Abstract
This study employed both short-read sequencing (SRS, Illumina) and long-read sequencing (LRS Oxford Nanopore Technologies) platforms to conduct a comprehensive analysis of the equid alphaherpesvirus 1 (EHV-1) transcriptome. The study involved the annotation of canonical mRNAs and their transcript variants, encompassing transcription start site (TSS) and transcription end site (TES) isoforms, in addition to alternative splicing forms. Furthermore, the study revealed the presence of numerous non-coding RNA (ncRNA) molecules, including intergenic and antisense transcripts, produced by EHV-1. An intriguing finding was the abundant production of chimeric transcripts, some of which potentially encode fusion polypeptides. Moreover, EHV-1 exhibited a greater incidence of transcriptional overlaps and splicing compared to related viruses. It is noteworthy that many genes have their unique TESs along with the co-terminal transcription ends, a characteristic scarcely seen in other alphaherpesviruses. The study also identified transcripts that overlap the replication origins of the virus. Moreover, a novel ncRNA, referred to as NOIR, was found to intersect with the 5′-ends of longer transcript isoform specified by the major transactivator genes ORF64 and ORF65, surrounding the OriL. These findings together imply the existence of a key regulatory mechanism that governs both transcription and replication through, among others, a process that involves interference between the DNA and RNA synthesis machineries.
Keywords: Equid alphaherpesvirus 1, EHV-1, Transcriptome, Replication origin, Long-read sequencing, Nanopore sequencing, Direct RNA sequencing, Illumina sequencing
1. Introduction
Equid alphaherpesvirus 1 (EHV-1) belongs to the Varicellovirus genus of herpesviruses [1,2]. EHV-1 is an important veterinary pathogen causing severe losses in equine industry throughout the world. The most common symptoms of EHV-1 infection include disease of upper respiratory tract, spontaneous abortion in pregnant mares, death in newborns, and life-threatening myeloencephalopathy [[3], [4], [5]]. The virus has an approximately 150 kbp linear double-stranded DNA genome, with 56.7% GC content [6]. The EHV-1genome is composed of two unique regions: the unique short (US) surrounded by a long inverted repeat region (IR); and the unique long (UL) flanked by a short IR [6]. The viral genome encompasses eighty open reading frames (ORFs) encoding 76 protein-coding genes; four genes are located in the IR region [7]. EHV-1 codes for 5 genes (ORF1, 2, 67, 71, and 75) of which no homologs can be found in other alphaherpesviruses with annotated genomes [8]. Similarly to other alphaherpesviruses, EHV-1 can productively infect the cells or enter latency in specific sensory nerve cells [9]. The virus can infect the susceptible cells through two different mechanisms, including endocytosis or fusion between the viral envelope and the membrane of the host cell [10]. Recognition of target cells by EHV-1 is a receptor-dependent process mediated by the viral glycoproteins gB, gC, and gD [11].
The expression of viral genes follows a well-organized cascade, which are controlled by transcription activators including ORF5, 12, 63, 64, and 65 [[12], [13], [14], [15]]. The immediate-early (IE) genes of herpesviruses are produced in the absence of de novo viral protein synthesis. Among the IE genes, ORF64 (homologous to the ie180 gene of pseudorabies virus, PRV, and the icp4 gene of herpes simplex virus 1, HSV-1) is the sole IE gene identified in EHV-1 [16]. The early (E) viral genes typically encode enzymes necessary for DNA replication, while the late (L) genes code for structural polypeptides of the virion, such as splike and capsid proteins. The L genes can be further categorized as leaky late (L1) and true late (L2) depending on whether their expression relies on DNA replication [16].
Short-read sequencing (SRS) and long-read sequencing (LRS) platforms have proved to be exceptionally successful in the analysis of the structural aspects of the transcriptomes. The Illumina technique has a high coverage and base accuracy, but due to its short-read-based assembly it is inefficient for the identification of the transcript ends, including transcription start sites (TSSs), transcription end sites (TESs), alternative splice sites, the embedded transcripts, the multigenic RNA molecule and also the parallel transcription overlaps [17]. LRS platforms developed by Oxford Nanopore Technologies (ONT) and Pacific Biosciences (PacBio) [18,19]. While these methods can accurately identify full-length cDNA and native RNA sequences, they come with the trade-off of having a lower output and higher rates of sequencing errors [[20], [21], [22], [23]]. LRS is superior to the SRS in the detection of multigenic RNA molecules and transcript isoforms. Despite the high error rate associated with the ONT-based approach, it doesn't pose a significant challenge in transcriptome research, as long as there is high genome coverage and well-annotated genomes are accessible.
Direct RNA sequencing (dRNA-Seq) has risen the prominence as the gold standard in RNA sequencing [24] due to its potential to avoid the generation of non-specific reads, which can result from reverse transcription (RT), second strand synthesis, or PCR amplification. Furthermore, In addition, dRNA-Seq maintains the orientation of read sequences and enables the identification of RNA modifications [[25], [26], [27]]. However, the dRNA-Seq technique has its limitations. For instance, it can't fully capture entire transcript lengths, as sequences from the 5'-termini (15–30 base pairs) and often also the poly(A)-tails are absent from the reads [28]. Another limitation of the dRNA-Seq technique is its comparatively lower throughput when juxtaposed with cDNA sequencing Additionally, we observed that dRNA-Seq produced certain transcripts, which were undetected by other techniques, and conversely, true transcripts detected by cDNA sequencing were unidentified using dRNA-Seq [29]. An integrated approach including SRS, LRS, and also the various library preparation techniques is able to circumvent the above problems, and to provide a highly efficient and reliable method in transcriptome research.
Besides SRS [30] (Oláh et al., 2015), herpesvirus transcriptomes have been analyzed by various LRS techniques, including synthesis-based sequencing (from PacBio) [PRV: Tombácz et al. [23]; Torma et al. [31] Epstein-Barr virus: O’Grady et al. [32]; human cytomegalovirus: Balázs et al. [33], 2017; HSV-1: Tombácz et al. [34], nanopore sequencing (from ONT) [varicella-zoster virus (VZV): Prazsák et al. [35]) and LoopSeq single-molecule synthetic long-read sequencing (from Loop Genomics) on Illumina platform (Bovine alphaherpesvirus 1 (BoHV-1):Moldován et al. [29] ].
EHV-1 transcriptome has already been sequenced using an SRS technique [36], however, in this study, only the transcriptional activity within the genomic regions is documented, with no accompanying annotation of viral transcripts. In addition, this work detected miRNAs in EHV-2 and EHV-5, but not in EHV-1. Our objective in this present study was to provide a comprehensive transcriptome annotation of a field isolate EHV-1 using ONT MinION and Illumina MiSeq platforms. We applied amplified cDNA sequencing (Illumina), direct cDNA sequencing (dcDNA-Seq, ONT), as well as direct RNA sequencing (dRNA-Seq, ONT).
2. Results
2.1. Decoding the architecture of the EHV-1 transcriptome
In this study we carried out RNA sequencing using a dual SRS-LRS (Illumina/ONT) approach for profiling the poly(A)+ fraction of the EHV-1 lytic transcriptome. We utilized various library preparation techniques, including methods based on cDNA and native RNA (Fig. 1). Libraries based on the Terminator enzyme were also prepared for both dcDNA-Seq and dRNA-Seq strategies. The LoRTIA pipeline, developed in our lab [37], was used for the analysis and annotation of mapped reads. We utilized the minimap2 alignment tool to map the reads for both the EHV-1 (NC_001491.2) and the host genome (GCF_000003625.3) using the minimap2 alignment tool. Read statistics is available in Table 1 and in Supplementary Table S1.
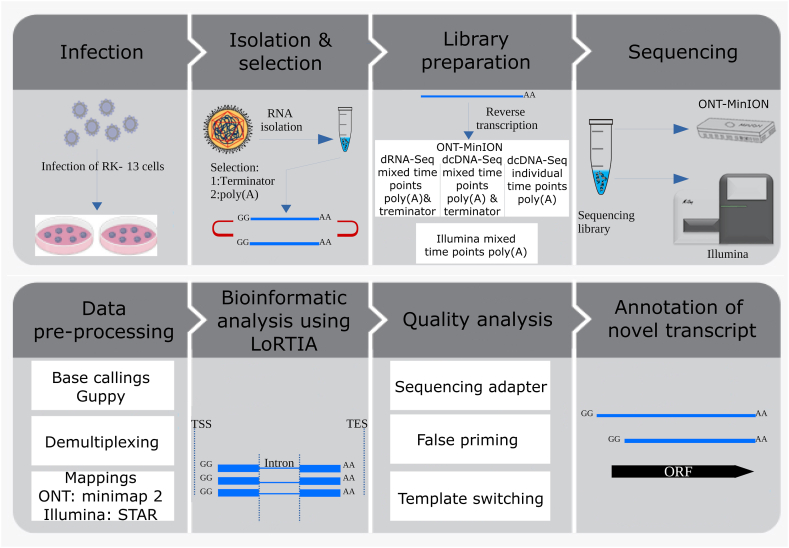
Fig. 1.
Workflow. This figure shows the steps of our analysis starting from the infection of RK-13 cells with a field isolate of EHV-1 and ends with the annotation of transcripts.
Table 1.
Statistics of EHV-1 reads.
| Samples | Read count | Mapped read count | Mean mapped read length | Stdev mapped length |
|---|---|---|---|---|
| ONT dcDNA | 740,128 | 133,838 | 910.084 | 861.209 |
| ONT dcDNA terminator | 1,817,784 | 319,379 | 1,019.045 | 1,048.886 |
| ONT dRNA | 449,468 | 53,625 | 722.195 | 399.855 |
| ONT dRNA terminator | 334,958 | 40,001 | 826.734 | 458.918 |
| ONT dcDNA A | 6,590,468 | 1,345,994 | 663.408 | 665.950 |
| ONT dcDNA B | 5,878,000 | 1,185,499 | 648.338 | 678.449 |
| ONT dcDNA C | 6,268,862 | 1,175,280 | 665.327 | 663.394 |
Native RNA sequencing is often viewed as the gold standard in transcriptome studies due to its ability to avoid the creation of false transcripts, a common issue found in the library preparation and sequencing stages of other methods. However, transcripts can be truncated by the viral and host RNase enzymes, or during the preparation of RNA molecules, therefore false TSSs can be produced. Indeed, in this and in previous studies, we obtained a large variety of 5′ transcript ends, of which a certain fraction is likely non-functional or even non-biological.
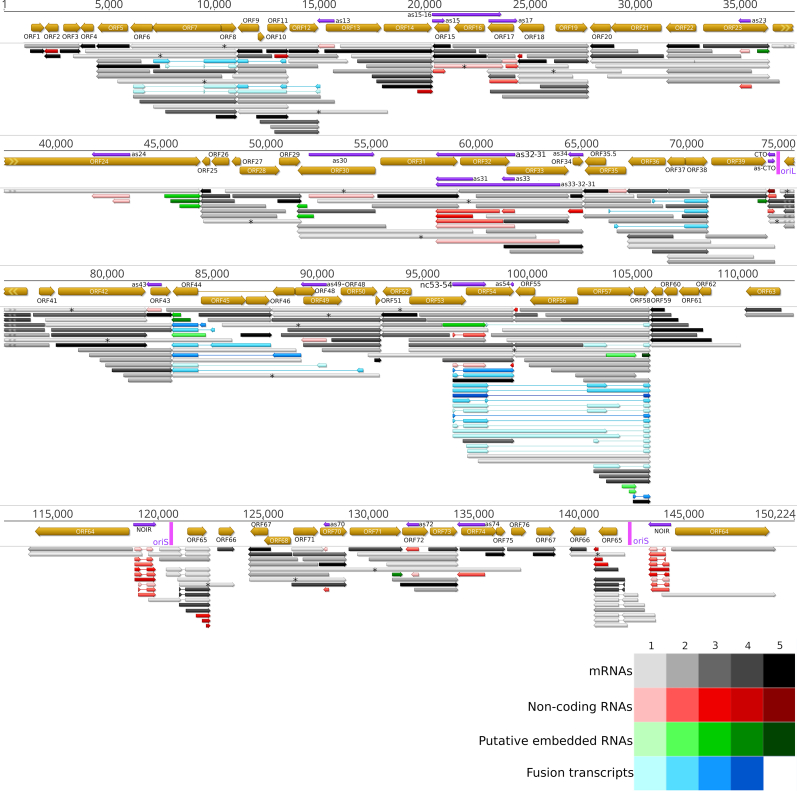
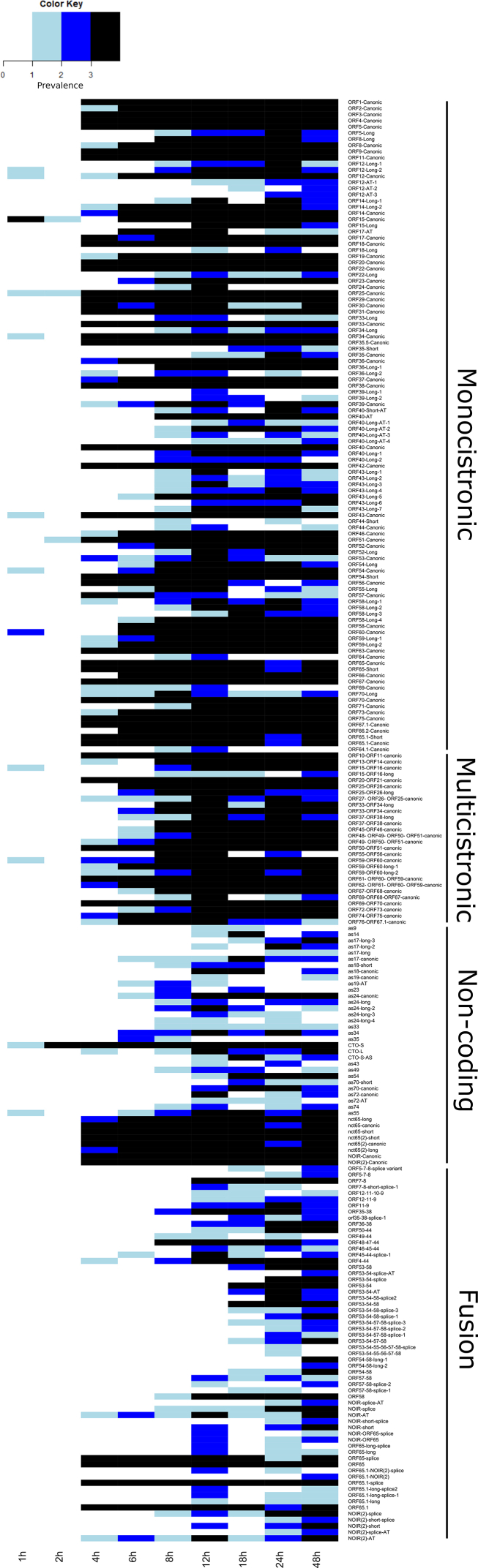
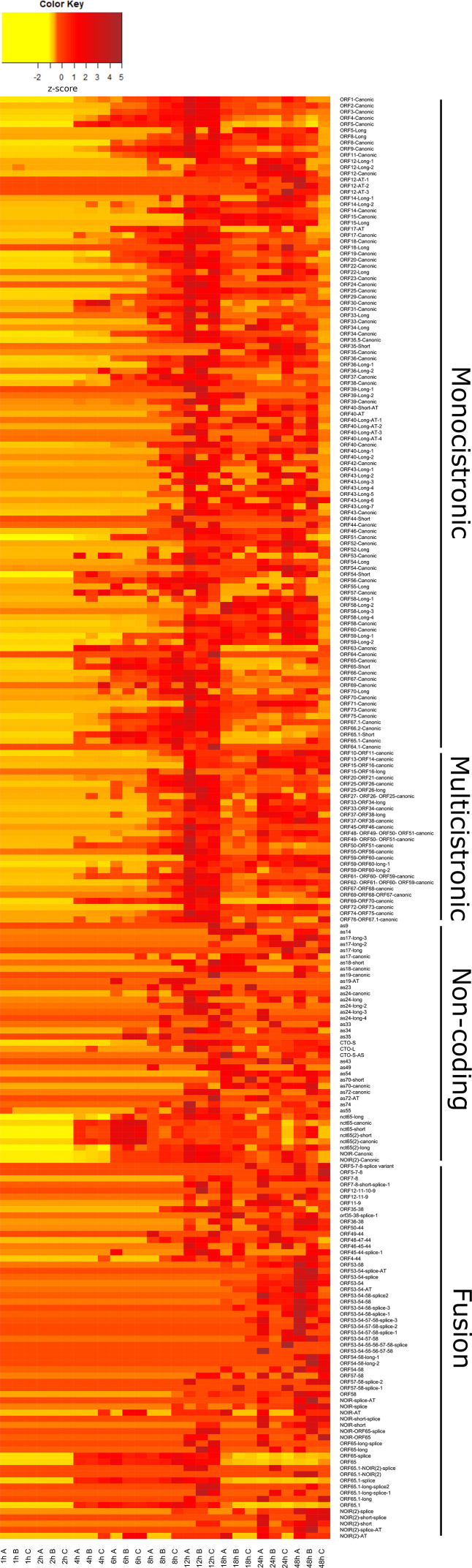
The LoRTIA software checks the quality of poly(A) sequences and sequencing adapters, while also eliminating incorrect TSSs, TESs, and splice sites that could be produced by RNA degradation, RT, second strand synthesis, PCR amplification, or incorrect priming during the sequencing process [37]. To further ensure the accuracy of the transcripts annotated by LoRTIA, more rigorous filtering standards were employed (refer for details to the Read Pre-Processing and Data Analysis part in the Materials and Methods section). The initial LoRTIA results yielded 2,338 transcripts, however the used stringent filtering procedure resulted in a final count of 376. Thus, the application of these very stringent criteria effectively filtered out potential spurious transcripts, but likely has led to a loss of several rare true transcripts of biological origin (Fig. 2, Supplementary Figs. S1 and S2, Supplementary Tables S2–S4). Supplementary Fig. S3 (Panel A to E) illustrates the presence or absence of the viral transcripts in the three replicates, while Supplementary Fig. S4 (Panel A to E) shows the kinetics of the normalized transcript counts.
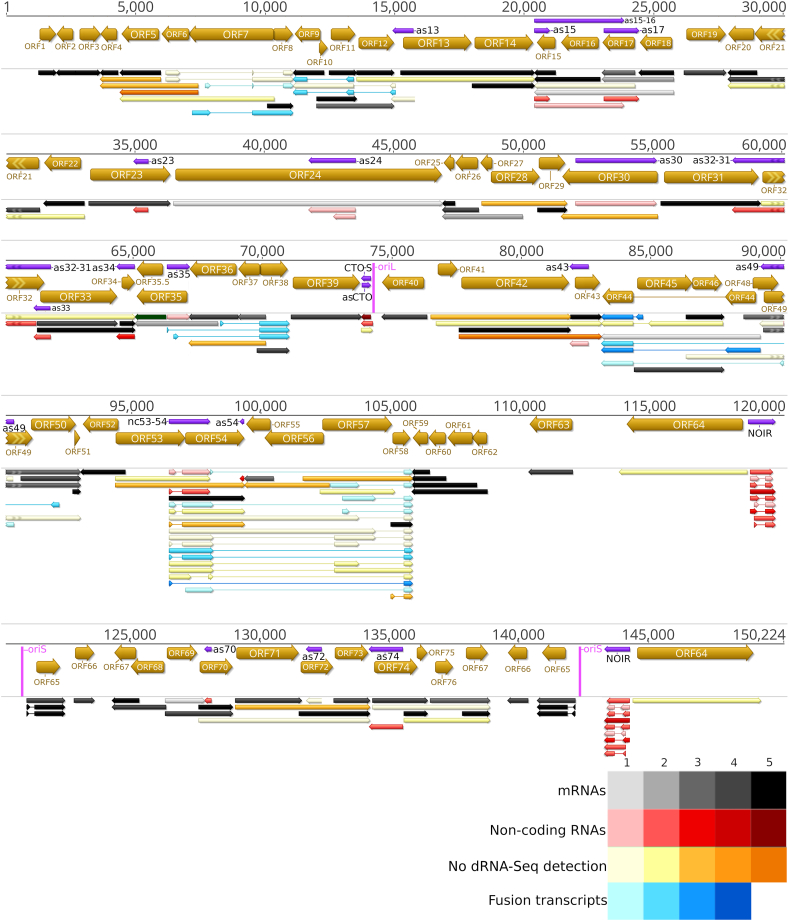
Fig. 2.
Canonical EHV-1 transcripts. This figure presents the canonical transcripts of EHV-1 identified using the LoRTIA program suit. Canonical mRNAs are defined as the most abundant transcript containing the same ORFs and the same exon-intron structure if spliced. All of the annotated non-coding RNAs and fusion transcripts are also depicted. Color code: black: mRNAs, red: ncRNAs, blue: fusion transcripts, yellow: mRNAs undetected by dRNA-Seq and fusion transcripts of which only the introns and TESs were detected by dRNA-Seq but not the TSS of the transcript. The shade of the colors corresponds to the abundance: 1: 1–9 reads, 2: 10–49 reads, 3: 50–199 reads, 4: 200–999 reads, 5: >1000 reads.
Although the total read count of transcripts showed a steady increase at the 48 hpi samples it decreased and there are no transcripts whose read count is the highest in this sample. The variety of transcript species also showed an increase (the number of different annotated transcripts) during the viral infection. The number of identified transcripts in the 1–12 hpi samples, in the 12–24 hpi samples and in the 48 hpi samples were 231, 263 and 248, respectively. Thus, even in the 48 hpi samples, the complexity, or the noise of transcription did not increase substantially, compared to the previous time-points. There were only 5 transcripts that could be detected in the 48 hpi samples, these are mainly splice variants (Supplementary Table S3).
Our pipeline also checks that the potential presence of A-rich regions upstream of the mapped regions, and discards these reads, as these are potentially the results of false priming events. Nevertheless, in order to ascertain that the annotated transcripts are not the results of possible internal priming events, we used the talon_label_reads submodule of the TALON software package [38] on the reads that were used by the LoRTIA program for transcript annotation. The results showed that out of the 503,506 LoRTIA reads, only 2,156 were labeled by this method. Supplementary Table S5A shows the summary of these results, while Supplementary Table S5B shows the per-transcript results. Filtering of these reads however didn’t lead to a loss of any LoRTIA transcript, as a read can support a transcript in a ± 10 nt window in the case of TSS, and ±20 in the case of TESs. Overall, this suggests that the annotated transcripts are indeed not the results of false priming events.
2.2. Promoter motifs, poly(A) signals, transcript start and end sites
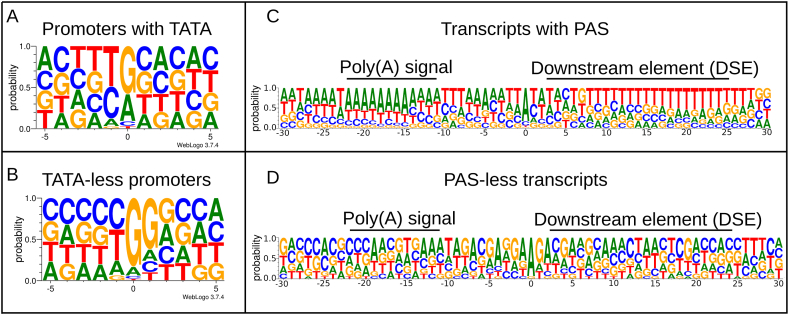
We identified 84 TATA boxes with an average distance of 30.86 bps from the TSSs, 195 GC boxes with an average distance of 60.35 bps from the TSSs, and 43 CAAT boxes with an average distance of 112.41 bps from the TSSs. We found that the +1 position of sequences containing TATA box are enriched in G bases, while in TATA-less sequences not only the +1, but also the +2 position is GC-rich (Fig. 3). Fig. 3A shows the TATA sequences, the TATA-less promoter sequences are shown in Fig. 3B. Fig. 3C and D shows the sequence motifs of transcripts with or without polyadenylation signals, respectively. The GC enrichment in HSV-1 VP5 promoter has already described [39]. The applied filtering criteria resulted in the annotation of 83 TESs (these were supported by dRNA-Seq data). Out of these, 77 had a PAS (92%), according to the reference annotation NC_001491.2. Five out of the six remaining PAS-less TESs are assigned with non-coding antisense transcripts, and one 3′-UTR isoform. These results are included in Supplementary Tables S2, S3 and S4, along with other information regarding the transcripts.
Fig. 3.
TATA boxes and poly(A) signals (A) Genomic surrounding of TSSs with TATA box within a ±5 bp interval. The first letter of TSSs (position 0) is enriched with G/A bases, while the − 1 position contains mainly C/T bases. (B) A) Genomic surrounding of TSSs without TATA box within a ±5 bp interval. The 0 and + 1 TSS positions are enriched with G letters (C) Sequence motifs of transcripts containing polyadenylation signals. (D) Sequence motifs of transcripts without polyadenylation signals.
The LoRTIA software also determines whether a read possess a poly(A) tail, by aligning the ‘softclip’ region to a poly(A) sequence, however to further validate that the accepted TES sites are supported by transcripts of biological origin with appropriate poly(A) tails, we estimated the poly(A)-tail lengths with the nanopolish software (https://github.com/jts/nanopolish). We found that 84.4% of the two dRNA sequencing library reads possess a poly(A)-tail, according to the nanopolish model (Supplementary Table S6). These reads validated 81 TES positions (in a ±20 nt window) out of the described 83 TESs. The remaining 2 TESs are associated with antisense transcripts that had very low abundance and could only be detected in dcDNA-Seq samples.
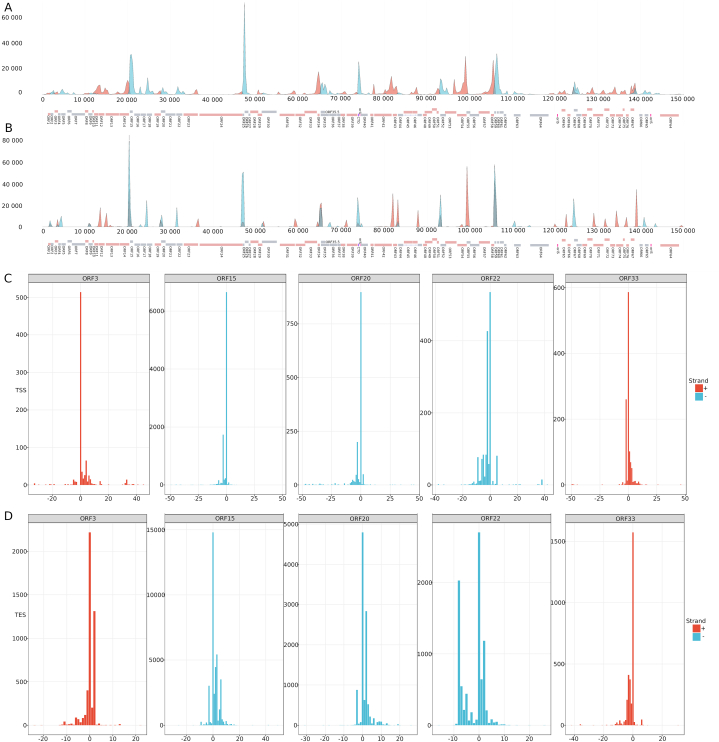
In accordance with the eukaryotic splice site consensus sequences, we identified A/C cleavage sites and U/G downstream elements at the transcripts containing PASs, while no such consensus sequences were detected in PAS-less transcripts. Fig. 4 illustrates the distribution of TSSs (Fig. 4A) and TES (Fig. 4B) along the EHV-1 genome. Here, we demonstrate that transcription starts and ends at multiple closely spaced points (typically within a ±25 bp interval), which are termed as TSS (Fig. 4C) or TES (Fig. 4D) clusters. A canonical TSS or TES is considered to be the most abundant transcript end. This study detected a high level of a TSS polymorphism in each viral transcript. The long 5′-UTR isoforms of EHV-1 appear to be longer on average and are produced in higher proportion than in other herpesviruses. In contrast to the poxviruses [40] and the baculoviruses [41], herpesvirus transcripts exhibit a low level of TES polymorphism. Our investigations confirmed this phenomenon also in EHV-1: except the case of premature termination of mRNAs and independent TESs of the upstream genes in tandem clusters (see below), the usage of alternative TSSs is also rare in EHV-1.
Fig. 4.
Transcription start and end sites. A. Genome-wide localization of TSSs. The relative amount of the TSSs is calculated from a mixed time-point sample. B. Genome-wide localization of TESs. The relative amount of the TSSs is calculated from a mixed time-point sample. C. TSS clusters illustrated by 5 examples D. TES clusters illustrated by 5 examples.
2.3. Canonical mRNAs
In this part of the work we annotated the canonical transcripts by identifying their TSS and TES clusters and splice sites (Supplementary Tables S2–S4). The canonical transcripts were defined as the highest abundance RNA isoform specified by a given protein-coding or a non-coding gene. Transcripts containing 5′-truncated ORFs were not considered in this calculation because the short RNA molecules are overrepresented due to the preference of LRS toward the sequences falling into this size range. We were able to identify canonical transcripts for every EHV-1 gene.
2.4. Identification of putative nested genes
The utilization of LRS has enabled us to distinctly differentiate between the larger host transcripts and the smaller embedded transcripts. In this phase of the study, we focused on identifying potential 5'-truncated mRNAs that lack specific upstream regions of the gene, including the canonical ATG start codon, but retain one or more downstream in-frame ATGs (refer to Fig. 2, Supplementary Figs. S1 and S2, Supplementary Tables S2–S4). These shorter open reading frames (ORFs), if translated, would result in the production of N-terminal truncated polypeptides. We refer to these ORFs as "in-frame ORFs" (ifORFs), while the genes harboring them are termed "putative nested genes," and the corresponding transcripts are referred to as "putative nested mRNAs."
The nested mRNAs also exhibit polymorphism concerning the length of their 5'-untranslated regions (UTRs), which are the coding portions of the larger host genes. The term "ifORF" is employed when the truncated in-frame ORF is detected within monocistronic genes. However, for ifORFs located within the 5'-UTR of RNAs encoded by downstream genes, it becomes challenging to determine whether these transcripts containing ifORFs represent simply long 5'-UTR isoforms of the downstream gene or if they undergo translation. This is a critical matter since, in the latter scenario, the downstream gene would not be translated. The true coding capacity of these 5'-truncated ORFs and their status as biological products are yet to be determined.
2.5. Non-coding transcripts
The non-coding RNA (ncRNA) molecules include those transcripts which do not contain functional ORFs (Supplementary Tables S2–S4). Most of the detected EHV-1 non-coding transcripts are long ncRNAs (lncRNAs) which, by definition, are made of more than 200 nucleotides. We also identified short ncRNAs (sncRNAs) although our approach is not optimal for the detection of this transcript type, especially in the case of molecules comprised of less than 50 nucleotides, such as microRNAs. The non-coding transcripts have their own promoters and are located in either intragenic, or in intergenic positions, or overlap the mRNAs in antiparallel manner.
Ten canonical antisense RNAs (asRNA) and their isoforms (altogether 27 asRNAs) were detected in this work. While asRNAs are controlled by their own promoters, antisense segments can also be part of mRNAs as a result of convergent or divergent transcriptional overlaps between adjacent or even distal antiparallel genes. We identified as70 transcript overlapping the ORF70, which is a homolog of PRV US4-AS, but we did not detect the PRV AZURE transcript that runs opposite to the us3 (homolog of ORF69) and us4 (homolog of ORF70) genes [31]. EHV-1 expresses a higher number of asRNAs than other alphaherpesviruses including PRV, its close relative. Most of the identified asRNAs have not been detected in other alphaherpesviruses. Due to the extensive transcriptional overlaps, practically, both DNA strands are transcriptionally active throughout the entire viral genome. Intriguingly, Coding Potential Calculator 2 [42] analysis gave the result that the small (average: 141.44 bp) ORFs of 9 asRNAs resemble to the coding sequences of the vertebrate organisms, therefore they might have coding potential (Supplementary Table S7).
We detected two very abundant groups of intergenic non-coding transcript, the NOIR and the CTO-S (the latter is discussed in the next section). Both transcripts have homologs in PRV (NOIR-1 and CTO-S, respectively), but not in other herpesviruses with annotated transcriptomes. Possibly, the NTO2-4 transcripts of VZV described by our research group [35] have a similar function as the NOIR transcripts, but in contrast to the NOIR, they are located within the canonical ORF62 (icp4 homolog). While in PRV, the NOIR-1 has a single splice variant, the EHV-1 homolog has two splice isoforms besides the unspliced RNA with a 88 nt difference in their splice donor site (Fig. 5A). The function of this RNA gene is completely unknown. No homolog of the low-abundance PRV noir-2 non-coding gene was detected in the EHV-1 genome.
Fig. 5.
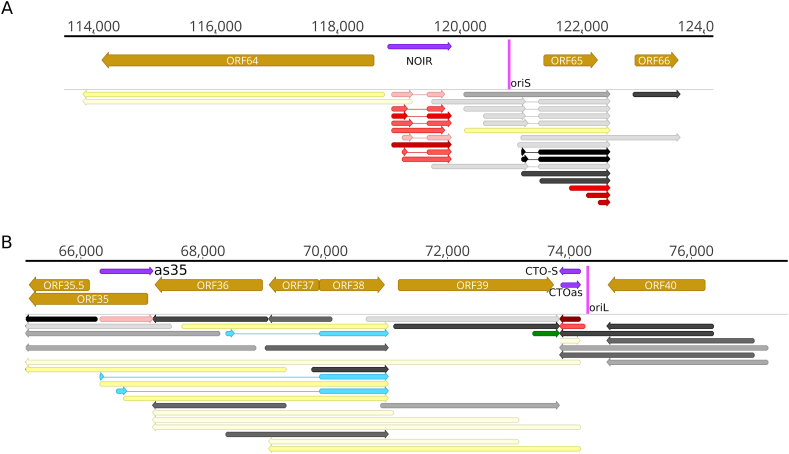
Transcription near the replication origins. In this figure, the presence of a transcript in the dRNA-Seq data was a prerequisite. A. OriS: ORF64-65 region. B. OriL: ORF35-41 region. The color code is defined in the figure. The shade of the colors corresponds to the abundance: 1: 1–9 reads, 2: 10–49 reads, 3: 50–199 reads, 4: 200–999 reads, 5: >1000 reads.
One type of intragenic ncRNAs are those ones which share their promoters with the mRNAs but lack the STOP codon due to the premature transcription termination. These transcripts are designated as ‘non-coding start’ (ncs). Similar to the mRNAs, the ‘ncs’ transcripts have the same alternative TSSs. One of the specialties of EHV-1 transcriptome is the presence of abundant ‘ncs’ transcripts, e.g., ORF13-ncs, ORF53-ncs, and ORF63-ncs. Additionally, intragenic ncRNAs without functional ORFs can be the result of 5′-truncation of mRNAs. This type of ncRNAs is termed as ‘non-coding coterminal’ (nct). Some of the monocistronic 5′-truncated transcripts discussed in the previous section may also be ‘nct’ transcripts. Since the frequency of false TSSs in short reads are higher than in longer reads [29], we accepted an ‘nct’ transcript as true if its abundance reached the 5% of the canonical mRNA into which it is embedded. The ‘non-coding start and stop’ (ncss) transcripts are also intragenic but lack both the TSS and TES of the host mRNA. An example of this transcript type is the ORF54-ncss.
2.6. Replication origin-associated transcripts
Replication origin-associated RNAs (raRNAs) are mapped near the genomic location of the replication origins (Oris). Such transcripts have been discovered in all viruses studied, including alphaherpesviruses [43]. Many raRNAs overlap the Oris, while others are terminated in their close vicinity [43]. In herpesviruses, these transcripts can either be ncRNAs, or they can be the longer TSS or TES versions of mRNAs, encoded by one or both neighboring genes [35]. Similar to PRV, EHV-1 contains a single OriL at the unique long region and two OriS at the repeat region. Like in other alphaherpesviruses, the raRNAs overlapping the OriS are the long 5′-UTR isoforms of ORF65 homologous to the us1 (icp22) gene of alphaherpesviruses (Fig. 5A). EHV-1 encodes the very abundant PRV homolog, the CTO-S transcripts from an RNA gene located near the OriL (Fig. 5B). The CTO-S transcript has very long TES isoforms being co-terminal with the ORF35, 36 and 37 genes. This transcript is a complex RNA (cxRNA) molecule because it contains genes (ORF38 and 39) with antiparallel orientation, and it is likely ncRNAs because its first ATG (of ORF 37) is too far from its TSS (Fig. 5B). A transcript, antiparallel to CTO-S, was also detected. Furthermore, the ul21 homolog of EHV-1 (ORF40) codes for a TES isoform (also termed as CTO-L), which is co-terminal with the canonical CTO-S. However, we could not detect the PRV homolog CTO-M for which the reason may be the relatively low data coverage at this region. Intriguingly, the TATA box of the longer CTO-S isoform is co-localized with the OriL. Likewise, we detected a TATA box within the OriS and identified the transcript which is likely to be controlled by this promoter element. It is possible that the NOIR transcripts have also a direct or indirect role in the regulation of replication (see Discussion for explanation).
2.7. Multigenic transcripts
Multigenic transcripts contain two or more genes on an RNA molecule. We used less stringent criteria for the identification of the TSSs of very long transcripts because of their low abundance resulted by the bias of LRS toward the short transcripts (or rather against long sequences), and therefore, the annotated TSSs of the long RNAs might be inaccurate.
2.8. Polycistronic transcripts
A characteristic feature of the organization of herpesvirus genomes is that tandem genes are transcribed as polycistronic transcripts in the following pattern: ‘abcd’, ‘bcd’, ‘cd’, ‘d’, where ‘a’ being the most upstream and ‘d’ the most downstream gene. However, in contrast to the prokaryotic polycistronic RNAs, in herpesviruses only the most upstream gene is translated. If an ifORF happens to be located within the most upstream gene of a polycistronic transcript, functional analysis is needed to determine whether it is only a long TSS isoform, or the 5′-truncated ORF is translated. A key discovery in this research is that, in EHV-1, numerous upstream genes within the tandem gene clusters possess individual TESs in addition to the shared co-terminal TESs.
2.9. Complex transcripts
Complex RNA molecules comprise more than one gene, at least one of which is oriented in a direction opposite to the others. Those transcripts in which the most upstream gene stands in antiparallel orientation are probably non-coding because of the long distance between the TSSs and the canonical ATGs. Despite this possibility we labeled them as coding transcripts with very long 5′-UTR in Fig. 2. Another distinctive feature of EHV-1 is the common occurrence of relatively abundant very long cxRNA molecules. Altogether, we identified 81 cxRNAs, which is obviously an underestimation of the real number.
2.10. Splicing, splice isoforms and fusion transcripts
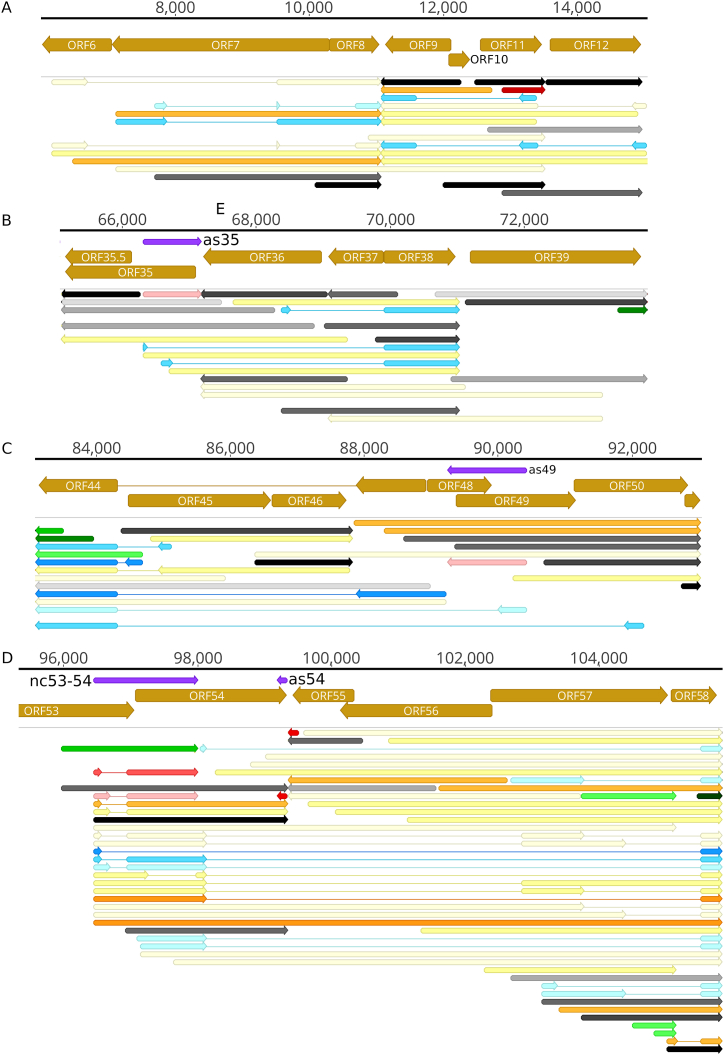
We used rigorous criteria for the identification of splice sites: besides their incidence in at least three distinct samples prepared by different techniques, we also requested the presence of splice consensus sites and the detection by dRNA-Seq. Alphaherpesviruses produce much less spliced transcripts and a lower variety of splice isoforms than other herpesviruses. However, in EHV-1, we detected complex splicing patterns even in those transcripts which are unspliced in the related viruses (Fig. 6). The most intriguing spliced transcripts are the fusion RNAs (fRNAs). One type of fRNAs utilizes the genomic segments from an 5′-UTR of adjacent or more distal genes standing in an opposite direction (e.g., ORF8 in Fig. 6A). The pre-mRNAs of these transcripts are complex transcripts. ORF8 (ul51) utilizes some parts of ORF6 and ORF7 as 5′-UTR in various combinations. Intriguingly, a truncated coding sequence of ORF8 is also produced. A similar complex splicing pattern is also observable in ORF9 (ul50). The pre-mRNAs of the fusion transcripts expressed from the ORF35-38 region (Fig. 6B) are cxRNAs because the first three genes stand in an antiparallel orientation relative to the ORF38. These transcripts contain the entire coding region of ORF38 and various 5′-UTR segments from the ORF35 and ORF36 genes. The ORF44 (ul15) gene is encoded in a special way in alphaherpesviruses: the continuity of its ORF is disrupted by two other genes (ORF45/ul17 and ORF46/ul16), which are spliced out from the mature ORF44 transcripts. The downstream exon is also expressed independently (Fig. 6C). The EHV-1 ORF44 is encoded an even more intricate manner: many of these transcripts contain a much longer intron which encompass the entire ORF48 and 49 genes and a large part of ORF50 gene. The ORF53-58 (ul9-4) region exhibits the highest complexity (Fig. 6D). In this region genes produce fusion proteins in various combinations. This genomic segment can also be used for exemplifying the 5′-truncated ORFs, the intragenic ncRNAs, the complex transcripts, and also the independent termination of the upstream members of tandem gene clusters. Many fusion results in-frame chimeric protein molecules, but in some cases the second or third exon is not at the same reading frame as the upstream exon. However, in this latter case, a close stop codon is located in the new reading frame.
Fig. 6.
Splicing and fusion transcripts. In this figure, the presence of a transcript in the dRNA-Seq data was a prerequisite. A. ORF6-12. B. ORF35-38. C. ORF44-50. D. ORF53-58. The color code is defined in the figure. The shade of the colors corresponds to the abundance: 1: 1–9 reads, 2: 10–49 reads, 3: 50–199 reads, 4: 200–999 reads, 5: >1000 reads.
2.11. Transcriptional overlaps
Gene pairs can have parallel (co-oriented; →→), convergent (→←), or divergent (←→) orientations. Transcriptional overlaps between convergent and co-oriented genes arise from transcriptional readthrough, while divergent overlaps result from the overlapping 5′-UTRs of divergently oriented gene products. Supplementary Fig. S5 demonstrates that nearly every divergent gene pair generates transcripts with extensive head-to-head overlaps. Another distinctive feature of the EHV-1 transcriptome is the presence of very long transcriptional overlaps that span multiple genes. Canonical transcripts encoded by convergent gene pairs typically do not overlap; however, they occasionally produce transcriptional read-throughs ('soft' overlap). The ORF 29/30 (ul31-30) gene pair is an exception, as their canonical transcripts overlap ('hard' overlap), which is consistent with other alphaherpesviruses.
3. Discussion
The past decade has seen a rapid progress of sequencing technologies. Third-generation LRS approaches led to a paradigm shift in genome and transcriptome research, especially in small-genome organisms [23,44]. The transcriptomic architecture of viruses proved to be much more complex than previously expected [17]. A large variety of overlapping transcripts have been discovered [31,34,35,45].
A recent study by Torma et al. [31] has revealed that the presence of nested genes within larger canonical genes is more prevalent in viruses than previously believed. The use of the SRS technique has proven to be ineffective in detecting these nested mRNAs, which is the reason they remained unnoticed in earlier studies. Ribosome profiling and other genome-wide translational analyses are needed to perform for confirming the translation of this transcript type. Despite the growing number of identified lncRNAs, their functional mechanisms remain poorly understood. These RNAs were previously dismissed as mere "transcriptional noise" due to their lack of protein-coding capacity. However, emerging evidence implies that they participate in a wide range of functions through distinct pathways, as indicated by Statello et al. [46].
In our study, we discovered various ncRNAs, including transcripts located within and between genes, as well as asRNAs. The asRNAs are encoded by the complementary DNA strands of protein-coding genes and are regulated by their own promoters. Additionally, we found that mono- and polycistronic transcripts, as well as cxRNAs, can also contain antisense regions. These regions are formed either through transcriptional read-through events between convergent gene pairs or through overlapping regions between divergent neighboring or distal genes. Notably, we identified 27 asRNAs, which is a higher number compared to what has been previously described in related viruses.
It has been shown that 72% of mammalian replication origin-associated transcripts are controlled by active promoters [47]. Similar raRNA molecules have also been detected in viruses [17]. In human BK polyomavirus, it has been demonstrated that the presence of an raRNA specimen leads to a substantial suppression of virus replication. This suppression is achieved by the raRNA binding to both the sense and antisense DNA strands within the Ori region, thereby interfering with the synthesis of RNA primers required for replication [48]. These Ori-associated transcripts undergo rapid evolution even within alphaherpesviruses. In alphaherpesviruses, the position of OriS remains conserved, residing between the icp4 and us1 genes. However, the presence of OriL varies among different viruses. For instance, in VZV and BoHV-1, OriL is either absent or not detected. In other alphaherpesviruses such as HSV-1, OriL is found between the ul29 and ul30 genes, while in PRV and EHV-1, it is located in different genomic position (between the ul21 and ul22 genes). The raRNAs of OriS include transcripts that are the long TSS isoforms of US1 transcripts, which overlap the origin of replication, or are initiated closely to it (in all alphaherpesviruses). The raRNAs of OriL include the long TES isoform of UL21 transcripts, and the CTO-S ncRNAs mapped downstream of the ul21 gene (in PRV and EHV-1). We observed that promoter elements of viral transactivator genes are co-localized with both OriS and OriL (such as in mammalian cells: Dellino et al. [47]), which suggests a co-regulation of the initiation of transcription and replication. The ori-overlapping RNAs might regulate the later phases of replication. The precise function of raRNAs of alphaherpesviruses, however, remains to be ascertained.
The EHV-1 NOIR represents an intriguing group of ncRNAs. Its homolog (NOIR-1) has been described in PRV [23], but no convergent partner (NOIR-2 in PRV) was detected in EHV-1. The canonical form of these EHV-1 transcripts overlaps both the long 5′-UTR isoforms of ORF64 (rs1/icp4 of HSV-1) and ORF65 (us1/icp22 of HSV-1). We can speculate that the process of transcription and/or the transcripts themselves might affect the activity of these transcription factor genes, which, if it does, would have a role in the control of genome-wide gene expression and also in DNA replication. In other words, the icp4-us1 genomic region of alphaherpesviruses might be the center for the viral regulatory mechanisms: the viral transcription factors and the ncRNAs at this locus might coordinate the onset and progression of both the replication and the global gene expression by physical interactions of their apparatuses, including collision, as well as competition for the promoters and Oris. The raRNAs are very likely not only by-products of an interference-based mechanism, but they also have function through e.g., forming a DNA-RNA hybrid at the Ori region [49].
Our results show that splicing events in EHV-1 are more frequent than in the related alphaherpesviruses. ORF44 (ul15), ORF65 (us1) and NOIR transcripts are also spliced in other alphaherpesviruses. However, splicing at other genomic regions (ORF6/12, ORF35/39, ORF53/58) is unique in EHV-1. Furthermore, the splicing events in ORF44 in EHV-1 are extended to the adjacent genomic regions including ORF49/50, which is also unique.
We detected relatively abundant fusion transcripts, some of which encode chimeric proteins in various exon combinations. Other fusion transcripts are the results of the combination of 5′-UTR sequences of one or more upstream genes with complete or 5′-truncated form of ORF of one or more downstream genes. The 5′-UTR sequences of the fusion transcripts in many cases are derived from the antiparallel strand of upstream genes. Low-abundance fusion genes in alphaherpesviruses have also been described by others [24].
Polycistronism, the phenomenon of encoding multiple genes within a single mRNA molecule, is widely seen in bacteria and viruses but is extremely uncommon in eukaryotic organisms. In prokaryotes and bacteriophages, the presence of the Shine-Dalgarno sequence in the mRNA allows for the translation of multiple genes within polycistronic RNA molecules. However, small-genome eukaryotic viruses have evolved various strategies to overcome this hurdle, including the use of ribosomal frameshifting, internal ribosome entry sites (IRES), or leaky ribosomal scanning, as detailed by Stacey et al. [50].
In herpesviruses, genes arranged in the same direction tend to be organized into gene clusters. These clusters produce transcripts that share common downstream sequences but possess distinct 5'-exons. The configuration of these transcripts adheres to a pattern: 'abcd', 'bcd', 'cd', and 'd', where 'a' signifies the most upstream gene and 'd' represents the most downstream gene. The precise function of multigenic transcripts in large DNA viruses remains uncertain, primarily because, with a few exceptions (such as uORFs, which are translated alongside the canonical ORFs, as described by Vilela et al., [51]; Kronstad et al. [52]), translation from the downstream genes has not been extensively documented.
Polycistronic EHV-1 RNAs encoded by tandem genes represent parallel overlaps. In other alphaherpesviruses, the majority of upstream genes of tandem gene clusters do not produce monocistronic transcripts, or if so, these RNA molecules are expressed in very low abundance. We found a more extensive use of alternative TESs by the upstream genes of EHV-1. Furthermore, EHV-1 produces bicistronic RNA molecules containing the two upstream genes of a tricistronic tandem gene cluster, which is also unique alphaherpesviruses.
Similar to PRV and HSV-1, we detected a ‘hard’ overlap between the ORF 29/30 (ul31/30), but not between ORF54/55 [such as in PRV: ul8/7 (Tombácz et al. [23])] or ORF58/60 [such as in HSV-1: ul4/3 (Tombácz et al. [34])]. We did not observe an increased extent of convergent overlaps and readthroughs, however, the divergent overlaps were found to be more extensive in EHV-1 than in other alphaherpesviruses. This genomic layout implies the possibility of a widespread transcriptional interference, resulting from the clash and/or rivalry of transcription machinery, which introduces a new layer of genetic regulation [53]. The influence of genomic context on the regulation of gene expression has recently been elucidated in yeast [54,55].
3.1. Limitations of the study
One of the limitations of this study is that although the sequencing reads cover the entire EHV-1 genome, at certain loci the coverage is in insufficient for the precise annotation of the given genomic segment. However, it is not a critical problem in transcript identification. Another limitation of our approach is that the applied method is not optimal for the identification of sncRNAs (especially of microRNAs), and also of very long lncRNAs, therefore it does not provide a complete atlas of EHV-1 transcriptome. Additionally, some of the low-abundance transcripts may have gone undetected due to the given level of read coverage. Finally, LRS is biased toward 200–600 bp transcription reads, which therefore produces relatively large read coverage at this size range. Although, LoRTIA software suit filters out false transcripts, we cannot exclude that at this size range some of the identified TSSs and transcripts are non-biological but represent mere technical artifacts. To avoid the identification of false TSSs, we implemented extremely rigorous criteria for the annotations. This led to a substantial decrease in the complexity of the transcriptome. The category of nested genes exists since many such RNA molecules encoded by them have already been detected. Each novel putative embedded mRNA has to be individually analyzed.
4. Materials and Methods
4.1. Cells and viruses
In this study we used a field isolate equid alphaherpesvirus 1 (EHV-1) strain MdBio (EHV-1-MdBio), which was isolated from the organs of an aborted colt fetus in the 1980’s at Marócpuszta (Hungary). The virus was cultured in confluent rabbit kidney (RK-13) epithelial cells (ECACC: 00021715). The cells were grown in DMEM supplemented with 10% fetal calf serum and 80 μg/ml of gentamycin at 37 °C with 5% CO2. To prepare the virus stock solution, cells were infected with an MOI (multiplicity of infection) of 0.1. The viral infection was let to continue until a full cytopathic effect was observed. Afterward, the infected cells underwent three consecutive cycles of freezing and thawing to destroy the structure of the cells and release the viruses. For the sequencing reactions, the same cell line was infected with EHV-1-MdBio using an MOI of 4. This process was performed using three technical replicates. To synchronize gene expressions, the infected cells were first incubated at 4 °C for 1 h. Next, the virus suspension was eliminated, and the cells were washed with phosphate-buffered saline. Subsequently, the infected cells were supplemented with fresh culture medium and then incubated for different durations. Once the incubation period was completed, the culture medium was removed, and the samples were frozen at −80 °C for subsequent use.
4.2. RNA isolation
To purify RNA from the cells, we utilized the NucleoSpin RNA kit (Macherey-Nagel). The infected cells were incubated with a lysis buffer provided in the kit. Subsequently, the RNA molecules were bound to a silica membrane. To eliminate any remaining genomic (g)DNA contaminants, DNase I treatment was employed. The purified RNA samples were then eluted using RNase-free water. Furthermore, to eliminate potential residual gDNA contamination, we carried out an extra DNase treatment using the TURBO DNA-free™ Kit (Invitrogen). The concentration of the RNA samples was measured using Qubit 4.0 fluorometer (Invitrogen) and Qubit Broad Range RNA Assay Kit (Invitrogen) were used for measuring the concentration of RNA solutions. For quality control purposes, the Agilent TapeStation 4150 was utilized. Samples with RIN (RNA Integrity Number) scores equal to or greater than 9.2 were selected for cDNA production in subsequent steps.
4.3. Purification of polyadenylated RNA
To isolate the poly(A)+ fraction of RNA fraction of the total RNA samples, we employed the Oligotex mRNA Mini Kit from Qiagen. Here's a summary of the procedure: Initially, 250 μL of diluted total RNA sample was combined with 50 μL of Oligotex suspension and 250 μL of OBB buffer, both provided in the Qiagen kit. The mixture was then heated to 70 °C and incubated for 3 min, followed by cooling to 25 °C for 10 min. Subsequently, centrifugation at 14,000×g for 2 min was performed, and the supernatant was carefully removed. To the remaining pellet, 400 μL of OW2 wash buffer (from the kit) was added. The solution was then loaded onto the spin columns provided in the Oligotex kit. After two centrifugation steps (at 14,000×g for 1 min each), the polyadenylated RNA fraction bound to the membrane was eluted by adding 50 μL of hot elution buffer (EB) from the Qiagen kit. The RNA was extracted and eluted using a volume of 60 μL of EB. In order to optimize the yield, we performed a second elution step.
4.4. rRNA removal
To identify the potential non-polyadenylated RNAs, we utilized the Ribo-Zero Magnetic Kit H/M/R from Epicentre/Illumina. The following steps were performed: First, 5 μg of a total RNA mixture of was combined with Ribo-Zero Reaction Buffer and Ribo-Zero rRNA Removal Solution. Next, the mixture was kept at 68 °C for 10 min, then cooled to room temperature (RT) and incubated for 5 min. Subsequently, the mixture was added to washed Magnetic Beads (225 μl, provided in the kit). After a brief vortexing and incubation at RT for 5 min, the sample was heated to 50 °C for 5 min. The mixture was then placed on a magnetic stand, allowing the Magnetic Beads to capture the rRNA and form a pellet while the supernatant containing the purified RNAs was collected, then further purified using the AMPure XP Bead washing method (Beckman Coulter).
4.5. Treatment with terminator enzyme
In order to enrich the full-length RNA molecules in a poly(A)+ RNA mixture, we employed Terminator™ 5'-Phosphate-Dependent Exonuclease (Lucigen). The following components were added to the RNA mixture: 10X Reaction Buffer A, RiboGuard RNase Inhibitor, and one unit from the Terminator Exonuclease. The mixture was then kept at 30 °C for 60 min. One μL 100 mM EDTA (pH 8.0) was added to terminate the reaction. Finally, the samples were purified using Agencourt RNAClean XP beads (Beckman Coulter).
4.6. Illumina MiSeq short-read sequencing
The complete transcriptome of EHV-1 was sequenced using a short-read sequencing method. To achieve this, a mixture derived from rRNA-depleted- and poly(A)+ RNA samples was utilized, along with the NEXTflex® Rapid Directional qRNA-Seq Kit (PerkinElmer). The sequencing protocol involved the following steps: (1) RNA fragmentation: The RNA samples were fragmented using the NEXTflex® RNA Fragmentation Buffer through an enzymatic reaction at 95 °C for 10 min (2) First cDNA strand synthesis: the primer provided by the company was used to generate the first cDNA strand. The mixture was kept at 65 °C for 5 min and then cooled down on ice. (3) RT: the buffer and the Rapid Reverse Transcriptase enzyme were added to the sample and then kept at RT. The reaction conditions included incubation at 25 °C for 10 min, followed by 50 °C for 50 min, and termination at 72 °C for 15 min (4) Second cDNA strand synthesis: We used the solution provided by the company for the synthesis of the second cDNA strand. The reaction was carried out at 16 °C for 60 min (5) Polyadenylation: The obtained double-stranded cDNAs were polyadenylated using the NEXTflex® Adenylation Mix. The adenylation reaction took place at 37 °C for 30 min and was terminated by warming the samples to 70 °C and incubated them for 5 min at this temperature. (6) Ligation of Molecular Index Adapters: The NEXTflex® Ligation Mix was used to ligate Molecular Index Adapters to the samples at 30 °C for 10 min (7) PCR amplification: The ligated cDNAs were subjected to PCR amplification. This involved the addition of PCR Master Mix, qRNA-Seq Universal forward primer, and qRNA-Seq Barcoded Primer (sequence: AACGCCAT, all from the NEXTflex® kit). The PCR protocol details can be found in Supplementary Table S8. AMPure XP Beads were used for purification after each enzymatic step, and the final elution was performed using the buffer from the NEXTflex® Kit. (8) Library loading and sequencing: A library mix with a concentration of 10 pM was loaded onto the reagent cassette for paired-end transcriptome sequencing. The sequencing was conducted using the MiSeq Reagent Kit v2 (300 cycles).
Quantification of the library was performed using a Qubit 4.0 fluorometer and the Qubit dsDNA HS Assay kit. Sample quality was evaluated using an Agilent TapeStation device and the Agilent High Sensitivity D1000 ScreenTape. The average size of the fragments was 420 bp.
4.7. ONT MinION – dcDNA sequencing
To perform direct cDNA sequencing, we utilized the ONT Direct cDNA Sequencing Kit (SQK-DCS109) following the recommendations provided in the kit's manual. The sequencing was conducted on both poly(A)+-enriched samples and poly(A)+-enriched samples treated with Terminator. Here is a summary of the protocol: (1) RNA preparation: The RNAs were mixed with the VN primer (VNP) from the ONT kit and 10 mM dNTPs. The mixtures were then incubated at 65 °C for 5 min (2) RT: 5x RT Buffer, RNaseOUT (Thermo Fisher Scientific), and Strand-Switching Primer (ONT) Kit were added To the RNA mixture. The samples were heated to 42 °C and kept for 2 min (3) First cDNA strand synthesis: Maxima H Minus Reverse Transcriptase enzyme (Thermo Fisher Scientific) was used to synthesize the first cDNA strands. The RT and strand-switching reactions were carried out at 42 °C for 90 min. The reactions were stopped by elevating the temperature to 85 °C for 5 min (4) RNA removal: RNase Cocktail Enzyme Mix (Thermo Fisher Scientific) was applied to remove RNA from the RNA-cDNA hybrids. The samples were incubated at 37 °C for 10 min (5) Second cDNA strand synthesis: The LongAmp Taq Master Mix from New England Biolabs (NEB) and PR2 Primer were used for the second cDNA strand synthesis. PCR reactions were performed according to the details provided in Supplementary Table S8. (6) End-repair and dA-tailing: The fragmented DNAs treated by the NEBNext End repair/dA-tailing Module (NEB). The reactions were carried out at 20 °C for 5 min followed by incubation at 65 °C for 5 min (7) Adapter ligation: The sequencing adapter was ligated to the samples at RT for 10 min using the NEB Blunt/TA Ligase Master Mix (NEB). ONT dcDNA libraries were labeled using barcodes (Supplementary Table S9) from the ONT Native Barcoding (12) Kit as recommended by the supplier. (8) Library preparation: The adapted and tethered cDNA libraries were purified and loaded onto ONT R9.4.1 SpotON Flow Cells. A total of five Flow Cells were used for dcDNA sequencing. To avoid potential ‘barcode hopping’, samples from earlier time points were sequenced separately from the later time points. (9) Purification and quantification: enzymatic treatments were followed by using AMPure XP Beads for sample purification. The samples were eluted in UltraPure™ nuclease-free water (Invitrogen). The concentration of the samples was measured as described in Supplementary Table S10.
4.8. ONT MinION– dRNA sequencing
To mitigate potential errors associated with RT and PCR, the direct RNA sequencing technique was employed for library preparation. This approach is considered the "gold standard" for detecting and validating novel splice variants and 3'-UTR isoforms. Two RNA mixtures were used: 1) standard Poly(A)+ RNA and 2) Poly(A)+ RNA treated with Terminator enzyme. For library preparation, the RNA mixtures were mixed with RT Adapter (oligo dT-containing T10 adapter), RNA CS (used for sequencing quality monitoring), NEBNext Quick Ligation Reaction Buffer, and T4 DNA ligase. The mixture was incubated at RT for 10 min. RT reactions were carried out using dNTPs, 5x first-strand buffer, DTT, and UltraPure™ DNase/RNase-Free water. SuperScript III enzyme was added to the samples, and the RTs were performed at 50 °C for 50 min, followed by termination at 70 °C for 10 min. Next, the samples were mixed with RNA adapter (RMX; ONT kit), ligation buffer, T4 DNA ligase, and nuclease-free water. The ligation was performed at RT for 10 min. To clean the RNA-cDNA hybrids, Agencourt RNAClean XP Beads were used, and AMPure XP Beads were applied after each additional enzymatic step. The samples were eluted in nuclease-free water between the reactions, and ONT's elution buffer was used for the last elution. Following library preparation, nucleic acid concentrations were measured using Qubit (as shown in Supplementary Table S11). Subsequently, 100 fmol of the library samples was loaded onto two MinION Flow Cells for sequencing.
4.9. Read Pre-Processing and Data Analysis
Raw data were first basecalled using Guppy v3.4.5. The reads were aligned to the reference genome (accession number: NC_001491.2) using the minimap2 program with the following option: Y -C5 -ax splice –cs. SeqTools was used for the identification of promoter elements and for the assembly of basic statistics (https://github.com/moldovannorbert/seqtools).
The LoRTIA tool, created by our research group, was used for the identification of TSS, TES and intron (‘features’) and for transcript annotation (https://github.com/zsolt-balazs/LoRTIA, v.0.9.9). The pipeline first searches for sequencing adapters and homopolymer A-s, and removes spurious reads generated by RNA degradation, template switching or false priming. This first part of the workflow was conducted using the following parameters: Samprocessor. py --five_adapter GCTGATATTGCTGGG --five_score 14 --check_in_soft 15 --three_adapter AAAAAAAAAAAAAAA --three_score 14 input output.
Next, the workflow identifies potential TESs and TSSs. Since TES positions are usually have a wobble, we increased this value to 20, while the wobble for TSSs was set to the default (10) value. The second part of the workflow was thus carried out using the following parameters for each ’sam’ file: Stats. py -r genome -f r5 -b 10 and Stats. py -r genome -f l5 -b 10 for the TSS detection and Stats. py -r genome -f r3 -b 20; while Stats. py -r genome -f r3 -b 20 and Stats. py -r genome -f l3 -b 20 for TES detection; and Stats. py -r genome -f in for intron detection.
The third part of the workflow, which summarizes the detected potential features and estimates their significance against the Poisson distribution (correcting the p-value is using the Bonferroni method) was carried out using the following parameters: Gff_creator.py -s poisson -o.
A splice site was considered valid if it contained the canonical GT/AG or GC/AG splice junction sequences and if it appeared in a minimum of four independent reads, with at least one of those reads originating from the dRNA-Seq results. In order to be classified as a genuine transcript, a sequencing read had to encompass previously annotated TSSs and TESs. Among the transcripts derived from a viral gene, the most prevalent one was regarded as the canonical RNA, while other transcript isoforms resulting from alternative TSS, TES, or splice variants were considered as lower abundance variants (Fig. 2, Supplementary Figs. S1 and S2, Supplementary Tables S2–S4). For the annotation of putative transcripts with a size of 2 kbps or less, we introduced the following additional criteria: the proportion of such transcripts had to reach at least 50% of the canonical RNAs and they had to be detected by both (regular and terminator-based) dRNA-Seq techniques. Moreover, putative TSSs within a ± 200 bps interval are not regarded as separate transcript ends for any putative transcripts independently of its size.
Finally, the transcript annotator module was performed, which annotates transcripts by assigning the validated features (TSSs, TESs and introns) to each read, with the following parameters: Transcript_Annotator_two_wobbles.py -z 20 -a 10.
In order to validate that the reads that were used for transcript annotation by LoRTIA are not the products of internal priming events, we used talon_label_reads from the TALON software package [38] (with default parameters). This script flags those reads wherein the percent of As are more than 50% in a stretch of 20 nt-s after the alignment on the reference genome.
The nanopolish (https://github.com/jts/nanopolish) software was used with default parameters to estimate poly(A)-tails on the dRNA and terminator enzyme-treated dRNA sequencing reads (direct cDNA data is not supported by this program). The primer used in dRNA sequencing is a sticky-ended double-stranded primer that can attach only to the poly(A) tail of the mRNA molecule. Consequently, only in the case where the mRNA is broken in an A-rich region, can mRNAs with false priming events be sequenced – the chance of which is very low indeed. Thus, this method can be used to validate TES sites.
Coding potential estimation on the annotated transcripts was carried out using Coding Potential Calculator 2 [42] using default parameters.
The Illumina reads were processed using the TrimGalore software using the following options: paired –length 20 –quality 30 (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/), which was followed by mapping them the reference genome using the STAR 2.7.10a software. BAM file were visualized using the Geneious Prime 2022.0.2 (https://www.geneious.com) and IGV [56] software.
Author contribution statement
Dóra Tombácz: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Gábor Torma, Gábor Gulyás, Ádám Fülöp, Balázs Kakuk: Analyzed and interpreted the data; Wrote the paper.
Ákos Dörmő, István Prazsák, Zsolt Csabai, Máté Mizik, Ákos Hornyák: Performed the experiments; Wrote the paper.
Zoltán Zádori: Conceived and designed the experiments; Performed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Zsolt Boldogkői: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Data availability statement
The sequencing datasets generated in this study are available at the European Nucleotide Archive under the accession: PRJEB52190.
Funding statement
The funding for this study was provided by the National Research, Development and Innovation Office through grants K 128247 and 142674 to ZB and FK 128252 and 142676 to DT. The publication fee (APC), was covered by the University of Szeged Open Access Fund under grant number 5758. The funders played no role in the design of the study, data collection and interpretation, or the decision to submit the work for publication.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e17716.
Contributor Information
Dóra Tombácz, Email: tombacz.dora@med.u-szeged.hu.
Gábor Torma, Email: torma.gabor@med.u-szeged.hu.
Gábor Gulyás, Email: gulyas.gabor@med.u-szeged.hu.
Ádám Fülöp, Email: fulop.adam@med.u-szeged.hu.
Ákos Dörmő, Email: dormo.akos@med.u-szeged.hu.
István Prazsák, Email: prazsak.istvan@med.u-szeged.hu.
Zsolt Csabai, Email: csabai.zsolt@med.u-szeged.hu.
Máté Mizik, Email: mizik.mate@med.u-szeged.hu.
Ákos Hornyák, Email: akos1526@gmail.com.
Zoltán Zádori, Email: zadori.zoltan@vmri.hu.
Balázs Kakuk, Email: kakuk.balazs@med.u-szeged.hu.
Zsolt Boldogkői, Email: boldogkoi.zsolt@med.u-szeged.hu.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
figs2.
figs3.
figs4.
figs5.
References
- 1.O’Callaghan DJ H.R. Academic Press, Harcourt Brace & Company; San Diego, CA: 1994. Encyclopedia of Virology. [Google Scholar]
- 2.Oladunni F.S., Horohov D.W., Chambers T.M. EHV-1: a constant threat to the horse industry. Front. Microbiol. 2019;10 doi: 10.3389/fmicb.2019.02668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carroll C.L., Westbury H.A. Isolation of equine herpesvirus 1 from the brain of a horse affected with paresis. Aust. Vet. J. 1985;62:345–346. doi: 10.1111/j.1751-0813.1985.tb07660.x. [DOI] [PubMed] [Google Scholar]
- 4.Allen G.P., Bryans J.T. Molecular epizootiology, pathogenesis, and prophylaxis of equine herpesvirus-1 infections. Prog. Vet. Microbiol. Immunol. 1986;2:78–144. http://www.ncbi.nlm.nih.gov/pubmed/2856183 [PubMed] [Google Scholar]
- 5.Patel J.R., Heldens J. Equine herpesviruses 1 (EHV-1) and 4 (EHV-4) – epidemiology, disease and immunoprophylaxis: a brief review. Vet. J. 2005;170:14–23. doi: 10.1016/j.tvjl.2004.04.018. [DOI] [PubMed] [Google Scholar]
- 6.Roizmann B., Desrosiers R.C., Fleckenstein B., Lopez C., Minson A.C., Studdert M.J. The familyHerpesviridae: an update. Arch. Virol. 1992;123:425–449. doi: 10.1007/BF01317276. [DOI] [PubMed] [Google Scholar]
- 7.Telford E.A.R., Watson M.S., McBride K., Davison A.J. The DNA sequence of equine herpesvirus-1. Virology. 1992;189:304–316. doi: 10.1016/0042-6822(92)90706-U. [DOI] [PubMed] [Google Scholar]
- 8.Allen G K., Kydd J., Slater J., Smith . Oxford University Press; Cape Town: 2004. Infectious Diseases of Livestock, Oxford. 228–232. [Google Scholar]
- 9.Paillot R., Case R., Ross J., Newton R., Nugent J. Equine herpes virus-1: virus, immunity and vaccines. Open Vet. Sci. J. 2008;2:68–91. doi: 10.2174/1874318808002010068. [DOI] [Google Scholar]
- 10.Frampton A.R., Stolz D.B., Uchida H., Goins W.F., Cohen J.B., Glorioso J.C. Equine herpesvirus 1 enters cells by two different pathways, and infection requires the activation of the cellular kinase ROCK1. J. Virol. 2007;81:10879–10889. doi: 10.1128/JVI.00504-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Osterrieder N. Construction and characterization of an equine herpesvirus 1 glycoprotein C negative mutant. Virus Res. 1999;59:165–177. doi: 10.1016/S0168-1702(98)00134-8. [DOI] [PubMed] [Google Scholar]
- 12.Caughman G.B., Staczek J., O’Callaghan D.J. Equine herpesvirus type 1 infected cell polypeptides: evidence for immediate early/early/late regulation of viral gene expression. Virology. 1985;145:49–61. doi: 10.1016/0042-6822(85)90200-4. [DOI] [PubMed] [Google Scholar]
- 13.Kim S.K., Jang H.K., Albrecht R.A., Derbigny W.A., Zhang Y., O’Callaghan D.J. Interaction of the equine herpesvirus 1 EICP0 protein with the immediate-early (IE) protein, TFIIB, and TBP may mediate the antagonism between the IE and EICP0 proteins. J. Virol. 2003;77:2675–2685. doi: 10.1128/JVI.77.4.2675-2685.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Derbigny W.A., Kim S.K., Jang H.K., O’Callaghan D.J. EHV-1 EICP22 protein sequences that mediate its physical interaction with the immediate-early protein are not sufficient to enhance the trans-activation activity of the IE protein. Virus Res. 2002;84:1–15. doi: 10.1016/S0168-1702(01)00377-X. [DOI] [PubMed] [Google Scholar]
- 15.Kim S.K., Ahn B.C., Albrecht R.A., O’Callaghan D.J. The unique IR2 protein of equine herpesvirus 1 negatively regulates viral gene expression. J. Virol. 2006;80:5041–5049. doi: 10.1128/JVI.80.10.5041-5049.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith R.H., Caughman G.B., O’Callaghan D.J. Characterization of the regulatory functions of the equine herpesvirus 1 immediate-early gene product. J. Virol. 1992;66:936–945. doi: 10.1128/jvi.66.2.936-945.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boldogkői Z., Moldován N., Balázs Z., Snyder M., Tombácz D. Long-read sequencing – a powerful tool in viral transcriptome research. Trends Microbiol. 2019;27:578–592. doi: 10.1016/j.tim.2019.01.010. [DOI] [PubMed] [Google Scholar]
- 18.Workman R.E., Tang A.D., Tang P.S., Jain M., Tyson J.R., Razaghi R., Zuzarte P.C., Gilpatrick T., Payne A., Quick J., Sadowski N., Holmes N., de Jesus J.G., Jones K.L., Soulette C.M., Snutch T.P., Loman N., Paten B., Loose M., Simpson J.T., Olsen H.E., Brooks A.N., Akeson M., Timp W. Nanopore native RNA sequencing of a human poly(A) transcriptome. Nat. Methods. 2019;16:1297–1305. doi: 10.1038/s41592-019-0617-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marx V. Long road to long-read assembly. Nat. Methods. 2021;18:125–129. doi: 10.1038/s41592-021-01057-y. [DOI] [PubMed] [Google Scholar]
- 20.Laver T., Harrison J., O’Neill P.A., Moore K., Farbos A., Paszkiewicz K., Studholme D.J. Assessing the performance of the Oxford nanopore technologies MinION. Biomol. Detect. Quantif. 2015;3:1–8. doi: 10.1016/j.bdq.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rhoads A., Au K.F. PacBio sequencing and its applications, genomics. Proteomics Bioinforma. 2015;13:278–289. doi: 10.1016/j.gpb.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Irimia M., Weatheritt R.J., Ellis J.D., Parikshak N.N., Gonatopoulos-Pournatzis T., Babor M., Quesnel-Vallières M., Tapial J., Raj B., O’Hanlon D., Barrios-Rodiles M., Sternberg M.J.E., Cordes S.P., Roth F.P., Wrana J.L., Geschwind D.H., Blencowe B.J. A highly conserved program of neuronal microexons is misregulated in autistic brains. Cell. 2014;159:1511–1523. doi: 10.1016/j.cell.2014.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tombácz D., Csabai Z., Oláh P., Balázs Z., Likó I., Zsigmond L., Sharon D., Snyder M., Boldogkői Z., Boldogkoi Z. Full-length isoform sequencing reveals novel transcripts and substantial transcriptional overlaps in a herpesvirus. PLoS One. 2016;11:1–29. doi: 10.1371/journal.pone.0162868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Braspenning S.E., Sadaoka T., Breuer J., G.M.G.M.G.M. Verjans, W.J.D.D. Ouwendijk. Depledge D.P., Depledge Are Co-Senior D.P., Shenk T. Decoding the architecture of the varicella-zoster virus transcriptome. mBio. 2020;11:1–19. doi: 10.1128/mBio.01568-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balázs Z., Tombácz D., Csabai Z., Moldován N., Snyder M., Boldogkői Z. Template-switching artifacts resemble alternative polyadenylation. BMC Genom. 2019;20:824. doi: 10.1186/s12864-019-6199-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo G.X., Taylor J. Template switching by reverse transcriptase during DNA synthesis. J. Virol. 1990;64:4321–4328. doi: 10.1128/jvi.64.9.4321-4328.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cocquet J., Chong A., Zhang G., Veitia R.A. Reverse transcriptase template switching and false alternative transcripts. Genomics. 2006;88:127–131. doi: 10.1016/j.ygeno.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 28.Workman R.E., Myrka A.M., Wong G.W., Tseng E., Welch K.C., Timp W. Single-molecule, full-length transcript sequencing provides insight into the extreme metabolism of the ruby-throated hummingbird Archilochus colubris. GigaScience. 2018;7 doi: 10.1093/gigascience/giy009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moldován N., Torma G., Gulyás G., Hornyák Á., Zádori Z., Jefferson V.A., Csabai Z., Boldogkői M., Tombácz D., Meyer F., Boldogkői Z. Time-course profiling of bovine alphaherpesvirus 1.1 transcriptome using multiplatform sequencing. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-77520-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oláh P., Tombácz D., Póka N., Csabai Z., Prazsák I., Boldogkői Z. Characterization of pseudorabies virus transcriptome by Illumina sequencing. BMC Microbiol. 2015;15:130. doi: 10.1186/s12866-015-0470-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Torma G., Tombácz D., Csabai Z., Göbhardter D., Deim Z., Snyder M., Boldogkői Z. An integrated sequencing approach for updating the pseudorabies virus transcriptome. Pathogens. 2021;10:242. doi: 10.3390/pathogens10020242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Grady T., Wang X., Höner zu Bentrup K., Baddoo M., Concha M., Flemington E.K., Höner zu Bentrup K., Baddoo M., Concha M., Flemington E.K. Global transcript structure resolution of high gene density genomes through multi-platform data integration. Nucleic Acids Res. 2016;44:1–17. doi: 10.1093/nar/gkw629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Balázs Z., Tombácz D., Szűcs A., Snyder M., Boldogkői Z. Long-read sequencing of the human cytomegalovirus transcriptome with the Pacific Biosciences RSII platform. Sci. Data. 2017;4 doi: 10.1038/sdata.2017.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tombácz D., Torma G., Gulyás G., Moldován N., Snyder M., Boldogkői Z. Meta-analytic approach for transcriptome profiling of herpes simplex virus type 1. Sci. Data. 2020;7:1–11. doi: 10.1038/s41597-020-0558-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prazsák I., Moldován N., Balázs Z., Tombácz D., Megyeri K., Szűcs A., Csabai Z., Boldogkői Z. Long-read sequencing uncovers a complex transcriptome topology in varicella zoster virus. BMC Genom. 2018;19:873. doi: 10.1186/s12864-018-5267-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zarski L.M., Weber P.S.D., Lee Y., Soboll Hussey G. Transcriptomic profiling of equine and viral genes in peripheral blood mononuclear cells in horses during equine herpesvirus 1 infection. Pathogens. 2021;10:43. doi: 10.3390/pathogens10010043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Balázs Z., Tombácz D., Csabai Z., Moldován N., Snyder M., Boldogkoi Z. Template-switching artifacts resemble alternative polyadenylation. BMC Genom. 2019;20:1–10. doi: 10.1186/s12864-019-6199-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wyman D., Balderrama-Gutierrez G., Reese F., Jiang S., Rahmanian S., Forner S., Matheos D., Zeng W., Williams B., Trout D., England W., Chu S.-H., Spitale R.C., Tenner A.J., Wold B.J., Mortazavi A. A technology-agnostic long-read analysis pipeline for transcriptome discovery and quantification. bioRxiv. 2020 https://www.biorxiv.org/content/10.1101/672931v2%0Ahttps://www.biorxiv.org/content/10.1101/672931v2.abstract [Google Scholar]
- 39.Huang L., Zhu Y., Anders D.G. The variable 3’ ends of a human cytomegalovirus oriLyt transcript (SRT) overlap an essential, conserved replicator element. J. Virol. 1996;70:5272–5281. doi: 10.1128/jvi.70.8.5272-5281.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tombácz D., Prazsák I., Torma G., Csabai Z., Balázs Z., Moldován N., Dénes B., Snyder M., Boldogkői Z. Time-Course transcriptome profiling of a poxvirus using long-read full-length Assay. Pathogens. 2021;10:919. doi: 10.3390/pathogens10080919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moldován N., Tombácz D., Szucs A., Csabai Z., Balázs Z., Kis E., Molnár J., Boldogkoi Z., Szűcs A., Csabai Z., Balázs Z., Kis E., Molnár J., Boldogkői Z. Third-generation sequencing reveals extensive polycistronism and transcriptional overlapping in a baculovirus. Sci. Rep. 2018;8:8604. doi: 10.1038/s41598-018-26955-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kang Y.-J., Yang D.-C., Kong L., Hou M., Meng Y.-Q., Wei L., Gao G. CPC2: a fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 2017;45:W12–W16. doi: 10.1093/nar/gkx428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boldogkői Z., Balázs Z., Moldován N., Prazsák I., Tombácz D. Novel classes of replication-associated transcripts discovered in viruses. RNA Biol. 2019;16:166–175. doi: 10.1080/15476286.2018.1564468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Depledge D.P., Breuer J. 2021. Varicella-Zoster Virus—Genetics, Molecular Evolution and Recombination; pp. 1–23. [DOI] [PubMed] [Google Scholar]
- 45.Balázs Z., Tombácz D., Szucs A., Csabai Z., Megyeri K., Petrov A.N., Snyder M., Boldogkoi Z. Long-read sequencing of human cytomegalovirus transcriptome reveals RNA isoforms carrying distinct coding potentials. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-16262-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Statello L., Guo C.-J., Chen L.-L., Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021;22:96–118. doi: 10.1038/s41580-020-00315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dellino G.I., Cittaro D., Piccioni R., Luzi L., Banfi S., Segalla S., Cesaroni M., Mendoza-Maldonado R., Giacca M., Pelicci P.G. Genome-wide mapping of human DNA-replication origins: levels of transcription at ORC1 sites regulate origin selection and replication timing. Genome Res. 2013;23:1–11. doi: 10.1101/gr.142331.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tikhanovich I., Liang B., Seoighe C., Folk W.R., Nasheuer H.P. Inhibition of human BK polyomavirus replication by small noncoding RNAs. J. Virol. 2011;85:6930–6940. doi: 10.1128/JVI.00547-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tai-Schmiedel J., Karniely S., Lau B., Ezra A., Eliyahu E., Nachshon A., Kerr K., Suárez N., Schwartz M., Davison A.J., Stern-Ginossar N. 2020. Human Cytomegalovirus Long Noncoding RNA4.9 Regulates Viral DNA Replication, Public Library of Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stacey S.N., Jordan D., Williamson A.J.K., Brown M., Coote J.H., Arrand J.R. Leaky scanning is the predominant mechanism for translation of human papillomavirus type 16 E7 oncoprotein from E6/E7 bicistronic mRNA. J. Virol. 2000;74:7284–7297. doi: 10.1128/JVI.74.16.7284-7297.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vilela C., McCarthy J.E.G. Regulation of fungal gene expression via short open reading frames in the mRNA 5′untranslated region. Mol. Microbiol. 2003;49:859–867. doi: 10.1046/j.1365-2958.2003.03622.x. [DOI] [PubMed] [Google Scholar]
- 52.Kronstad L.M., Brulois K.F., Jung J.U., a Glaunsinger B. Dual short upstream open reading frames control translation of a herpesviral polycistronic mRNA. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boldogköi Z. Transcriptional interference networks coordinate the expression of functionally related genes clustered in the same genomic loci. Front. Genet. 2012;3:1–17. doi: 10.3389/fgene.2012.00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gilet J., Conte R., Torchet C., Benard L., Lafontaine I. Additional layer of regulation via convergent gene orientation in yeasts. Mol. Biol. Evol. 2020;37:365–378. doi: 10.1093/molbev/msz221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brooks A.N., Hughes A.L., Clauder-Münster S., Mitchell L.A., Boeke J.D., Steinmetz L.M. Transcriptional neighborhoods regulate transcript isoform lengths and expression levels. Science. 2022;80(375):1000–1005. doi: 10.1126/science.abg0162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing datasets generated in this study are available at the European Nucleotide Archive under the accession: PRJEB52190.