Abstract
Objective
Transmission of antimicrobial resistant bacteria between people and household pets, such as dogs and cats, is an emerging global public health problem. This scoping review synthesized existing evidence of human-pet bacteria transmission to understand the magnitude and breadth of this issue.
Methods
The search included specific and generic terms for bacteria, resistance, transmission, pets, and humans. Searches were conducted through PubMed, Scopus, Web of Science, CABI Global Health, Networked Digital Library of Theses and Dissertations, Google Scholar. All studies published in English and Mandarin that isolated bacteria from pets (cats and dogs) and humans who had contact with the pets, and reported phenotypic or genotypic antimicrobial sensitivity test results, were included in this review. In cases of bacterial species that are commonly associated with pets, such as Staphylococcus pseudintermedius and Pasteurella multocida, we also included studies that only isolated bacteria from humans.
Results
After removing duplication, the search captured 9355 studies. A total of 1098 papers were screened in the full-text review, and 562 studies were identified as eligible according to our inclusion criteria. The primary reason for exclusion was the lack of sensitivity testing. The included studies were published between 1973 and 2021. The most common study location was the United States (n = 176, 31.3%), followed by the United Kingdom (n = 53, 9.4%), Japan (n = 29, 5.2%), and Canada (n = 25, 4.4%). Most of the included studies were case reports (n = 367, 63.4%), cross-sectional/prevalence studies (n = 130, 22.4%), and case series (n = 51, 8.8%). Only few longitudinal studies (n = 14, 2.4%), case-control studies (n = 12, 2.1%), and cohort studies (n = 5, 0.9%) were included in our review. Most studies focused on Pasteurella multocida (n = 221, 39.3%), Staphylococcus aureus (n = 81, 14.4%), and Staphylococcus pseudintermedius (n = 52, 8.9%). For the 295 studies that used strain typing methods to compare bacteria from humans and pets, most used DNA banding pattern-based methods (n = 133, 45.1%) and DNA sequencing-based methods (n = 118, 40.0%).
Conclusion
Transmission of bacteria could occur in both directions: pets to humans (e.g., S. pseudintermedius and P. multocida) and humans to pets (e.g., S. aureus). The majority of studies provided a low level of evidence of transmission (e.g., case reports), suggesting that more rigorous longitudinal, cohort, or case-control studies are needed to fully understand the risk of human-pet resistant bacterial transmission.
Keywords: Antimicrobial resistance, One health, Companion animals, Scoping review, Zoonotic disease
Highlights
-
•
Researchers paid more attention to One Health aspect of antimicrobial resistance issue in recent years, especially after 2010
-
•
Antimicrobial resistant bacteria transmission is expected to be a bidirectional pathway between humans and pets
-
•
P. multocida, S. aureus, and S.pseudintermedius were the most common reported bacteria transmitted between humans and pets
-
•
More longitudinal studies of high quality are required in this area
1. Introduction
Throughout history, the discovery and development of antibiotics have been one of the most important advancements in both human and veterinary medicine; however, antimicrobial resistance (AMR) has become a critical problem in modern medicine [1]. AMR occurs when bacteria, parasites, viruses or fungi develop an ability to survive in the presence of antimicrobials [2]. An estimated 700,000 people died as a result of AMR infections in 2014, and this number is projected to increase to 10 million in 2050 if there are no effective actions taken [3].
The transmission of antimicrobial resistant bacteria has become a One Health issue since AMR bacteria can be transmitted between humans, animals, and the environment [4,5]. Antimicrobial use in any of these sectors can increase the burden of AMR [6,7]. Due to the growing popularity of household pets [8], the probability of close contact between pet owners and pets is increasing. According to a recent survey, almost 90.5 million U.S. households, 70% of all American families, own at least one pet. Among them, 45.3 million homes have a cat and 69.0 million have a dog [9]. Veterinarians often prescribe medically important antimicrobials that are commonly used in humans to dogs and cats [10,11]. This drug use may select AMR bacteria, which may be transmitted between individuals and their pets [12] through both a direct and an indirect pathway [1,13]. Direct pathway refers to close contact between pets and humans, such as petting/touching or kissing/licking, while indirect pathway mainly refers to environmental transmission, as bacteria may transmit when humans and pets live in the same household [14].
Pet ownership is beneficial for human health, both physical and mental [15]. However, behaviors of pet owners such as allowing dogs to lick their faces and handling their dogs' feces have the potential to result in antimicrobial resistant bacterial transmission [14,16,17]. According to a survey of 260 dog-owning households in a community in the UK, most dogs (79%) were fed by their owners in the kitchen, also, nearly half of all dogs (42%) included in the study always or often slept in the kitchen [18]. Common dog-owner interactions also included the dogs sniffing or nudging their owners with their nose, jumping up on their owners, and licking their owners' hands; these contact behaviors were commonly reported as occurring “sometimes” or “often” [18]. Another survey among 108 German dog owners found that 88.9% of them shared their households with pets, 68.5% of pet owners allowed dogs to stay on sofas. Most of them (93.5%) let their pets lick their hands, and 52.8% of all participants let dogs lick their faces [14].
There has not been a systematic assessment of the risk of AMR bacterial transmission between humans and pets, although there are an increasing number of reports suggesting such transmission. A cohort study including samples routinely collected from 74 pets and 74 humans indicated that extended-spectrum beta-lactamase-producing Enterobacteriaceae could be transmitted between companion animals and humans in veterinary clinics and households [19]. Another study indicated that the most common antimicrobial resistant bacteria that could be transmitted from pets to humans included methicillin-resistant Staphylococcus aureus (MRSA), methicillin-resistant Staphylococcus pseudintermedius (MRSP), ESBL/AmpC-producing Enterobacteriaceae and MDR non-fermenting Gram-negative bacteria [20]. Some of these pathogens are found in both humans and pets (e.g., Enterobacteriaeceae) so the direction of transmission can be challenging to determine. However, other pathogens such as S. pseudintermedius and Pasteurella multocida, are adapted to domestic cats and dogs and are therefore likely to be transmitted from pets to humans rather than humans to pets [21,22]. Finally, pets can also be a reservoir for some human pathogenic bacteria, such as MRSA, vancomycin-resistant Enterococci (VRE) and multidrug-resistant Salmonella typhimurium DT104 [23]. It is important to assess the existing knowledge base of AMR transmission between pets and humans as it can help guide policies and identify studies or interventions needed to tackle AMR globally with a One Health approach. A scoping review is often used as a precursor for further studies, as it can provide a broad overview of existing literature and studies. This scoping review focused on mapping the extent of this issue to provide information for researchers, policymakers, veterinarians, and physicians to decrease AMR transmission between humans and dogs and cats.
2. Methods
The PICo framework [24] was used to formulate the research question of this scoping review, which is to identify the existing evidence of AMR transmission (the phenomenon of Interest) between people and pets (dogs and cats) (Population) in the world (Context).
The protocol of this scoping review, including background, rationale, inclusion and exclusion criteria, search strategy and review processes, was registered on August 31, 2020 (https://zenodo.org/record/3967957#.YobSfaiZNPa). The questions to guide inclusion and exclusion were refined on November 19, 2020, following a discussion with the review team, before the full-text screening.
2.1. Inclusion criteria
For the title/abstract screening stage, eligible studies were: 1) published in English or Mandarin; 2) primary research, narrative reviews, systematic reviews, or scoping reviews; 3) with a study population of domestic cats or dogs, and humans who have contact with cats or dogs; and 4) related to AMR bacteria transmission between humans and pets (cats and dogs). The bacteria must be transmitted from humans to pets or from pets to humans either directly or indirectly (e.g., via a shared environment).
For the full-text review stage, studies were included if they met the following criteria: 1) the full text of the study can be acquired; 2) the study is written in English or Mandarin; 3) the study is primary research; 4) this study includes cases of AMR transmission between pets (dogs and cats) and humans; 5) samples of both the pets and people who have had contact with the pets were analyzed OR only human samples were cultured, but pets are the primary source of infections with the bacteria in humans; 6) the bacteria were tested for antimicrobial resistance (phenotypic or genotypic).
2.2. Exclusion criteria
Studies that did not meet all of the inclusion criteria above were excluded from this scoping review. Studies that reported only metagenomic data were excluded from this review. Narrative reviews, systematic reviews, and scoping reviews were only used to identify additional relevant primary research and were excluded after the full-text review stage. Studies were also excluded if the full text could not be acquired.
2.3. Information sources
The following databases were searched to identify eligible studies using the designed search strategy: PubMed, Scopus, Web of Science, CABI Global Health, Networked Digital Library of Theses and Dissertations, Google Scholar. The initial search was performed on October 30, 2020, and the search strategy is available in the supplementary materials. Additional studies were identified from forward and backward reference screening of studies that meet the inclusion criteria following full-text review. Forward searching was done with Web of Science or Google Scholar cited reference searches. At the data extraction stage, references cited by included studies from the initial search and forward reference search were screened for relevance (backward screening). De-duplication was performed at each stage with EndNote (Clarivate, EndNote X8) and Covidence.
2.4. Review process and study selection
All studies found via the information sources were added to Covidence [25] and reviewed and assessed by a review team of seven reviewers, with each study assessed by two reviewers. Covidence is an online software platform designed to help reviewers to work together to manage the various stages of conducting a systematic review, including study selection, data extraction, and synthesis of results [25]. At the title and abstract screening stage, studies advanced to full-text review if both reviewers thought the study should be included or if they were unsure. Any disagreement among reviewers (e.g., one voted to include and the other voted to exclude) was resolved by discussion of the entire review team. A similar process was used for the full text review, with either a third reviewer or the entire review team assisting with disagreements from the two primary reviewers. The reason for exclusion at the full text review stage was recorded.
The review team tested the title and abstract screening process on 100 studies and refined the process for clarity and consistency, as measured by interrater reliability. The full text review and data charting process was tested on 5 studies.
2.5. Data extraction
The data from included studies was extracted using a standard data extraction form designed by the review team. All data extraction was done in Covidence, and each study was extracted by two reviewers. Conflicts were resolved by consensus, and the consensus data was recorded for the data analysis process.
The following seven components were recorded in the data extraction process: general information (year and location of publication, study design, study setting, etc.), pet species (dogs, cats, both), humans' relationship with pets (pet owners, veterinarians, veterinary nurses or technicians, or veterinary students, other or unknown), bacteria studied, antimicrobial resistance (type of antimicrobial susceptibility testing method, antimicrobials tested, percentage of multidrug resistant bacteria among all bacteria tested in the study, proportion of specific resistances for certain bacteria [e.g, the proportion of methicillin-resistant Staphylococcus aureus among all Staphylococcus aureus tested for each study]), evidence of transmission, and backward reference screening. The data extraction form is included in the supplemental materials.
2.6. Evidence of transmission assessment
Study design quality and bias for each included article were not assessed. Three questions in data extraction form were used to assess the quality of transmission evidence. The questions were as follows:
“Was the same bacterial species isolated from pets and humans? Yes, no, NA”.
“Did at least one pair of bacteria isolated from pets and humans have the same AMR phenotype or genotype? Yes, no, NA”.
“Was at least one bacterial species the same strain, as determined by strain typing (including DNA banding pattern-based methods, DNA sequencing-based methods, or DNA hybridization-based methods, see Supplement C for details), in pets and humans? Yes, no, unknown (strain typing not performed), NA”.
If none of the answers to these questions were “Yes”, the study was classified as a “presumptive study”. If one, two, or three of the answers was “yes”, the paper was listed as “low quality”,”medium quality”, or”high quality”, respectively. Due to the large number of strain typing methodologies used in the identified studies, we did not further divide or characterize the typing methodology. However, some typing methodologies, e.g., MLST and whole genome sequencing, provide stronger evidence for strain relatedness than others, e.g., PFGE.
2.7. Data analysis
Average Cohen's Kappa and average prevalence-adjusted bias-adjusted Kappa (PABAK) were computed to assess the inter-rater agreement in each study review stage.
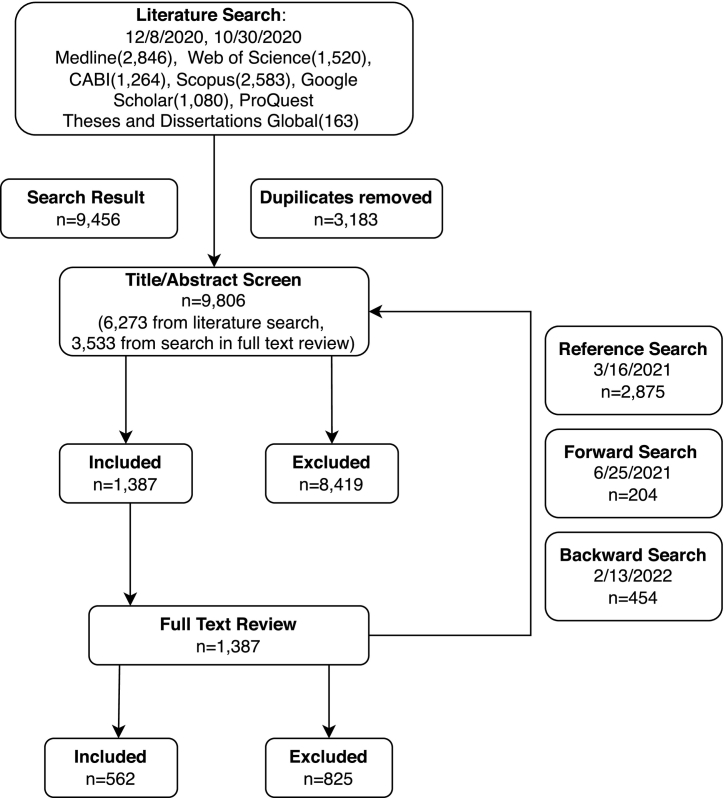
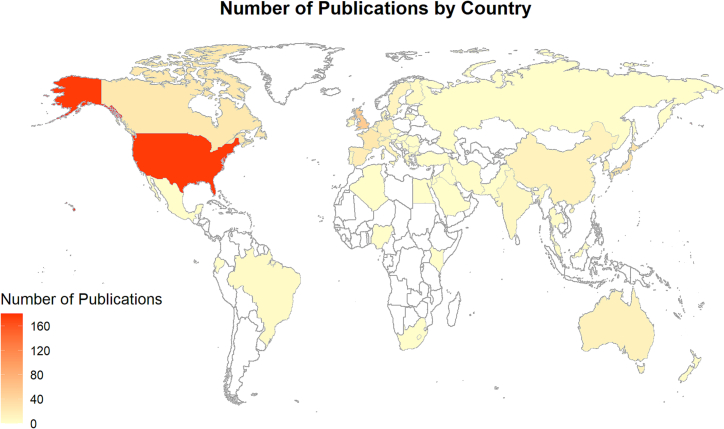
All data were recorded using Covidence and exported using Microsoft Excel. A PRISMA (preferred reporting items for systematic reviews and meta-analyses) flowchart was created to show the result of the literature search and study selection processes. Fig. 1, Fig. 2 were used to describe the studies included in our scoping review by location and year of publication. Cross-tabulations were used to show the relationship between two data items. All data analysis was conducted using R 4.2.1.
Fig. 1.
The PRISMA flowchart showing the search and screening process.
Fig. 2.
Map of the number of publications (n = 562) by the country of the studied population.
3. Results
3.1. General information
A total of 9456 studies were identified through a literature search in chosen databases, with 3183 duplicates being removed. Forward and backward searching plus scanning references of reviews yielded an additional 3533 studies before removal of duplicates. After title/abstract screening, 8419 studies were excluded, leaving 1387 eligible studies. Then 825 studies were identified as not eligible for full-text review criteria, so 562 studies were included in the data charting process (Fig. 1). During the full-text review, the most common reasons for exclusion were “No testing for antimicrobial resistance” (321, 39.0%) and “Not primary research” (158, 19.2%).
The included papers were from 52 countries, predominantly in Europe (n = 24 countries), Asia (n = 16), America (n = 5), Africa (n = 5), and Oceania (n = 2). The most common study locations (Fig. 2) were the United States (n = 176 studies, 31.3%), followed by the United Kingdom (n = 53, 9.4%), Japan (n = 29, 5.2%), and Canada (n = 25, 4.4%). A total of 279 studies (49.6%) were conducted in countries where English is the official language.
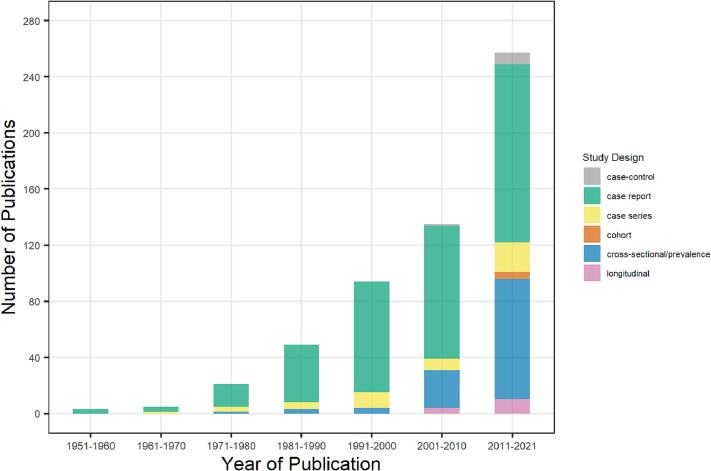
Among the 562 included studies, the date of publication was between 1953 and 2021. Only three and six studies included were published between 1951 and 1960 and 1961–1970, respectively. The majority of studies were published between 2010 and 2021 (n = 255, 45.4%) and 2001–2010 (n = 134, 23.8%) (Fig. 3).
Fig. 3.
Studies (n = 562) by decade of publication.
3.2. Study population
Most of the studies included did not sample the pets but reported humans infected with pet-associated bacteria [21,22] (e.g., S. pseudintermedius and P. multocida), so these studies were defined as “presumptive transmission” in our review (n = 331, 58.9%). Of other included studies, 109 sampled dogs and humans, 34 sampled cats and humans, and 88 studies included cats, dogs, and humans. Overall, over 16,000 dogs and nearly 4,000 cats were enrolled in these studies.
The majority of humans in eligible studies were pet owners (n = 370 studies), other or unknown (n = 66), and veterinarians, veterinary nurses or technicians, or veterinary students (n = 41). For the “other or unknown” category, most were people who fed stray dogs or cats, people who had contact with other people's pets, or animal shelter workers. Table 1 shows that most of the included studies were conducted in a human hospital/clinic (n = 399, 68.9%), household/community (n = 103, 17.8%), or veterinary clinic/veterinary school (n = 68, 11.7%). As for study design, case reports were the most common (n = 367, 63.4%), followed by cross-sectional/prevalence study (n = 130, 22.4%), case series (n = 51, 8.8%), longitudinal study without a control group (n = 14, 2.4%), case-control study (n = 12, 2.1%), and cohort study (n = 5, 0.9%). The majority of case reports occurred in human hospital/clinic, while most of the cross-sectional studies were conducted in household/community and veterinary clinic/veterinary school.
Table 1.
Summary of primary study settings of studies (n = 562) by study design. Note that some papers reported more than one study design or setting.
| Primary Study Settings |
||||||
|---|---|---|---|---|---|---|
| Study Design | Household/Community | Veterinary clinic/school | Human hospital/Clinic | Laboratory | Other/Unknown | Total |
| Case report | 20 (19.4) | 5 (7.4) | 341 (85.5) | 1 (16.7) | 0 (0.0) | 367 (63.4) |
| Case series | 6 (5.8) | 3 (4.4) | 41 (10.3) | 1 (16.7) | 0 (0.0) | 51 (8.8) |
| Cross-sectional/ prevalence | 56 (54.4) | 52 (76.5) | 15 (3.8) | 4 (66.7) | 3 (100.0) | 130 (22.5) |
| Longitudinal | 10 (9.7) | 4 (5.9) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 14 (2.4) |
| Case-control | 6 (5.8) | 4 (5.9) | 2 (0.5) | 0 (0.0) | 0 (0.0) | 12 (2.1) |
| Cohort | 5 (4.9) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 5 (0.9) |
| Total | 103 (17.8) | 68 (11.7) | 399 (68.9) | 6 (1.0) | 3 (0.5) | 579 |
We recorded the use of antimicrobials in humans after the isolation of the studied bacteria and found 334 studies in which the humans were given antimicrobials and recovered, 52 studies stated that the individuals were given antimicrobials and died, and 23 studies mentioned that the individuals were given antimicrobials, but the outcome was not recorded. There were also 151 studies without a clear statement of antimicrobial use and outcome, and only six studies mentioned that the individuals anywhere given no antimicrobials.
3.3. Antimicrobial resistance testing
Among all studies included in this review, 295 studies used strain typing methods to compare bacteria from humans and pets. Most of the 295 studies used DNA banding pattern-based methods (n = 133, 45.1%) and DNA sequencing-based methods (n = 118, 40.0%). Only 10 used DNA hybridization-based methods (3.4%), such as DNA macroarrays. There were 34 studies using strain typing methods other than these three types, such as mass spectrometry or serotyping. The pattern-based methods, including pulsed-field gel electrophoresis, which is the most common method used for strain typing (n = 100), generally provide a lower level of evidence for bacterial strain relatedness than sequencing-based methods. Only 54 studies used multilocus sequence typing, 40 studies used polymerase chain reaction, and 26 used whole genome sequencing.
The most common susceptibility testing methods used in our included studies were disc diffusion (n = 163, 24.4% of all reported methods), broth dilution or microdilution (n = 92, 13.8%), and PCR for specific AMR genes (n = 63, 9.4%). Among all studies included, 264 studies (39.6%) did not provide information about the susceptibility testing methods used but reported susceptibility results.
The most common antimicrobials included in susceptibility testing were penicillin (n = 254 studies, 45.2%), gentamicin (n = 219, 39.0%), ampicillin (n = 214, 38.1%), tetracycline (n = 208, 37.0%), and ciprofloxacin (n = 181, 32.2%). Among the 192 studies reporting AMR of Staphylococcus, 103 studies (53.9%) included Staphylococcus isolates with methicillin or oxacillin resistance and 10 studies (5.2%) included isolates with reduced vancomycin susceptibility or vancomycin resistance. Among 59 studies of Acinetobacter, Pseudomonas, or Enterobacterales, 39 papers (66.1%) reported isolates with resistance to ceftriaxone or cefotaxime and 10 studies (16.9%) reported isolates with resistance to ertapenem, meropenem, or imipenem.
3.4. Bacteria studied
Table 2 shows the most common bacteria sampled in our included studies: Pasteurella multocida (n = 221), Staphylococcus aureus (n = 81), Staphylococcus pseudintermedius (n = 50), Capnocytophaga canimorsus (n = 49), and Bordetella bronchiseptica (n = 46). These five most common bacteria were followed by Enterobacterales, which were included in 36 studies.
Table 2.
Summary of study characteristics by the five most common bacteria isolated.
| Characteristic |
P.multocida (n = 221) |
S.aureus (n = 81) |
S.pseudintermedius (n = 50) |
C.canimorsus (n = 49) |
B.bronchiseptica (n = 46) |
|---|---|---|---|---|---|
| Study Design | |||||
| Case report (n = 299) | 192 (86.9%) | 7 (8.6%) | 20 (40.0%) | 42 (85.7%) | 38 (82.6%) |
| Case series (n = 43) | 21 (9.5%) | 6 (7.4%) | 3 (6.0%) | 5 (10.2%) | 8 (17.4%) |
| Cross-sectional/Prevalence (n = 91) | 8 (3.6) | 59 (72.8%) | 22 (44.0%) | 2 (4.1%) | 0 (0.0%) |
| Longitudinal (n = 8) | 0 (0.0%) | 5 (6.2%) | 3 (6.0%) | 0 (0.0%) | 0 (0.0%) |
| Cohort (n = 3) | 0 (0.0%) | 2 (2.5%) | 1 (2.0%) | 0 (0.0%) | 0 (0.0%) |
| Case-control (n = 3) | 0 (0.0%) | 2 (2.5%) | 1 (2.0%) | 0 (0.0%) | 0 (0.0%) |
| Primary Study Setting | |||||
|
Household/ Community (n = 64) |
9 (4.1%) | 37 (41.6%) | 16 (31.4%) | 2 (4.1%) | 0 (0.0%) |
|
Veterinary clinic/ Veterinary school (n = 48) |
1 (0.5%) | 37 (41.6%) | 10 (19.6%) | 0 (0.0%) | 0 (0.0%) |
| Human hospital/Clinic (n = 334) | 209 (94.6%) | 12 (13.5%) | 22 (43.1%) | 45 (91.8%) | 46 (100.0%) |
| Laboratory (n = 6) | 1 (0.5%) | 1 (1.1%) | 2 (3.9%) | 2 (4.1%) | 0 (0.0%) |
| Other/unknown (n = 4) | 1 (0.5%) | 2 (2.2%) | 1 (2.0%) | 0 (0.0%) | 0 (0.0%) |
| Pet Species Sampled | |||||
| Dog (n = 74) | 15 (6.8%) | 34 (42.0%) | 20 (40.0%) | 3 (6.1%) | 2 (4.3%) |
| Cat (n = 28) | 20 (9.0%) | 6 (4.2%) | 0 (0.0%) | 2 (4.1%) | 0 (0.0%) |
| Both (n = 55) | 8 (3.6%) | 35 (43.2%) | 11 (22.0%) | 0 (0.0%) | 1 (2.2%) |
| None (n = 290) | 178 (80.5%) | 6 (7.4%) | 19 (38.0%) | 44 (89.8%) | 43 (93.5%) |
| Antimicrobial Use in Humans and Outcome | |||||
| Yes and died (n = 49) | 26 (11.8%) | 1 (1.2%) | 1 (2.0%) | 11 (22.4%) | 10 (20.8%) |
| Yes and recovered (n = 278) | 178 (80.5%) | 10 (12.3%) | 19 (38.0%) | 35 (71.4%) | 36 (75.0%) |
| Yes and unknown outcome (n = 17) | 10 (4.5%) | 3 (3.7%) | 1 (2.0%) | 1 (2.0%) | 2 (4.2%) |
| No antimicrobial use (n = 5) | 0 (0.0%) | 4 (4.9%) | 1 (2.0%) | 0 (0.0%) | 0 (0.0%) |
| Unknown use and outcome (n = 100) | 7 (3.2%) | 63 (77.8%) | 28 (56.0%) | 2 (4.1%) | 0 (0.0%) |
| Immunocompromise Status of Human | |||||
| Yes (n = 110) | 56 (25.3%) | 3 (3.7%) | 7 (14.0%) | 15 (30.6%) | 29 (63.0%) |
| No (n = 77) | 41 (18.6%) | 13 (16.0%) | 8 (16.0%) | 11 (22.4%) | 4 (8.7%) |
| Both (n = 21) | 7 (3.2%) | 4 (4.9%) | 5 (10.0%) | 2 (4.1%) | 3 (6.5%) |
| Unknown (n = 239) | 117 (52.9%) | 61 (75.3%) | 30 (60.0%) | 21 (42.9%) | 10 (21.7%) |
| Evidence of Transmission Assessment | |||||
| Presumptive Transmission (n = 302) | 180 (81.4%) | 9 (11.1%) | 20 (40.0%) | 47 (95.9%) | 46 (100.0%) |
| Low (n = 35) | 19 (8.6%) | 10 (12.3%) | 5 (10.0%) | 1 (2.0%) | 0 (0.0%) |
| Medium (n = 44) | 15 (6.8%) | 20 (24.7%) | 8 (16.0%) | 1 (2.0%) | 0 (0.0%) |
| High (n = 66) | 7 (3.2%) | 42 (51.9%) | 17 (34.0%) | 0 (0.0%) | 0 (0.0%) |
The majority of studies investigating P. multocida were case reports (n = 192, 86.9%) and were conducted in human hospital/clinic (n = 209, 94.6%), while most studies of S. aureus were cross-sectional/prevalence studies (n = 59, 72.8%). The mortality rate of C. canimorsus and B. bronchiseptica were the highest among these five bacteria, which were 22.4% (11/35) and 20.8% (10/46) of infected humans dying, respectively, while only 1 of 81 cases died after being infected with S. aureus (1.2%). Most humans with B. bronchiseptica were reported as immunocompromised (n = 29, 63.0%).
According to the evidence assessment criteria, the majority of studies (n = 347, 61.7%) were classified as presumptive studies (Table 2), which refer to studies without solid evidence of transmission between humans and pets. These studies either did not attempt to isolate the bacteria from pets but the authors stated that the infection in humans was presumptively from the pet, or they were not able to isolate the same bacterial species from pets and humans. There were 47 studies with evidence of low quality (8.4%), which isolated the same bacterial species from pets and humans but did not find the same AMR pattern in the bacterial isolates. There were 58 studies with evidence of medium quality (10.3%), which found bacteria of the same species and with the same AMR pattern in humans and pets. Finally, 110 studies had high-quality evidence of transmission (19.6%) with the study authors determining that humans and pets carried or were infected by the same bacterial strain through the use of strain typing. We did not further characterize the strength of the typing methodology, but note that studies using sequencing or genomic typing approaches likely provide higher-quality evidence of transmission than studies using pattern-based methods.
P. multocida was the most common bacteria isolated in our eligible studies (Table 2). However, most of these studies were case reports with no solid evidence of transmission between pets and humans (e.g., frequently the transmission was assumed because of reported pet contact and pets were not sampled). Of the 5 most commonly reported bacteria, only S. aureus was often transmitted from humans to pets. Generally, studies focusing on S. aureus provided a higher quality of transmission evidence (>50% of studies had high quality of evidence) than other four bacteria, which were all primarily pet to human transmission. Half the studies of S. pseudintermedius, a commensal staphylococcal species in dogs, had medium or high quality of evidence for transmission [26].
4. Discussion
We identified an increasing interest in AMR transmission between humans and pets. There were only three studies eligible for our review in the 1950s but 255 studies from 2010 to 2021, which could reflect increasing awareness of the One Health aspect of AMR, and/or increasing prevalence of transmission. A recent review of pet to human zoonosis from a One Health perspective suggested that the role of companion animals is usually underestimated in One Health communications, the authors thought that changing attitudes towards pets among humans lead to the changing modes of human-pet interaction, which can cause increasing risks of zoonosis, such as AMR transmission, but the frequency is difficult to estimate because the limited severity and non-specific symptoms [8]. However, fewer studies focused on human to pet AMR transmission compared to pet to human transmission; in addition, sometimes the direction of transmission can not be determined [27,28].
Of the five most commonly reported bacteria, P. multocida, S. pseudintermedius, C. canimorsus, and B. bronchiseptica were identified as pet-associated bacteria in our review with pet to human transmission. Only S.aureus was often transmitted from humans to companion animals. In our review, only 36 eligible papers studied antimicrobial resistant Enterobacteriaceae, although these bacteria caused the most human deaths in 2019 among all antimicrobial resistant pathogens [29]. Since Enterbacteriaceae are found in all mammals, it can be more challenging to study transmission between pets and humans; sampling both the pet and the human and using high-quality strain typing is required. More rigorous studies with high-quality evidence of human-pet transmission focusing on Enterobacteriaceae are needed to help determine the risk and direction of Enterobacteriaceae transmission between humans and pets. Most research focusing on pet-associated bacteria were case studies without strong evidence of transmission from the pets; the reason could be that human physicians usually do not take samples from pets and may not have easy access to veterinarians or other qualified personnel who can assist with collecting microbiological samples from pets. Additionally, sampling the pet may not add value to the physician's diagnostic and treatment plan for infected humans. However, taking bacterial cultures from pets when zoonotic transmission is suspected can have significant value to public health. We recommend that physicians, veterinarians, and public health officials consider developing guidelines for multi-species testing in cases of zoonotic or anthropozoonotic infection.
It is estimated that about 60% of all infectious diseases are zoonoses [30]. Pets might be blamed as potential reservoirs and sources of some dangerous zoonotic pathogens, which may even lead to abandonment or lack of responsible ownership [31]. However, the risk of anthropozoonosis (human to pet transmission) may be overlooked and additional studies are needed to understand the risk to pets from AMR bacteria originating in humans. In particular, pets living with people who work in human healthcare or who have been recently hospitalized could be at higher risk for becoming infected or colonized with AMR or MDR pathogens [30].
Due to the trends of increasing companion animal population and closer pet-human interactions in the world [32], it is important to think about the AMR issue from the One Health perspective, which aims to achieve optimal health outcomes by recognizing the interconnectedness between people, animals, plants, and their shared environment [33]. Studies showed that pet owners might have a limited understanding of antibiotic use among pets, and they are unwilling to change their affectionate behaviors with companion animals [34,35]. Therefore, antibiotic stewardship and judicious use are essential to protect companion animals and their owners from AMR, and veterinarians play an important role in this process. Specific guidelines for antibiotic use among pets [35] are an important tool for limiting harmful AMR and subsequent resistant bacterial zoonoses.
This scoping review has several limitations. According to our eligibility criteria, we only included studies in English and Mandarin Chinese. Nearly one-third of all studies were conducted in the US, and there could be a bias towards English-speaking countries as only studies published in English were included in our review. Although we aimed to also include studies in Mandarin, no eligible studies were identified. Excluding studies in languages other than English may limit the global generalizability of this review. Also, as only studies that cultured bacteria and tested for at least one phenotypic or genotypic resistance were eligible for this review, studies that used a metagenomic approach were excluded. Metagenomic data may lend some insight into the spread of AMR genes and this data type should be considered when designing future studies of human-pet AMR transmission.
Our quality assessment of the evidence for transmission was broad because of the diversity of methods used by the eligible studies. It is important to consider the specific strain typing methods and approaches used in a study before confidently determining that transmission has occurred. We encourage readers to critically evaluate the methods of each study of interest. We did not assess the study design quality and bias for each included article. However, scoping reviews do not always seek to provide a quality assessment of each study but aim to include an extensive range of literature on a broad question [36]. We synthesized the data extracted from eligible studies, whose literature provided different levels of quality and used different types of study design. The strain typing methods and antimicrobial susceptibility testing methods also varied and might use different criteria or techniques due to the limitations of technology at the time of publication. Therefore, the AMR rates cannot be synthesized across studies because of substantial differences in bacterial populations sampled (e.g., year, location) and methods of sampling. Additionally, some studies provided data at an isolate-level and others only provided overall percentages of resistance.
5. Conclusion
Our review provided evidence that transmission can occur in both directions: pets to humans (e.g., S. pseudintermedius and P. multocida) and humans to pets (e.g., S. aureus). This study has indicated a clearly increasing interest in AMR bacterial transmission between companion animals and humans, particularly in the last 20 years. However, >60% of our eligible studies presumed transmission between humans and pets and did not provide solid evidence of transmission, which requires culturing both pets and humans, applying the same type of susceptibility testing to the isolates, and strain typing. Also, the majority of studies were case reports or case series studies, suggesting that more rigorous primary studies, such as longitudinal, cohort, or case-control studies are needed to provide higher levels of evidence and help us fully understand the risk of human-pet bacterial transmission.
Funding
Marwan Osman is supported by the Atkinson Postdoctoral Fellowship (Cornell University).
Declaration of Competing Interest
The authors have no competing interests to disclose.
Acknowledgements
The review team is grateful for the input of Erin Eldermire, Head of Flower-Sprecher Veterinary Library at Cornell University College of Veterinary Medicine, on developing the search strategy.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.onehlt.2023.100593.
Appendix A. Supplementary data
Appendix A Searching strategy
Appendix B Research protocol
Appendix C Data extraction form
Appendix D Included studies
Data availability
I have shared the raw data (included paper of this review) at the Attached File step.
References
- 1.Blondeau J.J.M. Antimicrobial resistance & “Man’s best friend”: What they give to us we might be giving right back. Future Microbiol. 2017;12:549–553. doi: 10.2217/fmb-2017-0043. [DOI] [PubMed] [Google Scholar]
- 2.CDC About Antibiotic Resistance 2020, Centers for Disease Control and Prevention. 2021. https://www.cdc.gov/drugresistance/about.html
- 3.O’Neill J. 2016. Antimicrobial Resistance:Tackling a Crisis for the Health and Wealth of Nations. [Google Scholar]
- 4.Collignon P., McEwen S. One health—its importance in helping to better control antimicrobial resistance. Trop. Med. Infect. Dis. 2019;4:22. doi: 10.3390/tropicalmed4010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Velazquez-Meza M.E., Galarde-López M., Carrillo-Quiróz B., Alpuche-Aranda C.M. Antimicrobial resistance: One Health approach. Vet. World. 2022:743–749. doi: 10.14202/vetworld.2022.743-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Neill Jim. 2016. Tackling Drug-Resistant Infections Globally:Final Report and Recommendations the Review on Antimicrobial Resistance. [Google Scholar]
- 7.Laxminarayan R., Duse A., Wattal C., Zaidi A.K.M., Wertheim H.F.L., Sumpradit N., Vlieghe E., Hara G.L., Gould I.M., Goossens H., Greko C., So A.D., Bigdeli M., Tomson G., Woodhouse W., Ombaka E., Peralta A.Q., Qamar F.N., Mir F., Kariuki S., Bhutta Z.A., Coates A., Bergstrom R., Wright G.D., Brown E.D., Cars O. Antibiotic resistance—the need for global solutions. Lancet Infect. Dis. 2013;13:1057–1098. doi: 10.1016/S1473-3099(13)70318-9. [DOI] [PubMed] [Google Scholar]
- 8.Overgaauw P.A.M., Vinke C.M., van Hagen M.A.E., Lipman L.J.A. A one health perspective on the human-companion animal relationship with emphasis on zoonotic aspects. Int. J. Environ. Res. Public Health. 2020;17:1–29. doi: 10.3390/ijerph17113789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.APPA 2021–2022 National pet Owner's Survey, American Pet Products Association. 2022. https://www.americanpetproducts.org/press_industrytrends.asp
- 10.Lhermie G., La Ragione R.M., Weese J.S., Olsen J.E., Christensen J.P., Guardabassi L. Indications for the use of highest priority critically important antimicrobials in the veterinary sector. J. Antimicrob. Chemother. 2020;75:1671–1680. doi: 10.1093/jac/dkaa104. [DOI] [PubMed] [Google Scholar]
- 11.Frey E. The role of companion animal veterinarians in one-health efforts to combat antimicrobial resistance. J. Am. Vet. Med. Assoc. 2018;253:1396–1404. doi: 10.2460/javma.253.11.1396. [DOI] [PubMed] [Google Scholar]
- 12.Murphy C.P., Reid-Smith R.J., Boerlin P., Weese J.S., Prescott J.F., Janecko N., McEwen S.A. Out-patient antimicrobial drug use in dogs and cats for new disease events from community companion animal practices in Ontario. Can. Vet. J. 2012;53:291. /pmc/articles/PMC3280785/ (accessed January 16, 2022) [PMC free article] [PubMed] [Google Scholar]
- 13.Robinson T.P., Bu D.P., Carrique-Mas J., Fèvre E.M., Gilbert M., Grace D., Hay S.I., Jiwakanon J., Kakkar M., Kariuki S., Laxminarayan R., Lubroth J., Magnusson U., Ngoc P.T., Van Boeckel T.P., Woolhouse M.E.J. Antibiotic resistance is the quintessential one health issue. Trans. R. Soc. Trop. Med. Hyg. 2016;110:377. doi: 10.1093/TRSTMH/TRW048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walther B., Hermes J., Cuny C., Wieler L.H., Vincze S., Elnaga Y.A., Stamm I., Kopp P.A., Kohn B., Witte W., Jansen A., Conraths F.J., Semmler T., Eckmanns T., Lübke-Becker A. Sharing more than friendship — nasal colonization with coagulase-positive staphylococci (CPS) and co-habitation aspects of dogs and their owners. PLoS One. 2012;7 doi: 10.1371/JOURNAL.PONE.0035197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McNicholas J., Gilbey A., Rennie A., Ahmedzai S., Dono J.A., Ormerod E. Pet ownership and human health: a brief review of evidence and issues. BMJ. 2005;331:1252–1254. doi: 10.1136/BMJ.331.7527.1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naziri Z., Firouzi R., Derakhshandeh A., Tabrizi A.S. Comparative analysis of phylogenetic group and antimicrobial resistance pattern of fecal Escherichia coli isolates between healthy dogs and their owners. Comp. Clin. Pathol. 2015;24:1211–1220. doi: 10.1007/s00580-015-2062-7. [DOI] [Google Scholar]
- 17.Derakhshandeh A., Eraghi V., Boroojeni A.M., Niaki M.A., Zare S., Naziri Z. Virulence factors, antibiotic resistance genes and genetic relatedness of commensal Escherichia coli isolates from dogs and their owners. Microb. Pathog. 2018;116:241–245. doi: 10.1016/j.micpath.2018.01.041. [DOI] [PubMed] [Google Scholar]
- 18.Westgarth C., Pinchbeck G.L., Bradshaw J.W.S., Dawson S., Gaskell R.M., Christley R.M. Dog-human and dog-dog interactions of 260 dog-owning households in a community in Cheshire. Vet. Rec. 2008;162:436–442. doi: 10.1136/vr.162.14.436. [DOI] [PubMed] [Google Scholar]
- 19.Schmitt K., Kuster S.P., Zurfluh K., Jud R.S., Sykes J.E., Stephan R., Willi B. Transmission chains of extended-spectrum beta-lactamase-producing enterobacteriaceae at the companion animal veterinary clinic–household interface. Antibiotics. 2021;10:171. doi: 10.3390/antibiotics10020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pomba C., Rantala M., Greko C., Baptiste K.E., Catry B., van Duijkeren E., Mateus A., Moreno M.A., Pyörälä S., Ružauskas M., Sanders P., Teale C., Threlfall E.J., Kunsagi Z., Torren-Edo J., Jukes H., Törneke K. Public health risk of antimicrobial resistance transfer from companion animals. J. Antimicrob. Chemother. 2016:dkw481. doi: 10.1093/jac/dkw481. [DOI] [PubMed] [Google Scholar]
- 21.The Center for Food Security and Public Health at Iowa State University . 2013. Zoonotic Diseases of Companion Animals By Animal Species. 1–1. [Google Scholar]
- 22.Centers for Disease Control and Prevention Diseases That Can Spread Between Animals and People. 2020. https://www.cdc.gov/healthypets/diseases/index.html (accessed July 21, 2020)
- 23.Bhat A.H. Bacterial zoonoses transmitted by household pets and as reservoirs of antimicrobial resistant bacteria. Microb. Pathog. 2021;155 doi: 10.1016/j.micpath.2021.104891. [DOI] [PubMed] [Google Scholar]
- 24.Booth A., Noyes J., Flemming K., Moore G., Tunçalp Ö., Shakibazadeh E. Formulating questions to explore complex interventions within qualitative evidence synthesis. BMJ Glob. Health. 2019;4:1–7. doi: 10.1136/bmjgh-2018-001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Covidence systematic review software 2020. www.covidence.org
- 26.Lynch S.A., Helbig K.J. The complex diseases of Staphylococcus pseudintermedius in canines: where to next? Vet. Sci. 2021;8:11. doi: 10.3390/vetsci8010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rutland B.E., Weese J.S., Bolin C., Au J., Malani A.N. Human-to-dog transmission of methicillin-resistant Staphylococcus aureus. Emerg. Infect. Dis. 2009;15:1328–1330. doi: 10.3201/eid1508.081635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Duijkeren E., Wolfhagen M.J.H.M., Box A.T.A., Heck M.E.O.C., Wannet W.J.B., Fluit A.C. Human-to-dog transmission of methicillin-resistant Staphylococcus aureus. Emerg. Infect. Dis. 2004;10:2235–2237. doi: 10.3201/eid1012.040387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murray C.J.L., Ikuta K.S., Sharara F., Swetschinski L., Robles Aguilar G., Gray A., Han C., Bisignano C., Rao P., Wool E., Johnson S.C., Browne A.J., Chipeta M.G., Fell F., Hackett S., Haines-Woodhouse G., Kashef Hamadani B.H., Kumaran E.A.P., McManigal B., Achalapong S., Agarwal R., Akech S., Albertson S., Amuasi J., Andrews J., Aravkin A., Ashley E., Babin F.-X., Bailey F., Baker S., Basnyat B., Bekker A., Bender R., Berkley J.A., Bethou A., Bielicki J., Boonkasidecha S., Bukosia J., Carvalheiro C., Castañeda-Orjuela C., Chansamouth V., Chaurasia S., Chiurchiù S., Chowdhury F., Clotaire Donatien R., Cook A.J., Cooper B., Cressey T.R., Criollo-Mora E., Cunningham M., Darboe S., Day N.P.J., De Luca M., Dokova K., Dramowski A., Dunachie S.J., Duong Bich T., Eckmanns T., Eibach D., Emami A., Feasey N., Fisher-Pearson N., Forrest K., Garcia C., Garrett D., Gastmeier P., Giref A.Z., Greer R.C., Gupta V., Haller S., Haselbeck A., Hay S.I., Holm M., Hopkins S., Hsia Y., Iregbu K.C., Jacobs J., Jarovsky D., Javanmardi F., Jenney A.W.J., Khorana M., Khusuwan S., Kissoon N., Kobeissi E., Kostyanev T., Krapp F., Krumkamp R., Kumar A., Kyu H.H., Lim C., Lim K., Limmathurotsakul D., Loftus M.J., Lunn M., Ma J., Manoharan A., Marks F., May J., Mayxay M., Mturi N., Munera-Huertas T., Musicha P., Musila L.A., Mussi-Pinhata M.M., Naidu R.N., Nakamura T., Nanavati R., Nangia S., Newton P., Ngoun C., Novotney A., Nwakanma D., Obiero C.W., Ochoa T.J., Olivas-Martinez A., Olliaro P., Ooko E., Ortiz-Brizuela E., Ounchanum P., Pak G.D., Paredes J.L., Peleg A.Y., Perrone C., Phe T., Phommasone K., Plakkal N., Ponce-de-Leon A., Raad M., Ramdin T., Rattanavong S., Riddell A., Roberts T., Robotham J.V., Roca A., Rosenthal V.D., Rudd K.E., Russell N., Sader H.S., Saengchan W., Schnall J., Scott J.A.G., Seekaew S., Sharland M., Shivamallappa M., Sifuentes-Osornio J., Simpson A.J., Steenkeste N., Stewardson A.J., Stoeva T., Tasak N., Thaiprakong A., Thwaites G., Tigoi C., Turner C., Turner P., van Doorn H.R., Velaphi S., Vongpradith A., Vongsouvath M., Vu H., Walsh T., Walson J.L., Waner S., Wangrangsimakul T., Wannapinij P., Wozniak T., Young Sharma T.E.M.W., Yu K.C., Zheng P., Sartorius B., Lopez A.D., Stergachis A., Moore C., Dolecek C., Naghavi M. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet. 2022;399:629–655. doi: 10.1016/S0140-6736(21)02724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Littleton J., Karstens S., Busse M., Malone N. Human-animal interactions and infectious disease. Bioarchaeol. Int. 2022 doi: 10.5744/bi.2021.0002. [DOI] [Google Scholar]
- 31.Fatjó J., Bowen J., García E., Calvo P., Rueda S., Amblás S., Lalanza J. Epidemiology of dog and cat abandonment in Spain (2008–2013) Animals. 2015;5:426–441. doi: 10.3390/ani5020364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vercelli C., Gambino G., Amadori M., Re G. Implications of veterinary medicine in the comprehension and stewardship of antimicrobial resistance phenomenon. From the origin till nowadays. Vet. Anim. Sci. 2022;16 doi: 10.1016/j.vas.2022.100249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.CDC . 2023. One Helth, Centers for Disease Control and Prevention. [Google Scholar]
- 34.Smith M., King C., Davis M., Dickson A., Park J., Smith F., Currie K., Flowers P. Pet owner and vet interactions: exploring the drivers of AMR. Antimicrob. Resist. Infect. Control. 2018;7:46. doi: 10.1186/s13756-018-0341-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dickson A., Smith M., Smith F., Park J., King C., Currie K., Langdridge D., Davis M., Flowers P. Understanding the relationship between pet owners and their companion animals as a key context for antimicrobial resistance-related behaviours: an interpretative phenomenological analysis. Health Psychol. Behav. Med. 2019;7:45–61. doi: 10.1080/21642850.2019.1577738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arksey H., O’Malley L. Scoping studies: towards a methodological framework. Int. J. Soc .Res .Methodol .Theory Pract. 2005;8:19–32. doi: 10.1080/1364557032000119616. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix A Searching strategy
Appendix B Research protocol
Appendix C Data extraction form
Appendix D Included studies
Data Availability Statement
I have shared the raw data (included paper of this review) at the Attached File step.