Abstract
Vibrio vulnificus is a rod shape, Gram-negative bacterium that causes sepsis (with a greater than 50% mortality rate), necrotizing fasciitis, gastroenteritis, skin, and soft tissue infection, wound infection, peritonitis, meningitis, pneumonia, keratitis, and arthritis. Based on pathogenicity V. vulnificus is categorized into three biotypes. Type 1 and type 3 cause diseases in humans while biotype 2 causes diseases in eel and fish. Due to indiscriminate use of antibiotics V. vulnificus has developed resistance to many antibiotics so curing is dramatically a challenge. V. vulnificus is resistant to cefazolin, streptomycin, tetracycline, aztreonam, tobramycin, cefepime, and gentamycin. Subtractive genome analysis is the most effective method for drug target identification. The method is based on the subtraction of homologous proteins from both pathogen and host. By this process set of proteins present only in the pathogen and perform essential functions in the pathogen can be identified. The entire proteome of Vibrio vulnificus strain ATCC 27562 was reduced step by step to a single protein predicted as the drug target. AlphaFold2 is one of the applications of deep learning algorithms in biomedicine and is correctly considered the game changer in the field of structural biology. Accuracy and speed are the major strength of AlphaFold2. In the PDB database, the crystal structure of the predicted drug target was not present, therefore the Colab notebook was used to predict the 3D structure by the AlphaFold2, and subsequently, the predicted model was validated. Potent inhibitors against the new target were predicted by virtual screening and molecular docking study. The most stable compound ZINC01318774 tightly attaches to the binding pocket of bisphosphoglycerate-independent phosphoglycerate mutase. The time-dependent molecular dynamics simulation revealed compound ZINC01318774 was superior as compared to the standard drug tetracycline in terms of stability. The availability of V. vulnificus strain ATCC 27562 has allowed in silico identification of drug target which will provide a base for the discovery of specific therapeutic targets against Vibrio vulnificus.
Keywords: Vibrio vulnificus, Subtractive genomics, New drug target, Alphafold2, MD simulation
1. Introduction
Vibrio vulnificus is a rod shape, Gram-negative bacterium and is a member of family Vibrionaceae. Worldwide this lethal pathogen causes seafood-related deaths in humans [1]. Vibrio vulnificus causes sepsis greater than 50% mortality rate, necrotizing fasciitis, gastroenteritis, skin and soft tissue infection, wound infection, peritonitis, meningitis, pneumonia, keratitis, and arthritis. Chronic liver disease, kidney disease, hemochromatosis, immune deficiency and heart diseases are the predominant risk factors for Vibrio vulnificus [2]. Pili, Cytotoxin, hydrolytic enzyme, flagella, and capsule polysaccharide are the feasible virulent determinant for V. vulnificus. Based on pathogenicity V. vulnificus is categorized into three biotypes. Type 1 and type 3 cause diseases in humans, while biotype 2 causes diseases in eel and fishes [1]. The majority of microorganisms that cause infections are resistant to antibiotics. Due to indiscriminate use of antibiotics V. vulnificus has developed resistance to many antibiotics so curing is dramatically a challenge [1]. Pan et al. stated that V. vulnificus showed resistant to cefazolin, streptomycin, tetracycline, aztreonam, tobramycin, cefepime and gentamycin [3]. The mechanism by which V. vulnificus interact with different factors of host defense was investigated by researchers to understand its pathogenicity better. The first host defense encounter by V. vulnificus is the highly acidic environment of the stomach. To neutralize low-pH of the environment Gram-negative bacteria catabolize amino acids to amines and carbon dioxide, to encounter acidic environment V. vulnificus utilize the same strategy [4]. Target identification is the first step in drug discovery. Experimental methods are usually time-consuming, costly, and yield insufficient results whereas, subtractive genome analysis is a simple, efficient and less costly process therefore, computational genomics approaches are mostly preferred over experimental approaches [5]. This method can be used to predict both drug and vaccine target. With the help of subtractive genome analysis unique drug target can be identified in the pathogen proteome, as the target is unique to the pathogen so human proteins and drugs will not cross-bind d [6]. The approach of the subtractive genome is widely used to find drug target against the pathogenic bacteria. This approach compares pathogen proteome with humans and identifies a protein that is not present in humans and must be essential for pathogen survival. The target identified by this process is very fast and inexpensive [7]. The aim of the present study was to identify human non-homologous, virulent, essential, and non-gut flora drug target against V. Vulnificus and to develop 3D structure of the drug target. Further, molecular docking studies were carried out to find the effective drug against the new drug target. The best-docked complex was further subjected to molecular dynamic simulation to evaluate the stability of the complex.
2. Material and methods
2.1. Pathogen complete proteome
The total proteome of V. Vulnificus strain ATCC27562 was downloaded from the NCBI (https://www.ncbi.nlm.nih.gov) database.
2.2. Duplicate protein identification
For paralogous protein identification, the CD-hit suite was used. The duplicate proteins identified by CD-hit were removed [8]. Non-paralogs and those proteins having sequences >100 amino acids were further analyzed.
2.3. Humans’ non-homologous proteins identification
BLASTp was carried out with E value > 10−3 to identify the human non-homologs proteins of the pathogen [9]. The human homologous proteins were then removed.
2.4. Essential genes identification
To seek out essential genes of pathogen BLASTp against DEG (database of essential genes) was done. Essential genes are needed for pathogen survival and growth [10].
2.5. Pathogen unique pathways identification
Using the KEGG database comparative analysis of pathogen and human pathways was done. With the help of this database, specific pathways were identified [11]. Functional annotation of the genes was provided by the KAAS server which provided the KO codes that indicates a particular protein involved in the specific pathway of the pathogen [12].
2.6. Subcellular localization prediction
For appropriate drug target identification, it is important to identify the location of proteins within the cells. The localization of proteins was predicted via PSORTb [13]. Five important locations of proteins in microbes include cytoplasm, periplasm, plasma membrane, extracellular and outer membrane [12]. Membrane proteins may be referred to as vaccine targets and those found in the cytoplasm were referred to as drug targets [14].
2.7. Virulent proteins identification
Complete information of the virulent proteins was obtained by the use of PAID b server [15].
2.8. Proteins druggability potential
Proteins that were essential and human non-homologous were blasted with FDA-approved drug targets. Targets that revealed a favorable similarity with FDA-approved drug targets were described as druggable [16].
2.9. Screening of gut flora proteins
More than 1014 beneficial microbes reside in the gut flora of healthy individuals that may help in food digestion and prevent the growth of pathogenic bacteria in the gut. Inhibiting the intestinal gut flora proteins could cause an adverse effect on the human host. Protein BLAST was performed against gut flora proteins using 0.001 E value [17,18]. Proteins that revealed significant similarities with gut flora were removed.
2.10. Homology modeling by AlphaFold2
Computational methods are used for decades to predict the 3D structure in the absence of experimental structures. The AlphaFold is one of the applications of deep learning algorithms in biomedicine and is correctly considered the game changer in the field of structural biology [19]. The protein sequence, as well as the template structure, can be used as input by AlphaFold 2. The accuracy and speed are the major strength of alpha fold 2 approximately 400 residues structure can be predicted within a minute by a single GPU [20]. The sequence of the target was retrieved from the NCBI database and then the Colab notebook was used to predict the 3D structure of the target from the sequence.
2.11. Model validation
The 3D structure of the drug target developed by the AlphaFold2 server was validated by PROCHECK and ERRAT servers [21].
2.12. Virtual screening against the drug target
2.12.1. Chemical compounds collection for VS
The VS was carried out with a total of 12,000 drug-like chemical compounds retrieved from the ZINC15 database. Using the MOE-washed module the chemical structures were washed. Partial charges were assigned and hydrogen atoms were added to all the structures.
2.12.2. Molecular docking study
All the compounds were docked into the active site of receptor. The MOE software was used to perform the 3D protonation of the modeled 3D structure. Energy minimization using the default parameters i. e a gradient of 0.05 was carried out. The protein structure that has been reduced to its lowest energy form was further analyzed [22]. MOE virtual screening method with Triangle Matcher placement method, London dG scoring function, and force field (GBVI/WSA dG) refinement method were employed for docking. After screening the 11,000 chemical compounds against the receptor the docking score was arranged in ascending order and 10% of compounds were analyzed for interaction with the receptor.
2.13. Molecular dynamics simulation study
To evaluate the behavior of the receptor in the presence of water, GROMACS (version 5.18.3) was used to perform the MD simulations for the best-docked complex [23]. The GROMOS9643a1 force field was used to generate the topology of the receptor. A simple point-charge model (SPC/E) was used to solve all the systems in a cubic box [24]. The counter ions such as Na+ or Cl ions, were added to the system to neutralize the systems. The steepest descent and conjugate gradients were used to reduce the energy of systems (50,000 steps for each). For system equilibration, the volume (NVT) regulation and pressure (NPT) were operated. The NVT ensemble was used with a constant pressure of 1 bar and temperature of 300 K. The H-atoms were constrained at their equilibrium distances and periodic boundary conditions using the SHAKE algorithm. Finally, 100 ns of production runs were completed. At intervals of 10 ps, the energy, velocity, and trajectory were updated. The GROMACS utilities and MDTraj-based Python scripts were used to carry out the MD simulation analysis [25].
2.14. Principal component analysis
Principal Component Analysis (PCA), an unsupervised learning technique, was used to understand about the internal motion of the systems [26]. For PCA analysis gmx covar was used. The covariance matrix was generated for the eigenvector and its atomic coordinates. Using the orthogonal coordinate transformation, a diagonal matrix of eigenvalues was generated. Principal components were extracted using the eigenvectors and eigenvalues.
3. Results and discussion
The aim of the present study was to identify a drug target against V. Vulnificus. In this study, we subtracted the pathogen proteome from the human host and identified the essential and human non-homologous proteins of V. Vulnificus pathogenic bacteria. Table 1 described the systemic workflow of the whole work. V. Vulnificus genome comprising 1653 protein sequences was downloaded from the NCBI database. After running the CD-hit the number of proteins got reduced to 1634. After NCBI BLASTp against Homo sapiens 348 proteins were found non-homologous to humans. For further analysis, only non-homolog proteins were used while the homologous proteins were removed. Essential genes support the basic and vital functions of cells and help in pathogen survival [27]. Performing BLASTp against DEG 442 essential proteins were found in the proteome of V. Vulnificus. By the use of the KEGG database 13 pathways were found as unique to the pathogen. Fourteen proteins were involved in these unique pathways. Table 2 described the unique pathways of V. Vulnificus as well as pathways IDs while Table 3 describes the protein involved in unique pathways. Pathways that were present in pathogens as well as in humans were referred to as common pathways, while those present only in pathogens were considered unique [11]. Localization of proteins was predicted by the PSORTb server which indicated that six proteins were cytoplasmic, three were membrane, one was extracellular, and two were unknown Table 4.
Table 1.
Subtractive genomics steps for Vibrio Vulnificus Strain ATCC 27562.
| S. no | Step | No of proteins |
|---|---|---|
| 1 | Total proteome of ATCC 27562 strain retrieved from NCBI | 1653 |
| 2 | Duplicate proteins removed after found in CD-hit | 19 |
| 3 | Non duplicate proteins | 1634 |
| 4 | Human's non-homologs proteins | 1286 |
| 5 | Essential proteins found out in DEG | 442 |
| 6 | Unique metabolic pathways of Vibrio Vulnificus | 13 |
| 7 | Proteins involved in unique pathways | 14 |
| 8 | Druggable proteins | 12 |
| 9 | Cytoplasmic proteins identified via PSORTb server | 6 |
| 10 | Non gut flora proteins | 1 |
Table 2.
Unique metabolic pathways of V. Vulnificus.
| S.no | Entry ID | Pathway name |
|---|---|---|
| 1 | vvu00281 | Geraniol degradation |
| 2 | vvu00625 | Quorum sensing |
| 3 | vvu00626 | Beta lactam resistance |
| 4 | vvu00643 | Styrene degradation |
| 5 | vvu00633 | Nitrotoluene degradation |
| 6 | vvu00930 | Caprolactam degradation |
| 7 | vvu01120 | Microbial metabolism in diverse environments |
| 8 | vvu01501 | Naphthalene degradation |
| 9 | vvu01502 | Vancomycin resistance |
| 10 | vvu01503 | Cationic antimicrobial peptide (CAMP) resistance |
| 11 | vvu02024 | Chloroalkane and chloroalkene degradation |
| 12 | vvu02030 | Bacterial chemotaxis |
| 13 | vvu02060 | Phosphotransferase system (PTS) |
Table 3.
Proteins involved in V. Vulnificus unique metabolic pathways.
| S. No | Accession no | KO code | Pathways |
|---|---|---|---|
| 1 | WP_025302287.1 | K03632 | Geraniol degradation |
| 2 | WP_025302407.1 | K08300 | Caprolactam degradation |
| 3 | WP_025304679.1 | K15633 | Phosphotransferase system (PTS) |
| 4 | WP_025303907.1 | K17713 | Bacterial chemotaxis |
| 5 | WP_025305120.1 | K07091 | Quorum sensing |
| 6 | WP_025303371.1 | K02616 | beta-Lactam resistance |
| 7 | WP_025303718.1 | K07090 | Vancomycin resistance |
| 8 | WP_025304748.1 | K02029 | Microbial metabolism in diverse environments |
| 9 | WP_004937569.1 | K06204 | Two component system |
| 10 | WP_015379223.1 | K09823 | Bacterial chemotaxis |
| 11 | WP_004929672.1 | K00247 | Geraniol degradation |
| 12 | WP_004940959.1 | K07703 | Two component system |
| 13 | WP_025302798.1 | K22103 | Geraniol degradation |
| 14 | WP_004937507.1 | K09910 | Vancomycin resistance |
Table 4.
Virulency and subcellular localization of proteins.
| S. no | Accession no | Localization | Virulency |
|---|---|---|---|
| 1 | WP_025302287.1 | Extracellular | Virulent |
| 2 | WP_025302407.1 | cytoplasmic | Virulent |
| 3 | WP_025304679.1 | cytoplasmic | Virulent |
| 4 | WP_025303907.1 | Membrane | Virulent |
| 5 | WP_025305120.1 | cytoplasmic | Virulent |
| 6 | WP_025303371.1 | cytoplasmic | Virulent |
| 7 | WP_025303718.1 | Unknown | Non-virulent |
| 8 | WP_025304748.1 | Membrane | Virulent |
| 9 | WP_004937569.1 | cytoplasmic | Virulent |
| 10 | WP_015379223.1 | cytoplasmic | Non-virulent |
| 11 | WP_004929672.1 | unknown | Virulent |
| 12 | WP_004940959.1 | membrane | Virulent |
Identification of virulent proteins was done by PAIDb v2.5, which revealed that eight proteins were virulent while two were non-virulent. Drugbank database was used for the identification of the druggability potential of essential, virulent, and human non-homologous proteins of V. Vulnificus. A total of twelve proteins revealed similarities with the drug bank database. Druggable proteins and their respected drug bank IDs are shown in Table 5. To identify the non-gut flora proteins of V. Vulnificus Protein BLAST was performed against gut flora proteins using 0.001 E value. Inhibiting the intestinal gut flora proteins could cause an adverse effect on the human host so we discarded those proteins that showed similarity with gut flora proteins while the non-gut flora proteins were identified as the drug targets against V. Vulnificus. Only one protein bisphosphoglycerate-independent phosphoglycerate mutase was predicted as a non-gut flora protein. A drug target bisphosphoglycerate-independent phosphoglycerate mutase was predicted as the new drug target against V. Vulnificus via subtractive genome analysis. The predicted drug target bisphosphoglycerate-independent phosphoglycerate mutase was involved in the Quorum sensing pathway. Bacteria can respond to the environment and perform cell-to-cell communication via a two-component system. This pathway is also involved in bacterial pathogenesis. For developing new antibiotics, the proteins of TCS can be considered a potent target as this pathway is completely absent in humans as well as other mammals [28]. PTS maintain the virulency of many pathogenic bacteria [29]. Quorum sensing bacteria produced autoinducers chemical signals that help in bacterial gene expression [30].
Table 5.
Proteins Druggability potential.
| S. no | Accession | Drug bank target | Drug bank ID |
|---|---|---|---|
| 1 | WP_025302287.1 | drugbank_target|P15924 Desmoplakin |
|
| 2 | WP_025302407.1 | drugbank_target|P51788 Chloride channel protein 2 |
|
| 3 | WP_025304679.1 | drugbank_target|P0AES4 DNA gyrase subunit A |
|
| 4 | WP_025303907.1 | drugbank_target|P0AES4 DNA gyrase subunit A |
|
| 5 | WP_025305120.1 | drugbank_target|P49747 Cartilage oligomeric matrix protein |
|
| 6 | WP_025303371.1 | drugbank_target|P49776 MULTISPECIES: chromosome partition protein MukB |
|
| 7 | WP_025303718.1 | drugbank_target|P23008 Genome polyprotein |
|
| 8 | WP_025304748.1 | drugbank_target|Q99250 Sodium channel protein type 2 subunit alpha |
|
| 9 | WP_004937569.1 | drugbank_target|P51648 Fatty aldehyde dehydrogenase |
|
| 10 | WP_015379223.1 | drugbank_target|P11759GDP mannose 6-dehydrogenase |
|
| 11 | WP_004929672.1 | drugbank_target|Q9Y5Y4 Prostaglandin D2 receptor 2 |
|
| 12 | WP_004940959.1 | drugbank_target|P32239Gastrin/cholecystokinin type B receptor |
|
3.1. Homology modeling by AlphaFold2
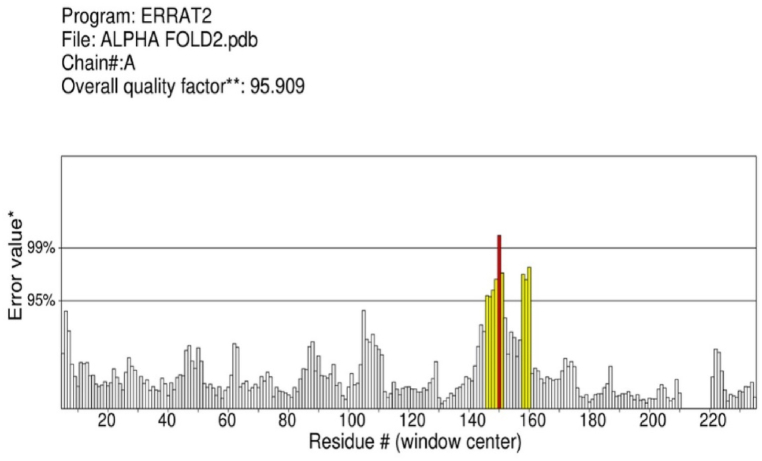
As the three-dimensional (3D) structure of the identified drug target was not present in the PDB database, homology modeling [20] was performed via the AlphaFold 2 server. The breakthrough with the AI-based tool AlphaFold2 (AF2), holds promise for achieving this goal, but the practical utility of AF2 remains to be explored. The majority of the residues in the developed model have very high confidence scores that are correlated with model accuracy (pLDDT > 90, Fig. 1A). Light green is bad (high error), while dark green is ideal (low error) as shown in expected position error Fig. 1B. Alpha fold generates a per-residue confidence value between 0 and 100. Some low PDDDT regions might be unstructured and disconnected Fig. 1A. The developed model was validated by PROCHECK and ERRAT tools [31]. According to Ramachandran plot, 95.5% of residues are in the most favored region, 4.1% residues in the additional allowed region, 0.5% in the generously allowed and 0% residues in the disallowed region in the structure of bisphosphoglycerate-independent phosphoglycerate mutase as described in Fig. 2. ERRAT plot indicates an overall quality factor of 95.90 for the developed model indicating the high quality of the model Fig. 3. The AlphaFold 2.0 has completely transformed our ability to deduce protein structures from sequences. This tool also unintentionally creates lots of new unexpected possibilities [32]. For more than 50 years, the ‘protein folding problems structure prediction component has been an incredible open scientific problem [33]. In spite of recent advances when no homologous structure is available existing methods lack atomic accuracy. AlphaFold is the first computational method that can predict protein structures with atomic precision on a regular basis, even if the template is unknown [34,35]. AlphaFold outperforms other methods and achieves accuracy comparable to experimental structures in most cases. The AlphaFold is based on a novel machine-learning approach that incorporates physical and biological knowledge about protein structure into the design of the deep-learning algorithm by utilizing multiple sequence alignments [20].
Fig. 1.
A).3D structure of 2, 3 bisphosphoglycerate-independent phosphoglycerate mutase developed by AlphaFold2. AlphaFold2 generates a per-residue confidence score (pLDDT) between 0 and 1n the model the region below 50 pLDDT score revealed the unstructured region. B) The expected aligned error of the model. The dark green color indicates a low error while the light green color indicates a high error. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 2.
Ramachandran plot of bisphosphoglycerate-independent phosphoglycerate mutase generated by PROCHECK server. 95.5% of the residues are in the most favored regions, 4.1% residues are in the additional allowed regions while 0.5% of the residues are in the generously allowed regions indicating a high quality of the model developed by AlphaFold 2.
Fig. 3.
ERRAT plot indicating overall quality factor of 95.90. Good quality structure yield values around 95% or higher.
3.2. Docking analysis
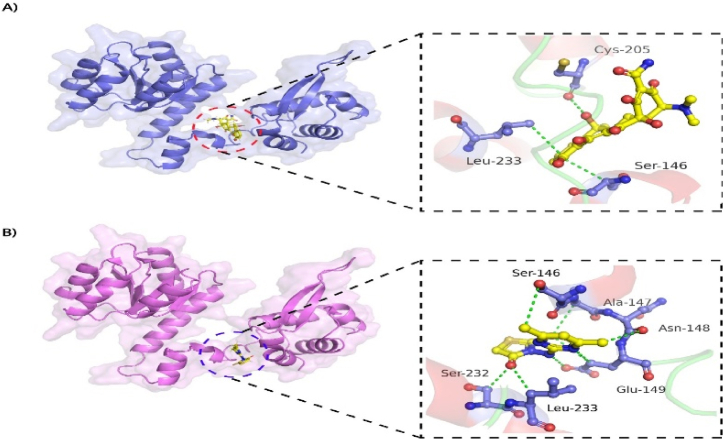
Molecular docking is frequently used to perform virtual screening of large libraries of compounds and explore the interactions of ligands with the targets [36]. The successful molecular docking technique employs scoring functions that precisely rank the hits [37]. Among all the 12,000 docked compounds ZINC01318774 showed the lowest docking score of −11.43 followed by ZINC05675633 with a docking score of −8.95. The compound ZINC01318774 made a total of three H-donor interactions, five H-acceptor interactions and three pi-H interactions with the active site residues including SER 146, ALA 147, ASN 148, GLU 149, SER 232 and LEU 233. Compound ZINC05675633 made a total of one H-donor, four H-acceptor and one H-pi interactions with GLU 149, LEU 233, THR 152, HIS 230 and HIS 206 active site residues. The docking score of standard drug tetracycline was predicted as −7.92 and the standard compound formed one H-donor interaction with Cys 205, two pi-H interactions with Ser 146 and Leu 233 active site residues. Tetracyclines are frequently used to treat vibrio infections [38]. In this study, tetracycline was taken as a control drug. Table 6 revealed the docking score and interaction of the best-scored compounds. Fig. 4A and B), showed the 3D interactions of the best-docked compound as well as the standard compound with the receptor.
Table 6.
The top best docking score along with their interactions and Zinc compounds IDs.
| Compound ID | Ligand | Receptor | Residues | Interaction | Distance | E (kcal/mol) | Docking score |
|---|---|---|---|---|---|---|---|
| ZINC05675633 |
|
|
|
|
|
|
−8.95 |
| ZINC72187409 |
|
|
|
|
|
|
−7.34 |
| ZINC01318774 |
|
|
|
|
|
|
−11.43 |
| ZINC04343551 |
|
|
|
|
|
|
−6.08 |
| ZINC04722102 |
|
|
|
|
|
|
−8.00 |
| ZINC05370287 |
|
|
|
|
|
|
−8.64 |
| ZINC67736629 |
|
|
|
|
|
|
−8.71 |
| Tetracycline |
|
|
|
|
|
|
−7.92 |
Fig. 4.
A) Interaction of reference compound (tetracycline) within the active site of receptor. B) Interaction of compound ZINC01318774 within the active site of receptor. The ligands are represented as blue sticks while the green dashed line represent the bond. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3. MD simulation analysis
MD simulations can reveal the stability and interaction mechanism between a ligand and receptor [39]. Each MD simulation was run at a time scale of 0–100 ns (ns). To measure the structural changes parameters like RMSD (Root mean square deviations) and RMSF (Root mean square fluctuations) were calculated.
3.3.1. RMSD and RMSF analysis
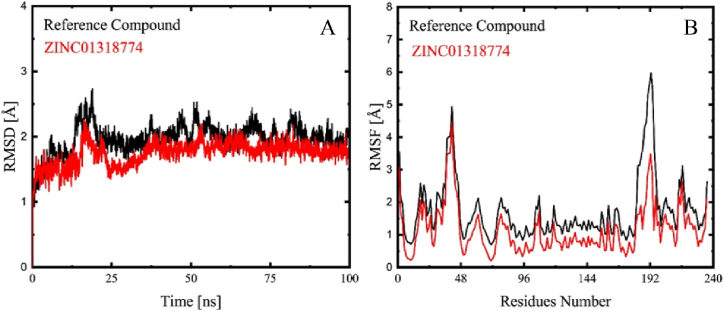
Average RMSD values of 2 Å and 2.5 Å were obtained from the MD simulation of bisphosphoglycerate-independent phosphoglycerate mutase and ZINC01318774 system and tetracycline in complex with bisphosphoglycerate-independent phosphoglycerate mutase respectively as in Fig. 5A. In contrast, the tetracycline in complex with the drug target system reached equilibrium in 75 ns with a substantially larger average RMSD value. The amplitudes of the fluctuations are inversely correlated with the system's stability. An increased RMSD value indicated higher fluctuations and a decrease in the stability of the systems [40]. Overall ZINC01318774 had a stable binding to bisphosphoglycerate-independent phosphoglycerate mutase as compared to the standard drug (tetracycline). The RMSF analysis indicates the flexibility of each residue of the protein. The increased mobility of the C alpha atoms due to relative fluctuations is indicated by the RMSF plots. The residues including Glu41, Ile 42, Ile 43, Phe 44, Asn 45, Ser 46, Gly 47, Ile 190, Ser 192, and Arg 193 revealed high flexibility during MD simulation while most of the binding pocket residues revealed stability during the 100ns MD simulation. The standard drug tetracycline with a slightly high degree of flexibility followed a like pattern Fig. 5B.
Fig. 5.
A). RMSD plots of reference compound tetracycline (Black) and RMSD plot of compound ZINC01318774 (Red). B) RMSF plots of reference compound tetracycline (Black) and compound ZINC01318774 (Red). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3.2. Principal component analysis
Principal component analysis (PCA) captures the high-amplitude principal motions in proteins. To evaluate the dynamic behavior of each system, the covariance matrix was calculated based on the Cartesian coordinates of Cα of the 5000 snapshots of each system for the whole trajectories. The covariance matrix shows the dynamic behavior of Cα atoms based on their average position. The eigenvectors and eigenvalues were extracted from the covariance matrix by diagonalization. The direction of high-amplitude motion and their mean square fluctuation were shown by the eigenvectors and eigenvalues, respectively. Fig. 6A shows the PCA plots of reference compound tetracycline and Fig. 6B showed the PCA plot of compound ZINC01318774 in complex with the receptor. For each system, the first two PCs such as PC1 and PC2 were calculated and plotted to monitor their motions. PCA analysis revealed that the compound ZINC01318774 showed the most cluster type of motions as compared to the reference compound.
Fig. 6.
A) PCA plots of reference compound tetracycline (B) and ZINC01318774. Each dot corresponds to a single frame.
4. Conclusion
In this study, various bioinformatics tools were used to predict a new drug target in the V. vulnificus proteome. The bisphosphoglycerate-independent phosphoglycerate mutase was identified as a drug target through the process of subtractive genome analysis. To our knowledge, this is the first study that identifies the new drug target in the Vibrio vulnificus strain ATCC 27562, and also new inhibitors against the drug target. This study will provide a base for the discovery of specific therapeutic targets against Vibrio vulnificus. Furthermore, the 3D structure was developed via AlphaFold 2 deep learning-algorithms and validated. Potent inhibitors against the new target were predicted by virtual screening and molecular docking study. The most stable compound ZINC01318774 perfectly binds to the binding pocket of bisphosphoglycerate-independent phosphoglycerate mutase. This potential candidate should be evaluated further for invitro study in the future to eradicate V. Vulnificus-associated infections.
Author contribution statement
Bader Saud Alotaibi; Amar Ajmal: Performed the experiments; Wrote the paper.
Mohammed Ageeli Hakami; Arif Mahmood: Analyzed and interpreted the data; Abdul Wadood; Junjian Hu: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data.
Data availability statement
No data was used for the research described in the article.
Additional information
No additional information is available for this paper.
Availability of data and materials
Dr. Abdul Wadood should be contacted for data and materials or other information.
Funding
The authors would like to thank the deanship of scientific research at Shaqra University for supporting this work.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
The authors thank Dr. Ajmal Khan, University of Nizwa for providing technical support for computational study.
Contributor Information
Abdul Wadood, Email: awadood@awkum.edu.pk.
Junjian Hu, Email: hujunjian79@163.com.
References
- 1.Heng S.P., Letchumanan V., Deng C.Y., et al. Vibrio vulnificus: an environmental and clinical burden. Front. Microbiol. 2017;8:997. doi: 10.3389/FMICB.2017.00997/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Horseman M.A., Surani S. A comprehensive review of Vibrio vulnificus: an important cause of severe sepsis and skin and soft-tissue infection. Int. J. Infect. Dis. 2011;15:e157–e166. doi: 10.1016/J.IJID.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 3.Pan J., Zhang Y., Jin D., et al. Molecular characterization and antibiotic susceptibility of Vibrio vulnificus in retail shrimps in hangzhou, People's Republic of China. J. Food Protect. 2013;76:2063–2068. doi: 10.4315/0362-028X.JFP-13-161. [DOI] [PubMed] [Google Scholar]
- 4.Jones M.K., Oliver J.D. Vibrio vulnificus: disease and Pathogenesis. Infect. Immun. 2009;77:1723–1733. doi: 10.1128/IAI.01046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sakharkar K.R., Sakharkar M.K., Chow V.T.K. A novel genomics approach for the identification of drug targets in pathogens, with special reference to Pseudomonas Aeruginosa. Silico Biol. 2004;4:355–360. [PubMed] [Google Scholar]
- 6.Musharaf Hossain M., Mosnaz A.T.M.J., Sajib A.M., et al. Identification of putative drug targets of Listeria monocytogenes F2365 by subtractive genomics approach. J. BioSci. Biotech. 2013:63–71. [Google Scholar]
- 7.Maurya S, Akhtar S, Siddiqui MH, et al Subtractive Proteomics for Identification of Drug Targets in Bacterial Pathogens: A Review.
- 8.Huang Y., Niu B., Gao Y., et al. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics. 2010;26:680–682. doi: 10.1093/BIOINFORMATICS/BTQ003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahmad S., Navid A., Akhtar A.S., et al. Subtractive genomics, molecular docking and molecular dynamics simulation revealed LpxC as a potential drug target against multi-drug resistant Klebsiella pneumoniae. Interdiscip. Sci. 2019;11:508–526. doi: 10.1007/S12539-018-0299-Y/FIGURES/11. [DOI] [PubMed] [Google Scholar]
- 10.Zhang R, research YL-N acids, 2009 undefined DEG 5.0, a database of essential genes in both prokaryotes and eukaryotes.academic.oup.com. [DOI] [PMC free article] [PubMed]
- 11.Moriya Y., Itoh M., Okuda S., et al. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:W182–W185. doi: 10.1093/NAR/GKM321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maurya S, Akhtar S, Siddiqui MH, et al Subtractive Proteomics for Identification of Drug Targets in Bacterial Pathogens: A Review.
- 13.Gardy J.L., Laird M.R., Chen F., et al. PSORTb v.2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics. 2005;21:617–623. doi: 10.1093/BIOINFORMATICS/BTI057. [DOI] [PubMed] [Google Scholar]
- 14.Hema K., Priyadarshini I.V., Swargam S., et al. 2015. 202 Subunit Vaccine Design against Pathogens Causing Atherosclerosis; pp. 135–136. 33. [DOI] [Google Scholar]
- 15.Yoon S.H., Park Y.K., Kim J.F. PAIDB v2.0: exploration and analysis of pathogenicity and resistance islands. Nucleic Acids Res. 2015;43:D624–D630. doi: 10.1093/NAR/GKU985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knox C., Law V., Jewison T., et al. DrugBank 3.0: a comprehensive resource for ‘Omics’ research on drugs. Nucleic Acids Res. 2011;39:D1035–D1041. doi: 10.1093/NAR/GKQ1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sears C.L. A dynamic partnership: celebrating our gut flora. Anaerobe. 2005;11:247–251. doi: 10.1016/J.ANAEROBE.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 18.Keely S., Talley N.J., Hansbro P.M. Pulmonary-intestinal cross-talk in mucosal inflammatory disease. Mucosal Immunol. 2011;5(15):7–18. doi: 10.1038/mi.2011.55. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.David A., Islam S., Tankhilevich E., Sternberg M.J.E. The AlphaFold database of protein structures: a biologist's guide. J. Mol. Biol. 2022;434 doi: 10.1016/J.JMB.2021.167336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jumper J., Evans R., Pritzel A., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:7873. doi: 10.1038/s41586-021-03819-2. 596:583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rafi M.O., Al-Khafaji K., Sarker M.T., et al. Design of a multi-epitope vaccine against SARS-CoV-2: immunoinformatic and computational methods. RSC Adv. 2022;12:4288–4310. doi: 10.1039/D1RA06532G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chenafa H., Mesli F., Daoud I., et al. 2021. In Silico Design of Enzyme α-amylase and α-glucosidase Inhibitors Using Molecular Docking, Molecular Dynamic, Conceptual DFT Investigation and Pharmacophore Modelling; pp. 6308–6329. 40. [DOI] [PubMed] [Google Scholar]
- 23.Abraham M.J., Murtola T., Schulz R., et al. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1(2):19–25. doi: 10.1016/J.SOFTX.2015.06.001. [DOI] [Google Scholar]
- 24.Mark P., Nilsson L. Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J. Phys. Chem. A. 2001;105:9954–9960. doi: 10.1021/JP003020W. [DOI] [Google Scholar]
- 25.McGibbon R.T., Beauchamp K.A., Harrigan M.P., et al. MDTraj: a modern open library for the analysis of molecular dynamics trajectories. Biophys. J. 2015;109:1528–1532. doi: 10.1016/J.BPJ.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ajmal A., Ali Y., Khan A., Wadood A., Rehman A.U. Identification of novel peptide inhibitors for the KRas-G12C variant to prevent oncogenic signaling. J. Biomol. Struct. Dyn. 2022:1–10. doi: 10.1080/07391102.2022.2138550. [DOI] [PubMed] [Google Scholar]
- 27.Judson N., Mekalanos J.J. TnAraOut, A transposon-based approach to identify and characterize essential bacterial genes. Nat. Biotechnol. 2000;18(7):740–745. doi: 10.1038/77305. [DOI] [PubMed] [Google Scholar]
- 28.Wang R., Mast Y., Wang J., et al. Identification of two-component system AfsQ1/Q2 regulon and its cross-regulation with GlnR in Streptomyces coelicolor. Mol. Microbiol. 2013;87:30–48. doi: 10.1111/MMI.12080. [DOI] [PubMed] [Google Scholar]
- 29.Deutscher J., Francke C., Postma P.W. How Phosphotransferase system-related protein Phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 2006;70:939–1031. doi: 10.1128/MMBR.00024-06/ASSET/A042D096-46C1-4F55-A469-056488BE7864/ASSETS/GRAPHIC/ZMR0040621380010.JPEG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller M.B., Bassler B.L. 2001. Quorum sensing in bacteria; p. 48. 12. [DOI] [PubMed] [Google Scholar]
- 31.Jin Y., Fayyaz A., Liaqat A., Khan A., Alshammari A., Wang Y., Gu R.X., Wei D.Q. Proteomics-based vaccine targets annotation and design of subunit and mRNA-based vaccines for Monkeypox virus (MPXV) against the recent outbreak. Comput. Biol. Med. 2023 doi: 10.1016/j.compbiomed.2023.106893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monzon V., Haft D.H., Bateman A. Folding the unfoldable: using AlphaFold to explore spurious proteins. Bioinf. Adv. 2022;2 doi: 10.1093/BIOADV/VBAB043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dill K.A., Ozkan S.B., Shell M.S., Weikl T.R. The protein folding problem. Annu. Rev. Biophys. 2008;37:289. doi: 10.1146/ANNUREV.BIOPHYS.37.092707.153558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Senior A.W., Evans R., Jumper J., et al. Improved protein structure prediction using potentials from deep learning. Nature. 2020;577:7792. doi: 10.1038/s41586-019-1923-7. 577:706–710. [DOI] [PubMed] [Google Scholar]
- 35.Zheng W., Li Y., Zhang C., et al. Deep-learning contact-map guided protein structure prediction in CASP13. Proteins: Struct., Funct., Bioinf. 2019;87:1149–1164. doi: 10.1002/PROT.25792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alam M, Malebari A, Syed N, et al Design, Synthesis and Molecular Docking Studies of Thymol Based 1, 2, 3-triazole Hybrids as Thymidylate Synthase Inhibitors and Apoptosis Inducers against Breast. Elsevier. [DOI] [PubMed]
- 37.Lindahl E.R. Molecular dynamics simulations. Methods Mol. Biol. 2008;443:3–23. doi: 10.1007/978-1-59745-177-2_1. [DOI] [PubMed] [Google Scholar]
- 38.Adesiyan I., Bisi-Johnson M., Reports A.O.-S. nature.com; 2022. Undefined Incidence of Antibiotic Resistance Genotypes of Vibrio Species Recovered from Selected Freshwaters in Southwest Nigeria. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khan A., Umbreen S., Hameed A., et al. In silico mutagenesis-based remodelling of SARS-CoV-1 peptide (ATLQAIAS) to inhibit SARS-CoV-2: structural-dynamics and free energy calculations. Interdiscip. Sci. 2021;13:521–534. doi: 10.1007/S12539-021-00447-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei-Ya L., Yu-Qing D., Yang-Chun M., et al. Exploring the cause of the inhibitor 4AX attaching to binding site disrupting protein tyrosine phosphatase 4A1 trimerization by molecular dynamic simulation. J. Biomol. Struct. Dyn. 2019;37:4840–4851. doi: 10.1080/07391102.2019.1567392. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.
Dr. Abdul Wadood should be contacted for data and materials or other information.