Abstract
Diagnosing biofilm infections has remained a constant challenge for the last 50 years. Existing diagnostic methods struggle to identify the biofilm phenotype. Moreover, most methods of biofilm analysis destroy the biofilm making the resultant data interpretation difficult. In this study we introduce Fourier Transform Infra-Red (FTIR) spectroscopy as a label-free, non-destructive approach to monitoring biofilm progression. We have utilised FTIR in a novel application to evaluate the chemical composition of bacterial biofilms without disrupting the biofilm architecture. S. epidermidis (RP62A) was grown onto calcium fluoride slides for periods of 30 min–96 h, before semi-drying samples for analysis. We report the discovery of a chemical marker to distinguish between planktonic and biofilm samples. The appearance of new proteins in biofilm samples of varying maturity is exemplified in the spectroscopic data, highlighting the potential of FTIR for identifying the presence and developmental stage of a single biofilm.
Keywords: Staphylococcus epidermidis, Biofilm, Infection, FTIR, Continuous monitoring, Chemical composition, Clinical diagnosis
1. Introduction
Biofilms are the preferred form of bacterial growth and are widespread both in nature and in human infection [[1], [2], [3]]. Despite our understanding of the health, industrial, environmental and economic impact of biofilms, there is much that remains unclear about biofilm development, including the temporal changes of the extracellular matrix (ECM) over time [4]. After attachment to a surface, bacteria become embedded within the ECM [5]. This hydrated matrix contains polysaccharides, lipids, proteins, phospholipids and eDNA and fulfils a number of critical roles including intercellular communication, resistance to desiccation and tolerance to antimicrobial treatment [[6], [7], [8]]. Once a biofilm infection has progressed, no current antimicrobial therapies will completely eradicate or disinfect them [9]. In the healthcare setting, such as in chronic wound infection, a key problem in clinical management is determining at what point a biofilm phenotype is present, which has significant implications on the effectiveness of antimicrobial treatment. Bacteria in a biofilm are at least 10 times more likely to resist available treatment options [10].
A major problem in investigating the transition from free-floating to adherent cells, onto the definitive biofilm phenotype and hence determining the best treatment options, is that current methods for quantitative analysis of biofilm requires the disruption of the ECM. This makes accurate temporal analysis of biofilm ECM inherently problematic. Diagnosis of infection is primarily rooted in culture plate assay or imaging techniques which remain unchanged since the 1980s [11,12]. These existing standardised clinical methods struggle to differentiate between planktonic and biofilm growing phenotypes [13]. Other methods of diagnosis are reliant on chemical labelling and total dehydration of the sample before analysis, which not only changes the chemical composition of the sample, but also requires a significant investment of time [14,15]. For example, in the culture plate methods, samples undergo multiple 24 h incubation periods, during which the patient's condition may worsen [13]. A rapid diagnostic solution is required where samples can be collected and analysed in the same day, to both determine which microorganism is causing an infection and whether the phenotype of the microbe has switched from planktonic to sessile. This would allow a more targeted approach to antimicrobial use [16,17].
The use of Fourier transform infrared spectroscopy (FTIR), a non-destructive method, has the potential to allow multiple analyses of the same biofilm, without damaging the encased bacteria [18]. FTIR is a vibrational spectroscopic technique that uses Fourier transformation, a mathematical method, to convert the information yielded by infra-red spectroscopy into an easily read format [19]. Infrared spectroscopy (IR) exposes the sample to IR light in a spectrometer. Photons are either absorbed or transmitted by the sample producing a spectrum which is unique to each chemical bond. FTIR spectroscopy is a widely used tool for structural and compositional analysis of natural materials [20]. Recently, spectroscopy has emerged as a major tool for biomedical applications and has made significant progress in the field of clinical evaluation [21].
Microbiology experienced an explosion in the use of FTIR for classifying bacteria in the late 20th century [22,23]. One of the first studies into the use of FTIR for microorganism analysis was conducted by Flemming et al. [24] This study evidenced the notion of identifying bacterial strains with spectroscopy by assigning chemical signatures to each strain and from then myriad studies have reported the use of FTIR to detect bacteria [25]. Research has since expanded to the spectroscopic study of a wide range of microbes and the biofilms they can form, incorporating real-time monitoring of cellular activity and how the surrounding environment influences biofilm composition [[26], [27], [28]]. Furthermore investigations have been carried out into the process by which sessile cells adhere within their environment as well as the preferred conformation which they adopt [29]. FTIR is gradually becoming more standardised, though it is often applied in conjunction with other analytical techniques to confirm the presence of chemical components. For instance, Serra et al. utilised FTIR to supplement their investigation into B. pertussis biofilm growth, using the spectroscopy to provide chemical information to accurately characterise the microorganism [30]. Despite the volume of research dedicated to optimising FTIR for biofilm analysis and characterisation, there is a lack of reports monitoring the growth of a single biofilm. It would be beneficial to understand chemical changes as microbes shift from the planktonic to the biofilm state. Ideally a chemical marker could be specifically associated with the irreversible attachment of microbial cells to a surface, strengthening the potential to explore methods to prevent or delay biofilm maturation in infected wound sites.
In this study we have used FTIR side by side with routine methods to examine temporal changes of the biofilm over a 96-h period in order to identify biomarkers of the switching to a biofilm phenotype. Here we show a number of differences in key areas of the FTIR spectrum that begin to develop as early as 30 min into biofilm development and remain consistent over the subsequent 24–96 h of biofilm growth.
2. Materials & methods
2.1. Characterisation of biofilm
Staphylococcus epidermidis RP62A (ATCC 35984) and ATCC 12228 were used throughout this study and was stored long-term in glycerol at −80 °C. S. epidermidis (RP62A) was revived on Luria Bertani (LB) agar (Sigma-Aldrich, UK) for 24 h at 37 °C. Following initial growth on solid media, bacteria were propagated in LB broth (Sigma-Aldrich, UK) overnight and washed by centrifugation. Bacterial cells were suspended in PBS and standardised to an optical density of 0.6 at 600 nm, equivalent to 1x108 cells/mL. Standardised cells were then adjusted to 1x107 cells/mL in nutrient broth (NB, Sigma-Aldrich, UK).
Biofilms were grown at over 96 h in nutrient broth on Thermanox coverslips (ThermoFisher, UK) at 37 °C. At early and late time points of 0.5, 1, 2, 4, 24, 48, 72 and 96 h biofilm media and any non-adherent cells were removed and biofilms were incubated with alamarBlue (ThermoFisher, UK), which was prepared in biofilm growth media as per the manufacturer's instructions, for 1.5 h at 37 °C. Fluorescence of alamarBlue incubated with each biofilm was measured in a plate reader at excitation/emission wavelengths of 544/590 nm. For colony forming unit (CFU) counting the biomass was removed by sonication in an ultrasonic water bath for 10 min before being serially diluted in PBS and plated on LB agar to quantify biofilm CFUs. Plates were subsequently incubated at 37 °C for 24 h. Biofilm characterisation data was plotted and analysed using GraphPad Prism (version 9). Means of biofilm viability, biomass and CFUs were compared using Kruskal-Wallis tests with Dunn's post-hoc test and statistical significance was determined where P < 0.05.
2.2. Biofilm sample preparation for FTIR
Cells for biofilms were prepared as described above and then grown in 20 mL of growth media (nutrient broth or brain heart infusion) on calcium fluoride windows (Galvoptics, UK) from 30 min to 96 h by incubating the slides in the bacterial culture. Following incubation, samples removed from solution and transferred to an isolated desiccant chamber for 30 min to partially dry, removing bulk hydration whilst maintaining the ECM. Chamber was a sealed Perspex box, containing fixed and regularly cleaned Perspex shelf with silica bead tray directly underneath. Biofilm samples analysed with FTIR immediately following drying. Planktonic samples analysed in a similar way by looping onto clean calcium fluoride slide and isolating for 30 min in desiccant chamber before spectra collection.
2.3. Fourier-transform infrared (FTIR) spectroscopy
A desktop Summit PRO FTIR spectrometer (Nicolet, Thermo Scientific, UK) with iD1 transmission sampling apparatus was used for all analysis. All data was collected using OMNIC Paradigm™ software (Thermo Scientific, UK). Data acquisition was performed at 4 cm−1 resolution, accumulating 64 scans over a spectral range of 4000-800 cm−1. For each sample, 3 locations were analysed, demonstrating varying biomass distribution on the calcium fluoride slides. Each sample was repeated in triplicate.
Data analysis was conducted using OMNIC software in the first instance. Each spectrum was processed in the same way, completing normalisation followed by a base line correction. Unscrambler X (CAMO, Sweden) was then used for the multivariant analysis (MVA) methods Cluster Analysis (CA), Principal Component Analysis (PCA) and Linear Discriminant Analysis (LDA). Every PCA was setup with a minimum of 12 orthogonal variables depending on the spectral region used, where the number of PCs chosen for each setup described >99% of the variation. All LDA models were setup over the full spectral range. In each LDA, three random spectra from the data set were left out of the training models at each run.
3. Results
3.1. Assessing biofilm growth kinetics
Firstly, to determine S. epidermidis biofilm dynamics, bacterial cells were grown in microtiter plates and the biofilm phenotype was assessed over 96 h (Fig. 1A). S. epidermidis (RP62A) was found to form biofilms after only 0.5 h of growth, but this became a more robust phenotype following 2 h of growth where biofilm viability and biomass increased 3.2 and 2.8-fold, respectively, compared to the previous observed time point (1 h). Biofilm viability continued to increase until 48 h (P < 0.05). From 72 h incubation, the biofilm thickness increased such that uptake of Alamar Blue dye was inhibited, resulting in apparent lowered cell viability, a phenomena which has previously been noted in the literature [31]. However, continued growth of the biofilm is evidenced by the S. epidermidis biomass increasing significantly following 96 h growth (P < 0.01). This significant increase in biomass was reflected in total biofilm CFUs (Fig. 1B) where total biofilm cells increased significantly to 1.6x107 and 2.4x107 CFU/mL after 48 and 96 h growth, respectively (P < 0.05 and 0.01, respectively).
Fig. 1.
Characterisation of Staphylococcus epidermidis biofilms.S. epidermidis biofilms were grown on coverslips for up to 96 h in nutrient broth at 37 °C. Throughout the growth period, biofilm biomass, viability and colony forming units (CFUs) were quantified. Biofilm viability (A; black squares) was measured by fluorescence of alamar blue after 1.5 h incubation and biomass was assessed by crystal violet staining (A; white squares). Biofilms were then removed from the coverslips via sonication, serially diluted and plated on LB agar for CFU quantification (B). Symbols denote statistical significance when compared to S. epidermidis following 0.5 h growth and error bars represent standard deviation of the mean, hash symbols relate to alamar blue data whereas Asterix is related to crystal violet data (#, *, P < 0.05; ##, **, P < 0.01).
3.2. Differentiating between planktonic and biofilm FTIR
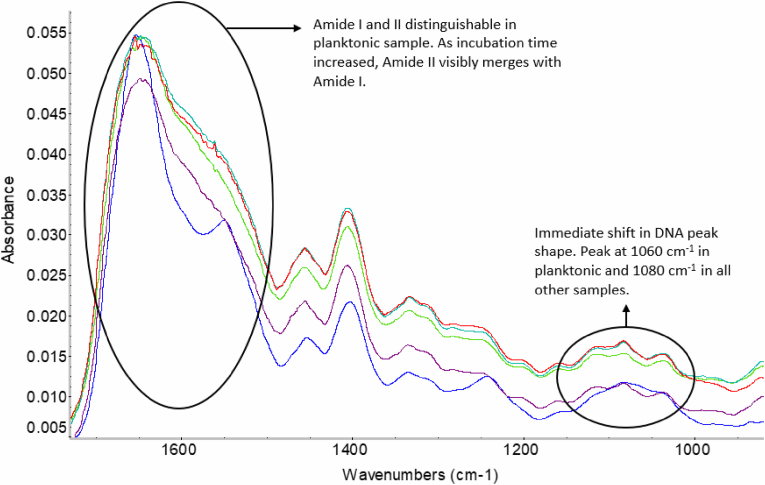
The FTIR spectra from a representative S. epidermidis (RP62A) planktonic sample and mature biofilm sample, incubated in Nutrient Broth (NB), are compared where the main areas of difference are annotated (Fig. 2). Firstly, significant spectral differences were observed in the Amide I and II regions at 1700-1500 cm−1. Two amide regions are definable in the planktonic sample, but once the biofilm has formed we observe the merging of these two regions, exhibited by the shoulder peak at 1600 cm−1. Secondly, changes are observed in the peak region relating to DNA and RNA structures at 1085-1060 cm−1. Here peak definition is shifted, as the peak at 1082 cm−1 becomes more defined in the biofilm sample. Further to this, the lipid region, 3500–2900 cm−1, is broadened in the biofilm sample. The changes found between planktonic and mature biofilms samples have been quantified across 5 separate experiments such that the average wavenumbers (in cm−1) of the: amide I region is 1653.6 and 1646.6; phospholipid peak is 1064.6 and 1082.6; and, key DNA peak is 934.6 and 919, for planktonic and matured biofilms, respectively for each of the three regions. For a complete peak analysis, detailing variability between repeat experiments, Supplementary Table 1.
Fig. 2.
FTIR spectrum of S. epidermidis planktonic cells (blue) overlaid with spectrum for semi-dry mature biofilm (red). All samples have been analysed on calcium fluoride slides. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
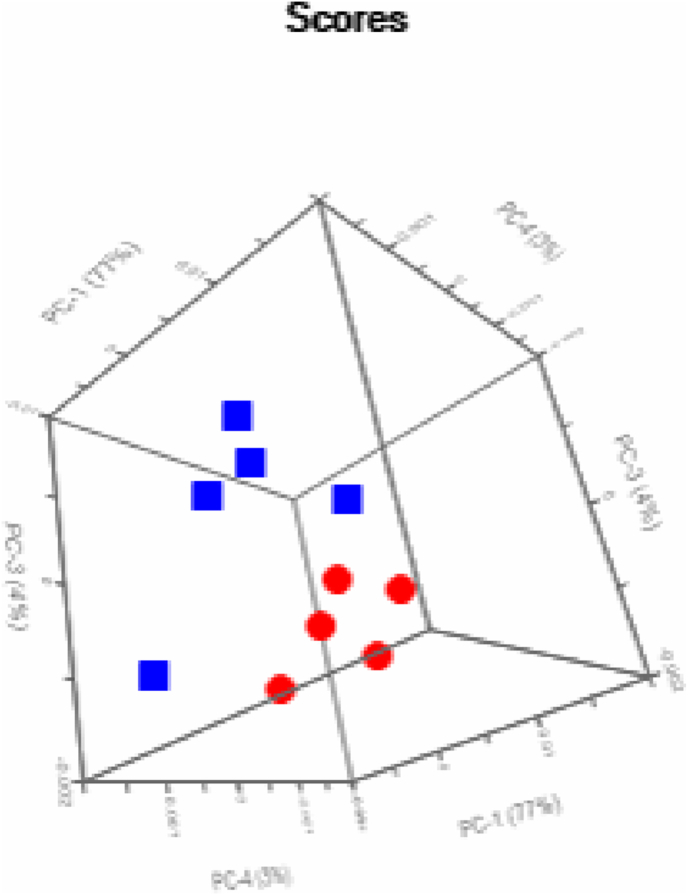
Spectra from the planktonic and mature biofilm were then compared by principal component analysis (PCA) with the results shown in Fig. 3. All data points remained consistently separated with zero outliers to the data set (Supplementary Fig. 1), validating the use of FTIR to distinguish the bacterial samples. Linear discriminant analysis (LDA) was also applied to demonstrate that the spectra can be predicted to 100% accuracy (Supplementary Fig. 2). This reinforced the notion that FTIR can be successfully used to differentiate between planktonic and biofilm samples.
Fig. 3.
Principal component analysis for biofilm (red) and planktonic cells (blue) computed in three-dimensions. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
To confirm that the FTIR spectral shifts relate solely to the production of a biofilm, a non-biofilm forming S. epidermidis strain, ATCC 12228, was incubated in NB for 72 h. The consequent FTIR spectra demonstrate negligible shift in the amide I peak between the planktonic and biofilm sample (spectra given in Supplementary Fig. 3). For further validation, to ensure observed spectral differences are not a result of the NB, the experiment was repeated in a different growth media, Brain Heart Infusion (BHI). In this case, the amide I peak shifted from 1653 cm−1 in the planktonic sample to 1640 cm−1 in the mature biofilm sample (complete peak analysis, Supplementary Table 2). This reaffirms the observation that FTIR can distinguish between a planktonic and biofilm sample.
3.3. Investigation of mature biofilms
Analysis of biofilms grown for 24, 48, 72 and 96 h show a clear shift between the planktonic and all biofilm samples in the lower wavenumber section, i.e. the “fingerprint region” (1700-900 cm−1) (Fig. 4). When comparing biofilm samples, spectral analysis was limited to the fingerprint region because differing hydration can cause variations within the spectra (exemplified in Supplementary Fig. 4). The broadening of the amide I and II regions is the most evident feature of the spectral change, with the two regions distinct in the planktonic sample but merging into a single peak in the 24-h biofilm, and then remaining unchanged for the 96-h duration of the experiment. There is also a shift of 7–10 cm−1 between the planktonic sample and the 24-h biofilm sample. However, between the 24-h sample and the 48 to 96-h samples, only small shifts are evident (Supplementary Table 3). To determine the statistical significance of the data set, a PCA was performed (Supplementary Fig. 5), from which we see separation between 24, 48- and 72-h samples, but as we move to 96-h samples the data becomes overlapped.
Fig. 4.
FTIR spectra from single analysis of S. epidermidis planktonic sample (blue) plotted against semi-dry biofilm on calcium fluoride after incubation for: 24 h (purple), 48 h (green), 72 h (teal) and 96 h (red). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.4. Early stage biofilms
Fig. 5 overlays the FTIR spectra collected for samples incubated from 30 min to 4 h, alongside the planktonic sample spectrum, seen in Fig. 2, Fig. 4. The amide I region (1700-1600 cm−1) is shifted to lower wavenumbers as the biofilm forms: planktonic (1652 cm−1), 30 min (1648 cm−1), 1 h (1647 cm−1), 2 h (1646 cm−1) and 4 h (1644 cm−1). This shift is complete by 24 h, with an overall shift of between 7 and 10 cm−1 repeatably reported. This was reproducible across three experiments (Supplementary Table 4). The amide II region (1600-1500 cm−1) is merging into one broad peak with the amide I region. The peak at 1550 cm−1 is only completely distinct in the planktonic sample. The phospholipid region is changed immediately between the planktonic sample and the sample incubated for 30 min.
Fig. 5.
FTIR spectra of representative S. epidermidis planktonic sample (blue) plotted against semi-dry biofilm forming samples, from one experiment, incubated for: 30 min (purple), 1 h (green), 2 h (teal) and 4 h (red). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
Biofilms are known to be important in the pathophysiology of infections, however diagnosing the presence of a biofilm in clinical samples has always presented problems as most methods used to evaluate the presence of biofilm disrupt the biofilm itself. This causes problems both in the diagnostic lab, but also in studying the development of biofilms as current techniques make longitudinal measurements difficult. We have used FTIR, a non-destructive method, to follow the development of the same biofilm over time. FTIR produces a spectrum by measuring the excitation and relaxation of biochemical groups (such as C O, CH2, CH3, C – O – C and O–P–O) or linkages, belonging to phospholipids, proteins, carbohydrates, collagen and amino acids. FTIR spectra from bacterial samples exhibit a complex peak distribution with prominent adsorption in almost all regions of the spectra. We have examined changes in wavenumber of specific spectral peaks related to different chemical signatures which are linked to the development of the biofilm phenotype. We have also seen differences in the intensity, height, of spectral peaks, but it is not reliable to use FTIR spectral measurements quantitatively.
In this study we chose to use S. epidermidis as a model, firstly because this organism is widely implicated in infections of implanted medical devices and intravascular cannulas [32]. For the work, calcium fluoride was utilised as the substrate material because it is transparent to infrared light, reducing interference with signals from the microorganism sample. To prevent sample and substrate damage, a simple transmission FTIR method was applied. Whilst an ATR approach may have optimised the spectroscopic result, using transmission FTIR permitted the repeated analysis of a single sample without direct contact with the sampling apparatus. By analysing with ATR, these samples would arguably be damaged and the risk of contamination would be increased. To repeat the work, without repeated analysis, using ATR-FTIR, aluminium foil could be explored as an alternative substrate. The strain of S epidermidis which we used (RP62A) develops features of a biofilm phenotype within the first 4 h of incubation, with an increase in biomass between 1- and 4-h of incubation that plateaus between 4 h and 24 h incubation. This allows us to examine the changes relating to early biofilm development, attachment and micro-colony formation, occurring in the first 4 h. Our initial experiments concentrated on the differentiation of planktonic cells from biofilm. When we compare planktonic and biofilm phenotypes a clear wavenumber shift occurs of 7–10 cm−1 in the Amide I region. We also report a shift of 14 cm−1 in the phospholipid region and a convergence of two peak at 935/895 cm−1 in the planktonic sample, to a single peak at 919 cm−1 in the biofilm samples. All of these spectral alterations, that remained the same in two different growth media, indicate the development of a different chemical composition in the sample once a biofilm has begun to form [33]. For comparison a non-biofilm forming S. epidermidis strain (ATCC 12228) was incubated in NB for 72 h [34]. From evaluating the FTIR spectra of the planktonic cells and the incubated sample, the amide I region remained unshifted. This strengthens the conclusion that the amide I shift relates directly to the formation of the biofilm.
When we examined spectral shifts between planktonic and biofilms over time the most noticeable feature is the shift in the amide region at 1650 cm−1. This change, which begins between 30 min and 1 h and is complete by 24 h of biofilm maturity is the clearest biomarker of the biofilm phenotype in this organism. This spectral shift of 10 cm−1 of the same band represents a significant change in the chemical composition of the biofilm and is related to changes in the chemical structures of proteins developing in the biofilms over this time frame. The spectra in Fig. 5 appear to have additional peaks within the defining peak at 1645 cm−1. This difference in curve smoothness originates from signal reflectance when collecting FTIR spectra and does not affect peak shift. Another consideration between the FTIR spectra presented in this study are the differing absorbance scales. This is a result of differing amounts of biomass present at the surface during the analysis. In the early stage of biofilm growth there are less bacteria, hence the spectral scale is 0.005–0.055, whereas the spectra relating to mature biofilms range from 0.02 to 0.20. This does not cause an issue when examining peak wavenumber shifts.
Further data analysis demonstrated that despite natural biological variance between sample sets, spectral differences remain generally consistent. We do however observe small peak shifts that arguably relate to biological differences between different experiments. This requires further exploration if this method is to become widely used and it will be important to standardise at which point peak shifts become significant and consistent between the data sets [35]. When small shifts, less than 2 cm−1, are reported, it is conceivable that the concentration of proteins has changed very slightly but this is often not consistent between experiments [36]. When peak shifts increase and become consistent between experiments, we can suggest a fundamental change in protein composition, as we see when comparing planktonic and biofilm FTIR spectra, Fig. 2. This is exemplified in our PCA where late-stage biofilms have overlapping data points, despite small variations in peak positions. Whereas, our planktonic and biofilms sample data points, which have larger peak shifts of more than 5 cm−1, definitively separate in multivariant analysis.
Another area of difference in the FTIR spectra was in the lipid region, 3500-2900 cm−1. We decided not to include this when comparing biofilms of varying maturity as changes could be related to the influence of differing sample hydration. While we controlled this by trying to standardise the level of hydration in all samples, in our opinion relying on analysis of this area would introduce too much uncertainty. Our method to standardise semi-drying involved isolating the sample in a sealed Perspex container containing a silica drying agent, for 30 min. Without this partial drying, the resultant FTIR spectra became overwhelmed with water signals, leading to biofilm samples of differing maturity, that clearly demonstrated the expected shift in amide I peak, but had confusing overlapping regions linked to water content. Despite our effort to control hydration, there was variation. If we had applied significantly longer drying times there was a risk of complete sample desiccation which would disrupt the biofilm architecture, arguably rendering the chemical composition inaccurate. Thus, we decided to concentrate on analysis of the fingerprint region, which is less influenced by water content and where the key spectral changes occur as the phenotype switches from planktonic to biofilm.
As the biofilm matures, we expect protein composition to change with the production of the ECM and subsequent development of the biofilm structure, evidenced by the amide I peak shift in the FTIR spectra. We observe minimal development after 24 h as this S. epidermidis biofilm is fully mature by 24 h so we are unable to identify different chemical signatures. As such there are protein signatures that remain constant between the planktonic and biofilm samples particularly proteins which exhibit peaks in the amide III region at 1333 cm−1. It is also important to note that changes co-incident with the shift in the amide peak also occur such as changes in the phospholipid peak, 1082 cm−1, but these secondary changes do not give us any additional information about the chemical evolution of the biofilm.
As we have shown the spectra between 24 and 96 h of biofilm maturity remain constant, but FTIR spectroscopy has shown substantial chemical changes in the early stage of biofilm development. We observe a clear merging of the amide I and II regions which is complete by 24 h of biofilm development. In terms of chemical changes these regions are linked to protein structures which are likely to change as the biofilm matures. The amide I region relates to C O stretching modes and amide II is the N–H bend and stretch. These regions come from the amino acids present in the sample. Amide II specifically relates to the secondary protein structure and is a defined individual peak in the planktonic sample. As the incubation period is increased, this amide II peak at 1550 cm−1 converges with the amide I peak, denoting a change in the structure of the proteins within the sample. The amide I peak simultaneously shifts as the biofilm develops with the planktonic average wavenumber at 1653.6 cm−1, steadily decreasing until we have a mature biofilm where the average wavenumber occurs at 1646.8 cm−1. These changes coincide with the community of bacteria irreversibly attaching to the surface of the calcium fluoride slide and the production of extracellular polymeric substances.
Not all changes in the spectra relate so closely to chemical changes when evaluating the presence of the biofilm phenotype compared to a planktonic sample. A further difference in the FTIR spectra occurs in the high wavenumber region, 3500-2900 cm−1. The peak in this area, centred at 3280 cm−1, is noticeably sharper in the planktonic sample than the 24-h biofilm sample. The primary factor affecting this region of the FTIR spectrum is water content and as biofilms are likely to be more hydrated than planktonic organisms, this results in a broadening of the peak unrelated to changes in the chemical composition of the biofilm [37]. This emphasises the importance of sample preparation to ensure that all samples are analysed in a similar state of hydration i.e. semi dried, to prevent the presence of water overwhelming other components contributing to the spectrum. Despite the semi-dry state of the biofilm samples, the lipid region of the FTIR spectra is consistently broadened suggesting the presence of the ECM. To further clarify this point, samples were rehydrated after analysis allowing repeated FTIR spectra collections, revealing comparable results. Partial drying of these samples for 30 min in a simple desiccant chamber was crucial because the FTIR signals of the sample could alter with complete drying potentially causing ECM and protein structures to break down, this would result in a different series of FTIR peaks. Similarly, analysing completely hydrated biofilm samples could result in an overwhelming water peak, preventing reliable spectral analysis. The standardised method we employed, using semi-dry samples, ensured both the reliability of analysis and the viability of microorganisms, proven by re-hydrating the samples with PBS and ensuring that the biofilms were still viable post sampling.
5. Conclusion
We have developed a potential spectroscopic approach to allow label-free, non-destructive testing of an intact biofilm. For a single strain of S. epidermidis (RP62A), the biofilm shows clear differences in the FTIR spectra compared to the planktonic phenotype, from as early as 30 min of biofilm maturity. The shift of amide I wavenumber position and the merging of the amide I and II regions is a consistent and early biomarker for the development of a biofilm phenotype in our S. epidermidis model.
This is a proof-of-concept study and further work is needed firstly to see if this spectral shift occurs in different bacterial species, as they form biofilms, to determine whether the same changes occur across a number of organisms, or if each species has its own unique biomarker. Secondly, we need to determine whether the cells within the biofilm or the ECM are the major component driving the chemical changes that we are observing as the biofilm develops. With further investigation and considerations for background signals that exist within realistic biopsy samples, this method has potential future applications within clinical diagnosis to differentiate between infections derived from planktonic microorganisms and those which have already switched to the biofilm phenotype.
Funding
This work has been supported by Greater Innovation for Smarter Materials Optimisation (GISMO) [DLUHC/03R18P02671].
CRediT authorship contribution statement
Amy R. Crisp: Methodology, Validation, Formal analysis, Investigation, Resources, Data curation, Writing – original draft, Visualization, Project administration. Bryn Short: Investigation, Data curation. Laurence Rowan: Investigation, Data curation. Gordon Ramage: Writing – review & editing, Project administration. Ihtesham U.R. Rehman: Conceptualization, Resources, Writing – review & editing, Supervision, Project administration, Funding acquisition. Robert D. Short: Supervision, Funding acquisition. Craig Williams: Conceptualization, Supervision, Project administration, Methodology, Resources, Writing – review & editing, Project administration.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioflm.2023.100141.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.De la Fuente-Núñez C., Reffuveille F., Fernández L., Hancock R.E.W. Curr Opin Microbiol. 2013;16:580–589. doi: 10.1016/j.mib.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 2.Lindsay D., von Holy A. J Hosp Infect. 2006;64:313–325. doi: 10.1016/j.jhin.2006.06.028. [DOI] [PubMed] [Google Scholar]
- 3.Srivastava S., Bhargava A. Biotechnol Lett. 2016;38:1–22. doi: 10.1007/s10529-015-1960-8. [DOI] [PubMed] [Google Scholar]
- 4.Flemming H.C., Neu T.R., Wozniak D.J. J Bacteriol. 2007;189:7945–7947. doi: 10.1128/JB.00858-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sauer K., Stoodley P., Goeres D.M., Hall-Stoodley L., Burmølle M., Stewart P.S., Bjarnsholt T. Nat Rev Microbiol. 2022;20:608–620. doi: 10.1038/s41579-022-00767-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hall-Stoodley L., Costerton J.W., Stoodley P. Nat Rev Microbiol. 2004;2(2) doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 7.Solano C., Echeverz M., Lasa I. Curr Opin Microbiol. 2014;18:96–104. doi: 10.1016/j.mib.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 8.Rabin N., Zheng Y., Opoku-Temeng C., Du Y., Bonsu E., Sintim H.O. Future Med Chem. 2015;7:493–512. doi: 10.4155/fmc.15.6. [DOI] [PubMed] [Google Scholar]
- 9.Gupta S., Andersen C., Black J., Fife C., Lantis J.I., Niezgoda J., Snyder R., Sumpio B., Tettelbach W., Treadwell T., Weir D. Wounds a Compend. Clin. Res. Pract. 2017;29(9) [PubMed] [Google Scholar]
- 10.Divakar S., Lama M., Asad U K. Antimicrob Resist Infect Control. 2019;8:76. doi: 10.1186/s13756-019-0533-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hassan A., Usman J., Kaleem F., Omair M., Khalid A., Iqbal M. Braz J Infect Dis. 2011;15:305–311. [PubMed] [Google Scholar]
- 12.Marrie T.J., Nelligan J., Costerton J.W. Circulation. 1982;66(6) doi: 10.1161/01.cir.66.6.1339. [DOI] [PubMed] [Google Scholar]
- 13.Harika K., Shenoy V., Narasimhaswamy N., Chawla K. J Global Infect Dis. 2020;12:129–134. doi: 10.4103/jgid.jgid_150_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lawrence J.R., Korber D.R., Hoyle B.D., Costerton J.W., Caldwell D.E. J Bacteriol. 1991;173:6558–6567. doi: 10.1128/jb.173.20.6558-6567.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorman S.P., Mawhinney W.M., Adair C.G., Issouckis M. J Med Microbiol. 1993;38:411–417. doi: 10.1099/00222615-38-6-411. [DOI] [PubMed] [Google Scholar]
- 16.Holmes A.H., Moore L.S.P., Sundsfjord A., Steinbakk M., Regmi S., Karkey A., Guerin P.J., Piddock L.J.V. Lancet. 2016;387:176–187. doi: 10.1016/S0140-6736(15)00473-0. [DOI] [PubMed] [Google Scholar]
- 17.MacLean R.C., Millan A.S. Science. 2019;365:1082–1083. doi: 10.1126/science.aax3879. [DOI] [PubMed] [Google Scholar]
- 18.Alvarez-Ordóñez A., Mouwen D.J.M., López M., Prieto M. J Microbiol Methods. 2011;84:369–378. doi: 10.1016/j.mimet.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 19.Ismail A.A., van de Voort F.R., Sedman J. In: Instrumental methods in food analysis. Pare J.R., Belanger J.M.R., editors. Elsevier Science; 1997. pp. 93–139. [Google Scholar]
- 20.Berthomieu C., Hienerwadel R. Photosynth Res. 2009;101:157–170. doi: 10.1007/s11120-009-9439-x. [DOI] [PubMed] [Google Scholar]
- 21.Petibois C., Desbat B. Trends Biotechnol. 2010;28:495–500. doi: 10.1016/j.tibtech.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 22.Ojeda J.J., Dittrich M. In: Microbial systems biology. Methods in molecular biology. Navid A., editor. Humana Press; Totowa, NJ: 2012. [Google Scholar]
- 23.Naumann D., Helm D., Labischinski H. Nature. 1991;351:81–82. doi: 10.1038/351081a0. [DOI] [PubMed] [Google Scholar]
- 24.Schmitt J., Flemming H.C. Int Biodeterior Biodegrad. 1998;41:1–11. [Google Scholar]
- 25.Burgula Y., Khali D., Kim S., Krishnan S.S., Cousin M.A., Gore J.P., Reuhs B.L., Mauer L.J. J Rapid Methods Autom Microbiol. 2007;15:146–175. [Google Scholar]
- 26.Nivens D.E., Ohman D.E., Williams J., Franklin M.J. J Bacteriol. 2001;183:1047–1057. doi: 10.1128/JB.183.3.1047-1057.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holman H.Y.N., Miles R., Hao Z., Wozei E., Anderson L.M., Yang H. Anal Chem. 2009;81:8564–8570. doi: 10.1021/ac9015424. [DOI] [PubMed] [Google Scholar]
- 28.Ariafar M.N., Iğci N., Akçelik M., Akçelik N. Arch Microbiol. 2019;201:1233–1248. doi: 10.1007/s00203-019-01681-5. [DOI] [PubMed] [Google Scholar]
- 29.Quilès F., Polyakov P., Humbert F., Francius G. Biomacromolecules. 2012;13:2118–2127. doi: 10.1021/bm300497c. [DOI] [PubMed] [Google Scholar]
- 30.Serra D., Bosch A., Russo D.M., Rodríguez M.E., Zorreguieta Á., Schmitt J., Naumann D., Yantorno O. Anal Bioanal Chem. 2007;387:1759–1767. doi: 10.1007/s00216-006-1079-9. [DOI] [PubMed] [Google Scholar]
- 31.Skogman M., Vuorela P., Fallarero A. J Antibiot (Tokyo) 2012;65:453–459. doi: 10.1038/ja.2012.49. [DOI] [PubMed] [Google Scholar]
- 32.Severn M.M., Horswill A.R. Nat Rev Microbiol. 2022 doi: 10.1038/s41579-022-00780-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie J., Yang F., Shi H., Yan J., Shen H., Yu S., Gan N., Feng B., Wang L. Int J Biol Macromol. 2022;207:358–364. doi: 10.1016/j.ijbiomac.2022.02.161. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y.Q., Ren S.X., Li H.L., Wang Y.X., Fu G., Yang J., Qin Z.Q., Miao Y.G., Wang W.Y., Chen R.S., Shen Y., Chen Z., Yuan Z.H., Zhao G.P., Qu D., Danchin A., Wen Y.M. Mol Microbiol. 2003;49:1577–1593. doi: 10.1046/j.1365-2958.2003.03671.x. [DOI] [PubMed] [Google Scholar]
- 35.Amiali N.M., Mulvey M.R., Sedman J., Louie M., Simor A.E., Ismail A.A. J Microbiol Methods. 2007;68:236–242. doi: 10.1016/j.mimet.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 36.Wenning M., Scherer S. Appl Microbiol Biotechnol. 2013;97:7111–7120. doi: 10.1007/s00253-013-5087-3. [DOI] [PubMed] [Google Scholar]
- 37.Yin W., Wang Y., Liu L., He J. Int J Mol Sci. 2019;20(4) doi: 10.3390/ijms20143423. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.