Abstract
X chromosome dosage compensation (XDC) refers to the process by which X-linked genes acquire expression equivalence between two sexes. Ohno proposed that XDC is achieved by two-fold upregulations of X-linked genes in both sexes and by silencing one X chromosome (X chromosome inactivation, XCI) in females. However, genes subject to two-fold upregulations as well as the underlying mechanism remain unclear. It’s reported that gene dosage changes may only affect X-linked dosage-sensitive genes, such as protein complex coding genes (PCGs). Our results showed that in human PCGs are more likely to escape XCI and escaping PCGs (EsP) show two-fold higher expression than inactivated PCGs (InP) or other X-linked genes at RNA and protein levels in both sexes, which suggest that EsP may achieve upregulations and XDC. The higher expressions of EsP possibly result from the upregulations of the single active X chromosome (Xa), rather than escaping expressions from the inactive X chromosome (Xi). EsP genes have relatively high expression levels in humans and lower dN/dS ratios, suggesting that they are likely under stronger selection pressure over evolutionary time. Our study also suggests that SP1 transcription factor is significantly enriched in EsP and may be involved in the up-regulations of EsP on the active X. Finally, human EsP genes in this study are enriched in the toll-like receptor pathway, NF-kB pathway, apoptotic pathway, and abnormal mental, developmental and reproductive phenotypes. These findings suggest misregulations of EsP may be involved in autoimmune, reproductive, and neurological diseases, providing insight for the diagnosis and treatment of these diseases.
Keywords: Autoimmune diseases, Dosage sensitivity, Mental developments, SP1, XCI escapees
1. Introduction
The male mammalians have one X chromosome, while females have two copies. The sex chromosomes of mammals (X and Y) originated from a pair of autosomes about 150 million years ago [1,2]. Then X and Y chromosomes began to diverge, possibly caused by recombination suppression events, such as inversions on the Y chromosome [3,4]. Except for the two small pseudo-autosomal regions (PARs), the Y chromosome did not recombine between chromosomes or undergo a different type of recombination in a single chromosome called gene conversion [5] and gradually degenerated and lost most of its genes [6]. This leads to between-sex gene content imbalance: male XY versus female XX. To maintain gene expression balance between sexes, XDC takes place, in which, according to Susumu Ohno, one random X chromosome in females is inactivated, and the expression of the active X chromosome in females and the single-copy X in males are doubled [7]. In this way, the expression of X-linked genes becomes equivalent between the two sexes, and at the same time, maintains the dosage balance between X-linked and autosomal genes [8]. This two-step model is established as a foundation of our understanding of the evolution of mammal sex chromosomes [9,10].
However, in general, no or only weak up-regulations of X-linked genes at both RNA and protein levels have been observed [[11], [12], [13]], questioning the two-fold upregulation of the entire X chromosome [14]. An alternative hypothesis is that only part of the X chromosome may be dosage compensated, as demonstrated in silkworm [15], chicken [15,[16], [17]], and platypus [18]. In this scenario, dosage-sensitive genes may be more likely to achieve dosage compensation [19]. PCGs are thus good representatives of dosage-sensitive genes [19], such as, in yeast most protein complex genes are likely to be dosage sensitive [20], and In multicellular organisms, PCGs were found overrepresented among dosage-balanced ohnologues [19] and many haploinsufficient genetic diseases are caused by PCGs [21]. Expression levels of X-linked genes coding large protein complexes are similar to autosomal genes and may achieve upregulations [19].
Over 20% of X-linked genes can escape XCI (termed as XCI escapees), which are expressed from both active X (Xa) and inactive X (Xi) chromosomes in females [22]. XCI escapees in humans are clustered on X and probably regulated at the chromatin domain level [23,24]. XCI escapee genes have been reported to exhibit higher expression than X-inactivated genes across tissues, [25] suggesting that XCI status may be associated with X upregulations and XDC, but the specific relationship as well as how XCI status affects XDC of dosage-sensitive genes (i.e. PCGs) remains unclear.
Moreover, upregulations and XDC of XCI escapees can be achieved by either upregulations of Xa or escaping expression from Xi. Allele-specific expression analyses of RNA-seq datasets revealed that expression from Xi reaches low levels, on average 33%, of expression from Xa [23,26]. These two mechanisms (Xa upregulations and Xi escaping) may contribute to the XDC differently and their contributions to XDC are still unknown.
In the study, we propose that only some genes on the X chromosome achieved upregulations and XDC. We want to find out X-linked genes that may achieve upregulations and XDC, measure the possible contributions of Xa and Xi in the upregulations, explore the underlying mechanism for upregulations of X-linked genes, estimate the selection pressure of X-linked genes using dN/dS values, and finally identify candidate genes that may underlie X chromosome inactivation-related diseases.
2. Results
2.1. PCGs are more likely to escape X chromosome inactivation (XCI)
Firstly, we focus on the relationship between protein complex coding genes (PCGs) and XCI escaping status. According to the statuses of XCI escaping and being PCGs, X-linked genes are divided into four groups: EsP, InP, EsNP (escaping non-PCGs), and InNP (inactive non-PCGs) (Supplementary Table 1). We found that 59.3% of PCGs can escape XCI, compared with only 45.2% of non-PCGs (Fig. 1A and 59.3% vs. 45.2%, chi-square test with Yates' correction, p = 0.0157). These results suggest that PCGs may have some sort of connection with XCI escaping.
Fig. 1.
Comparisons of gene expression at transcriptome and proteome levels between the X chromosome and autosomes in human tissues. (A) The relationship between X-linked protein complex coding genes (PCGs) and XCI escaping. Escapees and inactive genes refer to X-linked genes that can escape XCI or be subject to XCI. PCGs and non-PCGs refer to genes encoding protein complex coding genes or not. (B) Global expression value for each gene by taking the average of its expression values across tissues. The labels of the X-axis are AuP, EsP, InP, AuNP, EsNP, and InNP. EsNP and InNP refer to non-protein complex coding genes that can escape XCI or be subject to XCI, respectively. AuNP means non-protein complex coding genes in autosomes. (C) The median RNA expression of genes in 5 groups (AuP, EsP, InP, EsNP, InNP) across 23 tissues. (D) The corresponding RNA expressions of AuP, EsP, and InP in males and females. (E) Global protein expression value for each gene by taking the average of its expression values across tissues. (F) The X:AA ratios between EsP, InP, and AuP across 30 tissues. In all boxplots, the central horizontal line represents the median value and the top and bottom lines of the box are the 25th and 75th percentiles. “*” = p < 0.05, “**” = p < 0.01 and “***” = p < 0.001.
2.2. EsP but not InP show higher expression at transcriptome and proteome levels and may achieve total X chromosome dosage compensation
Next, we adopted a method, which used the expression of autosomal genes as the reference, to evaluate the role of XCI escaping statuses and being PCGs in XDC [19,27]. That is, X:AA = 1 means total dosage compensation, and X:AA = 0.5 indicates that no dosage compensation occurs. Autosomal protein complex genes and the rest are referred to as AuP and AuNP, respectively.
At the transcriptome level, we obtained RPKM values for all genes in each tissue from the Genotype-Tissue Expression project (GTEx) and found that the global expression levels of EsP are similar to that of AuP (Fig. 1B, p = 0.926, EsP:AuP = 0.900). By contrast, InP showed significantly lower expression than that of EsP and AuP (p = 0.005 and 0.0002, respectively), with only about half of the expression level of the latter groups (InP:EsP = 0.533 and InP:AuP = 0.478). These results indicate that EsP is twofold expression of InP at the mRNA level, which remains true for most tissues, as shown in Fig. 1C. In addition to females, the EsP also shows significantly higher expression than InP in males (Fig. 1D, p = 0.014), implying that the higher expressions of EsP mainly result from the upregulations of Xa, rather than escaping expressions from Xi. Moreover, EsP and InP genes, on average, are expressed at similar levels between two sexes (Fig. 1D, p = 0.791 and 0.922, respectively). To further test that escaping XCI alone can’t explain the higher expression of EsP than InP, we also compared the expression of EsNP and InNP, where these genes are less dosage-sensitive. For PCGs, the global expression of EsP is significantly higher than that of InP (Supplementary Fig. 1A, EsP:InP = 1.96, p = 0.0097), but no significant difference is observed for nonPCGs (Supplementary Fig. 1B, EsNP:InNP = 1.27, p = 0.3243). Moreover, there is a significant synergistic effect of being PCGs and XCI escaping status in the higher expression of EsP (Supplementary Table 2, two-way ANOVA, p = 0.0008) and XCI status is not significant separately (Supplementary Table 2, two-way ANOVA, p = 0.4364). These results suggest that the upregulation of active X, rather than escaping XCI, may mainly account for the higher expression of EsP – a group of dosage-sensitive genes.
Similar to the results at the transcriptome level, EsP also generally exhibited two-fold higher expression than InP at the protein level (Fig. 1E, p = 0.010). Moreover, while the InP:AuP ratio is 0.522 (p = 0.016), the EsP:AuP is close to 1 (p = 0.316), which still holds for most tissues as shown in Fig. 1F.
We can also ask whether the expression levels of EsP genes became higher in evolution. Z-score was used to measure expression changes, and positive Z-scores mean expression increases since the human/Chimpanzee common ancestor [28]. We found that Genes in group EsP have greater Z-scores than other X-linked genes (Supplementary Fig. 1C, p = 0.008). Moreover, the median Z-score of genes in the EsP group is 0.2, compared to other X-linked genes with a median Z-score close to 0 (i.e., no change). Several increased genes tend to occur in a cluster (data not shown). These data suggest that several EsP genes such as SH3KBP1 and HDAC6 may be slightly increasing more expression during the recent evolution of the X chromosome.
2.3. Transcription factor SP1 is enriched in genes of group EsP
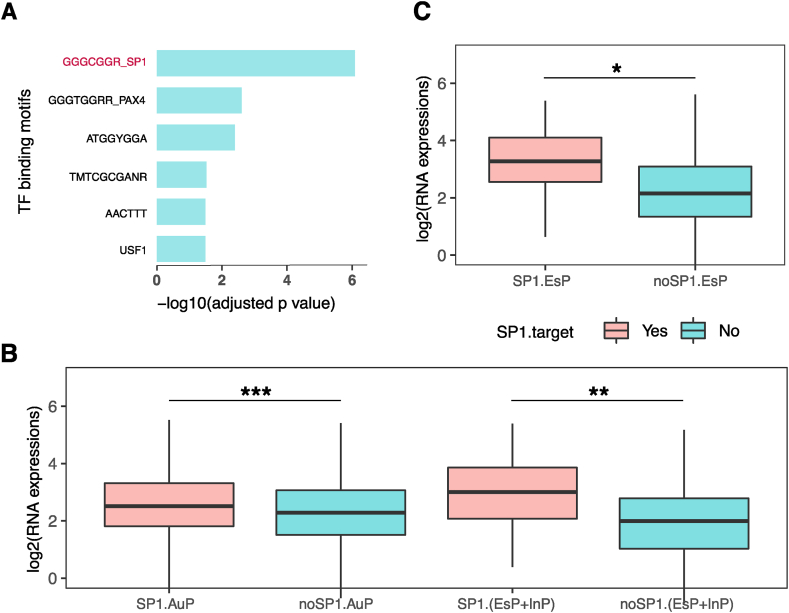
The specific mechanism for upregulations of X-linked genes is not fully understood. As EsP genes possibly achieve two-fold upregulations, we tried to explore the underlying expression regulation mechanisms of EsP, such as transcription factors, compared to InP, EsNP, and InNP using the Enrichr tool (see Methods) [29]. No significantly enriched motifs are found in the InP, EsNP, and InNP. By contrast, there are six significantly enriched motifs in EsP (Fig. 2A). Among them, SP1, PAX4, and USF are known specific transcription factors and SP1 ranks first (p = 2.175E-06). Then we extracted the SP1 binding sites of genes from RegNetwork database [30] and found that more than 43% (32/73) of EsP genes contain SP1 binding motifs around their promoter regions. SP1 can play parallel roles with YY1 and CTCF in regulating XIST transcription from the XIST promoter [31]. And the binding motif of SP1 was reported to be exclusively enriched in the regulatory elements on the Xa rather than Xi [32,33].
Fig. 2.
SP1 may be responsible for the dosage compensation of X-linked escaping PCGs. (A) Escaping protein complex coding genes (EsP) genes are significantly enriched with six binding motifs. X-axis indicates the −log10 adjusted p values and Y-axis indicates the motifs. (B) Autosomal PCGs (AuP) and X-linked PCGs (EsP + InP) with SP1 binding motifs are significantly higher than corresponding PCGs without SP1 binding motifs. (C) Within the EsP group, genes with SP1 binding motifs show higher expression than others. noSP1 means genes have no SP1 binding motifs, and SP1 means that genes have SP1 binding motifs. “*” = p < 0.05, “**” = p < 0.01 and “***” = p < 0.001.
Moreover, PCGs with SP1 binding sites have significantly higher protein expression than genes without SP1 binding sites both on the X chromosome (p = 0.008) and autosomes (p < 0.001) (Fig. 2B). Within the EsP class, genes with SP1 binding motifs also exhibit higher expressions than genes without SP1 binding motifs (Fig. 2C, p = 0.031). Expressions of EsP without SP1 binding motifs were marginally higher than those of InP (p = 0.0309), suggesting that there may be other factors besides SP1. These results suggest that SP1 may, at least in part, takes part in the upregulations of EsP on the Xa.
There have been no reports about including PAX4 and USF1 in XCI in the literature so far. PAX5 has been reported to be significantly enriched in XCI escapees via a secondary mechanism instead of a sequencing-dependent manner [34]. PAX4 may also participate in XDC similarly. USF belongs to basic helix-loop-helix leucine zipper family and can activate transcription through pyrimidine-rich initiator elements and E-box motifs.
2.4. EsP showed lower dN/dS ratios and may be under stronger purifying selection pressure
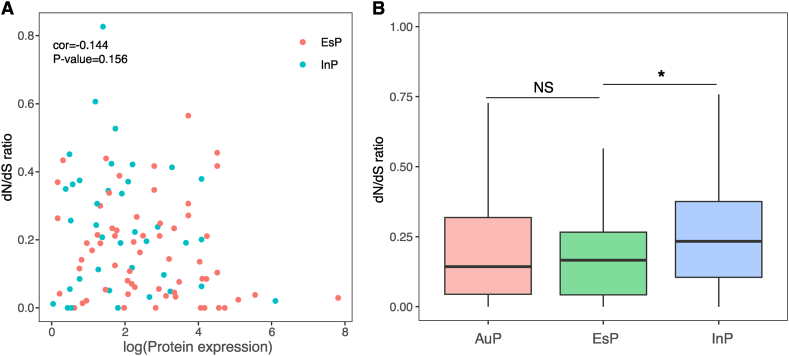
Next, we want to find out the differences in selection pressures between EsP and InP using dN/dS ratios. dN/dS < 1 suggests purifying selection and dN/dS > 1 indicates adaptive selection [35]. Expression levels are reported to be highly correlated with sequence evolutionary rates, and genes with higher expression levels tend to be under greater selection pressure [36,37]. We found that protein expressions of EsP and InP are not associated with dN/dS (Fig. 3A), and the dN/dS ratios of EsP are still significantly lower than that of InP after removing the effects of protein expression levels (Fig. 3B, ANCOVA test, p = 0.034), suggesting that EsP may suffer from a stronger purifying selection.
Fig. 3.
Escaping protein complex coding genes (EsP) showed lower dN/dS ratios and higher expression increases during evolution. (A) The relationship between dN/dS and protein expressions for escaping protein complex coding genes (EsP) and inactive protein complex coding genes (InP). (B) The distributions of dN/dS within PCGs. “NS” = p > 0.05, “*” = p < 0.05, “**” = p < 0.01 and “***” = p < 0.001.
2.5. EsP are enriched in important pathways and phenotypes and may be underlying mechanisms for XCI-related diseases
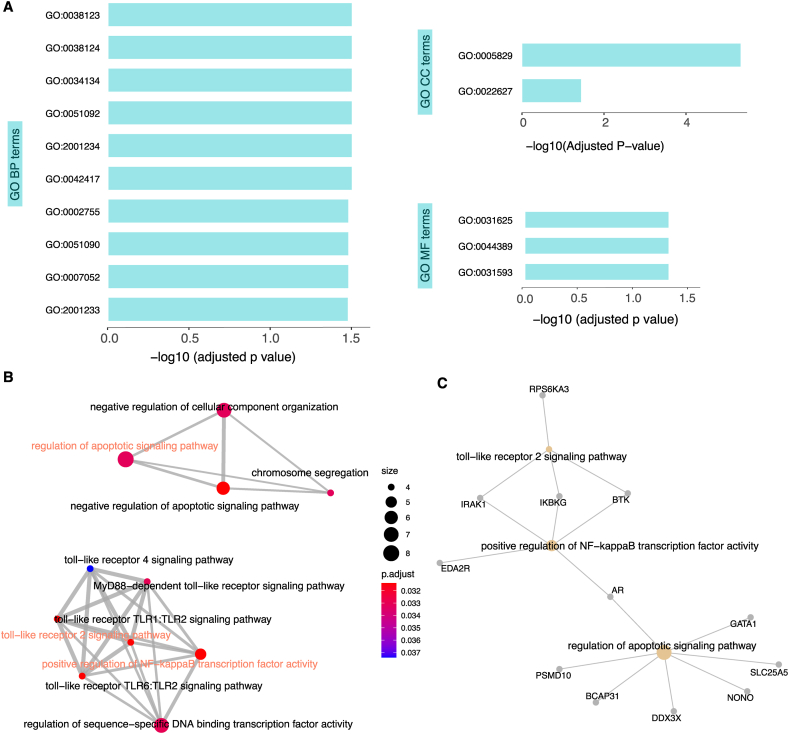
Then we tried to explore the possible downstream consequences of EsP genes having high dosages in females and males. We performed gene functional enrichment analysis (see Methods) and found that 22 GO terms (17 BP terms, 2 CC terms, and 3 MF terms) are significantly enriched with EsP genes (Fig. 4A, adjusted p < 0.05 for these 22 GO terms). Most enriched GO terms are associated with immune regulations, including NF-kappaB and toll-like receptor-related pathways, as well as apoptosis (GO: 2001234) and cell division (GO: 0007052 and GO: 0007059) (Fig. 4B). By contrast, InP genes were enriched only in one term (GO: 000609, pyruvate-related pathway, adjusted p = 0.02214). There were no significantly enriched terms for genes in EsNP and InNP, and even the top-ranking terms are independent of the above signaling pathways.
Fig. 4.
Several GO and KEGG terms are enriched by escaping protein complex coding genes (EsP). (A) Significantly enriched GO terms of genes in group EsP. The horizontal axis referred to the adjusted p values (in logs) for enriched GO terms. The top 10 in 17 BP terms are shown and they are toll−like receptor TLR1:TLR2 signaling pathway (GO:0038123), toll−like receptor TLR6:TLR2 signaling pathway (GO:0038124), toll−like receptor 2 signaling pathway (GO:0034134), positive regulation of NF−kappaB transcription factor activity (GO:0051092) negative regulation of apoptotic signaling pathway (GO:2001234), dopamine metabolic process (GO:0042417), MyD88−dependent toll−like receptor signaling pathway (GO:0002755) regulation of sequence−specific DNA binding transcription factor activity (GO:0051090) mitotic spindle organization (GO:0007052) and regulation of apoptotic signaling pathway (GO:2001233). Five MF and CC GO terms are cytosol (GO:0005829), cytosolic small ribosomal subunit (GO:0022627), ubiquitin protein ligase binding (GO:0031625), small conjugating protein ligase binding (GO:0044389) and polyubiquitin binding (GO:0031593). (B) The relationship between enriched GO BP terms. (C) The interaction network between genes and pathways.
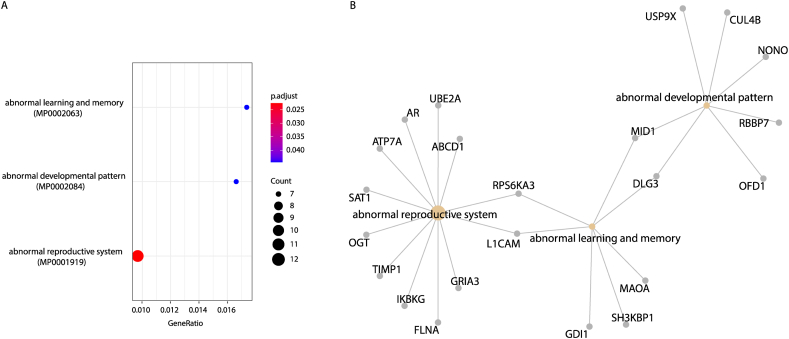
Similarly, we adopted the phenotypic information in the MGI phenotype database to analyze the phenotypic differences between EsP and three other groups using Enrichr (see Methods). There are three significantly enriched phenotype terms for EsP: abnormal learning and memory, abnormal developmental pattern, and abnormal reproduction system (Fig. 5). By contrast, there were no terms for genes in the other three groups. The X chromosome has accumulated a disproportionate number of genes associated with mental functions [38] and reproduction [39], and the enrichment results of EsP are consistent with previous studies [39], suggesting that dysregulations of EsP may partly underlie X-linked abnormal phenotypes.
Fig. 5.
The abnormal phenotype terms are enriched by escaping protein complex coding genes (EsP). (A) Three phenotype terms significantly enriched by genes in group EsP are shown. (B) The interaction network between phenotype terms and corresponding genes in EsP.
2.6. Overexpression of EDA2R may be associated with systemic lupus erythematosus
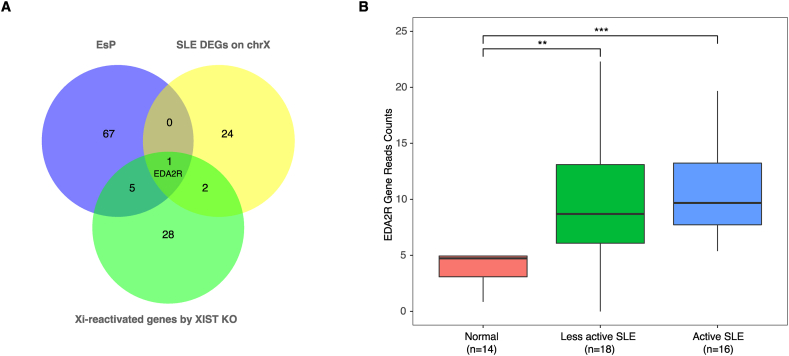
EsP may provide us with a collection of probable candidate genes, dysregulations of which may cause autoimmune diseases [40]. Taking systemic lupus erythematosus (SLE) for example, we built a PPI network between 71 SLE pathogenic genes (obtained from DisGeNET database [41]) and EsP genes and found that many EsP genes had direct protein-protein interactions with SLE pathogenic genes (Supplementary Fig. 2), suggesting these genes may be involved in SLE. Moreover, a recent study reveals that SLE is likely due to the partial reactivation of Xi resulting from the lack of XIST in rare CD11c+ atypical B cells. They identified 36 XIST-dependent reactivated X-linked genes [42]. So, it is reasonable to suppose that overexpression of X-linked dosage-sensitive genes resulting from Xi reactivation may participate in SLE. EDA2R is the only gene in EsP that belongs to Xi-reactivated genes from XIST knockout and is over-expressed in both SLE patients with active and less active diseases (Fig. 6). EDA2R encodes a lesser-known TNFR superfamily member and mediates the activation of the non-canonical NF-kappa-B pathway [43]. EDA2R requires the kinase activity of NIK and IKKα to bind TRAF2 and TRAF6. Although EDA2R has not been investigated in SLE, it is significantly upregulated in Sjogren’s syndrome (SS) compared to healthy individuals [44]. Sjogren’s syndrome is frequently associated with SLE and rheumatoid arthritis. These results highlight the potential functional implication of EDA2R in SLE and other autoimmune diseases.
Fig. 6.
The expressions of EDA2R in systemic lupus erythematosus (SLE) patients (A) The intersection between three gene sets, including escaping protein complex coding genes (EsP), XIST-dependent genes, and (differentially expressed genes) DEGs in one SLE cohort. (B) The expressions of EDA2R between healthy donors, SLE with less active disease, and SLE with active disease. “*” = p < 0.05, “*” = p < 0.01 and “***” = p < 0.001.
3. Discussion
3.1. Escaping but not inactive PCGs may achieve X upregulations and dosage compensation
Ohno hypothesized that XDC is achieved by random XCI in females and two-fold expression increases of X-linked genes in both sexes [7]. Previous research did not show evidence of global X up-regulation, which may be limited to dosage-sensitive genes, such as PCGs [12,19]. It has been reported that X:AA = 1 indicates possible dosage compensation, and X:AA = 0.5 means that there was possibly no dosage compensation [19,27]. From our study, we found that PCGs are more likely to escape XCI. And EsP shows similar expressions to autosomal PCGs, almost twice InP expressions, at both RNA and proteome levels, suggesting that EsP but not the InP may have achieved upregulations and XDC, which is partly consistent with the previous report that X-linked PCGs may achieve XDC [19]. The discrepancy may be due to the overrepresentation of XCI escapees in PCGs. As nearly 60% of PCGs can escape XCI, the overall distribution will deviate to EsP, and expression signals of InP may be shielded. When we put EsP and InP together (meaning all PCGs), the ratio (E + I)/A is 0.837 and near 1, which may partly account for this phenomenon. Our two-way ANOVA results revealed that PCGs and escaping XCI had a significant synergistic effect on higher expressions of EsP (Supplementary Table 2, p < 0.05 with Tukey post hoc test). Moreover, EsP genes have greater median Z-scores than other X-linked genes (Supplementary Fig. 1C), suggesting that they became higher in expression level in evolution during the recent evolution of the X chromosome. These results further support the hypothesis that EsP may achieve upregulations and XDC. Since our results are based on the bulk RNA-seq from GTEx, it will miss the cellular heterogeneity of gene expressions between different cell types.
3.2. Upregulations of EsP possibly result from higher expressions of Xa rather than escaping expression of Xi
In human, most EsP and InP genes exhibited similar expression levels between females (XaXi) and males (XaY) (Fig. 1D), suggesting that escaping expressions from Xi may have little influence on overall expressions [25]. We found EsP showed higher expression levels than InP in both females and males (Fig. 1D), implying that higher expressions of EsP relative to InP may mainly result from high expressions of Xa rather than escaping expression from Xi if assuming the same regulatory mechanism is used in both sexes. Since we have found EsP shows similar expressions to autosomal PCGs, almost twice of InP expressions (Fig. 1) and only EsP may have achieved two-fold upregulations, these above results suggested upregulations of EsP may result from higher expressions of Xa rather than escaping expressions of Xi, which were consistent with the report that expression level of escapee genes from the Xi is usually lower than from Xa [45]. For example, as a XCI escapee, TLR7 expression from Xi usually accounts for a small portion of Xa [42].
3.3. SP1 may be responsible for X up-regulation and dosage compensation of EsP
Several mechanisms participated in the X up-regulation process, including RNA stability, translational regulation, and increased ribosome density [[46], [47], [48], [49]]. Different mechanisms may have independently evolved to increase expression levels of corresponding X-linked genes [39]. We found SP1 motif is significantly enriched in the promoters of EsP rather than InP (Fig. 2A, p < 0.05), which is consistent with a previous report that SP1 was exclusively enriched in transcriptome regulation elements of expressed genes in Xa [32,33]. We further extracted SP1 binding information of genes from RegNetwork database [30]. PCGs with SP1 binding sites showed significantly higher protein expressions than PCGs without SP1 binding sites on autosomes and the X chromosome (Fig. 2B). These results indicate that SP1 may be involved in the up-regulations of EsP on the Xa.
Since we found EsP shows similar expressions to autosomal PCGs, almost twice of InP expressions, in box males and females, suggesting that only EsP (Escaping PCGs) but not the InP(Inactive PCGs) may have achieved two-fold upregulations on the Xa. If this assumption is true, there seem to be some common characteristics between XCI escaping on Xi and up-regulations on Xa. So, the mechanisms of X upregulation, including SP1, may help genes on Xi escape inactivation. CpG islands DNA (CGIs) methylation is associated with gene silencing in XCI [50]. SP1 can recruit RNA polymerase II, prevent the spreading of heterochromatin [51,52], protect CGIs from de novo methylation and maintain expression of downstream genes [53,54]. Mutations of SP1-binding sites fail to maintain the unmethylated state of the Aprt gene CGI [51]. These results SP1 showed that SP1 might also participate in XCI escaping on the Xi. And XCI escaping may serve as contingent effects of XDC due to some unknown transcriptional regulation mechanisms rather than the direct cause.
3.4. EsP genes are involved in several important pathways and abnormal phenotypes
The phenotypic consequences of sex chromosome aneuploidies highlight the importance of X-linked dosage-sensitive genes in autoimmune diseases, reproductive diseases, and neurodevelopmental disorders, but the underlying genes remain unclear [[55], [56], [57], [58]]. Genome-wide association studies (GWAS) studies often excluded the X chromosome because of genetic complexity, limiting our understanding of its roles in complex diseases [57]. Moreover, Variable escape genes recently were reported to be significantly enriched in female-biased diseases [59]. We found that EsP genes, rather than InP, EsNP, and InNP, are enriched with several important pathways, including apoptosis, NF-κB, Toll-like receptors, and abnormal phenotypes and are highly associated with X chromosome-related diseases. For example, IRAK1 in EsP acts as the MyD88 complex on/off switch to activate the NF-κB inflammatory pathway and is a susceptibility gene for SLE (Fig. 4C) [40]. Apoptosis is a programmed cell death process, which maintains tissue homeostasis. Defects of apoptosis may result in uncleared apoptotic cells and their accumulation in tissue and initiate systemic autoimmunity in several chronic inflammatory diseases, including SLE and Rheumatoid Arthritis [60]. NF-κB signaling pathway is reported to play crucial roles in the proper maturation and development of lymphocytes and dendritic cells. Abnormal NF-κB signaling will result in the secretions of autoreactive T-cells and has also been implicated in the pathogenesis of the autoimmune disease [61]. Toll-like receptors (TLRs) belong to the family of pattern recognition receptors that recognize a wide range of pathogen-associated molecular patterns (PAMPs). They are released by the host cells and trigger various intracellular pathways to protect against invading microorganisms. Several studies pointed out the essential role of TLRs in the pathogenesis of autoimmune diseases. TLR pathway-associated proteins, such as IRAK, TRAF, and SOCS, are often dysregulated in autoimmune diseases [62]. Women account for 80% of patients with autoimmune diseases [63]. Klinefelter syndrome patients (males, XXY) have a 14-fold higher risk to develop SLE than normal males (XY) [63], suggesting that the high risk of SLE is dominated by the increased expressions of X-linked genes, especially immune genes [42]. Genes escaping XCI may be expressed higher in females than males and thus cause female-biased immune reactions such as higher antibodies, misregulation of these escaping dosage-sensitive genes may cause serious autoimmune diseases [64].
We also discovered a significant enrichment of EsP genes with abnormal learning and memory, abnormal developmental patterns, and abnormal reproduction system (Fig. 5). Evidenced is emerging for the influence of X-linked genes upon intelligence. RPS6KA3 and MAOA in EsP are involved in brain development. The MAOA gene provides instructions for making the enzyme monoamine oxidase A, which is involved in the breakdown of the neurotransmitter’s serotonin, epinephrine, and dopamine and is associated with regulating mood, emotion, sleep, and appetite. Monoamine oxidase A appears to be involved in normal brain development and is a susceptibility gene for psychiatric disorders [65,66]. The RPS6KA3 gene encodes a member of the RSK (ribosomal S6 kinase) family of growth factor-regulated serine/threonine kinases. RSKs appear to have important roles in cell cycle progression, differentiation, and cell survival. Dysfunctions of RPS6KA3 are associated with intellectual disability [67]. It seems promising to explore the role of misregulations and mutations of RPS6KA3 in brain diseases. So, EsP may play important roles in immune, reproduction, and brain development and may need to compensate for haploinsufficiency during the evolution of sex chromosomes. Expression changes of EsP resulting from sex chromosome aneuploidies may underlie sex chromosome aneuploidies-related disorders.
We further found that the dN/dS ratios of EsP are still significantly lower than that of InP (Fig. 3B, ANCOVA test, p = 0.034) after removing the effects of differences of gene expression levels in the ANCOVA test, which indicate that EsP genes may be under strong functional constraints possibly because these significantly enriched pathways of EsP are highly conserved across different organisms. The inconsistent results between this study and Zhang, Y. et al. (2013) [68] at the dN/dS ratios of XCI escapees may be due to that the classifications of XCI status are based on different datasets.
3.5. Misregulations of EsP such as EDA2R are highly associated with autoimmune diseases
It’s recently reported that the fidelity of XCI maintenance was impaired in the CD11c+ atypical B cells, a rare population in healthy individuals but aberrantly expanded in autoimmune diseases [42]. Loss of XIST and partial reactivation of Xi in CD11c+ atypical B cells result in overexpression of X-linked immune genes including TLR7 [45], raising a possibility that certain autoimmune diseases seem to be due to abnormal reactivation of X-linked dosage-sensitive immune genes from Xi [64]. Overexpression of TLR7 plays an important role in the development and pathogenesis of SLE [69]. As a XCI escapee, TLR7 expression from Xi usually only accounts for a small portion of Xa and can revert to a similar expression as Xa when activated by XIST knockout [42]. Similarly, as EsP are more likely to be dosage-sensitive and enriched in important pathways, EsP genes are possibly predisposed to reactivate and overexpress in Xi and underlie autoimmune diseases, such as SLE. We found that EDA2R in EsP can be reactivated by XIST knockout and over-expressed in SLE patients with active and less active diseases (Fig. 6), suggesting the misregulations of EsP and the potential functional role of EDA2R in the SLE and other autoimmune diseases.
3.6. Future perspective
Given that bulk RNA-seq presents an average expression of genes among all cells in a cell pool, it is interesting to analyze the cellular heterogeneity of EsP expressions at the single-cell levels and perform functional experiments to explore their roles in autoimmune diseases.
4. Conclusion
Using transcriptome, proteome and other data, we demonstrated that EsP shows similar expressions to autosomal PCGs, almost twice of InP expressions in boxes female and males, implying that EsP may achieve two-higher upregulations and higher expressions of EsP relative to InP may mainly result from upregulations of Xa. SP1 motif is significantly enriched in EsP and EsP genes with SP1 binding sites showed significantly higher protein expressions than EsP without SP1 binding sites. These results suggest that SP1 may be involved in the up-regulations of EsP on the Xa. Moreover, EsP genes are involved in several important pathways and abnormal phenotypes. We also found that misregulations of EDA2R may be associated with SLE. We hope that our study could provide several candidate genes that may underlie XCI-related diseases.
5. Methods
5.1. Obtaining protein-complex coding genes (PCGs)
Human protein complex datasets were downloaded from the PCDq database (https://dbarchive.biosciencedbc.jp/data/pcdq/LATEST/pcdq_subunit_member.zip) [70]. PCDq database predicted human protein complexes based on six integrated PPI data (BIND, DIP, MINT, HPRD, IntAct, and GNP_Y2H) by finding densely connected regions in the PPI network and then manually annotating the predicted complexes to confirm that they are experimentally defined protein complex members in literature [70]. We used BioMart (v.GRCH37) (https://grch37.ensembl.org/biomart/martview/, Ensembl 75) to retrieve genomic coordinates of protein complex coding genes (PCGs) and 3463 autosomal and 143 X-chromosomal PCGs, respectively, were obtained by homemade Perl script. Other genes were classified as non-PCGs. In this study, we use PCGs as the representatives of dosage-sensitive genes.
5.2. Obtaining XCI escaping status of X-linked genes
The XCI status of X-linked genes on chromosome X was obtained from a prior study [25]. As the XCI status of most genes is highly consistent across tissues and variable escapees seem to be tissue specific [25], they classified X-linked genes into XCI escapees and X-inactivated genes and used an inclusive definition of XCI escapees: genes that showed significant evidence of escape in any number of tissues or individuals were classified as escape genes [25]. Genes without XCI status information were excluded. We chose the HGNC gene symbol as the standard, and other formats such as Ensembl ID are converted to gene symbols using the online Hyperlink Management System tool (http://biodb.jp/).
5.3. X-linked genes grouped by XCI status and PCGs
This study investigated the effect of dosage sensitivity and XCI status on X chromosome dosage compensation. We divided genes on chromosome X into four groups according to escape status and protein complex coding information to facilitate further comparison. The four groups are abbreviated as EsP, InP, EsNP, and InNP, respectively (Supplementary Table 1). To reduce the effects of other factors, we did not consider the genes without corresponding escape and protein complex information. Similarly, protein-complex coding genes on the autosome are referred to as group AuP (3463 genes), and other autosomal genes are referred to as AuNP.
5.4. Human transcription profiles in GTEx database
To assess the differences in transcription levels of different groups of genes, we downloaded the publicly available gene expression data from the GTEx database, which used RPKM (Reads per kilo base per million mapped reads) as the gene expression quantification method [71]. GTEx provided RPKM values of 56318 genes for 30 different tissue types of 78 American women and 138 American males (http://www.gtexportal.org/static/datasets/gtex_analysis_v6/rna_seq_data/GTEx_Analysis_v6_RNA-seq_RNA-SeQCv1.1.8_gene_median_rpkm.gct.gz).
We only consider the entire tissues for tissues with multiple subtypes, excluding the expression data of subtypes and excluded sex-specific tissues (cervix, fallopian tube, ovary, uterus, vagina, prostate, and testis) finally, leaving 23 tissue types. Then we used homemade scripts to extract the expression levels of the autosomal and X chromosome genes. 2 genes in EsNP and 7 in InNP had no corresponding expression data; others have one-to-one correspondence. Many genes in several tissues cannot be expressed or are expressed at a low level, which may affect the estimation of the average expression level of groups. Therefore, we adopted a fixed cutoff (0.1 RPKM) which has been used in GTEx [71] and other projects [25,72]. We calculate the average RPKM values of genes across all the tissues in which the gene is expressed.
5.5. Human proteome profiles in HPM database
We downloaded human proteome from the Human Proteome Map (HPM) portal. Analytical processes and methods can refer to the literature [73]. HPM is an interactive resource by integrating the massive peptide sequencing result from the draft map of the human proteome project. The HPM contains protein expressions of more than 17,000 human genes, covering over 84% of the annotated human protein-coding genes from individuals with clinically defined healthy tissues, which includes 17 adult tissues, 6 primary hematopoietic cells, and 7 fetal tissues. We obtained the RPKM values of 30057 proteins form HPM in RefSeq protein format and then converted them into HGNC format through the Hyperlink Management System tool (http://biodb.jp/). After excluding 2395 RefSeq proteins without corresponding gene symbols, we obtain protein expression of 27662 genes. The expression of autosomal and X genes was extracted by a homemade Perl script. Most genes have protein expression values: AuP (3134 genes, 90.5%), group EsP (66 genes, 90.4%), group InP (44 genes, 88%), EsNP group (184 genes, 88%) and InNP group (202 genes, 84.2%).
5.6. Exploring the role of XCI escaping statuses and being PCGs in XDC
If dosage compensation occurs, gene expression levels will revert to original levels as ancient autosomes. Otherwise, they will be about the same as half of the original levels. Because the dosage changes during sex chromosome evolution are thought to mainly affect the X-linked genes, we adopted a method used in previous research [19,27], which used the expression of autosomal genes as the reference. That is, X:AA = 1 means total dosage compensation, and X:AA = 0.5 indicates that no dosage compensation occurs. Autosomal protein complex genes and the rest are referred to as AuP and AuNP, respectively.
5.7. The evolution of expression of X-linked genes
We have adopted a method described previously [28], in which a Bayesian approach is employed to infer the ancestral expression state in the human/Chimpanzee common ancestor. Z-score was used to measure expression changes, and positive Z-scores mean expression increases since the common ancestor [28]. X-linked genes were divided into group EsP and other genes due to the small size of genes of some groups.
5.8. Evolutionary selection pressure analysis of X-link genes
We employ dN/dS ratios to measure the selection pressure of genes. And the corresponding dN and dS of human and rhesus one-to-one orthologous protein-coding genes were downloaded from the online Ensembl BioMart (v.GRCH37) database (https://grch37.ensembl.org/biomart/martview/, release 75). By necessity, we exclude genes without corresponding dN or dS values. Differences of dN/dS values between each group are measured by pairwise Mann–Whitney U tests.
5.9. Statistical analysis
In this study, the differences of RPKM values, protein levels and dN/dS ratios between groups were measured by Mann–Whitney U tests in the R language. ANCOVA and ANOVA tests were performed by aov function in R language. TukeyHSD package (v.4.0.3) in the R language was used to perform the ANOVA post hoc test.
5.10. Functional enrichment analysis of X-linked genes
The functional enrichment analysis of X-linked genes is performed on the Online Enrichr database with the default setting [29], which integrates various commonly used databases. Our research mainly involves three databases: GO, KGEE, MGI Mammalian Phenotype database, and Genome Browser PWMs [29]. Genome Browser PWMs is a TF binding motifs database, MGI Mammalian Phenotype is a commonly used mouse phenotype database. We extracted SP1 binding information of genes from RegNetwork database [30]. ClusterProfiler package (v.3.16) in R was used to perform the visualization of functional enrichment Results [74].
5.11. RNA-seq datasets of SLE patients
We wanted to find out the overexpressed X-linked genes in SLE patients. RNA-seq of healthy donors, less active SLE, and active SLE patients are obtained from the GEO database (accession number GSE97264). DESeq2 package (v.1.29.16) in R was used to retrieve differentially expressed genes (DEGs) between healthy individuals and SLE patients with |log2FC| > 1 and adjust p < 0.05.
6. List of abbreviations
The abbreviations and their full name are listed below: X chromosome dosage compensation (XDC); X chromosome inactivation (XCI); protein complex coding genes (PCGs); escaping PCGs (EsP); inactivated PCGs (InP); EsNP (escaping non-PCGs); InNP (inactive non-PCGs); active X chromosome (Xa); inactive X chromosome (Xi); pseudo-autosomal regions (PARs); Genotype-Tissue Expression project (GTEx); systemic lupus erythematosus (SLE); CpG islands DNA (CGIs); Genome-wide association studies (GWAS); pathogen-associated molecular patterns (PAMPs) and RPKM (Reads per kilo base per million mapped reads).
Author contribution statement
Zhihao Xing: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Yuchao Zhang, Weiwei Xiao, Chunqing Zhu: Performed the experiments.
Zhongyuan Tian, Songhui Zhao: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Meng Wang, Yufei Zhu: Analyzed and interpreted the data.
Landian Hu, Xiangyin Kong: Conceived and designed the experiments; Wrote the paper.
Data availability statement
Data included in article/supplementary material/referenced in article.
Funding statement
This work was funded by Guangdong Basic and Applied Basic Research Fund (2019A1515110665 and 2020A1515010246), Shenzhen Children's Hospital (ynkt2020-zz05) and National Key R&D Program of China (No. 2021YFA0805200).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We wish to acknowledge Dr. Zhenguo Zhang for his help in the scientific writing and interpretation of the significance of the results. We wish to thank Prof. Laurence D Hurst for his valuable comments.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e17721.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
The relationship between XCI escaping and expression levels of X-linked genes. (A, B)The effects of XCI escaping on expression increases of X-linked PCGs and non-PCGs at RNA and protein levels. EsP/InP ratio means the effects of escaping on the PCGs, while the EsNP/InNP ratio means the effects of escaping on the non-PCGs. (C) Changes of X-linked gene expression since the human-Chimpanzee common ancestor. Similar to the above boxplots, the central horizontal line represents the median value and the top and bottom of box are the 25th and 75th percentile.
The PPI networks between genes in EsP and SLE susceptibility genes. Blues rectangles are systemic lupus erythematosus (SLE) susceptibility genes retrieved in DisGeNET database, and red rectangles refer to genes in escaping protein complex coding genes (EsP).
References
- 1.Potrzebowski L., Vinckenbosch N., Marques A.C., Chalmel F., Jegou B., Kaessmann H. Chromosomal gene movements reflect the recent origin and biology of therian sex chromosomes. PLoS Biol. 2008;6:e80. doi: 10.1371/journal.pbio.0060080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Graves J.A. The origin and function of the mammalian Y chromosome and Y-borne genes--an evolving understanding. Bioessays. 1995;17:311–320. doi: 10.1002/bies.950170407. [DOI] [PubMed] [Google Scholar]
- 3.Ross M.T., V Grafham D., Coffey A.J., Scherer S., McLay K., Muzny D., Platzer M., Howell G.R., Burrows C., Bird C.P., Frankish A., Lovell F.L., Howe K.L., Ashurst J.L., Fulton R.S., Sudbrak R., Wen G., Jones M.C., Hurles M.E., Andrews T.D., Scott C.E., Searle S., Ramser J., Whittaker A., Deadman R., Carter N.P., Hunt S.E., Chen R., Cree A., Gunaratne P., Havlak P., Hodgson A., Metzker M.L., Richards S., Scott G., Steffen D., Sodergren E., Wheeler D.A., Worley K.C., Ainscough R., Ambrose K.D., Ansari-Lari M.A., Aradhya S., Ashwell R.I., Babbage A.K., Bagguley C.L., Ballabio A., Banerjee R., Barker G.E., Barlow K.F., Barrett I.P., Bates K.N., Beare D.M., Beasley H., Beasley O., Beck A., Bethel G., Blechschmidt K., Brady N., Bray-Allen S., Bridgeman A.M., Brown A.J., Brown M.J., Bonnin D., Bruford E.A., Buhay C., Burch P., Burford D., Burgess J., Burrill W., Burton J., Bye J.M., Carder C., Carrel L., Chako J., Chapman J.C., Chavez D., Chen E., Chen G., Chen Y., Chen Z., Chinault C., Ciccodicola A., Clark S.Y., Clarke G., Clee C.M., Clegg S., Clerc-Blankenburg K., Clifford K., Cobley V., Cole C.G., Conquer J.S., Corby N., Connor R.E., David R., Davies J., Davis C., Davis J., Delgado O., Deshazo D., Dhami P., Ding Y., Dinh H., Dodsworth S., Draper H., Dugan-Rocha S., Dunham A., Dunn M., Durbin K.J., Dutta I., Eades T., Ellwood M., Emery-Cohen A., Errington H., Evans K.L., Faulkner L., Francis F., Frankland J., Fraser A.E., Galgoczy P., Gilbert J., Gill R., Glockner G., Gregory S.G., Gribble S., Griffiths C., Grocock R., Gu Y., Gwilliam R., Hamilton C., Hart E.A., Hawes A., Heath P.D., Heitmann K., Hennig S., Hernandez J., Hinzmann B., Ho S., Hoffs M., Howden P.J., Huckle E.J., Hume J., Hunt P.J., Hunt A.R., Isherwood J., Jacob L., Johnson D., Jones S., de Jong P.J., Joseph S.S., Keenan S., Kelly S., Kershaw J.K., Khan Z., Kioschis P., Klages S., Knights A.J., Kosiura A., Kovar-Smith C., Laird G.K., Langford C., Lawlor S., Leversha M., Lewis L., Liu W., Lloyd C., Lloyd D.M., Loulseged H., Loveland J.E., Lovell J.D., Lozado R., Lu J., Lyne R., Ma J., Maheshwari M., Matthews L.H., McDowall J., McLaren S., McMurray A., Meidl P., Meitinger T., Milne S., Miner G., Mistry S.L., Morgan M., Morris S., Muller I., Mullikin J.C., Nguyen N., Nordsiek G., Nyakatura G., O’Dell C.N., Okwuonu G., Palmer S., Pandian R., Parker D., Parrish J., Pasternak S., Patel D., V Pearce A., Pearson D.M., Pelan S.E., Perez L., Porter K.M., Ramsey Y., Reichwald K., Rhodes S., Ridler K.A., Schlessinger D., Schueler M.G., Sehra H.K., Shaw-Smith C., Shen H., Sheridan E.M., Shownkeen R., Skuce C.D., Smith M.L., Sotheran E.C., Steingruber H.E., Steward C.A., Storey R., Swann R.M., Swarbreck D., Tabor P.E., Taudien S., Taylor T., Teague B., Thomas K., Thorpe A., Timms K., Tracey A., Trevanion S., Tromans A.C., d’Urso M., Verduzco D., Villasana D., Waldron L., Wall M., Wang Q., Warren J., Warry G.L., Wei X., West A., Whitehead S.L., Whiteley M.N., Wilkinson J.E., Willey D.L., Williams G., Williams L., Williamson A., Williamson H., Wilming L., Woodmansey R.L., Wray P.W., Yen J., Zhang J., Zhou J., Zoghbi H., Zorilla S., Buck D., Reinhardt R., Poustka A., Rosenthal A., Lehrach H., Meindl A., Minx P.J., Hillier L.W., Willard H.F., Wilson R.K., Waterston R.H., Rice C.M., Vaudin M., Coulson A., Nelson D.L., Weinstock G., Sulston J.E., Durbin R., Hubbard T., Gibbs R.A., Beck S., Rogers J., Bentley D.R. The DNA sequence of the human X chromosome. Nature. 2005;434:325–337. doi: 10.1038/nature03440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lemaitre C., V Braga M.D., Gautier C., Sagot M.F., Tannier E., Marais G.A.B. Footprints of inversions at present and past pseudoautosomal boundaries in human sex chromosomes. Genome Biol. Evol. 2009;1:56. doi: 10.1093/gbe/evp006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang C.H., Larracuente A.M. Heterochromatin-enriched assemblies reveal the sequence and organization of the drosophila melanogaster Y Chromosome. Genetics. 2019 doi: 10.1534/genetics.118.301765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Charlesworth B., Charlesworth D. The degeneration of Y chromosomes. Philos. Trans. R. Soc. London, A. 2011;355:1563. doi: 10.1098/rstb.2000.0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soukup S.W. In: Sex Chromosomes and Sex-Linked Genes. Ohno S., editor. Springer-Verlag; Berlin, Heidelberg, New York: 1971. p. 1967. 192. Teratology. [DOI] [Google Scholar]
- 8.Sex chromosomes and sex-linked genes. Ann. Intern. Med. 1968 doi: 10.7326/0003-4819-68-6-1375_2. [DOI] [PubMed] [Google Scholar]
- 9.Charlesworth B. The evolution of chromosomal sex determination and dosage compensation. Curr. Biol. 1996;6:149–162. doi: 10.1016/s0960-9822(02)00448-7. [DOI] [PubMed] [Google Scholar]
- 10.Payer B., Lee J.T. X chromosome dosage compensation: how mammals keep the balance. Annu. Rev. Genet. 2008;42:733–772. doi: 10.1146/annurev.genet.42.110807.091711. [DOI] [PubMed] [Google Scholar]
- 11.Lin F., Xing K., Zhang J., He X. Expression reduction in mammalian X chromosome evolution refutes Ohno’s hypothesis of dosage compensation. Proc. Natl. Acad. Sci. U. S. A. 2012;109:11752–11757. doi: 10.1073/pnas.1201816109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Julien P., Brawand D., Soumillon M., Necsulea A., Liechti A., Schutz F., Daish T., Grutzner F., Kaessmann H. Mechanisms and evolutionary patterns of mammalian and avian dosage compensation. PLoS Biol. 2012;10 doi: 10.1371/journal.pbio.1001328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiong Y., Chen X., Chen Z., Wang X., Shi S., Wang X., Zhang J., He X. RNA sequencing shows no dosage compensation of the active X-chromosome. Nat. Genet. 2010;42:1043–1047. doi: 10.1038/ng.711. [DOI] [PubMed] [Google Scholar]
- 14.Casci T. What dosage compensation? Nat. Rev. Genet. 2011;12:2. doi: 10.1038/nrg2921. [DOI] [PubMed] [Google Scholar]
- 15.Zha X., Xia Q., Duan J., Wang C., He N., Xiang Z. Dosage analysis of Z chromosome genes using microarray in silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 2009;39:315–321. doi: 10.1016/j.ibmb.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 16.Itoh Y., Melamed E., Yang X., Kampf K., Wang S., Yehya N., Van Nas A., Replogle K., Band M.R., Clayton D.F., Schadt E.E., Lusis A.J., Arnold A.P. Dosage compensation is less effective in birds than in mammals. J. Biol. 2007;6:2. doi: 10.1186/jbiol53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Itoh Y., Replogle K., Kim Y.H., Wade J., Clayton D.F., Arnold A.P. Sex bias and dosage compensation in the zebra finch versus chicken genomes: general and specialized patterns among birds. Genome Res. 2010;20:512–518. doi: 10.1101/gr.102343.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deakin J.E., Hore T.A., Koina E., Marshall Graves J.A. The status of dosage compensation in the multiple X chromosomes of the platypus. PLoS Genet. 2008;4 doi: 10.1371/journal.pgen.1000140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pessia E., Makino T., Bailly-Bechet M., McLysaght A., Marais G.A.B., Baillybechet M., McLysaght A., Marais G.A.B., Bailly-Bechet M., McLysaght A., Marais G.A.B. Mammalian X chromosome inactivation evolved as a dosage-compensation mechanism for dosage-sensitive genes on the X chromosome. Proc. Natl. Acad. Sci. U. S. A. 2012;109:5346–5351. doi: 10.1073/pnas.1116763109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Papp B., Pál C., Hurst L.D. Dosage sensitivity and the evolution of gene families in yeast. Nature. 2003;424:194. doi: 10.1038/nature01771. [DOI] [PubMed] [Google Scholar]
- 21.Kondrashov F.A., V Koonin E. A common framework for understanding the origin of genetic dominance and evolutionary fates of gene duplications. Trends Genet. Tig. 2004;20:287. doi: 10.1016/j.tig.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 22.Carrel L., Brown C.J. When the lyon(Ized chromosome) roars: ongoing expression from an inactive X chromosome. Philos. Trans. R. Soc. B Biol. Sci. 2017;372 doi: 10.1098/rstb.2016.0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tukiainen T., Villani A.C., Yen A., Rivas M.A., Marshall J.L., Satija R., Aguirre M., Gauthier L., Fleharty M., Kirby A., Cummings B.B., Castel S.E., Karczewski K.J., Aguet F., Byrnes A., Gelfand E.T., Getz G., Hadley K., Handsaker R.E., Huang K.H., Kashin S., Lek M., Li X.X., Nedzel J.L., Nguyen D.T., Noble M.S., Segrè A.V., Trowbridge C.A., Abell N.S., Balliu B., Barshir R., Basha O., Battle A., Bogu G.K., Brown A., Brown C.D., Chen L.S., Chiang C., Conrad D.F., Cox N.J., Damani F.N., Davis J.R., Delaneau O., Dermitzakis E.T., Engelhardt B.E., Eskin E., Ferreira P.G., Frésard L., Gamazon E.R., Garrido-Martín D., Gewirtz A.D.H., Gliner G., Gloudemans M.J., Guigo R., Hall I.M., Han B., He Y., Hormozdiari F., Howald C., Im H.K., Jo B., Kang E.Y., Kim Y., Kim-Hellmuth S., Lappalainen T., Li G., Li X.X., Liu B., Mangul S., McCarthy M.I., McDowell I.C., Mohammadi P., Monlong J., Montgomery S.B., Muñoz-Aguirre M., Ndungu A.W., Nicolae D.L., Nobel A.B., Oliva M., Ongen H., Palowitch J.J., Panousis N., Papasaikas P., Park Y.Y., Parsana P., Payne A.J., Peterson C.B., Quan J., Reverter F., Sabatti C., Saha A., Sammeth M., Scott A.J., Shabalin A.A., Sodaei R., Stephens M., Stranger B.E., Strober B.J., Sul J.H., Tsang E.K., Urbut S., Van De Bunt M., Wang G., Wen X., Wright F.A., Xi H.S., Yeger-Lotem E., Zappala Z., Zaugg J.B., Zhou Y.H., Akey J.M., Bates D., Chan J., Claussnitzer M., Demanelis K., Diegel M., Doherty J.A., Feinberg A.P., Fernando M.S., Halow J., Hansen K.D., Haugen E., Hickey P.F., Hou L., Jasmine F., Jian R., Jiang L., Johnson A., Kaul R., Kellis M., Kibriya M.G., Lee K., Li J.B., Li Q., Lin J., Lin S., Linder S., Linke C., Liu Y., Maurano M.T., Molinie B., Nelson J., Neri F.J., Park Y.Y., Pierce B.L., Rinaldi N.J., Rizzardi L.F., Sandstrom R., Skol A., Smith K.S., Snyder M.P., Stamatoyannopoulos J., Tang H., Wang L., Wang M., Van Wittenberghe N., Wu F., Zhang R., Nierras C.R., Branton P.A., Carithers L.J., Guan P., Moore H.M., Rao A., Vaught J.B., Gould S.E., Lockart N.C., Martin C., Struewing J.P., Volpi S., Addington A.M., Koester S.E., Little A.R., Brigham L.E., Hasz R., Hunter M., Johns C., Johnson M., Kopen G., Leinweber W.F., Lonsdale J.T., McDonald A., Mestichelli B., Myer K., Roe B., Salvatore M., Shad S., Thomas J.A., Walters G., Washington M., Wheeler J., Bridge J., Foster B.A., Gillard B.M., Karasik E., Kumar R., Miklos M., Moser M.T., Jewell S.D., Montroy R.G., Rohrer D.C., Valley D.R., Davis D.A., Mash D.C., Undale A.H., Smith A.M., Tabor D.E., Roche N.V., McLean J.A., Vatanian N., Robinson K.L., Sobin L., Barcus M.E., Valentino K.M., Qi L., Hunter S., Hariharan P., Singh S., Um K.S., Matose T., Tomaszewski M.M., Barker L.K., Mosavel M., Siminoff L.A., Traino H.M., Flicek P., Juettemann T., Ruffier M., Sheppard D., Taylor K., Trevanion S.J., Zerbino D.R., Craft B., Goldman M., Haeussler M., Kent W.J., Lee C.M., Paten B., Rosenbloom K.R., Vivian J., Zhu J., Regev A., Ardlie K.G., Hacohen N., MacArthur D.G. Landscape of X chromosome inactivation across human tissues. Nature. 2017;550:244–248. doi: 10.1038/nature24265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aguet F., Barbeira A.N., Bonazzola R., Brown A., Castel S.E., Jo B., Kasela S., Kim-Hellmuth S., Liang Y., Oliva M., Flynn E.D., Parsana P., Fresard L., Gamazon E.R., Hamel A.R., He Y., Hormozdiari F., Mohammadi P., Muñoz-Aguirre M., Park Y.S., Saha A., Segrè A.V., Strober B.J., Wen X., Wucher V., Ardlie K.G., Battle A., Brown C.D., Cox N., Das S., Dermitzakis E.T., Engelhardt B.E., Garrido-Martín D., Gay N.R., Getz G.A., Guigó R., Handsaker R.E., Hoffman P.J., Im H.K., Kashin S., Kwong A., Lappalainen T., Li X., MacArthur D.G., Montgomery S.B., Rouhana J.M., Stephens M., Stranger B.E., Todres E., Viñuela A., Wang G., Zou Y., Anand S., Gabriel S., Graubert A., Hadley K., Huang K.H., Meier S.R., Nedzel J.L., Nguyen D.T., Balliu B., Conrad D.F., Cotter D.J., deGoede O.M., Einson J., Eskin E., Eulalio T.Y., Ferraro N.M., Gloudemans M.J., Hou L., Kellis M., Li X., Mangul S., Nachun D.C., Nobel A.B., Park Y., Rao A.S., Reverter F., Sabatti C., Skol A.D., Teran N.A., Wright F., Ferreira P.G., Li G., Melé M., Yeger-Lotem E., Barcus M.E., Bradbury D., Krubit T., McLean J.A., Qi L., Robinson K., Roche N.V., Smith A.M., Sobin L., Tabor D.E., Undale A., Bridge J., Brigham L.E., Foster B.A., Gillard B.M., Hasz R., Hunter M., Johns C., Johnson M., Karasik E., Kopen G., Leinweber W.F., McDonald A., Moser M.T., Myer K., Ramsey K.D., Roe B., Shad S., Thomas J.A., Walters G., Washington M., Wheeler J., Jewell S.D., Rohrer D.C., Valley D.R., Davis D.A., Mash D.C., Branton P.A., Barker L.K., Gardiner H.M., Mosavel M., Siminoff L.A., Flicek P., Haeussler M., Juettemann T., Kent W.J., Lee C.M., Powell C.C., Rosenbloom K.R., Ruffier M., Sheppard D., Taylor K., Trevanion S.J., Zerbino D.R., Abell N.S., Akey J., Chen L., Demanelis K., Doherty J.A., Feinberg A.P., Hansen K.D., Hickey P.F., Jasmine F., Jiang L., Kaul R., Kibriya M.G., Li J.B., Li Q., Lin S., Linder S.E., Pierce B.L., Rizzardi L.F., Smith K.S., Snyder M., Stamatoyannopoulos J., Tang H., Wang M., Carithers L.J., Guan P., Koester S.E., Little A.R., Moore H.M., Nierras C.R., Rao A.K., Vaught J.B., Volpi S. The impact of sex on gene expression across human tissues. Science (80-.) 2020;369 doi: 10.1126/SCIENCE.ABA3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Slavney A., Arbiza L., Clark A.G., Keinan A. Strong constraint on human genes escaping X-inactivation is modulated by their expression level and breadth in both sexes. Mol. Biol. Evol. 2016;33:491–492. doi: 10.1093/molbev/msv225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cotton A.M., Ge B., Light N., Adoue V., Pastinen T., Brown C.J. Analysis of expressed SNPs identifies variable extents of expression from the human inactive X chromosome. Genome Biol. 2013 doi: 10.1186/gb-2013-14-11-r122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen X., Zhang J. No X-chromosome dosage compensation in human proteomes. Mol. Biol. Evol. 2015;32:1456. doi: 10.1093/molbev/msv036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hurst L.D., Ghanbarian A.T., Forrest A.R.R., Huminiecki L. The constrained maximal expression level owing to haploidy shapes gene content on the mammalian X chromosome. PLoS Biol. 2015;13 doi: 10.1371/journal.pbio.1002315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., McDermott M.G., Monteiro C.D., Gundersen G.W., Ma’ayan A. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Z.P., Wu C., Miao H., Wu H. RegNetwork: an integrated database of transcriptional and post-transcriptional regulatory networks in human and mouse. Database J. Biol. Databases Curation. 2015;2015 doi: 10.1093/database/bav095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shah R., Sharma A., Kelkar A., Sengupta K., Galande S. A novel cis regulatory element regulates human XIST in a CTCF-dependent manner. Mol. Cell Biol. 2021;41 doi: 10.1128/MCB.00382-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calabrese J.M., Sun W., Song L., Mugford J.W., Williams L., Yee D., Starmer J., Mieczkowski P., Crawford G.E., Magnuson T. Site-specific silencing of regulatory elements as a mechanism of X-inactivation. Cell. 2012;151:951–963. doi: 10.1016/j.cell.2012.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jégu T., Blum R., Cochrane J.C., Yang L., Wang C.Y., Gilles M.E., Colognori D., Szanto A., Marr S.K., Kingston R.E., Lee J.T. Xist RNA antagonizes the SWI/SNF chromatin remodeler BRG1 on the inactive X chromosome. Nat. Struct. Mol. Biol. 2019;26:96–109. doi: 10.1038/s41594-018-0176-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen C.Y., Shi W., Balaton B.P., Matthews A.M., Li Y., Arenillas D.J., Mathelier A., Itoh M., Kawaji H., Lassmann T. YY1 binding association with sex-biased transcription revealed through X-linked transcript levels and allelic binding analyses. Sci. Rep. 2016;6 doi: 10.1038/srep37324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Z., Nielsent R. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol. Biol. Evol. 2002 doi: 10.1093/oxfordjournals.molbev.a004148. [DOI] [PubMed] [Google Scholar]
- 36.Wall D.P., Hirsh A.E., Fraser H.B., Kumm J., Giaever G., Eisen M.B., Feldman M.W. Functional genomic analysis of the rates of protein evolution. Proc. Natl. Acad. Sci. U. S. A. 2005;102:5483–5488. doi: 10.1073/pnas.0501761102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Drummond D.A., Bloom J.D., Adami C., Wilke C.O., Arnold F.H. Why highly expressed proteins evolve slowly. Proc. Natl. Acad. Sci. U. S. A. 2005;102:14338–14343. doi: 10.1073/pnas.0504070102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skuse D.H. X-linked genes and mental functioning. Hum. Mol. Genet. 2005;14(Spec No):R27. doi: 10.1093/hmg/ddi112. [DOI] [PubMed] [Google Scholar]
- 39.Deng X., Berletch J.B., Nguyen D.K., Disteche C.M., Di K.N., Disteche C.M. X chromosome regulation: diverse patterns in development, tissues and disease. Nat. Rev. Genet. 2014;15:367–378. doi: 10.1038/nrg3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mousavi M.J., Mahmoudi M., Ghotloo S. Escape from X chromosome inactivation and female bias of autoimmune diseases. Mol. Med. 2020;26:127. doi: 10.1186/s10020-020-00256-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Piñero J., Ramírez-Anguita J.M., Saüch-Pitarch J., Ronzano F., Centeno E., Sanz F., Furlong L.I. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2020;48:D845–D855. doi: 10.1093/NAR/GKZ1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu B., Qi Y., Li R., Shi Q., Satpathy A.T., Chang H.Y. B cell-specific XIST complex enforces X-inactivation and restrains atypical B cells. Cell. 2021;184 doi: 10.1016/j.cell.2021.02.015. 1790-1803.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Verhelst K., Gardam S., Borghi A., Kreike M., Carpentier I., Beyaert R. XEDAR activates the non-canonical NF-κB pathway. Biochem. Biophys. Res. Commun. 2015 doi: 10.1016/j.bbrc.2015.08.019. [DOI] [PubMed] [Google Scholar]
- 44.Sisto M., Lorusso L., Lisi S. X-linked ectodermal dysplasia receptor (XEDAR) gene silencing prevents caspase-3-mediated apoptosis in Sjögren’s syndrome. Clin. Exp. Med. 2017 doi: 10.1007/s10238-015-0404-z. [DOI] [PubMed] [Google Scholar]
- 45.Wang J., Syrett C.M., Kramer M.C., Basu A., Atchison M.L., Anguera M.C. Unusual maintenance of X chromosome inactivation predisposes female lymphocytes for increased expression from the inactive X. Proc. Natl. Acad. Sci. U.S.A. 2016;113:E2029–E2038. doi: 10.1073/pnas.1520113113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yin S., Deng W., Zheng H., Zhang Z., Hu L., Kong X. Evidence that the nonsense-mediated mRNA decay pathway participates in X chromosome dosage compensation in mammals. Biochem. Biophys. Res. Commun. 2009;383:378–382. doi: 10.1016/j.bbrc.2009.04.021. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Z., Presgraves D.C. Drosophila X-linked genes have lower translation rates than autosomal genes. Mol. Biol. Evol. 2016;33:413–428. doi: 10.1093/MOLBEV/MSV227. [DOI] [PubMed] [Google Scholar]
- 48.Deng X., Berletch J.B., Ma W., Nguyen D.K., Hiatt J.B., Noble W.S., Shendure J., Disteche C.M. Mammalian X upregulation is associated with enhanced transcription initiation, RNA half-life, and MOF-mediated H4K16 acetylation. Dev. Cell. 2013;25:55–68. doi: 10.1016/j.devcel.2013.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Faucillion M.L., Larsson J. Increased expression of X-linked genes in mammals is associated with a higher stability of transcripts and an increased ribosome density. Genome Biol. Evol. 2015 doi: 10.1093/gbe/evv054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shen L., Kondo Y., Guo Y., Zhang J., Zhang L., Ahmed S., Shu J., Chen X., Waterland R.A., Issa J.P.J. Genome-wide profiling of DNA methylation reveals a class of normally methylated CpG island promoters. PLoS Genet. 2007;3:2023. doi: 10.1371/journal.pgen.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gebhard C., Benner C., Ehrich M., Schwarzfischer L., Schilling E., Klug M., Dietmaier W., Thiede C., Holler E., Andreesen R. General transcription factor binding at CpG islands in normal cells correlates with resistance to de novo DNA methylation in cancer cells. Cancer Res. 2010;70:1398. doi: 10.1158/0008-5472.CAN-09-3406. [DOI] [PubMed] [Google Scholar]
- 52.Ishii K., Laemmli U.K. Structural and dynamic functions establish chromatin domains. Mol. Cell. 2003;11:237–248. doi: 10.1016/s1097-2765(03)00010-8. [DOI] [PubMed] [Google Scholar]
- 53.Baron B. Intech; 2012. Breaking the Silence: the Interplay between Transcription Factors and DNA Methylation. [Google Scholar]
- 54.Straussman R., Nejman D., Roberts D., Steinfeld I., Blum B., Benvenisty N., Simon I., Yakhini Z., Cedar H. Developmental programming of CpG island methylation profiles in the human genome. Nat. Struct. Mol. Biol. 2009;16:564–571. doi: 10.1038/nsmb.1594. [DOI] [PubMed] [Google Scholar]
- 55.Natri H., Garcia A.R., Buetow K.H., Trumble B.C., Wilson M.A. The pregnancy pickle: evolved immune compensation due to pregnancy underlies sex differences in human diseases. Trends Genet. 2019 doi: 10.1016/j.tig.2019.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Neri G., Schwartz C.E., Lubs H.A., Stevenson R.E. X-linked intellectual disability update 2017. Am. J. Med. Genet. 2018 doi: 10.1002/ajmg.a.38710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Posynick B.J., Brown C.J. Escape from X-chromosome inactivation: an evolutionary perspective. Front. Cell Dev. Biol. 2019;7:241. doi: 10.3389/fcell.2019.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hong D.S., Reiss A.L. Cognitive and neurological aspects of sex chromosome aneuploidies. Lancet Neurol. 2014 doi: 10.1016/S1474-4422(13)70302-8. [DOI] [PubMed] [Google Scholar]
- 59.Sauteraud R., Stahl J.M., James J., Englebright M., Chen F., Zhan X., Carrel L., Liu D.J. Inferring genes that escape X-Chromosome inactivation reveals important contribution of variable escape genes to sex-biased diseases. Genome Res. 2021;31:1629–1637. doi: 10.1101/gr.275677.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mahajan A., Herrmann M., Muñoz L.E. Clearance deficiency and cell death pathways: a model for the pathogenesis of SLE. Front. Immunol. 2016;7:1–12. doi: 10.3389/fimmu.2016.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Open N., Mishra R.K. Corresponding author article history citation involvement of NF-κB signaling pathway in the pathogenesis of systemic lupus erythematosus. Nephrol Open J. 2016;2:9–13. doi: 10.17140/NPOJ-2-112. [DOI] [Google Scholar]
- 62.Chen J.Q., Szodoray P., Zeher M. Toll-like receptor pathways in autoimmune diseases. Clin. Rev. Allergy Immunol. 2016;50:1–17. doi: 10.1007/s12016-015-8473-z. [DOI] [PubMed] [Google Scholar]
- 63.Libert C., Dejager L., Pinheiro I. The X chromosome in immune functions: when a chromosome makes the difference. Nat. Rev. Immunol. 2010;10:594–604. doi: 10.1038/nri2815. [DOI] [PubMed] [Google Scholar]
- 64.Klein S.L., Flanagan K.L. Sex differences in immune responses. Nat. Rev. Immunol. 2016;16:626–638. doi: 10.1038/nri.2016.90. [DOI] [PubMed] [Google Scholar]
- 65.Tong J., Rathitharan G., Meyer J.H., Furukawa Y., Ang L.C., Boileau I., Guttman M., Hornykiewicz O., Kish S.J. Brain monoamine oxidase B and A in human parkinsonian dopamine deficiency disorders. Brain. 2017;140:2460–2474. doi: 10.1093/brain/awx172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Behl T., Kaur D., Sehgal A., Singh S., Sharma N., Zengin G., Andronie-Cioara F.L., Toma M.M., Bungau S., Bumbu A.G. Role of monoamine oxidase activity in alzheimer’s disease: an insight into the therapeutic potential of inhibitors. Molecules. 2021;26:1–21. doi: 10.3390/molecules26123724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tejada M.I., Ibarluzea N. Non-syndromic X linked intellectual disability: current knowledge in light of the recent advances in molecular and functional studies. Clin. Genet. 2020;97:677–687. doi: 10.1111/cge.13698. [DOI] [PubMed] [Google Scholar]
- 68.Zhang Y., Castillomorales A., Jiang M., Zhu Y., Hu L., Urrutia A.O., Kong X., Hurst L.D. Genes that escape X-inactivation in humans have high intraspecific variability in expression, are associated with mental impairment but are not slow evolving. Mol. Biol. Evol. 2013;30:2588. doi: 10.1093/molbev/mst148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fillatreau S., Manfroi B., Dörner T. Toll-like receptor signalling in B cells during systemic lupus erythematosus. Nat. Rev. Rheumatol. 2021 doi: 10.1038/s41584-020-00544-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kikugawa S., Nishikata K., Murakami K., Sato Y., Suzuki M., Altaf-Ul-Amin M., Kanaya S., Imanishi T. PCDq: human protein complex database with quality index which summarizes different levels of evidences of protein complexes predicted from H-Invitational protein-protein interactions integrative dataset. BMC Syst. Biol. 2012;6:S7. doi: 10.1186/1752-0509-6-S2-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lonsdale J., Thomas J., Salvatore M., Phillips R., Lo E., Shad S., Hasz R., Walters G., Garcia F., Young N. The genotype-tissue expression (GTEx) project. Nat. Genet. 2015;13:307–308. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hackett N.R., Butler M.W., Shaykhiev R., Salit J., Omberg L., Rodriguezflores J.L., Mezey J.G., Strulovicibarel Y., Wang G., Didon L. RNA-Seq quantification of the human small airway epithelium transcriptome. BMC Genom. 2012;13:82. doi: 10.1186/1471-2164-13-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim M.S., Pinto S.M., Getnet D., Nirujogi R.S., Manda S.S., Chaerkady R., Madugundu A.K., Kelkar D.S., Isserlin R., Jain S., Thomas J.K., Muthusamy B., Leal-Rojas P., Kumar P., Sahasrabuddhe N.A., Balakrishnan L., Advani J., George B., Renuse S., Selvan L.D., Patil A.H., Nanjappa V., Radhakrishnan A., Prasad S., Subbannayya T., Raju R., Kumar M., Sreenivasamurthy S.K., Marimuthu A., Sathe G.J., Chavan S., Datta K.K., Subbannayya Y., Sahu A., Yelamanchi S.D., Jayaram S., Rajagopalan P., Sharma J., Murthy K.R., Syed N., Goel R., Khan A.A., Ahmad S., Dey G., Mudgal K., Chatterjee A., Huang T.C., Zhong J., Wu X., Shaw P.G., Freed D., Zahari M.S., Mukherjee K.K., Shankar S., Mahadevan A., Lam H., Mitchell C.J., Shankar S.K., Satishchandra P., Schroeder J.T., Sirdeshmukh R., Maitra A., Leach S.D., Drake C.G., Halushka M.K., Prasad T.S., Hruban R.H., Kerr C.L., Bader G.D., Iacobuzio-Donahue C.A., Gowda H., Pandey A. A draft map of the human proteome. Nature. 2014;509:575–581. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu G., Wang L.G.G., Han Y., He Q.Y.Y. ClusterProfiler: an R package for comparing biological themes among gene clusters. OMICS A J. Integr. Biol. 2012 doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The relationship between XCI escaping and expression levels of X-linked genes. (A, B)The effects of XCI escaping on expression increases of X-linked PCGs and non-PCGs at RNA and protein levels. EsP/InP ratio means the effects of escaping on the PCGs, while the EsNP/InNP ratio means the effects of escaping on the non-PCGs. (C) Changes of X-linked gene expression since the human-Chimpanzee common ancestor. Similar to the above boxplots, the central horizontal line represents the median value and the top and bottom of box are the 25th and 75th percentile.
The PPI networks between genes in EsP and SLE susceptibility genes. Blues rectangles are systemic lupus erythematosus (SLE) susceptibility genes retrieved in DisGeNET database, and red rectangles refer to genes in escaping protein complex coding genes (EsP).
Data Availability Statement
Data included in article/supplementary material/referenced in article.