Abstract
Hydroxyapatite (HAp) is critical to health both as the main structural material of the skeleton and storage material of calcium and phosphate. Nano-sized HAp (nHAp) is naturally produced by mineralizing cells during bone formation and remodeling and is the main constituent of the skeleton. As such, HAp is currently being investigated as a therapeutic biomaterial for orthopedic and dental purposes. Recent studies have suggested that extracellular nHAp can influence osteoblast lineage commitment and cell function through changes in gene expression, however the mechanisms remain to be elucidated. Here, the cellular and molecular mechanism by which rod-shaped nHAp (10 × 100 nm) stimulates gene expression in pre-osteoblast bone marrow stromal cells was investigated. Electron microscopy detected a rapid and stable interaction of nHAp with the cell membrane, which correlated with a strong stimulation of the Erk1/2 signaling pathway. Results also identified the requirement of the Fgf receptor signaling and phosphate-transporters for nHAp regulated gene expression whereas a calcium-sensing receptor inhibitor had no effect. Collectively, the study uncovers novel signaling pathways and cellular events specifically stimulated by and required for the cellular response to free extracellular HAp. The results provide insight into the osteoblastic response to HAp relevant to functional mineralization and pathological calcification and could be used in the development of biomaterials for orthopedic purposes.
Keywords: hydroxyapatite nanomaterials, Erk1/2 signaling, Phosphate-transporter, Fgf receptor signaling, osteopontin
INTRODUCTION
Calcium and phosphate are critically important in human health with functions ranging from cell signaling, to DNA formation, to structural integrity of the body. They can exist as free ions or in different complexed forms of calcium and phosphate (CaP) in the body and are closely associated with functional mineralization of bone and dentition1 and pathological calcification of cartilage and vascular tissues.2-10 Hydroxyapatite (HAp), which is essentially identical to bone apatite, is crystalline in form and consists mainly of calcium and phosphate (Ca10(PO4)6(OH)2) at a stoichiometric Ca/P ratio of 1.67. HAp is one of the most stable forms of CaP and does not spontaneously develop at physiological temperatures and pHs. HAp is therefore actively produced in the skeleton by bone forming osteoblasts as well as during tooth development as the main constituent of tooth enamel by ameloblasts. Formation of nano-sized HAp (nHAp) during functional skeletal mineralization is a controlled process performed by differentiating osteoblasts.11 Although not completely understood, a leading hypothesis posits that the initiation of HAp occurs through an active process by which osteoblasts deposit 50-200 nm needle-like HAp crystals in the microenvironment.1, 12 In the earliest stages of initiation the individual crystals have been demonstrated to form round clusters in cartilage and bone.12-13 Following initiation, crystal growth occurs resulting in a more regularly bundled mineralized matrix. Due to the functional genesis of HAp during mineralization and the resulting physiological importance it is currently being investigated as a bioactive material for skeletal and dental repair.
Currently, HAp is being explored as a biomaterial for its potential use in orthopedic repair14-15 utilized as a coating or surface film to improve adhesion,16 osseointegration,17 and osteoinductivity.18 Additionally, HAp is being investigated in tissue engineering as a biomaterial that can be incorporated into more flexible polymeric networks (reviewed in19). Nano-sized CaP products, including nHAp, have only more recently begun to be investigated for direct effects on the osteoblast lineage. Results have been conflicting; with some studies finding beneficial effects on osteogenesis and others suggesting decreased proliferation and increased apoptosis and these differences may be due to size, shape and synthesis method.20-27 Although it appears clear that both HAp coatings and nHAp, either engineered or endogenous, influence cell function the cellular and molecular mechanisms by which it exerts these effects remain mostly unknown. Understanding the mechanisms by which nHAp influences cell behavior will provide insight into the natural process of bone mineralization and pathological calcification as well as allow for further development and optimization of HAp as a bioactive material.
Exposure of osteoblast lineage cells to increasing concentrations of rod-like nHAp (10 × 100 nm) for three and seven days has been demonstrated to strongly alter the RNA levels of a number of genes including; increased osteopontin (Opn) as well as a strong dose dependent suppression of the alkaline phosphatase (Alp).28 The cellular response to nHAp appears to be mainly the result of its intrinsic material property as 100 nm silica based nanoparticles did not alter gene expression in the same manner.28 Similarly, a 4 μm crystalline calcium based particle (calcium oxide; CaO) also did not produce the changes as seen with nHAp,28 suggesting that the similar shape and calcium composition of the CaO particle is not sufficient to mimic the effects of nHAp. Therefore, it is likely to be a relatively specific recognition of the Ca/P ratio in HAp that produces the biological effect. Collectively, these previous results identified HAp as a bioactive nanomaterial with the capacity to produce changes in gene expression. The idea that HAp can alter cell function has implications for human health since HAp and CaP are produced endogenously and associated with functional mineralization as well as with diseases such as hydroxyapatite crystal deposition disease29 and the mineral and bone disorders associated with chronic kidney disease.30 The mechanism(s) by which HAp influences cell behavior in health and disease remain to be elucidated.
The underlying mechanisms by which HAp is so carefully deposited to form the bones of the skeletal during formation, remodeling, and repair are not fully understood. Therefore, the general objective of the current study was to understand the cellular and molecular mechanisms by which nHAp influences gene expression in osteoblast lineage cells. Specifically, we hypothesized that extracellular nHAp is sensed by specific membrane based proteins resulting in the stimulation of specific signal transduction pathways leading to changes in gene expression. A better understanding of how osteoblast lineage cells respond to HAp will not only increase our knowledge of skeletal biology but might also provide insight into the design of biomaterials for skeletal repair.
MATERIALS AND METHODS
Synthesis and characterization of hydroxyapatite nanomaterials.
Synthesis and characterization have been described previously.28 Briefly, pure phase hydroxyapatite (HAp: Ca10(PO4)6(OH)2) was synthesized using a sonochemistry based precipitation method. HAp was obtained by filter press and lyophilization and characterized by powder X-ray diffractometry (XRD), Fourier Transfer Infrared spectroscopy (FT-IR), and Transmission Electron Microscopy (TEM).28 For the measurement of the size and surface charge by Dynamic Light Scattering (DLS) with a Zetasizer Nano ZS90 (Malvern Instruments Ltd., Malvern, UK), nHAp was dispersed in; Dulbecco’s Phosphate-Buffered Saline (DPBS) (1X) (#21-031-CV; without calcium and magnesium; Mediatech, Manassas, VA), culture medium; α-Modified Eagle’s Medium (α-MEM with nucleosides; Thermo (formerly Gibco;#32571-036)), or growth medium; α-MEM with 10% fetal bovine serum (FBS; Atlanta Biologicals, Lawrenceville, GA) at a concentration of 5 mg/ml and pH 7, adjusted using NaOH and HCl as needed.
Cell culture.
A bone marrow stromal cell (BMSC) line with osteoblastic potential was developed from repeated sub-culturing of primary BMSCs obtained from red marrow by centrifugation of the femur as described.31 These BMSCs are characterized as pre-osteoblast cells based on the ability to mineralize, induce expression to alkaline phosphatase and express osteoblastic marker genes in a time dependent factor when stimulated to differentiate.28 Stock cells (passage 5) were cultured in 10 cm plates and sub-cultured every three to four days for up to 10 passages. MC3T3-E1 (passage 22) cells have been described previously,32-33 were cultured as BMSCs and used to as a second model to validate results. All cells were cultured in α-MEM at 37°C in 5% CO2 and supplemented with 50 U/ml penicillin, 50mg/ml streptomycin, and 2mM L-glutamine (Thermo) and 10% FBS.34 Osteoblast differentiation medium consisted of growth medium with the addition of 50 μg/ml L-ascorbic acid (AA) (Sigma-Aldrich, St. Louis, MO) but not β-glycerophosphate. For RNA and protein experiments cells were plated in 6-well (seeded at ~5x105) or 10 cm plates (seeded at ~5x106) and experiments performed within 72 hours. nHAp was used at 25 μg/ml unless otherwise noted.
RNA isolation, cDNA synthesis, and qRT-PCR.
RNA was extracted using TRIzol reagent following the manufacturer’s protocol (Thermo). The RNA concentration was quantified by spectrophotometer (Nanodrop; Thermo) and complementary DNA (cDNA) was synthesized using QuantiTech Reverse Transcription kit (Qiagen, Valencia CA). qRT-PCR was performed using VeriQuest SYBR Green qPCR master mix (Affymetrix, Santa Clara CA) on a StepOnePlus thermocycler (Applied Biosystems, NY). Primers were designed using qPrimerDepot software (http://mouseprimerdepot.nci.nih.gov/) for mouse and synthesized by Integrated DNA Technologies, Inc. (Coralville, IA) with sequences described in Table S1. The results are normalized to 18S and expressed as fold change based on the ΔΔCT method +/− SD of 3 replicates.
Scanning Electron Microscopy.
BMSCs were grown on precut silicon chips (5 mm square x 0.5 mm thick) (Electron Microscopy Sciences; Hatfield, PA). Chips were first prepared by immersion in sulfuric acid and hydrogen peroxide solution (7:3 ratio) for one hour, washed with deionized water four times, coated with Rat tail type I collagen solution (Thermo) for one hour, washed with 1x PBS twice followed by addition of the cells. The cells were then treated with nHAp for 24 hr, 1 hr, 15 min, or untreated. Cells were thoroughly washed three times in 1x PBS for 5 minutes with rocking. All samples were simultaneously fixed in buffered glutaraldehyde (2.5% glutaraldehyde in 0.1 M cacodylate, pH 7.4) and post fixed in 1% osmium tetroxide (chemicals purchased from Electron Microscopy Sciences, Hatfield, PA) after which they were washed and dehydrated through an ethanol series to 100% dry ethanol. The ethanol was exchanged for liquid CO2 in a Polaron E3000 Critical Point Drying Unit (Quorum Technologies Ltd. East Sussex, UK) and the CO2 brought to its critical point of 1073 psi and 31°C. After the CO2 was slowly and gently released, the dry samples were secured to SEM stubs and sputter coated with ~8 nm chromium (Denton, Moorestown, NJ) using a Denton DV602 Magnetron Sputter Coater. The samples were imaged in the upper stage of a Topcon DS150 FE-SEM (Tokyo, Japan) at 5 kV.
Western Blot Analysis.
The nHAp treated (test) or control cells were rinsed with DPBS and total cell lysate was generated by lysis in p300 lysis buffer (250 mM NaCl, 0.1% NP-40, 20 mM sodium phosphate, 30 mM sodium pyrophosphate, 5 mM EDTA, and 10 mM sodium fluoride (Sigma), adjusted at pH 7.0) supplemented with Halt Protease & Phosphatase Single-Use Inhibitor Cocktail (Thermo). Total cell lysate (10 - 20 μg) was separated by polyacrylamide gel electrophoresis (12%) and electro-transferred to PVDF membrane Hybond-P (GE Health Sciences, Piscataway, NJ). Membranes were initially blocked in 1x TBST with 5% non-fat dry milk. Non-phospho antibodies were used in 5% milk in Tris-buffered saline/Tween 20 (20 mM Tris, 150 mM NaCl, pH 7.5) while phospho-specific antibodies were used in 5% Bovine Serum Albumin (Sigma) in the Tris-buffered saline/Tween 20 solution overnight at 4 °C. The anti- phospho-Erk1/2, phospho-Jnk, phospho-p38, and Erk1/2 antibodies were purchased from Cell Signaling Technologies Inc. (Beverly, MA), antibodies to p38 and Jnk from Santa Cruz Biotechnologies Inc. (Dallas, TX). The blots were visualized by enhanced chemiluminescence (ECL) (Thermo).
Inhibitors and concentrations.
Inhibitors targeting; Fibroblast growth factor (Fgf) receptor (iFgfr; PD173074-100 nM) was purchased from Selleckchem (Houston, TX), phosphate-transporter (iPiT; Foscarnet-1 mM), calcium-sensing receptor (iCaSR; NPS2143-100 nM), and methyl-β-cyclodextrin (MβCD) (5mM) from Sigma-Aldrich. Erk1/2 (iErk1/2; U0126-30 μM), p38 (iP38; SB203580-10 μM), phenylarsine oxide (50 nM) and amiloride hydrochloride (200 μM) from CalBiochem (San Diego, CA), and Jnk (iJnk; SP600125-10 μM) from Tocris (Ellisville, MO). Inhibitors were added for 1 hour prior to the start of the experiment.
Immunoprecipitation.
The nHAp treated (test) or control cells were lysed in p300 buffer (above) and the resulting lysate (500 μg) was incubated with primary antibody (total Frs2α, Santa Cruz) and Protein A/G PLUS-Agarose (Santa-Cruz) overnight at 4°C with rotation. The immunoprecipitates were collected at 2,000g, washed twice with IP lysis buffer, separated by SDS-PAGE, immunoblotted with phospho-Frs2α (Tyr-196) (Cell Signaling) and visualized by ECL.
Microarrays.
RNA was isolated from cells using Trizol (Thermo) according to manufactures protocol. RNA quality was determined by Agilent 2100 Bio- analyzer (Santa Clara, CA). Transcriptome wide analysis was performed on Affymetrix ® Mouse Gene 2.0 ST Array Plates (Affymetrix). These arrays contain >28,000 coding transcripts and >7,000 non-coding (include ~2,000) long intergenic non-coding transcripts. The raw data was imported in Partek Genomics suite v 6.6 with RMA background correction. Differentially expressed genes were identified using Analysis of variance. Differentially expressed genes were filtered with unadjusted p value of 0.05 or less and a ≥ 2 fold change.
Statistical Analysis.
For multiple comparisons a one-way ANOVA was performed with Tukey’s multiple comparison post hoc test on normally distributed data (normality determined by Shapiro-Wilk test) (Fig. 1). For simple comparisons a standard two-tailed unpaired Student’s t-test analysis was performed on the qRT-PCR data of other experiments.
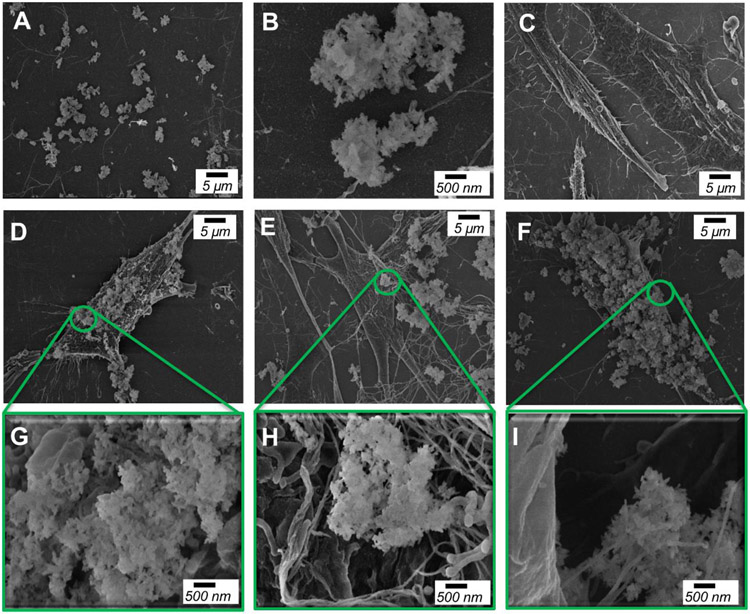
Figure 1. Scanning Electron Microscopy identifies nHAp on cell surface.
SEM analysis of; (A and B) nHAp on collagen-coated silicon chip, (C) untreated cells and (D and G) nHAp-treated BMSCs for 15 minutes, (E and H) 1 hour, and (F and I) 24 hours. G, H, and I are magnified areas of D, E, and F respectively. Scale bars as indicated in the figure.
RESULTS
Characterization of nHAp.
A recent study identified a significant change in gene expression in differentiating osteoblasts in response to exposure to medium dispersed nHAp.28 The nHAp particles were characterized by TEM analyses as rod-like in shape and approximately 10 nm in width and 100 nm in length and non-toxic at medium concentrations up to 100 μg/ml.28 To determine the effects of culture medium on size and charge of the nHAp particles we further characterized them by Dynamic Light Scattering (DLS) (Table 1). DPBS consists mainly of NaCl (137 mM) and Na2HPO4 (8 mM) and cell culture media (α-MEM) which contains phosphate (1.0 mM) and calcium (1.8 mM), among other minerals and amino acids, and growth medium, which is culture medium plus 10% fetal bovine serum (FBS). The size was in the range from 0.5 to 3 μm (Table 1). Zeta potential, representing the overall charge of nHAp, was negative under all the conditions (Table 1). Therefore, the cells in this study were generally exposed to negatively charged nHAp in the size range of 0.5 to 3 μm.
Table 1.
Characterization of nHAp by DLS
| 1x DPBS | Medium | Growth Medium | |
|---|---|---|---|
| Average size (nm) | 2484 ± 539 | 1879 ± 656 | 542 ± 296 |
| Zeta potential (mV) | −19.6 ± 3.4 | −12.5 ± 4.0 | −27.2 ± 3.9 |
nHAp stably interacts with the cell surface.
A primary mechanism by which external stimuli can stimulate molecular and cellular changes is through cell surface binding and interaction with one or more surface proteins such as a receptor. Scanning Electron Microscopy (SEM) was used to determine if nHAp directly contacts to or interacts with the surface of cells. Following 0, 15 min, 1 hour and 24 hours of cell exposure to nHAp cells were washed thoroughly three times before fixation and analysis by SEM. The aggregate (or size) of nHAp in cell culture medium is ~0.5 to 3 μm (Figure 1A and 1B), in agreement with the results obtained by DLS (Table 1). The untreated control samples showed a clean, smooth cell surface (Figure 1C) whereas cells treated for 15 minutes, 1 hour or 24 hours showed clusters of nHAp on the cell surface (Figure 1D to 1I). Images clearly identified nHAp bound to the surface of the cells within 15 minutes of exposure which increased through 24 hours, at which point most of the cell surface was covered (Figure 1).
nHAp induces rapid changes in gene expression.
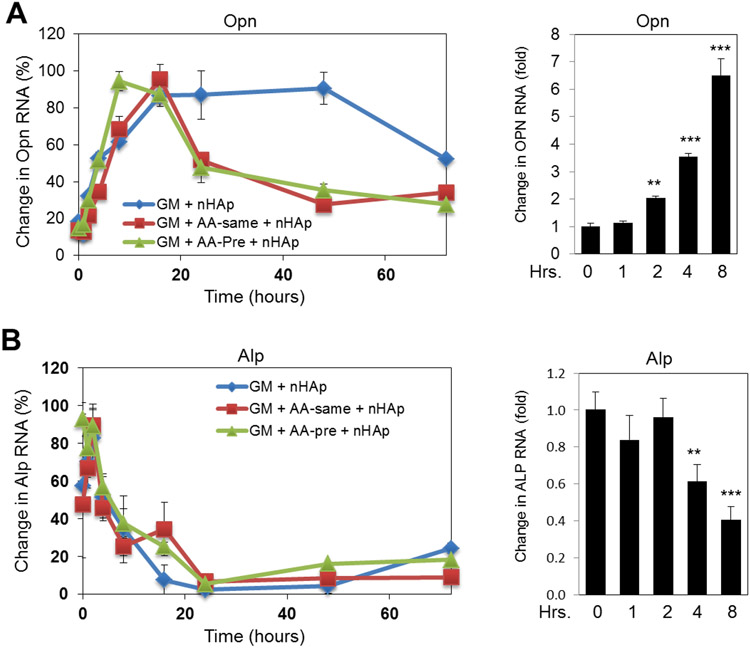
To determine if the changes in gene expression correlated with the interaction of nHAp and the cell surface and to better define the time frame for potential cellular signaling event(s) required for nHAp to regulate gene expression we performed a time course. RNA levels of Alp and Opn were analyzed in response to nHAp under different culture conditions; growth medium, pretreated for one day with the osteoblast promoting ascorbic acid, or ascorbic acid treatment added simultaneously with nHAp. The cells were harvested at time points ranging from 1 hour of exposure to 72 hours. An increase in Opn expression (Figure 2A) and decrease in Alp expression (Figure 2B) were detected regardless of culture conditions. The results also identified a specific time frame for the start of transcriptional regulation with the increase in Opn expression starting between 1 and 2 hours and the decrease in Alp RNA levels starting between 2 and 4 hours. HAp is very stable with pKsp 117.3 at 37°C, and its dissolution rate is 0.25%/day at pH 7.4, and requires at least two months to one year for complete dissolution35 and therefore the response was not likely due to an increase in free calcium or phosphate in the medium. Together, these results suggested that any signaling events generated by nHAp and necessary for regulation of gene expression generally occur within the first 2 hours.
Figure 2. Time course of nHAp stimulated osteopontin and repressed alkaline phosphatase gene expression;
BMSCs were cultured in; growth medium (GM), GM with ascorbic acid (AA) added at the same time as 25 μg/ml of nHAp (AA-same) or 24hrs before nHAp (AA-pre) and RNA was analyzed by qRT-PCR for (A) osteopontin (Opn) or (B) Alkaline phosphatase (Alp) expression levels. Right panels; an expanded graph of early time points calculated as fold change from untreated control. **p<0.005 and ***p<0.0005 (one-way ANOVA).
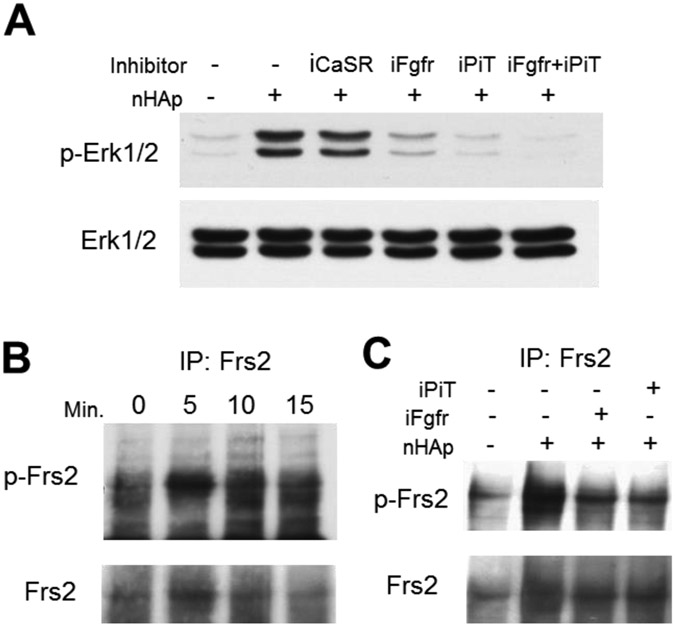
Phosphate-transporter (PiT) and Fibroblast growth factor receptor (Fgfr) signaling are required for nHAp regulated gene expression.
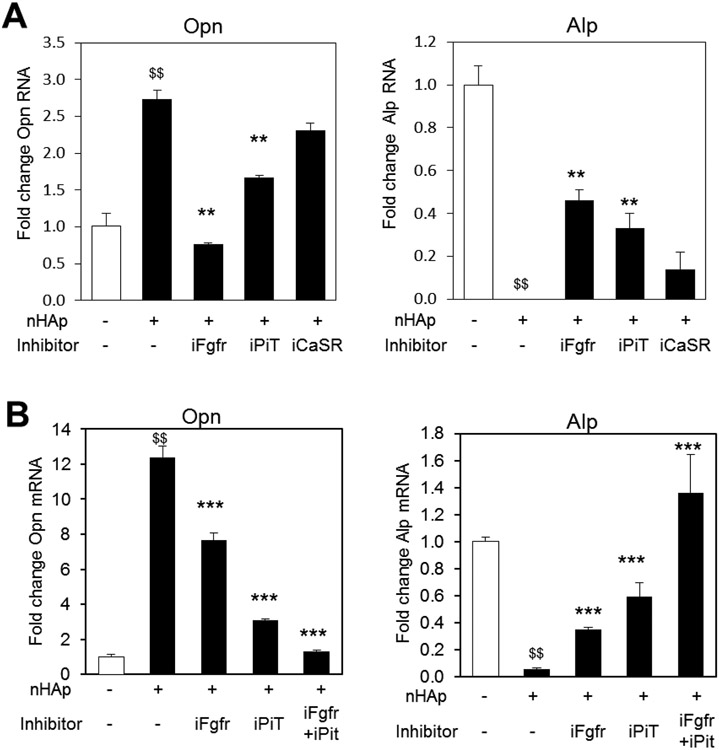
The main components of HAp are calcium and phosphate. These two elements are known to be recognized by cells by the calcium-sensing receptor (CaSR)36 and a family of sodium dependent phosphate transporters (PiT).37-38 Additionally, previous studies on gene regulation by extracellular inorganic phosphate identified the requirement of Fibroblast growth factor receptor (Fgfr) signaling as well as sodium-dependent phosphate co-transporters.31 To determine if the CaSR, PiT, or Fgfr are necessary for the nHAp response, cells were pretreated with pharmacologic inhibitors of the three membrane proteins; CaSR: NPS2143, PiT: phosphonoformic acid (Foscarnet), and Fgfr: PD173074 for 1 hour prior to the addition of nHAp. Quantification of Opn gene expression revealed that inhibition of the Fgfr resulted in an almost complete block of OPN expression, inhibition of Pi-transport resulted in a partial but significant block, and inhibition of the CaSR had only a minimal effect (Figure 3A). In the case of Alp regulation, both Fgfr and PiT inhibition resulted in a partial prevention of RNA suppression. We therefore addressed whether the combination of inhibitors would result in full inhibition, and in fact, the simultaneous inhibition of Fgfr and PiT resulted in complete inhibition of nHAp repressed Alp levels (Figure 3B).
Figure 3. Inhibition of FGF receptor and phosphate-transport blocks nHAp regulated gene expression and Erk1/2 phosphorylation.
(A) BMSCs were pretreated with inhibitors of the Fgf receptor inhibitor (iFgfr), Pi-transport inhibitor (iPiT) or calcium-sensing receptor inhibitor (iCaSR) and harvested after 24 hours. (B) BMSCs were treated as in (A) with iFgfr, iPiT and the combination of iFgfr and iPiT. $$p<0.005 compared to untreated control. **p<0.005 compared to the nHAp treated (Student’s t-test).
nHAp stimulates the Erk1/2 signaling pathway.
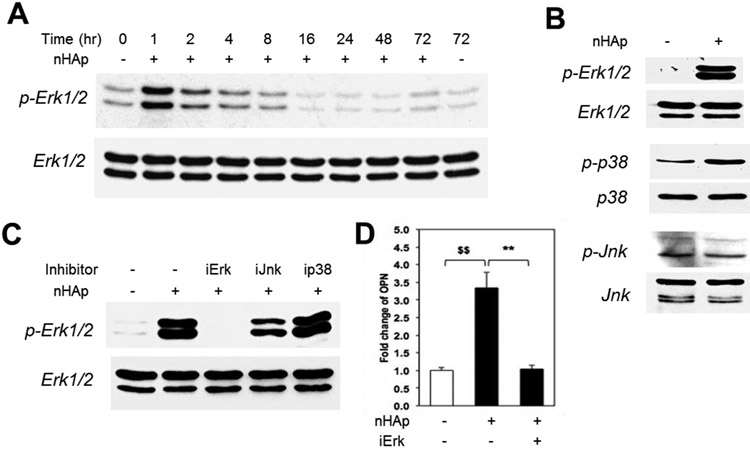
One mechanism by which external stimuli can influence gene expression is through the stimulation of intracellular signaling pathways. The Mitogen Activated Protein Kinases (MAPKs); Erk1/2 (p42/p44), p38, and Jun N-terminal Kinase (Jnk) are three common pathways by which many extracellular generated signals are transduced to alter gene expression. These proteins are key kinases in distinct signaling pathways and are activated by phosphorylation. A previous study, on the effects of elevated inorganic phosphate identified an increase in Erk1/2 signaling.39 To determine if nHAp stimulated Erk1/2 signaling pathway BMSCs were treated with nHAp for similar time points as our gene expression analysis (Figure 1) and analyzed for phosphorylation status using Western blotting. An increase in Erk1/2 phosphorylation was detected within 1 hour of cell exposure to nHAp (Figure 4A). To determine if nHAp specifically stimulates the Erk1/2 pathway the phosphorylation status of the other MAPKs, p38 and Jnk, was also analyzed. Results identified 15 minutes as the strongest response for Erk1/2 phosphorylation however no change in the Jnk pathway and only a slight increase in p38 were detected (Figure 4B). Similar results were obtained in serum free medium (Figure S1A) suggesting that nHAp is unlikely to stimulate signaling through the binding of medium proteins. The results therefore identify a strong and rapid stimulation of the Erk1/2 pathway by nHAp.
Figure 4. nHAp stimulates phosphorylation of Erk1/2.
(A) BMSCs were treated with nHAp (25 μg/ml) for indicated times and analyzed by Western blotting or phospho-Erk1/2 (pErk1/2) or total Erk1/2. (B) Cells were serum starved and treated with nHAp and samples analyzed by Western blotting as indicated; p-phosphorylated form. (C) Cells were pre-treated with inhibitors for Erk1/2 (iErk1/2), Jnk (iJnk), and p38 (ip38) and the resulting lysate was analyzed by Western blotting and probed as indicated. (D) Cells were pretreated with Erk inhibitor for 24 hours and samples analyzed by qRT-PCR for osteopontin expression. $$p<0.005 compared to untreated control; **p<0.005 compared to the nHAp treated (Student’s t-test).
The Erk1/2 signaling pathway is necessary for nHAp stimulated OPN expression.
To confirm the specificity of nHAp stimulated phosphorylation of Erk1/2, pharmacologic inhibitors of the three MAPK pathways were employed. In agreement with the time course data suggesting stimulation of Erk1/2 phosphorylation, U0126 (30 μM), an inhibitor of MEK, the upstream kinase of Erk1/2, blocked phosphorylation of Erk1/2 in response to nHAp treatment (Figure 4C). Similar results were obtained in the pre-osteoblast cell line MC3T3-E1 (Figure S1B), demonstrating that the findings are generally relevant to osteoblast lineage cells. To determine if the stimulation of Erk1/2 phosphorylation is required for nHAp regulated gene expression, cells were pretreated with the MEK-Erk1/2 inhibitor (U0126) prior to exposure to nHAp. Analysis of Opn gene expression by qRT-PCR revealed a complete inhibition of Opn stimulation by blocking Erk1/2 activation (Figure 4D). The expression level of Alp was decreased by U0126 alone and therefore could not be analyzed using this approach. Taken together, the results identify the Erk1/2 pathway as a functionally required upstream signaling event for nHAp regulated Opn gene expression.
The Erk1/2 signaling pathway including Frs2 is downstream of Fgfr and PiT.
To determine if the requirement of these membrane proteins in the response is upstream of Erk1/2 phosphorylation Western blot analyses were performed. In agreement with the gene expression data the Fgfr and phosphate-transport inhibitors inhibited Erk1/2 phosphorylation in response to nHAp, whereas the CaSR inhibitor had no effect (Figure 5A). Frs2α represents an intracellular signaling protein immediately downstream of the Fgfr family and upstream of Erk1/2 activation. To determine if Frs2α is activated in response to nHAp cells were treated with nHAp for 5, 10 and 15 minutes and immunoprecipitations were performed. The resulting Western blot was probed for phospho-Frs2α (Figure 5B). Results suggest that, in fact Frs2α is phosphorylated (activated) within 5 minutes of nHAp treatment. The Fgfr and PiT inhibitors were used to assess the requirement of these membrane proteins in the response and as expected the Fgfr inhibitor blocked phosphorylation and somewhat unexpected was the fact that the PiT inhibitor also blocked phosphorylation (Figure 5C) suggesting that both events are upstream of Frs2 activation.
Figure 5. Stimulation of Erk1/2 and Frs2 requires Fgf receptor and PiT signaling.
(A) BMSCs were pretreated with inhibitors as Fig. 3, followed by addition of nHAp for 15 minutes and harvested for Western Blotting. The resulting blots were probed with antibodies as indicated. (B) BMSCs were treated with nHAp for the indicated times followed by immunoprecipitation with Frs2 antibody and the resulting blot probed as indicated. (C) BMSCSs were treated with inhibitors as indicated followed by immunoprecipitation with Frs2 antibody. Representative of multiple experiments.
Endocytosis is not required for nHAp to influence cell signaling.
Endocytosis has been shown to be an important membrane event in the biological effects of a number of nanomaterials.40-41 To determine if the process of endocytosis is required for changes in cell signaling in response to nHAp, in addition to cell surface interactions, cells were pre-treated with the inhibitor of three different endocytic processes; caveolin mediated endocytosis; methyl-β-cyclodextrin (MβCD, 5 mM) which depletes and interferes with membrane cholesterol42 an inhibitor of Clathrin-mediated (Phenlyarsine oxide, 50 nM), and an inhibitor of macropinocytosis (Amiloride hydrochloride, 200 μM). The stimulation of Erk1/2 phosphorylation by nHAp was assessed following addition of inhibitors. These concentrations were determined as non-toxic to the cells (Figure S2A). None of the three endocytosis inhibitors altered the stimulation of phospho-Erk1/2 by nHAp (Figure S2B). The results therefore suggest that internalization of nHAp is not required for the initiation of cell signaling resulting in regulation of gene expression.
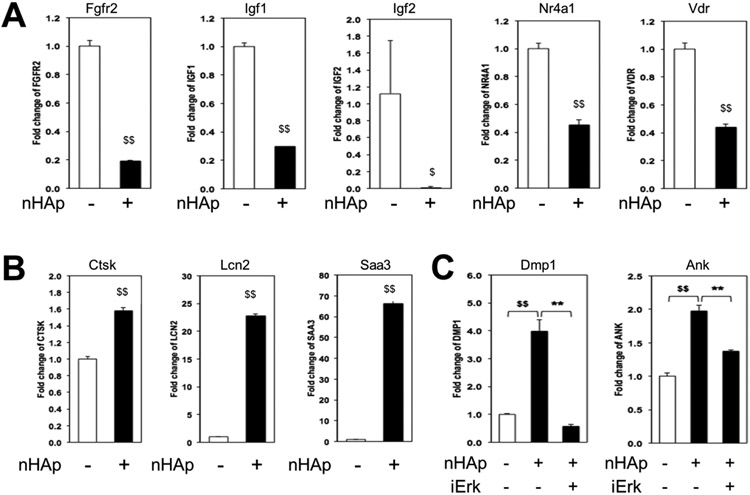
The transcriptional response to nHAp involves numerous genes.
To determine if the transcriptional response to nHAp was limited to a few osteoblast differentiation genes or whether a more extensive cell-wide response was generated we performed an exploratory transcriptome array analysis. Results identified hundreds of genes increased and hundreds decreased by 2-fold or greater in BMSCs exposed to nHAp for 3 days (Table S2). Some of the most interesting genes are shown in Table 2. The genes identified covered a broad range of functions from osteoblast lineage genes such as Dentin matrix protein 1 (Dmp1), Progressive ankylosis (Ank), osterix (Sp7), Bsp (Ibsp), type-1 collagen (Col1α1), and vitamin D receptor (Vdr), among others, to a number of secreted factors including Insulin-like growth factor 1 and 2 (Igf1, Igf2), Bone morphogenetic protein 4 (Bmp4), Lipocalin (Lcn2) and serum amyloid A3 (Saa3), to the membrane proteins integrin β7 (Itgb7) and α6 (Itga6), Fgfr2, and Epidermal growth factor receptor (Egfr). To validate the results, gene expression analysis was performed for a number of these genes and in fact found the results to be very representative of the array analysis (Figure 6A,B). The requirement of Erk1/2 signaling was confirmed for nHAp induced Dmp1 and Ank expression (Figure 6C) suggesting a common regulatory pathway. Taken together, the results identify extracellular nHAp as a stimuli capable of regulating numerous genes through specific signaling pathways.
Table 2.
nHAp regulated genes
| Down regulated to nHAp | Up regulated to nHAp | |||||
|---|---|---|---|---|---|---|
| Gene | Full name | Fold1) | Gene | Full name | Fold1) | |
| Pappa2 | pappalysin 2 | 17.21 | Saa3 | serum amyloid A3 | 21.77 | |
| Omd | osteomodulin | 13.50 | Cst6 | cystatin E/M | 4.99 | |
| Npnt | nephronectin | 11.54 | Hmga1 | high mobility group AT-hook 1 | 4.29 | |
| Mest | mesoderm specific transcript | 6.65 | Dmp1 | dentin matrix protein 1 | 3.99 | |
| Ibsp | integrin binding sialoprotein | 6.10 | Hmga2 | high mobility group AT-hook 2 | 3.44 | |
| Igf1 | insulin-like growth factor 1 | 5.22 | Gadd45α | growth arrest and DNA-damage-inducible 45α | 3.27 | |
| Sfrp2 | secreted frizzled-related protein 2 | 4.11 | Lcn2 | lipocalin 2 | 3.08 | |
| Fabp4 | fatty acid binding protein 4, adipocyte | 3.82 | Itgb7 | integrin beta 7 | 2.94 | |
| Aspn | Aspirin | 3.61 | Etv1 | ets variant 1 | 2.93 | |
| Fgfr2 | fibroblast growth factor receptor 2 | 3.45 | Adamts7 | Adamts7 | 2.84 | |
| Cyr61 | cysteine rich protein 61 | 3.42 | Ctsk | cathepsin K | 2.65 | |
| Postn | periostin, osteoblast specific factor | 3.35 | Kitl | kit ligand | 2.63 | |
| Igf2 | insulin-like growth factor 2 | 3.32 | Pgf | placental growth factor | 2.63 | |
| Fmod | fibromodulin | 3.17 | Ctsl | cathepsin L | 2.56 | |
| Nr4a1 | nuclear receptor subfamily 4, group A, member 1 | 3.15 | Ank | progressive ankylosis | 2.55 | |
| Fbn2 | fibrillin 2 | 3.12 | Itga6 | integrin alpha 6 | 2.54 | |
| Daam2 | dishevelled associated activator of morphogenesis 2 | 3.11 | Adamts5 | Adamts5 | 2.42 | |
| Nid2 | nidogen 2 | 2.98 | Mmp3 | matrix metallopeptidase 3 | 2.41 | |
| Bmp4 | bone morphogenetic protein 4 | 2.93 | Ostn | Osteocrin | 2.41 | |
| Egr2 | early growth response 2 | 2.79 | Vnn1 | vanin 1 | 2.37 | |
| Fabp5 | fatty acid binding protein 5 | 2.79 | Ifrd1 | interferon-related developmental regulator 1 | 2.35 | |
| Fzd2 | frizzled homolog 2 (Drosophila) | 2.76 | Vcan | Versican | 2.30 | |
| S1pr1 | sphingosine-1-phosphate receptor 1 | 2.73 | Cd38 | CD38 antigen | 2.19 | |
| Vdr | vitamin D receptor | 2.72 | Spry1 | sprouty homolog 1 (Drosophila) | 2.16 | |
| Ctgf | connective tissue growth factor | 2.52 | Etv5 | ets variant 5 | 2.16 | |
| Enpp2 | ectonucleotide pyrophosphatase/phosphodiesterase 2 | 2.45 | Timp3 | tissue inhibitor of metalloproteinase3 | 2.15 | |
| Sp7 | Sp7 transcription factor 7 (osterix) | 2.24 | Cp | Ceruloplasmin | 2.13 | |
| Col1a1 | collagen, type I, alpha 1 | 2.13 | Efnb2 | ephrin B2 | 2.08 | |
| Wnt10b | wingless-type MMTV integration site family, member 10B | 2.10 | Ereg | Epiregulin | 2.01 | |
Fold change (Fold) shown as Log2.
Figure 6. Validation of gene array data.
BMSCs were cultured in growth medium and ascorbic acid for 3 days +/− nHAp and harvested RNA samples were analyzed by qRT-PCR for downregulated genes (A) or upregulated (B) as indicated. (C) BMSCs were treated with the Erk1/2 inhibitor prior to harvesting samples for qRT-PCR as indicated. $p<0.05 and $$p<0.005 compared to untreated control and **p<0.005 compared to nHAp treated control (Student’s t-test).
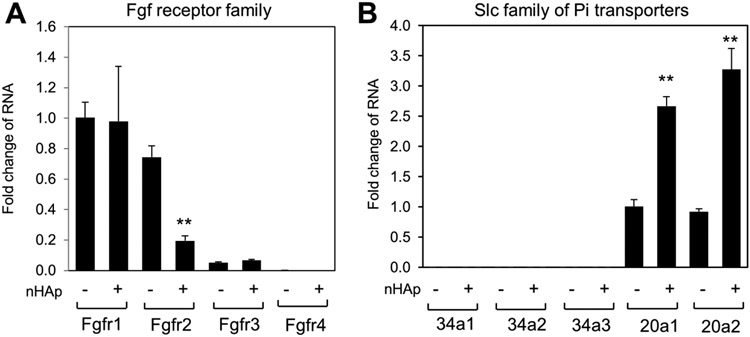
Expression and regulation of the Fgfr and PiT families of genes.
The above microarray results suggested the downregulation of Fgfr2. To determine if nHAp altered the expression of other FGFr family members, gene expression was analyzed in BMSCs treated with nHAp for 24 hours. Results confirmed the strong downregulation of Fgfr2 however Fgfr1, 3, and 4 were not as responsive (Figure 7A). Additionally, results are expressed as a fold change from Fgfr1 to allow a comparison of general expression levels which revealed that Fgfr1 and Fgfr2 were detected at similar levels and that Fgfr3 is expressed at lower levels while Fgfr4 is only minimally detected. Likewise, a comparison of the members of the two families of PiT (Slc34 and Slc20) revealed that both Slc20a1 and Slc20a2 are detected at similar levels and are stimulated by nHAp whereas the Slc34 family is not substantially detected (Figure 7B).
Figure 7. Analysis of Fgf receptor and PiT family member gene expression.
BMSCs were treated with nHAp for 24 hours and cells harvested for RNA analysis by qRT-PCR of Fgfr family members (A) of Slc family members (B). The results are calculated as fold change from untreated Fgfr1 (A) and Slc20a1 (B). Statistical analysis is nHAp treated compared to untreated control for each gene. **p<0.005 (Student’s t-test).
DISCUSSION
HAp and functional mineralization.
Mineral apatite is naturally produced by the cells of the skeleton and dentition and is thought to occur in two general steps; initiation and growth. Although not fully understood, during the initiation phase HAp is thought to be secreted by mineralizing cells as needle or rod-like nHAp in the 50-200 nm range that subsequently form round clusters.12-13, 43 During the growth phase the needle-like clusters are elongated into bundles of parallel crystallites likely guided by collagen and non-collagenous proteins. Rod-like nHAp clusters are also being investigated in the form of nanocrystalline hydroxyapatite paste used therapeutically as a bone substitute.44 Therefore, cells at sites of active mineralization, such as osteoblasts, are likely exposed to HAp similar to the nHAp as used herein (reviewed in1, 12, 45).
HAp and pathological calcification.
Results presented herein might also be relevant to disease related calcification. The deposition of calcium-phosphate crystals in non-mineralizing tissues such as cartilage and vascular smooth muscle can significantly decrease tissue performance as well as generating inflammatory and pain responses. Pathological calcification associated with vascular disease has been demonstrated to correlate with the presence of HAp in the range of hundreds of nanometers,2, 10, 46-49 including crystals isolated from human carotid atherosclerotic plaques.50 These studies also identify needle-like or globular clusters of nHAp generally in the in the 1 to 5 μm range during the early stages of disease. The pathogenic mechanisms remain to be completely defined however our results suggest potential membrane therapeutic targets to diminish the cellular phenotypic changes produced by exposure to nHAp and potentially reduce disease initiation or progression. We have also identified a number of novel genes responsive to nHAp and it will be interesting to determine if they might also be altered in these pathogenic states.
nHAp interacts with the cell surface.
Using SEM, the interaction of nHAp with the surface of BMSCs was identified. These studies identified clusters of rod shaped nHAp of approximately 0.5 to 3 μm interacting with the surface of cells which persisted even after extensive washing suggesting a relatively strong interaction with the cell surface. The SEM images did not suggest any evidence that the nHAp was internalized and this was further supported using pharmacological inhibitors of the three main endocytic mechanisms which did not block nHAp induced Erk1/2 phosphorylation. The internalization of nHAp has been suggested to be influenced by size and charge, with smaller and more positively charged HAp being internalized more readily.51 Our cell studies were performed in growth medium (+10% FBS) and interestingly DLS identified that the size was the smallest and zeta potential the highest relative to whether in DPBS or culture medium alone. The results suggest an influence of FBS which is rich in proteins. Proteins interact with nHAp and bind on the surface by multiple interactions such as charge and hydrophilic/hydrophobic interactions. The most abundant protein, albumin has a negative charge and hydrophobic pockets. These proteins, including albumin, would be able to make a protein shell, called protein corona. Because zeta potential measures overall charge, not specifically surface charge, bound proteins could influence the zeta potential to become more negative. Further, the proteins can act as a stabilizer, preventing aggregates from forming. As a result, the size in the growth medium showed less aggregation than the non-protein containing buffers (Table 1).
Collectively, our data support the idea that the intracellular signaling events are generated by cell surface-nHAp interaction and not the internalization of the particles. Moreover, the fact that nHAp stimulated Erk1/2 phosphorylation in the absence of FBS suggested that the any protein corona did significantly not influence the response. Therefore, the interpretation of the result is that the cell has the ability to recognize the unique composition of HAp. Neither our free particle nHAp nor film/surface HAp is internalized but in both cases the cell membrane is exposed. However, as discussed above, surfaces coated with HAp generally generate an increase in proliferation and an increase in expression of osteoblast related genes such as Alp. The differences in the cellular response could be due to the cells ability to recognize differences in size, shape, or crystallinity of the materials or differences in the protein content of the apical or basolateral surfaces due to cell polarity.52 Understanding these interactions and the mechanism underlying the different responses will be important in optimizing HAp as a biomaterial.
The cellular and molecular effects of nHAp are inhibited by phosphonoformic acid (Foscarnet).
The potential cell surface sensing mechanism(s) by which nHAp might initiate these internal signaling events was also investigated. Foscarnet was identified as a compound capable of significantly inhibiting nHAp stimulated gene expression. One function identified for Foscarnet is the inhibition of sodium-dependent phosphate co-transporters.53 Phosphate transporters are classified into three families (types) (reviewed in37, 54). Type 1 transporters (current nomenclature Slc17a-1 to 7) are not thought to be phosphate specific and may serve as general anion channels. Type 2 transporters (current nomenclature Slc34a-1,2 &3) are thought to be responsible mainly for absorption in the intestine and resorption in the kidney. Type 3 transporters (current nomenclature Slc20a-1&2) are expressed more ubiquitously and until recently have been thought to perform “house-keeping” functions. Slc20a1 (Glvr-1, PiT-1), in particular, has been suggested to be involved in functional mineralization,55 enamel formation,56 and pathological calcification.57-58 Further, Slc20a1 has been identified in a large-scale identification and characterization screen of human genes that activate the NF-κB and MAPK signaling pathways59 supporting its potential role as an originator of signal transduction. A recent study investigating the effects of calcium-phosphate mineralized matrix on osteogenesis of human mesenchymal stem cells identified the requirement of Slc20a1 for the promotion of osteogenic differentiation.60 Although functional studies have not investigated the role of Slc20a2 in bone biology, a recent study has suggested decreased bone mineral density and content in Slc20a2 knockout mice.61
nHAp induced changes in gene expression require Fgf receptor signaling.
The full downstream gene regulatory response to nHAp was also found to require a second membrane event, Fgf receptor signaling. The Fgf receptor family consists of 4 family members, Fgfr1, 2, 3, and 4, with a number of splice variants, and function as membrane receptors that for fibroblast growth factors. These are transmembrane proteins that function, upon ligand binding, as intracellular tyrosine kinases to initiate a number of downstream signaling cascades including activation of the mitogen activated protein kinases (MAPKs). Interestingly, elevated levels of inorganic phosphate have been previously demonstrated to also require Fgfr signaling to induce changes in gene expression as well as activation of Erk1/2 signaling.31 The requirement of Fgfr signaling for both upregulated Opn and downregulated Alp by nHAp suggests that this receptor family is critical for most if not all the nHAp generated cellular signaling. Further, the differential effect of both increased and decreased gene expression presents the possibility that more than one receptor is involved. To our knowledge, this is one of the first examples of a requirement of a growth factor receptor for a nanomaterial to influence cell function.
nHAp induced gene expression requires the Erk1/2 signaling pathway.
Studies herein identified the specific intracellular signal transduction pathway encompassing Erk1/2 as required for nHAp regulated gene expression. The Erk1/2 pathways represents one of three major MAPK signaling pathways the others being Jnk and p38. Activation of the Erk1/2 signaling pathway is a common downstream event for growth factor receptor signaling and has been established as an essential step for proper osteoblast differentiation and skeletal development.62 In regards to osteoblast differentiation, at least two other nanoparticles (NPs) have been demonstrated to activate the Erk1/2 signaling pathway. Gold nanoparticles (20 and 40 nm) and silica (50 nm) NPs have been identified to stimulate Erk1/2.63-64 Although nHAp stimulates Erk1/2 similar to gold and silica NPs the biological effect is different. Silica and gold NPs stimulate Alp expression, whereas nHAp downregulated Alp. This suggests that the cell has the ability to recognize different compositions and/or that cell surface interactions relative to internalization generate secondary signals resulting in a differential response. Taken together, although a range of nanomaterials have been demonstrated to stimulate the Erk1/2 signaling pathway in osteoblast like cells, the downstream effects are substantially different, identifying the likely substantial influence of secondary and tertiary signaling pathways required for the full biological response.
nHAp represents a novel external stimulus capable of regulating a range of genes.
This study identified the upregulation or downregulation of hundreds of genes in BMSCs in response to stimulation with nHAp. Genes such as Dmp1, Ank, Hmga1, Fgfr2, Omd, Postn and Npht, among others, have been previously identified in osteoblast-like cells as responsive to long-term exposure to high (4 - 10 mM) extracellular levels of inorganic phosphate,31, 65 some of which may be due to the generation of octacalcium phosphate nanocrystals, a precursor of HAp.66 A number of novel genes were identified as nHAp responsive and downregulated such as; the Vdr, Igf-1 and Igf-2 and the important osteoblast transcription factor osterix. Other upregulated genes linked to bone homeostasis include the secreted factors, serum amyloid A367 and Lipocalin.68 The down regulation of two osteoblast transcription factors, Vdr and osterix, important in the early stages of osteoblast commitment supports the notion that the process of mineralization (HAp generation) is a signal to transition the osteoblast to a terminally differentiated phenotype.28, 69 The change in expression of circulating factors such as Igf-1/2, Lcn2 and Saa3 represents a potentially novel mechanism by which osteoblast cells exposed to HAp during mineralization might alter the micro-environment niche as paracrine factors or even distant sites as endocrine factors. Interestingly, increased expression of Dmp1, Dusp1, and Tenascin C were also identified in an array analysis of osteoblasts grown on a HAp discs however other genes showed no overlap.70 The result raises the possibility that surface/film HAp and free nHAp generate overlapping but also distinct signaling modules.
CONCLUSIONS
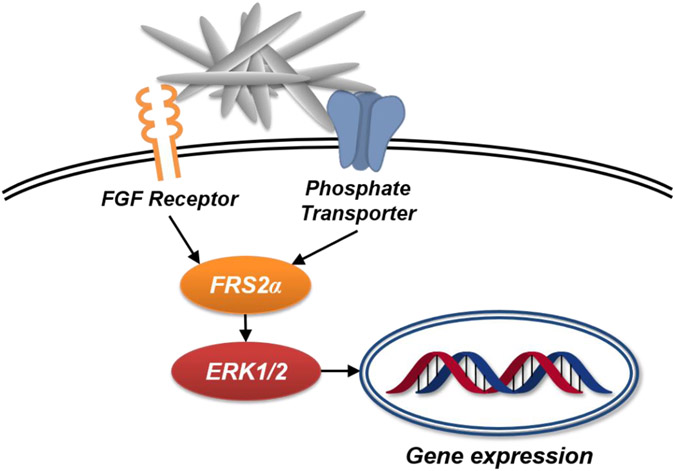
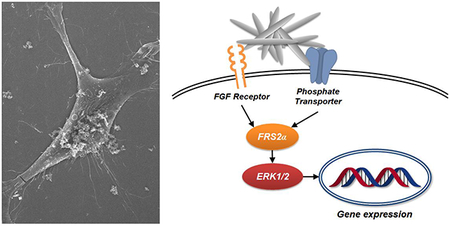
The current study investigated the mechanisms by which rod-shaped nHAp acts as an external stimulus capable of generating specific changes in the expression of a range of genes in osteoblast lineage cells. Understanding the mechanisms by which osteoblast-like cells recognize and respond to the generation of HAp in the extracellular environment is relevant to understanding the process of functional mineralization required for bone homeostasis. At the cell surface, the need for two specific HAp sensors in phosphate-transporters and Fgf receptors was identified (Scheme 1) as well as the stimulation of specific signaling proteins, Frs2α and Erk1/2, downstream of the membrane proteins. Taken with a previous study suggesting that nHAp enhances the differentiation of osteoblast towards terminally differentiated osteocytes, the newly identified signaling framework might be modulated for future health benefits to enhance terminal differentiation by stimulation or to keep cells in a less differentiated state by inhibition. A number of non-osteoblast specific genes that are regulated by nHAp were also identified suggesting the possibility that other cell types may also respond by changes in cell behavior. This finding may be relevant to better understanding the etiology of pathological calcification. Finally, with the advances provided from nanochemistry, findings from this study might be used to modify or engineer HAp to maximize specific behavioral changes in osteoblast lineage cells, improving its potential as a biomaterial for orthopedic repair.
Scheme 1.
Schematic of the upstream signaling events generated by nHAp leading to changes in gene expression.
Supplementary Material
SYNOPSIS.
ACKNOWLEDGMENT
This study was supported by grants to G.R Beck Jr. from Biomedical Laboratory Research & Development Service Award Number I01BX002363 from the VA Office of Research and Emory University Research Committee grant (00067461). The content is solely the responsibility of the authors and does not represent the official views of the Department of Veterans Affairs, National Institutes of Health, or the United States Government. We thank Jeannette Taylor of the Emory University Robert P. Apkarian Integrated Electron Microscopy Core for expert assistance with scanning electron microscopy. The array studies were supported in part by the Emory Integrated Genomics Core (EIGC), which is subsidized by the Emory University School of Medicine and is one of the Emory Integrated Core Facilities. Additional support was provided by the National Center for Advancing Translational Sciences of the National Institutes of Health under Award Number UL1TR000454. The content is solely the responsibility of the authors and does not necessarily reflect the official views of the National Institutes of Health.
Footnotes
Conflict of Interest
The authors declare no competing financial interests.
Supporting Information Available:
Western blots of MAPK signaling proteins, viability studies, Western blots with endocytosis inhibitors, and primers for qRT-PCR studies. This materials is available free of charge via the internet at http://pubs.acs.org
REFERENCES
- (1).Wu LN; Genge BR; Dunkelberger DG; LeGeros RZ; Concannon B; Wuthier RE Physicochemical Characterization of the Nucleational Core of Matrix Vesicles. J. Biol. Chem 1997, 272 (7), 4404–4411. [DOI] [PubMed] [Google Scholar]
- (2).Bertazzo S; Gentleman E; Cloyd KL; Chester AH; Yacoub MH; Stevens MM Nano-Analytical Electron Microscopy Reveals Fundamental Insights into Human Cardiovascular Tissue Calcification. Nat. Mater 2013, 12 (6), 576–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Reynolds JL; Joannides AJ; Skepper JN; McNair R; Schurgers LJ; Proudfoot D; Jahnen-Dechent W; Weissberg PL; Shanahan CM Human Vascular Smooth Muscle Cells Undergo Vesicle-Mediated Calcification in Response to Changes in Extracellular Calcium and Phosphate Concentrations: A Potential Mechanism for Accelerated Vascular Calcification in Esrd. J. Am. Soc. Nephrol 2004, 15 (11), 2857–2867. [DOI] [PubMed] [Google Scholar]
- (4).Lee SY; Kim HY; Gu SW; Kim HJ; Yang DH 25-Hydroxyvitamin D Levels and Vascular Calcification in Predialysis and Dialysis Patients with Chronic Kidney Disease. Kidney Blood Press. Res 2012, 35 (5), 349–354. [DOI] [PubMed] [Google Scholar]
- (5).Villa-Bellosta R; Millan A; Sorribas V Role of Calcium-Phosphate Deposition in Vascular Smooth Muscle Cell Calcification. Am. J. Physiol. Cell Physiol 2011, 300 (1), C210–220. [DOI] [PubMed] [Google Scholar]
- (6).Hunter LW; Charlesworth JE; Yu S; Lieske JC; Miller VM Calcifying Nanoparticles Promote Mineralization in Vascular Smooth Muscle Cells: Implications for Atherosclerosis. Int. J. Nanomedicine 2014, 9, 2689–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Dai XY; Zhao MM; Cai Y; Guan QC; Zhao Y; Guan Y; Kong W; Zhu WG; Xu MJ; Wang X Phosphate-Induced Autophagy Counteracts Vascular Calcification by Reducing Matrix Vesicle Release. Kidney Int. 2013, 83 (6), 1042–1051. [DOI] [PubMed] [Google Scholar]
- (8).Shanahan CM; Crouthamel MH; Kapustin A; Giachelli CM Arterial Calcification in Chronic Kidney Disease: Key Roles for Calcium and Phosphate. Circ. Res 2011, 109 (6), 697–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Kirsch T; Nah HD; Demuth DR; Harrison G; Golub EE; Adams SL; Pacifici M Annexin V-Mediated Calcium Flux across Membranes Is Dependent on the Lipid Composition: Implications for Cartilage Mineralization. Biochemistry 1997, 36 (11), 3359–3367. [DOI] [PubMed] [Google Scholar]
- (10).Belem LC; Zanetti G; Souza AS Jr.; Hochhegger B; Guimaraes MD; Nobre LF; Rodrigues RS; Marchiori E Metastatic Pulmonary Calcification: State-of-the-Art Review Focused on Imaging Findings. Respir. Med 2014, 108 (5), 668–676. [DOI] [PubMed] [Google Scholar]
- (11).Stein GS; Lian JB; Owen TA Relationship of Cell Growth to the Regulation of Tissue-Specific Gene Expression During Osteoblast Differentiation. FASEB J. 1990, 4 (13), 3111–3123. [DOI] [PubMed] [Google Scholar]
- (12).Anderson HC; Garimella R; Tague SE The Role of Matrix Vesicles in Growth Plate Development and Biomineralization. Front. Biosci 2005, 10, 822–837. [DOI] [PubMed] [Google Scholar]
- (13).Bonucci E The Locus of Initial Calcification in Cartilage and Bone. Clin. Orthop. Relat. Res 1971, 78, 108–139. [DOI] [PubMed] [Google Scholar]
- (14).Balasundaram G; Webster TJ Nanotechnology and Biomaterials for Orthopedic Medical Applications. Nanomedicine (Lond) 2006, 1 (2), 169–176. [DOI] [PubMed] [Google Scholar]
- (15).Zakaria SM; Sharif Zein SH; Othman MR; Yang F; Jansen JA Nanophase Hydroxyapatite as a Biomaterial in Advanced Hard Tissue Engineering: A Review. Tissue Eng. Part B Rev 2013, 19 (5), 431–441. [DOI] [PubMed] [Google Scholar]
- (16).Balasundaram G; Sato M; Webster TJ Using Hydroxyapatite Nanoparticles and Decreased Crystallinity to Promote Osteoblast Adhesion Similar to Functionalizing with Rgd. Biomaterials 2006, 27 (14), 2798–2805. [DOI] [PubMed] [Google Scholar]
- (17).Breding K; Jimbo R; Hayashi M; Xue Y; Mustafa K; Andersson M The Effect of Hydroxyapatite Nanocrystals on Osseointegration of Titanium Implants: An in Vivo Rabbit Study. Int. J. Dent 2014, 2014, 171305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Hu J; Zhou Y; Huang L; Liu J; Lu H Effect of Nano-Hydroxyapatite Coating on the Osteoinductivity of Porous Biphasic Calcium Phosphate Ceramics. BMC Musculoskelet. Disord 2014, 15, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Swetha M; Sahithi K; Moorthi A; Srinivasan N; Ramasamy K; Selvamurugan N Biocomposites Containing Natural Polymers and Hydroxyapatite for Bone Tissue Engineering. Int. J. Biol. Macromol 2010, 47 (1), 1–4. [DOI] [PubMed] [Google Scholar]
- (20).Cai YR; Liu YK; Yan WQ; Hu QH; Tao JH; Zhang M; Shi ZL; Tang RK Role of Hydroxyapatite Nanoparticle Size in Bone Cell Proliferation. J. of Mater. Chem 2007, 17 (36), 3780–3787. [Google Scholar]
- (21).Remya NS; Syama S; Gayathri V; Varma HK; Mohanan PV An in Vitro Study on the Interaction of Hydroxyapatite Nanoparticles and Bone Marrow Mesenchymal Stem Cells for Assessing the Toxicological Behaviour. Colloids Surf. B Biointerfaces 2014, 117, 389–397. [DOI] [PubMed] [Google Scholar]
- (22).Xu Z; Liu C; Wei J; Sun J Effects of Four Types of Hydroxyapatite Nanoparticles with Different Nanocrystal Morphologies and Sizes on Apoptosis in Rat Osteoblasts. J. Appl. Toxicol 2012, 32 (6), 429–435. [DOI] [PubMed] [Google Scholar]
- (23).Kalia P; Vizcay-Barrena G; Fan JP; Warley A; Di Silvio L; Huang J Nanohydroxyapatite Shape and Its Potential Role in Bone Formation: An Analytical Study. J. R. Soc. Interface 2014, 11 (93), 20140004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Xu JL; Khor KA; Sui JJ; Zhang JH; Chen WN Protein Expression Profiles in Osteoblasts in Response to Differentially Shaped Hydroxyapatite Nanoparticles. Biomaterials 2009, 30 (29), 5385–5391. [DOI] [PubMed] [Google Scholar]
- (25).Wang L; Zhou G; Liu H; Niu X; Han J; Zheng L; Fan Y Nano-Hydroxyapatite Particles Induce Apoptosis on Mc3t3-E1 Cells and Tissue Cells in Sd Rats. Nanoscale 2012, 4 (9), 2894–2899. [DOI] [PubMed] [Google Scholar]
- (26).Qing F; Wang Z; Hong Y; Liu M; Guo B; Luo H; Zhang X Selective Effects of Hydroxyapatite Nanoparticles on Osteosarcoma Cells and Osteoblasts. J. Mater. Sci. Mater. Med 2012, 23 (9), 2245–2251. [DOI] [PubMed] [Google Scholar]
- (27).Shi Z; Huang X; Cai Y; Tang R; Yang D Size Effect of Hydroxyapatite Nanoparticles on Proliferation and Apoptosis of Osteoblast-Like Cells. Acta Biomater. 2009, 5 (1), 338–345. [DOI] [PubMed] [Google Scholar]
- (28).Ha SW; Jang HL; Nam KT; Beck GR Jr. Nano-Hydroxyapatite Modulates Osteoblast Lineage Commitment by Stimulation of DNA Methylation and Regulation of Gene Expression. Biomaterials 2015, 65, 32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Garcia GM; McCord GC; Kumar R Hydroxyapatite Crystal Deposition Disease. Semin. Musculoskelet. Radiol 2003, 7 (3), 187–193. [DOI] [PubMed] [Google Scholar]
- (30).Martin KJ; Gonzalez EA Metabolic Bone Disease in Chronic Kidney Disease. J. Am. Soc. Nephrol 2007, 18 (3), 875–885. [DOI] [PubMed] [Google Scholar]
- (31).Camalier CE; Yi M; Yu LR; Hood BL; Conrads KA; Lee YJ; Lin Y; Garneys LM; Bouloux GF; Young MR; Veenstra TD; Stephens RM; Colburn NH; Conrads TP; Beck GR Jr. An Integrated Understanding of the Physiological Response to Elevated Extracellular Phosphate. J. Cell. Physiol 2013, 228 (7), 1536–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Sudo H; Kodama HA; Amagai Y; Yamamoto S; Kasai S In Vitro Differentiation and Calcification in a New Clonal Osteogenic Cell Line Derived from Newborn Mouse Calvaria. J. Cell Biol 1983, 96 (1), 191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Beck GR Jr.; Sullivan EC; Moran E; Zerler B Relationship between Alkaline Phosphatase Levels, Osteopontin Expression, and Mineralization in Differentiating MC3T3-E1 Osteoblasts. J. Cell. Biochem 1998, 68 (2), 269–280. [DOI] [PubMed] [Google Scholar]
- (34).Beck GR Jr.; Ha SW; Camalier CE; Yamaguchi M; Li Y; Lee JK; Weitzmann MN Bioactive Silica-Based Nanoparticles Stimulate Bone-Forming Osteoblasts, Suppress Bone-Resorbing Osteoclasts, and Enhance Bone Mineral Density in Vivo. Nanomedicine: NBM 2012, 8 (6), 793–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Uskokovic V; Desai TA Phase Composition Control of Calcium Phosphate Nanoparticles for Tunable Drug Delivery Kinetics and Treatment of Osteomyelitis. I. Preparation and Drug Release. J. Biomed. Mater. Res. A 2013, 101 (5), 1416–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Goltzman D; Hendy GN The Calcium-Sensing Receptor in Bone--Mechanistic and Therapeutic Insights. Nat. Rev. Endocrinol 2015, 11 (5), 298–307. [DOI] [PubMed] [Google Scholar]
- (37).Werner A; Dehmelt L; Nalbant P Na+-Dependent Phosphate Cotransporters: The Napi Protein Families. J. Exp. Biol 1998, 201 (Pt 23), 3135–3142. [DOI] [PubMed] [Google Scholar]
- (38).Biber J; Hernando N; Forster I Phosphate Transporters and Their Function. Annu. Rev. Physiol 2013, 75, 535–550. [DOI] [PubMed] [Google Scholar]
- (39).Beck GR Jr.; Knecht N Osteopontin Regulation by Inorganic Phosphate Is Erk1/2-, Protein Kinase C-, and Proteasome-Dependent. J. Biol. Chem 2003, 278 (43), 41921–41929. [DOI] [PubMed] [Google Scholar]
- (40).Oh N; Park JH Endocytosis and Exocytosis of Nanoparticles in Mammalian Cells. Int. J. Nanomedicine 2014, 9 Suppl 1, 51–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Sahay G; Alakhova DY; Kabanov AV Endocytosis of Nanomedicines. J. Control. Release 2010, 145 (3), 182–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Rothberg KG; Ying YS; Kamen BA; Anderson RG Cholesterol Controls the Clustering of the Glycophospholipid-Anchored Membrane Receptor for 5-Methyltetrahydrofolate. J. Cell Biol 1990, 111 (6 Pt 2), 2931–2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Porter AE; Botelho CM; Lopes MA; Santos JD; Best SM; Bonfield W Ultrastructural Comparison of Dissolution and Apatite Precipitation on Hydroxyapatite and Silicon-Substituted Hydroxyapatite in Vitro and in Vivo. J. Biomed. Mater. Res. A 2004, 69 (4), 670–679. [DOI] [PubMed] [Google Scholar]
- (44).Huber FX; Belyaev O; Hillmeier J; Kock HJ; Huber C; Meeder PJ; Berger I First Histological Observations on the Incorporation of a Novel Nanocrystalline Hydroxyapatite Paste Ostim in Human Cancellous Bone. BMC Musculoskelet. Disord 2006, 7, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Farbod K; Nejadnik MR; Jansen JA; Leeuwenburgh SC Interactions between Inorganic and Organic Phases in Bone Tissue as a Source of Inspiration for Design of Novel Nanocomposites. Tissue Eng. Part B Rev 2014, 20 (2), 173–188. [DOI] [PubMed] [Google Scholar]
- (46).Azari F; Vali H; Guerquin-Kern JL; Wu TD; Croisy A; Sears SK; Tabrizian M; McKee MD Intracellular Precipitation of Hydroxyapatite Mineral and Implications for Pathologic Calcification. J. Struct. Biol 2008, 162 (3), 468–479. [DOI] [PubMed] [Google Scholar]
- (47).Hutcheson JD; Goettsch C; Bertazzo S; Maldonado N; Ruiz JL; Goh W; Yabusaki K; Faits T; Bouten C; Franck G; Quillard T; Libby P; Aikawa M; Weinbaum S; Aikawa E Genesis and Growth of Extracellular-Vesicle-Derived Microcalcification in Atherosclerotic Plaques. Nat. Mater 2016, 15 (3), 335–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Cui L; Houston DA; Farquharson C; MacRae VE Characterisation of Matrix Vesicles in Skeletal and Soft Tissue Mineralisation. Bone 2016, 87, 147–158. [DOI] [PubMed] [Google Scholar]
- (49).Kapustin AN; Davies JD; Reynolds JL; McNair R; Jones GT; Sidibe A; Schurgers LJ; Skepper JN; Proudfoot D; Mayr M; Shanahan CM Calcium Regulates Key Components of Vascular Smooth Muscle Cell-Derived Matrix Vesicles to Enhance Mineralization. Circ. Res 2011, 109 (1), e1–12. [DOI] [PubMed] [Google Scholar]
- (50).Ewence AE; Bootman M; Roderick HL; Skepper JN; McCarthy G; Epple M; Neumann M; Shanahan CM; Proudfoot D Calcium Phosphate Crystals Induce Cell Death in Human Vascular Smooth Muscle Cells: A Potential Mechanism in Atherosclerotic Plaque Destabilization. Circ. Res 2008, 103 (5), e28–34. [DOI] [PubMed] [Google Scholar]
- (51).Chen L; McCrate JM; Lee JC; Li H The Role of Surface Charge on the Uptake and Biocompatibility of Hydroxyapatite Nanoparticles with Osteoblast Cells. Nanotechnology 2011, 22 (10), 105708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Zhao H Membrane Trafficking in Osteoblasts and Osteoclasts: New Avenues for Understanding and Treating Skeletal Diseases. Traffic 2012, 13 (10), 1307–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Szczepanska-Konkel M; Yusufi AN; VanScoy M; Webster SK; Dousa TP Phosphonocarboxylic Acids as Specific Inhibitors of Na+-Dependent Transport of Phosphate across Renal Brush Border Membrane. J. Biol. Chem 1986, 261 (14), 6375–6383. [PubMed] [Google Scholar]
- (54).Tenenhouse HS Phosphate Transport: Molecular Basis, Regulation and Pathophysiology. J. Steroid Biochem. Mol. Biol 2007, 103 (3-5), 572–577. [DOI] [PubMed] [Google Scholar]
- (55).Yoshiko Y; Candeliere GA; Maeda N; Aubin JE Osteoblast Autonomous Pi Regulation Via Pit1 Plays a Role in Bone Mineralization. Mol. Cell. Biol 2007, 27 (12), 4465–4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Yoshioka H; Yoshiko Y; Minamizaki T; Suzuki S; Koma Y; Nobukiyo A; Sotomaru Y; Suzuki A; Itoh M; Maeda N Incisor Enamel Formation Is Impaired in Transgenic Rats Overexpressing the Type III NaPi Transporter Slc20a1. Calcif. Tissue Int 2011, 89 (3), 192–202. [DOI] [PubMed] [Google Scholar]
- (57).El Husseini D; Boulanger MC; Fournier D; Mahmut A; Bosse Y; Pibarot P; Mathieu P High Expression of the Pi-Transporter Slc20a1/Pit1 in Calcific Aortic Valve Disease Promotes Mineralization through Regulation of Akt-1. PLoS One 2013, 8 (1), e53393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Li X; Yang HY; Giachelli CM Role of the Sodium-Dependent Phosphate Cotransporter, Pit-1, in Vascular Smooth Muscle Cell Calcification. Circ. Res 2006, 98 (7), 905–912. [DOI] [PubMed] [Google Scholar]
- (59).Matsuda A; Suzuki Y; Honda G; Muramatsu S; Matsuzaki O; Nagano Y; Doi T; Shimotohno K; Harada T; Nishida E; Hayashi H; Sugano S Large-Scale Identification and Characterization of Human Genes That Activate Nf-Kappab and Mapk Signaling Pathways. Oncogene 2003, 22 (21), 3307–3318. [DOI] [PubMed] [Google Scholar]
- (60).Shih YR; Hwang Y; Phadke A; Kang H; Hwang NS; Caro EJ; Nguyen S; Siu M; Theodorakis EA; Gianneschi NC; Vecchio KS; Chien S; Lee OK; Varghese S Calcium Phosphate-Bearing Matrices Induce Osteogenic Differentiation of Stem Cells through Adenosine Signaling. Proc. Natl. Acad. Sci. U. S. A 2014, 111 (3), 990–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Bezerra DP; Oliveira JR New Studies on Knockout Mouse for the Slc20a2 Gene Show Much More Than Brain Calcifications. J. Mol. Neurosci 2016, 59 (4), 565–566. [DOI] [PubMed] [Google Scholar]
- (62).Ge C; Xiao G; Jiang D; Franceschi RT Critical Role of the Extracellular Signal-Regulated Kinase-Mapk Pathway in Osteoblast Differentiation and Skeletal Development. J. Cell Biol 2007, 176 (5), 709–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Zhang D; Liu D; Zhang J; Fong C; Yang M Gold Nanoparticles Stimulate Differentiation and Mineralization of Primary Osteoblasts through the Erk/Mapk Signaling Pathway. Mater. Sci. Eng. C Mater. Biol. Appl 2014, 42, 70–77. [DOI] [PubMed] [Google Scholar]
- (64).Ha SW; Sikorski JA; Weitzmann MN; Beck GR Jr. Bio-Active Engineered 50 Nm Silica Nanoparticles with Bone Anabolic Activity: Therapeutic Index, Effective Concentration, and Cytotoxicity Profile in Vitro. Toxicol. In Vitro 2014, 28 (3), 354–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Beck GR Jr.; Moran E; Knecht N Inorganic Phosphate Regulates Multiple Genes During Osteoblast Differentiation, Including Nrf2. Exp. Cell Res 2003, 288 (2), 288–300. [DOI] [PubMed] [Google Scholar]
- (66).Sage AP; Lu J; Tintut Y; Demer LL Hyperphosphatemia-Induced Nanocrystals Upregulate the Expression of Bone Morphogenetic Protein-2 and Osteopontin Genes in Mouse Smooth Muscle Cells in Vitro. Kidney Int. 2011, 79 (4), 414–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Thaler R; Sturmlechner I; Spitzer S; Riester SM; Rumpler M; Zwerina J; Klaushofer K; van Wijnen AJ; Varga F Acute-Phase Protein Serum Amyloid A3 Is a Novel Paracrine Coupling Factor That Controls Bone Homeostasis. FASEB J. 2015, 29 (4), 1344–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Rucci N; Capulli M; Piperni SG; Cappariello A; Lau P; Frings-Meuthen P; Heer M; Teti A Lipocalin 2: A New Mechanoresponding Gene Regulating Bone Homeostasis. J. Bone Miner. Res 2015, 30 (2), 357–368. [DOI] [PubMed] [Google Scholar]
- (69).Beck GR Jr. Inorganic Phosphate as a Signaling Molecule in Osteoblast Differentiation. J. Cell. Biochem 2003, 90 (2), 234–243. [DOI] [PubMed] [Google Scholar]
- (70).Xie J; Baumann MJ; McCabe LR Osteoblasts Respond to Hydroxyapatite Surfaces with Immediate Changes in Gene Expression. J. Biomed. Mater. Res. A 2004, 71 (1), 108–117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.