded tissue pieces mounted on slicing plate.
(C) Slicing procedure.
(D) Mouse tissue slice with islets indicated by white arrows.
Slice procedure
Timing: 45 min–1 h
This section describes how to prepare pancreatic tissue slices using a vibratome.
-
15.
Glue tissue blocks by placing them upside down on the mounting plate and allow glue to solidify for a moment (Figure 2B and Methods video S1).
-
16.
Mount the plate into the tray.
-
17.
Fill tray with HEPES buffer (containing baseline glucose concentration) until agarose blocks are completely covered with solution.
-
18.
Carefully place the blade in the blade holder.
Note: Lower blade to start shortly above tissue and ensure the blade is covered by solution to avoid tissue rupture.
-
19.
Set parameters for slicing.
Note: Set parameters for continuous slicing by marking start and end points.
Slice parameters
| Amplitude | 8 (0.8 mm) |
|---|---|
| Speed | 2–4 (0.2–0.4 mm/s) |
| Angle (if adjustable) | 15° |
| Interval (mouse) | 150 μm |
| Interval (human) | 120 μm |
-
20.
Start cutting.
-
21.
Collect slices carefully using curved forceps or a small brush (Figures 2C and 3D).
Note: Start cutting very slowly and increase speed if tissue quality allows it.
CRITICAL: Using inhibitors (e.g. aprotinin) is essential for tissue viability if kept static in a dish.
CRITICAL: Do not overcrowd the dish with slices (not more than 30 slices in 60 mm dish). Use multiple dishes for larger amounts.
CRITICAL: Continue with perifusion or imaging in a timely manner to ensure tissue viability. Slices can be kept in HEPES buffer (containing baseline glucose concentration and aprotinin) at 22°C for up to 1 h. If more time is needed place slices in an incubator at 37°C (in HEPES buffer with glucose and aprotinin). For culture of tissue slices use appropriate culture conditions (see MMF Qadir for a detailed culture protocol3).
Note: Place dish with slices on a slowly moving shaker to allow wash out of digestive enzymes and improve viability if kept longer.
Perifusion
Timing: 3–6 h
This section describes how to set up and perform dynamic perifusion using an automated perifusion system (Biorep Technologies Inc.). Step 22–25 should be performed prior or in parallel to slicing for optimal timing.
-
22.
Turn on perifusion machine and select number of samples/chambers and choose/program a protocol.
Note: Add a flushing step of 90 min in baseline HEPES buffer to allow proper wash out and resting of the slices to ensure a stable baseline for hormone release.
Standard perifusion protocol (for 2 chambers)
| Step # | Step | Repetition | Seconds | Flow rate | A | B | C | D | Pause |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Flushing | 48 | 112 | 100 | 1 | 1 | 0 | 0 | no |
| 2 | Baseline (5.5 or 7 mM glucose) | 10 | 60 | 100 | 1 | 1 | 0 | 0 | no |
| 3 | Stimulation (high glucose, 16.7 mM) | 20 | 60 | 100 | 2 | 2 | 0 | 0 | no |
| 4 | Stimulation (low glucose, 1 mM) | 30 | 60 | 100 | 3 | 3 | 0 | 0 | no |
| 5 | Membrane depolarization (30 mM KCl) | 5 | 60 | 100 | 4 | 4 | 0 | 0 | no |
| 6 | Baseline (5.5 or 7 mM glucose) | 10 | 60 | 100 | 1 | 1 | 0 | 0 | no |
Note: Repetitions and seconds of flushing step are calculated to fit a single 96 Well plate (90 min/ (96 wells/ 2 chambers) × 60 s).
Note: Protocols can be designed more complex, but are limited to 8 solutions, 4 outlets and maximum of 12 chambers (for the Peri4) or 12 solutions and 12 outlets (for the Peri5).
-
23.
Place necessary solutions in the tray and connect inflow and pump tubing’s.
-
24.
Prime the machine using the automated priming protocol.
-
25.
Once priming is completed, close the machine and switch on heating before continuing.
-
26.
Choose 3–5 slices and trim slices under the stereomicroscope using forceps and scalpel.
Note: Pre-select slices by identifying slices rich in islets using transmitted light microscopy to allow maximal hormone detection (Figure 2D).
-
27.
Gently place each slice on a separate grid of the slice chamber (Figure 4A and Methods video S2).
Note: Assemble each section of the slice chamber and add a drop of baseline HEPES buffer to place slice easier.
-
28.
Assemble slice chamber from the top to bottom (Figure 4B).
-
29.
Connect slice chamber to perifusion machine and immediately start the protocol (Figures 4C and 4D).
Note: Connect slice chambers to machine one by one to avoid drying out.
Note: If many chambers must be connected, protocol can be stopped and re-started to allow full 90 min flushing for all chambers.
Note: Place a collection container in tray for 90 min flushing period to avoid spilling (empty tip boxes fit very well).
-
30.
Connect ice-water bucket to cooling pump any time during 90 min flushing protocol.
-
31.
Change 96 Well plates when necessary, during the protocol. For the protocol above plate needs to be changed once after 90 min and once after 48 min.
Note: Plate 1 will always be for flushing and does not have to be collected or measured for hormone release. If calculated correctly the first plate need to be placed after 90 min.
Note: The timing to change the plate depends on the number of chambers and seconds chosen to collect samples.
-
32.
If hormones are measured directly after perifusion, store plates at 4°C. Otherwise store at −80°C to avoid degradation of hormones.
-
33.
After protocol is finished, stop heater and cooling pump, disconnect chambers from machine and place in chamber holder.
-
34.
Disassemble chambers and collect slices from each chamber. To measure total hormone content, collect slices in 500 μL ice cold acidic ethanol (3% HCl in pure ethanol, lysate buffer).
Note: Slices can be collected in any chosen buffer for lysis or be reused for perifusion/imaging by placing them back in HEPES buffer (containing baseline glucose concentration and aprotinin).
-
35.
Generate an experiment report and save it.
-
36.
Clean the machine with preset protocol and disassemble tubing’s after cleaning (follow on screen instructions)
-
37.
Return tray to park position and switch off the machine.
Note: Cleaning procedure takes 32 min and requires ddH2O and 10% household bleach solution.
Figure 4.
Slice perifusion procedure related to step 4
(A) Single slice mounted on metal plate of slice chamber.
(B) Assembled slice chamber with 3 slices.
(C) Slice chambers connected to perifusion machine.
(D) Setup for slice perifusion with 4 chambers and 4 solutions in a Peri4 machine.
Hormone detection
Timing: 3 h
This part describes how to measure and analyze hormones released from perfusates.
-
38.
Thaw perifusion plates.
Note: If dilutions are not clear, perform a dilution test with a limited number of samples first.
-
39.
Perform ELISA according to user manual and read plate(s).
-
40.
For data analysis, interpolate values with the standard curve using four parameter logistic regression.
Note: As slices have unknown number of islets, absolute values should be normalized.
-
41.
Normalize values to baseline (stimulation index), KCl peak and /or total hormone content (if available).
-
42.
Plot data as perifusion trace and indicate switch of solution.
Note: The system has a dead volume time of roughly 2 min. That means this time will have to be added to the switch time.
Orientation for hormone concentration ranges
| Step | Tissue source | Insulin concentration range | Glucagon concentration range (pmol/L) | Insulin stimulation index (fold) | Glucagon stimulation index (fold) |
|---|---|---|---|---|---|
| Baseline (5.5 or 7 mM glucose) | Human slices | 5–50 mU/L | 1–10 | 0.5–2 | 0.5–2 |
| Human islets | 50–400 mU/L | 10–60 | 0.8–1.2 | 0.6–1.5 | |
| Mouse islets | 2–5 ng/mL | 30–100 | 0.8–1.2 | 0.8–1.2 | |
| Stimulation (high glucose, 16.7 mM) | Human slices | 50–200 mU/L | 0.2–5 | 2–20 | 0.2–1 |
| Human islets | 300–600 mU/L | 20–50 | 2–5 | 0.2–0.6 | |
| Mouse islets | 4–10 ng/mL | 10–60 | 2–8 | 0.4–0.8 | |
| Stimulation (low glucose, 1 mM) | Human slices | 1–20 mU/L | 2–30 | 0.5–1 | 2–20 |
| Human islets | 20–100 mU/L | 40–150 | 0.1–0.8 | 1–4 | |
| Mouse islets | 1–4 ng/mL | 70–200 | 0.5–1.2 | 1.2–3 | |
| Membrane depolarization (30 mM KCl) | Human slices | 50–200 mU/L | 3–50 | 2–20 | 2–20 |
| Human islets | 300–1000 mU/L | 150–250 | 2–8 | 4–10 | |
| Mouse islets | 5–15 ng/mL | 50–200 | 5–20 | 2–5 |
Values shown in this table are based on healthy tissue from 3 human slices/ 100 human islets/ 150 mouse islets. Please note that concentrations are heterogeneous between samples and highly depend on multiple factors including tissue size, islet number, donor characteristics, etc. This table only serves as orientation for hormone assessment.
Labeling for imaging
Timing: 1 h
This step describes how to label slices to perform functional imaging. We labeled with a Calcium reporter; however other dyes might work equally well.
-
43.
Use a 24 well plate and add 500 μL of baseline HEPES buffer with inhibitor to one well.
-
44.
Add dye to the well. We use Fluo4-AM at a final concentration of 6 μM.
-
45.
Cover the plate with aluminum foil.
-
46.
Incubate slices for 1 h at 22°C on a shaker.
Note: Some dyes might label sufficiently already after 30 min incubation time.
-
47.
Wash slice with baseline buffer by transferring slice to another well.
CRITICAL: Wash out of the dye is critical to not contaminate the imaging chamber or objective with loading dye.
Functional imaging
Timing: 1–3 h
This section describes how to perform functional imaging on tissue slices.
-
48.
Connect all tubing’s and switch on heater (Figures 5A and 5B).
-
49.
Fill and prime all syringes with their respective solutions.
-
50.
Assemble imaging chamber.
Note: First 3 steps can be done while slices are incubating.
-
51.
Add baseline buffer to the imaging chamber and gently transfer a single slice.
-
52.
Gently press down the slice using the harp.
-
53.
Connect the chamber to in and out flow and place it under the scope (Figures 5C and 5D).
-
54.
Find islets using a low magnification objective (10× works well).
-
55.
Focus on the islet and switch to higher magnification objective for imaging (20× / 40×).
-
56.
Define top and bottom of the islet by setting the Z-stack.
-
57.
Use a Z-step size of 5–15 μM depending on islet size and labeling quality.
Note: Avoid imaging the surface layer of the slice as it might show unspecific, bright labeling.
-
58.
Capture a xyz image with reflected light and dye wavelength, in this case 488 laser.
-
59.
Switch on pump and start perfusing with baseline glucose solution.
-
60.
Define parameters for xyzt imaging as follows:
Imaging parameters
| z-step size | 5–15 μM |
|---|---|
| Total z-stack size | 50–100 μM |
| interval | 5 s |
| resolution | 512 × 512 |
| pinnhole | 1–2 airy units |
-
61.
Start imaging and switch solutions according to protocol. The protocol we used to study alpha cells in mouse tissue slices is as follows:
Imaging protocol
| Buffer | Time | Frames | Total frames |
|---|---|---|---|
| 7 mM glucose | 5 | 60 | 60 |
| 16.7 mM glucose | 10 | 120 | 180 |
| 1 mM glucose | 10 | 120 | 300 |
| 7 mM glucose | 5 | 60 | 360 |
| 100 μM glutamate | 3 | 36 | 396 |
| 7 mM glucose | 5 | 60 | 456 |
| 10 μM epinephrine | 3 | 36 | 492 |
| 7 mM glucose | 5 | 60 | 552 |
| 100 μM kainate | 3 | 36 | 588 |
| 7 mM glucose | 5 | 60 | 648 |
| 30 mM KCl | 3 | 36 | 684 |
| 7 mM glucose | 5 | 60 | 744 |
Note: All stimuli are diluted in baseline buffer (7 mM glucose HEPES buffer).
-
62.
Perfuse slices continuously while acquiring.
-
63.
Once completed stop imaging.
-
64.
Stop the pump.
-
65.
Save images.
-
66.
Remove the slice from the chamber and transfer to a 24 well plate in baseline buffer.
-
67.
Fix the slice with 4% PFA for 30 min for immunohistochemistry.
-
68.
Repeat steps 51–67 with a slice for the same or a different protocol as desired.
Figure 5.
Imaging procedure related to step 4
(A) Pump and perifusion setup for microscope.
(B) Complete imaging setup with perifusion.
(C) Imaging chamber connected to in- and outflow.
(D) Setup for slice imaging with dipping lens.
Expected outcomes
This protocol describes how to generate pancreatic tissue slices and use them to perform physiological measurements of hormone secretion and Ca2+ imaging. The first 3 steps describe in detail how to generate slices to be utilized for in situ experiments. If correctly performed a single mouse pancreas can be cut in 50–100 slices, depending on tissue piece size. Not every slice necessarily contains islets so pre-screening is recommended for optimal outcome. Slices then can be used for functional studies as described above but are not limited to these readouts. For dynamic hormone secretion we typically measure insulin and glucagon, however other hormones (including secretion from exocrine compartments, e.g., amylase) can be measured in parallel, if detection kits are available. If detection is too low, number of slices can be increased and/or flow rate reduced to reach higher yields. Usually, perfusates need dilution for insulin detection, while glucagon levels are lower and need no dilution. In case of functional imaging, multiple dyes are commercially available to study different aspects of metabolism and cell signaling and can be applied as recommended by supplier. The described protocol on how to image slices can be used no matter the dye.
Although this protocol is focused on alpha cell function, exocrine cell function can be studied in parallel. If cells are labeled with using general calcium indicators like Calbryte or Flou-4AM, responses from any cell type can be analyzed. This has been proven successful for a variety of studies already studying immune cells, innervation or blood vessels.4,5,6,7,8
Quantification and statistical analysis
Quantification of hormone measurements
Quantification of hormone release was measured using commercially available ELISA kits (Mercodia). For insulin perfusates had to be diluted 1:1 and 1:2 to remain within detection limits, glucagon was measured undiluted. Concentrations of insulin/glucagon were obtained by plotting the absorbance of calibrators versus their concentration using sigmoidal four-parameter logistic with Prism 9 software (GraphPad software, La Jolla, CA). Calculated concentrations are absolute values and need to be normalized as number of islets within the slices is unknown. Data can be normalized to average baseline secretion (fold), percent of KCl peak or as percent of total hormone content (%). Hormone content was measured from lysates obtained from all slices used after the experiment and diluted 1:500 or 1:1000.
Quantification of cytosolic Ca2+ levels
To quantify changes in intracellular Ca2+ levels, we first identify the endocrine area by using a backscatter image. We then drew regions of interest around individual islet cells and measured changes in mean GCaMP3/Fluo4-AM/Calbryte fluorescence intensity using ImageJ. Regions of interest were drawn using single planes. Maximum Intensity is measured for each region/frame and exported to Excel sheets for every islet individually. Changes in fluorescence intensity were then calculated as percentage change over baseline (ΔF/F). The baseline was defined as the mean intensity of 3 min baseline prior to each stimulus. To ensure viability cells we preselect cells based on their response to KCl depolarization. We used a threshold of 2xSD of baseline activity for Calcium dye and 4xSD over baseline activity for slices from transgenic mice. Data can then be presented either as heatmaps using MATLAB R2019a (MathWorks software, Natick, MA) or as average traces using Prism 9 (GraphPad software, La Jolla, CA).
Statistical analysis
For statistical comparisons we used Prism 9 (GraphPad software, La Jolla, CA) and performed Student’s t tests (unpaired) or one-way analysis of variance (ANOVA) corrected for multiple comparisons (using Tukey’s Multiple Comparison Test; each row represented matched data). p-values < 0.05 were considered statistically significant.
Limitations
Pancreatic tissue slices preserve pancreas morphology and thus cell-cell contacts of different cell types present in the pancreas. This communication is of advantage studying pancreas physiology but leads to specific limitations and must be considered when interpreting results. As tissue slices contain both endocrine and exocrine cell types, stimulus application might not only influence the targeted cell. Thus, certain stimuli could cause indirect effects by other cell types and might lead to feedback loops. Particularly when substances inhibit or activate certain receptors, this might target other cell types present in slices containing the same receptors. For specific cell targeting traditional in vitro protocols might be more appropriate. Another aspect to be considered is that acinar cells contain pancreatic enzymes that lead to the breakdown of proteins and thus digestion of the slice itself. Consequently, trypsin inhibitors need to be used at all times when slices are static to ensure viability. Utilization of these inhibitors might interfere with the successful transfer of viruses that might be used for labeling or other experiments.
For the study of human samples, reduced availability is a major limitation. Furthermore, human samples are variable in general, it is challenging to obtain consistent results. Consequently, timely and consistent handling of pancreatic tissue slices with adequate controls for viability is essential.
Troubleshooting
Problem 1
Difficulty with agarose injection if pancreas is not inflating properly (related to step 1).
Potential solution
-
•
Agarose has higher viscosity and therefore more effort is necessary to infuse the pancreas. If injections for e.g., isolations are usually successful you might just need more pressure or longer injection time compared to water based injections.
-
•
Unsuccessful cannulation of the duct. It helps to remove fatty and fibrotic tissue to visualize the duct and allow proper cannulation. Use forceps to stabilize the needle and avoid pinching through the duct. Try to put some tension on the duct by pulling carefully on one end to allow easier cannulation.
-
•
Multiple attempts to cannulate might leave incisions in the duct. If that happens, try to cannulate closer towards the intestine to avoid leaking.
-
•
If agarose inflates the intestine or stomach, make sure that the common bile duct is clamped correctly.
-
•
Low melting point agarose solidifies quickly and can clot the needle within very short time. Keep a little pressure on the syringe constantly to allow agarose flowing and try to be a quick as possible.
Problem 2
Difficulty cutting pancreatic tissue slices (related to step 3).
Potential solution
-
•
If the pancreas is poorly injected, it will be very hard to slice. Only use tissue pieces that are inflated with agarose.
-
•
Adipose tissue, ducts and fibrotic tissue are difficult to slice. Try to clean the tissue very well before embedding.
-
•
Make sure to use a sharp razor blade. Use a fresh blade every time.
-
•
Blot dry tissue pieces well before embedding to avoid them from breaking out of the agarose. If pancreas tissue falls out of the agarose as a whole, it can be dried and re-embedded.
Problem 3
Leaking perifusion chambers (related to step 4 perifusion).
Potential solution
-
•
Make sure chambers are closed properly.
-
•
Trim slices to fit the chambers. If agarose overlaps, chamber might to close properly.
-
•
Check O-rings and replace if necessary.
-
•
Make sure all tubing’s are connected properly.
Problem 4
Chamber runs dry (related to step 4 perifusion)
Potential solution
-
•
Make sure all tubing’s are properly connected.
-
•
Ensure tubing’s are fully immersed in solution.
-
•
Make sure chamber and nozzles are clean and solution can flow.
-
•
Ensure number of channels matches with the protocol to avoid pumping from a closed channel.
Problem 5
Air bubbles in the chamber (related to step 4 perifusion).
Potential solution
-
•
Make sure all tubing’s are properly connected and dead ends are used on all channels that are not in use.
-
•
Ensure tubing’s are fully immersed in solution.
-
•
If air bubbles accumulate in islet chambers, gently remove air either using a syringe or opening the chamber and adding a drop of solution.
-
•
Any manipulation of the chamber might cause a change in hormone kinetics so it is essential to be very careful and only to remove air bubbles if absolutely necessary (when accumulation might cause drying out).
-
•
We highly recommend using the newly developed slice chambers from Biorep as they cause less problems with air bubbles due to a vertical flow and minimal volume.
Problem 6
Weak or no hormone release (related to step 4 perifusion).
Potential solution
-
•
If samples were diluted, use lower dilution factor.
-
•
Make sure slices contain islets by screening them under a stereo microscope. Use more slices if possible.
-
•
If there is no response to membrane depolarization with KCl slices were not viable. In that case injection/slicing procedure was not successful. Try with a new sample and make sure steps are performed in a timely manner and slices are incubated in the presence of inhibitors when static. Might be helpful to perform live/dead assay with 2 slices to check viability prior to experiment.
Problem 7
No labeling of the slices (related to step 5 imaging).
Potential solution
-
•
Incubate slices a little longer (∼30 min) and check again.
-
•
Make sure dye is not expired.
-
•
Use a higher concentration of the dye.
-
•
Try with another slice.
Problem 8
Weak Ca2+ responses (related to step 5 imaging).
Potential solution
-
•
Stimulate slice with KCl to check viability. If there is no response use another slice. If slice does not respond to KCl, start over with a fresh sample and make sure Step 1–3 are performed in a timely manner and slices are incubated with inhibitors when static.
-
•
Make sure solutions contain glucose and/or other stimuli.
-
•
Make sure pump is running and solutions are flowing.
-
•
Use another slice and try again.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Alejandro Caicedo, acaicedo@med.miami.edu.
Materials availability
This study did not generate new unique reagents.
Data and code availability
This study did not generate/analyze datasets/code.
Acknowledgments
This work was funded by NIH grants R56DK084321 (A.C.), R01DK084321 (A.C.), R01DK111538 (A.C.), R01DK113093 (A.C.), U01DK120456 (A.C.), R33ES025673 (A.C.), R21ES025673 (A.C.), and R01DK130328 (A.C.); the Leona M. and Harry B. Helmsley Charitable Trust grants G-2018PG-T1D034 (A.C.) and G-1912-03552 (A.C.); and the American Diabetes Association 4-22-PDFPM (J.K.P.).
We thank Alexander Kao and Guillermo Camarena from Biorep Technologies, Inc. for developing the tissue slice chambers used in this paper.
Author contributions
J.K.P. and A.C. wrote the paper.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2023.102399.
Contributor Information
Julia K. Panzer, Email: jpanzer@coh.org.
Alejandro Caicedo, Email: acaicedo@med.miami.edu.
References
- 1.Panzer J.K., Tamayo A., Caicedo A. Restoring glutamate receptor signaling in pancreatic alpha cells rescues glucagon responses in type 1 diabetes. Cell Rep. 2022;41:111792. doi: 10.1016/j.celrep.2022.111792. [DOI] [PubMed] [Google Scholar]
- 2.Schindelin J., Arganda-Carreras I., Frise E., et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. https://www.nature.com/articles/nmeth.2019#citeas. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qadir M.M.F., Álvarez-Cubela S., Weitz J., Panzer J.K., Klein D., Moreno-Hernández Y., Cechin S., Tamayo A., Almaça J., Hiller H., et al. Long-term culture of human pancreatic slices as a model to study real-time islet regeneration. Nat. Commun. 2020;11:3265. doi: 10.1038/s41467-020-17040-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohrs C.M., Chen C., Jahn S.R., Stertmann J., Chmelova H., Weitz J., Bähr A., Klymiuk N., Steffen A., Ludwig B., et al. Vessel network architecture of adult human islets promotes distinct cell-cell interactions in situ and is altered after transplantation. Endocrinology. 2017;158:1373–1385. doi: 10.1210/en.2016-1184. [DOI] [PubMed] [Google Scholar]
- 5.Weitz J.R., Makhmutova M., Almaça J., Stertmann J., Aamodt K., Brissova M., Speier S., Rodriguez-Diaz R., Caicedo A. Mouse pancreatic islet macrophages use locally released ATP to monitor beta cell activity. Diabetologia. 2018;61:182–192. doi: 10.1007/s00125-017-4416-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huber M.K., Drotar D.M., Hiller H., Beery M.L., Joseph P., Kusmartseva I., Speier S., Atkinson M.A., Mathews C.E., Phelps E.A. Observing islet function and islet-immune cell interactions in live pancreatic tissue slices. J. Vis. Exp. 2021 doi: 10.3791/62207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Makhmutova M., Weitz J., Tamayo A., Pereira E., Boulina M., Almaça J., Rodriguez-Diaz R., Caicedo A. Pancreatic beta-cells communicate with vagal sensory neurons. Gastroenterology. 2021;160:875–888.e11. doi: 10.1053/j.gastro.2020.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Almaca J., Weitz J., Rodriguez-Diaz R., Pereira E., Caicedo A. The pericyte of the pancreatic islet regulates capillary diameter and local blood flow. Cell Metab. 2018;27:630–644.e634. doi: 10.1016/j.cmet.2018.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate/analyze datasets/code.